Abstract
Genome editing has proven to be highly potent in the generation of functional gene knockouts in dividing cells. In the CNS however, efficient technologies to repair sequences are yet to materialize. Reprogramming on the mRNA level is an attractive alternative as it provides means to perform in situ editing of coding sequences without nuclease dependency. Furthermore, de novo sequences can be inserted without the requirement of homologous recombination. Such reprogramming would enable efficient editing in quiescent cells (e.g., neurons) with an attractive safety profile for translational therapies. In this study, we applied a novel molecular-barcoded screening assay to investigate RNA trans-splicing in mammalian neurons. Through three alternative screening systems in cell culture and in vivo, we demonstrate that factors determining trans-splicing are reproducible regardless of the screening system. With this screening, we have located the most permissive trans-splicing sequences targeting an intron in the Synapsin I gene. Using viral vectors, we were able to splice full-length fluorophores into the mRNA while retaining very low off-target expression. Furthermore, this approach also showed evidence of functionality in the mouse striatum. However, in its current form, the trans-splicing events are stochastic and the overall activity lower than would be required for therapies targeting loss-of-function mutations. Nevertheless, the herein described barcode-based screening assay provides a unique possibility to screen and map large libraries in single animals or cell assays with very high precision.
Keywords: trans-splicing, barcoding, viral vectors, plasmid library
INTRODUCTION
By delivering DNA/RNA via viral vectors it is possible to repair or disrupt disease causing genes and also to introduce novel genes (Puttaraju et al. 1999; Mansfield et al. 2000; Jinek et al. 2012; Naldini 2015). Through synthetic modifications of the viruses’ binding properties or by using cell-specific promoters to regulate the transgene expression, it is conceivable to target specific cell types in desired areas and have a defined expression of the delivered gene (Weeratna et al. 2001; Boulaire et al. 2009; Deverman et al. 2016). However, to replace a gene, the viral vector must deliver a complete gene together with the entire expression machinery (promoter, poly-adenylation sequence, etc.). Considering the restricted loading capacity in viral vectors, usually 4–8 kb depending of the viral strain (Kantor et al. 2014), there is a limitation to which genes can be replaced or which promoters can be utilized. Therefore, correction of genes in situ is an attractive approach and may allow for the delivery of shorter coding sequences in the viral vectors, thereby increasing the pool of possible target genes significantly. One possible approach for in situ gene correction is spliceosome mediated trans-splicing (Puttaraju et al. 2001; Tahara et al. 2004; Nakayama et al. 2005; Berger et al. 2015).
Trans-splicing has many properties, making its application in viral vectors very interesting and with great potential for both experimental research and clinical therapy. In trans-splicing, two physically separate pre-mRNAs are joined together to form a mature mRNA (Garcia-Blanco 2003). The trans-splicing normally occurs within an intron, and the end product becomes a combination of the upstream exons from one pre-mRNA and the downstream exons from the other (Fig. 1A).
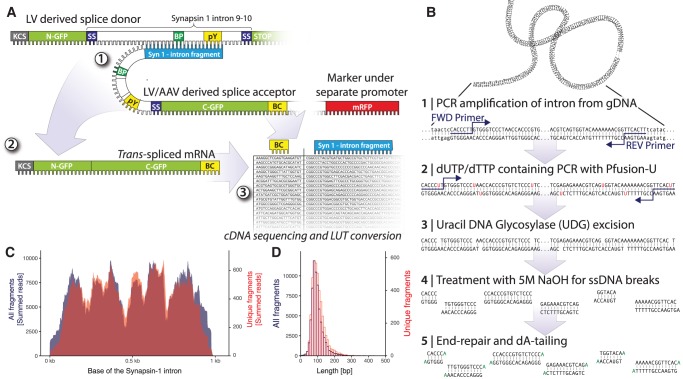
FIGURE 1.
Generation and validation of the splice acceptor plasmid library. (A) Schematics showing the split-GFP trans-splicing screening approach. LV derived splice donor expressing N-terminal GFP and full Synapsin I intron 9–10 and splice acceptor expressing intron fragment, C-terminal GFP and DNA barcode (BC). The trans-spliced mRNA is a hybrid between donor and acceptor and the result is the full open reading frame of GFP. By deep sequencing of molecular barcodes and intron fragments from the plasmid library prior to the cell culture or in vivo screening assay, a look-up table can be created to create a link between the two. (B) A schematic overview of the process used for the preparation of intron fragments for library cloning by using dUTP-based fragmentation via Uracil DNA Glycosylase and NaOH followed by end-repair and A-tailing. (C) Plot of the overall coverage of the Synapsin I intron with the negative (active) strand in red and positive (inactive) strand orientation in blue. (D) Length distribution of all inserted intron fragments. (See Supplemental Fig. S1 for additional information.)
