Abstract
Using 13C-NMR, we demonstrate that [13C]methanol readily entered sycamore (Acer pseudoplatanus L.) cells to be slowly metabolized to [3-13C]serine, [13CH3]methionine, and [13CH3]phosphatidylcholine. We conclude that the assimilation of [13C]methanol occurs through the formation of 13CH3H4Pte-glutamate (Glu)n and S-adenosyl-methionine, because feeding plant cells with [3-13CH3]serine, the direct precursor of 13CH2H4Pte-Glun, can perfectly mimic [13CH3]methanol for folate-mediated single-carbon metabolism. On the other hand, the metabolism of [13C]methanol in plant cells revealed assimilation of label into a new cellular product that was identified as [13CH3]methyl-β-d-glucopyranoside. The de novo synthesis of methyl-β-d-glucopyranoside induced by methanol did not require the formation of 13CH3H4Pte-Glun and was very likely catalyzed by a “transglycosylation” process.
Most plants produce and emit methanol, especially during the early stages of leaf expansion, because of pectin demethylation (for review, see Fall and Benson, 1996), and this volatile organic compound exits leaves via stomata (Nemecek-Marshall et al., 1995). The proportion of methanol production that is recycled in plants is not known, but it is obvious that plant tissues metabolize methanol. Although higher plants likely do not possess methanol oxidase (this enzyme has so far only been found in microorganisms), it has been observed that they can convert [14C]methanol to 14CO2 (Cossins, 1964). A mitochondrial NAD-dependent formate dehydrogenase has been found in higher plants (Halliwell and Butt, 1974; Oliver, 1981; Colas des Francs-Small et al., 1993; Hourton-Cabassa et al., 1998; Suzuki et al., 1998), and isolated mitochondria from various tissues can oxidize formate with a tight coupling to an electrogenic translocation of protons across the inner membrane (Hourton-Cabassa et al., 1998). Formate is also a potential single-carbon source in higher plants, and several authors have shown that externally added formate is readily metabolized into Gly and Ser in a process involving the ATP-dependent synthesis of 10-formyl 5,6,7,8-tetrahydropteroylpolyglutamate (H4Pte-Glun) (Shingles et al., 1984; Prabhu et al., 1996). Indeed, a formyl H4Pte-Glun synthetase has been purified from spinach leaves (Nour and Rabinowitz, 1991); it is mainly associated with the cytosolic fraction (Kirk et al., 1994), but some activity was also detected in mitochondria (Clandinin and Cossins, 1972).
Furthermore, in higher plants 5,10-methylene H4Pte-Glun, 5,10-methenyl H4Pte-Glun, and 10-formyl H4Pte-Glun are readily interconvertible, thus providing an equilibrium between the pool of C1 units at the formyl and methylene levels of oxidation. These interconversions are catalyzed by a bi-functional methylene H4Pte-Glun dehydrogenase/methenyl H4Pte-Glun cyclohydrolase (Nour and Rabinowitz, 1991; Kirk et al., 1995; Chen et al., 1997). This bi-enzyme complex is present in various cell compartments, including the cytosol (Kirk et al., 1995; Chen et al., 1997), the mitochondria (Suzuki and Iwai, 1974; Neuburger et al., 1996; Chen et al., 1997), and the plastids (Neuburger et al., 1996), and this distribution matches that of Ser hydroxymethyltransferase (Besson et al., 1995). Higher plants therefore possess all of the enzymatic machinery necessary to catalyze the incorporation of the carbon deriving from formate (and perhaps from methanol) into methyl groups of various organic compounds, such as the polar head group of phosphatidylcholine.
Using 13C-NMR, we demonstrate that the carbon atom of [13C]methanol given to higher plant cells is readily incorporated into the methyl groups of numerous molecules, including Met and phosphatidylcholine. We also demonstrate that methanol induces the de novo synthesis of methyl-β-d-glucopyranoside.
RESULTS
Methanol Transport
13C-NMR spectroscopy was performed on intact sycamore cells to follow methanol import. After a few minutes of incubation in the presence of various concentrations (0.2–5 mm) of [13C]methanol, we observed an accumulation of methanol in the cells characterized by its unique resonance at 49.7 ppm (Fig. 1). At all the concentrations tested up to 5 mm, methanol was not transported via a carrier because substrate saturation was not observed. At 5 mm this alcohol entered very rapidly in sycamore cells, because in less than 20 s the intracellular methanol concentration equilibrated with the concentration of methanol added to the perfusion medium (data not shown). These results indicate that the plasmalemma membrane did not constitute an obstacle to the diffusion of methanol. Interestingly, comparison of the increase in the cell number between control culture and cultures in the presence of 5 mm methanol indicated that methanol at this concentration did not affect the initial growth rates or maximum density of the cells. In both cases the cell number doubling time was 40 to 48 h after a lag phase of approximately 2 d, and the maximum cell density of sycamore cells was attained after 6 to 7 d of growth, when the stationary phase was attained, indicating that 5 mm methanol did not affect cell growth parameters.
Figure 1.
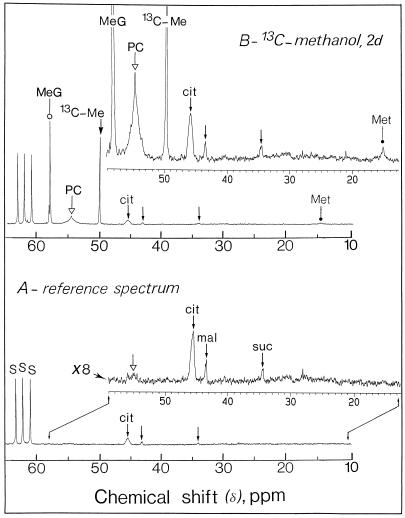
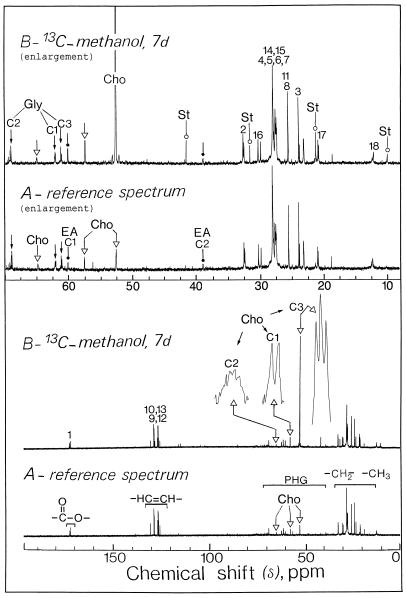
Representative in vivo 13C-NMR spectra (expanded scale from 10–65 ppm) obtained from sycamore cells. The spectra, recorded at 20°C, are the results of 3,600 transients (60 min). Cells (9 g wet weight) were taken from a standard exponentially growing suspension culture, packed in a 25-mm NMR tube as described previously (Aubert et al., 1996), and perfused with a Mn2+-free culture medium containing 5 mm Suc. A, Control cells at pH 6.0; B, cells incubated for 2 d with 5 mm [13C]methanol at pH 6.0. Note that the methyl carbons resonance of phosphatidylcholine is broadened because this polar lipid is not free to move. In addition, signals from Ser (C-3, 13C-enriched) and methyl-β-d-glucopyranoside (CH3, 13C-enriched) overlapped in 13C-NMR spectra of intact cells (compare with Fig. 2). Part of the amino acid and organic acid methylene groups are shown on expanded scales (magnification, ×8). Peak assignments are as follows: MeG, [13C]methyl-β-d-glucopyranoside; PC, [13CH3]phosphatidylcholine; 13C-Me, [13C]methanol; cit, citrate; mal, malate; suc, succinate; Met, [13CH3]Met; S, Suc.
