Abstract
High-isoelectric-point (pI) α-glucosidase was purified 7,300-fold from an extract of barley (Hordeum vulgare) malt by ammonium sulfate fractionation, ion-exchange, and butyl-Sepharose chromatography. The enzyme had high activity toward maltose (kcat = 25 s−1), with an optimum at pH 4.5, and catalyzed the hydrolysis by a retaining mechanism, as shown by nuclear magnetic resonance. Acarbose was a strong inhibitor (Ki = 1.5 μm). Molecular recognition revealed that all OH-groups in the non-reducing ring and OH-3 in the reducing ring of maltose formed important hydrogen bonds to the enzyme in the transition state complex. Mass spectrometry of tryptic fragments assigned the 92-kD protein to a barley cDNA (GenBank accession no. U22450) that appears to encode an α-glucosidase. A corresponding sequence (HvAgl97; GenBank accession no. AF118226) was isolated from a genomic phage library using a cDNA fragment from a barley cDNA library. HvAgl97 encodes a putative 96.6-kD protein of 879 amino acids with 93.8% identity to the protein deduced from U22450. The sequence contains two active site motifs of glycoside hydrolase family 31. Three introns of 86 to 4,286 bp interrupt the coding region. The four exons vary from 218 to 1,529 bp. Gene expression analysis showed that transcription reached a maximum 48 h after the start of germination.
α-Glucosidases (EC 3.2.1.20) release α-d-Glc from the non-reducing ends of α-glucosides, oligosaccharides, and starch. Type I enzymes prefer aryl glucosides and Suc, type II prefer maltose and isomaltose, and type III resemble type II but also attack starch (Chiba, 1988, 1997; Frandsen and Svensson, 1998). Plant α-glucosidases have been purified to homogeneity from buckwheat (Kanaya et al., 1976), corn (Chiba and Shimomura, 1975), pea (Sun et al., 1995), rice (Takahashi et al., 1971), spinach (Sugimoto et al., 1995), sugar beet (Chiba et al., 1978), and broccoli (Monroe et al., 1999), and genes were cloned from barley (Hordeum vulgare) (Tibbot and Skadsen, 1996), sugar beet (Matsui et al., 1997), spinach (Sugimoto et al., 1997), potato (Taylor et al., 1998), and Arabidopsis (GenBank accession no. AF014806; Monroe et al., 1999). The sequences belong to glycoside hydrolase family 31, which includes fungal α-glucosidases; mammalian sucrase-isomaltase, maltase-glucoamylase, and lysosomal α-glucosidase; and α-glucosidase II in N-linked sugar biosynthesis (Henrissat, 1991).
In conjunction with β-amylase, limit dextrinase, and α-amylase, α-glucosidase in germinating seeds was proposed to mobilize endosperm starch (MacGregor, 1987). Early work addressed the purification and specificity of these enzymes in malt (Jørgensen, 1963, 1964; Jørgensen and Jørgensen, 1963, 1967). α-Glucosidase I and II (50 and 130 kD, respectively) have high activity on p-nitrophenyl α-d-glucoside, and maltase I and II (14 and 66 kD, respectively) were described subsequently (Stark and Yin, 1987). Malt α-glucosidase was found to exert an initial attack on and, with barley α-amylase, to make a synergistic 11-fold enhanced digestion of starch granules (Sun and Henson, 1990). A similar study showed a 2-fold synergy (Sissons and MacGregor, 1994), suggesting that the former α-glucosidase preparation or perhaps both preparations may have contained traces of amylolytic enzymes.
Cloning of a gibberellin-induced putative barley α-glucosidase cDNA provided new knowledge on this enzyme (Tibbot and Skadsen, 1996). The encoded 97-kD protein belonged to glycoside hydrolase family 31 (Tibbot and Skadsen, 1996) and was larger than the 33-kD enzyme reported previously (Im and Henson, 1995). Recombinant inactive and active α-glucosidases were produced in Escherichia coli and Pichia pastoris (Tibbot et al., 1998); however, the active form had approximately 300 times lower specific activity than the malt enzyme. The reason for this discrepancy is unknown, but the recombinant enzyme lacked an N-terminal region. Moreover, immunoblotting with antibodies raised against the inactive E. coli form indicated that the full-length protein was processed in germinating seeds (Tibbott et al., 1998).
In the present study, a high-pI α-glucosidase was purified 7,300-fold from 6-d old barley malt. Matrix-assisted laser desorption ionization (MALDI) mass spectrometry (MS) of tryptic fragments of this 92-kD protein showed that its sequence and that deduced from the cDNA (Tibbot and Skadsen, 1996) were very similar. This enzyme had the highest activity reported for the high-pI α-glucosidase from barley. Acarbose and 5′-thio-4-N-α-maltoside (Svensson and Sierks, 1992; Sigurskjold et al., 1994; Andrews et al., 1995) were strong inhibitors. Glucoside bonds were hydrolyzed with retention of the anomeric configuration characteristic of family 31 enzymes (Frandsen and Svensson, 1998). Recognition of deoxy maltosides indicated that all OH-groups of the non-reducing ring and OH-3 of the reducing ring contributed to transition state stabilization. The sequence of a corresponding barley α-glucosidase encoding DNA (cv Igri) of 7.1 kb contained four exons.
RESULTS
Purification and Identification of Barley Malt High-pI α-Glucosidase
A protocol was established for the purification of a highly active high-pI α-glucosidase from barley malt. The progress of purification was monitored by SDS-PAGE (Fig. 1), and the degree of purification and yields at individual steps are given in Table I. The protocol took advantage of the fact that high-pI α-glucosidase, in contrast to low-pI α-glucosidase, did not bind to DEAE-Fractogel at pH 7.5. The ratio of high- to low-pI enzymes in the ammonium sulfate precipitate of malt extract was approximately 0.25, based on the activity for maltose of the eluted low-pI (not shown) and high-pI α-glucosidases.
Figure 1.
SDS-PAGE of the purification of high-pI α-glucosidase. Lane 1, Ammonium sulfate precipitate (640 μg); lane 2, DEAE Fractogel (135 μg); lane 3, COO− Fractogel, pH 5.5 (20 μg); lane 4, rechromatography (12.4 μg); lane 5, butyl-Sepharose (1.3 μg); and lane 6, COO− Fractogel, pH 7.3 (1.2 μg). M, Marker proteins.
Table I.
Purification of high-pI α-glucosidase from 5.7 kg of barley malt flour
| Fraction | Activity with Maltose | Protein | Specific Activity | Yielde | Purification |
|---|---|---|---|---|---|
| units | E280 | U/E280 | % | -fold | |
| Extract | 14,400 | 936,000 | 0.015 | 100 | 1 |
| 20%–70% Ammonium sulfate | 8,300 | 72,530 | 0.11 | 58 | 8 |
| Fractogel DEAEa | 1,630 | 16,850 | 0.1 | 11 (58) | 6 |
| First Fractogel COO−b | 1,030 | 660 | 1.6 | 7 (36) | 103 |
| Second Fractogel COO−c | 580 | 155 | 3.8 | 4 (20) | 250 |
| Butyl-Sepharose | 150 | 6 | 25.0 | 1 (5) | 1,590 |
| Third Fractogel COO−d | 55 | 0.5 | 110.0 | 0.4 (2) | 7,300 |
Activity in the pass-through and wash in 20 mm HEPES and 5 mm CaCl2, pH 7.5.
NaCl gradient elution in 50 mm sodium acetate and 5 mm CaCl2, pH 5.5.
Rechromatography.
NaCl gradient elution in 20 mm HEPES and 5 mm CaCl2, pH 7.3.
Values in parentheses indicate the recovery corrected for the presence of low-pI α-glucosidase in the first two steps (see text).
