Abstract
The crisis of antimicrobial resistance (AMR) is one of the most serious issues facing us today. The scale of the problem is illustrated by the recent commitment of Heads of State at the UN to coordinate efforts to curb the spread of AMR infections. In this review, we explore the biochemistry behind the headlines of a few stories that were recently published in the public media. We focus on examples from three different issues related to AMR: (i) hospital-acquired infections, (ii) the spread of resistance through animals and/or the environment and (iii) the role of antimicrobial soaps and other products containing disinfectants in the dissemination of AMR. Although these stories stem from three very different settings, the underlying message in all of them is the same: there is a direct relationship between the use of antimicrobials and the development of resistance. In addition, one type of antimicrobial could select for cross-resistance to another type and/or for multidrug resistance. Therefore, we argue the case for increased stewardship to not only cover clinical use of antibiotics, but also the use of antimicrobials in agriculture and stewardship of our crucially important biocides such as chlorhexidine.
Keywords: antimicrobial resistance, antiseptic, cross-resistance, hospital acquired infections, last resort antibiotic, spread of resistance
Introduction
The serendipitous discovery of penicillin by Alexander Fleming, and subsequent development of the fungal metabolite into a viable treatment for infections by Howard Florey and his group, marked the beginning of the golden age of antibiotics. Huge optimism reigned during the 1960s and into the 1970s due to the defeat of smallpox and polio. However, less than 50 years later the WHO published its first Global Report on Antimicrobial Resistance and concluded that, without intervention, we are heading for a post-antibiotic era, where minor infections and small injuries will once again be fatal [1]. Later that year the first economic report on the impact of antimicrobial resistance (AMR), commissioned by David Cameron (the then Prime Minister of Britain) and headed by economist Lord O’Neill, was published [2]. According to the O’Neill report, if nothing is done, antibiotic resistance-related deaths would increase from 700000 annually to 10 million annually by 2050, overtaking cancer as the main cause of mortality and costing the healthcare industry trillions of USD [2].
Microorganisms are incredibly adaptable, and considering that Escherichia coli can divide every 20 min under ideal conditions, this gives them a huge evolutionary advantage over humans with an average lifespan of 75 years. They can respond and resist any treatments we might challenge them with. Most of our antibiotics come from other microorganisms [3]. This is hardly surprising as they excrete these compounds to fight each other in their continuous competition for resources. This also provides an explanation for the discovery of resistance genes in 30000-year-old permafrost [4], making the development of resistance against antimicrobials unsurprising. What is astounding though, is the immense speed with which resistance is developing. There is no doubt that human intervention plays a substantial role in the development of AMR, as there is a linear correlation between the use of antibiotics and the development of resistance [5,6]. Moreover, if the use and abuse of antibiotics in healthcare settings is alarming, it is worth pausing to consider the fact that animals in the U.S.A. consume more than twice as many medically important antibiotics as humans [7]. This alarm is compounded by the fact that in many countries, several tonnes of last-resort antibiotics, such as colistin, are used in animal feed every year [8–10]. Although antibiotics are used therapeutically in food animals to treat clinical disease, they are also applied prophylactically to prevent common disease outbreaks (particularly in intensively farmed animals) and subtherapeutically to enhance animal growth [11,12]. As for the use of antimicrobials in general household items, there is no limitation (or even quantification) of the amounts used, and the ability of these antimicrobials to hasten the development of resistance has gone largely unnoticed.
In this review, we will look at some of the stories on AMR that have made headlines recently. An analysis of the development of the resistance will be provided and the biochemical mechanism underlying the observed resistance will be explored. The current state of our preventative measures to curb the threat of AMR will also be evaluated.
Hospital-acquired infections
Hospital-acquired infections frequently make news headlines and gain considerable public interest. These infections are caused by a range of opportunistic pathogens (organisms that only cause disease in immunocompromised individuals); many of them multidrug resistant [13]. Device-associated infections such as ventilator-associated pneumonia (VAP) and urinary tract infections (UTI) account for approximately 60% of all hospital-associated infections [13,14]. A meta-analysis found that in the United States, the treatment of hospital-acquired infections cost an estimated $10 billion annually [15].
We will focus on the most notorious and widespread hospital-acquired infection [16], responsible for nearly 20000 in-hospital deaths every year in the U.S.A. alone [17], namely methicillin-resistant Staphylococcus aureus (MRSA), and explore the biochemistry behind the development of resistance in this pathogen.