Methanol Metabolism
The rate of methanol disappearance in the growth medium measured by 13C-NMR was linear with time, approximately 0.2 μmol h−1 g−1 wet weight at a fixed concentration of added methanol (5 mm).
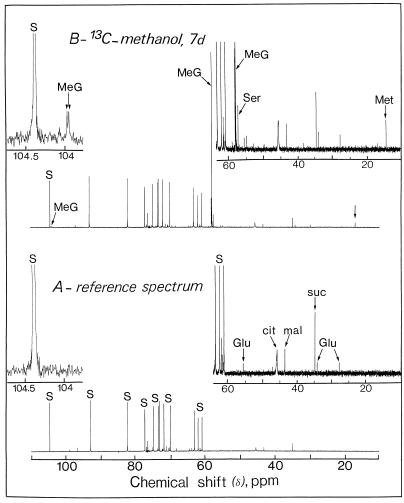
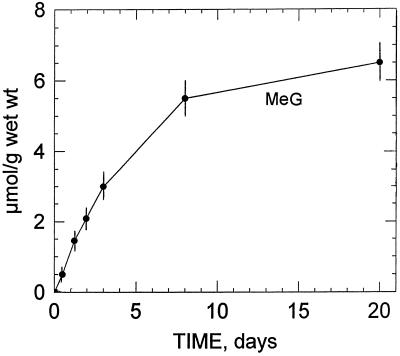
Figure 2 illustrates the changes that occurred in the 13C-NMR spectra of perchloric acid extracts obtained from sycamore cells maintained at pH 6.0 for 7 d in a nutrient medium containing 5 mm [13C]methanol. In the 13C-NMR reference spectra (in which methanol was omitted from the nutrient solution), the strongest signals were from glucosyl and fructosyl moieties of Suc and corresponded to an intracellular concentration of approximately 75 to 80 μmol g−1 wet weight (Rébeillé et al., 1985). In addition, the major resonances (13C-natural abundance) in the chemical shift range of 10 to 80 ppm arose from Glu (at 27.8, 34.4, and 55.6 ppm), citrate (at 45.7 ppm, C-2 + C-4), malate (at 43.2 ppm, C-3), and succinate (at 34.9 ppm, C-2 + C-3). Under these conditions, resonances from Ser and Met were undetectable (threshold of detection around 1–2 μmol/g wet weight). This relatively low sensitivity can be explained by the fact that peaks present in this spectrum result from the 1.1% naturally occurring 13C of cellular metabolites normally present at high concentrations in the cell. On the other hand, cells maintained in a medium containing [13C]methanol (5 mm for 7 d at pH 6.0) permitted visualization in the chemical range from 10 to 60 ppm, C-3 (13C-enriched) of Ser at 57.3 ppm, and the methyl carbon (13C-enriched) of Met at 14.7 ppm. [13C]Methanol also triggered the appearance of a unique resonance centered at 58.0 ppm. To our knowledge, this resonance has not been reported in plant extracts. We identified it as originating from the methyl group of methyl-β-d-glucopyranoside, based on the following observations. First, when adding methyl-β-d-glucopyranoside (13C-natural abundance) to the extract, seven resonances centered at 58.0 (methyl carbon), 61.6, 70.5, 73.9, 76.5, 76.7, and 104.0 ppm were observed. Under these conditions the unique resonance at 58.0 ppm (13C-enriched; pH 7.5) was enhanced. Second, at higher resolution the resonance centered at 104.0 ppm (C-1, actually 1.1% natural abundance 13C), which is specific to methyl-β-d-glucopyranoside, appeared as two distinct peaks due to the homonuclear spin-spin carbon interaction (the 2JCC coupling constant, 0.2 Hz, of this doublet is typical of -C-O-CH3; inset of Fig. 2). Interestingly, this resonance was not characterized in the reference spectra (methanol was omitted from the nutrient solution), suggesting that methanol induces the de novo synthesis of this methyl glucoside. Third, we verified that the considerable accumulation of [13C]methyl-β-d-glucopyranoside (resonance at 58.0 ppm) was not attributable to the methyl group of methyl-α-d-glucopyranoside, 3-O-methyl-Glc, methyl-α-d-galactopyranoside, or methyl-α-d-mannopyranoside. Fourth, the presence of methyl-β-d-glucopyranoside was confirmed unambiguously by mass spectrometry. Indeed, the mass spectrum of the trimethylsilyl derivative of this methyl glucoside from sycamore cell extracts incubated with methanol was identical to the mass spectrum of the trimethylsilyl derivative of an authentic sample of methyl-β-d-glucopyranoside and previously published by Dejongh et al. (1969; result not shown). Independently of the incubation time, the 13C-enrichment at the methyl position of methyl-β-d-glucopyranoside was found to be 100%. Free methyl-β-d-glucopyranoside accumulated steadily during the first 3 d of incubation with methanol (Fig. 3). After a 6-d incubation with 5 mm methanol the concentration of methyl-β-d-glucopyranoside attained in the cell was approximately 6 μmol/g cell wet weight.
Figure 2.
