Abstract
Emerging evidence points to a strong association between the gut microbiota and the risk, development and progression of gastrointestinal cancers such as colorectal cancer (CRC) and hepatocellular carcinoma (HCC). Bile acids are produced in the liver and are metabolized by enzymes derived from intestinal bacteria, and are critically important for maintenance of a healthy gut microbiota, balanced lipid and carbohydrate metabolism, insulin sensitivity and innate immunity. Given the complexity of bile acid signalling and the direct biochemical interactions between the gut microbiota and the host, a systems biology perspective is required to understand the liver–bile acid– microbiota axis and its role in gastrointestinal carcinogenesis in order to reverse the microbiota-mediated alterations in bile acid metabolism that occur in disease states. An examination of the recent progress of research in this area is urgently needed. In this Review, we discuss the mechanistic links between bile acids and gastrointestinal carcinogenesis in CRC and HCC, which involve two major bile acid-sensing receptors, farnesoid X receptor (FXR) and G protein-coupled bile acid receptor (TGR5). We also highlight the strategies and cutting-edge technologies to target the gut microbiota-dependent alterations in bile acid metabolism in the context of the cancer therapy.
ETOC BLURB
Bile acids link the gut microbiota to both hepatic and intestinal metabolism, and this tripartite relationship has been implicated in gastrointestinal disease. In this Review, the authors outline the mechanistic links between bile acid–microbiota crosstalk and gastrointestinal inflammation and carcinogenesis, with specific emphasis on the major bile acid sensing receptors.
Introduction
The pathogenesis of human gastrointestinal cancers involves extensive and complex gene–environment interactions1. Cancers of the gastrointestinal tract represent the leading cause of mortality worldwide2. In the United States, colorectal cancer (CRC) is the third most common cancer and the second most common cause of cancer-related deaths3. Hepatocellular carcinoma (HCC), the most common primary cancer of the liver, is the fifth most common cancer in men (~554,000 cases) and the ninth most common cancer in women (~228,000 cases) globally4 and is the third leading cause of cancer-related death5. In the United States, it is estimated that there will be ~135,430 new cases of CRC and ~40,710 new cases of liver cancer (~90% of primary liver cancers in the United States are HCC) in 20173,6.
The human gastrointestinal tract is colonized by a diverse range of symbiotic bacteria and other microorganisms that are collectively known as the gut microbiota7,8. The host genome and microbiome (that is, the collective genomes of the gut microbiota) co-produce a large array of metabolites that serve as important signalling factors and energy substrates. Such metabolites include: bile acids such as deoxycholic acid (DCA) and lithocholic acid (LCA), which are involved in digestive processes, or represent etiological agents and mediators of the pathogenic process9,10; choline, which is involved in transportation of lipids, methylation reactions and neurotransmitter synthesis11; neurotransmitters such as glycine, glutamate, γ-aminobutyric acid (GABA)12; and short-chain fatty acids (SCFAs) such as acetate and butyrate, which shape the composition of the gut microbiota and thus influence colon physiology and serve as energy sources by host cells and the intestinal microbiota13,14. The cross-talk between this dynamic profile of small molecule metabolites — known as the metabolome — and the host modulates the immune system, regulates metabolic phenotypes, and influences risk factors for disease and response to therapy15,16.
Mounting evidence obtained from studies in humans and animal models has revealed the tumor-promoting effects of the gut microbiota, and has thus linked the microbiota to gastrointestinal carcinogenesis17–19. An example of bacterial pathogen-induced carcinogenesis is Helicobacter pylori–induced gastric cancer20; H. pylori triggers gastric atrophy and hypochlorhydria through the secretion of a virulence factor, cytotoxin-associated gene A (CagA), which renders the stomach susceptible to the overgrowth of other pH-sensitive bacterial species such as Acinetobacter, and subsequently leads to an increase in the rate at which bacteria convert dietary nitrates into carcinogens21. Another example is gallbladder cancer, which is associated with chronic infection with Salmonella enterica subsp. enterica serovar Typhi and Salmonella enterica subsp. enterica serovar Paratyphi22. S. Typhi has a mutagenic effect on the gallbladder epithelium by metabolizing bile salts into carcinogenic secondary bile acids and other genotoxins23. In addition, enterotoxigenic bacteria, such as Fusobacterium nucleatum, Escherichia coli NC101 and Bacteroides fragilis promote CRC development24,25. These bacteria can either: induce chronic inflammation and increase reactive oxygen species (ROS)-mediated genotoxicity; or directly promote carcinogenesis by secreting toxins that induce DNA damage responses, such as adhesin protein FadA from F. nucleatum, and polyketide synthases from E. coli26. Indeed, suppression of intestinal bacteria by antibiotic treatment in mice reduced the development of colon and liver cancer18,27.
Among the diverse array of endogenous metabolites derived from host–gut microbiota co-metabolism, bile acids are currently receiving increased attention due to their known tumor-promoting properties18,28–30. Bile acids are synthesized in the liver and have direct or indirect antimicrobial effects; bile acids can modulate the composition of the microbiota, which in turn regulates the size and composition of the bile acid pool. Bile acids are recognized as the endogenous ligands31,32 for a number of ligand-activated transcription factors of the nuclear hormone receptor family including farnesoid X receptor (FXR),31 G-protein-coupled bile acid receptor (TGR5),33,34 pregnane X receptor (PXR),35 vitamin D3 receptor (VDR)36 and constitutive androstane receptor (CAR)35. Accumulating evidence has demonstrated a crucial role for bile acids in gastrointestinal carcinogenesis.The gut bacterial metabolite DCA has been shown to promote the development of obesity-associated HCC in a mouse model pretreated with 7,12-dimethylbenzanthracene18. In addition, exposure of hepatocytes to DCA induced expression of inflammatory genes which is closely associated with the development of cancer37.
There has been a recent surge in research interest in the role of bile acid–microbiota crosstalk in gastrointestinal cancer, and the field has quickly advanced, and will undoubtedly continue to progress at a fast pace. In this Review, we discuss the biosynthesis, metabolism and transport of bile acids, with focus on biosynthetic pathways, amidation and key bile acid transporters. We will also highlight the mechanistic links between the gut microbiota, bile acids, and gastrointestinal carcinogenesis through the major bile acid sensing receptors, FXR, TGR5, PXR, CAR and VDR. Finally, the mechanistic roles of bile acids and microbiota in the development of HCC and CRC will be reviewed, and opportunities for control and prevention of gastrointestinal cancers by targeting the liver–bile acid–microbiota metabolic axis will be presented.
Enterohepatic circulation of bile acids
The processes of bile acid synthesis, transport metabolism and regulation has been extensively reviewed elsewhere34,38,39. In this section, we will therefore focus the elemental knowledge of bile acid enterohepatic circulation to provide context for later discussion of their role in gastrointestinal inflammation and carcinogenesis.
Bile acid synthesis, transport, and metabolism
Bile acids are synthesized in hepatocytes via cytochrome P450-mediated oxidation of cholesterol, which can occur through two biosynthetic pathways; the ‘classical’ and ‘alternative’ pathways (FIG. 1)40. The ‘classical’ pathway produces the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA), through the enzymatic actions of three cholesterol hydroxylase enzymes; CYP7A1, CYP8B1, and CYP27A133. The ‘alternative’ pathway yields CDCA via the hydroxylation of the cholesterol side chain by CYP27A1, followed by 7-α hydroxylation (that is, addition of a 7α-hydroxy group) by CYP7B141 to form the oxysterol intermediates.
Figure 1. Bile acid biosynthesis, transport and metabolism.
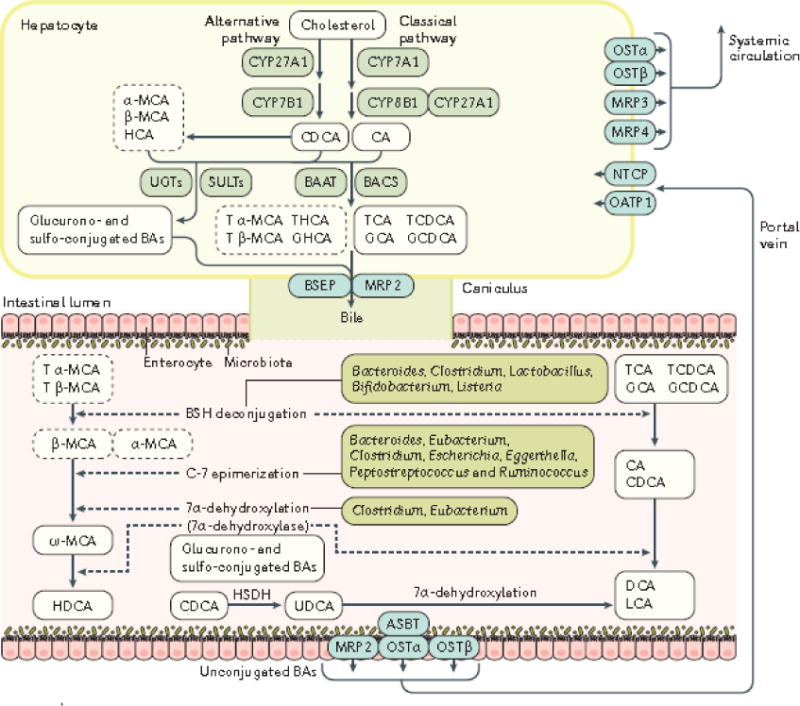
The most abundant bile acids in mammals are the primary bile acids CA and CDCA, and the secondary bile acids DCA and LCA. Bile acids are synthesized in hepatocytes via cytochrome P450-mediated oxidation of cholesterol, which occurs through two pathways: the ‘classical’ and ‘alternative’ pathways. The ‘classical’ pathway is initiated by CYP7A1 and produces the primary bile acids cholic acid (CA) and chenodeoxycholic acid (CDCA) through the subsequent enzymatic action of CYP8B1, and CYP27A1, The ‘alternative’ pathway is initiated by CYP27A1 and yields CDCA via CYP7B. Expression of CYP8B1 determines the ratio of cholic acid (CA) to chenodeoxycholic acid (CDCA) by promoting CA biosynthesis via the ‘classical’ pathway. In the rodent liver, the majority of CDCA is converted to α-muricholic acid (MCA and β-MCA); in the pig, CDCA is primarily converted to hyocholic acid (HCA), whereas in humans it remains as CDCA56. In hepatocytes, most bile acids are conjugated to glycine (glyco-, G) or taurine (tauro-, T) through the action of bile acid:CoA synthetase (BACS) and bile acid-CoA:amino acid N-acyltransferase (BAAT) prior to their secretion into bile via bile salt export pump (BSEP). Differences in the types of congugated bile acids produced exist between human and rodents; the solid line and dashed line rectangles list the dominant bile acids in humans and rodents, respectively. At the same time, sulphated (sulpho-) or glucuronidated (glucurono-) bile acids produced by the liver via sulfotransferases (SULTs) and UDP-glucuronosyltransferases (UGTs) amidated with a taurine or a glycine are taken up by the multidrug resistance-associated protein 2 (MRP2). In the intestine, microbial enzymes from gut bacteria (dashed arrows) metabolize bile acids34; glyco-conjugated and tauro-conjugated CA and CDCA are deconjugated via bile salt hydrolases (BSH) and 7α -dehydroxylated to form secondary bile acids (DCA and LCA). Tα-MCA and Tβ-MCA are deconjugated via BSH to form α-MCA and β-MCA. β-MCA is C-6 epimerized to form ω-MCA and then ω-MCA is 7α-dehydroxylated to from HDCA203. CDCA is transformed to UDCA using the hydroxysteroid dehydrogenase (HSDH)204. Glucurono and sulpho-conjugated BAs are mainly excreted into urine by MRP2205,206. The main bacterial genera of the gut microbiota involved in bile acid metabolism include: Bacteroides, Clostridium, Lactobacillus, Bifidobacterium and Listeria in bile acid deconjugation; Bacteroides, Eubacterium, Clostridium, Escherichia, Egghertella, Eubacterium, Peptostreptococcus and Ruminococcus in oxidation and epimerization of hydroxyl groups at C3, C7 and C12; Clostridium and Eubacterium in 7-dehydroxylation; Bacteroides, Eubacterium and Lactobacillus in esterification; and Clostridium, Fusobacterium, Peptococcus and Pesudomonas in desulfatation53,67. At the terminal ileum, most of the unconjugated bile acids including CA, CDCA, DCA, UDCA, HDCA, α-MCA, β-MCA, ω-MCA are reabsorbed by apical sodium-dependent BA transporter (ASBT) into the enterocytes, and secreted into the portal circulation via basolateral bile acid transporters organic solute transporter α (OSTα), OSTβ, MRP2 and MRP3. Bile acids are then taken up by sodium-dependent taurocholate cotransporting polypeptide (NTCP) and organic anion-transporting polypeptide 1 (OATP1) into hepatocytes. Hepatic MRP3, MRP4 and OST-α/β also provide alternative excretion routes for bile acids into the systemic circulation42,43.
CA, cholic acid; CDCA, chenodeoxycholic acid; α-MCA, α-muricholic acid; β-MCA, β-muricholic acid; ω-MCA, ω-muricholic acid; HCA, hyocholic acid; DCA, deoxycholic acid; HDCA, hyodeoxycholic acid; GCA, glycocholic acid; GCDCA, glycochenodeoxycholic acid; TCA, taurocholic acid; TCDCA, taurochenodeoxycholic acid; T α-MCA, tauro α-muricholic acid; T β-MCA, tauro β-muricholic acid; LCA, Lithocholic acid; UDCA, ursodeoxycholic acid; CYP7A1, cholesterol 7 alpha-hydroxylase; CYP8B1, sterol 12α-hydroxylase; CYP7B1, oxysterol 7α-hydroxylase; CYP27A1, sterol 27-hydroxylase; BACS, bile acid, CoA synthetase; BAAT, bile acid-CoA, amino acid N-acyltransferase; BSEP, bile salt export pump; ASBT, apical sodium-dependent bile acid transporter; OSTα/β, organic solute transporter alpha/beta; MRP2, multidrug resistance-associated protein 2; MRP3, multidrug resistance-associated protein 3; MRP4, multidrug resistance-associated protein 4; NTCP, sodium-dependent taurocholate cotransporting polypeptide; OATP1, organic anion transporter 1; HSDH, hydroxysteroid dehydrogenase
Following the conjugation of the primary bile acids CA and CDCA to either taurine (predominantly in mice) or glycine (mainly in humans) by bile acid:CoA synthetase (BACS) and bile acid-CoA:amino acid N-acyltransferase (BAAT) to form taurocholic acid (TCA), taurochenodeoxycholic acid (TCDCA), glycocholic acid (GCA) and glycochenodeoxycholic acid (GCDCA), primary bile acids are secreted from the liver into the bile canaliculus via the canalicular bile salt export pump (BSEP)34. Concurrently, sulphated (sulpho-; catalyzed by enzymes such as sulfotransferase family 2A member 1 (SULT2A1)) or glucuronidated (glucurono-; catalyzed by UDP-glucuronosyltransferase (UGT) enzymes) bile acids (that are amidated with either taurine or glycine) are secreted from the liver into the bile via the multidrug resistance-associated protein 2 (MRP2)42,43. When the flow of bile is impaired under cholestatic conditions44 (that is, when the flow of bile from the liver is slowed or blocked), excessive amounts of hepatic bile acids and bilirubin can be excreted into the systemic circulation through basolateral export transport systems that are mediated by members of the multidrug resistance-associated protein (MRP) family including MRP3 and MRP4, as well as the organic anion transporting polypeptide 2 (OATP2) and the organic solute transporter-α/β (OST-α/β)42,43. In the postprandial state, the gallbladder contracts and releases bile acids (in the form of mixed micelles containing bile acids, cholesterol and phospholipids) into the intestinal lumen45, thus facilitating the emulsification and absorption of lipids in the small intestine46 Following lipid absorption, both conjugated and unconjugated bile acids are then reabsorbed in the distal ileum by the apical sodium-dependent BA transporter (ASBT), which is located in the brush border (that is, the microvillus border of intestinal epithelial cells at the small intestinal villi membrane)38,47. These bile acids are then bound to the ileal bile acid carrier protein (IBABP), which mediates bile acid transport across the enterocyte to the basolateral membrane, where the enteric OST-α/β facilitates transport into the portal vein48.