Methicillin-resistant S. aureus
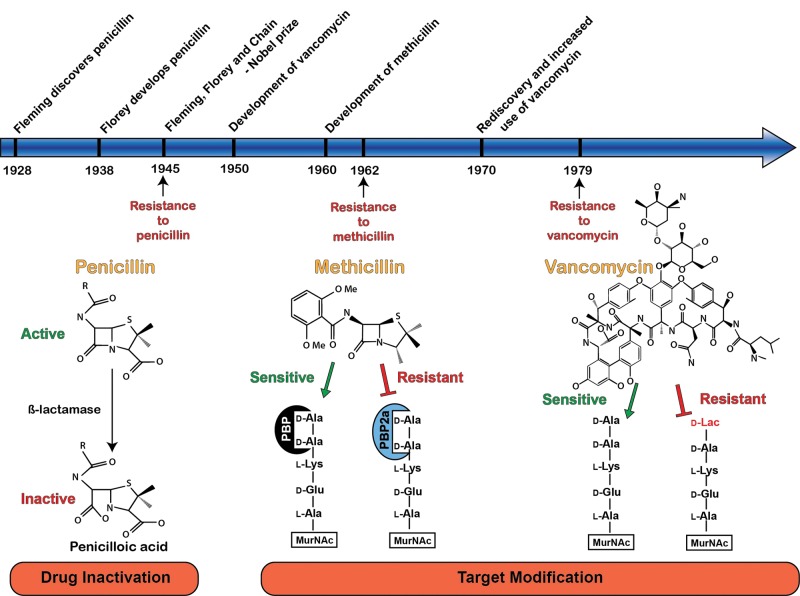
S. aureus is a Gram-positive bacterium that causes skin and wound infections, bacteraemia and toxic shock syndrome [18–20]. Historically, S. aureus infections have been associated with severe morbidity and mortality, particularly in the wounds of soldiers, which were highly prevalent in the early 20th century [21]. During World War II, however, S. aureus wound infections were treated successfully for the first time with the newly developed antimicrobial, penicillin (Figure 1) [22]. Unfortunately, by the end of the war and in the same year that Alexander Fleming, Howard Florey and Ernst Chain received their Nobel Prize for the discovery and development of penicillin, the first strains of S. aureus resistant to penicillin started to emerge (Figure 1) [19,21]. Resistance to penicillin is through the acquisition of the blaZ gene, which encodes β-lactamase [23]. This gene product enzyme hydrolyses the generic β-lactam structure of penicillin, which is core to all β-lactam antibiotics, rendering them ineffective as a clinical treatment (Figure 1). The transmission of the blaZ gene is achievable through conjugal transfer, which resulted in widespread S. aureus penicillin resistance [23]. Resistance towards penicillin necessitated the development of methicillin, a narrow spectrum β-lactam alternative to penicillin. Methicillin and the penicillins that followed (oxacillin, nafcillin, cloxacillin and dicloxacillin) were designed with bulky side groups so that they would not fit in the active site of the β-lactamase and hence could not be inactivated by these enzymes (Figure 1) [24]. However, resistance towards methicillin was first documented just 2 years after its clinical introduction, giving rise to MRSA. Methicillin resistance occurs through the acquired alternative penicillin-binding protein, penicillin-binding protein 2a (PBP2a), encoded by the mecA gene that is regulated by the blaZ-blaI-blaR1 and mecA-mecI-mecRI systems [23,25]. In methicillin-sensitive S. aureus, the antimicrobial activity of methicillin is mediated through high-affinity interaction with the bacterial PBPs. This binding event ultimately results in inhibition of the PBP enzymes, preventing cross-linking of bacterial peptidoglycan and leading to cell lysis [25]. However, the PBP2a variant has a much lower binding affinity for methicillin, allowing its continued activity even in the presence of methicillin and therefore, allowing bacterial survival [24]. MRSA strains are highly resistant against most β-lactams and many other classes of antibiotics. Vancomycin is the drug of choice for the treatment of infections caused by methicillin-resistant staphylococci [26,27]; however, S. aureus strains with increased MICs against vancomycin are also emerging now.
Figure 1. Timeline of selected antibiotic development and reported resistance.
A mechanism of resistance is illustrated for each antibiotic. Penicillin is commonly inactivated by bacterial β-lactamases, which cleave the β-lactam ring, forming the inactive penicilloic acid. Subsequent development of methicillin utilized a larger aryl side chain that was largely resistant to hydrolytic cleavage by β-lactamases. Instead, resistance to methicillin is driven by the expression of the alternative transpeptidase, PBP2a, which has a lower affinity for methicillin and can catalyse peptidoglycan cross-linking despite methicillin intervention. Resistance to vancomycin is driven by structural alteration of the terminal dipeptide that is modified from d-alanyl-d-alanine (d-Ala-d-Ala) to d-alanyl-d-lactate (d-Ala-d-Lac), reducing the affinity of the dipeptide for vancomycin and preventing disruption of peptidoglycan cross-linking.
Vancomycin intermediate S. aureus (VISA) and vancomycin-resistant S. aureus (VRSA)
Just like the β-lactam antibiotics, vancomycin also inhibits cell wall synthesis. However, it has a different target. Vancomycin binds to the D-Ala-D-Ala residues that are part of the building blocks of the peptidoglycan cell wall, and so prevent the formation of the essential peptide cross-links used to connect the units of the cell wall [28]. This mechanism of inhibition was thought to represent a major breakthrough in the field of antibiotics, as resistance towards vancomycin was absent for many years after its introduction. However, a strain of S. aureus with reduced vancomycin susceptibility (VISA) was first reported in 1997 [29]. Since then the incidence of VISA has been steadily increasing so that VISA is currently an immediate concern in the treatment of infections caused by S. aureus [30,31]. VISA strains have thicker cell walls compared with vancomycin-sensitive strains. These conditions restrict vancomycin to the outer layers of bacterium and prevent it from inducing its antimicrobial effects [32,33]. VRSA strains have also emerged [29]. MRSA obtains resistance to vancomycin through the conjugal transfer of the plasmid borne transposon Tn1546 from the vancomycin-resistant Enterococcus faecalis. The specific biochemical mechanism conferred by the Tn1546 transposon was shown to alter the dipeptide residue D-Ala-D-Ala to D-Ala-D-Lac, a dipeptide with substantially lower affinity for vancomycin (Figure 1) [34]. Only a few cases of VRSA have been reported so far and therefore it does not represent an urgent public health threat.
The worldwide concern about the prevalence of MRSA and VISA meant that considerable effort went into developing alternatives to vancomycin for the treatment of MRSA. Currently, the advanced generation cephalosporin ceftaroline, the lipopeptide daptomycin, the vancomycin analogues telavancin, oritavancin and dalbavancin, and the oxazolidinones linezolid and tedizolid, can still be used against MRSA [35–37]. However, the development of resistance has been observed against these drugs already [38–40]. Therefore, the significance of MRSA and VISA should not be underestimated, due to both its significance regarding global mortality and its historical ability to develop resistance mechanisms in the presence of antibiotic stress.
