Abstract
Previous studies have demonstrated the involvement of sulfoconjugation in the metabolism of catecholamines and serotonin. The current study aimed to clarify the effects of single nucleotide polymorphisms (SNPs) of human SULT1A3 and SULT1A4 genes on the enzymatic characteristics of the sulfation of dopamine, epinephrine, norepinephrine and serotonin by SULT1A3 allozymes. Following a comprehensive search of different SULT1A3 and SULT1A4 genotypes, twelve non-synonymous (missense) coding SNPs (cSNPs) of SULT1A3/SULT1A4 were identified. cDNAs encoding the corresponding SULT1A3 allozymes, packaged in pGEX-2T vector were generated by site-directed mutagenesis. SULT1A3 allozymes were expressed, and purified. Purified SULT1A3 allozymes exhibited differential sulfating activity toward catecholamines and serotonin. Kinetic analyses demonstrated differences in both substrate affinity and catalytic efficiency of the SULT1A3 allozymes. Collectively, these findings provide useful information relevant to the differential metabolism of dopamine, epinephrine, norepinephrine and serotonin through sulfoconjugation in individuals having different SULT1A3/SULT1A4 genotypes.
Keywords: Single nucleotide polymorphisms, cytosolic sulfotransferase, SULT1A3, sulfation, catecholamines, serotonin
Graphical abstract
![]()
1. Introduction
Catecholamines, including dopamine (DA), epinephrine (EP) and norepinephrine (NE), and serotonin (5-hydroxytryptamine; 5-HT) constitute an important group of monoamine neurotransmitters/hormones that play key roles in the regulation of physiological processes such as mood, body temperature, heart rate, blood pressure, gastrointestinal motility and secretions, as well as the development of various neurological, psychiatric, endocrine, and cardiovascular diseases [1–4]. They are synthesized centrally and peripherally in, for example, the adrenal glands, gastrointestinal tract, kidneys, pancreas, and immune system [5–12]. For 5-HT, its sulfated derivative, 5-HT sulfate, has also been detected in blood and cerebrospinal fluids [13,14]. Studies have shown that more than 98% of DA and approximately 80% of total EP and NE are present in the sulfated form in circulation [15,16]. Thus, sulfation has been proposed to play important roles in the regulation and biotransformation of catecholamines as well as 5-HT [17–19].
In mammals, sulfation as catalyzed by the cytosolic sulfotransferases (SULTs) has been shown to be involved in the metabolism and elimination of xenobiotics as well as the homeostasis of key endogenous compounds such as catecholamines, 5-HT, steroid/thyroid hormones, cholesterol, and bile acids [20–22]. The SULTs mediate the transfer of a sulfonate group from the donor co-substrate, 3´-phosphoadenosine 5´-phosphosulfate (PAPS), to the hydroxyl or amino group of an acceptor substrate compound [23]. While there are occasionally exceptions, sulfate-conjugated compounds generally become inactive and more hydrophilic and thus can be eliminated more easily from the body [21,22]. Of the 13 human SULTs, SULT1A3 has been identified as the main enzyme responsible for sulfating catecholamines and 5-HT [24,25]. It is noted that SULT1A3 is found only in humans and closely related primates [26]. Interestingly, genomic studies have revealed the duplication of the gene coding for SULT1A3 during the evolutionary process [27,28]. Two genes, designated SULT1A3 and SULT1A4, located on chromosome 16 have been identified and shown to encode identical protein products, collectively called SULT1A3 [28,29]. Importantly, ethnic-specific inherited alterations in catecholamine sulfation in humans have been reported [30]. An intriguing question is whether the genetic polymorphisms of SULT1A3 and SULT1A4 may affect the enzymatic activity of the resulting SULT1A3 protein product and thus the homeostasis of catecholamines and 5-HT and possibly pathophysiological conditions associated with these latter compounds.
The current study was based on the hypothesis that nonsynonymous missense single nucleotide polymorphisms (SNPs) of SULT1A3 and SULT1A4 genes could lead to SULT1A3 allozymes with differential sulfating activities toward DA, EP, NE and 5-HT. We report in this paper a systematic database search for human SULT1A3 and SULT1A4 SNPs. Twelve missense SNPs were identified and the cDNAs and the corresponding SULT1A3 allozymes were generated, expressed, and purified. Purified SULT1A3 allozymes were analyzed for their enzymatic characteristics toward catecholamines and 5-HT.
2. Materials and methods
2.1. Materials
Adenosine 5′-triphosphate (ATP), dithiothreitol (DTT), dimethyl sulfoxide (DMSO), N-2-hydroxylpiperazine-N′-2-ethanesulfonic acid (HEPES), DA, EP and 5-HT were products of Sigma Chemical Company (St. Louis, MO, USA). NE was purchased from Cayman Chemical Company (Ann Arbor, MI, USA). Cellulose thin-layer chromatography (TLC) plates were from EMD Millipore Corporation (Burlington, MA, USA). 3′-Phosphoadenosine-5′-phospho[35S]sulfate (PAP[35S]) was prepared using ATP and carrier-free [35S]sulfate based on a previously established protocol [31]. Oligonucleotide primers were synthesized by Eurofins Genomics (Louisville, KY, USA). X-Ray films were products of Research Products International Corporation (Mt Prospect, IL, USA). Prime STAR GXL DNA Polymerase was purchased from Clontech Laboratories, Inc. (Mountain View, CA, USA). PCR kit was from G Biosciences (St. Louis, MO, USA). Protein molecular weight markers were from Bioland Scientific LLC (Paramount, CA, USA). QIAprep Spin Miniprep Kit was a product of QIAGEN (Germantown, MD, USA). Ecolume scintillation cocktail was from MP Biomedical LLC (Irvine, CA, USA). Glutathione Sepharose was purchased from GE Healthcare Life Sciences (Pittsburgh, PA, USA). All other chemicals were of the highest grades commercially available.
2.2. Database search
A comprehensive search was performed for SULT1A3 and SULT1A4 SNP clones deposited in two databases located at, respectively, the U.S. National Center for Biotechnology Information (NCBI) and the UniProt Knowledgebase (UniProtKB). Moreover, the NCBI PubMed was searched for previous studies (for example, Thomae et al. 2003 [30] and Hildebrandt et al. 2004 [28]) describing SNPs of these two genes.
2.3. Generation, expression, and purification of SULT1A3 allozymes
Site-directed mutagenesis was performed to generate cDNAs encoding SULT1A3 allozymes packaged in pGEX-2TK vector. Briefly, mutagenic primers (cf. Table 1) corresponding to specific SULT1A3/SULT1A4 missense cSNPs were designed and synthesized. Wild-type SULT1A3 cDNA packaged in pGEX-2TK prokaryotic vector was used as the template in conjunction with specific mutagenic primers to amplify mutated SULT1A3 cDNAs coding for different SULT1A3 allozymes. The sequences of the “mutated” cDNAs were verified by nucleotide sequencing [32]. The pGEX-2TK vector harboring individual mutated SULT1A3 cDNA was transformed into BL21 E. coli cells. After induction of recombinant SULT1A3 expression by IPTG, affinity chromatography using glutathione-Sepharose was performed, followed by thrombin digestion to release purified recombinant SULT1A3 allozyme. The purity of purified recombinant allozymes was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) [33,34]. The concentrations of these allozymes were measured based on Bradford protein assay [35].
Table 1.