High-pI α-glucosidase was purified 250-fold after COO−-Fractogel rechromatography at pH 5.5 concomitant with removal of abundant proteins (Fig. 1, lanes 2–4): α-amylase/subtilisin inhibitor (21 kD; Svendsen et al., 1986), (1,3;1,4)-β-glucanase (33 kD; Woodward and Fincher, 1982), β-d-glucan exohydrolase (69 kD; Hrmova et al., 1996), and lipoxygenase 2 (90 kD; Doderer et al., 1992), as identified by N-terminal sequencing, western blotting using specific antibodies, or in-gel trypsin digestion and MALDI-MS peptide mapping. The N-terminal sequence AXPKTVGVYELTKGDFSAKVTNLGATVTDD of a 38-kD protein (Fig. 1, lanes 3 and 4) has 54% identity to aldose-1-epimerase-like protein from tobacco (Nicotiana tabacum; GenBank G2739168). The 21-, 33-, and 38-kD proteins could be removed by gel filtration (Sephacryl S-200 HR), but butyl-Sepharose chromatography separated both these proteins and lipoxygenase 2 (90 kD) from α-glucosidase. The butyl-Sepharose eluate contained high-pI α-glucosidase and β-d-glucan exohydrolase, which were partially separated on COO−-Fractogel at pH 7.3, resulting in further 4.5-fold purification of the α-glucosidase (Fig. 1, lane 6). The high-pI α-glucosidase of 92 kD was thus purified 7,300-fold from malt extract (Table I) in 2% yield taking into account that the low-pI enzyme possessed 80% of the activity in the extract. The modest recovery is considered to stem from the large number of purification steps, the hydrophobic nature, and the small quantities present of the 92-kD high-pI α-glucosidase, which was reported to be processed to a predominant 81-kD form after 5 to 7 d of germination (Tibbott et al., 1998).
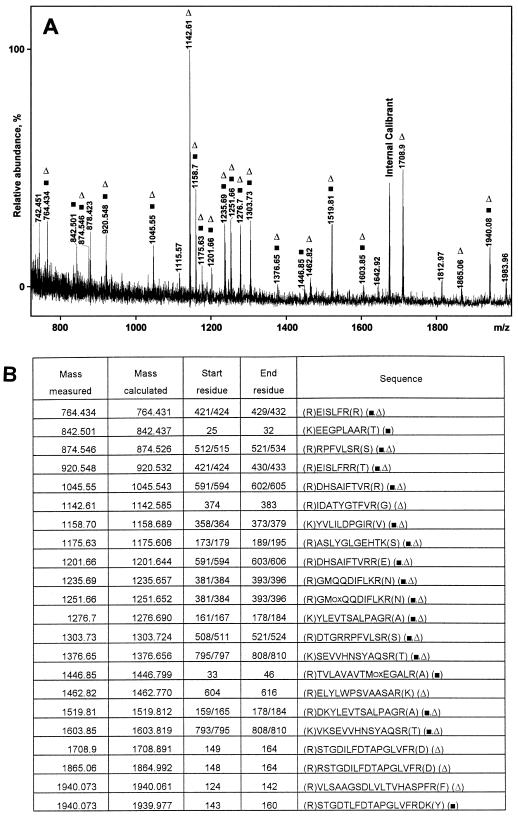
N-terminal sequencing identified β-d-glucan exohydrolase (69 kD; Hrmova et al., 1996) in the highly purified α-glucosidase. The 92-kD protein was N-terminally blocked. MALDI-MS of the mixture of tryptic fragments generated by in-gel digestion of the 92-kD protein indicated that it was likely encoded by a putative barley α-glucosidase gene for which a cDNA was cloned (Tibbot and Skadsen, 1996). Protein alignment showed 93% sequence identity between the two deduced protein sequences. Peptide masses determined by MALDI-MS were used in database searches, and the pattern (Fig. 2A) contained little noise information in the form of significant unmatched components. The matches gave (Fig. 2B) about 20% coverage of the sequence deduced from the α-glucosidase cDNA of cv Morex (Fig. 2A; Tibbot and Skadsen, 1996) and cv Igri (Fig. 2A; the present work; GenBank AF118226). A majority (14 of 22) of peptides (Fig. 2) matched both the Igri and Morex sequences, while five matched only Igri and three only Morex. This variation was expected as evident from the alignment of the Igri and Morex sequences comprising 46 differences and eight gaps. Five unmatched peaks in Figure 2A (two of medium and three of lower intensity) probably stem from parts of the Alexis α-glucosidase that vary from the known sequences. MALDI-MS gives accurate masses but provides no sequence information, so peptides from Alexis with a single amino acid substitution relative to known sequences will escape identification in the search for a match.
Figure 2.
A, MALDI-TOF MS of peptide mixture from in-gel tryptic digestion of high-pI α-glucosidase (cv Alexis). Marked signals match the deduced α-glucosidase sequence (▪, cv Morex; Δ, cv Igri). B, Peptides are marked as in A. Measured and calculated molecular masses are given as protonated mono-isotopic masses.
Organization of the Gene Encoding Barley α-Glucosidase
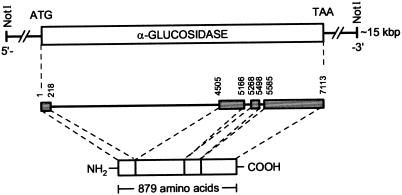
A single clone hybridizing to the cDNA clone AGL2790 (Tibbot and Skadsen 1996) was identified by rescreening two primary isolated clones from a barley genomic library at high stringency with a 2.3-kb cDNA from a barley EST library (cv Himalaya). This 15- to 18-kb genomic clone was characterized by restriction mapping, subcloned for sequencing, and found to contain the entire AGL97 gene flanked by non-coding regions. The organization of the α-glucosidase gene (HvAgl, cv Igri) and the localization of the protein coding regions by identification of exon/intron boundaries are shown in Figure 3. The gene from start to stop codon has 7,113 bases and four exons separated by introns of varying length. Exons have from 218 to 1,529 bp, whereas introns range from 86 to 4286 bp. All nucleotide sequences at exon/intron boundaries were consistent with the consensus GT/AG sequence at the donor and acceptor sites of RNA splicing. The nucleotide sequence was named HvAgl97 (GenBank accession no. AF118226). The four exons encode a protein of 879 amino acids with a calculated molar mass of 96.558 D. The theoretical pI is 6.93 (http://www.expasy.ch/tools/pi_tool.html), whereas isoelectric focusing gave an experimental value of ≥8.
Figure 3.
Genomic organization of the barley high-pI α-glucosidase gene (cv Igri).
Expression of the Barley α-Glucosidase Gene during Germination
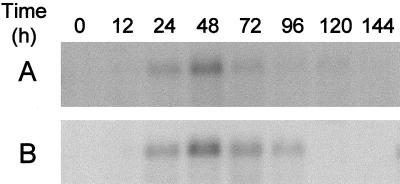
The transcription of the α-glucosidase gene in germinating seeds was followed from the grain (0 h) to 144 h (7 d) after the start of germination in the malt house. While dry grain has no detectable transcript, a very weak α-glucosidase transcription signal was detected 12 h after steeping. The signal increased during the next 12 h and expression reached a maximum after 48 h. The transcript decreased at d 3 to 4 and disappeared at d 5 to 6. In a parallel experiment, seeds (cv Alexis) were subjected to micromalting and hybridized to RNA prepared from samples taken at the same time points as those from the industrial malting. Micromalting and industrial malting gave similar expression profiles (Fig. 4), with a transcription maximum at 48 h. This demonstrated excellent agreement for a particular gene expression between performance in the malthouse and during micromalting.
Figure 4.
Northern-blot steady-state analysis of high-pI α-glucosidase mRNA (cDNA probe Lok-PS333) from industrial malt (A) and micromalt (B) 0 to 144 h after the start of steeping.
Enzymic Properties of the Barley High-pI α-Glucosidase
The pH optimum for hydrolysis of the preferred substrate maltose was at pH 4.0 to 4.5, and no activity was detected at a pH ≥ 8.0. The low amount of highly purified α-glucosidase allowed determination of kinetic parameters for only selected substrates. Maltose was the best substrate, with a kcat of 24.8 s−1 and a Km of 2.2 mm. For the α-1,6-linked isomaltose, kcat was 5 s−1 and Km was 11.0 mm, resulting in a 25-fold reduction in kcat/Km compared with maltose. p-Nitrophenyl α-d-glucoside was hydrolyzed with a kcat of 2.2 s−1 and a Km of 0.6 mm, resulting in a 3-fold decrease of kcat/Km. Preliminary tests indicated that the activity was lower on longer substrates, which agrees with results from other studies (Im and Henson, 1995). The α-glucosidase was free from contamination by α-amylase, β-amylase, and limit dextrinase in assays specific for these malt enzymes performed at a level of sensitivity allowing detection of amounts corresponding to 0.1% of the purified α-glucosidase.