Representative in vitro 13C-NMR spectra (scale from 10–110 ppm) of sycamore cells (perchloric extract). The spectra, recorded at 20°C, are the results of 900 transients (90 min). Perchloric extracts were prepared from a standard exponentially growing suspension culture (9 g wet weight) according to the procedure described in “Materials and Methods.” A, Control cells at pH 6.0; B, cells incubated for 2 d with 5 mm [13C]methanol at pH 6.0. Note that the C-3 resonance of Ser is well separated from the resonance of the methyl group of methyl-β-d-glucopyranoside (compare with Fig. 1). The resonance frequency associated with methanol was not observed in the perchloric extract (compare with Fig. 1, for an explanation see text). Inset, 0050 Expanded scale (magnification, ×20) from 103.75 to 104.75 ppm showing the C-1 resonance of methyl-β-d-glucopyranoside (this resonance appeared as two distinct peaks, for an explanation see text). The 13C-enrichment at the methyl position of methyl-β-d-glucopyranoside was found to be 100%. Part of the amino acid and organic acid methylene groups are shown on expanded scales (magnification, ×8). Peak assignments are as follows: MeG, [13C]methyl-β-d-glucopyranoside; cit, citrate; mal, malate; suc, succinate; Glu, Glu; Met, [13CH3]Met; S, Suc.
Figure 3.
Evolution of intracellular methyl-β-d-glucopyranoside in sycamore cells incubated with 5 mm [13C]methanol at pH 6.0, determined from in vivo 13C-NMR spectra. Note that methyl-β-d-glucopyranoside was not detected at the commencement of the incubation period indicating that methanol triggered the de novo synthesis of this methyl-glucoside (MeG). Results are the average of three measurements (expressed as μmol/g cell wet weight) and bars represent sd.
In vivo 13C-NMR spectra (Fig. 1) obtained from sycamore cells maintained at pH 6.0 for 7 d in a nutrient medium containing 5 mm [13C]methanol showed the resonances frequencies associated with methanol (methanol was not observed in the perchloric extract owing to its evaporation during lyophilization, see “Materials and Methods”), Met (CH3, 13C-enriched), and methyl-β-d-glucopyranoside (CH3, 13C-enriched). Signals from Ser (C-3, 13C-enriched) and methyl-β-d-glucopyranoside (CH3, 13C-enriched) overlapped in 13C-NMR spectra of intact cells (Fig. 1), whereas they were clearly distinguishable in 13C-NMR spectra obtained from cell extracts (Fig. 2). No free formaldehyde and formate were detected. Figure 1 also shows a broad resonance centered at 54.5 ppm, which was not observed in 13C-NMR spectra obtained from cell extracts and is very likely attributable to the polar head group of phosphatidylcholine, the major polar lipid of plant cells. This resonance was almost undetectable in the control cells (in which methanol was omitted from the nutrient solution). To characterize further this unknown compound, a lipid extract (see “Materials and Methods”) was prepared from 9 g of sycamore cells maintained for 7 d in a nutrient medium containing 5 mm [13C]methanol.
31P- and 13C-NMR Studies of Polar Lipids
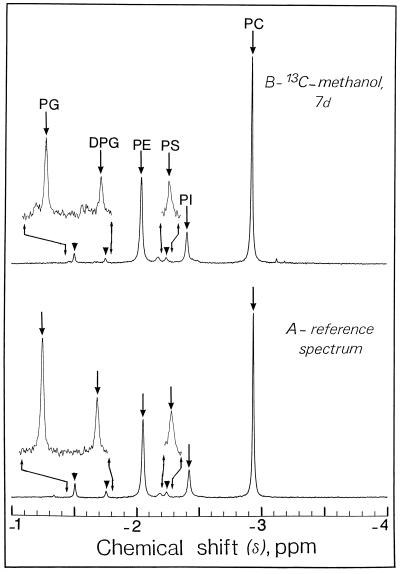
31P-NMR spectroscopy of a cell lipid extract (for review, see Pearce et al., 1991) gave signals from phosphatidylcholine (−2.95 ppm), phosphatidylethanolamine (−2.05 ppm), and phosphatidylinositol (−2.41 ppm) (Fig. 4). Minor signals centered at −1.50, −1.76, and −2.24 ppm clearly distinguishable from the background noise, coincide with the phosphate group of phosphatidylglycerol, diphosphatidylglycerol (cardiolipin), and phosphatidyl-Ser, respectively. Figure 4 also indicates that the phospholipid composition (percentage of total polar lipids) of intact cells was not modified by the presence of [13C]methanol in the growth medium. The phospholipid composition measured using 31P-NMR (see “Materials and Methods”) is in very good agreement with the phospholipid compositions of a number of different non-green cells measured by using previous fastidious biochemical determinations (Bligny and Leguay, 1987).
Figure 4.
Representative in vitro 31P-NMR spectra (expanded scale from −1 to 4 ppm) of a lipid extract from sycamore cells. The spectra, recorded at 20°C, are the results of 256 transients (9 min). Lipid extracts were prepared from a standard, exponentially growing suspension culture (9 g wet weight) according to the procedure described in “Materials and Methods.” A, Control cells at pH 6.0; B, cells incubated for 7 d with 5 mm [13C]methanol at pH 6.0. Note that the phospholipid composition of cells was not modified by the presence of [13C]methanol in the growth medium. Peak assignments are as follows: PG, phosphatidylglycerol; DPG, diphosphatidylglycerol (cardiolipin); PE, phosphatidylethanolamine; PS, phosphatidyl-Ser; PI, phosphatidylinositol; PC, phosphatidylcholine. The signal at 2.18 ppm next to PS was not ascribed to a known phospholipid.
13C-NMR spectroscopy was then performed on a cell lipid extract prepared in 100% 12C-chloroform. Figure 5 illustrates the changes that occurred when the cells were incubated for 7 d in a nutrient medium containing 5 mm [13C]methanol. In the absence of exogenous methanol, we observed a multitude of sharp signals deriving from methylene and methyl carbons of esterified fatty acids (10–40 ppm), all of the olephinic carbons belonging to mono- or polyunsaturated chains (120–135 ppm), and carbonyl carbons of esterified fatty acids (165–175 ppm). We also observed carbons associated with the polar head groups of lipids, including carbons of the glycerol backbone (peaks at 61.8, 67.5, and 60.9 ppm from C-1, C-2, and C-3, respectively), carbons of ethanolamine (peaks at 59.9 and 38.8 ppm from C-1 [carbon bound to a hydroxyl group] and C-2, respectively), and carbons of choline moiety of phosphatidylcholine (peaks at 57.3, 64.9, and 52.4 ppm from C-1, C-2, and the three methyl carbons of choline, respectively) (40–80 ppm). Some general rules for the assignment of the spectra of polar lipids were found in Pollesollo et al. (1996).
