Abstract
We isolated pgi1-1, an Arabidopsis mutant with a decreased plastid phospho-glucose (Glc) isomerase activity. While pgi1-1 mutant has a deficiency in leaf starch synthesis, it accumulates starch in root cap cells. It has been shown that a plastid transporter for hexose phosphate transports cytosolic Glc-6-P into plastids and expresses restricted mainly to the heterotrophic tissues. The decreased starch content in leaves of the pgi1-1 mutant indicates that cytosolic Glc-6-P cannot be efficiently transported into chloroplasts to complement the mutant's deficiency in chloroplastic phospho-Glc isomerase activity for starch synthesis. We cloned the Arabidopsis PGI1 gene and showed that it encodes the plastid phospho-Glc isomerase. The pgi1-1 allele was found to have a single nucleotide substitution, causing a Ser to Phe transition. While the flowering times of the Arabidopsis starch-deficient mutants pgi1, pgm1, and adg1 were similar to that of the wild type under long-day conditions, it was significantly delayed under short-day conditions. The pleiotropic phenotype of late flowering conferred by these starch metabolic mutations suggests that carbohydrate metabolism plays an important role in floral initiation.
Most plants synthesize starch in their chloroplasts during photosynthesis and degrade it during the subsequent night. The regulation of transitory starch metabolism in photosynthetic tissues is clearly different from the long-term, reserve starch metabolism in non-photosynthetic tissues (Caspar, 1994). Many mutations that affect the starch of certain cereal seeds, potato, pea, and Chlamydomonas reinhardtii have been isolated and characterized (Hannah, 1997). Although studies of these mutants have greatly added to our knowledge of starch metabolism, these mutants studied are relatively specific for the reserve and reproductive organs, and do not affect starch metabolism in the vegetative parts of plants. Mutants that affect starch metabolism in the vegetative parts of Arabidopsis would be useful to extend our understanding on starch metabolism and its role in the plant.
Previously, several nuclear-encoded, recessive mutants of Arabidopsis, pgm1, adg1, and adg2, were isolated and characterized for their low starch content or lack of starch in leaves (Caspar et al., 1985; Wang et al., 1997, 1998). Phosphoglucomutase (PGM, EC 2.7.5.1) catalyzes the conversion of Glc-6-P to Glc-1-P, while ADP-Glc pyrophosphorylase (ADGase, EC 2.7.7.27) catalyzes Glc-1-P and ATP to ADP-Glc. The plastid isozymes of PGM and ADGase are synthesized in the cytosol and transported into plastids. Along with the plastid form of PGM, there are cytosolic forms of PGM isozymes in Arabidopsis. The deficiency of starch in the pgm1 mutant, which has no detectable plastidial PGM activity, shows that the cytosolic Glc-1-P synthesized by PGM isozymes is not transported into plastids for starch synthesis. The deficiency of starch in the adg1 mutant, which does not contain ADGase activity in the plastids, suggests that ADP-Glc is apparently the major substrate for starch synthetase and that the nucleotide sugars synthesized in the cytosol are likely not transported into plastids for starch synthesis.
Phospho-Glc isomerase (PGI, EC 5.3.1.9) is a dimeric enzyme that catalyzes the reversible isomerization of Fru-6-P and Glc-6-P (Smith and Doolittle, 1992). Two isozymes of PGI exist in Arabidopsis, one in the plastids and the other in the cytosol (Caspar et al., 1985). Several plant cytosolic PGIs (Thomas et al., 1992, 1993; Nowitzki et al., 1998) and a plastidial PGI of spinach (Nowitzki et al., 1998) were cloned and characterized mainly to investigate the evolutionary history of eukaryotic PGI genes.