Acarbose, a pseudotetrasaccharide strongly inhibiting glucoamylase, α-amylase, and other retaining or inverting α-glucoside-specific hydrolases and transferases (Legler, 1990; Sinnott, 1990; Svensson and Sierks, 1992; Sigurskjold et al., 1994; Svensson et al., 1995; Frandsen and Svensson, 1998), was a competitive inhibitor with a Ki of 1.5 μm for the high-pI α-glucosidase. This Ki value is 2- to 3-fold lower than that for barley α-amylase (Søgaard et al., 1993). Surprisingly, the maltose analog methyl 5′-thio-4-N-α-maltoside inhibited the high-pI α-glucosidase with Ki = 0.9 μm, which is extremely efficient compared with the Km of 2.5 mm for maltose. A similar relation was found for this inhibitor in maltose hydrolysis by A. niger glucoamylase (Andrews et al., 1995). The two α-glucosidase inhibitors were thus roughly 100-fold more potent than castanospermine, which was reported to give a Ki of 0.11 mm for barley high-pI α-glucosidase (Henson and Sun, 1995). Kinetic parameters and substrate specificity of different plant α-glucosidases have been reviewed (Frandsen and Svensson, 1998). Very recently, a recombinant potato enzyme was found to have a Km of 17 mm for maltose and a Vmax of 3.25 nm Glc formed h−1 μg−1 protein (Taylor et al., 1998), which was recalculated to a kcat of 0.09 s−1. It is remarkable that this catalytic constant was as low as the value determined for the recombinant barley enzyme produced in P. pastoris (Tibbot et al., 1998). Although kcat was not reported for the natural enzyme from potato, this coincidence may indicate that these large recombinant proteins were not obtained in proper functional form or that the sequence from potato corresponds to an α-glucosidase with a different functional role, possibly in the biosynthesis of glycoconjugates.
Stereochemistry of Methyl β-Maltoside Hydrolysis
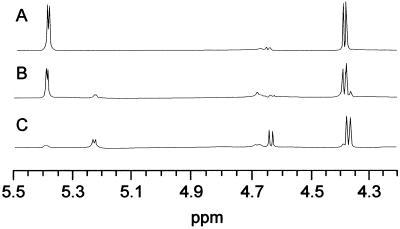
The stereochemistry of the catalytic hydrolysis of the glucoside bond was determined by 1H-NMR spectroscopy. The NMR spectra of methyl β-maltoside before (Fig. 5A) and 7 or 120 min after the addition of enzyme (Figs. 5, B and C) showed a doublet centered at 5.22 μL L−1 and assigned to H-1 of free α-Glc. The doublets at 4.37 and 4.64 μL L−1 were assigned to H-1 of the products, β-methyl Glc and β-Glc, respectively. The spectrum recorded 7 min after addition of the enzyme (Fig. 5B) indicated a ratio of 65% α-Glc to 35% β-Glc. The latter arose by mutarotation of initially formed α-Glc. Because high-pI α-glucosidase released α-Glc from the maltoside, it catalyzed glycoside bond hydrolysis with retention of the anomeric configuration, in agreement with the stereospecificity of the mechanism in glycoside hydrolase family 31 (Frandsen and Svensson, 1998).
Figure 5.
Methyl β-maltoside hydrolysis by high-pI α-glucosidase followed by 1H-NMR. Spectra were recorded before (A), 7 min after (B), and 120 min after (C) enzyme addition.
Molecular Recognition of Maltose Analogs
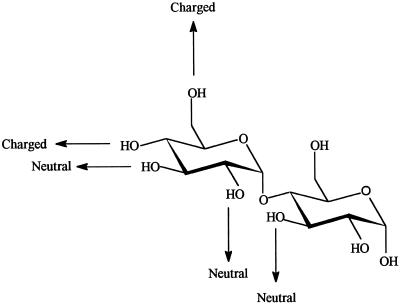
The specificity constant was determined for the α-glucosidase catalyzed hydrolysis of β-methyl maltoside and its seven monodeoxy analogs. The results made it possible to identify and assess the strength of individual intermolecular enzyme-substrate OH-hydrogen bonds implicated in stabilization of the transition state in the hydrolysis. Deoxygenation at OH-4′ or -6′ thus resulted in about 1.2 × 103-fold reduction in Vmax/Km, and at OH-3′, -2′, and -3 in modest 14- to 60-fold reductions (Table II), but no significant effect occurred when OH-1 or -2 were replaced by hydrogen (Table II). The difference in the transition state stabilization energy (ΔΔG‡) was calculated from the Vmax/Km values determined for analog and parent substrates. This molecular recognition approach is an established procedure for probing binding energy contributed by individual carbohydrate OH-groups to transition-state stabilization. ΔΔG‡ values of approximately 19 kJ mol−1 were correlated with the elimination of OH-4′ and -6′, thus indicating that these OH-groups participate in strong charged hydrogen bonds to the enzyme (Fersht et al., 1985). The OH-2′ and -3′ at the non-reducing, and the OH-3 at the reducing end ring each contributed 7 to 11 kJ mol−1 to transition-state stabilization (Table II), which is compatible with the formation of neutral hydrogen bonds between the OH-groups and the enzyme.
Table II.
Specificity constants and ΔΔG‡ values for high-pI α-glucosidase catalyzed hydrolysis of monodeoxy maltose analogs
| Substrate | Vmax/Km | ΔΔG‡ |
|---|---|---|
| kJ mol−1 | ||
| Methyl β-maltoside | 9.51 × 10−3 ± 1.77 × 10−3 | — |
| Methyl 1-deoxy-β-maltoside | 1.05 × 10−2 ± 1.02 × 10−3 | −0.3 |
| Methyl 1,2-dideoxy-β-maltoside | 1.17 × 10−2 ± 1.14 × 10−3 | −0.3 |
| Methyl 3-deoxy-β-maltoside | 6.73 × 10−4 ± 1.09 × 10−5 | 7.0 |
| 2′-Deoxy maltose | 2.83 × 10−4 ± 3.00 × 10−5 | 9.3 |
| 3′-Deoxy maltose | 1.66 × 10−4 ± 2.62 × 10−5 | 10.7 |
| 4′-Deoxy maltose | 7.53 × 10−6 ± 1.47 × 10−7 | 18.9 |
| 6′-Deoxy maltose | 8.09 × 10−6 ± 1.45 × 10−7 | 18.7 |
Assays were performed at 37°C in 50 mm sodium acetate, pH 4.5. ΔΔG‡ was calculated as −RTln[Vmax/Km)a/(Vmax/Km)b], where a and b designate analog and parent compound, respectively.
DISCUSSION
Barley high-pI α-glucosidase was purified 7,300-fold from an extract of 6-d-old malt, resulting in a specific activity of 16.5 units mg−1, corresponding to a kcat = 25 s−1. A cDNA clone encoding barley α-glucosidase was identified previously (Tibbot and Skadsen, 1996), and a corresponding recombinant protein was produced in Pichia pastoris (Tibbot et al., 1998). The Vmax of this enzyme toward maltose was 0.054 μmol min−1 mg−1 (Tibbot et al., 1998), which is equal to a kcat of 0.08 s−1, as recalculated using the theoretical size of the recombinant enzyme of 89 kD. However, this protein was not purified prior to characterization. Earlier kinetic analysis of a malt α-glucosidase resulted in a kcat of 12.6 μmol min−1 mg−1 (Im and Henson, 1995), recalculated to kcat = 20 s−1 using 97 kD, the theoretical enzyme molar mass, rather than the 33 kD reported by the authors. These authors determined the enzyme concentration by active site titration with the inhibitor castanospermine, and thus got a correct value for kcat even though the preparation contained other proteins.
The recombinant enzyme from P. pastoris was constructed to lack part of the N-terminal sequence, which perhaps resulted in low activity (Tibbot et al., 1998). However, adverse post-translational modification or misfolding may also be the cause. This comparison makes it clear that highly active, high-pI α-glucosidase of the correct size was purified in the present work. The low yield of the 92-kD α-glucosidase may in part stem from important processing to smaller forms during germination, as indicated by immunoblotting of aleurone and seed extracts using antibodies directed against the inactive recombinant enzyme produced in E. coli (Tibbot et al., 1998). The larger form was claimed to be a minor form in malt (Tibbott et al., 1998). It is not known if the forms have the same activity.