FXR is the nuclear receptor that is most dedicated to signalling by bile acids. Activation of ileal FXR by unconjugated bile acid downregulates the expression of ASBT and induces the expression of IBABP and OST-α/β49. Ileal bile acids are recycled to the liver where they are taken up primarily by the sodium-dependent taurocholate cotransporting polypeptide (NTCP), and to a lesser extent by OATP1 and OATP422,23, and are negatively regulated by nuclear FXR through a feedback mechanism to limit the increase of bile acid levels in liver50. In humans, NTCP largely accounts for the uptake of conjugated bile acids (>80%)51, whereas the members of the OATP family efficiently transport unconjugated or sulfated bile acids52 to the liver. The process of bile acid biosynthesis, transport and metabolism, along with the associated change in the intestinal bile acid profile, is termed enterohepatic circulation. Optimally, a bile acid pool of about 3 grams (~90-95%) is recycled between the gut and the liver approximately eight times per day, with only ~0.2-0.6 grams of de novo synthesized bile acids being produced per day to maintain a stable pool of bile acids38,53.
Signalling regulation of bile acid synthesis and transport
Hepatic FXR is the primary regulator of bile acid biosynthesis and enterohepatic circulation. Under normal physiological conditions, the activity of hepatic FXR regulates bile acid levels in the biliary tree and intestine, while limiting bile acid accumulation in the liver (FIG. 2a). Normal intestinal FXR activity maintains a good efflux of bile acids back into the portal vein, and a controlled reuptake of bile acids into the enterocyte that limits intracellular bile acid levels54. Activation of intestinal FXR upregulates the expression of fibroblast growth factor 15 (FGF15) in mice, and its orthologue FGF19 in humans, which inhibits bile acid synthesis in hepatocytes via activation of hepatic fibroblast growth factor receptor 4 (FGFR4)49. FIG. 2 illustrates the signalling pathway for bile acid synthesis and distribution.
Figure 2. Enterohepatic circulation of bile acids under normal physiological conditions (a) and during dysbiosis and inflammation (b).
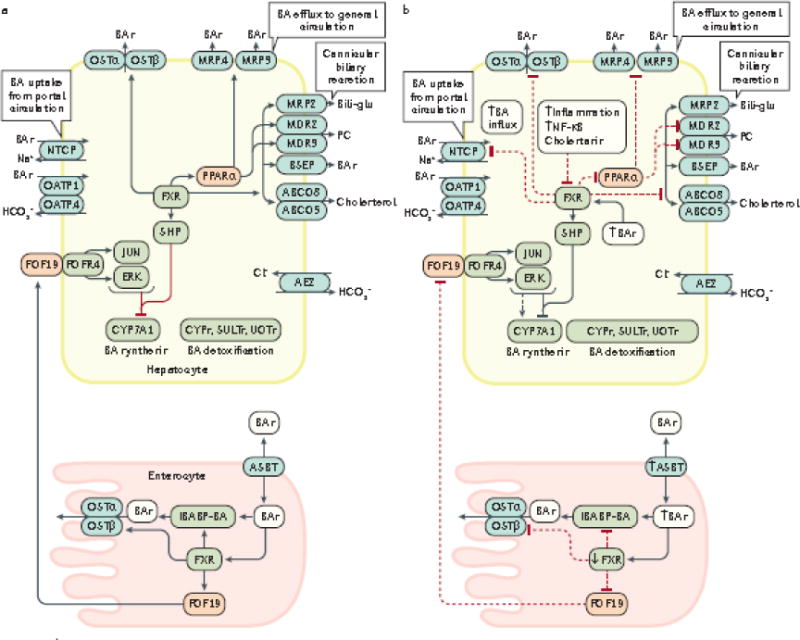
The enterohepatic circulation of bile acids between the intestine (enterocytes) and liver (hepatocytes) under normal physiological conditions (a, solid black arrows), and the alterations that occur in this process during dysbiosis of the gut microbiota and inflammation (b, dashed black arrows), are shown. During intestinal inflammation, which occurs as a result of intestinal barrier dysfunction, expression of intestinal FXR is downregulated207, which results in: reduced FGF19-FGFR4 signalling; downregulation of intestinal bile acid transport protein (IBABP); downregulation of the bile acid efflux transporters OSTα an OSTβ54,208; and upregulation of the apical sodium-dependent BA transporter (ASBT). Reduced expression of IBABP carrier protein results in a reduction in the transfer of bile acids across the enterocyte to the OSTα/β efflux transporters for entry into the portal vein, thus disrupting enterohepatic circulation. The combination of these events permit increased influx of bile acids into the enterocyte and prevention of bile acid efflux back into the portal vein. Collectively, increased influx and decreased efflux of bile acids might increase inflammation in the intestinal mucosa49. Diminished signalling via the FGF19-FGFR4 axis also increases bile acid synthesis in the liver via reduced activation of c-JUN/ERK/CYP7A1 axis209–211. During hepatic inflammation caused by bile acid perturbation of hepatocyte membranes and subsequent activation of pro-inflammatory PKC pathways leads to the activation NF-κB which inhibits transcription of hepatic FXR, and also prevents the subsequent activation of SHP that normally inhibits the synthesis of the rate-limiting enzyme for BA synthesis, CYP7A1. Therefore, decreased FXR expression promotes increased bile acid synthesis in combination with increased influx of bile acids98 due to the fact that OSTα/β, MRP3/4 and all of the cannicular transporters (depicted in grey ovals) are under transcriptional control by FXR.98 Decreased FXR transcription also leads to decreased expression of NTCP transporters which are involved in bile acid influx; however, OATP transporters are not affected by FXR212. FXR also controls bile acid detoxification; decreased transcription of FXR leads to decreased expression of PPARα and its target genes which encode Cytochrome P450 enzymes (CYPs), sulfotransferases (SULTs) and UDP-glucuronosyltransferases (UGTs).208 Decreased PPARα expression also induces additional decreases in expression of the MDR2/3, MRP3 and MRP4 transporters. Under these conditions, both cholestasis and inflammation are intensified which can lead to the development of liver cancer. Cholestasis causes inflammation via various mechanisms which leads to upregulation and activation of NF-κB which in turn, binds directly to the FXR promoter to inhibit its transcription124.
BSEP, bile salt export pump; CYP7A1, cholesterol-7α-hydroxylase; MDR3/4, multidrug resistance protein 3/4; MRP2/3/4, multidrug resistance-associated protein 2/3/4; NTCP, sodium taurochlorate co-transporting polypeptide; Ostα/β, organic solute transporter α/β; PC, phosphatidylcholine; PPARα, peroxisome proliferator-activated receptor α; SHP, small heterodimer partner; IBABP, ileal bile acid binding protein; FGF19, fibroblast growth factor 19; OATP, multi-specific orgnanic anion transporters; FGFR4, fibroblast growth factor receptor 4; ABCG5/8 ATP-binding cassette subfamily G, members 5/8; ASBT, apical sodium-dependent bile acid transporter; SULTs, sulfotransferases; UGTs, UDP-glucuronosyltransferases
Species-specific differences in bile acid synthesis, transport and metabolism occur, particularly with respect to the composition of the bile acid pool. In the rodent liver, the majority of CDCA is converted to β-muricholic acid (βMCA) through the action of a 6α/β-hydroxylase55. Conversely, in the pig, CDCA is largely converted to hyocholic acid (HCA)56 in liver via C-6α-hydroxylation by CYP3A456,57; however, some studies report that HCA might be formed from α -muricholic acid (α-MCA) in the intestine58. The majority of human hepatic bile acids tend to be glycine-conjugated, whereas a higher proportion of rodent bile acids are taurine-conjugated34. A study that compared serum bile acid composition between mice, rats, and humans reported that unconjugated bile acids account for the majority of total bile acids in mice and rats (84% and 92%, respectively), including CA (24-50%) and MCAs (22-43%); however, glycine-conjugated bile acids (42%) were most abundant in human serum, followed by deoxycholic acid (DCA) (19%), CA (16%) and CDCA (15%)59. In mice, taurine-amidated βMCA (tauro-βMCA) antagonizes FXR signalling in the intestine, and, when present at low concentrations in the gut, creates a more hydrophobic bile acid profile that activates intestinal FXR60. Inhibition of intestinal FXR by tauro-βMCA downregulated FGF15 in mice and induced FGFR4-dependent activation of CYP7A1 via inhibition of JUN transcriptional activity61, leading to elevated hepatic bile acid synthesis.
Signalling by bile acids
The ability of the gut microbiota to biotransform intestinal bile acids to their unconjugated forms is central to the metabolic homeostasis of the gastrointestinal tract; these unconjugated bile acids can activate bile acid signalling receptors such as FXR, pregnane X receptor (PXR), constitutive androstane receptor (CAR), vitamin D receptor (VDR) and G-protein coupled receptor TGR533,34. The main bacterial genera of the gut microbiota involved in bile acid metabolism include: Bacteroides, Clostridium, Lactobacillus, Bifidobacterium and Listeria in bile acid deconjugation; Bacteroides, Eubacterium, Clostridium, Escherichia, Egghertella, Eubacterium, Peptostreptococcus and Ruminococcus in oxidation and epimerization of hydroxyl groups at C3, C7 and C12; Clostridium and Eubacterium in 7-dehydroxylation; Bacteroides, Eubacterium and Lactobacillus in esterification; and Clostridium, Fusobacterium, Peptococcus and Pesudomonas in desulfatation53,62. Intestinal anaerobic bacteria of the genera Bacteroides, Eubacterium and Clostridium deconjugate taurine-conjugated and glycine-conjugated bile acids to their respective unconjugated free forms through the action of bile salt hydrolase (BSH)39. Subsequently, anaerobes of the genera Bacteroides, Clostridium, Eubacterium, Lactobacillus and Escherichia convert the unconjugated primary bile acids CDCA and CA, into the secondary bile acids lithocholic acid (LCA) and DCA, through 7α-dehydroxylation by CYP7A153. The majority of CA, CDCA, DCA and LCA are then reabsorbed in the intestine and transported back to the liver, whereas most of the LCA is excreted in faeces53.
Bile acids, FXR and TGR5
Unconjugated bile acids (such as CDCA, LCA, DCA and CA) act as high affinity ligand agonists of FXR63–67. Many in vitro studies in human liver and/or colon cell lines, or in cell lines from monkey or human, that were transfected to overexpress human FXR have consistently shown a rank order for the ability of bile acids to activate FXR: CDCA > DCA > LCA > CA. However, an in vivo mouse study indicated that this rank order may be different under physiological, non-hepatotoxic conditions66; the repression of the FXR target gene Cyp7a1 at a bile acid dose of 0.1% of their dietary intake showed a rank order: DCA = ursodeoxycholic acid (UDCA) > CA > CDCA > LCA, whereas at a dose of 0.3%, the order changed to UDCA > DCA > CDCA > CA > LCA (thus, at higher doses of bile acid, CDCA precedes CA as an FXR activator). Although there is some controversy regarding the rank order of FXR activating potential between studies performed in vitro and in vivo, there is a consensus that unconjugated bile acids have greater potential to activate FXR than conjugated bile acids36,67,68.
TGR5, a bile acid-specific G-protein coupled receptor (GPCR) that is ubiquitously expressed in humans and animals, activates various intracellular signalling pathways upon interaction with bile acids69. In vitro studies have shown that the bile acid rank order for activation of TGR5 is: LCA (EC50, the concentration of a drug that gives half-maximal response: 35±5 nM) > DCA (EC50 = 575 ±75 nM) > CDCA (EC50 = 4.00 ± 0.66 μM) > CA (EC50 > 10 μM) for unconjugated bile acids70. The rank order of both conjugated and unconjugated BAs for activation of TGR5 in Chinese hamster ovary cells (CHO) transfected with human TGR5 has also been determined: TLCA > LCA > GLCA > TDCA > DCA > GDCA > TCDCA > CDCA > GCDCA > TCA > CA > GCA71. Interestingly, the microbiota-induced secondary bile acids LCA and DCA, and their conjugated forms, were the best activators of TGR571. Therefore, these data infer that the gut microbiota can potentiate bile acid signalling through both bile acid signalling receptors, TGR5 and FXR.
Bile acids, PXR, CAR and VDR
PXR is primarily expressed in the liver and intestine, whereas CAR is mainly expressed in hepatic tissues35. Both receptors are principal regulators of drug metabolism in the liver72. PXR activates the bile acid hydroxylation enzyme CYP3A, bile acid–hydroxylating enzymes CYP2B, bile acid conjugation enzymes SULT2A1 and UGTs, the canalicular transporter MRP2, and the basolateral transporter OATP273–75. CAR also activates CYP3A, CYP2B and SULT75. CAR and PXR might play a synergistic role in maintaining bile acid homeostasis in vivo, presumably through the combined induction of CYP3A4 and SULT2A176, as evidenced by the observation that combined loss of both receptors induced aggravated LCA-induced liver damage in mice77,78. PXR-dependent activation of intestinal FGF19 only occurred in colon cancer cells but not in normal intestinal crypt cells, and thus PXR has been associated with the promotion of colon tumor growth79. Interestingly, Pxr−/− mice fed a lithogenic diet had a higher susceptibility for the development of cholesterol gallstones than wild-type mice, owing to decreased Cyp7a1 gene expression, reduced biliary bile acid secretion, and higher intestinal FGF15 expression due to increased activation of enteric FXR80. In addition, LCA activates PXR, and indirectly activates CAR, which inhibit CYP7A1 transcription by interacting with HNF4α53.