Resistance through the use of antibiotics in veterinary science and agriculture
Apocalypse pig and the demise of a last resort antibiotic
‘Apocalypse pig’ refers to the first pig reported to harbour a Gram-negative organism resistant to the last resort antibiotic, colistin [41]. The term ‘last resort antibiotic’ refers to an antibiotic that still has activity against resistant pathogens and are therefore used as a last line of treatment when other antibiotics fail. Once organisms have developed resistance against a last resort antibiotic, hardly any treatment options remain [42]. Colistin has an interesting history. It is a polymyxin type of antibiotic (also known as polymyxin E) that act by permeabilizing the outer membrane of Gram-negative organisms [43]. Electrostatic interactions between colistin and the lipid A subunits present in the lipopolysaccharide (LPS) of the outer membrane [43–45] alter the structural integrity of LPS, leading to cellular membrane permeability and resulting in bacterial death (Figure 2) [46,47]. Colistin is characterized by remarkable antimicrobial activity against hard to treat Gram-negative organisms such as multidrug-resistant Pseudomonas aeruginosa, Acinetobacter baumannii and Klebsiella pneumoniae [48–50]. It was first developed in the 1950s but was shelved in the 1970s due to its nephrotoxicity [45]. When widespread resistance developed against other antibiotics, Li et al. [51] revisited colistin and provided the pharmacokinetic and pharmacodynamic data that enabled the development of colistin for new clinical applications. Although colistin can be dosed as the parent compound, more commonly it is administered as a prodrug, colistin methanesulfonate, leading to some confusion in dosing regimes.
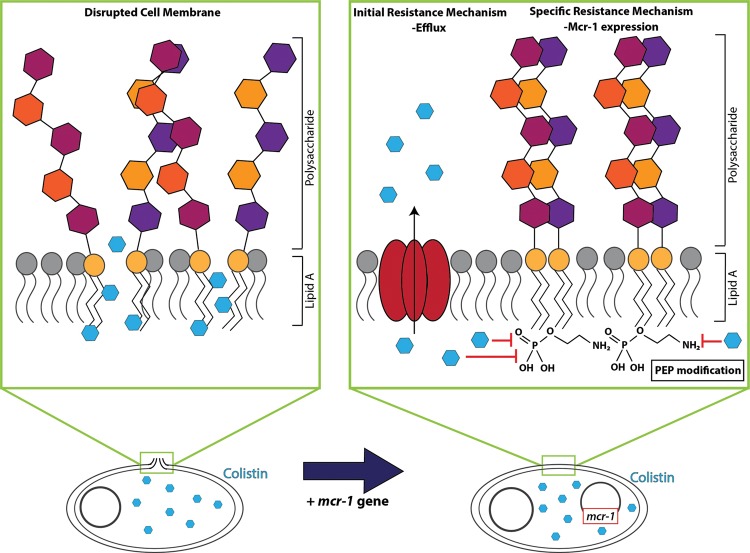
Figure 2. Selected mechanisms of colistin resistance.
The bactericidal activity of colistin relies on disruption of the bacterial cell membrane, initiated by electrostatic interaction between colistin and the lipid A portion of bacterial LPS. Immediate, albeit non-specific, resistance to colistin is mediated through transcriptional up-regulation of drug efflux pumps. Specific resistance to colistin is facilitated by the plasmid-mediated mcr-1 gene, encoding a phosphoethanolamine transferase, which modifies lipid A with a phosphoethanolamine (PEP) group, preventing interaction between colistin and lipid A.
Since colistin was not used clinically for approximately 30 years, resistance against colistin was very rare. Early research indicated that colistin resistance in E. coli and Salmonella isolates could be conferred by a mutation in the two-component PhoP-PhoQ and/or PmrA-PmrB systems [46]. These mutations produce structural modifications in the lipid A subunit that reduce electrostatic interactions between the positively charged amino groups of colistin and negatively charged phosphate groups of the lipid A subunits, preventing disruption of the cell membrane [47]. However, recently, the plasmid-encoded mcr-1 gene was discovered in colistin-resistant, commensal E. coli strains from food animals in China [10]. This non-chromosomal mechanism of colistin resistance raised fears for the rapid spread of resistance, similar to the dissemination of the β-lactamase coding antibiotic resistance genes blaKPC and blaNDM-1 [52–54]. In addition, colistin is also subject to efflux by drug efflux pumps that could give initial resistance to this antibiotic (Figure 2) [55–57]. Even though colistin was not used clinically for a long time, many countries have been using colistin in agriculture [9]. China currently represents the world’s largest user of colistin in livestock, dedicating 11942 tonnes to livestock agricultural feeds annually [10]. Colistin-resistant organisms have been isolated from people with no prior exposure to colistin, indicating the possibility for the mcr-1 gene to spread from animals to humans. The detection of the mcr-1 gene in multiple countries makes it difficult to pinpoint the origin of AMR. Retrospective analysis indicated that the mcr-1 gene has been present since the 1980s in E. coli in China without causing serious issues [58]. However, it is clear that a global reassessment of the agricultural use of this crucially important, last resort antibiotic, is now imperative. To this extent, the use of colistin as a feed additive for animals has been banned in China effective from 1 November 2016 [12].
Resistance from farm to fork
Several other outbreaks of infectious disease caused by multidrug-resistant organisms, acquired through food sources, have brought the issue of the use of antibiotics in agriculture firmly into public attention. In 2014, a multistate outbreak of multidrug-resistant Salmonella heidelberg in the U.S.A. was linked to consumption of chicken meat from one supplier [59]. Prophylactic use of antimicrobials in factory farmed chickens is huge, currently estimated at a global annual consumption of 148 mg of antibiotic per kg of animal produced [60], to prevent outbreaks in the crowded and unhygienic conditions. Incidences like these have prompted many public calls for ‘antibiotic-free meat’. However, it is not residual antibiotics in the meat that poses a problem, the real issue is the selection of multidrug-resistant superbugs in animals raised on antimicrobials. These animals could act as reservoirs of resistant organisms that could eventually find their way to human consumers, either through the environment or through direct contact [61–63].