Purification of barley high-pI malt α-glucosidase of 92 kD was extremely difficult, and resulted in about 30 μg enzyme kg−1 malt. Peptide mapping by MALDI-MS of tryptic fragments showed that the sequence of this protein was very similar to the deduced cDNA sequences from cv Morex (Tibbot and Skadsen, 1996; GenBank U22450) and cv Igri (the present study) of mutual identity of 93.8% (Fig. 6). The mass of a few tryptic fragments did not match the cv Morex sequence, but did match the cv Igri sequence and vice versa. Some did not match either of these sequences, as expected because the protein was from a third cultivar, cv Alexis.
Figure 6.
Alignment of deduced protein sequences from the high-pI α-glucosidase genomic clone (AF118226, cv Igri) and cDNA clone (GenBank U22450, cv Morex). Identical residues are marked with asterisks (*). Conserved signature regions I and II in glucoside hydrolase family 31 are underlined (Frandsen and Svensson, 1998).
The DNA region encoding high-pI α-glucosidase spanned 7.1 kb, of which 37.2% is protein coding sequence. This is comparable to genes of barley β-amylase and limit dextrinase in which coding regions occupied 42.6% and 29.2%, respectively (GenBank accession nos. AF061203 and AF022725). In contrast, the coding region of barley α-amylases covered 87.3% and 73.7% of the Amy1/6-4 and Amy32b genes (GenBank accession nos. K02637 and X05166). The α-glucosidase cDNA encodes a putative 96.9-kD polypeptide (Tibbot and Skadsen, 1996), which agrees with the 92 kD found for the present malt enzyme by SDS-PAGE.
Previous expression analysis showed that transcription from an α-glucosidase gene increased during laboratory germination to a maximum at 3 to 5 d after imbibition. Increasing α-glucosidase activity was found up to d 7 in a series covering 10 d (Tibbot and Skadsen, 1996). The present study on gene expression during d 0 to 6 revealed that transcription of the gene encoding the high-pI enzyme reached a maximum at 48 h, both in micromalting and industrial malting. Because we found that the low-pI α-glucosidase was predominant in malt, it was not meaningful to attempt to correlate the present temporal transcription with the increase in enzyme activity in extracts made during germination. Recently, transcription of the barley limit dextrinase gene in the same samples used here was found to reach a maximum 72 to 96 h after imbibition (Kristensen et al., 1999). Thus, the α-glucosidase transcript preceded that of limit dextrinase and followed or coincided with that of α-amylase (Tibbot and Skadsen, 1996).
Barley high-pI α-glucosidase of glycoside hydrolase family 31 catalyzes glucoside hydrolysis with retention of the anomeric configuration resulting from a double-displacement mechanism through transition states with substantial oxocarbenium ion character (Davies and Henrissat, 1995; Frandsen and Svensson, 1998). Confirmation of this stereochemistry of methyl β-maltoside hydrolysis by 1H-NMR adds further support to the identification of the 92-kD protein. In this mechanism one carboxylic acid acts as a general acid/base catalyst and a second as a catalytic nucleophile (Sinnott, 1990; Svensson and Søgaard, 1993; McCarter and Withers, 1994; Tanaka et al., 1994; Davies and Henrissat, 1995). The pH activity dependence for the high-pI α-glucosidase (see also Henson and Sun, 1995; Tibbott et al., 1998) is compatible with a mechanism suggesting that the enzyme in the pH range around 4.5 of optimal activity has a deprotonated and a protonated carboxylic acid group at the active site. Mechanism-based active site labeling of sugar beet α-glucosidase by the inhibitor conduritol B epoxide led to identification of an essential Asp (Iwanami et al., 1995). The prominent sequence similarity with sugar beet α-glucosidase (Quaroni and Semenza, 1976; Chiba, 1997; Frandsen and Svensson, 1998) suggests that Asp-437 is the catalytic nucleophile in the barley enzyme. This agrees with site-directed mutagenesis to Asn of Asp-481 in α-glucosidase from Schizosaccharomyces pombe, leading to inactivation (Mori et al., 1999), and mechanism-based labeling of Asp-214 as a catalytic nucleophile in α-glucosidase from S. cerevisiae (McCarter and Withers, 1996a, 1996b).
Enzyme-substrate interactions through hydrogen bonds are essential in specificity and for transition state stabilization. Deoxygenated sugar analogs are widely used to identify the critical sugar OH groups and to quantitate the associated individual energies in complexes of carbohydrate-binding enzymes (Bundle and Young, 1992; Sierks and Svensson, 1992; Sierks et al., 1992; Frandsen et al., 1996; Lemieux et al., 1996). The transition-state stabilization energy (ΔΔG‡) was calculated from hydrolysis of a series of deoxy maltosides by α-glucosidase to be 19 kJ mol−1, which was attributed to charged hydrogen-bonds with OH-4′ and -6′. Neutral hydrogen bonds, contributing 7 to 11 kJ mol−1, were proposed to form with OH-2′, -3′, and -3 in maltose (Fig. 7). Remarkably, all four OH-groups of the non-reducing sugar ring participate in intermolecular hydrogen bonds in the enzyme-substrate transition state complex, whereas in the reducing end ring only OH-3 contributed to stabilization and did so less strongly than each of the OH-groups from the other ring. This pattern indicates that the enzyme is of type II, having poor activity on oligosaccharide substrates and starch. The reasonable activity on 4-nitrophenyl α-d-glucopyranoside, which lacks a hydrogen bond donor equivalent to OH-3, was explained by 4-nitrophenol being a good leaving group that compensated for the lack of the OH-3-mediated hydrogen bond to the enzyme.
Figure 7.
Schematic representation of proposed charged or neutral hydrogen bonds from maltose to high-pI α-glucosidase.
In summary, highly active, high-pI α-glucosidase was purified from barley malt and classified by peptide mapping and nucleotide sequencing to glycoside hydrolase family 31. This work contributes to the knowledge about genes and molecular properties of plant α-glucosidases, and forms a basis for future studies of the role of these enzymes during germination.
MATERIALS AND METHODS
Materials
Isomaltose, maltose, p-nitrophenyl α-d-glucopyranoside, Glc oxidase (Aspergillus niger), and the Glc oxidase kit were from Sigma-Aldrich, St. Louis). Red-Pullulan was from Megazyme (Ireland). Insoluble Blue starch, butyl-Sepharose 4 FF, SDS-PAGE, and isoelectric focusing gels were from Amersham-Pharmacia Biotech (Uppsala), and the β-amylase assay kit was from Behring Diagnostics (American Hoechst, Charlotte, NC). Fractogels EMD DEAE 650 (S) and EMD COO− 650 (S) were from Merck (Darmstadt, Germany). Chemicals for Tricine-SDS-polyacrylamide gels, Ready gels, and Bio-Gel P-6 DG were from Bio-Rad Laboratories (Hercules, CA). A barley (Hordeum vulgare, cv Igri) genomic library in λFIX II was obtained from Stratagene (La Jolla, CA). Nylon Hybond-N+ and nitrocellulose membranes were from Amersham (Buckinghamshire, UK), and Schleicher & Schuell (Dassel, Germany), respectively. [32P]dCTP was from DuPont-NEN (Stevenage, UK), and the XAR5 x-ray film was from Eastman-Kodak (Rochester, NY). The DNA sequencing kit was from Perkin-Elmer Applied Biosystems (Foster City, CA). The ProtoBlot kit, restriction enzymes, and sequencing grade trypsin were from Promega Biotechnology (Madison, WI). Ultrapure water was from a Milli-Q system (Millipore, Bedford, MA). Other chemicals were of analytical grade. Monodeoxy maltosides (Bock and Pedersen, 1987; Sierks et al., 1992), methyl 5′-thio-4-N-α-maltoside (Andrews et al., 1995), and acarbose were the kind gifts of Dr. K. Bock (Carlsberg Laboratory), Dr. B.M. Pinto (Simon Fraser University, Burnaby, British Columbia, Canada), and Dr. E. Möller (Bayer AG, Wuppertal, Germany), respectively. Antibodies against barley (1,3;1,4)-β-glucanase and lipoxygenase 1/2 were kindly provided by Drs. O. Olsen and J. Rouster (Carlsberg Research Laboratory).