VDR is primarily activated by 1α,25-dihydroxyvitamin D3, the biologically active form of vitamin D, and functions as a ligand-regulated transcription factor that is highly expressed in the large intestine, particularly in immune cells36. The activation of VDR directly interferes with dendritic cell differentiation and the production of pro-inflammatory cytokines36. VDR primarily functions as an intestinal bile acid sensor that can be activated by LCA to induce expression of CYP3A and SULT2A1, and therefore protect the gut from bile acid toxicity81. In addition, VDR activated Asbt gene expression in male Sprague-Dawley rats, which enhanced ileal bile acid transport82. It was reported that Vdr−/− mice expressed lower intestinal levels of FGF15, whereas treatment with 1α,25-dihydroxyvitamin D3 increased intestinal FGF15 in wild-type mice83. Activation of VDR might also activate the FGF19–FGFR4 axis in the intestine, and might thus contribute to normalization of enterohepatic circulation. Activation of VDR upregulates FGF19 expression in the intestine, leading to decreased activation of CYP7A1 in the liver and decreased bile acid synthesis in mice — a potentially critical bypass pathway for FXR signalling, which is downregulated in the intestine in CRC tumours83. Moreover, the vitamin D–VDR axis modulates fibrogenesis via epigenetic regulation of the TGFβ–SMAD pathway84, which can result in a variety of human diseases ranging from autoimmunity to fibrosis and cancer,85.
Bile acids, liver inflammation and HCC
Signalling via bile acid receptors results in the detoxification of bile acids and therefore anti-inflammatory effects86. In addition to its role in regulation of bile acid synthesis and transport, FXR is also a bile acid-modulated transcription factor and protective sensor that induces protective cellular responses in hepatic and gastrointestinal tissues, and that regulates inflammation, immune responses, and liver regeneration86,87. Dysregulated bile acid synthesis, transport, and metabolism is a common etiological factor for almost all human liver diseases37,88–90. In this section, the role of both hepatic FXR and dysbiosis of the gut microbiota in liver inflammation and carcinogenesis will be discussed.
Hepatic FXR in liver inflammation
As previously discussed, the canalicular export of bile acids from the liver is mediated by BSEP; this process is a major determinant of bile acid secretion and is therefore tightly regulated at transcriptional and post-transcriptional levels by several liver–enriched transcription factors, including FXR91. Sustained inhibition of BSEP expression occurs during hepatic inflammation; decreased FXR gene expression in response to inflammation reduced the flow of bile, and led to liver injury38. In conditions associated with hepatic accumulation of bile acids, activation of FXR also induced the expression of OST-α/β at the sinusoidal membrane, and promoted the efflux of hepatic bile acids to the systemic circulation (Fig. 2b)92,93. Diminished activity of BSEP and OST-α/β during inflammation directly related to decreased bile acid–dependent activation of FXR signaling (Fig 2b)38,91,92. In addition, NTCP expression is down-regulated at the mRNA and protein levels in response to increased hepatic levels of bile acids, and occurs through the FXR–small heterodimer partner (SHP) pathway94, which has an important role in maintaining bile acid and cholesterol levels by inhibiting the transcription of cholesterol CYP7A1 (Fig. 2b)95,96. Therefore, the decreased function of hepatic transporters that occurs as a result of reduced FXR signalling during inflammation leads to enhanced hepatic bile acid sequestration and sustained inflammation, which can promote the development of HCC97–99. Bile acids can directly disrupt the plasma membrane and cause activation of protein kinase C (PKC), which in turn activates the p38 MAPK pathway resulting in increased activation of p53 and nuclear factor kappa-B (NF-κB), ultimately leading to increased apoptosis and inflammation100. Activated NF-κB is translocated to the nucleus where it transcribes genes encoding proinflammatory cytokines such as TNFα, IL-1β and IL-6101,102. IL-6 can activates the Janus kinase–signal transducer and activator of transcription-3 (JAK–STAT3) pathway, which leads to decreased apoptosis and progression of HCC103. IL-1β activates the phosphoinositide 3-kinase (PI3K)–MDM2 pathway to negatively regulate p53, thus increasing survival of DNA damaged cells, which might lead to the development of HCC104. The NF-κB p50/p65 heterodimer binds directly to the promoter of FXR and inhibit its transcription, promoting the loss of bile acid transporters such as OSTα/β, increased biosynthesis of bile acids, and ultimately accumulation of bile acids in the liver86. Membrane perturbation by bile acids can also activate cytosolic phospholipase A2 (PLA2) which causes the release of arachidonic acid (AA) from the cell membrane via the activity of cyclooxygenase (COX) and lipoxygenase (LOX), which ultimately results in increased levels of reactive oxygen species (ROS) in the hepatocyte.105,106 ROS can directly activate NF-κB and can also induce direct DNA damage in cells which might lead to HCC (FIG. 3).107
Figure 3. Bile acid–induced hepatic inflammation and carcinogenesis.
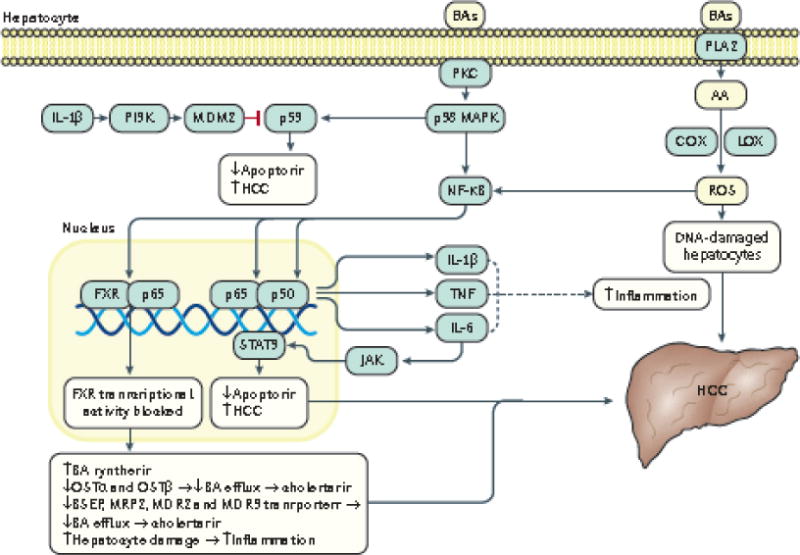
Owing to their lipophilic, detergent properties, bile acids can directly disrupt the plasma membrane and cause activation of PKC which in turn activates the p38 MAPK pathway. The resultant activation of p53 and NF-κB lead to induction of apoptosis and increased inflammation. Activated NF-κB translocates to the nucleus and promotes transcription of genes that encode pro-inflammatory cytokines such as TNFα, IL-1β and IL-6, which can positively regulate NF-κB activation and thus promote a continued cycle of inflammation. IL-6 also activates the Janus Kinase (JAK)–Signal Transducer and Activator of Transcription 3 (STAT3) pathway which leads to decreased apoptosis and progression of HCC103. IL-1β also activates the PI3K–MDM2 pathway to negatively regulate p53, thus increasing survival of DNA damaged cells which might lead to HCC104. The NF-κB p50/p65 heterodimer has also been shown to bind directly to the promoter of FXR and inhibits its transcription, resulting in: reduced expression of bile acid transporters such as OSTα/β, BSEP, MRP2, MDR2/3 which leads to decreased bile acid efflux and cholestasis and increased biosynthesis of bile acids124. Inhibition of FXR transcription collectively leads to increased amounts of bile acids in the liver which causes inflammation that can lead to HCC. Membrane perturbation by bile acids can also activate PLA2 which causes release of arachidonic acid (AA) from the cell membrane via cyclooxygenase (COX) and lipoxygenase (LOX), ultimately resulting in increased levels of reactive oxygen species (ROS) in the hepatocyte105,106. ROS can directly activate NF-κB and can also induce direct DNA damage in cells which might lead to HCC.107
PKC, protein kinase C; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor kappa B; PI3K, phosphoinositide 3-kinase; TNFα, tumor necrosis factor; JAK, Janus kinase; STAT3, signal transducer and activator of transcription-3; PLA2, phospholipase A2; COX, cyclooxygenase; LOX, lipoxygenase; ROS, reactive oxygen species; AA, arachidonic acid
Mice with whole body FXR deficiency develop liver tumors spontaneously96. In the absence of hepatic FXR, the re-expression of constitutively active FXR in the enterocytes of these Fxr-null mice restored bile acid homeostasis through activation of the FGF15 axis, and protected against the development of HCC108. However, hepatocyte-specific FXR deficiency (Fxrhep−/−) in mice did not promote spontaneous liver tumorigenesis, suggesting that hepatic FXR deficiency may only serve as a tumor initiator, and that further increased levels of bile acids are required for tumor development109. In addition, it has also been demonstrated that BSEP deficiency, inherited mutations in the ABCB11 gene cause impairment of bile salt export from hepatocytes into bile110 — which leads to progressive familial intrahepatic cholestasis and eventually pediatric HCC — was associated with marked increases in intracellular concentrations of bile acids in Children110,111.
An important determinant of the biological effects of a particular bile acid is its hydrophobicity. The ranking of hydrophobicity, based on chromatographic behavior, for important bile acids is: UDCA < CA < CDCA < DCA < LCA34. Several studies of bile acid-induced hepatocyte injury indicated that the hydrophobic bile acids LCA, DCA and CDCA were cytotoxic, and that the hydrophilic bile acid UDCA, along with its taurine-conjugated derivative TUDCA, were cytoprotective112. Intrahepatic accumulation of hydrophobic bile acids such as DCA and CDCA has been proposed as a mechanism of cholestatic liver injury113. Increased concentrations of hydrophobic bile acids caused mitochondrial damage and disruption of the cell membrane in mouse or human hepatocytes, which ultimately led to the development of HCC via increased levels of reactive oxygen species (ROS) and activation of RAS and NF-κB114. In particular, NF-κB activation induced by TNF-α in mouse liver inhibited hepatic FXR expression115, which subsequently downregulated the expression of SHP — an important tumor suppressor in the development of liver cancer that inhibits cell proliferation and induces apoptosis116,117. In stage I HCC tissues, FXR expression levels have been shown to be reduced to 40% of levels in normal liver tissues, and are further decreased progressively at later stages of disease118. Furthermore, Fxr−/− Shp−/− mice developed liver tumors spontaneously when exposed to chronically elevated intrahepatic concentrations of bile acids119. Bile acid accumulation in mouse hepatocytes was sufficient for Hippo pathway activation, which caused the nuclear translocation of transcription factor yes-associated protein (YAP), and increased cell proliferation119. This induction of proliferation mechanistically involved bile acid–induced overexpression of a key scaffolding protein, IQ motif containing GTPase activating protein 1 (IQGAP1); IQGAP1 decreased cell-cell adhesion via dissociation of α-catenin from the E-cadherin-β-catenin complex, thus causing the translocation of YAP to the nucleus119.
Bile acids and dysbiosis of gut microbiota
In addition, several bile acids, including DCA, LCA, CDCA, and TCDCA, have been individually demonstrated to have cytotoxic and cancer-promoting properties120. DCA was first shown to be a carcinogen that promoted CRC development in mice in 1940121. Administration of a high fat diet (HFD) in mice resulted in changes in the gut microbiota, and consequent increases in hepatic levels of DCA18; elevated DCA levels induced a senescence-associated secretory phenotype (SASP) in hepatic stellate cells and secretion of various inflammatory and tumor-promoting factors that facilitated the development of HCC in mice exposed to chemical carcinogen, 7,12-dimethylbenzanthracene (DMBA)18. DCA has also been implicated in the development and progression of colon and oesophageal cancer, suggesting that the microbiota might also influence colonic and oesophageal carcinogenesis through the production of DCA, particularly in the context of obesity28,29. In addition, male mice exposed to low-dose streptozotoxin, a chemical used for the destruction of insulin–producing cells and for the generation of Type 1 diabetes mellitus (T1DM) phenotypes in mice, in combination with a HFD for 16 weeks developed liver tumors122; the gut microbiota profile was markedly altered in mice that developed liver tumours compared with control mice exposed to a HFD alone for 58 weeks123, and a panel of bile acids — including DCA, TCA, TCDCA and TLCA — collaboratively contributed to liver tumor development122. Therefore, sequential steps in a possible mechanism of bile acid–induced liver carcinogenesis include: HFD–induced alteration of the gut microbiota and increased production of secondary bile acids in the lower intestine; increased intestinal reabsorption and return of bile acids to the liver; increased liver inflammation and consequent downregulation of hepatic FXR, which causes inhibition of several major bile acid transporters (in particular, BSEP); increased hepatic accumulation of bile acids, and subsequently, sustained hepatocyte DNA damage, apoptosis, and inflammation. In the gastrointestinal tract, disrupted enterohepatic circulation of bile acids from the liver can cause inflammation; activation of NF-κB results in increased expression of pro-inflammatory cytokines such as TNFα and IL-1β, which reduce transcription of FXR target genes88. This leads to reduced transport of bile acids from the enterocyte back into the portal vein via IBABP and OSTα/β, (FIG. 2b)124, resulting in bile acid accumulation in the intestinal mucosa.
Patients with liver disorders such as fatty liver disease, fibrosis, cirrhosis and HCC often exhibit gut dysbiosis that is characterized by a major elevation of aerobic, pro-inflammatory, BSH-rich bacteria of the genera Enterobacter, Enterococcus and Clostridium that are capable of producing increased amounts of secondary bile acids53. Conversely, individuals with intestinal conditions, such as inflammatory bowel disease (IBD), often suffer from gut dysbiosis that is characterized by diminished microbial diversity and decreased abundance of bacteria of the phylum Firmicutes; Faecalibacterium prausnitzii is particularly under-represented, which leads to lowered levels of secondary bile acids and elevated levels of conjugated bile acids in the intestine125–127. These microbiota-induced changes in bile acid profiles might therefore result in diminished activity of FXR, downregulation of the expression of hepatic BSEP, hepatic OST-α/β and intestinal OST-α/β, and consequently, increased intracellular retention of bile acids in hepatocytes and intestinal mucosa (Fig 2b)124,128. In addition, modification of gut microbiota profiles using probiotics enhanced faecal excretion of bile acids and promoted hepatic bile acid synthesis in mice, the mechanism of which involved decreased bile acid re-absorption from ileum and repression of the enterohepatic FXR–FGF15 axis129, which suggests that dysbiosis and abnormal bile acid metabolism can be reversed with probiotics. Therefore, bile acid and gut microbiota profiles influence each other, and when disrupted, jointly produce a gastrointestinal disease phenotype.
Bile acids, colon inflammation and CRC
In addition to the effect of hepatic accumulation of bile acids in liver carcinogenesis, mounting evidence suggests that patients who are diagnosed with colonic carcinomas who consume a Western diet (high in saturated fats, red meats and processed carbohydrates display elevated levels of faecal secondary bile acids, mostly DCA and LCA130,131. Elevated levels of secondary bile acids exert detrimental effects on the architecture and function of the colonic epithelium through multiple mechanisms including oxidative damage to DNA, inflammation, activation of NF-κB and enhanced cell proliferation131. Accordingly, bile acids have been considered tumor-promoting factors in the development of CRC29,105. In this section, bile acid-dependent action of TGR5 and the links between bile acids, inflammation and CRC will be discussed.