Figure 5.
Representative in vitro 13C-NMR spectra of a lipid extract from sycamore cells. The spectra, recorded at 20°C, are the results of 900 transients (90 min). Lipid extracts were prepared from a standard exponentially growing suspension culture (9 g wet weight) according to the procedure described in “Materials and Methods.” A, Control cells at pH 6.0; B, cells incubated for 7 d with 5 mm [13C]methanol at pH 6.0. Part of the lipid methylene groups are shown on expanded scales (magnification, ×8). Note the massive label in the methyl carbons of choline associated with phosphatidylcholine. Peak assignments are as follows: Gly, C1, C2, C3, carbons of the sn glycerol backbone (small arrow); Cho, C1, C2, C3, carbons of choline (▵); EA, C1, C2, carbons of ethanolamine (dot); St, 24-methyl, and 24-ethyl sterols such as camposterol and sitosterol (○); 1 to 18, carbon atoms of esterified fatty acids; COO−, carbonyl carbon of esterified fatty acids; –CH=CH–, olephinic carbons belonging to mono- or polyunsaturated esterified fatty acids; PHG, carbons associated with the polar head groups of lipids; –CH2–, –CH3–, methylene and methyl carbons of esterified fatty acids.
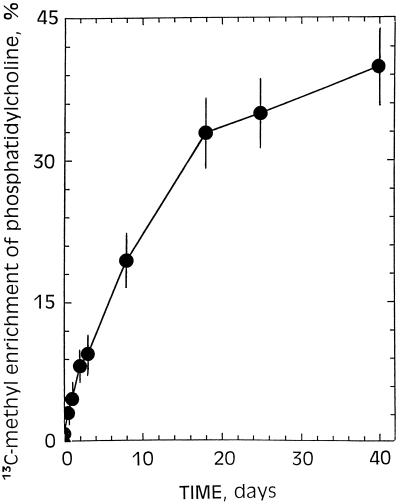
After 7 d of incubation in the presence of 5 mm [13C]methanol, significant changes in the spectra of cell lipid extract were evident (Fig. 5). Of particular interest was the massive label in the methyl carbon of choline associated with phosphatidylcholine. A close examination of an expanded scale of the spectra between 52.45 and 52.35 ppm showed three distinct peaks at approximately 52.40, 52.36, and 52.32 ppm, corresponding to the three methyl carbons of choline coupled to 14N. 13C-Enrichments (percent) in the methyl carbon of choline in phosphatidylcholine from sycamore cells grown with [13C]methanol increased slowly with time up to a steady-state equilibrium (45%) attained after 1 month of cell culture (Fig. 6). This indicates that at least half of the methyl carbons of phosphatidylcholine were derived from [13C]methanol. It is clear that the broad resonance observed at 54.5 ppm in 13C-NMR spectra obtained from intact sycamore cells maintained at pH 6.0 for 2 d in a nutrient medium containing 5 mm [13C]methanol (Fig. 1) was definitively attributable to the methyl carbons of phosphatidylcholine deriving from methanol.
Figure 6.
13C-Enrichment as a function of time in the methyl carbons of phosphatidylcholine from sycamore cells grown in the presence of 5 mm [13C]methanol. Relative 13C-enrichment was obtained for methyl carbon atoms from 13C-NMR spectra by the standard approach (phosphatidylcholine sample from sycamore cells with natural 13C-abundance versus phosphatidylcholine from sycamore cells grown with [13C]methanol). Cells were subcultured every 7 d in the presence of 5 mm methanol. The rate constant for choline enrichment roughly corresponds to the rate of membrane/cell doubling.
The 2.1-ppm shift observed in comparison with lipid extracts (Fig. 5) originates from the solvent (water versus chloroform) specific effect. Figure 5 also indicates a 50% 13C-enrichment in the C-1 carbon of choline in phosphatidylcholine, whereas no enrichment (natural abundance) was observed in the C-2 carbon. Likewise, there was a 50% enrichment in the C-1 carbon of ethanolamine in phosphatidylethanolamine, whereas no enrichment was observed in the C-2 carbon. In other words, carbon bound to a hydroxyl group in polar heads of phospholipids derived partly from [13C]methanol via Ser and ethanolamine (Mouillon et al., 1999). Finally, the four resonances at 10.1, 21.2, 31.3, and 41.3 ppm coincided with those of 24-methyl and 24-ethyl sterols such as camposterol and sitosterol. This alkylation of the side chain is catalyzed by S-adenosyl-Met-dependent C-methyltransferases, which have been well studied in higher plants (Bach and Benveniste, 1997).
These results altogether strongly suggest that [13C]methanol added to plant cells is utilized via folate-mediated single-carbon metabolism, in which 5-13CH3H4Pte-Glun and S-adenosyl-Met play a key role in a myriad of transmethylation reactions, including the methylation of homo-Cys to yield Met and the progressive methylation of ethanolamine to yield choline. However, the specific labeling of methyl-β-d-glucopyranoside by [13C]methanol remains obscure.
Accumulation and Metabolism of [3-13C]Ser in Plant Cells
To verify that label incorporation into the methyl groups derived directly or indirectly from 13CH2 H4Pte-Glun, sycamore cells were incubated with [3-13C]Ser. It is well established that the major source of one-carbon units is the 3-carbon of Ser, derived from glycolytic intermediates in eukaryotic cells (Schirch, 1984). The one-carbon unit is transferred to H4Pte-Glun in a reaction catalyzed by Ser hydroxymethyltransferase to generate CH2 H4Pte-Glun.
Amino acids, including Ser, are transported into plant cells by proton-coupled symporters that link translocation across the plasma membrane to the electrochemical potential generated by the H+-pumping ATPase (Bush, 1998). 13C-NMR spectroscopy was performed on intact sycamore cells incubated for 48 h in a nutrient medium containing 200 μm [3-13C]Ser (results not shown). The spectra showed the resonance frequency associated with Met (CH3 group, 13C-enriched) and phosphatidylcholine (methyl carbons of choline, 13C-enriched). Examination of the resonances in the chemical shift range from 60 to 50 ppm indicated that feeding [3-13C]Ser to these cells did not lead to any detectable accumulation of methyl-β-d-glucopyranoside (CH3, 13C-enriched).