Plant Material
Six-day-old barley (Hordeum vulgare cv Alexis; Carlsberg Maltings) malt was dried (30 kg) in a kiln (5 d) at 28°C (residual weight 17 kg) to 8% (w/w) water content. Industrial malt samples were from Carlsberg Maltings (batch 339), and micromalt samples of the same variety were prepared in a micromalt apparatus (Automated Malting System, Phoenix-Biosystems, Australia) using European Brewery Convention standard conditions.
Enzyme Purification
Malt Extraction and Ammonium Sulfate Precipitation
Dried malt (17 kg) was milled using 4-mm plate spacing (Carlsberg Technology Development), and the flour was stirred for 1 h in 200 L of 20 mm N-(2-hydroxyethyl)piperazine-N′-(2-ethanesulfonic acid) (HEPES), 5 mm CaCl2, pH 7.5, at room temperature, followed by settling for 16 h at 4°C. The pH was kept at 7.5 by adding 1 m NaOH. The supernatant was concentrated to 10 L by ultrafiltration (DDS-RO GR81P membrane, 20-kD cutoff, Danisco, Copenhagen), and adjusted to pH 6.7. Ammonium sulfate was added to 20% (w/v) saturation, and the extract was centrifuged at 4,550g for 45 min at 4°C. Ammonium sulfate was added to the supernatant to 70% (w/v) saturation. The precipitate, collected by centrifugation in six portions of approximately 185 g each, was stored at 4°C.
Chromatographic Separations
All steps were at 4°C. Ammonium sulfate precipitate (370 g) was stirred (1 h) into 700 mL of 20 mm HEPES and 0.5 m CaCl2, pH 7.5, centrifuged (4,550g, 30 min), and the supernatant was desalted on Bio-Gel P-6 (10 × 38 cm) in 10 mm HEPES and 2.5 mm CaCl2, pH 7.5 at a flow rate of 840 mL h−1. Fractions with high activity were pooled and applied at a flow rate of 108 mL h−1 to DEAE-Fractogel (2.6 × 30 cm) in 20 mm HEPES and 5 mm CaCl2, pH 7.5. The pass-through and the first wash (1 column volume) containing high-pI α-glucosidase were then pooled. Following continued washing (10 volumes), low-pI α-glucosidase (used in a separate study) was eluted by a linear gradient (0−0.8 m NaCl) in the buffer (2 × 900 mL; rate: 120 mL h−1). The high-pI α-glucosidase pool was dialyzed against 50 mm sodium acetate and 5 mm CaCl2, pH 5.5, applied to a COO−-Fractogel (2.6 × 25 cm; rate: 130 mL h−1) equilibrated in the buffer, and washed (10 volumes) and eluted using a linear gradient (0−0.4 m NaCl; 2 × 900 mL at 130 mL h−1). Fractions with activity were pooled, dialyzed, and rechromatographed as above on COO−-Fractogel (1.6 × 5 cm; linear gradient: 2 × 200 mL at 51 mL h−1). The pool with activity was mixed with 1 volume of 50 mm sodium acetate, 5 mm CaCl2, and 4 m NaCl, pH 5.5, and applied to butyl-Sepharose (1.6 × 2 cm) equilibrated in the same buffer containing 2 m NaCl. After a buffer wash (10 volumes), the buffer was made to 1.5 m, 1.25 m, and 1 m NaCl to be used for stepwise elution (6 column volumes each; rate: 51 mL h−1). The 1.25 and 1 m NaCl eluates containing activity were pooled, dialyzed against 20 mm HEPES and 5 mm CaCl2, pH 7.3, and applied to COO−-Fractogel (1.6 × 1 cm; rate: 60 mL h−1). After a buffer wash (10 volumes), the high-pI α-glucosidase was eluted by a linear gradient (0−0.2 m NaCl) in the buffer (2 × 20 mL at 60 mL h−1), pooled, and dialyzed against 0.1 m sodium acetate, pH 4.5. The enzyme preparation (E280 = 0.12) was stored at 4°C.
Enzymatic Procedures
Enzyme Activity Assays
Activity on maltose (15 mm) was assayed by adding 10-μL aliquots of fractions to 190 of μL 0.1 m of sodium acetate, pH 4.5, in 300-μL PCR tubes at 37°C. After a suitable reaction time (5–15 min) and enzyme inactivation (7 min, 100°C), the mixture was centrifuged. The supernatant was mixed in a microtiter plate with an enzyme-color solution (100 μL) containing 5 units mL−1 Glc oxidase, 1 unit mL−1 peroxidase, and 0.23 mg mL−1 o-dianisidine in 1 m Tris, pH 7.6. The A450 after 1 h at room temperature was read in a microtiter plate reader (Ceres Uv900Hdi, Bio-Tek) and standardized using d-Glc. One activity unit was considered to be the amount of enzyme that releases 2 μmol Glc from maltose min−1 at 37°C. Vmax and Km were obtained by fitting initial hydrolysis rates at 10 substrate concentrations (0.125 × Km − 8 × Km) to the Michaelis-Menten equation. The reaction was started by the addition of enzyme (10 μL) to substrate, and aliquots (35 μL) were transferred at intervals to Eppendorf tubes, heat inactivated, centrifuged, and added to enzyme-color solution (200 μL). The kcat = Vmax/[E0], where [E0] is the enzyme concentration determined from amino acid analysis. Ki was calculated using Ki = (100 − % inhibition) [I]/% inhibition (1 + [S]/Km). Activity for maltose (15 mm) was determined at 20 pH values (pH 2.5–7.3 in 0.1 m citrate-phosphate; pH 7.6–9.2 in 0.1 m boric acid/sodium tetraborate).
α-Amylase and β-amylase assays used Insoluble Blue Starch in 20 mm sodium acetate, 5 mm CaCl2, pH 5.0, at 37°C (Juge et al., 1995) and a mixture of 4-nitrophenyl α-d-maltopentaoside (0.85 mm) and 4-nitrophenyl α-d-maltohexaoside (0.65 mm) in the presence of ≥800 units L−1 microbial α-glucosidase in 47 mm sodium phosphate, pH 6.0, at 40°C (Mathewson and Seabourn, 1993), respectively. A test for limit dextrinase used 2% (w/v) Red Pullulan in 0.2 m sodium acetate and 5 mm CaCl2, pH 5.0, at 40°C (Kristensen et al., 1998).
Activity toward Deoxy-Maltose Analogs
Energetics of the enzyme transition state complex were mapped using seven deoxy maltose analogs. Because these were sparse, we did not determine Vmax and Km. Instead, the second-order rate constants Vmax/Km = vo/EoSo, where vo is the initial rate of hydrolysis and Eo and So are enzyme and substrate concentrations were measured. Activity was assayed at two So values near 0.1 × Km to confirm second-order conditions. Hydrolysis was started by adding enzyme (1.4−57 units mL−1) in 50- to 100-μL assays in 0.1 m sodium acetate, pH 4.5. A standard developing solution (see above) was used for deoxy analogs in the reducing ring, while the solution contained 60 units mL−1 Glc oxidase, 1 unit mL−1 peroxidase, and 0.10 mg mL−1 o-dianisidine for analogs at the non-reducing ring. The absorbance was read after 4 h at room temperature, and quantitated using the relevant deoxy sugar or d-Glc. The increase in activation energy for hydrolysis by removal of a substrate OH-group was calculated using the equation ΔΔG‡ = −RT ln[(Vmax/Km)a/(Vmax/Km)b] (Wilkinson et al., 1983), where a and b refer to the analog and parent substrates, respectively.
Stereochemistry of Glucoside Bond Hydrolysis
α-Glucosidase (62 units) from COO−-Fractogel rechromatography at pH 5.5 was passed on Sephacryl S-200 (1.6 × 85 cm) in 0.2 m sodium acetate, pH 5.5, 5 mm CaCl2. The sample (same specific activity as after butyl-Sepharose) was mixed with 10 volumes of 0.1 m sodium acetate, pH 4.5, in D2O (99%), concentrated 10-fold in a 10-kD cutoff ultrafiltration unit (Centricon, Amicon, Beverly, MA) at 4°C, and the solvent exchange was repeated. A substrate (45 mm methyl-β-maltoside, 600 μL) 1H-NMR spectrum was recorded at 310 K (AMX-600 spectrometer at 600 MHz, Bruker Instruments, Billerica, MA), and the stereochemistry of hydrolysis was followed by recording spectra at intervals after addition of enzyme.