Trans-splicing is a conserved event that has been reported in many different species, from nematodes to rodents and humans (Caudevilla et al. 1998; Flouriot et al. 2002; Fischer et al. 2008). There are three known variants of trans-splicing: 3′ trans-splicing, 5′ trans-splicing, and internal exon replacement. In 3′ trans-splicing, the splice-acceptor is provided in the exogenous RNA, and all exons downstream from the targeted intron are replaced. Whereas in 5′ trans-splicing, a de novo upstream exon is introduced into the endogenous RNA and the splice-donor site is provided in the exogenous transcript. Those two strategies can be merged to allow for the replacement of a single, internal exon but then require two trans-splicing events to occur (Mansfield et al. 2004; Koller et al. 2011). In this paper, we have focused only on the 3′–5′ trans-splicing reaction as this has been found to be most efficient (Yang and Walsh 2005).
When trans-splicing is utilized in a gene therapy setting, only the mRNA being expressed at a certain time and location can be subjected to trans-splicing. This means that if the DNA containing the trans-splicing acceptor sequence is delivered to a cell not expressing the target pre-mRNA, the delivered gene would have a negligible effect. Through trans-splicing, the expression level from the delivered DNA can thus be linked to the level of the endogenous target pre-mRNA, allowing cell-type selectivity in the absence of unwanted effects due to constitutive overexpression. As a therapeutic alternative, trans-splicing has the advantage over other editing techniques (e.g., CRISPR/Cas9) in that it is nuclease independent which provides a more attractive safety profile for long-term expression. Several seminal studies have been performed on trans-splicing and the method has been used in a wide variety of preclinical proof-of-concept studies. The therapeutic applications range from delivery of toxins within cancer cells to correcting hereditary disorders such as Huntington's disease, tauopathies, and immunodeficiencies (Tahara et al. 2004; Nakayama et al. 2005; Rodriguez-Martin et al. 2005; Rindt et al. 2012).
Since viral vector-mediated trans-splicing is a fairly unexplored technique, there are still many unknown factors and parameters that can potentially be improved to increase the efficacy of the method. For example, a thorough investigation into how the binding domain (the part of the construct providing pre-mRNA selectivity) should be designed to give the highest trans-splicing efficacy (trans-splicing compared to cis-splicing) has yet to be performed. While several factors exert an effect on the splicing efficacy (spacer, vector design, etc.), the least studied factor is how well the binding domain can hybridize with the endogenous intron to induce trans-splicing (Mansfield et al. 2004). In this study, we have characterized this component using a screening assay based on DNA barcoding.
DNA barcoding is a novel and extremely powerful technique to individually label sequences, viruses or various mutants of a protein and allows for assessment in parallel within the same assay, in cell culture or in vivo. A DNA barcode is a variable stretch of nucleotides (normally 4–20 bp) that can be used in a subsequent sequencing to determine the origin of the DNA or as a unique identifier of a longer, more complex sequence expressed elsewhere in the viral vector genome (Parameswaran et al. 2007; Patwardhan et al. 2009; Chen et al. 2012; Adachi et al. 2014; Davidsson et al. 2016). Barcodes can be extremely useful and even crucial in studies where the functional unit is not present following mRNA processing. Examples include promoter activity (Patwardhan et al. 2009), viral capsid functions (Adachi et al. 2014) and, as utilized here, for the assessment of trans-splicing.
In this study, we have developed a novel and unbiased screening assay for trans-splicing efficacy based on deep sequencing of molecular barcodes. By labeling each individual vector with a unique barcode we have been able to map trans-splicing efficacy both in cell culture and in vivo. We have validated these findings both in cell culture and in vivo in the mouse brain using both lentiviral (LV) and adeno-associated viral (AAV) vectors. This novel library generation and screening assay can be utilized to study a wide variety of biological functions stretching far beyond RNA reprogramming in cells or in the brain.
RESULTS
In order to investigate RNA reprogramming efficacy and the factors governing this process, we first established a synthetic in situ modeling and reporter system. The first version of this system was targeted toward monitoring on-target events and efficacy. It was thus built on a split-GFP approach. Centered over base 274 of the eGFP sequence, the sequence AG|GC is suitable for the insertion of a U2 type intron (spliced by the major spliceosome) (Sharp and Burge 1997). Through the insertion of an intron at this point, we generated two de novo exons of the eGFP protein (hereafter referred to as N-GFP and C-GFP, respectively), without affecting the fluorescent function of the protein after cis-splicing (Fig. 1A).