There is a widespread belief that antimicrobials that are not currently in clinical use are fine to use as growth promoters in feed animals, as resistance to these compounds would not lead to resistance to clinically used antimicrobials. However, this argument does not hold true, as pathogens can express drug efflux pumps that can expel many different classes of compounds, including the antimicrobials used as feed additives [64–66]. Organisms that express these efflux pumps will subsequently be resistant against a multitude of antimicrobial compounds, including those used in healthcare. This efflux-mediated resistance would confer a fitness advantage to organisms, sufficient to allow survival in the presence of clinical antibiotics until specific resistance mechanisms have been acquired [67,68]. Animals that are ill should undoubtedly be treated with antimicrobials, even medically important ones. However, the use of antibiotics and other antimicrobials both prophylactically and as growth-enhancers needs to be re-considered.
These facts are slowly being recognized and acted upon. In 2015, the WHO released their ‘Global action plan’ on AMR. One of the objectives in this report is to ‘optimize the use of antimicrobial medicines in human and animal health’ (Objective 4) by, among other things, curbing the ‘inappropriate or unregulated use of antimicrobial agents in agriculture’ [69]. In June, 2015, the Australian government adopted a ‘One Health’ approach, where one of their objectives is ‘Surveillance of antimicrobial resistance and antimicrobial usage in human health and animal care’ [70]. These measures are timely and much needed. It would be ideal though if monitoring programmes were extended not only to the use of antibiotics, but also to include biocides as addressed in the following section.
The issue with antimicrobial soaps
The FDA (U.S.A.) ban the inclusion of triclosan in antibacterial soaps
Our society is obsessed with a sense of cleanliness and (understandably) infection control, as is evident by the widespread use of antibacterial soaps and the inclusion of antiseptics/biocides and antimicrobial nanosilver in many household cleaning and personal care products; from soaps to socks that prevent smelly feet. In September 2016, the FDA in the U.S.A. banned the use of the antibacterial agent triclosan as well as 18 other compounds from soaps [71]. There were several reasons for this ban. Firstly, triclosan containing soaps are not more efficient in preventing the spread of infection compared with normal soap [72]. Secondly, there are concerns regarding the safety and health effect of long-term exposure to triclosan [73]. Thirdly and very importantly, the inclusion of triclosan in soap could lead to the development of AMR [74]. Unlike antibiotics, biocides, antiseptics and nanosilver particles do not have specific targets, but act rather non-selectively on microbial cells. For this reason, it was believed that resistance should not easily develop against these types of compounds [75]. However, bacteria have a natural defence mechanism to protect them from toxic compounds: drug efflux pumps. These efflux pumps are membrane proteins that expel antibiotics and other toxic compounds from the microbial cells, thereby lowering their concentration inside the cell to sub-toxic levels (Figure 3) [68,72,73]. Drug efflux pumps are the main contributors to multidrug resistance by virtue of their ability to expel a wide variety of structurally and functionally distinct antibiotics. The substrate range of drug efflux proteins is not limited to antibiotics, but include biocides, dyes, detergents, heavy metals and endogenous compounds such as virulence factors [76–82]. Therefore, efflux pumps have the ability to give widespread resistance against these compounds, and simultaneously render the organisms resistant against the antibiotics that are currently in clinical use to treat infections [83–92].
Figure 3. Biocide usage and antibiotic resistance.
Biocides such as triclosan and chlorhexidine exert their antimicrobial activity through non-specific interactions with cellular targets. An innate bacterial defence to toxic compounds, such as these, is up-regulation of multidrug efflux pumps, such as qacA in the Gram-positive organism S. aureus and mexAB-oprM in the Gram-negative organism P. aeruginosa. Once expressed, these efflux pathways will not only export biocides, but also antibiotics, antiseptics, heavy metals and dyes – hence resulting in the development of multidrug resistance.
Use of chlorhexidine selects for resistance against the last resort antibiotic colistin
In contrast with triclosan, the biocide chlorhexidine is highly efficient in the prevention and control of the spread of infectious organisms [93]. It is widely used in various clinical applications that require decolonization and infection control. For example, chlorhexidine gluconate (CHG) is used as an antiseptic to decolonize skin before operations [94], as a mouthwash to prevent plaque formation and sterilization of the mouth before dental implants [95], as a daily bath solution to prevent hospital-acquired infections in e.g. burn patients [96] or as a rinse to prevent infections associated with indwelling devices such as VAP or UTI [94]. Resistance to CHG will therefore have dire consequences for infection control in hospital settings. Although CHG is mostly still very effective, and resistance to the high concentrations (1–2%) used in hospital disinfectants has not been reported for Gram-positive organisms, increased tolerance (in some cases up to 100-fold increase in MIC to CHG) has been reported in Gram-negative organisms, such as P. aeruginosa and K. pneumoniae [97–99]. More worryingly, a pan drug resistant strain of K. pneumoniae has been identified that was able to multiply in a 1% CHG disinfectant solution [100]. Resistance to CHG usually arise through the expression of drug efflux proteins that can pump out CHG such as QacA/QacB from S. aureus and the MexAB-OprM system from P. aeruginosa [88]. This also poses the question as to the possible relationship between CHG tolerance and antibiotic resistance. Very recently, links between the use of CHG and resistance to both vancomycin and the last line antibiotic, colistin, have been established [97,101]. These worrying observations clearly indicate that in addition to antibiotic stewardship, we also need stewardship of our biocides especially the critically useful ones such as CHG.