Analysis of the Gene
Screening of Genomic Library
The barley genomic library was screened by plaque hybridization using a cDNA fragment from a barley expressed sequence tag (EST) library as a probe. This probe (LOK-PS333; cv Himalaya) corresponded to cDNA clone pAGL.2737 (GenBank U22450, cv Morex; Tibbot and Skadsen, 1996). Single plate lifts of approximately 0.5 × 106 plaques were prehybridized and hybridized at 42°C (16 h) in 5× SSC, 5× Denhart's solution, 50% (w/v) formamide, 0.2% (w/v) SDS, 10 μg mL−1 poly(A+), and 10 μL mL−1 sheared salmon sperm DNA (9.7 mg mL−1). Filters were washed three times for 15 min at 22°C in 2× SSC and 0.2% (w/v) SDS, then three times for 5 min at 65°C in 0.2× SSC and 0.1% (w/v) SDS, and exposed to XAR5 film (Eastman-Kodak) in cassettes for 16 h at −80°C. Two positive primary plaque areas were re-screened. A single confirmed positive plaque was subjected to a final screening.
Sequencing the Coding Region for the Barley α-Glucosidase
The α-glucosidase λ-clone was digested by restriction enzymes, and the DNA fragments were subcloned into pBlueScript SK−. Subcloned fragments encoding protein were sequenced using a dideoxynucleotide cycle sequencing kit (DyeDeoxy, Perkin-Elmer Applied Biosystems, Foster City, CA) and a DNA sequencer (model 373, Perkin-Elmer Applied Biosystems). Each nucleotide in the sequence was analyzed in at least two sequencing passes on both DNA strands. The samples were prepared as described by Rasmussen (1994). The DNA contig assembly was performed with ABI Prism sequencing software 1.0 for Apple Macintosh and Microgenie 6.0 (Beckman Instruments, Fullerton, CA). Analysis of nucleotide sequences was done using GCG 9.0 (Genetics Computer Group, Madison, WI) and DNA Tools 5.0 (S. Rasmussen, Carlsberg Laboratory).
Northern Analysis
Total RNA was isolated from germinating seeds (cv Alexis) as described previously (Leah and Mundy, 1989). Seed samples were taken 0 to 144 h after imbibition. For northern blots, 10 μg of RNA was separated in formaldehyde agarose gels, blotted onto Hybond-N+ membranes, and the blots were developed by a 32P−labeled DNA probe made using the Multiprime Labeling System (Amersham). RNA blots were hybridized and washed as described previously (Leah and Mundy, 1989). Electronic images of 32P-labeled blots were made and visualized using phosphor imager cassettes and ImagesQuant version 3.3 software (Molecular Dynamics, Sunnyvale, CA).
Malting and Micromalting of Barley
Uniform samples of standard malt were collected at 0 (dry grain), 12, 24, 48, 72, 96, 120, and 144 h after the start of steeping (i.e. the addition of water during an industrial malting of 65 tons of barley) (cv Alexis; Carlsberg Maltings). Samples of the same variety were prepared at the same time points using a microscale malting system on an 80-g scale (Carlsberg Technical Service). All samples were kept in liquid nitrogen and processed simultaneously.
Analytical Techniques
SDS-PAGE, Western Blotting, and Isoelectric Focusing
N-Tris(hydroxymethyl) methyl-Gly (Tricine) SDS-PAGE gels comprised a separation gel (16 × 12 × 0.075 cm) and a stacking gel (16 × 3 × 0.075 cm) of 8.25% (w/v) and 4% (w/v) acrylamide, respectively. Both were 1.5% (w/v) in bisacrylamide. Protein was denatured by boiling for 3 min in sample buffer (0.1 m Tris-HCl, pH 6.8, 8% [w/v] SDS, 21% [w/v] glycerol, and 0.02% [w/v] Coomassie Brilliant Blue G 250). The electrophoresis was run at 13°C for 16 h at 20 mA and 100 V (Bio-Rad II xi cell and power supply model 100/500; Schägger and von Jagow, 1987).
Proteins were blotted after SDS-PAGE (Phast gel, Pharmacia) to nitrocellulose by diffusion (15 min) at 70°C (Phast system, Pharmacia). The membranes were soaked for 30 min at room temperature in TBST (10 mm Tris-HCl, 0.15 m NaCl, and 0.05% [w/v] Tween 20, pH 8.0) containing 1% (w/v) bovine serum albumin, followed by TBST containing 0.1% (w/v) primary rabbit antibody (30 min), a wash (three times for 10 min), and TBST containing alkaline phosphatase-coupled goat IgG against rabbit IgG for 30 min. After washing three times for 10 min, the blot was developed in 5 mL of TBST with 21 μL of nitroblue tetrazolium (50 mg mL−1) and 16 μL of 5-bromo-4-chloro-3-indolyl-phosphate (50 mg mL−1).
Isoelectric focusing was performed with Phast gels (43 × 37 × 0.45 mm, 5% [w/v] acrylamide, 1.5% [w/v] bisacrylamide; Pharmacia), pH 3.0 to 9.0, and proteins were visualized by Coomassie Brilliant Blue R 350 or silver staining according to the instructions of the manufacturer. The gel was overlaid with 25 mm maltose, 0.1 m sodium acetate, pH 4.5, and 1% (w/v) agarose (8.5 mL) mixed with 60 mm Tris-HCl, pH 7.5, 115 units mL−1 Glc oxidase, 0.5 unit mL−1 peroxidase, 21% (w/v) glycerol (1 mL), and 2.5 mg mL−1 o-dianisidine (0.5 mL) at 37°C for 15 min to give a brown-colored zymogram.
In Situ Gel Plug Trypsin Digestion
Coomassie-stained protein bands were cut from SDS polyacrylamide (8.25% [w/v] Tricine) gels and kept in water at −18°C. Prior to digestion, the gel plug was washed three times for 20 min in 40% (w/v) acetonitrile/60% (w/v) 50 mm NH4HCO3, pH 7.8, at 37°C, and dried for 20 min in a vacuum centrifuge. Trypsin (5 μL; 0.33 μg μL−1) was added to the gel (sample:trypsin, approximately 25:1 [w/w]). After reswelling of the gel, 15 μL of 50 mm NH4HCO3, pH 7.8, was added and digestion continued at 37°C for 16 h. Peptides were extracted three times in 100 μL of 60% (w/v) acetonitrile, lyophilized, and analyzed by MALDI time of flight (TOF) MS.
MALDI-MS and Protein Identification by Database Searches
Mass spectra were acquired on a MALDI-TOF mass spectrometer (Voyager-Elite, Perseptive Biosystems, Framingham, MA) equipped with delayed ion extraction technology. Lyophilized samples were dissolved in 20 μL of 0.1% (w/v) trifluoroacetic acid and prepared for MALDI-MS analysis by placing a 0.8-μL sample on a probe tip followed by 0.4 μL of matrix solution. The MALDI matrices were 2,5-dihydroxybenzoic acid (Aldrich, Milwaukee, WI) dissolved in a mixture of 0.1% (v/v) trifluoroacetic acid and acetonitrile 2:1 (v/v) (25 g L−1), and α-cyano-4-hydroxy-cinnamic acid (Sigma-Aldrich) in 70% (v/v) acetonitrile (20 g L−1). All mass spectra were obtained in reflector mode and calibrated using internal calibration. Data processing was carried out using GRAMS/386 software (Galactic Industries, Salem, NH). Protein identification was performed by searching the European Molecular Biology Laboratory comprehensive non-redundant protein sequence database (nrdb) (ftp://ftp.embl-heidelberg.de/pub/databases/nrdb/) using the PeptideSearch software (http://www.mann.embl-heidelberg.de/Services/PeptideSearch/) (Mann et al., 1993).