Borchert et al. (1989) showed that there is a plastid transporter for hexose phosphate that transports cytosolic Glc-6-P into plastids but does not transport Glc-1-P. However, this hexose phosphate transporter may not exist in chloroplasts (Borchert et al., 1989; Kammerer et al., 1998), thus making a plastidial PGI activity essential for starch synthesis. A plastid pgi mutant of Clarkia xantiana , which accumulates about 60% as much starch in leaves as the wild type, has also been isolated previously (Jones et al., 1986). As the C. xantiana plastid pgi mutant has about 50% of wild-type plastidial PGI activity, it is difficult to ascertain whether cytosolic Glc-6-P can be imported to chloroplasts for starch synthesis. In this study, we isolated and characterized pgi1, an Arabidopsis mutant with a highly reduced level of plastidial PGI enzyme activity. While the leaf starch synthesis is impaired by the pgi1-1 mutation, starch synthesis in the root cap cells is not affected.
Several reports have suggested that carbohydrate metabolism may play a role in floral initiation (Bernier et al., 1993; Levy and Dean, 1998). The Arabidopsis pgm1 mutant has a flowering time similar to that of the wild type under long-day conditions; however, under short-day conditions, the pgm1 mutant delays its floral initiation. We found that the Arabidopsis pgi1 and adg1 mutants exhibit a delayed floral initiation phenotype similar to that of the pgm1 mutant. These monogenic mutations will be valuable in the analysis of the correlation between carbohydrate metabolism and floral initiation.
RESULTS
Isolation of the Arabidopsis pgi1 Mutant
To isolate starch-metabolic mutants of Arabidopsis, we screened ethyl methanesulfonate-mutagenized M2 plants by examining their leaf starch levels using iodine staining. Starch was present in the leaves and root cap cells of the wild type (Fig. 1A) and absent in those of the pgm1 mutant (Fig. 1C). A mutant line, TSY254, was isolated because its leaves turned a pale yellow color upon iodine staining (Fig. 1B), suggesting a deficiency of starch accumulation in the leaves. However, starch was present in root cap cells of the TSY254 mutant (Fig. 1B) at a level similar to that of the wild type (Fig. 1A).
Figure 1.
Leaves and roots of the wild type, pgi1-1, and pgm1-1 mutants stained for starch with iodine. Plants of the wild type (A), pgi1-1 (B), and pgm1-1 (C) were de-pigmented and stained with an iodine solution. Starch can be seen to be present in the wild-type leaves and in root cap cells (arrowheads) of the wild type and the pgi1-1 mutant.
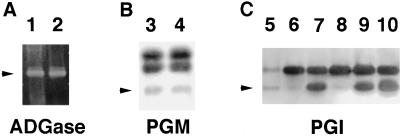
We measured the starch content in leaves from the TSY254 mutants and the wild type. The wild type, heterozygous TSY254, and homozygous TSY254 plants contained 2.64 ± 0.21 (mean ± se of five replica samples), 1.31 ± 0.16, and 0.04 ± 0.01 mg starch g−1 fresh weight, respectively. These results confirmed that the mutant TSY254 plants were indeed deficient in leaf starch accumulation. To identify the biochemical lesion of this mutant, we examined the PGI, PGM, and ADGase activities in leaf extracts from each of the plant types. Figure 2 shows that the mutant TSY254 (lanes 6 and 8) expressed approximately 2% of the wild-type plastidial PGI activity (lane 5). The plastidial PGI activity of the TSY254 heterozygotes (lane 9) had approximately 50% of the wild-type activity (lane 7), which was confirmed by the spectrophotometric enzyme assay. The plastidial PGI activity in the roots of the mutant TSY254 was reduced to the same level as in the leaves (data not shown).
Figure 2.
Activity gel assay for ADGase, PGM, and PGI in wild-type and mutant leaf extracts. Leaf extracts from the wild type (20 μL in lanes 1, 3, and 10; 10 μL in lane 7; and 0.4 μL in lane 5), TSY254 (20 μL in lanes 2, 4, and 8; 10 μL in lane 6), and TSY254/+ (20 μL in lane 9) were separated in a 7% (w/v) native PAGE and assayed for the ADGase, PGM, and PGI activity. ADGase activity was detected by calcium pyrophosphate precipitation; PGM and PGI activity were detected as the colored formazan formation in enzymatic coupling reactions. Arrowheads indicate the positions of the plastidial forms of the enzymes (Caspar et al., 1985).