Activation of TGR5 in macrophages
In chronic or acute inflammatory conditions such as Crohn’s disease (CD) or ulcerative colitis (UC), there is chronic activation of the innate immune system and dysregulated secretion of pro-inflammatory and anti-inflammatory cytokines132. Macrophages are major regulators of cytokine production in the gastrointestinal tract; their actions here are primarily through the activation of their bile acid–responsive, membrane-bound GPCR, TGR5. Indeed, bile acid–dependent activation of TGR5 inhibits the activation of NF-κB induced by lipopolysaccharide injection in wild-type mice compared with lipopolysaccharide–injected TGR5-deficient mice133. In addition, a series of elegant imaging studies and bioluminescence resonance energy transfer (BRET) assays showed that TGR5 remains membrane–bound and accumulates in lipid rafts in response to treatment with DCA, TLCA and the selective TGR5 agonists oleanolic acid and 3-(2-chlorophenyl)-N-(4-chlorophenyl)-N, 5-dimethylisoxazole-4-carboxamide in HEK293 cells transfected with TGR5-RLuc8 and colonocytes (NCM460) that endogenously express TGR5133, where it can transactivate epidermal growth factor receptor (EGFR) (FIG. 4)134.
Figure 4. Bile acid-induced TGR5 signalling pathways in macrophages.
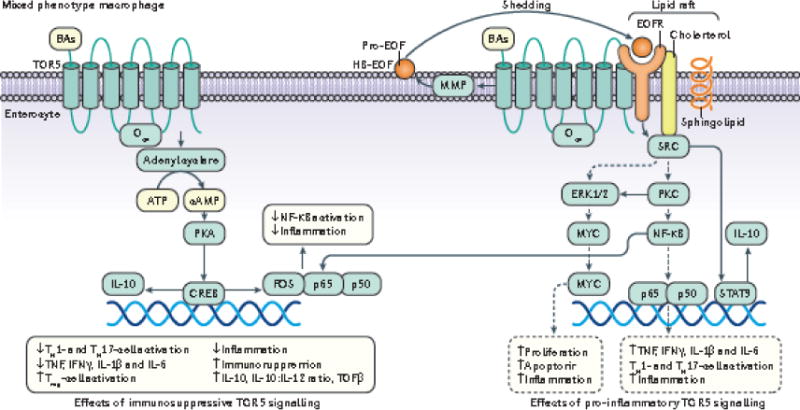
The M1 macrophage phenotype is pro-inflammatory and M2 phenotype is immunosuppressive. Although bile acids are not able to induce complete macrophage polarization to either the M1 or M2 phenotype, they have been shown to activate TGR5 and produce a ‘mixed phenotype’ that exhibits dominant immunosuppressive behavior as evidenced by an increased IL-10:IL-12 ratio141. In response to bile acids, TGR5 can activate both the adenyl cyclase–cAMP and EGFR–SRC pathways133. In the pro-inflammatory (dotted arrows) M1-like pathways, TGR5-dependent transactivation of EGFR can induce pro-inflammatory signalling via activation of SRC, which in turn activates ERK1/2 and PKC133. This transactivation has been shown to occur when EGFR and a relevant GPCR such as TGR5 co-exist side by side in a lipid raft (signified by cholesterol and sphingolipid). Upon TGR5 activation, metalloproteinase-dependent cleavage of the EGFR ligand HB-EGF occurs213. TGR5–dependent activation of PKC results in the activation of NF-κB, which causes increased expression of the pro-inflammatory cytokines IL-1β, IL-6 and TNFα, along with further autocrine activation of ERK1/2 and continued cycles of pro-inflammatory signaling. SRC-dependent activation of ERK1/2 in the M1-like phenotype results in increased expression of c-MYC which increases cell apoptosis214. Pro-inflammatory M1-like signalling also regulates innate immunity by increasing activation of pro-inflammatory responsive Th17 cells (increased IL-1β, IL-6) and Th1 cells (increased IL-12, IFNγ) with concurrent suppression of immunosuppressive Treg cells (decreased IL-10) via the pro-inflammatory cytokines it produces141. The pro-inflammatory M1-like phenotype is not dominant during bile acid-dependent activation of TGR5; concomitantly, SRC activation also activates STAT3, which promotes anti-inflammatory effects (solid arrows) including decreased activation of Th17 and Th1-cells, decreased production of TNFα, IFNβ, IL-6, IL-12, and increased production of IL-10 and TGFβ, which corresponds with an increase in the IL-10:IL-12 ratio73,143. Bile acid–activated TGR5 also activates the cAMP pathway independently from EGFR transactivation; cAMP activates PKA which leads to the upregulation of cAMP response element binding protein (CREB) expression and activity. cFos, an important target gene of CREB, binds to the p65 subunit of activated NF-κB to inhibit its translocation to the nucleus, representing an anti-inflammatory effect of TGR5. Another important target gene of CREB is IL-10 which exerts immunosuppressive and therefore anti-inflammatory effects73,141,143. Bold solid arrows represent M2-like immunosuppressive signaling pathways and dotted arrow represent M1-like signaling pathways.
cAMP, cyclic adenosine monophosphate; PKA, protein kinase A; CREB, cAMP response element binding protein; ERK, extracellular receptor kinase; STAT3, signal transducer and activator of transcription-3; Gαs, G-protein alpha-s; ROS, reactive oxygen species; PKC, protein kinase C; EGFR, epidermal growth factor receptor; HB-EGF, heparin-binding epidermal growth factor; MMP, matrix metalloproteinase; SRC, sarc kinase; IL-6,10,12, interleukin-6,10,12; TNFα, tumor necrosis factor-α; TGFβ, transforming growth factor-β; cFos, cFos gene/protein
Although the role of TGR5 in gastrointestinal carcinogenesis is unclear, strong immunohistochemical staining of TGR5 was reported in 12% of human intestinal metaplasia135. Studies have shown that there are two types of tumor associated macrophages: M1 macrophages, which are pro-inflammatory; and M2 macrophages, which are anti-inflammatory and immunosuppressive136,137. The M1 phenotype is characterized by the production of pro-inflammatory cytokines such as IL-1β, IL-6, IL-12, IL-23 and TNFα, that collectively activate Th17 and Th1-cells138. IL-12 is required to induce production of IFNγ in M1 macrophages in order to activate Th1-cells138. M2 macrophages are immunosuppressive and mainly produce IL-10, which induces the activation of Regulatory T (Treg) cells136. An M2-like phenotype, an intermediate form, is induced in response to TGR5 activation using the TGR5 agonist INT-767139, and is defined by increased production of IL-10, which causes immunosuppression and further differentiation of M2-like macrophages to the M2 state140. However, bile acid-dependent activation of TGR5 in macrophages cannot completely switch the polarization of a macrophage from the M1 to the M2 state. Instead, bile acid–activated TGR5 induces a ‘mixed phenotype’ whereby the immunosuppressive M2 phenotype predominates, as evidenced by an increase in the IL-10:IL-12 ratio141. Depending on the polarization state of the macrophage, activation of TGR5 can promote pro-inflammatory responses (in M1 macrophages), or anti-inflammatory responses (in M2 macrophages) that are immunosuppressive in the context of CRC (FIG. 4). The major pro-inflammatory responses that occur following TGR5 activation in M1 macrophages include: increased activation of c-Myc, which induces apoptosis, proliferation and inflammation in macrophages in response to surrounding DNA-damaged cells; and activation of NF-κB, which stimulates production of pro-inflammatory cytokines such as IL-6 and TNFα, that promote inflammation and inactivate FXR142. The major anti-inflammatory, immunosuppressive response that results from TGR5 activation in M2 macrophages is production of IL-10, which: decreases production of pro-inflammatory cytokines such as TNFα, IFNβ, and IL-6; increases TGFβ expression; and increases the population of Treg cells, which decrease the activities of Th17 and Th1-cells73,143. Therefore, bile acid–TGR5 signalling controls the intricate balance between the production of pro-inflammatory and anti-inflammatory cytokines in the intestine. As IL-10 induces the differentiation of T-cells into immunosuppressive Treg cells, and IL-12 promotes the differentiation of T-cells into pro-inflammatory Th1 cells, the ratio of IL-10:IL-12 is an indicator of the inflammatory status of the intestinal mucosa (FIG. 4)144,145.
Bile acid dysregulation in the development of CRC
Bile acids influence the intestinal environment by controlling the growth and maintenance of commensal microbiota, maintaining barrier integrity and modulating the immune system146,147. The control of intestinal inflammation through activation of intestinal FXR in response to secondary bile acids, which passively diffuse into the enterocyte, has an important role in the normal physiological function of the colon where the deconjugation and dehydroxylation of bile acids occurs39. Evidence for the influence of commensal bacteria in bile acid synthesis originated in vivo; mice that were raised in the absence of commensal bacteria, or that were treated with antibiotics to deplete commensal microbiota, produced fewer secondary bile acids, and more conjugated bile acids, in the faeces, than conventional counterparts that were raised with normal commensal bacterial profiles in the gut148,149. A study of human faecal samples revealed a substantial decrease in abundance of bacteria from the Firmicutes phylum, a marked decrease in the Faecalibacterium prausntizii:E. coli ratio, considerably higher levels of conjugated bile acids, and lower levels of secondary bile acids in patients with active IBD compared with healthy individuals149. Interestingly, increased amounts of 3-OH-sulphated secondary bile acids were also detected in patients with active IBD. The sulphation of hepatic bile acids increases their hydrophilicity, thus reducing their cytotoxicity and enhancing their excretion in faeces and urine compared with unsulfated bile acids150. Therefore, sulphation is an important mechanism of bile acid detoxification and homeostasis in the gastrointestinal tract. However, sulphation of hydrophobic secondary bile acids hampers their ability to activate the anti-inflammatory effector FXR149,151. In patients with IBD, the presence of high levels of sulphated bile acids in faeces also implies that there might be a lower abundance of specific microbiota in the gut that are capable of desulfating sulphonated bile acids, such as Clostridium, Fusobacterium, Peptococcus, and Pseudomonas152. Collectively, gut dysbiosis can result in a diminished ability of the microbiota to produce unconjugated and secondary bile acids, which are high affinity ligands for activation of FXR, as well a reduced ability to promote sulphation of these bile acids to prevent FXR activation.
In addition, many of the bacterial species of phylum Firmicutes (for example, Clostridium cluster XIVa bacteria Eubacterium rectale and Roseburia spp.) produce butyrate153, and a decreased abundance of these bacteria was observed in patients with CRC154. In vitro studies showed that butyrate can inhibit colorectal carcinogenesis by suppressing the growth of tumour cells, inducing differentiation and apoptosis and inhibiting cell proliferation155,156. Moreover, there is an inverse relationship between the levels of butyrate in the human colon and the incidence of CRC157,158, possibly owing to the observation that there is a reduced ability of gut microbiota from patients with CRC to produce butyrate87. Bacteria from Clostridium cluster XIVa have also been reported to produce secondary bile acids via 7α-dehydroxylation of primary bile acids39. A decreased level of secondary bile acids was observed in patients with IBD compared with healthy individuals149; this effect was due to a decrease in the abundance of bacteria of the Firmicutes phylum (for example, Clostridium species) which contributed to the loss of anti-inflammatory effects of secondary bile acids on intestinal epithelial cells, thus enhancing chronic inflammation, Although the observations regarding loss of butyrate-producing bacteria87 and loss of secondary bile acid–producing (BSH-rich) bacteria in patients with IBD39 were made separately, the butyrate-producing bacteria and bile acid-producing bacteria overlap to some extent, and both are depleted during chronic inflammation of the gastrointestinal tract39,53,149,154.
Inflammation promotes the development and progression of IBD to CRC115,159. Pharmacological activation of intestinal FXR reduced inflammation and preserved intestinal barrier function in two established murine models of experimental colitis induced by dextran sodium sulphate (DSS) and trinitrobenzenesulfonic acid (TNBS)124; FXR agonist INT-747 reduced the loss of goblet cells, lowered inflammatory immune cell infiltrates and decreased the permeability of intestinal epithelium in both models. In addition, INT-747 also decreased expression of pro-inflammatory genes including IL-1β, IL-6 and monocyte chemoattractant protein-1 (MCP-1/CCL2). However, FXR-null mice did not show improvements in colitis with INT-747 treatment124. Mechanistically, the anti-inflammatory effect of FXR agonism occurred by inhibiting the activation of the pro-inflammatory transcription factor NF-κB124,160. By contrast, inhibition of FXR activity by cytokines TNF and IL-1β, which are upregulated in response to NF-κB activation, reduced transcription of FXR target genes124; furthermore, the p65 subunit of activated NF-κB was found to interact with FXR directly. Therefore, there are bile acid-mediated bidirectional interactions between FXR and components of the inflammation process161. Overall, unconjugated bile acids, which are detrimental to hepatocytes at high concentrations, are important for controlling microbial populations in the gut, activating intestinal FXR, and modulating innate immunity to preserve intestinal barrier function124,162.
The pathogenesis of CRC is considered a multistep process that often includes a sequence of cell mutations during progression from adenoma to carcinoma163. The most common mutations in CRC are in the genes encoding APC (adematous poliposis coli), KRAS, p53, phosphoinositide 3-kinase (PI3K), and transforming growth factor β (TGFβ) (FIG. 5)164. Enteral bacterial species that are most frequently associated with CRC include Fusobacterium nucleatum, colibactin–producing E. coli and Bacteroides fragilis, all of which can activate the WNT–β-catenin signalling pathway165. Interestingly, FXR expression has been found to be decreased at the mRNA level in colonic polyps and even more in colonic adenocarcinoma166. In normal murine intestinal tissues, FXR mRNA expression was highest in fully differentiated epithelial cells at the apical surface, and FXR–deficient mice exhibited compromised intestinal barriers with increased infiltration of immune cells166. Consistent with these findings, progressive decreases in FXR mRNA expression was reported from early to advanced stages of human colitis–associated neoplasia, and was undetectable in four CRC cell lines167,168. Immunohistochemical evaluation of 159 stage I-IV CRC samples, 32 polyps, and 238 normal samples revealed loss of FXR expression in the majority (94%) of CRC samples, and markedly reduced FXR expression in precancerous polyps, compared with normal colon tissues169. The incidence of intestinal cancer is also associated with FXR expression; the ileum, which normally has a higher level of FXR expression than the distal colon, had a lower cancer incidence than the distal colon169,170.
Figure 5. Effects of intestinal bile acids on colorectal carcinogenesis.