Conclusions
We grew up in the golden age of antibiotics. A world without effective antimicrobials – where a simple scratch could cost you your life and where most modern medical procedures would no longer be possible – is unthinkable. Yet, we are bombarded on a daily basis with reports of AMR superbugs impervious to our best treatments. In this review, we explored the biochemistry behind reports on AMR in healthcare, agriculture and the environment. The underlying, unifying, factor of all these case studies is the fast development and global spread of AMR regardless of the geographical location or community of origin. This is a truly global problem that will only be solved by a global response.
The recent commitment by Heads of State at the UN general assembly to adopt a broad, coordinated approach to tackle AMR (http://www.un.org/pga/71/2016/09/21/press-release-hl-meeting-on-antimicrobial-resistance/) is therefore a much needed and a very timely global incentive. Good stewardship is needed not only in the medical use of antimicrobials, but also for the use of antimicrobials in animal health, the abundant use of antimicrobials in agriculture, and the widespread use of biocides and antiseptics in common household products. Only once these issues have been suitably addressed can we hope to slow the ever-increasing development of AMR.
“There is probably no chemotherapeutic drug to which in suitable circumstances the bacteria cannot react by in some way acquiring ‘fastness’ [resistance].”
—Alexander Fleming, 1946
Summary
Hospital-acquired infections remain a serious threat as increased resistance reduces or eliminates treatment options and costs billions of dollars per year to manage.
The use of antibiotics in agriculture results in the development of multidrug-resistant organisms that act as reservoirs of resistance. These organisms or their genes can spread to humans either through direct contact or through the environment.
The excessive use of antiseptics and biocides leads to resistance against these compounds and cross-resistance to antibiotics.
Good stewardship programmes are needed, not only for clinically used antibiotics, but also for antimicrobials used in agriculture and for critically important antiseptics such as chlorhexidine.
Abbreviations
- AMR
antimicrobial resistance
- CHG
chlorhexidine gluconate
- LPS
lipopolysaccharide
- MIC
Minimum inhibitory concentration
- MRSA
methicillin-resistant Staphylococcus aureus
- PBP
penicillin-binding protein
- UTI
urinary tract infections
- VAP
ventilator-associated pneumonia
- VISA
vancomycin intermediate Staphylococcus aureus
- VRSA
vancomycin-resistant Staphylococcus aureus
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.World Health Organisation. (2014) Antimicrobial Resistance: Global Report on Surveillance, http://www.who.int/drugresistance/documents/surveillancereport/en/ (accessed 2 February 2017) [Google Scholar]
- 2.O’Neill J. (2014) Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations, https://amr-review.org/ [Google Scholar]
- 3.Korzybski T., Kowszyk-Gindifer Z. and Kurylowicz W. (1967) Antibiotics: Origin, Nature and Properties, Pergamon Press, Poland [Google Scholar]
- 4.D’Costa V.M., King C.E., Kalan L., Morar M., Sung W.W., Schwarz C. et al. (2011) Antibiotic resistance is ancient. Nature 477, 457–461 [DOI] [PubMed] [Google Scholar]
- 5.Goossens H., Ferech M., Vander Stichele R., Elseviers M. and ESAC Project Group (2005) Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet (North Am. Ed.) 365, 579–587 [DOI] [PubMed] [Google Scholar]
- 6.Sun L., Klein E.Y. and Laxminarayan R. (2012) Seasonality and temporal correlation between community antibiotic use and resistance in the United States. Clin. Infect. Dis. 55, 687–694 [DOI] [PubMed] [Google Scholar]
- 7.O’Neill J. (2015) Antimicrobials in agriculture and the environment: reducing unnecessary use and waste, https://amr-review.org/Publications [Google Scholar]
- 8.Cuong N., Nhung N.T., Nghia N.H., Mai Hoa N.T., Trung N.V., Thwaites G. et al. (2016) Antimicrobial consumption in medicated feeds in vietnamese pig and poultry production. EcoHealth 13, 490–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempf I., Jouy E. and Chauvin C. (2016) Colistin use and colistin resistance in bacteria from animals. Int. J. Antimicrob. Agents 48, 598–606 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J. et al. (2016) Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168 [DOI] [PubMed] [Google Scholar]
- 11.Shen Z., Wang Y., Shen Y., Shen J. and Wu C. (2016) Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect. Dis. 16, 293. [DOI] [PubMed] [Google Scholar]
- 12.Walsh T.R. and Wu Y. (2016) China bans colistin as a feed additive for animals. Lancet Infect. Dis. 16, 1102–1103 [DOI] [PubMed] [Google Scholar]
- 13.Saint S., Kowalski C.P., Kaufman S.R., Hofer T.P., Kauffman C.A., Olmsted R.N. et al. (2008) Preventing hospital-acquired urinary tract infection in the United States: a national study. Clin. Infect. Dis. 46, 243–250 [DOI] [PubMed] [Google Scholar]
- 14.Koeman M., van der Ven A.J., Hak E., Joore H.C., Kaasjager K., de Smet A.G. et al. (2006) Oral decontamination with chlorhexidine reduces the incidence of ventilator-associated pneumonia. Am. J. Respir. Crit. Care Med. 173, 1348–1355 [DOI] [PubMed] [Google Scholar]
- 15.Zimlichman E., Henderson D., Tamir O., Franz C., Song P., Yamin C.K. et al. (2013) Health care-associated infections: a meta-analysis of costs and financial impact on the US health care system. JAMA Intern. Med. 173, 2039–2046 [DOI] [PubMed] [Google Scholar]