We also performed spectrophotometric enzyme assays for PGI activity based on the differential temperature stability of the plastidial and cytosolic PGI isozymes (Jones et al., 1986). Heating extracts of purified Arabidopsis chloroplasts to 50°C for 10 min completely inactivated the plastidial PGI but had no effect on the cytosolic PGI activity. While the PGI activity of the mutant was reduced by 3% upon a heat treatment, the PGI activity of the wild type was reduced by 46%, indicating that the plastidial PGI activity in the mutant is approximately 6.5% of the wild type. These data suggest that plastidial PGI enzyme activity is positively correlated with the leaf starch levels of plants. The activities of ADGase, PGM, and cytosolic PGI were apparently normal in the mutant (Fig. 2, lanes 2, 4, and 8). Therefore, TSY254 is specifically defective in the plastidial PGI activity and we designated this mutated allele present in TSY254 as pgi1-1.
Cloning and Sequencing of the Arabidopsis PGI1 Gene
The cytosolic PGI gene of Arabidopsis has been previously cloned and sequenced (Thomas et al., 1993). By BLAST searching the Arabidopsis EST sequence databank, we identified a clone (198H11T7; GenBank accession no. H76567) with homology to the Synechocystis PGI gene and to spinach plastidial PGI. However, the sequence of this EST differs from that of the Arabidopsis cytosolic PGI. We performed RFLP analysis of selfed F2 progenies of pgi1 (TSY254, in the Columbia ecotype) crossed with the wild type (in the Landsberg erecta ecotype). Southern-blot analysis was performed with DNA samples digested with BstU I, and probed with the EST clone 198H11T7. In a population of 50 pgi1 homozygous plants, which were identified by assaying for PGI activity and leaf starch content, the starch-deficient phenotype cosegregated with the RFLP of Columbia detected by the EST probe (data not shown). This result suggested that this clone corresponded to the gene mutated in pgi1-1.
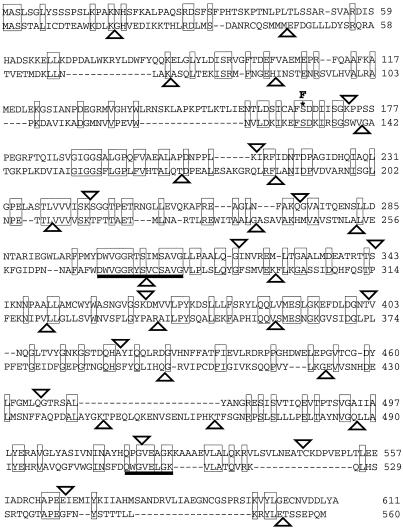
The PGI1 gene was further mapped by RFLP analysis using recombinant inbred lines (Lister and Dean, 1993) and probing with the EST clone to chromosome 4 to 67.3 centiMorgans. With this EST clone as a probe, we isolated genomic clones and cDNA clones. Primers for PCR were designed from the sequence of the genomic clones to obtain the full-length cDNA (GenBank accession no. AF120494), which encodes an open reading frame of 611 amino acids with a predicted transit peptide of 48 amino acids (Emanuelsson et al., 1999). The protein sequence, excluding the putative transit peptide, showed approximately 40% similarity to the Arabidopsis cytosolic PGI, and 70% similarity to the Synechocystis PGI and the spinach plastidial PGI, suggesting that the Arabidopsis PGI1 encodes the plastidial PGI. While the cytosolic PGI gene of Arabidopsis contains 21 introns, the Arabidopsis plastidial PGI gene contains 14 exons interrupted by 13 introns (Fig. 3). None of the introns inserted within these two genes can be aligned at the same position. Our finding supports the endosymbiotic origin of the plastidial PGI genes (Nowitzki et al., 1998).