In the second step, we selected a suitable modeling system that would allow for studying of RNA reprogramming events exclusively in post-mitotic cells in vivo. For this purpose, the abundant synaptic protein Synapsin I was chosen as the target. Through sequence analysis of the full Synapsin I gene, intron 9–10 was selected as it fulfilled the criteria of consensus 5′ and 3′ splice site (SS) sequences, identifiable poly-pyrimidine tract and putative branch point, and was sufficiently short to be inserted into an AAV or LV genome. From DNA extracted from tail biopsies, we then isolated and amplified the genomic sequence spanning the intron 9–10 from three commonly used mouse strains, C57bl/6, NMRI and Swiss to confirm that this intron is well preserved between strains. In all three strains, the sequence was verbatim to that of the ensemble reference sequence (ENSMUSG00000037217).
Generation of splice donor and splice acceptor constructs
To allow for cis-splicing in the donor construct (the part containing N-GFP and the Synapsin I 9–10 intron), but avoid fluorescence originating from the cis-splicing construct, we truncated the C-GFP exon to only 19 amino acids. This rendered the GFP protein nonfluorescent after cis-splicing events. The N-GFP|Synapsin I 9–10 intron|truncated C-GFP sequence was inserted into an expression plasmid, driven by the CMV promoter. This construct is hereafter referred to as the splice donor.
The splice acceptor was designed within a second-generation LV expression vector where two promoters, placed in line, allow for independent expression of two reporters (Pan et al. 2008). The CMV promoter controls the splice acceptor sequence and the downstream promoter (PGK) the mRFP gene. The splice acceptor sequence was de novo synthesized based on a sequence found in Tahara et al. (2004) with slight modifications.
To allow for unbiased, random and efficient insertion of binding domains 5′ of the splice acceptor region, we utilized the zero-background cloning technique described previously (Müller et al. 2005; Davidsson et al. 2016), where the ccdB toxin gene is flanked by cloning sites for fragment insertion.
Fragmentation of the Synapsin I intron, and generation of the screening library
To date, little is known about the requirements with regards to length and placement of the binding domain for efficient trans-splicing. Therefore, we used an approach of DNA fragmentation, ligation, and barcoding, which we have described elsewhere in detail (Davidsson et al. 2016). Briefly, this process allows for random fragmentation of a defined genetic sequence (here the Synapsin I intron 9–10) into varying lengths, and insertion into the splice acceptor LV vector in both directions with equal probability (Fig. 1A,C,D; Supplemental Fig. S1).
After successful trans-splicing with the splice donor construct, the binding domain and the splice acceptor region are spliced, and the mature mRNA contains only the full GFP sequence. To be able to monitor and map the RNA editing efficacy in situ, we therefore had to implement a molecular barcoding approach, inserting a degenerate 20-nucleotide (nt) sequence in the 3′ UTR of the C-GFP sequence of the splice acceptor construct (Fig. 1A, also described in detail in Davidsson et al. 2016). After the generation of a lookup table, (generated through NGS sequencing of the splice acceptor plasmid linking binding domain sequence and the barcode), the readout of barcodes from mRNA can be used to map the trans-splicing efficacy back to each binding domain. This cloning resulted in a highly diverse library of 34,640 bacterial colonies, which were pooled and grown together to form the plasmid library for LV production.
Before LV production, the plasmid library was sequenced using the PCR-free consensus circular sequencing (CCS) approach on the Pacific Biosciences RS II sequencer. This approach allows for sequencing of the entire region of the LV spanning the binding domain, splice acceptor, C-GFP, and barcode from a single molecule. The circular reading oversamples the reads 4–5 times at each base, improving the read accuracy significantly. The resulting high-quality sequences were used to generate a lookup table linking the unique barcodes to their respective intron fragment sequence (the binding domain). Using our previously described analysis workflow (Davidsson et al. 2016), we then analyzed the Synapsin I intron 9–10 splice acceptor library in depth (complete and executable workflow available as a Docker image on Docker Hub as Bjorklund/RNA-edit, and the raw sequencing are available as SRA [PRJNA403798]). (See Supplemental Fig. S1 for additional details.)
In trans-splicing, only binding domains with a sequence inserted in the reverse orientation to the endogenous intron are expected to work, since only they have the capacity to make RNA/RNA Watson–Crick base pairing. However, binding domains in the forward orientation serve as an optimal internal control. Using our unbiased cloning technique (Davidsson et al. 2016), regardless of orientation, the fragments were inserted into the plasmid with equal efficacy (Supplemental Fig. S1). The fragments spanned the entire intronic sequence (Fig. 1C) with a mean length of each binding domain of around 150 bp (Fig. 1D).