Amino Acid and Amino-Terminal Sequence Analyses
Amino acid analysis was performed on 200 to 500 pmol of protein after hydrolysis (6 m HCl; 24 h in sealed, evacuated tubes; 110°C) on a Pharmacia LKB Alpha Plus equipped with an ion-exchange column. N-terminal sequence analysis was performed on 50 to 500 pmol of protein on an sequenator (model 470A or 477A, Applied Biosystems) and a PTH analyzer (model 120, Applied Biosystems).
ACKNOWLEDGMENTS
We are grateful to Sidsel Ehlers, Suksawad Vongvisuttikun, and Aksel Englyst for excellent technical assistance, and to Bodil Corneliussen, Lone Soerensen, and Dr. Ib Svendsen for N-terminal sequencing and amino acid analysis. Jean Sage and Dr. Søren W. Rasmussen are thanked for advice on DNA sequencing. Bent Ole Pedersen is acknowledged for performing the NMR spectroscopy experiments.
Footnotes
This work was supported by the Danish Research Councils' Committee for Biotechnology (grant no. 95–02014 to P.R. and B.S.).
LITERATURE CITED
- Andrews JS, Weimar T, Frandsen TP, Svensson B, Pinto BM. Novel disaccharides containing sulfur in the ring and nitrogen in the interglycosidic linkage: conformation of methyl 5′-thio-4-N-α-maltoside bound to glucoamylase and its activity as a competitive inhibitor. J Am Chem Soc. 1995;117:10799–10804. [Google Scholar]
- Bock K, Pedersen H. The substrate specificity of the enzyme amyloglucosidase (AMG): part 1. Deoxy derivatives. Acta Chem Scand. 1987;B41:617–628. doi: 10.3891/acta.chem.scand.41b-0617. [DOI] [PubMed] [Google Scholar]
- Bundle DR, Young NM. Carbohydrate-protein interactions in antibodies and lectins. Curr Opion Struct Biol. 1992;2:666–673. [Google Scholar]
- Chiba S Amylase Research Society of Japan, editors. Handbook of Amylases and Related Enzymes. Oxford: Pergamon Press; 1988. α-Glucosidases; pp. 104–105. [Google Scholar]
- Chiba S. Molecular mechanism in α-glucosidase and glucoamylase. Biosci Biotechnol Biochem. 1997;61:1233–1239. doi: 10.1271/bbb.61.1233. [DOI] [PubMed] [Google Scholar]
- Chiba S, Inomata S, Matsui H, Shimomura T. Purification and properties of an α-glucosidase (glucoamylase) in sugar beet seeds. Agric Biol Chem. 1978;42:241–245. [Google Scholar]
- Chiba S, Shimomura T. Purification and some properties of flint corn α-glucosidase. Agric Biol Chem. 1975;39:1033–1040. [Google Scholar]
- Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- Doderer A, Kokkelink I, Veen van der S, Valk BE, Schram AW, Douma AC. Purification and characterization of two lipoxygenase isoenzymes from germinating barley. Biochim Biophys Acta. 1992;1120:97–104. doi: 10.1016/0167-4838(92)90429-h. [DOI] [PubMed] [Google Scholar]
- Fersht AR, Shi J-P, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MMY, Winter G. Hydrogen bonding and biological specificity analyzed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Frandsen TP, Stoffer BB, Palcic MM, Hof S, Svensson B. Structure and energetics of the glucoamylase-isomaltose transition-state complex probed by using modeling and deoxygenated substrates coupled with site-directed mutagenesis. J Mol Biol. 1996;263:79–89. doi: 10.1006/jmbi.1996.0557. [DOI] [PubMed] [Google Scholar]
- Frandsen TP, Svensson B. Plant α-glucosidases of the glucoside hydrolase family 31: molecular properties, substrate specificity, reaction mechanism, and comparison with family members of different origin. Plant Mol Biol. 1998;37:1–13. doi: 10.1023/a:1005925819741. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarity. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson CA, Sun Z. Barley seed α-glucosidases: their characteristics and roles in starch degradation. ACS Symp Ser. 1995;618:51–58. [Google Scholar]
- Hrmova M, Harvey AJ, Wang, Shirley NJ, Jones GP, Stone BA, Høj PB, Fincher GB. Barley β-d-glucan exohydrolase with β-d-glucosidase activity: purification, characterization, and determination of primary structure from a cDNA clone. J Biol Chem. 1996;271:5277–5286. doi: 10.1074/jbc.271.9.5277. [DOI] [PubMed] [Google Scholar]
- Im H, Henson CA. Characterization of high pI α-glucosidase from germinated barley seeds: substrate specificity, subsite affinities and active-site residues. Carbohydr Res. 1995;277:145–159. [Google Scholar]
- Iwanami S, Matsui H, Kimura A, Ito H, Mori H, Honma M, Chiba S. Chemical modification and amino acid sequence of active site in sugar beet α-glucosidase. Biosci Biotechnol Biochem. 1995;59:459–463. doi: 10.1271/bbb.59.459. [DOI] [PubMed] [Google Scholar]
- Iwanami S, Nishimoto Y, Murata S, Ito H, Matsui H, Honma M, Chiba S. Identification of essential ionizable groups of rice α-glucosidase II. J Appl Glucosci. 1996;43:67–71. [Google Scholar]
- Jørgensen BB, Jørgensen OB. Barley malt α-glucosidase with isomaltase activity. Acta Chem Scand. 1963;17:1765–1770. [Google Scholar]
- Jørgensen BB, Jørgensen OB. Inhibition of barley malt α-glucosidase by tris (hydroxymethyl) aminomethane and erythritol. Biochim Biophys Acta. 1967;146:167–172. doi: 10.1016/0005-2744(67)90083-6. [DOI] [PubMed] [Google Scholar]
- Jørgensen OB. Barley malt α-glucosidase: II. Studies on the substrate specificity. Acta Chem Scand. 1963;17:2471–2478. [Google Scholar]
- Jørgensen OB. Barley malt α-glucosidase: IV. Studies on the kinetics and the active groups. Acta Chem Scand. 1964;18:1115–1124. [Google Scholar]
- Juge N, Rodenburg KW, Guo XJ, Chaix JC, Svensson B. Isozyme hybrids within the protruding third loop domain of the barley α-amylase (β/α)8-barrel: implication for BASI sensitivity and substrate affinity. FEBS Lett. 1995;363:299–303. doi: 10.1016/0014-5793(95)00291-g. [DOI] [PubMed] [Google Scholar]
- Kanaya KI, Chiba S, Shimomura T, Nishi K. Improved method for purification of buckwheat α-glucosidase and some kinetic properties. Agric Biol Chem. 1976;40:167–172. [Google Scholar]
- Kristensen M, Lok F, Planchot V, Svendsen I, Leah R, Svensson B. Isolation and characterization of the gene encoding the starch debranching enzyme limit dextranase from germinating barley. Biochem Biophys Acta. 1999;1431:538–546. doi: 10.1016/s0167-4838(99)00077-1. [DOI] [PubMed] [Google Scholar]
- Kristensen M, Planchot V, Abe J, Svensson B. Large-scale purification and characterization of barley limit dextrinase, a member of the α-amylase structural family. Cereal Chem. 1998;75:473–479. [Google Scholar]
- Leah R, Mundy J. The bifunctional α-amylase/ subtilisin inhibitor of barley: nucleotide sequence and patterns of seed-specific expression. Plant Mol Biol. 1989;12:673–682. doi: 10.1007/BF00044158. [DOI] [PubMed] [Google Scholar]
- Legler G. Glycoside hydrolases: mechanistic information from studies with reversible and irreversible inhibitors. Adv Carbohydr Chem Biochem. 1990;71:319–384. doi: 10.1016/s0065-2318(08)60034-7. [DOI] [PubMed] [Google Scholar]
- Lemieux RU, Spohr U, Bach M, Cameron DR, Frandsen TP, Stoffer BB, Svensson B, Palcic MM. Chemical mapping of the active site of glucoamylase of Aspergillus niger. Can J Chem. 1996;74:319–335. [Google Scholar]
- MacGregor AW. α-Amylase, limit dextrinase, and α-glucosidase enzymes in barley and malt. CRC Crit Rev Biotechnol. 1987;5:117–128. doi: 10.3109/07388558709086972. [DOI] [PubMed] [Google Scholar]