Mutagenic primer sets used in the PCR-amplification of the cDNAs encoding human SULT1A3 allozymes
| SULT1A3 Allozymes | MAF1 | Primers |
|---|---|---|
| SULT1A3-T7P | 0.00035322 | 5′- ATGGAGCTGATCCAGGA
CCTCCCGCCCGCCACTGG -3′ 5′- CCAGTGGCGGGCGGGAGGGGTCCTGGATCAGCTCCAT -3′ |
| SULT1A3-S8P | 0.00001122 | 5′- GAGCTGATCCAGGACACC
CCCGCCCGCCACTGGAGT -3′ 5′- ACTCCAGTGGCGGGCGGGGGGTGTCCTGGATCAGCTC -3′ |
| SULT1A3-R9C | 0.000066452 | 5′- CTGATCCAGGACACCTCC
GCCCGCCACTGGAGTACG -3′ 5′- CGTACTCCAGTGGCGGGCAGGAGGTGTCCTGGATCAG -3′ |
| SULT1A3-P10L | 0.000009142 | 5′- TCCAGGACACCTCCCGCC
GCCACTGGAGTACGTGAA -3′ 5′- TTCACGTACTCCAGTGGCAGGCGGGAGGTGTCCTGGA -3′ |
| SULT1A3-V15M | 0.000040582 | 5′- CGCCCGCCACTGGAGTAC
TGAAGGGGGTCCCGCTCA -3′ 5′- TGAGCGGGACCCCCTTCATGTACTCCAGTGGCGGGCG -3′ |
| SULT1A3-V18F | 0.00022 | 5′- CTGGAGTACGTGAAGGGG
TCCCGCTCATCAAGTACT -3′ 5′- AGTACTTGATGAGCGGGAACCCCTTCACGTACTCCAG -3′ |
| SULT1A3-P101L | 0.0254 | 5′- CTCTGAAAGACACACCGC
CCCACGGCTCATCAAGTC -3′ 5′- GACTTGATGAGCCGTGGGAGCGGTGTGTCTTTCAGAG -3′ |
| SULT1A3-P101H | 0.0044 | 5′- CTCTGAAAGACACACCGC
CCCACGGCTCATCAAGTC -3′ 5′- GACTTGATGAGCCGTGGGTGCGGTGTGTCTTTCAGAG -3′ |
| SULT1A3-R144C | 0.0254 | 5′- TCCTACTACCATTTCCAC
GTATGGAAAAGGCGCACC -3′ 5′- GGTGCGCCTTTTCCATACAGTGGAAATGGTAGTAGGA -3′ |
| SULT1A3-K234N | 0.0424 | 5′- GTTCAAGGAGATGAAGAA
AACCCTATGACCAACTAC -3′ 5′- GTAGTTGGTCATAGGGTTATTCTTCATCTCCTTGAAC -3′ |
| SULT1A3-N235T | – | 5′- TCAAGGAGATGAAGAAGA
CCCTAT GACCAACTACAC -3′ 5′- GTGTAGTTGGTCATAGGGGTCTTCTTCATCTCCTTGA -3′ |
| SULT1A3-S290T | – | 5′- TGGCAGGCTGCAGCCTCA
CTTCCGCTCTGAGCTGTG -3′ 5′- CACAGCTCAGAGCGGAAGGTGAGGCTGCAGCCTGCCA -3′ |
2.4. Enzymatic assay
To analyze the sulfating activity of SULT1A3 allozymes toward DA, EP, NE and 5-HT, an established assay protocol was employed [36]. Two different concentrations were tested for each of the four substrates. Radioactive PAP[35S] was used as the sulfate donor. The assays were performed at pH 7.4 and allowed to proceed at 37°C for 10 min, followed by TLC separation of the reaction mixtures. Upon completion of TLC, autoradiography was performed using an X-ray film to locate the position of sulfated product. The radioactive sulfated product spot was then cut out and the sulfated product therein eluted for the quantitative measurement of [35S]-radioactivity using a liquid scintillation counter. Specific activity of the SULT1A3 allozymes was calculated based on the [35S]-radioactivity determined. In kinetic experiments, different substrate concentrations, 0, 0.25, 0.5, 1, 2.5, 5, 10, 25, 50, 75 and 100 μM for DA and EP, 0, 0.001, 0.1, 0.5, 1, 5, 10, 25, 50, 100, 150 and 200 μM for NE, and 1, 10, 50, 100, 125, 250, 500, 750 and 1000 μM for 5-HT.
2.5. Statistical analysis
Data obtained from the kinetic experiments were analyzed based on the non-linear regression of the Michaelis-Menten equation to calculate the kinetic constants. GraphPad Prism 7 software was used in data analysis.
3. Results
3.1. Analysis of human SULT1A3 and SULT1A4 single nucleotide polymorphisms
SNPs of SULT1A3 and SULT1A4 genes, as identified in above-mentioned database search, were categorized according to the location of variations in each of the two genes. For the SULT1A3 gene, a total of 38 SNPs, including 2 in the 3′-untranslated region (3′-UTR), 19 in the intron regions, 6 synonymous cSNPs, one nonsense cSNP, and 10 missense cSNPs, were identified. For the SULT1A4 gene, a total of 39 SNPs, including 1 in the 3′-UTR, 25 in the intron regions, 7 synonymous cSNPs, and 6 missense cSNPs, were identified. Of the 16 missense cSNPs found for both SULT1A3 and SULT1A4 genes, 13 distinct amino acid alterations were found. The designated names and their SNP ID number for these 13 SNPs are: SULT1A3-T7P (Reference SNP (rs)776817009/ rs754600221), SULT1A3-S8P (rs767263838), SULT1A3-R9C (rs762151655/ rs752303630), SULT1A3-P10L (rs757573592), SULT1A3-V15M (rs750575779/ rs758881470), SULT1A3-V18F (rs553050853), SULT1A3-P19L (rs747088850), SULT1A3-P101L (rs751527244), SULT1A3-P101H, SULT1A3-R144C and SULT1A3-K234N [28], SULT1A3-N235T (UniProt P0DMM9) and SULT1A3-S290T (UniProt P0DMM9). Figure 1 illustrates the locations of amino acid variations associated with above-mentioned cSNPs, together with previously reported sequences/residues involved in PAPS-binding, substrate-binding, and/or catalysis. Examination of the reported crystal structures of SULT1A3 [37] revealed that these amino acid residues are positioned close to the surface of the molecule (Figure 2). Interestingly, many of them are associated with the three important loops/segments, Asp66-Met77, Ser228-Gly259, and Lys85-Pro90, which have been proposed to be involved in the formation of a gate that governs the substrate selectivity [38] (cf. Figure 2).
Figure 1.
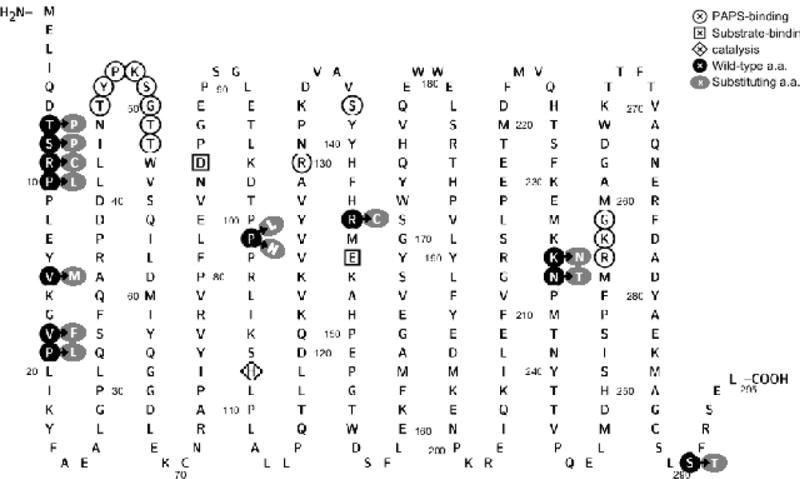
Amino acid sequence of the human SULT1A3 showing the locations of amino acid residues involved in the SULT1A3/SULT1A4 cSNPs and sequences/residues reported to be involved in PAPS-binding, substrate-binding, and/or catalysis. Residues circled with white background are involved in PAPS-binding. Residues enclosed in square are involved in substrate-binding. Residue enclosed in diamond is involved in catalysis. Residues circled with black background refer to the locations of amino acid substitutions in the polypeptide chain of the SULT1A3 molecule. Residues circled with gray background refer to the substituting amino acids. The figure was generated using Protter, a web tool for interactive protein feature visualization [63].
Figure 2.
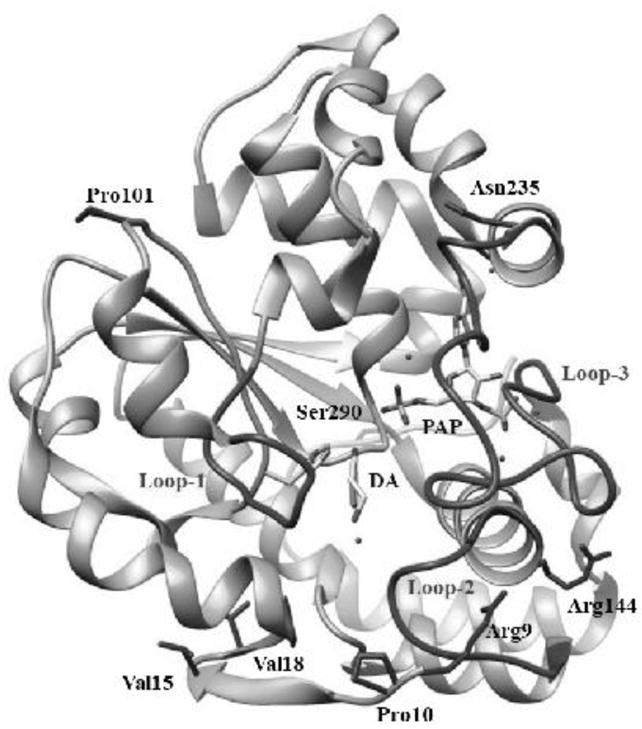
Ribbon diagram of the structure of human SULT1A3-dopamine-PAP complex showing the locations of amino acid residues involved in the SULT1A3/SULT1A4 cSNPs. The structure of SULT1A3 (Protein Data Bank code: 2A3R [37]) was edited using USCF Chimera, a molecular modeling software [64]. DA and PAP molecules in the structure are shown by bond structures. Loops 1, 2, and 3 refer to Asp66-Met77, Ser228-Gly259, and Lys85-Pro90 segments previously reported to form a gate for substrate entry. Side chains of the amino acid residues involved in the SULT1A3/SULT1A4 cSNPs, Arg9, Pro10, Val15, Val18, Pro101, Arg144, Asn235, Ser290, are indicated by bond structures.