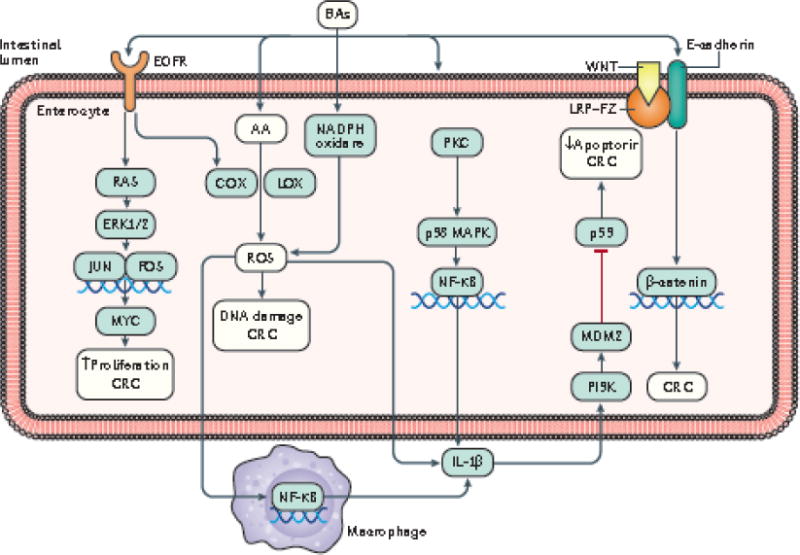
Secondary bile acids, in particular deoxycholic acid (DCA), influence several signaling pathways in enterocytes that can lead to the development of colorectal cancer (CRC). Bile acids can activate EGFR which in turn leads to activation of RAS–ERK1/2 signalling. ERK1/2 induces activation and nuclear translocation of c-JUN and c-FOS, which together form the Activator protein 1 (AP-1) transcription factor complex, which promotes the transcription of the AP-1 target gene c-MYC215. Expression of c-MYC increases cell proliferation and stemness. Activation of EGFR also leads to the upregulation of cyclooxygenase (COX) and lipoxygenase (LOX) which promote the production of reactive oxygen species (ROS). Bile acids can disrupt the plasma membrane to release arachidonic acid (AA) into the cytoplasm where it is rapidly used as a substrate for the biosynthesis of both COX and LOX enzymes105,177. NADP(H) oxidase can be stimulated by bile acids to further increase ROS production177. In the enterocyte, ROS can cause DNA damage and inflammation, leading to upregulated expression of the pro-inflammatory cytokine IL-1β177. All of these events contribute to the development and progression of CRC. Perturbation of the plasma membrane by bile acids also activates PKC, which subsequently activates p38 MAPK and NF-κB. Activated NF-κB translocates into the nucleus where it transcribes the gene encoding IL-1β. IL-1β can then signal in an autocrine and paracrine fashion to activate the PI3K–MDM2 axis which results in blockage of p53 activity. Decreased p53 activity leads to decreased apoptosis and increased survival of DNA-damaged cells, thus contributing to the potential development of CRC105,177. Bile acids also stimulate the binding of Wnt to the low-density lipoprotein receptor-related protein (LRP)–frizzled receptor (Fzr)–E-cadherin-β-catenin complex, which causes release of β-catenin from E-cadherin and into the cytoplasm where it translocates to the nucleus and stimulates transcription factors such as LEF/TCF to increase cell proliferation and cell stemness, thus contributing to the development and progression of CRC216.
DCA, deoxycholic acid; CRC, colorectal cancer; EGFR, epidermal growth factor; Ras, Ras oncoprotein; ERK1/2, extracellular signal-regulated kinase 1/2; C-Jun, C-Jun protein; C-Fos, C-Fos protein; c-Myc, myc oncoprotein; AA, arachidonic acid; COX, cyclooxygenase; LOX, lipoxygenase; ROS, reactive oxygen species; PKC, protein kinase C; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor kappa B; IL-1β, interleukin-one-beta; PI3K, phosphatidylinositol 3-kinase; MDM2, mouse double minute chromosome 2; Wnt, Wingless-related integration site; LRP, low density receptor related protein; Fzr, frizzled receptor; APC, adenomatous polypopsis coli; LEF, lymphoid enhancer factor; TCF, T-cell factor
Loss of FXR expression in CRC has been hypothesized to be due to hypermethylation of the FXR promoter168; however, hypermethylation was found to depend on the CRC cell line, whereby treatment with the DNA methyltransferase antagonist 5-Azacytidine caused dose dependent increases in FXR mRNA levels in the HCT-116 and SW480 cell lines, but not in the Caco-2 and HT-29 cell lines. Two mechanisms for FXR silencing have been proposed in human CRC; methylation of the FXR promoter in microsatellite instability (MSI)-high tumours; and activation of KRAS signalling in MSI-low tumours169.
In studies of obesity-related CRC, a high-fat diet (HFD) increased the risk of CRC in humans157. In addition, FXR−/− APCmin/+ mice — which are susceptible to CRC — exhibited an expanded bile acid pool due to decreased negative regulation of the FXR-FGF19 axis167,171. The increased risk for CRC in response to a HFD might be due to the increased production of secondary bile acids in the colon. Indeed, the overstimulation of bile acid discharge into the intestine in response to a HFD increases the production of secondary bile acids, which function as tumour promoters172. The hydrophobicity of DCA causes membrane perturbations that activate protein kinase C (PKC) and NADPH oxidase, which induce downstream activation of NF-κB173 (FIG. 5). Such membrane perturbations also facilitate the release of arachidonic acid — a substrate involved in the synthesis of the pro-inflammatory enzymes lipoxygenase and Cyclooxygenase (COX), which promote the production of ROS and the consequent induction of DNA-damage173,174. DCA also induces β-catenin signalling and also increases the degradation of p53 (FIG. 5), which together results in increased cell proliferation and invasiveness of colon cancer cells175. However, new evidence suggests that HFD-induced increases in intestinal levels of secondary bile acids might be a transient process, and that unconjugated bile acids (including DCA) might be gradually depleted from the intestine under chronic inflammatory conditions, presumably due to the depletion of BSH-rich bacteria in the intestine149,176. In addition, treatment with cholestyramine, a bile acid sequestrant, did not modify the tumor susceptibility of FXR−/− APCmin/+ mice171.
Secondary bile acids help to maintain a viable epithelial layer of cells by inducing apoptosis and promoting cell turnover. Therefore, exposure to high levels of secondary bile acids in response to HFD intervention and then to gradually decreased levels of secondary bile acids due to inflammation might have two important consequences: increased apoptosis of fully differentiated epithelial cells at the apical surface that express high levels of FXR; and continued renewal and increased selective growth of less differentiated cells that are resistant to apoptosis and express low levels of FXR167,177. Taken together, the carcinogenic effects of intestinal bile acids on the development and progression of CRC might be different from that of HCC; the gradual depletion of unconjugated secondary bile acids in response to gut dysbiosis, and subsequently, the emergence of less differentiated and highly proliferative colon epithelial cells with loss of FXR function, might be key factors contributing to colorectal carcinogenesis.
Targeting bile acid signalling
Based on the discussions outlined in this Review, the most important therapeutic considerations for the treatment of gastrointestinal diseases such as HCC and CRC are: restoring FXR signalling by correcting gut dysbiosis; re-establishing normal enterohepatic circulation of bile acids; and controlling inflammation of the gastrointestinal tract. Inflammation promotes the development of both CRC and HCC. Patients with inflammatory diseases such as IBD discussed in this Review are more likely to develop gastrointestinal cancer. Therapeutic targeting of FXR and TGR5 signalling might be important in restoring the normal enterohepatic circulation and controlling gastrointestinal inflammation. TABLE 1 summarizes the progress that has been made to date in targeting bile acid signalling in gastrointestinal diseases.
Table 1.
Clinical trials and ongoing studies of targeting bile acid signalling in gastrointestinal diseases
| Agent | Gastrointestinal Condition | Type of study | Status | Phase of development | Reference |
|---|---|---|---|---|---|
| FXR agonist | |||||
| OCA | Patients with PBC who have inadequate response to UDCA | Randomized, Double Blind, Placebo Controlled | Still recruiting | Phase III | 217 |
| Px-104 | NAFLD | Open label | Complete | phase II | 218 |
| OCA | PSC | Randomized, Double Blind, Placebo Controlled | Ongoing, but not recruiting participants | phase II | 219 |
| LJN452 | PBC | Randomized, Double-blind, Placebo-controlled | Still recruiting | phase II | 220 |
| OCA | NASH and fibrosis | Randomized, Double Blind, Placebo Controlled | Still recruiting | Phase III | 221 |
| OCA | Moderately severe alcoholic hepatitis | Randomized, Double Blind, Placebo Controlled | Still recruiting | phase II | 222 |
| OCA | NAFLD, NASH | Randomized, Double Blind, Placebo Controlled | Complete | Phase II | 223 |
| TGR5 agonist | |||||
| INT-767 (23-sulphate derivative of OCA) | PSC | – | – | phase I | 224 |
| INT-777 (6α-ethyl-23(S)-methylcholic acid) | NASH | – | – | preclinical | 225 |
| VDR related | |||||
| Vitamin D3 | IBD and Hypovitaminosis D | Randomized, Parallel Assignment, Double Blind | Complete | Phase III | 226 |
| Vitamin D3 | IBD, Ulcerative Colitis, Crohn Disease | Interventional, Single Group Assignment | Not yet recruiting | Phase III | 227 |
| UDCA related | |||||
| norUDCA | PSC | Randomized, Parallel Assignment, Double Blind | Complete | Phase II | 228 |
| UDCA | Polycystic Liver Disease | Interventional, Single Group Assignment, Open Label | Complete | Phase III | 229 |
| UDCA | Functional Dyspepsia | Randomized, Double Blind, Placebo Controlled | Not yet recruiting | Phase IV | 230 |
| TUDCA | PBC | Randomized, Parallel Assignment, Double Blind | Complete | Phase III | 231 |
| UDCA | FAP, Duodenal Neoplasms, Duodenal Polyps | Randomized, Parallel Assignment, Double Blind | Complete | Phase III | 232 |
Abbreviations:
CD, Crohn Disease; FAP, Familial Adenomatous Polyposis; FXR, farnesoid X receptor; IBD, inflammatory bowel disease; INT-767, 23-sulphate derivative of OCA; INT-777, 6α-ethyl-23(S)-methylcholic acid; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; Nor UDCA, norursodeoxycholic acid; OCA, obeticholic acid; UDCA, ursodeoxycholic acid; PBC, primary biliary cirrhosis; PSC, primary sclerosing cholangitis; TGR5, G-protein-coupled bile acid receptor 1; TUDCA, tauroursodeoxycholic acid; UC, Ulcerative Colitis; UDCA, ursodeoxycholic acid; VDR, Vitamin D receptor.
Gut dysbiosis is one of the most probable inducers of IBD. Patients with ulcerative colitis are six times more likely to develop CRC than the general population178. Although IBD-associated CRC accounts for only 1–2% of all cases of CRC, IBD with colon involvement is among the top three high-risk conditions for CRC178. CRC accounts for approximately 10–15% of all deaths among patients with IBD178. Probiotics — live microorganisms which, when administered in adequate amounts, confer a health benefit on the host179 — are ingested to normalize the gut microbiota and improve mucosal barrier function through different mechanisms including the production of short-chain fatty acids (SCFAs), which activate GPCRs such as TGR5 on enterocytes and immune cells to increase the amounts of anti-inflammatory cytokines (such as IL-10) and TReg cells180. A systematic Review of 14 studies in patients with Crohn’s Disease and 21 studies in patients with Ulcerative Colitis revealed that patients with Crohn’s Disease that had taken probiotics in conjunction with conventional therapy did not show a substantial clinical improvement in disease symptoms; however, addition of probiotics to conventional treatment with mesalamine improved both induction and maintenance of remission for patients with ulcerative colitis181. A probiotic formulation, VSL#3 (produced by VSL Pharmaceuticals), also showed promising results for mild to moderately severe ulcerative colitis, but not in Crohn’s Disease182,183.
Bile acid receptors — such as FXR, PXR, CAR, and VDR — are therapeutic targets for the treatment of cholestatic liver disease, fatty liver disease, diabetes mellitus, gallstones, obesity and metabolic syndrome34,184,185. Almost all of the proteins involved in the production, uptake, metabolism and transport of bile acids can alter the size and composition of the bile acid pool and bile acid signalling, and thus, represent potential therapeutic targets. For example, 24-norursodeoxycholic acid (norUDCA) — a derivative of ursodeoxycholic acid (UDCA) formed by removal of a methylene group — is more hydrophilic and less toxic than UDCA68,186, resistant to conjugation and can be absorbed by cholangioctyes by passive diffusion187. NorUDCA stimulates the secretion of bicarbonate (HCO3−) from biliary cells, which forms an alkaline (HCO3−) ‘umbrella’ on the apical cholangiocyte surface, leading to deprotonation of apolar hydrophobic bile acids and therefore preventing apolar hydrophobic bile acids from entering into cholangiocytes and/or hepatocytes188. FXR agonists have been developed that have a higher affinity for FXR than endogenous bile acids, and are being evaluated as drugs for the the clinical management of primary biliary cirrhosis (PBC) and primary sclerosing cholangitis (PSC)189,190. Both UDCA therapy and the synthetic FXR agonist PX-102 are under current clinical assessment in phase II and phase III clinical trials, respectively191. The bile acid derivative obeticholic acid (OCA), has been approved by the FDA for the treatment of PBC, both in combination with UDCA in adults with an inadequate response to UDCA or as monotherapy in adults who are unable to tolerate UDCA192.
TGR5 agonists have shown promise for the control of inflammation in human cell-based models and animal models of both CD and UC73,193; however, these agents are not being tested in clinical trials. Several TGR5 agonists have been designed to confer specificity for the TGR5 receptor over FXR in the treatment of TGR5-mediated disorders, which limits adverse effects linked to an inadvertent activation of FXR194. Methylation at the C-23 position of endogenous bile acids promoted selectivity for TGR5195, which led to the development of INT-777, a semisynthetic bile acid196 derived from CA. Nonsteroidal TGR5 agonists include naturally occurring compounds such as oleanolic acid, which is derived from olive leaves, and synthetic compounds such as 3-Aryl-4-isoxazolecarboxamides that are derived from chemical libraries193. Although TGR5 agonists have shown promise in control of hepatic and gastrointestinal inflammation73,193, additional studies are necessary before translation to the clinical setting.
Recently bile acid-binding resins (or bile acid sequestrants) such as cholestyramine and colestimide have been used to remove bile acids from the intestine via chelating with them and promoting their excretion in faeces, leading to decreased enterohepatic circulation of bile acids and the accelerated conversion of cholesterol to bile acids via increased bile acid synthesis due to decreased bile acid reabsorption in the intestine197. Due to the metabolic interactions between the intestinal bile acid pool and the gut microbiota, the administration of bile acid-binding resins might readily alter the composition of intestinal microbial species, and might also have therapeutic value in the treatment of gastrointestinal cancers, particularly HCC. In addition, inhibition of enterohepatic circulation using ASBT inhibitor SC-435 improved both hepatic and systemic symptoms of nonalcoholic fatty liver disease (NAFLD) in mice fed a HFD198. Another ASBT inhibitor, A4250, ameliorated liver cholestasis and bile duct injury in a mouse model of PSC199.
The use of intestinal microbial strains that are engineered to increase the direct consumption of pathogenic metabolites or exogenous chemicals also represents a novel strategy to target the metabolic crosstalk between the liver, bile acids and gut microbiota for the prevention of gastrointestinal cancers. It has been shown that hyperammonemia can be effectively ameliorated in mouse models by administration of a strain of Lactobacillus plantarum that was genetically engineered to consume ammonia, compared with a wild-type L. plantarum strain200 ; the ammonia hyperconsuming L. plantarum strain lowered ammonia levels in the blood and faeces and improved the survival of hyperammonemic mice compared with hyperammonemic mice treated with the wild-type strain. Similarly, L. plantarum 80 (pCBH1), a genetically engineered strain overexpressing BSH, efficiently degraded GDCA and TDCA in vitro201. Such intervention strategies are advantageous over other strategies based on modulation of the gut microbiota; genetically engineered microbial strains can be directed specifically to the pathogenic molecule or metabolic pathways of interest, rather than targeting the entire enteral microbial community — a ‘silver-bullet’ approach.