Clonal mapping of trans-splicing efficacy
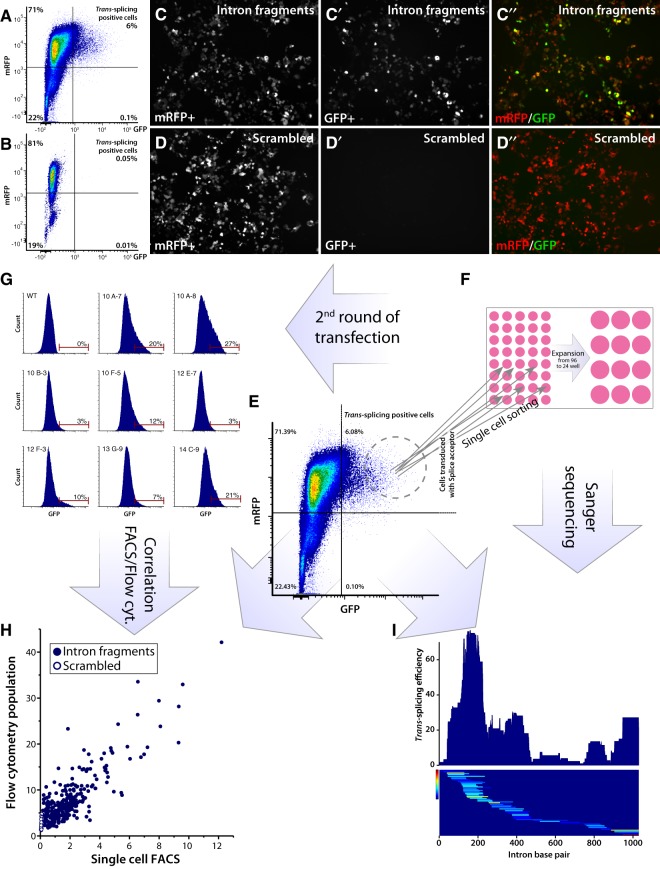
The LV splice acceptor library was used to make a low multiplicity of infection (MOI) transduction of HEK293T cells resulting in less than 10% of mRFP+ cells to maximize the chances of a single integration event per cell. The stably transduced cells were then enriched by fluorescence-activated cell sorting (FACS) based on mRFP expression and expanded (Fig. 2A). As a negative control, a similar cell line was generated using a splice acceptor containing a scrambled binding domain with a very low sequence homology with the Synapsin I intron (Fig. 2B). When transiently transfected with the N-GFP splice donor plasmid (containing the full Synapsin I intron 9–10), the cells containing the active splice acceptor library displayed a subset of cells with strong GFP fluorescence (Fig. 2C′,C″,A), which was completely absent in the cell line containing the scrambled binding domain (Fig. 2D′,D″,B).
FIGURE 2.
trans-splicing screening assay in cell culture based of split-GFP fluorescence. (A,B) FACS plot showing trans-splicing positive cells expressing GFP and mRFP of cells from C (A) and from D (B). (C–D″) Confocal image of stable cell lines expressing splice acceptor either from the LV intron fragment library (C–C″) or the negative control containing the scrambled sequence (D–D″) (both expressing C-GFP), transfected with splice donor (expressing N-GFP). Successful trans-splicing is indicated by GFP expression and both constructs constitutively express mRFP for assessment of transduction efficacy and FACS enrichment of transduced cells. (E–I) Flowchart of the single cell assessment assay of trans-splicing efficacy based on fluorescence intensity. (E) FACS plot showing the distribution of GFP+ cells used for single cell sorting. Cells were sorted and analyzed in FACS based on GFP/mRFP double fluorescence. (F) Expansion of single sorted cells from 96-well to 24-well. (G) Single sorted cells were after expansion subjected to a second round of transfection with splice donor, and GFP fluorescence was analyzed in a flow cytometer. (H) Correlation between first (FACS) and second (flow cytometer) round of transfection for each single sorted and expanded cell. Fluorescence was quantified using the MESF standard beads. Dots with full circle are cells with splice acceptors containing intron fragments, and dots with white center are cells expressing a splice acceptor containing only the scrambled sequence. (I) DNA from cells in F was extracted and PCR amplified and sent for Sanger sequencing. The sequenced intron fragment was then mapped to trans-splicing efficacy based on the fluorescence data in H. Top part shows trans-splicing efficacy and bottom part shows each fragment's position in the Synapsin I intron, color-coded based on the achieved fluorescence intensity.