3.2. Expression and purification of recombinant human SULT1A3 allozymes
cDNAs corresponding to the twelve chosen SULT1A3 missense genotypes packaged in pGEX-2TK prokaryotic expression vector were individually transformed into BL21 E. coli host cells for the expression of SULT1A3 allozymes. To purify the SULT1A3 allozymes, glutathione-Sepharose was used to fractionate the GST-SULT1A3 proteins from the transformed E. coli cell homogenates, followed by thrombin digestion to release SULT1A3 allozymes. Purified SULT1A3 allozymes appeared to be highly homogeneous as judged by SDS-PAGE. As shown in Figure 3, the apparent molecular weights of SULT1A3 allozymes were similar to the predicted molecular weight (34,196) of wild-type SULT1A3.
Figure 3.
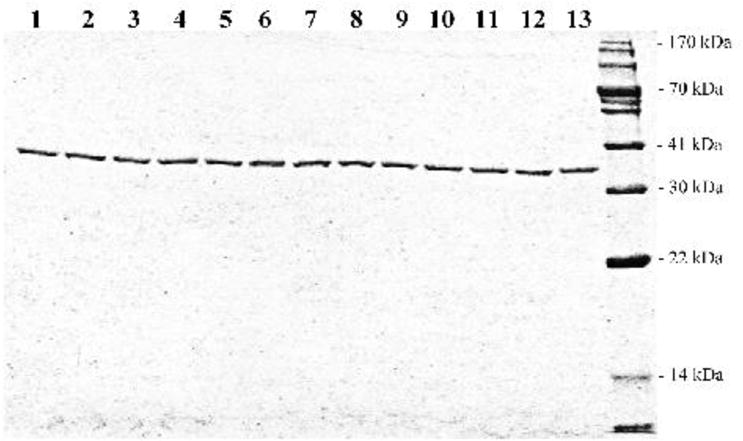
SDS gel electrophoretic pattern of the purified human SULT1A3 allozymes. SDS-PAGE was performed on a 12% gel, followed by Coomassie Blue staining. Samples analyzed in lanes 1 through 13 correspond to SULT1A3-WT (wild-type), SULT1A3-T7P, SULT1A3-S8P, SULT1A3-R9C, SULT1A3-P10L, SULT1A3-V15M, SULT1A3-V18F, SULT1A3-P101L, SULT1A3-P101H, SULT1A3-R144C, SULT1A3-K234N, SULT1A3-N235T and SULT1A3-S290T. Positions of protein molecular weight markers are indicated on the right.
3.3. Enzymatic characterization of the SULT1A3 allozymes
Purified wild-type and SULT1A3 allozymes were characterized with regard to their sulfating activity with catecholamines and 5-HT as substrates. In the initial experiments, the specific activity of wild-type and SULT1A3 allozymes was determined using two different concentrations, one considerably below and the other close to the reported Km [6.46 ± 0.59, 9.16 ± 1.81, 10.65 ± 1.14 and 71.38 ± 7.99 μM], of each of the four substrates (DA, EP, NE and 5-HT). Results obtained are shown in Figures 4–7.
Figure 4.
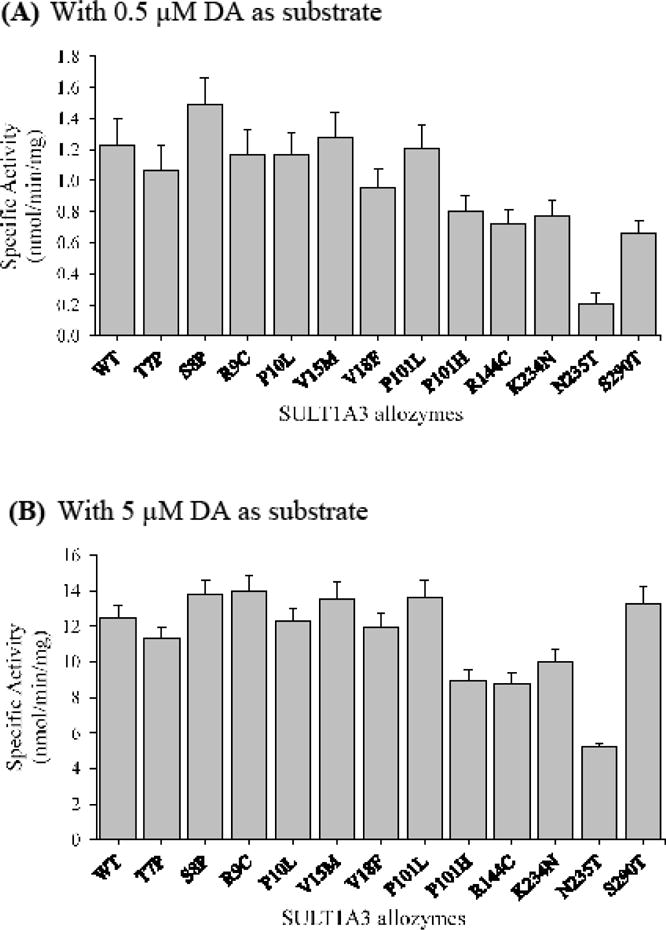
Specific activities of the sulfation of dopamine (DA) by human SULT1A3 allozymes. Concentrations of dopamine used in the enzymatic assays were 0.5 μM (A) and 5.0 μM (B). Specific activity refers to nmol dopamine sulfated/min/mg of purified allozyme. Data shown represent mean ± standard deviation derived from three determinations. WT refers to wild-type SULT1A3.
Figure 7.
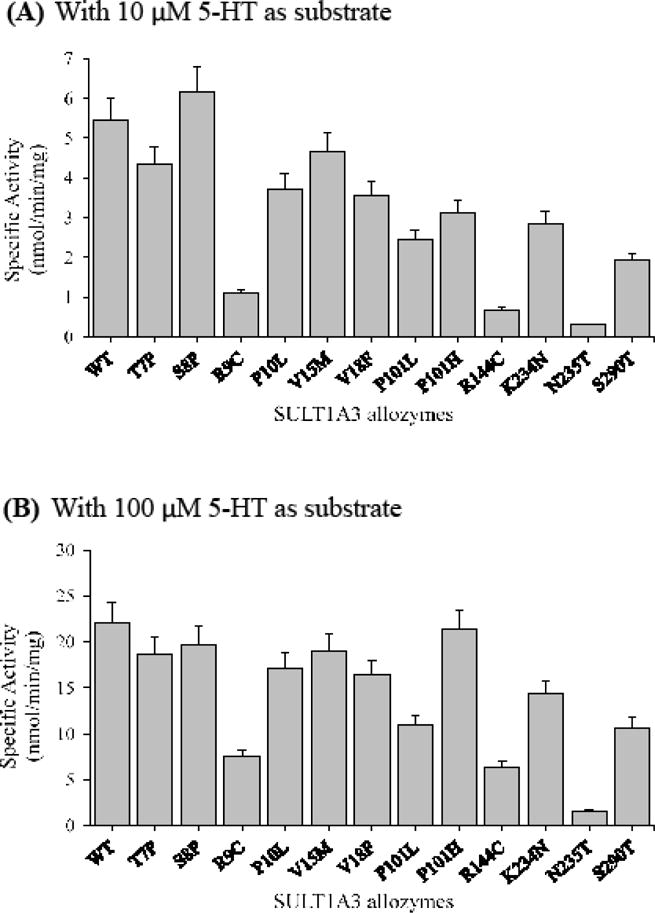
Specific activities of the sulfation of serotonin (5-HT) by human SULT1A3 allozymes. Concentrations of serotonin used in the enzymatic assays were 10 μM (A) and 100 μM (B). Specific activity refers to nmol serotonin sulfated/min/mg of purified allozyme. Data shown represent mean ± standard deviation derived from three determinations. WT refers to wild-type SULT1A3.
With DA as the substrate
At low substrate (un-saturating) concentration (0.5 μM), SULT1A3-S8P showed a higher specific activity than the wild-type enzyme, while seven other SULT1A3 allozymes (SULT1A3-T7P to SULT1A3-P101L) displayed comparable specific activities with the wild-type (Figure 4A). In contrast, the specific activities determined for the other five allozymes (SULT1A3-P101H, SULT1A3-R144C, SULT1A3-K234N, SULT1A3-N235T and SULT1A3-S290T) were considerably lower than that of the wild-type enzyme. Among these five latter SULT1A3 allozymes, SULT1A3-N235T exhibited much lower activity (16.4 % of that of the wild-type enzyme) than the other four. At higher substrate concentration (5 μM), eight SULT1A3 allozymes showed comparable or slightly higher specific activities, compared with the wild-type enzyme, whereas four allozymes (SULT1A3-P101H, SULT1A3-R144C, SULT1A3-K234N and SULT1A3-N235T) showed lower DA-sulfating activity than the wild-type enzyme, with SULT1A3-N235T showing the lowest specific activity (Figure 4B).