Finally, 13C-NMR spectroscopy performed on lipid extracts from cells maintained in the presence of [3-13C]Ser in place of [13C]methanol indicated considerable 13C-enrichment in the methyl carbons of choline in phosphatidylcholine and in the methyl and ethyl carbons at C-24 in plant sterols (results not shown). These results therefore demonstrated that [3-13C]Ser can perfectly mimic [13C]methanol for folate-mediated single-carbon metabolism. These results also demonstrated that the accumulation of methyl-β-d-glucopyranoside (CH3, 13C-enriched) previously observed with methanol is not associated with the classical C1 metabolism involving H4Pte-Glun.
DISCUSSION
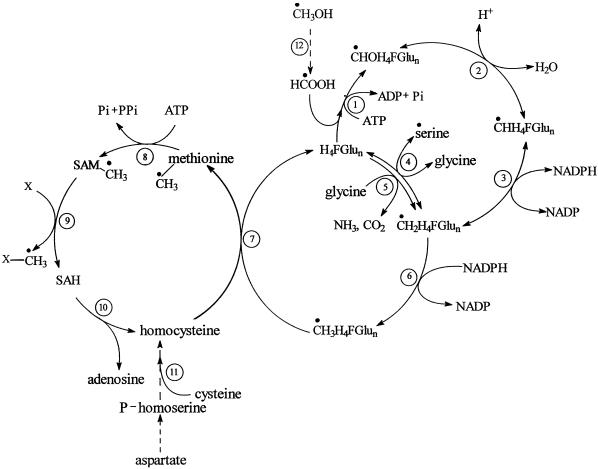
Our results demonstrate that 5 mm methanol did not decrease the initial growth rates and did not limit the maximum density of the sycamore cells. We also show that [13C]methanol readily entered sycamore cells (likely through aquaporin-like protein) (Maurel, 1997) to be slowly metabolized to [3-13C]Ser, [13CH3]Met, and [13CH3]phosphatidylcholine (proposed metabolic pathway in methanol assimilation see Fig. 7). Cossins (1964), using various plant tissues, demonstrated that carbon from methanol is also incorporated into Ser and Met. We conclude that the assimilation of [13CH3]methanol by plant cells occurs through the formation of 13CH2H4Pte-Glun, 13CH3H4Pte-Glun, S-adenosyl-Met (a methyl donor in a myriad of transmethylation reactions), and their subsequent utilization to yield [3-13C]Ser, [13CH3]Met, and [13CH3]phosphatidylcholine (Figs. 1, 2, and 5; for review, see Rébeillé and Douce, 1999). Assimilation of methanol would therefore require its prior oxidation, although the exact mechanism by which this oxidation occurs is unclear. A reasonable possibility would be formation of formaldehyde followed by its oxidation to formate, a reaction in which methanol oxidase and formaldehyde dehydrogenase operate in concert. Indeed, formate is a potential single-carbon source in higher plants (the ATP-dependent synthesis of 10-formyl H4Pte-Glun from formate involves the enzyme formyl H4Pte-Glun synthetase). However, the pathway for formaldehyde production would have to be efficiently regulated to prevent this lethal metabolite from accumulating.
Figure 7.
Proposed metabolic pathway involved in methanol assimilation by sycamore cells (the origin of methyl-β-d-glucopyranoside is not indicated). Enzymes: 1, Formyltetrahydrofolate synthetase; 2, methenyl H4Pte-Glun cyclohydrolase; 3, methylenetetrahydrofolate dehydrogenase; 4, Ser hydroxymethyltransferase; 5, Gly decarboxylase; 6, methylenetetrahydrofolate reductase; 7, cobalamin-independent Met synthase; 8, S-adenosyl-Met synthetase; 9, methyltransferase; 10, S-adenosylhomo-Cys hydrolase; 11, cystathionine γ-synthase and cystathionine β-lyase; 12, methanol oxidase and formaldehyde dehydrogenase. H4FGlun, 5,6,7,8 tetrahydropteroylpoly-Glu (tetrahydrofolate); CHOH4F-Glun, 10-formyltetrahydrofolate; CHH4F-Glun, 5,10-methenyltetrahydrofolate; CH2H4F-Glun, 5,10-methylenetetrahydrofolate; CH3H4FGlun, 5-methyltetrahydrofolate; SAM, S-adenosyl-Met; SAH, S-adenosylhomo-Cys. ●, (13C) carbon.
Our results also indicate that sycamore cells metabolize methanol rather slowly (approximately 0.2 μmol h−1 g−1 wet weight), and that some of the carbons from methanol (30%–60%) are channeled toward 13C-compound synthesis, including Ser, Met, 24-methyl and 24-ethyl sterols, phosphatidylcholine, and methyl-β-d-glucopyranoside (NMR-visible compounds). Therefore, in sycamore cells an important fraction of the carbon flow from formate goes into the formyl H4Pte-Glun synthetase pathway rather than through the mitochondrial oxidation pathway. However, several studies have characterized a NAD-dependent formate dehydrogenase in plant mitochondria, an enzyme that catalyzes the oxidation of formate to CO2. For example, Oliver (1981) and Hourton-Cabassa et al. (1998) showed that spinach mitochondria can oxidize formate rather rapidly. In support of this observation, preliminary results carried out in our laboratory indicated that intact sycamore cells can oxidize formate more rapidly than they metabolize methanol (approximately 1 μmol h−1 g−1 wet weight versus 0.2 μmol h−1 g−1 wet weight). Uncertainty therefore arises concerning the partitioning of formate between the oxidation pathway via formate dehydrogenase and the assimilation pathway via 10-formyl H4Pte-Glun. Apparently, according to the tissue, the relative flux of formate toward oxidation and assimilation can fluctuate considerably (Cossins, 1964), because there are strong indications for a control of formate oxidation in plants at the level of gene expression; furthermore, the major site for such a regulation is probably NAD-dependent formate dehydrogenase (Hourton-Cabassa et al., 1998). The poor affinity of formate dehydrogenase for its substrate (Km formate ≈ 1.7 mm; Halliwell, 1974), in contrast with the situation observed for formyl H4Pte-Glun synthetase (Km formate ≈ 35 μm; Kirk et al., 1995), could explain the fact that part of the carbon deriving from methanol is diverted toward C1 metabolism.