In order to assess which of the binding domains resulted in the most efficient trans-splicing, we subsequently performed single cell FACS based on GFP fluorescence (Fig. 2E). Each cell was sorted into a unique well of a 96-well plate and expanded into populations of around 200,000 cells (Fig. 2F). These monoclonal populations (containing the same binding domain in all cells of the clone) were then split into two fractions; the first was re-transfected with the splice donor plasmid and re-analyzed by flow cytometry for GFP fluorescence (Fig. 2G); and the second subjected to PCR for amplification of the binding domain, followed by parallel Sanger sequencing of amplicons. The re-assessment with flow cytometry showed excellent test-retest correlation between the single cell fluorescence data and the expanded population (Fig. 2H), confirming the correct sorting and monoclonal expansion.
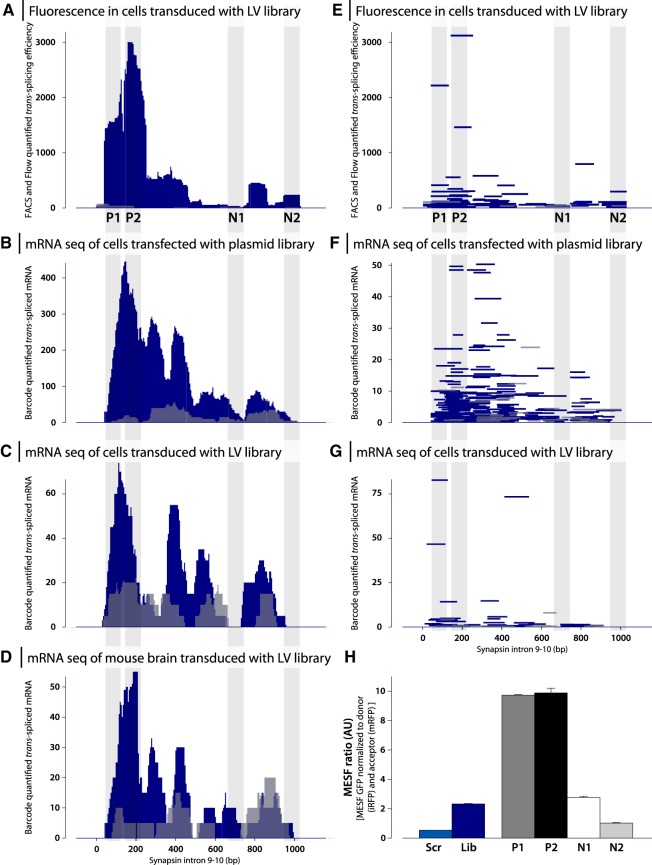
The Sanger sequencing recovered binding domains from 171 clones, of which 163 contained a single integration event representing 95 unique fragments. Of these, 89 were found in the reverse orientation as expected, and only 6 fragments in forward orientation. Using R-based bioinformatics workflow (also available in the Bjorklund/RNA_edit Docker image), we then re-aligned the recovered fragments with the Synapsin I intron 9–10, and included the recorded trans-splicing efficacy as assessed through the fluorescence at the clonal level (Fig. 2I). This analysis revealed a very distinct pattern of trans-splicing efficacy, with most of the efficient events occurring in the 5′ end of the intron, and very few events occurring in the 3′ half of the intron. The region containing the branch point, the poly(Y) sequence (the region previously targeted for trans-splicing) at the end of the intron was also permissive for trans-splicing, but to a lesser extent than the 5′ region. As a validation step, to ensure that this distribution pattern was not due to a bias in the fragment library, we then normalized the trans-splicing efficacy to the relative abundance of each fragment in the library based on the PacBio CCS sequencing (Fig. 3A,E). In this figure, the fragments’ orientation is visualized as well.
FIGURE 3.
Results from three trans-splicing screening assays based both in cell culture and in vivo. (A–D) Trans-splicing efficacy over Synapsin I intron 9–10 plotted as trans-splicing efficacy for each base in the intron. (A) The collected results from Figure 2, i.e., the screening based on fluorescence in HEK293T cells transduced with LV-intron fragment library and transfected with splice donor. Data now normalized based on the relative distribution of each fragment in the complete library to allow for comparison to the mRNA base screening assays below. (B–D) Results from screening based on mRNA sequencing of barcodes. Efficacy calculated from fragments in reverse orientation are shown in blue, and fragments in forward orientation are shown in gray. Fragments selected for further validations P1, P2, N1, and N2 are shown by vertical gray lines. (B) Screening in HEK293T cells transfected with intron fragment library and splice donor. (C) Screening in HEK293T cells were transduced with intron fragment library and transfected with splice donor. (D) Screening in C57BL/6 mice injected in striatum with LV-intron fragment library. (E–G) Trans-splicing efficacy over Synapsin I intron 9–10 plotted as individual fragments. Plots E–G correspond to A–C. Recovered fragments in reverse orientation are again shown in blue and recovered fragments in forward orientation are shown in gray. (H) Validation of trans-splicing efficacy for selected fragments P1, P2, N1, and N2 as well as intron fragment library and scrambled sequence. HEK293T cells were transfected with splice acceptor and splice donor, and trans-splicing efficacy (GFP expression) was assessed by flow cytometry. GFP expression was normalized to both iRFP (splice donor) and mRFP (splice acceptor).