With EP as the substrate
At low substrate (un-saturating) concentration (1 μM), eight SULT1A3 allozymes displayed higher specific activities than the wild-type enzyme, with SULT1A3-P10L and SULT1A3-P101L showing the highest specific activities (Figure 5A). The other four SULT1A3 allozymes showed lower specific activities than the wild-type, with SULT1A3-N235T displaying the lowest specific activity. At higher substrate concentration (10 μM), all SULT1A3 allozymes showed specific activities that were close to that of the wild-type enzyme except SULT1A3-N235T which displayed a specific activity nearly 10 times lower than the wild-type (Figure 5B).
Figure 5.
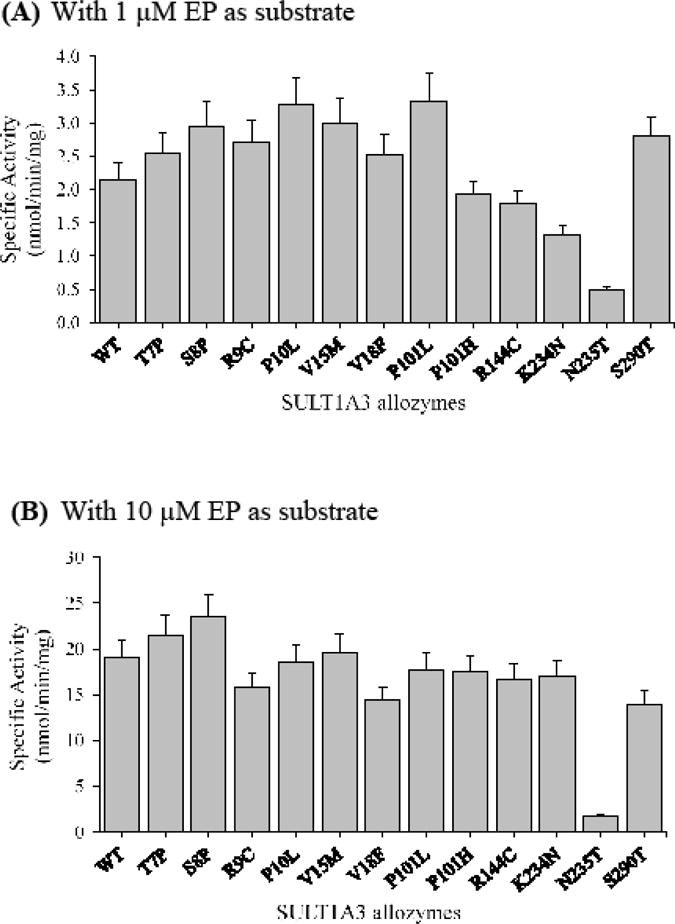
Specific activities of the sulfation of epinephrine (EP) by human SULT1A3 allozymes. Concentrations of epinephrine used in the enzymatic assays were 1 μM (A) and 10 μM (B). Specific activity refers to nmol epinephrine sulfated/min/mg of purified allozyme. Data shown represent mean ± standard deviation derived from three determinations. WT refers to wild-type SULT1A3.
With NE as the substrate
At low substrate (un-saturating) concentration (1 μM), all SULT1A3 allozymes exhibited lower NE-sulfating activities than the wild-type enzyme (Figure 6A). Among them, SULT1A3-R9C and SULT1A3-N235T showed much lower activities (~8 and 10 times, respectively) than the other ten SULT1A3 allozymes. At higher substrate concentration (10 μM), all SULT1A3 allozymes displayed similarly lower specific activities than the wild-type enzyme, with SULT1A3-R9C and SULT1A3-N235T again showing much lower specific activities than the rest (Figure 6B).
Figure 6.
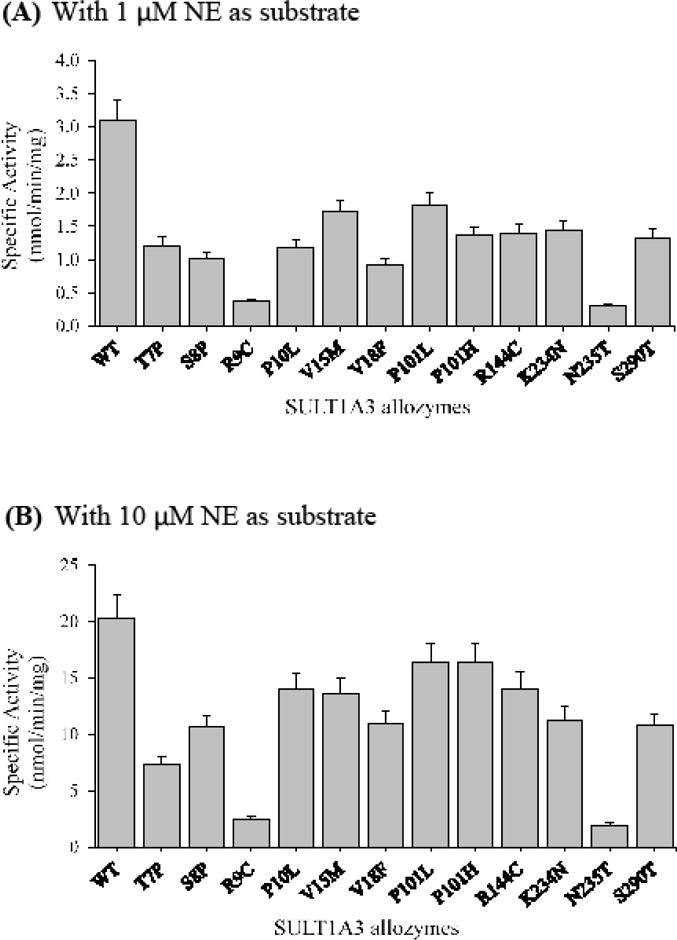
Specific activities of the sulfation of norepinephrine (NE) by human SULT1A3 allozymes. Concentrations of norepinephrine used in the enzymatic assays were 1 μM (A) and 10 μM (B). Specific activity refers to nmol norepinephrine sulfated/min/mg of purified allozyme. Data shown represent mean ± standard deviation derived from three determinations. WT refers to wild-type SULT1A3.
With 5-HT as the substrate
At low substrate (un-saturating) concentration (10 μM), SULT1A3-S8P displayed a slightly higher activity than the wild-type enzyme, while the other eleven SULT1A3 allozymes all showed lower specific activities than the wild-type enzyme, with SULT1A3-R9C, SULT1A3-R144C and SULT1A3-N235T showing the lowest 5-HT-sulfating activity than the rest (Figure 7A). At higher substrate concentration (100 μM), four SULT1A3 allozymes, SULT1A3-T7P, SULT1A3-S8P, SULT1A3-V15M and SULT1A3-P101H, displayed specific activities comparable to that of the wild-enzyme. The other eight SULT1A3 allozymes showed considerably lower specific activities than the wild-type enzyme, with SULT1A3-R9C, SULT1A3-R144C and SULT1A3-N235T again displaying much lower specific activities than the other five allozymes (Figure 7B).
3.4. Kinetic Analyses
To examine further the impact of the amino acid variations on the catecholamine/5-HT-sulfating activity of the SULT1A3 allozymes, kinetic studies were performed. Assays were carried out using varying concentrations of each of the four substrates (DA, EP, NE and 5-HT) at pH 7.4. Figure 8 shows the concentration-dependent sulfation of DA, EP, NE and 5-HT, respectively, by the wild-type SULT1A3. The arrow signs indicated the concentrations at which substrate inhibition was observed. Data obtained from the kinetic experiments using the wild-type and each of the twelve SULT1A3 allozymes were processed to generate Michaelis-Menten saturation curves and Lineweaver-Burk plots using GraphPad Prism 7 software for the determination of kinetic constants (Km, Vmax, and Vmax/Km).
Figure 8.
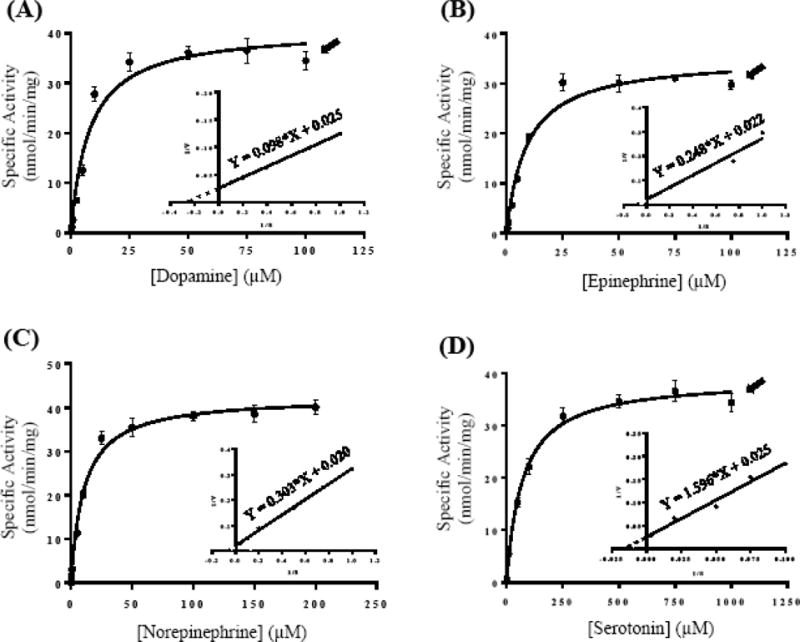
Kinetic analysis for the sulfation of catecholamines and serotonin by wild-type human SULT1A3. Panels (A), (B), (C) and (D) illustrate the Michaelis–Menten saturation curves for the sulfation of dopamine (DA), epinephrine (EP), norepinephrine (NE), and serotonin (5-HT), respectively. The insets show the Lineweaver-Burk plots generated based on the data shown in each of the four panels. Arrow signs indicate the concentrations at which substrate inhibition started taking place. Data shown represent calculated mean ± standard deviation derived from three experiments.