- 16.Loffler C.A. and MacDougall C. (2007) Update on prevalence and treatment of methicillin-resistant Staphylococcus aureus infections. Expert Rev. Anti Infect. Ther. 5, 961–981 [DOI] [PubMed] [Google Scholar]
- 17.Boucher H.W. and Corey G.R. (2008) Epidemiology of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 46, S344–S349 [DOI] [PubMed] [Google Scholar]
- 18.Lowy F.D. (1998) Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 19.Wertheim H.F., Melles D.C., Vos M.C., van Leeuwen W., van Belkum A., Verbrugh H.A. et al. (2005) The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 5, 751–762 [DOI] [PubMed] [Google Scholar]
- 20.Gould F.K., Brindle R., Chadwick P.R., Fraise A.P., Hill S., Nathwani D. et al. (2009) Guidelines (2008) for the prophylaxis and treatment of methicillin-resistant Staphylococcus aureus (MRSA) infections in the United Kingdom. J. Antimicrob. Chemother. 63, 849–861 [DOI] [PubMed] [Google Scholar]
- 21.Tong M.J. (1972) Septic complications of war wounds. JAMA 219, 1044–1047 [PubMed] [Google Scholar]
- 22.Swann J.P. (1983) The search for synthetic penicillin during World War II. Brit. J. Hist. Sci. 16, 154–190 [DOI] [PubMed] [Google Scholar]
- 23.Hao H., Dai M., Wang Y., Huang L. and Yuan Z. (2012) Key genetic elements and regulation systems in methicillin-resistant Staphylococcus aureus. Future Microbiol. 7, 1315–1329 [DOI] [PubMed] [Google Scholar]
- 24.Stapleton P.D. and Taylor P.W. (2002) Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci. Prog. 85, 57–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers H.F. (1997) Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens D.L. (2006) The role of vancomycin in the treatment paradigm. Clin. Infect. Dis. 42, S51–S57 [DOI] [PubMed] [Google Scholar]
- 27.Moellering R.C. (2006) Vancomycin: a 50-year reassessment. Clin. Infect. Dis. 42, S3–S4 [DOI] [PubMed] [Google Scholar]
- 28.Arthur M. and Courvalin P. (1993) Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 37, 1563–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiramatsu K., Hanaki H., Ino T., Yabuta K., Oguri T. and Tenover F. (1997) Methicillin-resistance Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 135–136 [DOI] [PubMed] [Google Scholar]
- 30.Tenover F.C., Biddle J.W. and Lancaster M.V. (2001) Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7, 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhand A. and Sakoulas G. (2012) Reduced vancomycin susceptibility among clinical Staphylococcus aureus isolates (‘the MIC Creep’): implications for therapy. F1000 Med. Rep. 4, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardete S. and Tomasz A. (2014) Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 124, 2836–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rishishwar L., Kraft C.S. and Jordan I.K. (2016) Population genomics of reduced vancomycin susceptibility in Staphylococcus aureus. mSphere 1, 4, e00094–16, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handwerger S. and Skoble J. (1995) Identification of chromosomal mobile element conferring high-level vancomycin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 39, 2446–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burnett Y.J., Echevarria K. and Traugott K.A. (2016) Ceftaroline as salvage monotherapy for persistent MRSA bacteremia. Ann. Pharmacother. 50, 1051–1059 [DOI] [PubMed] [Google Scholar]
- 36.Miller W.R., Bayer A.S. and Arias C.A. (2016) Mechanism of action and resistance to daptomycin in Staphylococcus aureus and Enterococci. CSH Perspect. Med. 6, 11, a026997, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeng D., Debabov D., Hartsell T.L., Cano R.J., Adams S., Schuyler J.A. et al. (2016) Approved glycopeptide antibacterial drugs: mechanism of action and resistance. CSH Perspect. Med. 6, 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan L.C., Basuino L., Diep B., Hamilton S., Chatterjee S.S. and Chambers H.F. (2015) Ceftobiprole-and ceftaroline-resistant methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 59, 2960–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skiest D.J. (2006) Treatment failure resulting from resistance of Staphylococcus aureus to daptomycin. J. Clin. Microbiol. 44, 655–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Syed Y.Y. and Scott L.J. (2015) Oritavancin: a review in acute bacterial skin and skin structure infections. Drugs 75, 1891–1902 [DOI] [PubMed] [Google Scholar]
- 41.McKenna M. (2015) Apocalypse pig: the last antibiotic begins to fail. Germination, National Geographic, http://phenomena.nationalgeographic.com/2015/11/21/mcr-gene-colistin/ [Google Scholar]
- 42.Cai Y., Chai D., Wang R., Liang B. and Bai N. (2012) Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J. Antimicrob. Chemother. 67, 1607–1615 [DOI] [PubMed] [Google Scholar]
- 43.Gallardo-Godoy A., Muldoon C., Becker B., Elliott A.G., Lash L.H., Huang J.X. et al. (2016) Activity and predicted nephrotoxicity of synthetic antibiotics based on polymyxin B. J. Med. Chem. 59, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gurjar M. (2015) Colistin for lung infection: an update. J. Intensive Care 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biswas S., Brunel J.-M., Dubus J.-C., Reynaud-Gaubert M. and Rolain J.-M. (2012) Colistin: an update on the antibiotic of the 21st century. Expert Rev. Anti Infect. Ther. 10, 917–934 [DOI] [PubMed] [Google Scholar]