Utilizing RNA barcoding to map RNA reprogramming in situ
To enable a higher resolution mapping of the trans-splicing efficacy, and to select the best candidate binding domains, we then utilized the presence of the molecular barcode located in the 3′ UTR of the C-GFP in the splice acceptor plasmid (see Fig. 1A). We first performed a double transient transfection of HEK293T cells with both the splice donor plasmid and the fragment containing the splice acceptor plasmid library, as this retains a higher diversity of binding domains than viral transduction. Forty-eight hours post-transfection, we extracted RNA from the cells, and performed targeted NGS sequencing of spliced mRNA (containing both N-GFP, and the barcode containing 3′UTR). The extracted barcodes were then translated back to the binding domain sequence using the lookup table, and their relative quantities (copies of mRNA) were plotted along the Synapsin I intron 9–10. Strikingly, the efficacy pattern of the transient transfection, and barcode readout were very similar to the overall patterns displayed from the single cell sorted stable clones in Figure 2 (Fig. 3B,F). Again, most of the fragments recovered were oriented in the reverse orientation, with only scattered events occurring in the forward orientation (Fig. 3B,F) with very low mRNA quantities.
To investigate trans-splicing mediated reprogramming in a living mammal, the splice acceptor library was moved into a lentiviral (LV) transduction system. Transduction was first tested in HEK293T cells which resulted in a similar trans-splicing efficiency plot, albeit with fewer unique fragments as compared to transfection (Fig. 3C,G). The LV library was then injected into the striatum of wt C57bl/6 mice, and RNA extracted from the striatal tissue 4 wk later. Targeted NGS sequencing was performed using a forward primer targeting the endogenous upstream exon of Synapsin I to recover trans-spliced reprogrammed products. While the in vivo approach recovered the fewest RNA reprogramming events, the efficiency of sequences that drive reprogramming was strikingly similar to that observed in transiently transfected and stably transduced tissue culture cells (Fig. 3D).
In order to assess single binding domains, and compare efficacy, we then selected two sequences that mediated the highest trans-splicing activity; one from the single cell sorting approach (P1), and one from the barcoding approach (P2). As a negative control, we designed one similarly sized fragment spanning the region of least effective trans-splicing (N1). For comparison with earlier studies targeting the 3′ region of intron, we generated one fragment covering that part of the intron (N2). Applying the above described transient transfection assay in HEK293T cells with a stably inserted splice donor, we compared these fragments individually with our complete library, and the scrambled sequence using flow cytometry. Somewhat surprisingly, the fragment N2 performed equally inefficiently as the scrambled sequence, while fragment N1 performed on par with the total library (Fig. 3H). In contrast, the positively selected fragments P1 and P2 performed with much higher efficacy, confirming the predictive nature of both the single cell sorting approach, and the barcoding-based approach.