With DA as the substrate
As shown in Table 2, the Km value (6.46 ± 0.59 μM) determined for the wild-type SULT1A3 was lower than those of all SULT1A3 allozymes. Of the twelve SULT1A3 allozymes, SULT1A3-P101H, SULT1A3-R144C and SULT1A3-N235T, displayed Km values (12.72 ± 3.29, 13.26 ± 2.99 and 12.91 ± 1.29 μM, respectively) that were approximately two times that (6.46 ± 0.59 μM) of the wild-type. In terms of the Vmax, most SULT1A3 allozymes showed comparable values to that of the wild-type enzyme, except SULT1A3-K234N. The Vmax/Km values, reflecting the catalytic efficiency, of all SULT1A3 allozymes were all lower than that of the wild-type enzyme. Notably, three of them, SULT1A3-P101H, SULT1A3-R144C and SULT1A3-N235T, showed calculated values (3.55, 3.38 and 3.33, respectively) that were nearly half of that (6.32) of the wild-type enzyme.
Table 2.
Kinetic parameters of the wild-type human SULT1A3 and allozymes with dopamine as a substrate.
| SULT1A3 allozymes | Km (μM) | Vmax (nmol/min/mg) | Vmax/Km |
|---|---|---|---|
| 1A3-WT1 | 6.46 ± 0.59 | 40.82 ± 1.14 | 6.32 |
| 1A3-T7P | 8.50 ± 2.06 | 36.76 ± 2.28 | 4.32 |
| 1A3-S8P | 8.86 ± 1.70 | 44.15 ± 2.19 | 4.98 |
| 1A3-R9C | 9.06 ± 2.07 | 45.48 ± 2.69 | 5.02 |
| 1A3-P10L | 8.56 ± 2.35 | 42.82 ± 3.00 | 5.00 |
| 1A3-V15M | 8.84 ± 2.11 | 45.26 ± 2.78 | 5.12 |
| 1A3-V18F | 8.81 ± 2.41 | 41.32 ± 2.91 | 4.69 |
| 1A3-P101L | 9.06 ± 1.39 | 43.18 ± 1.72 | 4.77 |
| 1A3-P101H | 12.72 ± 3.29 | 45.10 ± 3.32 | 3.55 |
| 1A3-R144C | 13.26 ± 2.99 | 44.77 ± 2.90 | 3.38 |
| 1A3-K234N | 7.54 ± 2.09 | 30.74 ± 2.12 | 4.08 |
| 1A3-N235T | 12.91 ± 1.29 | 42.99 ± 1.21 | 3.33 |
| 1A3-S290T | 6.56 ± 1.77 | 35.03 ± 2.29 | 5.34 |
Wild-type human SULT1A3.
With EP as the substrate
As shown in Table 3, with small fluctuations, the Km values determined for all SULT1A3 allozymes were comparable to that of the wild-type enzyme, except for SULT1A3-N235T which showed a Km (200.1 ± 64.51 μM) more than twenty times that of the wild-type. A similar situation was found with the Vmax, with all SULT1A3 allozymes showing values comparable to that of the wild-type SULT1A3, except for SULT1A3-N235T. Based on these data, the calculated Vmax/Km of SULT1A3-N235T (0.13) was near 30 times lower than that of the wild-type (3.85). Of the rest of the SULT1A3 allozymes, only SULT1A3-S290T showed considerable lower value of Vmax/Km than the wild-type.
Table 3.
Kinetic parameters of the wild-type human SULT1A3 and allozymes with epinephrine as a substrate.
| SULT1A3 allozymes | Km (μM) | Vmax (nmol/min/mg) | Vmax/Km |
|---|---|---|---|
| 1A3-WT1 | 9.16 ± 1.81 | 35.30 ± 1.82 | 3.85 |
| 1A3-T7P | 8.90 ± 0.89 | 33.72 ± 0.87 | 3.79 |
| 1A3-S8P | 9.73 ± 0.47 | 35.60 ± 0.45 | 3.66 |
| 1A3-R9C | 9.44 ± 0.40 | 30.65 ± 1.34 | 3.25 |
| 1A3-P10L | 10.86 ± 0.48 | 38.66 ± 0.46 | 3.56 |
| 1A3-V15M | 9.05 ± 1.22 | 34.38 ± 1.20 | 3.80 |
| 1A3-V18F | 9.62 ± 1.08 | 30.69 ± 0.91 | 3.19 |
| 1A3-P101L | 8.83 ± 1.23 | 34.97 ± 1.26 | 3.96 |
| 1A3-P101H | 9.67 ± 1.71 | 33.95 ± 1.58 | 3.51 |
| 1A3-R144C | 11.19 ± 1.69 | 34.70 ± 1.42 | 3.10 |
| 1A3-K234N | 8.58 ± 1.24 | 29.30 ± 1.08 | 3.41 |
| 1A3-N235T | 200.1 ± 64.51 | 25.85 ± 6.05 | 0.13 |
| 1A3-S290T | 11.12 ± 0.45 | 29.60 ± 1.27 | 2.66 |
Wild-type human SULT1A3.
With NE as the substrate
As shown in Table 4, all SULT1A3 allozymes showed higher Km values than the wild-type enzyme, with SULT1A3-R9C and SULT1A3-N235T displaying much higher Km values (9.4 and 5.0 times that of the wild-type) than the rest. The Vmax values determined for all SULT1A3 allozymes were all lower than that of the wild-type except SULT1A3-P101L, whereas SULT1A3-N235T showed the lowest Vmax value which was only 30% that of the wild-type enzyme. Accordingly, while all SULT1A3 allozymes showed lower Vmax/Km than the wild-type, SULT1A3-R9C and SULT1A3-N235T showed much lower values (~14 and 16 times lower, respectively) than the wild-type.
Table 4.
Kinetic parameters of the wild-type human SULT1A3 and allozymes with norepinephrine as a substrate.
| SULT1A3 allozymes | Km (μM) | Vmax (nmol/min/mg) | Vmax/Km |
|---|---|---|---|
| 1A3-WT1 | 10.65 ± 1.14 | 42.53 ± 0.99 | 3.99 |
| 1A3-T7P | 18.78 ± 2.91 | 27.34 ± 1.07 | 1.36 |
| 1A3-S8P | 15.80 ± 3.02 | 32.62 ± 1.51 | 2.06 |
| 1A3-R9C | 99.98 ± 15.44 | 27.91 ± 3.24 | 0.28 |
| 1A3-P10L | 17.10 ± 1.85 | 37.23 ± 0.99 | 2.18 |
| 1A3-V15M | 13.17 ± 2.12 | 36.13 ± 1.34 | 2.74 |
| 1A3-V18F | 12.49 ± 3.53 | 27.82 ± 1.79 | 2.23 |
| 1A3-P101L | 13.41 ± 2.10 | 42.86 ± 1.55 | 3.20 |
| 1A3-P101H | 11.52 ± 2.54 | 35.92 ± 1.77 | 3.12 |
| 1A3-R144C | 13.49 ± 2.71 | 36.77 ± 1.71 | 2.73 |
| 1A3-K234N | 14.32 ± 2.70 | 32.43 ± 1.44 | 2.26 |
| 1A3-N235T | 53.46 ± 4.76 | 12.88 ± 0.41 | 0.24 |
| 1A3-S290T | 10.99 ± 1.62 | 22.92 ± 0.74 | 2.09 |
Wild-type human SULT1A3.
With 5-HT as the substrate
As shown in Table 5, all SULT1A3 allozymes showed higher Km values than the wild-type (at 71.38 ± 7.99 μM), with SUT1A3-R9C, SULT1A3-P10L, SULT1A3-P101L, SULT1A3-R144C, SULT1A3-N235T and SULT1A3-S290T displaying much higher Km values (216.6 ± 27.32, 102.40 ± 15.99, 213.9 ± 33.26, 281.0 ± 9.16, 8112.0 ± 866 and 165.6 ± 21.58 μM, respectively) than the rest. With respect to Vmax, the fluctuations were smaller among SULT1A3 allozymes, except for SULT1A3-N235T which displayed a Vmax which was three times that of the wild-type. Based on these data, six of the twelve SULT1A3 allozymes (SULT1A3-R9C, SULT1A3-P10L, SULT1A3-P101L, SULT1A3-R144C, SULT1A3-N235T and SULT1A3-S290T) showed notably lower Vmax/Km than the wild-type enzyme.
Table 5.