Conclusions
Apart from facilitating the absorption of dietary fats, bile acids act as strong signalling molecules through different nuclear receptors and cell signalling pathways to regulate lipid, glucose and energy metabolism. Research in the past three decades has contributed substantially to our understanding of the mechanisms of bile acid metabolism and also to the pathogenesis of gastrointestinal cancers. In this Review, we discussed the mechanisms of bile acid synthesis, transport and metabolism, and highlighted the signalling regulation of bile acid synthesis and transport in normal and disease states. We discussed the mechanistic links between bile acids, gut microbiota and gastrointestinal inflammation and cancers, with a focus on CRC and HCC. Bile acids, bile acid-activated nuclear receptors FXR, PXR, CAR and VDR, as well as TGR5, are key therapeutic targets for the development of drugs to treat gastrointestinal cancers.
Our knowledge of the functional relevance of the human gut microbiota in gastrointestinal carcinogenesis is still in its infancy. With the advent of new profiling tools and strategies, including metagenomics and metabolomics, we are rapidly gaining mechanistic insights into the metabolic interactions between the gut and the liver, and into the signalling pathways that regulate the development of gastrointestinal cancers. Modulation of the gut microbiota and bile acid profile represent novel therapeutic approaches for the treatment of gastrointestinal cancers, including CRC and HCC; this strategy holds much promise and clearly represents the next frontier for gastrointestinal cancer research. Given the complexity of the processes involved in bile acid signalling, as well as the complex metabolic interactions between the gut microbiota and the host, a systems biology perspective is required to understand the complexities of liver–bile acid–microbiota cross-talk, and its influence in gastrointestinal carcinogenesis. The individual components of the liver–bile acid–microbiota axis cannot be isolated, and the successful treatment of gastrointestinal diseases will involve therapeutic interventions that modulate all of these components. Future research to inform the control and prevention of gastrointestinal cancers might include identifying clusters of beneficial commensal microbiota, and their critical metabolites, which modulate the inflammation that drives gastrointestinal carcinogenesis. Multifaceted approaches that integrate metagenomic and metabolomic profiling from well-characterized gnotobiotic animal models will be essential in determining the role of microbiota-mediated alterations in bile acids in the development of gastrointestinal cancers. In addition, distinct profiles of gut microbiota and bile acids have been reported in males and females, and these might contribute to the sex-based disparity in liver carcinogenesis202. A better understanding of the hormonal actions of bile acids on their receptors will reveal opportunities for prevention and control of gastrointestinal cancers that often exhibit gender differences. The prevention of bile acid toxicity in the intestine and liver by control of the quantity and types of bile acids that are present will require a greater understanding of the regulation of the cellular signalling pathways involved in synthesis, transport and metabolism of bile acids.
Key points.
Bile acids are critical components of the gastrointestinal tract that link the gut microbiota to hepatic and intestinal metabolism, and thus influence gastrointestinal motility, intestinal permeability, and carcinogenesis
The gut microbiota regulate bile acid production and signalling via biotransformation of intestinal bile acids to unconjugated and secondary forms that readily activate the bile acid receptors
Bile acids are ligands for G protein-coupled bile acid receptor (TGR5) and for the nuclear hormone receptor farnesoid X receptor (FXR)
The profiles of bile acids and gut microbiota influence each other; bile acids can modulate microbiota composition, which in turn regulates the size and composition of the bile acid pool
Disruption bile acid–microbiota cross-talk promotes inflammation and a gastrointestinal disease phenotype, which can contribute to carcinogenesis of gastrointestinal cancers including colorectal cancer and hepatocellular carcinoma
Modulation of gut microbiota and bile acid profiles holds promise as a novel therapeutic approach for the treatment of gastrointestinal cancers, and represents the next frontier for gastrointestinal cancer research
Acknowledgments
W.J. and G.X.X. are supported by grants from the NIH (1U01CA188387-01A1).
Glossary Terms
- Hypochlorhydria
a deficiency of hydrochloric acid in the stomach
- Enterotoxigenic bacteria
bacteria that cause disease in humans and domestic animals by producing enterotoxin
- Bile canaliculus
a thin tube that collects bile secreted by hepatocytes
- Enterohepatic circulation
the circulation of bile salts, bilirubin, drugs or other substances from the liver to the bile, followed by entry into the small intestine, absorption by the enterocyte and return to the liver via the portal circulation
- Lithogenic diet
a diet designed to increase the likelihood of stone formation, particularly gallstones
- Primary biliary cirrhosis
a type of liver disease caused by damage to the bile ducts in the liver
- Primary sclerosing cholangitis
a chronic liver disease characterized by a progressive course of cholestasis with inflammation and fibrosis of the intrahepatic and extrahepatic bile ducts
- Hyperammonemia
a metabolic disturbance characterized by an excess of ammonia in the blood
- Senescence-associated secretory phenotype
cells induced to senesce by genotoxic stress secrete proinflammatory cytokines, chemokines, and proteases associated with inflammation and malignancy
Biographies
Dr. Wei Jia is a Professor and Associate Director at the University of Hawaii Cancer Center (UHCC), Honolulu, USA. Previously, he spent 5 years as a Professor at University of North Carolina at Greensboro, USA, and as Co-Director of the UNCG Center for Translational Biomedical Research. Prior to his appointment with UNCG, Dr. Jia worked in China for 10 years as a Professor and Vice Dean at Tianjin University and Shanghai Jiao Tong University. Dr. Jia received his Ph.D. in Radiopharmaceutical Chemistry in 1996 from the University of Missouri, Columbia, USA. He has published over 260 papers in peer-reviewed journals including in top journals such as Science, Science Translational Medicine and Nature Reviews Drug Discovery. Dr. Jia’s research interest involves defining the molecular mechanisms that link metabolic disruptions in gut microbial-host co-metabolism to metabolic disorders and gastrointestinal cancer.
Dr. Guoxiang Xie is presently an assistant specialist (professor) at the University of Hawaii Cancer Center (UHCC), Honolulu, USA. He worked as a research scientist at Center for Translational Biomedical Research at the University of North Carolina at Greensboro before joining UHCC. In 2007, Dr. Xie received his M.D. degree in Pharmaceuticals from Shanghai Jiao Tong University, Shanghai, China. He has published over 80 peer-reviewed papers on metabolomics and biomedical research. His current research focuses on conducting comprehensive metabolomic studies to identify the metabolite markers for disease diagnosis and stratification, and to define the molecular mechanisms that link metabolic disruptions in gut microbial-host co-metabolism to metabolic disorders and gastrointestinal cancer.
Dr. Weiping Jia is the Director of Shanghai Clinical Center for Diabetes and Shanghai Diabetes Institute, Shanghai, China, and the President of Chinese Diabetes Society. She received her PhD and MD from Shanghai Jiao Tong University, Shanghai, China. She has published over 300 papers in international journals and received numerous awards for her diabetes research. She served as an advisory board member of Lancet Diabetes Endocrinology and Associate Editor of Diabetes Research and Clinical Practice, and the Journal of Diabetes Investigation. Her research focuses on the epidemiology, prevention, diagnosis, and therapy of metabolic diseases including diabetes and liver diseases in the Chinese population.
Footnotes
Author Contributions
Wei J. and G.X. researched data for the article, and wrote the article. Wei J., G.X. and Weiping J. made substantial contributions to discussion of content, and reviewed/edited the manuscript before submission.
Competing interests statement
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Subject Categories
Health sciences/Pathogenesis/Oncogenesis [URI /692/420/755]
Health sciences/Gastroenterology/Gastrointestinal diseases/Gastrointestinal cancer/Liver cancer/Hepatocellular carcinoma [URI /692/4020/1503/1504/1610/4029]
Health sciences/Diseases/Gastrointestinal diseases/Gastrointestinal cancer/Colorectal cancer/Colon cancer [URI /692/699/1503/1504/1885/1393]
Health sciences/Gastroenterology/Gastrointestinal system/Microbiota [URI /692/4020/2741/2135]
References
- 1.Mucci LA, Wedren S, Tamimi RM, Trichopoulos D, Adami HO. The role of gene-environment interaction in the aetiology of human cancer: examples from cancers of the large bowel, lung and breast. J Intern Med. 2001;249:477–493. doi: 10.1046/j.1365-2796.2001.00839.x. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society. About Colorectal Cancer. 2017 https://www.cancer.org/content/dam/CRC/PDF/Public/8604.00.pdf.
- 4.International Agency for Research on Cancer. GLOBOCAN 2012: Estimated Incidence, Mortality and Prevalence Worldwide in 2012. 2012 http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- 5.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 6.American Cancer Society. Key Statistics About Liver Cancer. 2017 https://www.cancer.org/cancer/liver-cancer/about/what-is-key-statistics.html.
- 7.Jia W, Li H, Zhao L, Nicholson JK. Gut microbiota: a potential new territory for drug targeting. Nat Rev Drug Discov. 2008;7:123–129. doi: 10.1038/nrd2505. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson JK, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 9.Monte MJ, Marin JJG, Antelo A, Vazquez-Tato J. Bile acids: Chemistry, physiology, and pathophysiology. World J Gastroenterol. 2009;15:804–816. doi: 10.3748/wjg.15.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swann JR, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumas ME, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes E, et al. Therapeutic modulation of microbiota-host metabolic interactions. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004244. 137rv136. [DOI] [PubMed] [Google Scholar]
- 13.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 14.Corrêa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MAR. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology. 2016;5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, et al. Melamine-induced renal toxicity is mediated by the gut microbiota. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005114. 172ra122. [DOI] [PubMed] [Google Scholar]
- 17.Ahn J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 19.Sheh A, Fox JG. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes. 2013;4:505–531. doi: 10.4161/gmic.26205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss SF. The Clinical Evidence Linking Helicobacter pylori to Gastric Cancer. Cell Mol Gastroenterol Hepatol. 2017;3:183–191. doi: 10.1016/j.jcmgh.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carboni M, et al. Chronic atrophic gastritis and risk of N-nitroso compounds carcinogenesis. Langenbecks Arch Chir. 1988;373:82–90. doi: 10.1007/BF01262769. [DOI] [PubMed] [Google Scholar]
- 22.Caygill CP, Hill MJ, Braddick M, Sharp JC. Cancer mortality in chronic typhoid and paratyphoid carriers. Lancet. 1994;343:83–84. doi: 10.1016/s0140-6736(94)90816-8. [DOI] [PubMed] [Google Scholar]
- 23.Prieto AI, Ramos-Morales F, Casadesús J. Bile-Induced DNA Damage in Salmonella enterica. Genetics. 2004;168:1787–1794. doi: 10.1534/genetics.104.031062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boleij A, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy AN, et al. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:15. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen GY, Shaw MH, Redondo G, Nunez G. The innate immune receptor Nod1 protects the intestine from inflammation-induced tumorigenesis. Cancer Res. 2008;68:10060–10067. doi: 10.1158/0008-5472.CAN-08-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quante M, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21:36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernstein C, et al. Carcinogenicity of deoxycholate, a secondary bile acid. Arch Toxicol. 2011;85:863–871. doi: 10.1007/s00204-011-0648-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lozano E, et al. Cocarcinogenic effects of intrahepatic bile acid accumulation in cholangiocarcinoma development. Mol Cancer Res. 2014;12:91–100. doi: 10.1158/1541-7786.MCR-13-0503. [DOI] [PubMed] [Google Scholar]
- 31.Forman BM, et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 32.Seol W, Choi HS, Moore DD. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 33.Hylemon PB, et al. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 35.Zollner G, Wagner M, Trauner M. Nuclear receptors as drug targets in cholestasis and drug-induced hepatotoxicity. Pharmacol Ther. 2010;126:228–243. doi: 10.1016/j.pharmthera.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol. 2008;8:685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen K, Jaeschke H, Copple BL. Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol. 2011;178:175–186. doi: 10.1016/j.ajpath.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res. 2009;50:2340–2357. doi: 10.1194/jlr.R900012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Axelson M, et al. Bile acid synthesis in cultured human hepatocytes: support for an alternative biosynthetic pathway to cholic acid. Hepatology. 2000;31:1305–1312. doi: 10.1053/jhep.2000.7877. [DOI] [PubMed] [Google Scholar]
- 41.Hylemon PB, et al. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 43.Meier PJ, Stieger B. Bile salt transporters. Annu Rev Physiol. 2002;64:635–661. doi: 10.1146/annurev.physiol.64.082201.100300. [DOI] [PubMed] [Google Scholar]
- 44.Zollner G, Marschall HU, Wagner M, Trauner M. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: pathogenetic and therapeutic considerations. Mol Pharm. 2006;3:231–251. doi: 10.1021/mp060010s. [DOI] [PubMed] [Google Scholar]
- 45.Lemoy MJMF, Westworth DR, Ardeshir A, Tarara RP. Reference Intervals for Preprandial and Postprandial Serum Bile Acid in Adult Rhesus Macaques (Macaca mulatta) J Am Assoc Lab Anim Sci. 2013;52:444–447. [PMC free article] [PubMed] [Google Scholar]
- 46.Holm R, Mullertz A, Mu H. Bile salts and their importance for drug absorption. Int J Pharm. 2013;453:44–55. doi: 10.1016/j.ijpharm.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Geyer J, Wilke T, Petzinger E. The solute carrier family SLC10: more than a family of bile acid transporters regarding function and phylogenetic relationships. Naunyn Schmiedebergs Arch Pharmacol. 2006;372:413–431. doi: 10.1007/s00210-006-0043-8. [DOI] [PubMed] [Google Scholar]
- 48.Dawson PA. Role of the Intestinal Bile Acid Transporters in Bile Acid and Drug Disposition. Handb Exp Pharmacol. 2011:169–203. doi: 10.1007/978-3-642-14541-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gadaleta RM, Cariello M, Sabba C, Moschetta A. Tissue-specific actions of FXR in metabolism and cancer. Biochim Biophys Acta. 2015;1851:30–39. doi: 10.1016/j.bbalip.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Matsubara T, Li F, Gonzalez FJ. FXR signaling in the enterohepatic system. Mol Cell Endocrinol. 2013;368:17–29. doi: 10.1016/j.mce.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kullak-Ublick GA, Stieger B, Meier PJ. Enterohepatic bile salt transporters in normal physiology and liver disease. Gastroenterology. 2004;126:322–342. doi: 10.1053/j.gastro.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Hata S, et al. Substrate specificities of rat oatp1 and ntcp: implications for hepatic organic anion uptake. Am J Physiol Gastrointest Liver Physiol. 2003;285:G829–839. doi: 10.1152/ajpgi.00352.2002. [DOI] [PubMed] [Google Scholar]