- Mann M, Højrup P, Roepstorff P. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- Mathewson PR, Seabourn BW. A new procedure for specific determination of β-amylase in cereals. J Agric Food Chem. 1993;31:1322–1326. [Google Scholar]
- Matsui H, Iwanami S, Ito H, Mori H, Honma M, Chiba S. Cloning and sequencing of a cDNA encoding α-glucosidase from sugar beet. Biosci Biotechnol Biochem. 1997;61:875–880. doi: 10.1271/bbb.61.875. [DOI] [PubMed] [Google Scholar]
- McCarter JD, Withers SG. Mechanisms of enzymatic glycoside hydrolysis. Curr Opin Struc Biol. 1994;4:885–892. doi: 10.1016/0959-440x(94)90271-2. [DOI] [PubMed] [Google Scholar]
- McCarter JD, Withers SG. 5-Fluoro glucosides: a new class of mechanism-based inhibitors of both α- and β-glucosidases. J Am Chem Soc. 1996a;118:241–242. [Google Scholar]
- McCarter JD, Withers SG. Unequivocal identification of Asp-214 as the catalytic nucleophile of Saccharomyces cerevisiae α-glucosidase using 5-fluoro glycosyl fluorides. J Biol Chem. 1996b;271:6889–6894. doi: 10.1074/jbc.271.12.6889. [DOI] [PubMed] [Google Scholar]
- Monroe JD, Gough CM, Chandler LE, Loch CM, Ferrante JE, Wright PW. Structure, properties, and tissue localization of apoplastic α-glucosidase in crucifers. Plant Physiol. 1999;119:385–397. doi: 10.1104/pp.119.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Okuyama M, Kimura A, Chiba S (1999) Carboxyl groups of Asp 481, Glu 484, and Asp 647 are essential for activity of α-glucosidase from Schizosaccharomyces pombe (abstract). Third Carbohydrate Bioengineering Meeting, April 11–14, Newcastle upon Tyne, UK
- Quaroni A, Semenza G. Partial amino acid sequences around the essential carboxylate in the active sites of the intestinal sucrase-isomaltase complex. J Biol Chem. 1976;251:3250–3253. [PubMed] [Google Scholar]
- Rasmussen SW. Sequencing of a 28.6 kb region of yeast chromosome XI includes FBA1 and TO A2 genes, an open reading frame (ORF) similar to a translationally controlled tumour protein, one ORF containing motifs also found in plant storage proteins and 13 ORFs with weak or no homology to known proteins. Yeast. 1994;10:63–68. doi: 10.1002/yea.320100008. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kD. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sierks MR, Bock K, Refn S, Svensson B. Active site similarities of glucose dehydrogenase, glucose oxidase and glucoamylase probed by deoxygenated substrates. Biochemistry. 1992;31:8972–8977. doi: 10.1021/bi00152a038. [DOI] [PubMed] [Google Scholar]
- Sierks MR, Svensson B. Kinetic identification of a hydrogen bonding pair in the glucoamylase/maltose transition state complex. Protein Eng. 1992;5:185–188. doi: 10.1093/protein/5.2.185. [DOI] [PubMed] [Google Scholar]
- Sigurskjold BW, Berland CR, Svensson B. Thermodynamics of inhibitor binding to the catalytic site of glucoamylase from Aspergillus niger determined by displacement titration calorimetry. Biochemistry. 1994;33:10191–10199. doi: 10.1021/bi00199a048. [DOI] [PubMed] [Google Scholar]
- Sinnott ML. Catalytic mechanisms of enzymic glycosyl transfer. Chem Rev. 1990;90:1171–1202. [Google Scholar]
- Sissons MJ, MacGregor AW. Hydrolysis of barley starch granules by α-glucosidases from malt. J Cereal Sci. 1994;19:161–169. [Google Scholar]
- Søgaard M, Kadziola A, Haser R, Svensson B. Site-directed mutagenesis of histidine 93, aspartic acid 180, glutamic acid 205, histidine 290, and aspartic acid 291 at the active site and tryptophan 279 at the raw starch binding site in barley α-amylase 1. J Biol Chem. 1993;268:22480–22484. [PubMed] [Google Scholar]
- Stark JR, Yin XS. Evidence for the presence of maltase and α-glucosidase isoenzymes in barley. J Inst Brew. 1987;93:108–112. [Google Scholar]
- Sugimoto M, Furui S, Suzuki Y. Multiple molecular forms of α-glucosidase from spinach seeds, Spinacia oleracea L. Biosci Biotechnol Biochem. 1995;59:673–677. [Google Scholar]
- Sugimoto M, Furui S, Suzuki Y. Molecular cloning and characterization of a cDNA encoding α-glucosidase from spinach. Plant Mol Biol. 1997;33:765–768. doi: 10.1023/a:1005766003923. [DOI] [PubMed] [Google Scholar]
- Sun Z, Duke SH, Henson CA. The role of pea chloroplast α-glucosidase in transitory starch degradation. Plant Physiol. 1995;108:211–217. doi: 10.1104/pp.108.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Henson CA. Degradation of native starch granules by barley α-glucosidases. Plant Physiol. 1990;94:320–327. doi: 10.1104/pp.94.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen I, Hejgaard J, Mundy J. Complete amino acid sequence of the α-amylase/subtilisin inhibitor from barley. Carlsberg Res Commun. 1986;51:43–50. [Google Scholar]
- Svensson B, Sierks MR. Role of the aromatic side chains in the binding of substrates, inhibitors, and cyclomalto-oligosaccharides to the glucoamylase from Aspergillus niger probed by perturbation difference spectroscopy, chemical modification, and mutagenesis. Carbohydr Res. 1992;227:29–44. doi: 10.1016/0008-6215(92)85059-9. [DOI] [PubMed] [Google Scholar]
- Svensson B, Søgaard M. Mutational analysis of glycosylase function. J Biotechnol. 1993;29:1–37. [Google Scholar]
- Svensson B, Stoffer B, Frandsen TP, Søgaard M, Sierks MR, Rodenburg KW, Sigurskjold BW, Dupont C. Basic molecular features, mechanism and specificity of protein–carbohydrate interactions in amylolytic enzymes. In: Bock K, Clausen H, editors. Proceedings of 36th Alfred Benzon Symposium. Copenhagen: Munksgaard; 1995. pp. 202–213. [Google Scholar]
- Takahashi N, Shimomura T, Chiba S. Studies on α-glucosidase in rice: part I. Isolation and some properties of α-glucosidase I and α-glucosidase II. Agric Biol Chem. 1971;35:2015–2024. [Google Scholar]
- Tanaka Y, Tao W, Blanchard JS, Hehre EJ. Transition state structures for the hydrolysis of α-d-glucopyranosyl fluoride by retaining and inverting reactions of glycosylases. J Biol Chem. 1994;269:32306–32312. [PubMed] [Google Scholar]
- Taylor MA, George LA, Ross HA, Davies HV. cDNA cloning and characterization of an α-glucosidase gene from potato (Solanum-tuberosum L.) Plant J. 1998;13:419–425. doi: 10.1046/j.1365-313x.1998.00051.x. [DOI] [PubMed] [Google Scholar]
- Tibbot BK, Henson CA, Skadsen RW. Expression of enzymatically active, recombinant barley α-glucosidase in yeast and immunological detection of α-glucosidase from seed tissue. Plant Mol Biol. 1998;38:379–391. doi: 10.1023/a:1006006203372. [DOI] [PubMed] [Google Scholar]
- Tibbot BK, Skadsen RW. Molecular cloning and characterization of a gibberellin-inducible, putative α-glucosidase gene from barley. Plant Mol Biol. 1996;30:229–241. doi: 10.1007/BF00020110. [DOI] [PubMed] [Google Scholar]
- Wilkinson AJ, Fersht AR, Blow DM, Winter G. Site-directed mutagenesis as a probe of enzyme structure and catalysis: tyrosyl-tRNA synthase cysteine-35 to glycine-35 mutation. Biochemistry. 1983;22:3581–3586. doi: 10.1021/bi00284a007. [DOI] [PubMed] [Google Scholar]
- Woodward JR, Fincher GB. Purification and chemical properties of two 13;1,4-β-glucan endohydrolases from germinating barley. Eur J Biochem. 1982;121:663–669. doi: 10.1111/j.1432-1033.1982.tb05837.x. [DOI] [PubMed] [Google Scholar]