Kinetic parameters of the wild-type human SULT1A3 and allozymes with serotonin as a substrate.
| SULT1A3 allozymes | Km (μM) | Vmax (nmol/min/mg) | Vmax/Km |
|---|---|---|---|
| 1A3-WT1 | 71.38 ± 7.99 | 38.99 ± 0.98 | 0.55 |
| 1A3-T7P | 72.38 ± 6.29 | 32.22 ± 2.68 | 0.45 |
| 1A3-S8P | 72.75 ± 10.97 | 39.48 ± 1.45 | 0.54 |
| 1A3-R9C | 216.60 ± 27.32 | 26.19 ± 1.15 | 0.12 |
| 1A3-P10L | 102.40 ± 15.99 | 34.01 ± 1.46 | 0.33 |
| 1A3-V15M | 83.18 ± 11.32 | 34.51 ± 1.20 | 0.41 |
| 1A3-V18F | 78.08 ± 10.24 | 29.07 ± 0.95 | 0.37 |
| 1A3-P101L | 213.90 ± 33.26 | 34.49 ± 1.87 | 0.16 |
| 1A3-P101H | 76.49 ± 6.82 | 37.90 ± 2.90 | 0.50 |
| 1A3-R144C | 281.00 ± 9.16 | 24.17 ± 1.30 | 0.09 |
| 1A3-K234N | 73.41 ± 7.92 | 25.38 ± 1.66 | 0.35 |
| 1A3-N235T | 8112.0 ± 866 | 117.50 ± 14.3 | 0.014 |
| 1A3-S290T | 165.60 ± 21.58 | 26.98 ± 1.13 | 0.16 |
Wild-type human SULT1A3.
4. Discussion
Previous studies have demonstrated sulfoconjugation as an important pathway in the biotransformation of catecholamines and 5-HT in humans, and SULT1A3 was identified as the major enzyme responsible for the sulfation of these monoamine neurotransmitters/hormones [24,25,39,40]. The current study aimed to systematically evaluate the effects of the genetic polymorphisms on the sulfating activity of SULT1A3 allozymes. Genetic polymorphism of human SULT1A3 was first reported in a study of a group of 232 individuals in which considerable variations in catecholamine-sulfating activity in the platelet samples prepared from these subjects were found [41]. In a later study using DNA samples from 60 African-American and 60 Caucasian-American subjects, a non-synonymous cSNP of the SULT1A3 gene was detected [30]. In a similar study, three additional non-synonymous cSNPs of SULT1A3 were found, and the corresponding SULT1A3 allozymes expressed were shown to exhibit varying DA-sulfating activity [28]. It therefore appears that the genetic polymorphisms may play a critical role in affecting the functional activity of SULT1A3 protein products, and such differences may possibly impact on the metabolism of catecholamine/5-TH through sulfation in individuals with different SULT1A3 and SULT1A4 genotypes. In the current study, we performed a systematic database search for human SULT1A3 and SULT1A4 SNPs. Thirteen missense cSNPs were identified. Site-directed mutagenesis was used to generate cDNAs for the expression of corresponding SULT1A3 allozymes. Twelve of the thirteen SULT1A3 allozymes were successfully expressed and purified, with one being present in inclusion body form and could not be purified.
The twelve SULT1A3 allozymes prepared were first analyzed for their catecholamine/5-HT-sulfating activity in comparison with the wild-type enzyme. Activity data shown in Figures 4–7 revealed differential DA-, EP-, NE-, and 5-HT-sulfating activity among all twelve SULT1A3 allozymes. It was noted that the variations in DA-sulfating activities of SULT1A3 allozymes were markedly smaller than the variations in their sulfating activities toward EP, NE, and 5-HT. It should be pointed out that of the twelve SULT1A3 allozymes examined, four, SULT1A3-P101L, SULT1A3-P101H, SULT1A3-R144C and SULT1A3-K234N, had been studied previously [28,30]. While the results reported in the previous studies showed some minor differences in comparison with the results obtained in the current study, the trend of the variations in their DA-sulfating activity appeared the same. It is noted that different DA and PAPS concentrations were used in the assays performed in the previous and current studies. Moreover, purified SULT1A3 allozymes were used in the current study, as compared with transfected COS-1 cell lysates used in the previous studies [28,30].
Kinetic constants shown in Tables 2–5 revealed distinct substrate affinity (as reflected by the Km) and catalytic activity (as reflected by the Vmax) of different SULT1A3 allozymes in catalyzing the sulfation of DA, EP, NE, and 5-HT. It was noted that despite their considerable differences in Km and Vmax with each of the four substrates, variations in catalytic efficiency (as reflected by Vmax/Km) were found to be smaller with DA than with any of the other three compounds as substrate. With DA as substrate, variations in Vmax/Km were less than 47.3 % for all twelve SULT1A3 allozymes in comparison with the wild-type enzyme. With EP, NE, or 5-HT as substrate, variations in Vmax/Km were found to be as high as 99.96, 99.94, and 99.84 times, compared with the wild-type. For example, SULT1A3-N235T, which showed the lowest specific activities (cf. Figures 4–7) and catalytic efficiencies (Tables 2–5) among all SULT1A3 allozymes, while displaying a catalytic efficiency approximately 52.6 % that of the wild-type enzyme toward DA, exhibited much lower catalytic efficiencies (3.3 %, 6 %, and 2.5 %, respectively) toward EP, NE and 5-HT. Whether the lower degree of variations in DA-sulfating activity of SULT1A3 allozymes indicates the critical importance of the maintenance of the homeostasis of dopamine, in comparison with other monoamine compounds, remains to be clarified. As noted earlier, studies have shown that more than 98% of DA in circulation is present in sulfoconjugated form [15], and sulfoconjugation has been reported as a high capacity (not easily saturated) pathway involved in the biotransformation of DA [42,43]. Moreover, DA, among other monoamines, has been demonstrated to induce its own metabolism by SULT1A3, possibly for protecting neurons from elevated DA levels, and that abnormal SULT1A3 activity may pose as a risk factor for DA-associated neurotoxicity [44]. In relation to this latter point, it has been reported that abnormal levels of catecholamines and 5-HT and/or their conjugates correlated with certain pathological conditions, including neurodegenerative diseases [45–47] attention deficit hyperactivity disorder (ADHD) [48,49], migraine [50,51], atrial fibrillation and blood pressure changes [51], and myocardial infarction [52,53].
Several crystal structures of SULT1A3 have been reported [26,37,54]. Some of the structural elements involved in the functioning of the enzyme include a catalytically important residue His108, the PAPS interacting regions (residues 45TYPKSGTT52, Arg130, Ser138, and residues 257RKG259), the substrate binding regions (including particularly, residues Asp86 and Glu146) which were proposed to play crucial roles in substrate specificity and sulfating activity [37], the C-terminal dimerization motif (residues Lys265-Glu274) with the sequence KXXXTVXXXE [55], as well as the N-terminal region that contains βA- and βB-sheets which were thought to be structural components important for the SULT folding [56]. Moreover, three loop segments, Asp66-Met77, Ser228-Gly259, and Lys85-Pro90, have been proposed to be involved in the formation of a gate that governs the substrate selectivity (cf. Figure 2) [38]. Of the SULT1A3 allozymes investigated, SULT1A3-T7P, SULT1A3-S8P and SULT1A3-R9C contain amino acid substitutions in the N-terminal region. It was noted that the substitutions of a polar amino acid (Thr, Ser, or Arg) with a non-polar or turn-inducing amino acid (Pro or Cys) in these there SULT1A3 allozymes resulted in lower catalytic efficiencies toward the four substrates tested (DA, EP, NE and 5-HT). From the perspective of the substrate, the differences in the chemical structures of the four monoamine compounds may also contribute to the differential catalytic efficiencies of these SULT1A3 allozymes. In the case of SULT1A3-R9C allozyme, the replacement of Arg, a positively charged amino acid, may possibly abolish H-bonding, salt-bridge, and/or van der Waals interactions [57–59]. Among the four monoamine substrates, NE and 5-HT were more highly affected by the R9C substitution, which might be related to the differences in the positions of their constituent functional groups. The substitution of a non-polar or turn-inducing amino acid (Pro or Val) with a non-polar or an aromatic amino acid (Leu, Met, or Phe) in SULT1A3-P10L, SULT1A3-V15M and SULT1A3-V18F also resulted in lower catalytic efficiencies. It appeared that Leu that replaces Pro in SULT1A3-P10L allozyme might have resulted in conformational changes that rendered the decrease in catalytic efficiency of this allozyme. Specifically, the substituted Val residue in SULT1A3-V15M and SULT1A3-V18F carries a more bulky side chain, which originally may compel the enzyme to adopt a relatively more restricted conformation. The results obtained with these two allozymes appeared to be compatible with the postulation that βA- and βB-sheets present in the N-terminal region represent important structural components of the SULT1A3 molecule [56]. It should be pointed out that the three N-terminal residues (Thr7, Ser8, and Arg9) as well as Val15, Val18, and Arg144 (as discussed below) are located near the proposed gate through which substrates must pass to enter the active site [38]. SULT1A3-P101L and SULT1A3-P101H, both with a turn-inducing proline residue replaced by either a non-polar Leu or a basic His residue, also showed differential sulfating efficiencies toward the four monoamine substrates. A previous study indicated that amino acid residues 84–104, together with residues 145–154, are involved in reshaping the substrate binding pocket, narrowing its cavity volume, and enabling the substrate binding [37]. SULT1A3-R144C showed lower catalytic efficiencies toward the four substrates tested (particularly, DA and 5-HT). Substitution of a basic Arg residue with a non-polar Cys residue is considered non-conservative, which could in part explain the impairment in the catalytic efficiency of this allozyme. Additionally, Arg144 is located within the amino acid residues 143–148 segment, which has been proposed to contribute to substrate-binding and catalysis of both human SULT1A1 and SULT1A3 [60]. Spatially, Arg144 residue is very close to the amino acid segment spanning residues 145–154, which has been proposed to be involved in the substrate binding pocket [37]. SULT1A3-K234N and SULT1A3-N235T carry amino acid substitutions close to the C-terminal region. Both these two SULT1A3 allozymes showed lower catalytic efficiencies than the wild-type enzyme toward the four substrates tested, with SULT1A3-N235T displaying the lowest specific activity and catalytic efficiency with DA, EP, NE and 5-HT in fact among all 12 SULT1A3 allozymes analyzed. Lys234 and Asn235 residues in the wild-type enzyme are located in the α15 sheet as revealed in the SULT1A3 crystal structure. This region has been proposed to be involved indirect binding of PAPS. These two residues also may contribute to restricting the conformations that allow for substrate binding when the c-substrate (PAPS) is bound [56]. Moreover, Lys234 and Asn235 residues are associated with a segment (Loop 2; Ser228-Gly259) that constitutes the substrate access gate [38] that governs the substrate selectivity. In the case of SULT1A3-N235T, the bulky side chain of the substituting Thr may have more difficulty in fitting into the alpha-helical element as revealed in the SULT1A3 crystal structure. Previous studies have shown that a common SULT1A1 allozyme (SULT1A1-Asn235Thr) displayed a substantially higher Km toward 4-nitrophenol [61,62]. For SULT1A3-S290T, although the substitution of Ser with Thr does not seem to be dramatic from the chemistry standpoint, it showed differential catalytic efficiencies toward monoamines, with a bigger variation with 5-HT and lower variation with DA. Whether these variations are due to its close proximity (position 290) to the conserved KTVE motif as revealed in the SULT1A3 crystal structure [26,55] remains to be clarified.