Figure 3.
Protein sequences of Arabidopsis PGIs. The top sequence is the plastid form and the bottom sequence is the cytosolic form. Identical amino acids are boxed, and the missense mutation (S166F) of pgi1-1 allele is labeled by an asterisk (*). Two conserved domains are underlined. The triangles indicate the positions of introns.
To identify the mutation site in the pgi1-1 allele, we cloned and sequenced the PGI1 genes from the wild type and the pgi1-1 mutant. In the pgi1-1 allele, a single nucleotide substitution at position 834 (C–T) caused a Ser-to-Phe (S166F) transition (Fig. 3) located downstream of the predicted transit peptide. Two conserved motifs present in the PGI proteins (Smith and Doolittle, 1992) have been identified in Arabidopsis plastidial PGI. One of the conserved motifs is located in the central region of the Arabidopsis plastidial PGI (D301–G314) and the other is located in the C-terminal region (Q517–K524). It was suggested that these two domains might be involved in the catalytic function of this enzyme (Meng et al., 1998).
PGI1-1 Mutation Reduced Plastidial PGI Protein Level
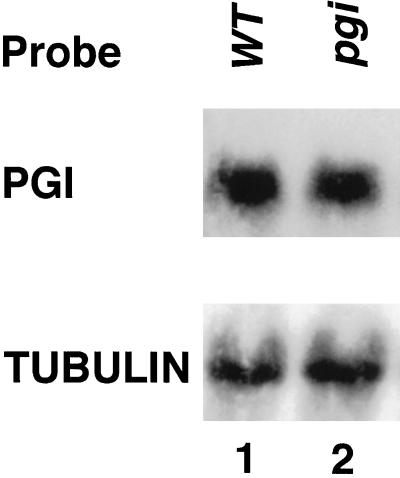
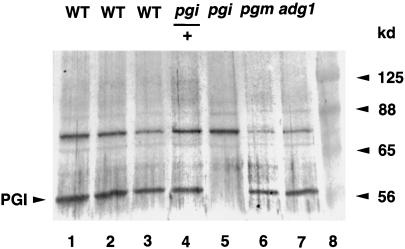
To determine the possible lesion of the pgi1 mutant, we examined PGI1 gene expression by northern- and western-blot analyses. The amount of PGI1 mRNA present in the mutant leaves was similar to that in the wild type (Fig. 4). However, immunoblot analysis of leaf proteins probed with an antibody raised against an Escherichia coli-expressed Arabidopsis plastidial PGI antigen indicated that the plastidial PGI protein present in the pgi1-1 mutant was below a detectable level (Fig. 5, lane 5). The plastidial PGI protein was detected in the heterozygous plant (Fig. 5, lane 4). These results suggest that the pgi1-1 mutant may contain a lesion at the translational or post-translational level, and that the S166F missense mutation may affect the stability of mutated plastidial PGI in plastids. The amount of plastidial PGI protein present in the pgm1 (Fig. 5, lane 6) and adg1 (Fig. 5, lane 7) mutants was similar to that of the wild type (Fig. 5, lane 1), suggesting that PGI1 expression is not affected by the downstream mutations.
Figure 4.
Northern-blot analysis of transcripts of plastidial PGI and tubulin in the wild type and pgi mutant. Northern blot of total leaf RNA (20 μg) isolated from the wild-type (WT) and pgi1-1 plants was probed with radioactive labeled Arabidopsis PGI1 and tubulin cDNA probes.
Figure 5.
Immunoblot analysis of the plastidial PGI protein in wild-type (WT) and mutant leaf extracts. Leaf extracts from the wild type (containing 24, 18, and 12 μg of proteins in lanes 1–3, respectively), pgi1/+ (24 μg of proteins in lane 4), pgi1, pgm1, adg1 (24 μg of proteins in lanes 5–7), and prestained molecular mass markers (lane 8) were separated in a 10% (w/v) SDS PAGE, electroblotted onto a nylon membrane, and probed with the antiserum against Arabidopsis plastidial PGI protein. Bands of the expected sizes were detected by the antiserum, as indicated. There were cross-reactive bands detected by the antibodies. These may be related proteins sharing antigenic sites with the Arabidopsis plastidial PGI protein or E. coli-derived proteins.