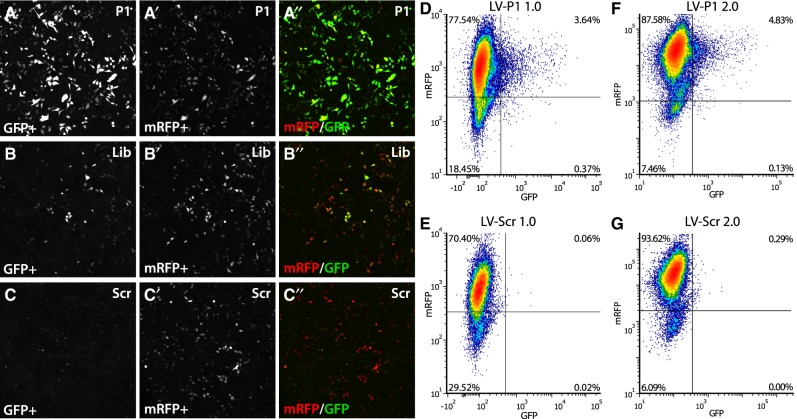
Validation of trans-splicing using lentiviral vectors reveals aberrant cis-splicing events
With the positively identified fragments P1 and P2, we then generated lentiviral splice acceptor vectors, and transduced HEK293T cells stably expressing the splice donor. Both P1 and P2 fragments displayed a significantly lower GFP fluorescence in transduced cells compared to that in transfected cells. In fact, the pattern of expression was also strikingly different, with very few scattered cells being GFP+. However, those GFP+ cells had reasonably high fluorescence (Fig. 4A,D). GFP fluorescence was negligible in cells transduced with the scrambled sequence (Fig. 4C,E). In order to assess the origin of this dramatic reduction, we set out to sequence the inserted LV derived splice acceptor sequence from the DNA of the stably transduced cell line from the left LTR and into the C-GFP. When running the amplicons on a gel, we observed both the correctly sized amplicon, and a significantly smaller band both in the positively selected fragments, and in the complete library (Supplemental Fig. S2). We found that the very strong 3′ splice sequence in the splice acceptor modified from Tahara et al. (2004) had recruited an earlier silent putative 5′ splice site in the LV vector 1564 bp from the 5′LTR, and that the sequence between them (including the binding domain) had been spliced out during the LV production. To circumvent this aberrant splicing, we generated a novel LV backbone, named 2.0 (see Supplemental Fig. S2). Using this novel LV vector, we again transduced the HEK293T cells with the stably integrated splice donor. However, the removal of the aberrant cis-splicing of the acceptor was not sufficient to improve the trans-splicing efficacy significantly, and the GFP expression was still restricted to a very small proportion, of 4.8%, of all cells (Fig. 4F,G).
FIGURE 4.
Validation of trans-splicing in cell culture and removal of aberrant cis-splicing in lentiviral vectors. (A–C) Validation of trans-splicing efficacy for the fragment P1, selected in the screening assay, compared to library and scrambled sequence. HEK293T cells stably expressing splice donor were transfected with P1 (A–A″), library (B–B″), and scrambled sequence (C-C″), respectively, and analyzed 48 h post-transfection by confocal microscopy. A–C shows trans-spliced GFP, A′–C″ shows the transfection control mRFP, and A″–C″ shows the pseudo colored overlay image. (D,E) Validation of LV-P1 (D) compared to LV-Scr (E) in HEK293T cells. Cells were transduced with LV, enriched by FACS and expanded, and then transfected with splice donor. Analysis was done using flow cytometry plotting trans-spliced GFP fluorescence against the transduction control mRFP. (F,G) Validation of LV-2.0 vectors after PCR confirmation. LV-P1 2.0 and LV-Scr 2.0 were used to transduce HEK293T cells. After enrichment and expansion, cells were transfected with splice donor and analyzed by flow cytometry. No improvement on trans-splicing efficacy was observed.
Development of a bidirectional LV vector for intron expression
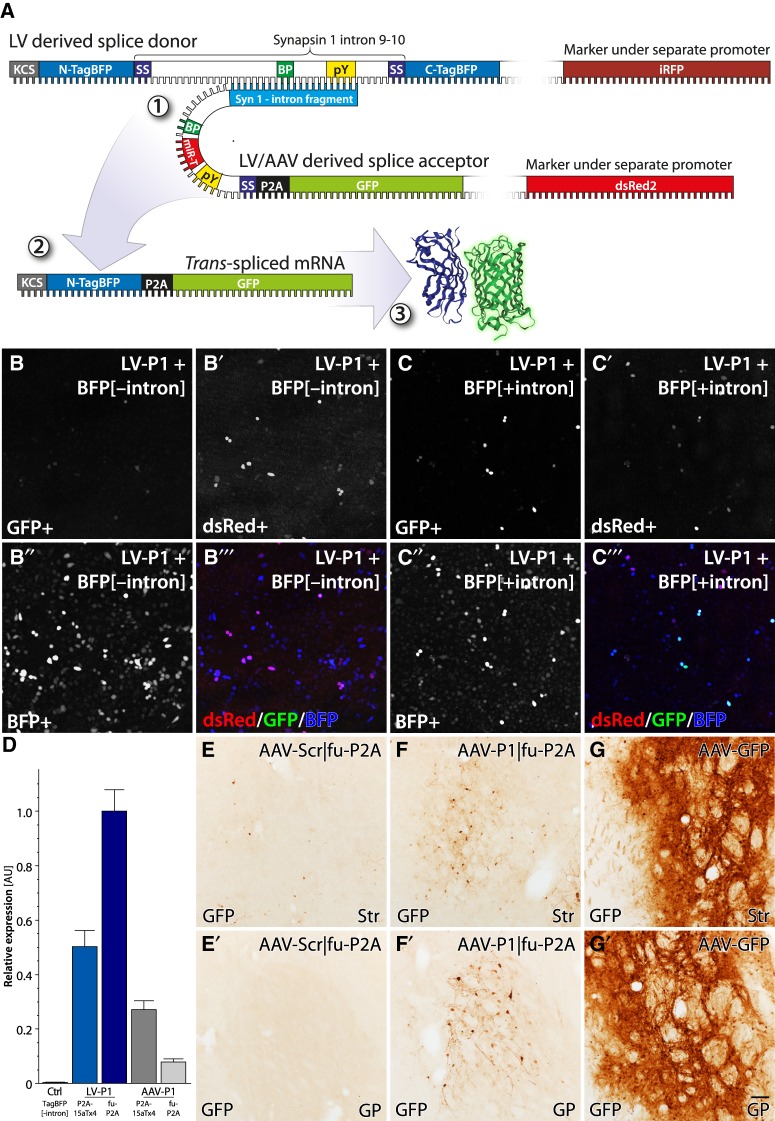