To summarize, we have generated, expressed, and purified twelve of the thirteen known human SULT1A3 allozymes. Enzymatic characterization of the purified SULT1A3 allozymes revealed differential substrate binding affinity and catalytic activity toward the four monoamines tested as substrates. These results may have implications in the differential metabolism of monoamine neurotransmitters in individuals with distinct SULT1A3/ SULT1A4 genotypes that code for different SULT1A3 allozymes. Moreover, pending further studies, the results obtained may provide clues to the link of particular SULT1A3/SULT1A4 genotype s to certain neuropathological disorders associated with abnormal levels of the monoamines that are used as substrates by SULT1A3.
Acknowledgments
This work was supported in part by a grant from National Institutes of Health (Grant # R03HD071146).
Abbreviations
- DA
dopamine
- EP
epinephrine
- NE
norepinephrine
- 5-HT
serotonin
- PAPS
3′-phosphoadenosine-5′-phosphosulfate
- SULT
cytosolic sulfotransferase
- TLC
thin-layer chromatography
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldstein DS, Swoboda KJ, Miles JM, Coppack SW, Aneman A, Holmes C, Lamensdorf I, Eisenhofer G. Sources and physiological significance of plasma dopamine sulfate. J Clin Endocrinol Metab. 1999;84:2523–2531. doi: 10.1210/jcem.84.7.5864. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein DS, Eisenhofer G, Kopin IJ. Sources and significance of plasma levels of catechols and their metabolites in humans. J Pharmacol Exp Ther. 2003;305:800–811. doi: 10.1124/jpet.103.049270. [DOI] [PubMed] [Google Scholar]
- 3.Sandyk R. Serotonergic mechanisms in amyotrophic lateral sclerosis. Int J Neurosci. 2006;116:775–826. doi: 10.1080/00207450600754087. [DOI] [PubMed] [Google Scholar]
- 4.Alex KD, Pehek EA. Pharmacologic mechanisms of serotonergic regulation of dopamine neurotransmission. Pharmacol Ther. 2007;113:296–320. doi: 10.1016/j.pharmthera.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schultz W. Multiple dopamine functions at different time courses. Annu Rev Neurosci. 2007;30:259–288. doi: 10.1146/annurev.neuro.28.061604.135722. [DOI] [PubMed] [Google Scholar]
- 6.Eisenhofer G, Kopin IJ, Goldstein DS. Catecholamine metabolism: a contemporary view with implications for physiology and medicine. Pharmacol Rev. 2004;56:331–349. doi: 10.1124/pr.56.3.1. [DOI] [PubMed] [Google Scholar]
- 7.Buttarelli FR, Fanciulli A, Pellicano C, Pontieri FE. The dopaminergic system in peripheral blood lymphocytes: from physiology to pharmacology and potential applications to neuropsychiatric disorders. Curr Neuropharmacol. 2011;9:278–288. doi: 10.2174/157015911795596612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lieberman M, Marks A, Peet A. Marks’ Basic Medical Biochemistry, A Clinical Approach. 4th. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. p. 175. [Google Scholar]
- 9.Moreira CG, Russell R, Mishra AA, Narayanan S, Ritchie JM, Waldor MK, Curtis MM, Winter SE, Weinshenker D, Sperandio V. Bacterial Adrenergic Sensors Regulate Virulence of Enteric Pathogens in the Gut. MBio. 2016;7:e00826–16. doi: 10.1128/mBio.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397–414. doi: 10.1053/j.gastro.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Mawe GM, Coates MD, Moses PL. Review article: intestinal serotonin signalling in irritable bowel syndrome. Aliment Pharmacol Ther. 2006;23:1067–1076. doi: 10.1111/j.1365-2036.2006.02858.x. [DOI] [PubMed] [Google Scholar]
- 12.Camilleri M. Serotonin in the gastrointestinal tract. Curr Opin Endocrinol Diabetes Obes. 2009;16:53–59. doi: 10.1097/med.0b013e32831e9c8e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lozda R, Purviņš I. The serotonin-O-sulfate as a potential plasma surrogate biomarker. Neurotransmitter. 2014;1:e281. [Google Scholar]
- 14.Tyce GM, Messick JM, Yaksh TL, Byer DE, Danielson DR, Rorie DK. Amine sulfate formation in the central nervous system. Fed Proc. 1986;45:2247–2253. [PubMed] [Google Scholar]
- 15.Johnson GA, Baker CA, Smith RT. Radioenzymatic assay of sulfate conjugates of catecholamines and DOPA in plasma. Life Sci. 1980;26:1591–1598. doi: 10.1016/0024-3205(80)90362-8. [DOI] [PubMed] [Google Scholar]
- 16.Falany CN. Enzymology of human cytosolic SULTs. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 17.Renskers KJ, Feor KD, Roth JA. Sulfation of dopamine and other biogenic amines by human brain phenol sulfotransferase. J Neurochem. 1980;34:1362–1368. doi: 10.1111/j.1471-4159.1980.tb11216.x. [DOI] [PubMed] [Google Scholar]
- 18.Costa JL, Launay JM, Kirk KL. Exploration of the role of phenolsulfotransferase in the disposition of serotonin in human platelets: implications for a novel therapeutic strategy against depression. Med Hypotheses. 1983;10:231–246. doi: 10.1016/0306-9877(83)90113-5. [DOI] [PubMed] [Google Scholar]
- 19.Reiter C, Mwaluko G, Dunnette J, Van Loon J, Weinshilboum R. Thermolabile and thermostable human platelet phenol sulfotransferase. Substrate specificity and physical separation. Naunyn Schmiedebergs Arch Pharmacol. 1983;324:140–147. doi: 10.1007/BF00497020. [DOI] [PubMed] [Google Scholar]
- 20.Mulder GJ, Jakoby WB. Sulfation in conjugation reactions. In: Mulder GJ, Jakoby WB, editors. Drug Metabolism. Taylor and Francis; London: 1990. pp. 107–161. [Google Scholar]
- 21.Falany C, Roth JA. Properties of human cytosolic sulfotransferases involved in drug metabolism. In: Jeffery EH, editor. Human Drug Metabolism; From Molecular Biology to Man. CRC Press; Boca Raton, FL: 1993. pp. 101–115. [Google Scholar]
- 22.Weinshilboum R, Otterness D. Sulfotransferase enzymes. In: Kaufmann FC, editor. Conjugation-Deconjugation Reactions in Drug Metabolism and Toxicity. Springer-Verlag; Berlin: 1994. pp. 45–78. [Google Scholar]
- 23.Lipmann F. Biological sulfate activation and transfer. Science. 1958;128:575–580. doi: 10.1126/science.128.3324.575. [DOI] [PubMed] [Google Scholar]
- 24.Dousa MK, Tyce GM. Free and conjugated plasma catecholamines, DOPA and 3-O-methyldopa in humans and in various animal species. Proc Soc Exp Biol Med. 1988;188:427–434. doi: 10.3181/00379727-188-42755. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda S, Liu MY, Suiko M, Sakakibara Y, Liu MC. Hydroxylated serotonin and dopamine as substrates and inhibitors for human cytosolic SULT1A3. J Neurochem. 2007;103:2679–2689. doi: 10.1111/j.1471-4159.2007.04948.x. [DOI] [PubMed] [Google Scholar]
- 26.Dajani R, Cleasby A, Neu M, Wonacott AJ, Jhoti H, Hood AM, Modi S, Hersey A, Taskinen J, Cooke RM, Manchee GR, Coughtrie MW. X-ray crystal structure of human dopamine sulfotransferase, SULT1A3. Molecular modeling and quantitative structureactivity relationship analysis demonstrate a molecular basis for sulfotransferase substrate specificity. J Biol Chem. 1999;274:37862–37868. doi: 10.1074/jbc.274.53.37862. [DOI] [PubMed] [Google Scholar]
- 27.Dooley TP. Cloning of the human phenol sulfotransferase gene family: three genes implicated in the metabolism of catecholamines, thyroid hormones and drugs. Chem Biol Interact. 1998;109:29–41. doi: 10.1016/s0009-2797(97)00118-x. [DOI] [PubMed] [Google Scholar]