Our studies of the metabolism of [13C]methanol in sycamore cells revealed assimilation of label into a new cellular product over several hours of exposure. This compound, characterized as methyl-β-d-glucopyranoside, has been identified in rose petals (Ichimura et al., 1997) and in a variety of species belonging to the genus Geum (Rosacae family) (Bonnier, 1934). In the present study, the assimilation of label from [13C]methanol into the methyl group of methyl-β-d-glucopyranoside did not require the formation of 13CH2H4Pte-Glun, because feeding sycamore cells with [3-13C]Ser, the direct precursor of 13CH2H4Pte-Glun, did not lead to the formation of [13C]methyl-β-d-glucopyranoside. In addition, a 13C-enrichment value close to 100% was obtained, indicating that the methyl group of methyl-β-d-glucopyranoside derives exclusively from methanol. This is in contrast to the situation observed with other compounds synthesized from [13C]methanol, such as phosphatidylcholine, in which 13C-enrichments (percent) in the methyl carbon of choline increase slowly with time up to a steady-state equilibrium (45%). This is because [13C]methanol and endogenous unlabeled Ser compete with one another to feed C1 metabolism.
The results of the present study indicate that a specific methyltransferase using S-adenosyl-Met is not involved in methyl-β-d-glucopyranoside synthesis. It is not clear whether the methanol-induced accumulation of methyl-β-d-glucopyranoside in sycamore cells has physiological significance (e.g. in osmotic stress tolerance). However, the existence of this metabolite in a variety of species that have not been fed with methanol speaks for its potential significance. It could be synthesized via an unspecific transglycosylation process catalyzed by a hydrolase in the presence of high concentrations of glycosyl acceptor (alcohol):
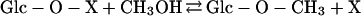 |
In support of this last suggestion, we have observed that the substitution of ethanol or glycerol for methanol led to the formation of an ethyl-β-d-glucopyranoside or glyceryl-β-d-glucopyranoside (results not shown). It is very likely that this transglycosylation reaction predominates over hydrolysis only when the concentration of suitable alcohol acceptor in the vicinity of the enzyme is considerable. For example, the action of a β-d-glucosidase (for example, see Crombie et al., 1998) is exactly what might be predicted for an enzyme involved in such a transglycosylation reaction. Obviously, this putative hydrolase remains to be characterized; however, this question was not further pursued in the present study. Finally, preliminary results carried out in our laboratory indicate that methyl-β-d-glucopyranoside accumulates in the vacuolar compartment and, once synthesized, exhibits a remarkable stability.
MATERIALS AND METHODS
Cell Preparation
Cell suspensions were chosen in preference to dense tissues to improve the homogeneity of the incubation conditions (particularly the extracellular pH and the oxygen supply). Sycamore (Acer pseudoplatanus L.) cells were grown at 20°C as a suspension in liquid nutrient medium according to the method of Bligny and Leguay (1987). The culture medium, well aerated and containing various concentrations of [13C]methanol (0.2–5 mm), was kept at a volume of 0.3 L and stirred continuously at 60 rpm. Under these conditions the cell number doubling time was 40 to 48 h, after a lag phase of approximately 2 d, and the maximum cell density was attained after 7 to 8 d of growth, when the stationary phase was attained. The cell suspensions were maintained in exponential growth by subcultures every 7 d. Anaerobiosis resulted in a failure of the cells to utilize methanol. The cell wet weight was measured after straining culture aliquots onto a glass-fiber filter.
Chemicals
[13C]Methanol (99%) and [3-13C]Ser (99%) were purchased from Leman (Saint Quentin-en-Yvelines, France). To simplify the quantification of intracellular [13C]-metabolites, the above chemicals were added in the external medium as 99% labeled. Methyl-β-d-glucopyranoside was purchased from Sigma-Aldrich (St. Louis).
In Vitro NMR Measurements
Perchloric Acid Extract Preparation
For perchloric acid extraction, cells (9 g wet weight) were quickly frozen in liquid nitrogen and ground to fine powder with a mortar and pestle with 1 mL of 70% (v/v) perchloric acid. The frozen powder was then placed at −10°C and thawed. The thick suspension thus obtained was centrifuged at 15,000g for 10 min to remove particulate matter, and the supernatant was neutralized with 2 m KHCO3 to approximately pH 5.0. The supernatant was then centrifuged at 10,000g for 10 min to remove KClO4, and the resulting supernatant was lyophilized and stored in liquid nitrogen. This freeze-dried material containing non-volatile compounds was redissolved in 2.5 mL of water containing 10% (v/v) 2H2O, neutralized to pH 7.5, and buffered with 50 mm 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES). Divalent cations (particularly Mn2+ and Mg2+) were chelated by the addition of sufficient amounts of 1,2-cyclohexylenedinitrilotetraacetic acid (CDTA) ranging from 50 to 100 μmol depending on the samples. The elimination of paramagnetic cations is a prerequisite for obtaining sharp resonance signals.
Polar Lipid Extraction
Cells (9 g wet weight) were fixed in boiling water for 5 min to destroy phospholipase D, and therefore prevent the formation of phosphatidic acid and phosphatidylmethanol (Douce et al., 1966). Cells were then lyophilized and ground to a fine powder. Polar lipids were extracted from this freeze-dried material according to the method of Folch et al. (1957). The final chloroform layer was evaporated to dryness in a stream of nitrogen, and the residue thus obtained was dissolved in 2.5 mL of pure 12C-deuterated chloroform (lipid extract) to avoid the 13C-resonance (natural abundance) of chloroform. To get sharp NMR signals (for review, see Meneses and Glonek, 1988), it was necessary to include in the chloroform extract (final volume 1 mL) small amounts of methanol (150 μL) and water (25 μL).
NMR Measurements
Spectra of neutralized perchloric acid extracts or lipid extract (see above) were recorded on a NMR spectrometer (AMX 400, Bruker, Billerica, MA) equipped with a 10-mm multinuclear probe tuned at 162 or 100.6 Mhz for 31P- or 13C-NMR studies, respectively. The deuterium resonance of 2H2O was used as a lock signal.
31P-NMR acquisition conditions used were 70° radiofrequency pulses (15 μs) at 3.6-s intervals; spectral width 8,200 Hz; 1,024 scans; Waltz-16 1H decoupling sequence (with two levels of decoupling: 1 W during acquisition time, 0.5 W during delay). Free induction decays were collected as 8K data points, zero filled to 16K, and processed with a 0.2-Hz exponential line broadening. 31P-NMR spectra are referenced to methylene diphosphonic acid, pH 8.9, at 16.38 ppm.