- 46.Olaitan A.O., Morand S. and Rolain J.-M. (2014) Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Needham B.D. and Trent M.S. (2013) Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11, 467–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zavascki A.P., Goldani L.Z., Li J. and Nation R.L. (2007) Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60, 1206–1215 [DOI] [PubMed] [Google Scholar]
- 49.Bergen P.J., Landersdorfer C.B., Lee H.J., Li J. and Nation R.L. (2012) ‘Old’ antibiotics for emerging multidrug-resistant bacteria. Curr. Opin. Infect. Dis. 25, 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Velkov T., Roberts K.D., Nation R.L., Thompson P.E. and Li J. (2013) Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol. 8, 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Rayner C., Nation R., Deans R., Boots R., Widdecombe N. et al. (2005) Pharmacokinetics of colistin methanesulfonate and colistin in a critically ill patient receiving continuous venovenous hemodiafiltration. Antimicrob. Agents Chemother. 49, 4814–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhouma M., Beaudry F. and Letellier A. (2016) Resistance to colistin: what is the fate for this antibiotic in pig production? Int. J. Antimicrob. Agents 48, 119–126 [DOI] [PubMed] [Google Scholar]
- 53.Hu Y., Liu F., Lin I.Y., Gao G.F. and Zhu B. (2016) Dissemination of the mcr-1 colistin resistance gene. Lancet Infect. Dis. 16, 146–147 [DOI] [PubMed] [Google Scholar]
- 54.Johnson A.P. and Woodford N. (2013) Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J. Med. Microbiol. 62, 499–513 [DOI] [PubMed] [Google Scholar]
- 55.Davies J. and Davies D. (2010) Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ni W., Li Y., Guan J., Zhao J., Cui J., Wang R. et al. (2016) Effects of efflux pump inhibitors on colistin resistance in multidrug-resistant Gram-negative bacteria. Antimicrob. Agents Chemother. 60, 3215–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goli H.R., Nahaei M.R., Rezaee M.A., Hasani A., Kafil H.S. and Aghazadeh M. (2016) Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iranian J. Microbiol. 8, 62–69 [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarz S. and Johnson A.P. (2016) Transferable resistance to colistin: a new but old threat. J. Antimicrob. Chemother. 71, 2066–2070 [DOI] [PubMed] [Google Scholar]
- 59.Gieraltowski L., Higa J., Peralta V., Green A., Schwensohn C., Rosen H. et al. (2016) National outbreak of multidrug resistant Salmonella heidelberg infections linked to a single poultry company. PLoS ONE 11, 9, e016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Boeckel T.P., Brower C., Gilbert M., Grenfell B.T., Levin S.A., Robinson T.P. et al. (2015) Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U.S.A. 112, 5649–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aarestrup F.M., Wegener H.C. and Collignon P. (2008) Resistance in bacteria of the food chain: epidemiology and control strategies. Expert Rev. Anti Infect. Ther. 6, 733–750 [DOI] [PubMed] [Google Scholar]
- 62.Verraes C., Van Boxstael S., Van Meervenne E., Van Coillie E., Butaye P., Catry B. et al. (2013) Antimicrobial resistance in the food chain: a review. Int. J. Environ. Res. Pub. Health 10, 2643–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshall B.M. and Levy S.B. (2011) Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poole K. (2001) Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 3, 255–264 [PubMed] [Google Scholar]
- 65.Ohene-Agyei T., Lea J.D. and Venter H. (2012) Mutations in MexB that affect the efflux of antibiotics with cytoplasmic targets. FEMS Microbiol. Lett. 333, 20–27 [DOI] [PubMed] [Google Scholar]
- 66.Sarmah A.K., Meyer M.T. and Boxall A.B. (2006) A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65, 725–759 [DOI] [PubMed] [Google Scholar]
- 67.Webber M. and Piddock L. (2003) The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 51, 9–11 [DOI] [PubMed] [Google Scholar]
- 68.Venter H., Mowla R., Ohene-Agyei T. and Ma S. (2015) RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front. Microbiol. 6, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.World Health Organisation. (2015) Global Action Plan on Antimicrobial Resistance, Geneva, Switzerland [Google Scholar]
- 70. Australian Government (2015) Responding to the threat of antimicrobial resistance: National antimicrobial resistance strategy 2015-2019, June (2015)
- 71.McNamara P.J. and Levy S.B. (2016) Triclosan: an instructive tale. Antimicrob. Agents Chemother. 2105–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giuliano C.A. and Rybak M.J. (2015) Efficacy of triclosan as an antimicrobial hand soap and its potential impact on antimicrobial resistance: a focused review. Pharmacotherapy: J. Hum. Pharmacol. Drug Ther. 35, 328–336 [DOI] [PubMed] [Google Scholar]
- 73.Yee A.L. and Gilbert J.A. (2016) Is triclosan harming your microbiome? Science 353, 348–349 [DOI] [PubMed] [Google Scholar]
- 74.Webber M.A., Whitehead R.N., Mount M., Loman N.J., Pallen M.J. and Piddock L.J. (2015) Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J. Antimicrob. Chemother. 70, 2241–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suller M. and Russell A. (2000) Triclosan and antibiotic resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 46, 11–18 [DOI] [PubMed] [Google Scholar]
- 76.Chuanchuen R., Karkhoff-Schweizer R.R. and Schweizer H.P. (2003) High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control. 31, 124–127 [DOI] [PubMed] [Google Scholar]
- 77.Silver S. and Phung L.T. (1996) Bacterial heavy metal resistance: new surprises. Ann. Rev. Microbiol. 50, 753–789 [DOI] [PubMed] [Google Scholar]
- 78.Piddock L.J. (2006) Multidrug-resistance efflux pumps – not just for resistance. Nat. Rev. Microbiol. 4, 629–636 [DOI] [PubMed] [Google Scholar]