Flowering Time of Starch-Deficient Mutants
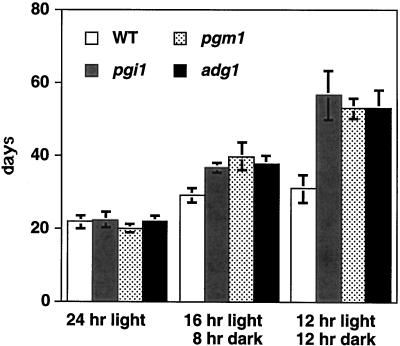
It has been previously reported that the flowering time of the Arabidopsis pgm1 mutant is similar to the wild type under long-day conditions, but is delayed under short-day conditions (Caspar et al., 1985). We compared the flowering time of pgi1, pgm1, adg1, and the wild type under different growth conditions (Fig. 6). All mutants had flowering times similar to the wild type when grown under long-day conditions. However, the flowering times of mutants grown under short-day conditions were significantly delayed compared with that of the wild type. The delay in flowering time was obviously not due to the slightly slower growth rate of the mutants, because mutants were found to bear more rosette leaves than the wild type at the time of flowering.
Figure 6.
Flowering time of the wild type and mutants under different growth conditions. Ten plants each of the wild type (WT; white columns), pgi1 (gray columns), pgm1 (stippled columns), and adg1 (black columns) mutants (all in Columbia ecotype) were grown under continuous light, 16-h-light, or 12-h-light conditions. Flowering time was recorded as the time the first floral stem appeared.
It is possible that the late floral initiation of these mutants was due to their lacking starch, which is normally degraded in the dark phase to provide small carbohydrate metabolites that serve as a signal for the floral initiation. To test this hypothesis, we examined the flowering time of the wild type and the pgi1-1 mutant grown under short-day conditions with a supplement of 1% sugar in the growth medium. The flowering time of the pgi1-1 mutant (69.4 ± 9.3 d) was significantly delayed compared with that of the wild type (44.0 ± 1.4 d) grown under short-day conditions on medium without Suc. Upon the addition of 1% (w/v) Suc in the growth medium, the flowering time of the pgi1-1 mutant (21.0 ± 2.4 d) was similar to that of the wild type (21.2 ± 2.2 d) grown under the same short-day conditions. Similarly, the addition of 1% (w/v) Glc or Fru in the growth medium also reversed the late-flowering phenotype of the pgi1-1 mutant grown under short-day conditions to that of the wild type.
DISCUSSION
In the present study we isolated and characterized an Arabidopsis pgi1 mutant. This pgi1 mutant affects the plastidial PGI activity in a manner similar to the Clarkia xantiana plastid pgi mutant (Jones et al., 1986), which contains a reduced level of starch in leaves. In these mutants, the cytosolic PGI isozymes are functional and can catalyze the formation of Glc-6-P in the cytosolic compartment. Biochemical analysis indicated that there is a hexose phosphate transporter supporting Glc-6-P but not Glc-1-P across the amyloplast envelope (Borchert et al., 1993). As the C. xantiana plastid pgi mutant has about 50% of wild-type plastidial PGI activity (Jones et al., 1986), it is difficult to ascertain whether cytosolic Glc-6-P can be imported to chloroplasts. The Arabidopsis pgi1-1 mutant has only a small fraction (2%–6.5%) of wild-type plastidial PGI activity, and the pgi1/+ heterozygote has approximately 50% of wild-type activity, with the corresponding amount of starch accumulation.