- 28.Hildebrandt MA, Salavaggione OE, Martin YN, Flynn HC, Jalal S, Wieben ED, Weinshilboum RM. Human SULT1A3 pharmacogenetics: gene duplication and functional genomic studies. Biochem Biophys Res Commun. 2004;321:870–878. doi: 10.1016/j.bbrc.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 29.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 30.Thomae BA, Rifki OF, Theobald MA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catecholamine sulfotransferase (SULT1A3) pharmacogenetics: Functional genetic polymorphism. J Neurochem. 2003;87:809–819. doi: 10.1046/j.1471-4159.2003.02027.x. [DOI] [PubMed] [Google Scholar]
- 31.Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, Takayanagi K, Natori Y, Liu MC. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5′-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem. 1998;62:1037–1040. doi: 10.1271/bbb.62.1037. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro AL, Viñuela E, Maizel JJ. Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem Biophys Res Commun. 1967;28:815–20. doi: 10.1016/0006-291x(67)90391-9. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 36.Kurogi K, Chen M, Lee Y, Shi1 B, Yan T, Liu M. Sulfation of buprenorphine, pentazocine, and naloxone by human cytosolic sulfotransferases. Drug Metab Lett. 2012;6:109–115. [PubMed] [Google Scholar]
- 37.Lu JH, Li HT, Liu MC, Zhang JP, Li M, An XM, Chang WR. Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3′-phosphoadenosine 5′-phosphate. Biochem Biophys Res Commun. 2005;335:417–423. doi: 10.1016/j.bbrc.2005.07.091. [DOI] [PubMed] [Google Scholar]
- 38.Cook I, Wang T, Almo SC, Kim J, Falany CN, Leyh TS. The gate that governs sulfotransferase selectivity. Biochemistry. 2013;52:415–424. doi: 10.1021/bi301492j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritchter D. The inactivation of adrenaline in vivo in man. J Physiol. 1940;98:361–374. doi: 10.1113/jphysiol.1940.sp003854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinshilboum RM. Sulfate conjugation of neurotransmitters and drugs. Introduction Fed Proc. 1986;45:2220–2222. [PubMed] [Google Scholar]
- 41.Price RA, Cox NJ, Spielman RS, Van Loon JA, Maidak BL, Weinshilboum RM. Inheritance of human platelet thermolabile phenol sulfotransferase (TL PST) activity. Genet Epidemiol. 1988;5:1–15. doi: 10.1002/gepi.1370050102. [DOI] [PubMed] [Google Scholar]
- 42.Claustre J, Pequignot JM, Bui-xuan B, Muchada R, Cottet-Emard RM, Peyrin L. Conjugation and deamination of circulating dopamine: relationship between sulfated and free dopamine in man. J Auton Nerv Syst. 1990;29:175–181. doi: 10.1016/0165-1838(90)90183-j. [DOI] [PubMed] [Google Scholar]
- 43.Eisenhofer G, Coughtrie MW, Goldstein DS. Dopamine sulphate: an enigma resolved. Clin Exp Pharmacol Physiol Suppl. 1999;26:S41–53. [PubMed] [Google Scholar]
- 44.Sidharthan NP, Minchin RF, Butcher NJ. Cytosolic sulfotransferase 1A3 is induced by dopamine and protects neuronal cells from dopamine toxicity: role of D1 receptor-NMDA receptor coupling. J Biol Chem. 2013;288:34364–34374. doi: 10.1074/jbc.M113.493239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S, Kulhara P. What is schizophrenia: A neurodevelopmental or neurodegenerative disorder or a combination of both? A critical analysis. Indian J Psychiatry. 2010;52:21–27. doi: 10.4103/0019-5545.58891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ungless MA, Grace AA. Are you or aren’t you? Challenges associated with physiologically identifying dopamine neurons. Trends Neurosci. 2012;35:422–430. doi: 10.1016/j.tins.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stokes AH, Hastings TG, Vrana KE. Cytotoxic and genotoxic potential of dopamine. J Neurosci Res. 1999;55:659–665. doi: 10.1002/(SICI)1097-4547(19990315)55:6<659::AID-JNR1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 48.Wu J, Xiao H, Sun H, Zou L, Zhu LQ. Role of dopamine receptors in ADHD: a systematic meta-analysis. Mol Neurobiol. 2012;45:605–620. doi: 10.1007/s12035-012-8278-5. [DOI] [PubMed] [Google Scholar]
- 49.Eagle K. ADHD impacted by sulfotransferase (SULT1A) inhibition from artificial food colors and plant-based foods. Physiol Behav. 2014;135:174–179. doi: 10.1016/j.physbeh.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Jones AL, Roberts RC, Colvin DW, Rubin GL, Coughtrie MW. Reduced platelet phenolsulphotransferase activity towards dopamine and 5-hydroxytryptamine in migraine. Eur J Clin Pharmacol. 1995;49:109–114. doi: 10.1007/BF00192368. [DOI] [PubMed] [Google Scholar]
- 51.Eagle K. Toxicological effects of red wine, orange juice, and other dietary SULT1A inhibitors via excess catecholamines. Food Chem Toxicol. 2012;50:2243–2249. doi: 10.1016/j.fct.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Benedict CR, Grahame-Smith DG. Plasma adrenaline and noradrenaline concentrations and dopamine-beta-hydroxylase activity in myocardial infarction with and without cardiogenic shock. Br Heart J. 1979;42:214–220. doi: 10.1136/hrt.42.2.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nadeau RA, de Champlain J. Plasma catecholamines in acute myocardial infarction. Am Heart J. 1979;98:548–554. doi: 10.1016/0002-8703(79)90278-3. [DOI] [PubMed] [Google Scholar]
- 54.Bidwell LM, McManus ME, Gaedigk A, Kakuta Y, Negishi M, Pedersen L, Martin JL. Crystal structure of human catecholamine sulfotransferase. J Mol Biol. 1999;293:521–530. doi: 10.1006/jmbi.1999.3153. [DOI] [PubMed] [Google Scholar]
- 55.Petrotchenko EV, Pedersen LC, Borchers CH, Tomer KB, Negishi M. The dimerization motif of cytosolic sulfotransferases. FEBS Letters. 2001;490:39–43. doi: 10.1016/s0014-5793(01)02129-9. [DOI] [PubMed] [Google Scholar]
- 56.Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, Loppna P, Martin F, Thonton J, Edwards AM, Bochkarev A, Plotnikov AN, Vedadi M, Arrowsmith CH. Structural and Chemical Profiling of the Human Cytosolic Sulfotransferases. PLoS Biol. 2007;5:e97. doi: 10.1371/journal.pbio.0050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flocco MM, Mowbray SL. Planar stacking interactions of arginine and aromatic side-chains in proteins. J Mol Biol. 1994;235:709–717. doi: 10.1006/jmbi.1994.1022. [DOI] [PubMed] [Google Scholar]
- 58.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Armstrong CT, Mason PE, Anderson JL, Dempsey CE. Arginine side chain interactions and the role of arginine as a gating charge carrier in voltage sensitive ion channels. Sci Rep. 2016;6:21759. doi: 10.1038/srep21759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen G, Chen X. Arginine Residues in the Active Site of Human Phenol Sulfotransferase (SULT1A1) J Biol Chem. 2003;278:36358–36364. doi: 10.1074/jbc.M306045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coughtrie MW. Sulfation through the looking glass-recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2:297–308. doi: 10.1038/sj.tpj.6500117. [DOI] [PubMed] [Google Scholar]
- 62.Zhu X, Veronese ME, Iocco P, McManus ME. cDNA cloning and expression of a new form of human aryl sulfotransferase. Int J Biochem Cell Biol. 1996;28:565–571. doi: 10.1016/1357-2725(95)00164-6. [DOI] [PubMed] [Google Scholar]
- 63.Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]