13C-NMR acquisition conditions used were 90° radiofrequency pulses (19 μs) at 6-s intervals; spectral width 20,000 Hz; 900 scans; Waltz-16 1H decoupling sequence (with two levels of decoupling: 2.5 W during acquisition time, 0.5 W during delay). Free induction decays were collected as 16K data points, zero filled to 32K, and processed with a 0.2-Hz exponential line broadening. 13C-NMR spectra are referenced to hexamethyldisiloxane at 2.7 ppm.
Identification and Quantification of Metabolites and Polar Lipids
Spectra of standard solutions of Ser, Met, methyl-β-d-glucopyranoside, and various polar lipids at pH 7.5 were compared with the spectra of extracts (perchloric acid or lipid extracts) of sycamore cells. The definitive assignments were made after running a series of spectra obtained by the addition of authentic compounds to the extracts, according to the methods described in previous publications (for 31P-NMR, see Roby et al., 1987; for 13C-NMR, see Gout et al., 1993). To determine accurately the total amount of metabolites and various polar lipids in extracts, a 20-s recycling time was used to obtain fully relaxed spectra, and calibration of the peak intensities by the addition of known amounts of the corresponding authentic compounds was performed. The possible errors in the measurements caused by perchloric acid extraction were estimated by adding known amounts of authentic compounds to frozen cells before grinding. For all of the compounds studied, the overall yield of recovery was about 80%. Intracellular concentrations were calculated on the following basis: 1 g of cell wet weight corresponds to 1 mL of cell volume and roughly 0.13 mL of cytoplasm and 0.78 mL of vacuole volume.
In Vivo NMR Measurements
To get a better signal-to-noise ratio, an experimental arrangement was devised to analyze the maximum cell volume and to optimize the homogeneity of the cell incubation conditions (Aubert et al., 1996). Such a system prevents gas bubbles within the cell mass and subsequent magnetic field inhomogeneity. The perfusion medium contained 5 mm Suc, 10 mm KNO3, 1 mm KCl, 0.5 mm MgSO4, 0.5 mm Ca(NO3)2, and 100 μm Pi. Spectra were recorded on a spectrometer (AMX 400, Bruker) equipped with a 25-mm probe tuned at 100.6 Mhz for 13C-NMR.
Acquisition conditions used for 13C-NMR were carried out according to a previous publication (Aubert et al., 1996). The assignments of resonance peaks were carried out according to the methods described in previous publications (Roberts and Jardetzky, 1981; Gout et al., 1993; Aubert et al., 1996) and from the spectra of the perchloric acid extracts that contained the soluble, low-Mr constituents (see above).
Mass Spectrometry
Methyl-β-d-glucopyranoside was identified by mass spectrometry (Quadripolar, Nermag R1010C, Paris, France) after separation of silylated sugars by HPLC using a capillary column (OV17, Spiral, Dijon, France).
ACKNOWLEDGMENT
Provision of various sterol compounds by Prof. Pierre Benveniste is gratefully acknowledged.
LITERATURE CITED
- Aubert S, Bligny R, Douce R. NMR studies of metabolism in cell suspensions and tissue cultures. In: Shachar-Hill Y, Pfeffer P, editors. Nuclear Magnetic Resonance in Plant Physiology. Rockville, MD: American Society of Plant Physiologists; 1996. pp. 109–154. [Google Scholar]
- Bach TJ, Benveniste P. Cloning of cDNAs of genes encoding enzymes of sterol biosynthesis from plants and other eukaryotes: heterologous expression and complementation analysis of mutations for functional characterization. Prog Lipid Res. 1997;36:197–226. doi: 10.1016/s0163-7827(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Besson V, Neuburger M, Rébeillé F, Douce R. Evidence for three serine hydroxymethyltransferases in green leaf cells: purification and characterization of the mitochondrial and chloroplastic isoforms. Plant Physiol Biochem. 1995;33:665–673. [Google Scholar]
- Bligny R, Leguay JJ. Techniques of cell suspension cultures. Methods Enzymol. 1987;148:3–16. [Google Scholar]
- Bonnier G. Grande Flore Complète. Paris: Belin; 1934. [Google Scholar]
- Bush DR. Amino acids transport. In: Singh BK, editor. Plant Amino Acids. New York: Marcel Dekker; 1998. pp. 305–318. [Google Scholar]
- Chen L, Chan S, Cossins EA. Distribution of folate derivatives and enzymes for synthesis of 10-formyltetrahydrofolate in cytosolic and mitochondrial fractions of pea leaves. Plant Physiol. 1997;115:299–309. doi: 10.1104/pp.115.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin MT, Cossins EA. Localization and interconversion of tetrahydropteroylpolyglutamates in isolated pea mitochondria. Biochem J. 1972;128:29–40. doi: 10.1042/bj1280029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Ambard-Bretteville F, Small ID, Rémy R. Identification of a major soluble protein in mitochondria from nonphotosynthetic tissues as NAD-dependent formate dehydrogenase. Plant Physiol. 1993;102:1171–1177. doi: 10.1104/pp.102.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossins EA. The utilization of carbon-1 compounds by plants: I. The metabolism of methanol-14C and its role in amino acid biosynthesis. Can J Biochem. 1964;42:1793–1802. [Google Scholar]
- Crombie HJ, Chengappa S, Hellyer A, Reid JS. A xyloglucan oligosaccharide-active, transglycosylating β-d-glucosidase from the cotyledons of nasturtium (Tropaeolum majus L.) seedlings-purification, properties and characterization of a cDNA clone. Plant J. 1998;15:27–38. doi: 10.1046/j.1365-313x.1998.00182.x. [DOI] [PubMed] [Google Scholar]
- Dejongh DC, Radford T, Hribar JD, Hanessian S, Bieber M, Dawson G, Sweeley CC. Analysis of trimethylsilyl derivatives of carbohydrates by gas chromatography and mass spectrometry. J Am Chem Soc. 1969;91:1728–1740. [Google Scholar]
- Douce R, Faure M, Maréchal J. Le phosphatidylméthanol: un phospholipide isolé d'une souche de crown-gall de Vigne. C R Acad Sci Paris. 1966;262:1549–1552. [Google Scholar]