The reduced plastidial PGI activity and leaf starch accumulation of the Arabidopsis pgi1-1 mutant strongly supports the idea that the chloroplastic hexose phosphate transporter either does not exist or does not function efficiently to complement the deficiency of chloroplastic PGI for starch synthesis. Indeed, a recent study demonstrated that the hexose phosphate transporter is expressed mainly in heterotrophic tissues and not in the chloroplasts (Kammerer et al., 1998). On the contrary, in the root cap cells of the pgi1 mutant, cytosolic Glc-6-P can be transported into amyloplasts to support starch synthesis (Fig. 1B). In roots, PGM, ADGase, and starch synthetase are not expressed except in the root cap cells (J. Chen, unpublished results). Thus, starch is synthesized only in the root cap cells of roots. From extensive mutant screening in the cereal breeding program, none of the mutants affecting starch accumulation in seeds was shown to be deficient in the PGI enzyme activities. In addition to the existence of hexose phosphate transporter in heterotrophic tissues (Kammerer et al., 1998), our results indicate that hexose phosphate transport for starch synthesis is different in chloroplasts and root amyloplasts in Arabidopsis.
While the majority of genes affecting starch biosynthesis show a dosage effect at the enzyme level, one copy of the wild-type gene is generally sufficient to confer a wild-type phenotype (Hannah, 1997). PGI has not been shown to be an allosteric enzyme, and thus has not been considered important in controlling the rate of starch synthesis. However, in the Arabidopsis pgi1-1 homozygous and heterozygous mutants, the decrease in starch accumulation was correlated with the reduction in plastidial PGI activity. This suggests that PGI does play a role in the control of starch synthesis. In addition, studies of C. xantiana pgi mutants showed that plastidial PGI controls the rate of starch synthesis and photosynthesis in saturating light intensity and CO2 (Kruckeberg et al., 1989). However, it is not known whether the starch content will increase further in plants with a higher PGI activity than the wild type. We intend to construct such transgenic plants with ectopic expression of PGI to evaluate this possible rate-limiting role of PGI .
Mutants deficient in PGI, PGM, and ADGase, pgi1, pgm1, and adg1, respectively, were isolated. These mutations are well characterized, and it is known that only a defined enzyme involved in the starch synthesis pathway is affected. All of these mutants had a flowering time similar to the wild type grown under long-day conditions. However, under short-day conditions, floral initiation of these mutants was delayed. An obvious reason for this late-flowering phenotype is that these mutants cannot accumulate assimilatory starch as a carbon source used for growth during the dark phase. However, we have isolated several mutants that can suppress the late-flowering phenotype without starch accumulation (T.-S. Yu and J. Chen, unpublished results). A plausible hypothesis is that these mutants lack starch, which is normally degraded in the dark phase to provide small carbohydrate metabolites that serve as a signal for floral initiation, along with other environmental and developmental cues (Bernier et al., 1993). This hypothesis is supported by the analysis of carbohydrate mobilization and floral induction of the starchless pgm mutant (Corbesier et al., 1998). It was shown that the late-flowering phenotype of the pgm mutant is due to the impossibility of mobilizing carbohydrate reserves in conditions in which floral induction is not accompanied by increased photosynthesis (Corbesier et al., 1998). Furthermore, our results show that the late-flowering phenotype under short-day conditions of the pgi1-1 mutant can be reversed to wild type by the addition of sugars (Glc, Fru, and Suc) in the growth medium. The exact identity of specific carbohydrate metabolites, and the interactive mechanisms between carbohydrate metabolism and floral initiation are key issues to be addressed in future studies.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The M2 seeds of ethyl methanesulfonate-mutagenized Arabidopsis (Columbia ecotype) were obtained from Lehle Seeds (Round Rock, TX). Plants were grown in soil at 23°C under 100 μmol quanta m−2 s−1 continuous fluorescent light or with different photoperiods, as indicated. Mutant screening was carried out as described by Caspar et al. (1985). Quantitative starch assays were conducted according to the method of Wang et al. (1998). Arabidopsis mutants, mapping lines, RFLP markers, and expressed sequence tag (EST) clones were obtained from the Arabidopsis Biological Resource Center at Ohio State University (Columbus).