- 79.Du D., Venter H., Pos K.M. and Luisi B.F. (2013) The machinery and mechanisms of multidrug efflux in Gram-negative bacteria. Micrbial Efflux Pumps (Yu E., Zhang Q., Brown M., eds), pp. 35–49, Caister Academic Press, Norfolk, U.K. [Google Scholar]
- 80.Wang Y., Venter H. and Ma S. (2016) Efflux pump inhibitors: a novel approach to combat efflux-mediated drug resistance in bacteria. Curr. Drug Targets 17, 702–719 [DOI] [PubMed] [Google Scholar]
- 81.Blanco P., Hernando-Amado S., Reales-Calderon J.A., Corona F., Lira F., Alcalde-Rico M. et al. (2016) Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms 4, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Abuzaid A., Hamouda A. and Amyes S. (2012) Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE efflux pump genes and antibiotic resistance. J. Hosp. Infect. 81, 87–91 [DOI] [PubMed] [Google Scholar]
- 83.Mc Cay P.H., Ocampo-Sosa A.A. and Fleming G.T. (2010) Effect of subinhibitory concentrations of benzalkonium chloride on the competitiveness of Pseudomonas aeruginosa grown in continuous culture. Microbiology 156, 30–38 [DOI] [PubMed] [Google Scholar]
- 84.Baker-Austin C., Wright M.S., Stepanauskas R. and McArthur J. (2006) Co-selection of antibiotic and metal resistance. Trends Microbiol. 1, 176–182 [DOI] [PubMed] [Google Scholar]
- 85.Kovacevic J., Ziegler J., Wałecka-Zacharska E., Reimer A., Kitts D.D. and Gilmour M.W. (2016) Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82, 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ho C.-M., Li C.-Y., Ho M.-W., Lin C.-Y., Liu S.-H. and Lu J.-J. (2012) High rate of qacA-and qacB-positive methicillin-resistant Staphylococcus aureus isolates from chlorhexidine-impregnated catheter-related bloodstream infections. Antimicrob. Agents Chemother. 56, 5693–5697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson R.C., Schlett C.D., Crawford K., Lanier J.B., Merrell D.S. and Ellis M.W. (2015) Recurrent methicillin-resistant Staphylococcus aureus cutaneous abscesses and selection of reduced chlorhexidine susceptibility during chlorhexidine use. J. Clin. Microbiol. 53, 3677–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kampf G. (2016) Acquired resistance to chlorhexidine – is it time to establish an ‘antiseptic stewardship’ initiative? J. Hosp. Infect. 94, 213–227 [DOI] [PubMed] [Google Scholar]
- 89.Kawamura-Sato K., Wachino J.-I., Kondo T., Ito H. and Arakawa Y. (2010) Correlation between reduced susceptibility to disinfectants and multidrug resistance among clinical isolates of Acinetobacter species. J. Antimicrob. Chemother. 65, 1975–1983 [DOI] [PubMed] [Google Scholar]
- 90.McNeil J.C., Kok E.Y., Vallejo J.G., Campbell J.R., Hulten K.G., Mason E.O. et al. (2016) Clinical and molecular features of decreased chlorhexidine susceptibility among nosocomial Staphylococcus aureus isolates at Texas Children’s Hospital. Antimicrob. Agents Chemother. 60, 1121–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu D., Lu R., Chen Y., Qiu J., Deng C. and Tan Q. (2016) Study of cross-resistance mediated by antibiotics, chlorhexidine and Rhizoma coptidis in Staphylococcus aureus. J. Glob. Antimicrob. Resist. 7, 61–66 [DOI] [PubMed] [Google Scholar]
- 92.Zhang M., O’Donoghue M., Ito T., Hiramatsu K. and Boost M. (2011) Prevalence of antiseptic-resistance genes in Staphylococcus aureus and coagulase-negative staphylococci colonising nurses and the general population in Hong Kong. J. Hosp. Infect. 78, 113–117 [DOI] [PubMed] [Google Scholar]
- 93.McDonnell G. and Russell A.D. (1999) Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Swan J.T., Ashton C.M., Bui L.N., Pham V.P., Shirkey B.A., Blackshear J.E. et al. (2016) Effect of chlorhexidine bathing every other day on prevention of hospital-acquired infections in the surgical ICU: a single-center, randomized controlled trial. Crit. Care Med. 44, 1822–1832 [DOI] [PubMed] [Google Scholar]
- 95.Supranoto S., Slot D., Addy M. and Van der Weijden G. (2015) The effect of chlorhexidine dentifrice or gel versus chlorhexidine mouthwash on plaque, gingivitis, bleeding and tooth discoloration: a systematic review. Int. J. Dent. Hyg. 13, 83–92 [DOI] [PubMed] [Google Scholar]
- 96.Lewis S.R., Butler A.R., Evans D.J., Alderson P. and Smith A.F. (2016) Chlorhexidine bathing of the critically ill for the prevention of hospital‐acquired infection. The Cochrane Library [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wand M.E., Bock L.J., Bonney L.C. and Sutton J.M. (2016) Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob. Agents Chemother. 60, e01162–e01216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bock L., Wand M. and Sutton J. (2016) Varying activity of chlorhexidine-based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J. Hosp. Infect. 93, 42–48 [DOI] [PubMed] [Google Scholar]
- 99.Saleem H.G.M., Seers C.A., Sabri A.N. and Reynolds E.C. (2016) Dental plaque bacteria with reduced susceptibility to chlorhexidine are multidrug resistant. BMC Microbiol. 16, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brooks S.E., Walczak M.A., Hameed R. and Coonan P. (2002) Chlorhexidine resistance in antibiotic-resistant bacteria isolated from the surfaces of dispensers of soap containing chlorhexidine. Infect. Cont. Hosp. Epidemiol. 23, 692–695 [DOI] [PubMed] [Google Scholar]
- 101.Bhardwaj P., Ziegler E. and Palmer K.L. (2016) Chlorhexidine induces VanA-type vancomycin resistance genes in enterococci. Antimicrob. Agents Chemother. 60, 2209–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]