- 53.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanimirov B, Stankov K, Mikov M. Bile acid signaling through farnesoid X and TGR5 receptors in hepatobiliary and intestinal diseases. Hepatobiliary Pancreat Dis Int. 2015;14:18–33. doi: 10.1016/s1499-3872(14)60307-6. [DOI] [PubMed] [Google Scholar]
- 55.Botham KM, Boyd GS. The metabolism of chenodeoxycholic acid to beta-muricholic acid in rat liver. Eur J Biochem. 1983;134:191–196. doi: 10.1111/j.1432-1033.1983.tb07550.x. [DOI] [PubMed] [Google Scholar]
- 56.Bergstrom S, Danielsson H, Goransson A. On the bile acid meataboism in the pig. Acta Chem Scand. 1959;13:776–783. [Google Scholar]
- 57.Martin FP, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao J, et al. Association between serum bile acid profiles and gestational diabetes mellitus: A targeted metabolomics study. Clin Chim Acta. 2016;459:63–72. doi: 10.1016/j.cca.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 59.García-Cañaveras JC, Donato MT, Castell JV, Lahoz A. Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method. J Lipid Res. 2012;53:2231–2241. doi: 10.1194/jlr.D028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sayin SI, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Stanimirov B, Stankov K, Mikov M. Pleiotropic functions of bile acids mediated by the farnesoid X receptor. Acta Gastroenterol Belg. 2012;75:389–398. [PubMed] [Google Scholar]
- 62.Gérard P. Metabolism of Cholesterol and Bile Acids by the Gut Microbiota. Pathogens. 2014;3:14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parks DJ, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 64.Makishima M, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 65.Vaquero J, Monte MJ, Dominguez M, Muntane J, Marin JJ. Differential activation of the human farnesoid X receptor depends on the pattern of expressed isoforms and the bile acid pool composition. Biochem Pharmacol. 2013;86:926–939. doi: 10.1016/j.bcp.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 66.Song P, Rockwell CE, Cui JY, Klaassen CD. Individual bile acids have differential effects on bile acid signaling in mice. Toxicol Appl Pharmacol. 2015;283:57–64. doi: 10.1016/j.taap.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanda T, et al. Regulation of expression of human intestinal bile acid-binding protein in Caco-2 cells. Biochem J. 1998;330(Pt 1):261–265. doi: 10.1042/bj3300261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Beuers U, Trauner M, Jansen P, Poupon R. New paradigms in the treatment of hepatic cholestasis: from UDCA to FXR, PXR and beyond. J Hepatol. 2015;62:S25–37. doi: 10.1016/j.jhep.2015.02.023. [DOI] [PubMed] [Google Scholar]
- 69.Duboc H, Taché Y, Hofmann AF. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig Liver Dis. 2014;46:302–312. doi: 10.1016/j.dld.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maruyama T, et al. Identification of membrane-type receptor for bile acids (M-BAR) Biochem Biophys Res Commun. 2002;298:714–719. doi: 10.1016/s0006-291x(02)02550-0. [DOI] [PubMed] [Google Scholar]
- 71.Kawamata Y, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 72.Stanley LA, Horsburgh BC, Ross J, Scheer N, Wolf CR. PXR and CAR: nuclear receptors which play a pivotal role in drug disposition and chemical toxicity. Drug Metab Rev. 2006;38:515–597. doi: 10.1080/03602530600786232. [DOI] [PubMed] [Google Scholar]
- 73.Yoneno K, et al. TGR5 signalling inhibits the production of pro-inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn’s disease. Immunology. 2013;139:19–29. doi: 10.1111/imm.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J Lipid Res. 2002;43:359–364. [PubMed] [Google Scholar]
- 75.Wagner M, et al. CAR and PXR agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology. 2005;42:420–430. doi: 10.1002/hep.20784. [DOI] [PubMed] [Google Scholar]
- 76.Zhou J, Liu M, Zhai Y, Xie W. The antiapoptotic role of pregnane X receptor in human colon cancer cells. Mol Endocrinol. 2008;22:868–880. doi: 10.1210/me.2007-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Uppal H, et al. Combined loss of orphan receptors PXR and CAR heightens sensitivity to toxic bile acids in mice. Hepatology. 2005;41:168–176. doi: 10.1002/hep.20512. [DOI] [PubMed] [Google Scholar]
- 78.Stedman CA, et al. Nuclear receptors constitutive androstane receptor and pregnane X receptor ameliorate cholestatic liver injury. Proc Natl Acad Sci U S A. 2005;102:2063–2068. doi: 10.1073/pnas.0409794102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang H, et al. Pregnane X receptor activation induces FGF19-dependent tumor aggressiveness in humans and mice. J Clin Invest. 2011;121:3220–3232. doi: 10.1172/JCI41514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.He J, Nishida S, Xu M, Makishima M, Xie W. PXR prevents cholesterol gallstone disease by regulating biosynthesis and transport of bile salts. Gastroenterology. 2011;140:2095–2106. doi: 10.1053/j.gastro.2011.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chatterjee B, Echchgadda I, Song CS. Vitamin D receptor regulation of the steroid/bile acid sulfotransferase SULT2A1. Methods Enzymol. 2005;400:165–191. doi: 10.1016/S0076-6879(05)00010-8. [DOI] [PubMed] [Google Scholar]
- 82.Chen X, et al. Transactivation of rat apical sodium-dependent bile acid transporter and increased bile acid transport by 1alpha,25-dihydroxyvitamin D3 via the vitamin D receptor. Mol Pharmacol. 2006;69:1913–1923. doi: 10.1124/mol.105.020792. [DOI] [PubMed] [Google Scholar]
- 83.Schmidt DR, et al. Regulation of bile acid synthesis by fat-soluble vitamins A and D. J Biol Chem. 2010;285:14486–14494. doi: 10.1074/jbc.M110.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding N, et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell. 2013;153:601–613. doi: 10.1016/j.cell.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gadaleta RM, et al. Activation of bile salt nuclear receptor FXR is repressed by pro-inflammatory cytokines activating NF-kappaB signaling in the intestine. Biochim Biophys Acta. 2011;1812:851–858. doi: 10.1016/j.bbadis.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 87.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- 88.Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lian JS, et al. A serum metabonomic study on the difference between alcohol- and HBV-induced liver cirrhosis by ultraperformance liquid chromatography coupled to mass spectrometry plus quadrupole time-of-flight mass spectrometry. Chin Med J. 2011;124:1367–1373. [PubMed] [Google Scholar]
- 90.Wang XN, et al. Urinary Metabolite Variation Is Associated with Pathological Progression of the Post-Hepatitis B Cirrhosis Patients. J Proteome Res. 2012;11:3838–3847. doi: 10.1021/pr300337s. [DOI] [PubMed] [Google Scholar]
- 91.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 92.Landrier JF, Eloranta JJ, Vavricka SR, Kullak-Ublick GA. The nuclear receptor for bile acids, FXR, transactivates human organic solute transporter-alpha and -beta genes. Am J Physiol Gastrointest Liver Physiol. 2006;290:G476–485. doi: 10.1152/ajpgi.00430.2005. [DOI] [PubMed] [Google Scholar]
- 93.Boyer JL, et al. Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTalpha-OSTbeta in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1124–1130. doi: 10.1152/ajpgi.00539.2005. [DOI] [PubMed] [Google Scholar]
- 94.Lu TT, et al. Molecular Basis for Feedback Regulation of Bile Acid Synthesis by Nuclear Receptors. Mol Cell. 6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 95.Sinal CJ, et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 96.Yang F, et al. Spontaneous development of liver tumors in the absence of the bile acid receptor farnesoid X receptor. Cancer Res. 2007;67:863–867. doi: 10.1158/0008-5472.CAN-06-1078. [DOI] [PubMed] [Google Scholar]
- 97.Cuperus FJ, Claudel T, Gautherot J, Halilbasic E, Trauner M. The role of canalicular ABC transporters in cholestasis. Drug Metab Dispos. 2014;42:546–560. doi: 10.1124/dmd.113.056358. [DOI] [PubMed] [Google Scholar]
- 98.Halilbasic E, Claudel T, Trauner M. Bile acid transporters and regulatory nuclear receptors in the liver and beyond. J Hepatol. 2013;58:155–168. doi: 10.1016/j.jhep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zollner G, et al. Hepatobiliary transporter expression in human hepatocellular carcinoma. Liver Int. 2005;25:367–379. doi: 10.1111/j.1478-3231.2005.01033.x. [DOI] [PubMed] [Google Scholar]
- 100.Anwer MS. Intracellular Signaling by Bile Acids. Journal of Bio-Science. 2014;20:23. doi: 10.3329/jbs.v20i0.17647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pikarsky E, et al. NF-[kappa]B functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 102.Tom L, Robert FS. NF-κB in the liver—linking injury, fibrosis and hepatocellular carcinoma. Nature Reviews Gastroenterology & Hepatology. 2011;8:108. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jung IH, et al. Predominant Activation of JAK/STAT3 Pathway by Interleukin-6 Is Implicated in Hepatocarcinogenesis. Neoplasia. 2015;17:586–597. doi: 10.1016/j.neo.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nag S, Qin J, Srivenugopal KS, Wang M, Zhang R. The MDM2-p53 pathway revisited. J Biomed Res. 2013;27:254–271. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bernstein H, Bernstein C, Payne CM, Dvorakova K, Garewal H. Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res. 2005;589:47–65. doi: 10.1016/j.mrrev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 106.Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. 2009;15:3329–3340. doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ouyang X, Ghani A, Mehal WZ. Inflammasome biology in fibrogenesis. Biochim Biophys Acta. 2013;1832:979–988. doi: 10.1016/j.bbadis.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 108.Degirolamo C, et al. Prevention of spontaneous hepatocarcinogenesis in farnesoid X receptor-null mice by intestinal-specific farnesoid X receptor reactivation. Hepatology. 2015;61:161–170. doi: 10.1002/hep.27274. [DOI] [PubMed] [Google Scholar]
- 109.Kong B, et al. Mice with hepatocyte-specific FXR deficiency are resistant to spontaneous but susceptible to cholic acid-induced hepatocarcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2016;310:G295–302. doi: 10.1152/ajpgi.00134.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Knisely AS, et al. Hepatocellular carcinoma in ten children under five years of age with bile salt export pump deficiency. Hepatology. 2006;44:478–486. doi: 10.1002/hep.21287. [DOI] [PubMed] [Google Scholar]
- 111.Iannelli F, et al. Massive gene amplification drives paediatric hepatocellular carcinoma caused by bile salt export pump deficiency. Nat Commun. 2014;5:3850. doi: 10.1038/ncomms4850. [DOI] [PubMed] [Google Scholar]
- 112.Amaral JD, Viana RJ, Ramalho RM, Steer CJ, Rodrigues CM. Bile acids: regulation of apoptosis by ursodeoxycholic acid. J Lipid Res. 2009;50:1721–1734. doi: 10.1194/jlr.R900011-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Woolbright BL, Jaeschke H. Novel insight into mechanisms of cholestatic liver injury. World Journal of Gastroenterology. 2012;18:4985–4993. doi: 10.3748/wjg.v18.i36.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pean N, Doignon I, Tordjmann T. Bile acids and liver carcinogenesis: TGR5 as a novel piece in the puzzle? Clin Res Hepatol Gastroenterol. 2013;37:226–229. doi: 10.1016/j.clinre.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 115.Gadaleta RM, et al. Bile acids and their nuclear receptor FXR: Relevance for hepatobiliary and gastrointestinal disease. Biochim Biophys Acta. 2010;1801:683–692. doi: 10.1016/j.bbalip.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Y, Wang L. Nuclear Receptor Small Heterodimer Partner in Apoptosis Signaling and Liver Cancer. Cancers. 2011;3:198–212. doi: 10.3390/cancers3010198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li G, et al. Small heterodimer partner overexpression partially protects against liver tumor development in farnesoid X receptor knockout mice. Toxicol Appl Pharmacol. 2013;272:299–305. doi: 10.1016/j.taap.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wolfe A, et al. Increased activation of the Wnt/beta-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J Pharmacol Exp Ther. 2011;338:12–21. doi: 10.1124/jpet.111.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anakk S, et al. Bile acids activate YAP to promote liver carcinogenesis. Cell Rep. 2013;5:1060–1069. doi: 10.1016/j.celrep.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. 2009;15:3329–3340. doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cook JW, Kennaway EL, Kennaway NM. Production of Tumours in Mice by Deoxycholic Acid. Nature. 1940;145:627–627. [Google Scholar]
- 122.Xie G, et al. Dysregulated hepatic bile acids collaboratively promote liver carcinogenesis. Int J Cancer. 2016;139:1764–1775. doi: 10.1002/ijc.30219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xie G, et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget. 2016;7:19355–19366. doi: 10.18632/oncotarget.8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gadaleta RM, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut. 2011;60:463–472. doi: 10.1136/gut.2010.212159. [DOI] [PubMed] [Google Scholar]
- 125.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sokol H, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 127.Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 128.Dawson PA, Karpen SJ. Intestinal transport and metabolism of bile acids. J Lipid Res. 2015;56:1085–1099. doi: 10.1194/jlr.R054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota Modification with Probiotics Induces Hepatic Bile Acid Synthesis via Downregulation of the Fxr-Fgf15 Axis in Mice. Cell Rep. 7:12–18. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 130.Nagengast FM, Grubben MJ, van Munster IP. Role of bile acids in colorectal carcinogenesis. Eur J Cancer. 1995;31a:1067–1070. doi: 10.1016/0959-8049(95)00216-6. [DOI] [PubMed] [Google Scholar]
- 131.Hofmann AF, Cravetto C, Molino G, Belforte G, Bona B. Simulation of the metabolism and enterohepatic circulation of endogenous deoxycholic acid in humans using a physiologic pharmacokinetic model for bile acid metabolism. Gastroenterology. 1987;93:693–709. doi: 10.1016/0016-5085(87)90430-6. [DOI] [PubMed] [Google Scholar]
- 132.Huang Y, Chen Z. Inflammatory bowel disease related innate immunity and adaptive immunity. Am J Transl Res. 2016;8:2490–2497. [PMC free article] [PubMed] [Google Scholar]
- 133.Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jensen DD, et al. The bile acid receptor TGR5 does not interact with beta-arrestins or traffic to endosomes but transmits sustained signals from plasma membrane rafts. J Biol Chem. 2013;288:22942–22960. doi: 10.1074/jbc.M113.455774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cao W, et al. Expression of bile acid receptor TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver Physiol. 2013;304:G322–G327. doi: 10.1152/ajpgi.00263.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: Potential therapeutic targets for anti-cancer therapy. Adv Drug Deliv Rev. 2016;99:180–185. doi: 10.1016/j.addr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 137.Algars A, et al. Type and location of tumor-infiltrating macrophages and lymphatic vessels predict survival of colorectal cancer patients. Int J Cancer. 2012;131:864–873. doi: 10.1002/ijc.26457. [DOI] [PubMed] [Google Scholar]