General Molecular Analysis
Standard cloning, DNA-blot, and RNA-blot techniques were used as described by Sambrook et al. (1989). DNA sequencing was performed with double-stranded plasmids using Sequenase (United States Biochemical, Cleveland). RFLP mapping was performed according to the method of Chang et al. (1988), and the data were analyzed using the JoinMap computer program (Stam, 1993).
Enzyme Assays
The assays for PGI, PGM, and ADGase were conducted according to previously described methods using 7% (w/v) PAGE (Wang et al., 1998). Spectrophotometric enzyme assays for the PGI activities based on the differential temperature stability of the plastid and cytosolic PGI isozymes was performed as described by Jones et al. (1986).
Isolation of Genomic and cDNA Clones
To isolate the corresponding genomic clones of Arabidopsis PGI, a genomic library of Arabidopsis ecotype Landsberg (supplied by the Arabidopsis Biological Resource Center) and a cDNA library (constructed with whole-plant mRNA; Stratagene, La Jolla, CA) were screened with the 198H11T7 cDNA insert. The positive cDNA λ-clone was in vivo excised to obtain a plasmid clone (pZpgi2) carrying an insert of 1.7 kb. A 14-kb SalI fragment of the PGI genomic λ-clone (λpgi5) was further subcloned into pBluescript SK+.
Immunoblot Analysis
Standard immunology techniques were used as described by Harlow and Lane (1988). Leaf extracts were electrophoresed in 10% (w/v) SDS-PAGE and electroblotted to nylon membranes (Nytran, Schleicher & Schull, Keene, NH). Rabbit antisera were prepared against the E. coli-expressed Arabidopsis PGI antigen and used as probes for the immunoblot analyses. The Arabidopsis PGI protein was isolated from an E. coli strain carrying a plasmid with an EcoRI-XhoI 1.7-kb fragment of the PGI cDNA clone (pZpgi2) inserted into pET30A (Novagen, Madison, WI). The primary antibody was detected with the Vectastain ABC kit (Vector Laboratories, Burlingame, CA).
ACKNOWLEDGMENTS
We thank H. Sun, N.S. Yang, S.M. Lee, and S. Zeeman for reading the manuscript prior to publication.
Footnotes
This work was supported by the National Science Council (Taiwan, Republic of China; grant nos. NSC 88–2311–B–002–030 to S.-M.W. and NSC 87–2311–B–001–074 to J.C.) and by Academia Sinica (to J.C.).
LITERATURE CITED
- Bernier G, Havelange A, Houssa C, Petitjean A, Lejeune P. Physiological signals that induce flowering. Plant Cell. 1993;5:1147–1155. doi: 10.1105/tpc.5.10.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert S, Grosse H, Heldt HW. Specific transport of inorganic phosphate, glucose 6-phosphate, dihydroxyacetone phosphate and 3-phosphoglycerate into amyloplasts from pea roots. FEBS Lett. 1989;253:183–186. [Google Scholar]
- Borchert S, Harborth J, Schunemann D, Hoferichter P, Heldt HW. Studies of the enzymic capacities and transport properties of pea root plastids. Plant Physiol. 1993;101:303–312. doi: 10.1104/pp.101.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T. Genetic dissection of the biosynthesis, degradation, and biological functions of starch. In: Meyerowitz EM, Somerville CR, editors. Arabidopsis. New York: Cold Spring Harbor Laboratory Press; 1994. pp. 913–936. [Google Scholar]