- Fall R, Benson AA. Leaf methanol: the simplest natural product from plants. Trends Plant Sci. 1996;1:296–301. [Google Scholar]
- Folch J, Lees M, Stanley GHS. A simple method for isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–505. [PubMed] [Google Scholar]
- Gout E, Bligny R, Pascal N, Douce R. 13C nuclear magnetic resonance studies of malate and citrate synthesis and compartmentation in higher plant cells. J Biol Chem. 1993;268:3986–3992. [PubMed] [Google Scholar]
- Halliwell B. Oxidation of formate by peroxisomes and mitochondria from spinach leaves. Biochem J. 1974;138:77–85. doi: 10.1042/bj1380077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B, Butt VS. Oxidative decarboxylation of glycolate and glyoxylate by leaf peroxisomes. Biochem J. 1974;138:217–224. doi: 10.1042/bj1380217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourton-Cabassa C, Ambard-Bretteville F, Moreau F, Davy de Virville J, Rémy R, Colas des Francs-Small C. Stress induction of mitochondrial formate dehydrogenase in potato leaves. Plant Physiol. 1998;116:627–635. doi: 10.1104/pp.116.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Kohata K, Koketsu M, Yamaguchi Y, Yamaguchi H, Suto K. Identification of methyl β-glucopyranoside and xylose as soluble sugar constituents in roses (Rosa hybrida L.) Biosci Biotech Biochem. 1997;61:1734–1735. doi: 10.1271/bbb.61.1734. [DOI] [PubMed] [Google Scholar]
- Kirk CD, Chen L, Imeson HC, Cossins EA. A 5,10-methylenetetrahydofolate dehydrogenase: 5,10-methenyltetrahydrofolate cyclohydrolase protein from Pisum sativum. Phytochemistry. 1995;39:1309–1317. [Google Scholar]
- Kirk CD, Imeson HC, Zheng L, Cossins EA. The affinity of pea cotyledon 10-formyltetrahydrofolate synthetase for polyglutamate substrates. Phytochemistry. 1994;35:291–296. [Google Scholar]
- Maurel C. Aquaporins and water permeability of plant membranes. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:27–50. doi: 10.1146/annurev.arplant.48.1.399. [DOI] [PubMed] [Google Scholar]
- Meneses P, Glonek T. High resolution 31P NMR of extracted phospholipids. J Lip Res. 1988;29:679–689. [PubMed] [Google Scholar]
- Mouillon JM, Aubert S, Bourguignon J, Gout E, Douce R, Rébeillé F. Glycine and serine catabolism in non-photosynthetic higher plant cells: their role in C1 metabolism. Plant J. 1999;20:197–205. doi: 10.1046/j.1365-313x.1999.00591.x. [DOI] [PubMed] [Google Scholar]
- Nemecek-Marshall M, MacDonald RC, Franzen JJ, Wojciechowski CL, Fall R. Methanol emission from leaves: enzymic detection of gas-phase methanol and relation of methanol fluxes to stomatal conductance and leaf development. Plant Physiol. 1995;108:1359–1368. doi: 10.1104/pp.108.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuburger M, Rébeillé F, Jourdain A, Nakamura S, Douce R. Mitohondria are a major site for folate and thymidylate synthesis in plants. J Biol Chem. 1996;271:9466–9472. doi: 10.1074/jbc.271.16.9466. [DOI] [PubMed] [Google Scholar]
- Nour JM, Rabinowitz JC. Isolation, characterization and structural organization of 10-formyltetrahydrofolate synthetase from spinach leaves. J Biol Chem. 1991;266:18363–18369. [PubMed] [Google Scholar]
- Oliver DJ. Formate oxidation and oxygen reduction by leaf mitochondria. Plant Physiol. 1981;68:703–705. doi: 10.1104/pp.68.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce JM, Shifman MA, Pappas AA, Komoroski RA. Analysis of phospholipids in human fluid by 31P NMR. Magn Reson Med. 1991;21:107–116. doi: 10.1002/mrm.1910210113. [DOI] [PubMed] [Google Scholar]
- Pollesollo P, Eriksson O, Höckersted K. Analysis of total lipid extracts from human liver by 13C and 1H nuclear magnetic resonance spectroscopy. Anal Biochem. 1996;236:41–48. doi: 10.1006/abio.1996.0129. [DOI] [PubMed] [Google Scholar]
- Prabhu V, Chatson KB, Abrams GD, King J. 13C nuclear magnetic resonance detection of interactions of serine hydroxymethyltransferase with C1-tetrahydrofolate synthase and glycine decarboxylase complex activities in Arabidopsis. Plant Physiol. 1996;112:207–216. doi: 10.1104/pp.112.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F, Bligny R, Martin JB, Douce R. Effect of sucrose starvation on sycamore (Acer pseudoplatanus) cell carbohydrate and Pi status. Biochem J. 1985;226:679–684. doi: 10.1042/bj2260679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F, Douce R. Folate synthesis and compartmentation in higher plants. In: Kruger NJ, Hill SA, Ratcliffe RG, editors. Regulation of Primary Metabolic Pathways in Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. pp. 53–99. [Google Scholar]
- Roberts JKM, Jardetzky O. Monitoring of cellular metabolism by NMR. Biochim Biophys Acta. 1981;639:53–76. doi: 10.1016/0304-4173(81)90005-7. [DOI] [PubMed] [Google Scholar]
- Roby C, Martin J-B, Bligny R, Douce R. Biochemical changes during sucrose deprivation in higher plant cells: II. Phosphorus-31 nuclear magnetic resonance studies. J Biol Chem. 1987;262:5000–5007. [PubMed] [Google Scholar]
- Schirch LV. Folates in glycine and serine metabolism. In: Blakley RL, Benkovic SJ, editors. Folates and Pterins. Vol. 1. New York: Wiley (Interscience); 1984. pp. 399–431. [Google Scholar]
- Shingles R, Woodrow L, Grodzinski B. Effects of glycolate pathway intermediates on glycine decarboxylation and serine synthesis in pea (Pisum sativum L) Plant Physiol. 1984;74:705–710. doi: 10.1104/pp.74.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Itai R, Suzuki K, Nakanishi H, Nishizawa N-K, Yoshimura E, Mori S. Formate dehydrogenase, an enzyme of anaerobic metabolism, is induced by iron deficiency in barley roots. Plant Physiol. 1998;116:725–732. doi: 10.1104/pp.116.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Iwai K. The occurrence and properties of methylenetetrahydrofolate dehydrogenase in pea seedlings, and intracellular distribution of some folate-linked enzymes in the plant. J Nutr Sci Vitaminol. 1974;20:89–96. doi: 10.3177/jnsv.20.89. [DOI] [PubMed] [Google Scholar]