- 138.Cook J, Hagemann T. Tumour-associated macrophages and cancer. Curr Opin Pharmacol. 2013;13:595–601. doi: 10.1016/j.coph.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 139.McMahan RH, et al. Bile Acid Receptor Activation Modulates Hepatic Monocyte Activity and Improves Nonalcoholic Fatty Liver Disease. J Bio Chem. 2013;288:11761–11770. doi: 10.1074/jbc.M112.446575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang Y, Sime W, Juhas M, Sjolander A. Crosstalk between colon cancer cells and macrophages via inflammatory mediators and CD47 promotes tumour cell migration. Eur J Cancer. 2013;49:3320–3334. doi: 10.1016/j.ejca.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 141.Calmus Y, Poupon R. Shaping macrophages function and innate immunity by bile acids: mechanisms and implication in cholestatic liver diseases. Clin Res Hepatol Gastroenterol. 2014;38:550–556. doi: 10.1016/j.clinre.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 142.Calmus Y, Poupon R. Shaping macrophages function and innate immunity by bile acids: Mechanisms and implication in cholestatic liver diseases. Clin Res Hepatol Gastroenterol. 2014;38:550–556. doi: 10.1016/j.clinre.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 143.Haselow K, et al. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J Leukoc Biol. 2013;94:1253–1264. doi: 10.1189/jlb.0812396. [DOI] [PubMed] [Google Scholar]
- 144.Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 145.Littman DR, Rudensky AY. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 2010;140:845–858. doi: 10.1016/j.cell.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 146.Shapiro H, Thaiss CA, Levy M, Elinav E. The cross talk between microbiota and the immune system: metabolites take center stage. Curr Opin Immunol. 2014;30:54–62. doi: 10.1016/j.coi.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 147.Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol Gastrointest Liver Physiol. 2013;304:29. doi: 10.1152/ajpgi.00267.2012. [DOI] [PubMed] [Google Scholar]
- 148.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Duboc H, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- 150.Attili AF, Angelico M, Cantafora A, Alvaro D, Capocaccia L. Bile acid-induced liver toxicity: relation to the hydrophobic-hydrophilic balance of bile acids. Med Hypotheses. 1986;19:57–69. doi: 10.1016/0306-9877(86)90137-4. [DOI] [PubMed] [Google Scholar]
- 151.Radominska A, et al. Human liver steroid sulphotransferase sulphates bile acids. Biochem J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gerard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2013;3:14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Van den Abbeele P, et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. Isme J. 2013;7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang T, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Goncalves P, Araujo JR, Pinho MJ, Martel F. In vitro studies on the inhibition of colon cancer by butyrate and polyphenolic compounds. Nutr Cancer. 2011;63:282–294. doi: 10.1080/01635581.2011.523166. [DOI] [PubMed] [Google Scholar]
- 156.Hague A, Elder DJ, Hicks DJ, Paraskeva C. Apoptosis in colorectal tumour cells: induction by the short chain fatty acids butyrate, propionate and acetate and by the bile salt deoxycholate. Int J Cancer. 1995;60:400–406. doi: 10.1002/ijc.2910600322. [DOI] [PubMed] [Google Scholar]
- 157.Bingham SA, et al. Dietary fibre in food and protection against colorectal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC): an observational study. Lancet. 2003;361:1496–1501. doi: 10.1016/s0140-6736(03)13174-1. [DOI] [PubMed] [Google Scholar]
- 158.Clausen MR, Bonnen H, Mortensen PB. Colonic fermentation of dietary fibre to short chain fatty acids in patients with adenomatous polyps and colonic cancer. Gut. 1991;32:923–928. doi: 10.1136/gut.32.8.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang YD, et al. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fiorucci S, et al. Protective effects of 6-ethyl chenodeoxycholic acid, a farnesoid X receptor ligand, in estrogen-induced cholestasis. J Pharmacol Exp Ther. 2005;313:604–612. doi: 10.1124/jpet.104.079665. [DOI] [PubMed] [Google Scholar]
- 162.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- 163.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 164.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic Alterations in Colorectal Cancer. Gastrointest Cancer Res. 2012;5:19–27. [PMC free article] [PubMed] [Google Scholar]
- 165.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.De Gottardi A, et al. The bile acid nuclear receptor FXR and the bile acid binding protein IBABP are differently expressed in colon cancer. Dig Dis Sci. 2004;49:982–989. doi: 10.1023/b:ddas.0000034558.78747.98. [DOI] [PubMed] [Google Scholar]
- 167.Modica S, Murzilli S, Salvatore L, Schmidt DR, Moschetta A. Nuclear bile acid receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008;68:9589–9594. doi: 10.1158/0008-5472.CAN-08-1791. [DOI] [PubMed] [Google Scholar]
- 168.Torres J, et al. Farnesoid X receptor expression is decreased in colonic mucosa of patients with primary sclerosing cholangitis and colitis-associated neoplasia. Inflamm Bowel Dis. 2013;19:275–282. doi: 10.1097/MIB.0b013e318286ff2e. [DOI] [PubMed] [Google Scholar]
- 169.Bailey AM, et al. FXR silencing in human colon cancer by DNA methylation and KRAS signaling. Am J Physiol Gastrointest Liver Physiol. 2014;306:G48–58. doi: 10.1152/ajpgi.00234.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lax S, et al. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer. 2012;130:2232–2239. doi: 10.1002/ijc.26293. [DOI] [PubMed] [Google Scholar]
- 171.Maran RR, et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J Pharm Exp Ther. 2009;328:469–477. doi: 10.1124/jpet.108.145409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164–164. doi: 10.1186/1477-7819-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Payne CM, Bernstein C, Dvorak K, Bernstein H. Hydrophobic bile acids, genomic instability, Darwinian selection, and colon carcinogenesis. Clin Exp Gastroenterol. 2008;1:19–47. doi: 10.2147/ceg.s4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Washo-Stultz D, et al. Role of mitochondrial complexes I and II, reactive oxygen species and arachidonic acid metabolism in deoxycholate-induced apoptosis. Cancer Lett. 2002;177:129–144. doi: 10.1016/s0304-3835(01)00786-8. [DOI] [PubMed] [Google Scholar]
- 175.Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm Sin B. 2015;5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Labbe A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in Crohn’s, ulcerative colitis and type 2 diabetes metagenomes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Barrasa JI, Olmo N, Lizarbe MA, Turnay J. Bile acids in the colon, from healthy to cytotoxic molecules. Toxicol In Vitro. 2013;27:964–977. doi: 10.1016/j.tiv.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 178.Mattar MC, Lough D, Pishvaian MJ, Charabaty A. Current Management of Inflammatory Bowel Disease and Colorectal Cancer. Gastrointest Cancer Res. 2011;4:53–61. [PMC free article] [PubMed] [Google Scholar]
- 179.Morelli L, Capurso L. FAO/WHO Guidelines on Probiotics: 10 Years Later. J Clin Gastroenterol. 2012;46:S1–S2. doi: 10.1097/MCG.0b013e318269fdd5. [DOI] [PubMed] [Google Scholar]
- 180.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 181.Ghouri YA, et al. Systematic review of randomized controlled trials of probiotics, prebiotics, and synbiotics in inflammatory bowel disease. Clin Exp Gastroenterol. 2014;7:473–487. doi: 10.2147/CEG.S27530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Scaldaferri F, et al. Gut microbial flora, prebiotics, and probiotics in IBD: their current usage and utility. Biomed Res Int. 2013;2013:435268. doi: 10.1155/2013/435268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mallon P, McKay D, Kirk S, Gardiner K. Probiotics for induction of remission in ulcerative colitis. Cochrane Database Syst Rev. 2007:CD005573. doi: 10.1002/14651858.CD005573.pub2. [DOI] [PubMed] [Google Scholar]
- 184.Boyer JL. Nuclear receptor ligands: rational and effective therapy for chronic cholestatic liver disease? Gastroenterology. 2005;129:735–740. doi: 10.1016/j.gastro.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 185.Zollner G, Trauner M. Nuclear receptors as therapeutic targets in cholestatic liver diseases. Br J Pharmacol. 2009;156:7–27. doi: 10.1111/j.1476-5381.2008.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Hohenester S, et al. A biliary HCO3- umbrella constitutes a protective mechanism against bile acid-induced injury in human cholangiocytes. Hepatology. 2012;55:173–183. doi: 10.1002/hep.24691. [DOI] [PubMed] [Google Scholar]
- 187.Fickert P, et al. 24-norUrsodeoxycholic Acid Is Superior to Ursodeoxycholic Acid in the Treatment of Sclerosing Cholangitis in Mdr2 (Abcb4) Knockout Mice. Gastroenterology. 2006;130:465–481. doi: 10.1053/j.gastro.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 188.Hohenester S, et al. A biliary HCO3− umbrella constitutes a protective mechanism against bile acid-induced injury in human cholangiocytes. Hepatology. 2012;55:173–183. doi: 10.1002/hep.24691. [DOI] [PubMed] [Google Scholar]
- 189.Ali AH, Carey EJ, Lindor KD. Recent advances in the development of farnesoid X receptor agonists. Ann Transl Med. 2015;3:5. doi: 10.3978/j.issn.2305-5839.2014.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.US National Library of Medicine. ClinicalTrials.gov, h. c. g. c. s. N. 2017 https://clinicaltrials.gov/ct2/show/NCT02516605.
- 191.Nevens F, et al. A Placebo-Controlled Trial of Obeticholic Acid in Primary Biliary Cholangitis. N Engl J Med. 2016;375:631–643. doi: 10.1056/NEJMoa1509840. [DOI] [PubMed] [Google Scholar]
- 192.De Magalhaes Filho CD, Downes M, Evans R. Bile Acid Analog Intercepts Liver Fibrosis. Cell. 166:789. doi: 10.1016/j.cell.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 193.Perino A, Schoonjans K. TGR5 and Immunometabolism: Insights from Physiology and Pharmacology. Trends Pharmacol Sci. 2015;36:847–857. doi: 10.1016/j.tips.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 194.Festa C, et al. Exploitation of Cholane Scaffold for the Discovery of Potent and Selective Farnesoid X Receptor (FXR) and G-Protein Coupled Bile Acid Receptor 1 (GP-BAR1) Ligands. J Med Chem. 2014;57:8477–8495. doi: 10.1021/jm501273r. [DOI] [PubMed] [Google Scholar]
- 195.Pellicciari R, et al. Nongenomic actions of bile acids. Synthesis and preliminary characterization of 23- and 6,23-alkyl-substituted bile acid derivatives as selective modulators for the G-protein coupled receptor TGR5. J Med Chem. 2007;50:4265–4268. doi: 10.1021/jm070633p. [DOI] [PubMed] [Google Scholar]
- 196.Pellicciari R, et al. Discovery of 6alpha-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. J Med Chem. 2009;52:7958–7961. doi: 10.1021/jm901390p. [DOI] [PubMed] [Google Scholar]
- 197.Kobayashi M, et al. Prevention and treatment of obesity, insulin resistance, and diabetes by bile acid-binding resin. Diabetes. 2007;56:239–247. doi: 10.2337/db06-0353. [DOI] [PubMed] [Google Scholar]
- 198.Rao A, et al. Inhibition of ileal bile acid uptake protects against nonalcoholic fatty liver disease in high-fat diet-fed mice. Sci Transl Med. 2016;8 doi: 10.1126/scitranslmed.aaf4823. 357ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Baghdasaryan A, et al. Inhibition of intestinal bile acid absorption improves cholestatic liver and bile duct injury in a mouse model of sclerosing cholangitis. J Hepatol. 2016;64:674–681. doi: 10.1016/j.jhep.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 200.Nicaise C, et al. Control of acute, chronic, and constitutive hyperammonemia by wild-type and genetically engineered Lactobacillus plantarum in rodents. Hepatology. 2008;48:1184–1192. doi: 10.1002/hep.22445. [DOI] [PubMed] [Google Scholar]
- 201.Jones ML, Chen H, Ouyang W, Metz T, Prakash S. Microencapsulated Genetically Engineered Lactobacillus plantarum 80 (pCBH1) for Bile Acid Deconjugation and Its Implication in Lowering Cholesterol. J Biomed Biotechnol. 2004;2004:61–69. doi: 10.1155/S1110724304307011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xie G, et al. Sex-dependent effects on gut microbiota regulate hepatic carcinogenic outcomes. Sci Rep. 2017:45232. doi: 10.1038/srep45232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Martin FPJ, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Molecular Systems Biology. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zheng MM, Wang RF, Li CX, Xu JH. Two-step enzymatic synthesis of ursodeoxycholic acid with a new 7β-hydroxysteroid dehydrogenase from Ruminococcus torques. Process Biochemistry. 2015;50:598–604. [Google Scholar]
- 205.Palmer RH. Bile acid sulfates. II. Formation, metabolism, and excretion of lithocholic acid sulfates in the rat. Journal of lipid research. 1971;12:680–687. [PubMed] [Google Scholar]
- 206.Lee J, et al. Adaptive regulation of bile salt transporters in kidney and liver in obstructive cholestasis in the rat. Gastroenterology. 2001;121:1473–1484. doi: 10.1053/gast.2001.29608. [DOI] [PubMed] [Google Scholar]
- 207.Nijmeijer RM, et al. Farnesoid X Receptor (FXR) Activation and FXR Genetic Variation in Inflammatory Bowel Disease. PLoS One. 2011;6:e23745. doi: 10.1371/journal.pone.0023745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Holt JA, et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 2003;17:1581–1591. doi: 10.1101/gad.1083503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Inagaki T, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 211.Mellor HR. Targeted inhibition of the FGF19-FGFR4 pathway in hepatocellular carcinoma; translational safety considerations. Liver Int. 2014;34:e1–9. doi: 10.1111/liv.12462. [DOI] [PubMed] [Google Scholar]
- 212.Halilbasic E, Baghdasaryan A, Trauner M. Nuclear receptors as drug targets in cholestatic liver diseases. Clin Liver Dis. 2013;17:161–189. doi: 10.1016/j.cld.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Prenzel N, et al. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 214.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 215.Lee HY, Crawley S, Hokari R, Kwon S, Kim YS. Bile acid regulates MUC2 transcription in colon cancer cells via positive EGFR/PKC/Ras/ERK/CREB, PI3K/Akt/IkappaB/NF-kappaB and p38/MSK1/CREB pathways and negative JNK/c-Jun/AP-1 pathway. Int J Oncol. 2010;36:941–953. doi: 10.3892/ijo_00000573. [DOI] [PubMed] [Google Scholar]
- 216.Pai R, Tarnawski AS, Tran T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell. 2004;15:2156–2163. doi: 10.1091/mbc.E03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02308111.
- 218.US National Library of Medicin. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01999101.
- 219.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02177136.
- 220.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02516605.
- 221.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02548351.
- 222.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02039219.
- 223.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01265498.
- 224.Intercept Pharmaceuticals Initiates Phase 1 Study of INT-767, a Dual FXR and TGR5 Agonist. 2015 [Google Scholar]
- 225.Pellicciari R, et al. Discovery of 6alpha-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. Journal of medicinal chemistry. 2009;52:7958. doi: 10.1021/jm901390p. [DOI] [PubMed] [Google Scholar]
- 226.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01877577.
- 227.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT03162432.
- 228.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01755507.
- 229.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT02021110.
- 230.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT03004118.
- 231.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT01857284.
- 232.US National Library of Medicine. ClinicalTrials.gov. 2017 https://clinicaltrials.gov/ct2/show/NCT00808743.