- Caspar T, Huber SC, Somerville C. Alterations in growth, photosynthesis, and respiration in a starchless mutant of Arabidopsis thaliana L. deficient in a chloroplast phosphoglucomutase activity. Plant Physiol. 1985;79:11–17. doi: 10.1104/pp.79.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Bowman JL, DeJohn AW, Lander ES, Meyerowitz EM. Restriction fragment length polymorphism map for Arabidopsis thaliana. Proc Natl Acad Sci USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, Lejeune P, Bernier G. The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: comparison between the wild type and a starchless mutant. Planta. 1998;206:131–137. doi: 10.1007/s004250050383. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. doi: 10.1110/ps.8.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah LC. Starch synthesis in the maize seed. In: Larkins BA, Vasil IK, editors. Cellular and Molecular Biology of Plant Seed Development. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 375–405. [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Jones TWA, Gottlieb LD, Pichersky E. Reduced enzyme activity and starch level in an induced mutant of chloroplastic phosphoglucose isomerase. Plant Physiol. 1986;81:367–371. doi: 10.1104/pp.81.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer B, Fischer K, Hilpert B, Schubert S, Gutensohn M, Weber A, Flugge U. Molecular characterization of a carbon transporter in plastids from heterotrophic tissues: the glucose 6-phosphate/phosphate antiporter. Plant Cell. 1998;10:105–117. doi: 10.1105/tpc.10.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruckeberg AL, Neuhaus HE, Feil R, Gottlieb LD, Stitt M. Decreased-activity mutants of phosphoglucose isomerase in the cytosol and chloroplast of Clarkia xantiana: impact on mass-action ratios and fluxes to sucrose and starch, and estimation of flux control coefficients and elasticity coefficients. Biochem J. 1989;261:457–467. doi: 10.1042/bj2610457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Dean C. The transition to flowering. Plant Cell. 1998;10:1973–1989. doi: 10.1105/tpc.10.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister C, Dean C. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 1993;4:745–750. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- Meng M, Chen Y-T, Hsiao Y-Y, Itoh Y, Bagdasarian M. Mutational analysis of the conserved cationic residues of Bacillus stearothermophilus 6-phosphoglucose isomerase. Eur J Biochem. 1998;257:500–505. doi: 10.1046/j.1432-1327.1998.2570500.x. [DOI] [PubMed] [Google Scholar]
- Nowitzki U, Flechner A, Kellermann J, Hasegawa M, Schnarrenberger C, Martin W. Eubacterial origin of nuclear genes for chloroplast and cytosolic glucose-6-phosphate isomerase from spinach: sampling eubacterial gene diversity in eukaryotic chromosomes through symbiosis. Gene. 1998;214:205–213. doi: 10.1016/s0378-1119(98)00229-7. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Smith MW, Doolittle RF. Anomalous phylogeny involving the enzyme glucose-6-phosphate isomerase. J Mol Evol. 1992;34:544–545. doi: 10.1007/BF00160467. [DOI] [PubMed] [Google Scholar]
- Stam P. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 1993;3:739–744. [Google Scholar]
- Thomas BR, Ford VS, Pichersky E, Gottlieb LD. Molecular characterization of duplicated cytosolic phosphoglucose isomerase genes in Clarkia and comparison to the single gene in Arabidopsis. Genetics. 1993;135:895–905. doi: 10.1093/genetics/135.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BR, Laudencia-Chingcuanco D, Gottlieb LD. Molecular analysis of the plant gene encoding cytosolic phosphoglucose isomerase. Plant Mol Biol. 1992;19:745–757. doi: 10.1007/BF00027071. [DOI] [PubMed] [Google Scholar]
- Wang SM, Chu B, Lue WL, Yu TS, Eimert K, Chen J. adg2-1 represents a missense mutation in the ADPG pyrophosphorylase large subunit gene of Arabidopsis thaliana. Plant J. 1997;11:1121–1126. doi: 10.1046/j.1365-313x.1997.11051121.x. [DOI] [PubMed] [Google Scholar]
- Wang SM, Lue WL, Yu TS, Long JH, Wang CN, Eimert K, Chen J. Characterization of ADG1, and Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J. 1998;13:63–70. doi: 10.1046/j.1365-313x.1998.00009.x. [DOI] [PubMed] [Google Scholar]