To further dissect the reasons for the stochastic and inefficient trans-splicing events after stable integration, we created a bidirectional LV vector that allow for the expression of intron containing genes, and retention of the introns through the LV production cycle (see Supplemental Fig. S3 for details). To ensure accurate quantification and normalization for integration-related modulation, we also designed the construct to express an independent fluorophore in the cis (forward) direction. In the new splice donor design, we expressed iRFP in cis under the PGK promoter and TagBFP containing the Synapsin I intron 9–10 in trans (reverse direction) under the CMV promoter (Fig. 5A). The bidirectional approach also enabled us to express a full-length GFP in the splice acceptor which still contains a marker gene (dsRed2) to select and normalize for transduction efficacy and silencing of integrated LV genomes. In this new approach, we also included a ribosome skipping sequence to allow for separation of the N-terminal TagBFP from the GFP after trans-splicing. (See Supplemental Fig. S3 for details.)
FIGURE 5.
Validation of a splice acceptors expressing full-length GFP trans-splicing in vivo in the mouse brain. (A) Schematic of the improved approach to the trans-splicing event between TagBFP[+intron] as splice donor and LV/AAV with full-length GFP as splice acceptor (1). After trans-splicing, this again forms a mature mRNA (2), but in this version, the full GFP is inserted in the reading frame of the N-TagBFP. Through the insertion of the P2A ribosome skipping sequence, the N-TagBFP is split from the GFP at the ribosomal translation into protein (3). (B,C) Transfection of stable cell lines expressing TagBFP[−intron] (B) and TagBFP[+intron] (C). Cell lines were transfected with LV-P1-fuP2A and analyzed using confocal microscopy. Some scattered cells in TagBFP[+intron] were positive for GFP meaning successful trans-splicing (C). dsRed was used as a transfection control (B′–C″). (D) Quantification of LV-P1 and AAV-P1 in HEK293 cells. Cells were transduced with either LV or AAV and trans-splicing efficacy was assessed by RT-qPCR with forward primer targeting TagBFP and reverse primer targeting GFP. Control sample was AAV transduction of cells expressing TagBFP[−intron]. (E–G). Trans-splicing in vivo. WT mice were injected with scrambled (Scr) acceptor vector with a furin-P2A (fu-P2A) cleavage site AAV-Scr|fu-P2A (E), an active construct AAV-P1|fu-P2A (F), or AAV-GFP (G) in striatum. Sections were stained for GFP using immunohistochemistry developed into a brown precipitation staining using the DAB-peroxidase reaction. The figure shows representative images from striatum (Str) (E–G) and globus pallidus (GP) (E′–G′). Scale bar in G′ represents 50 µm in E–G′.
Comparison of splice acceptors in LV and AAV vectors
With the bidirectional splice donor constructs we could now easily generate stable reporter cell lines through LV transduction. FACS sorting was conducted on both iRFP and TagBFP to ensure expression of both transgenes in the reporter cell line. We generated stable HeLa cell lines, either including or excluding the Synapsin I intron in the TagBFP gene. We avoided HEK293T cells in this step, as reports have shown them to be of a peripheral neuronal origin with low, but detectable levels of Synapsin I expression (Wang et al. 2009).
Once expanded, the reporter cells were transduced with the bidirectional splice acceptor with full GFP under the control of binding domains P1 or P2. When expressing a full-length, functional protein in the splice acceptor construct, off-target expression, i.e., when the acceptor (GFP) is expressed without being spliced into the donor (TagBFP), is of concern. For assessment of off-target expression, we evaluated the full-length splice acceptors in combination with the splice donor without the Synapsin I intron. The active fragments P1 and P2 in the absence of the Synapsin I intron displayed very low off-target expression in the screening system (Fig. 5B). (See also Supplemental Fig. S3 for the implemented secondary defense against leakiness.)
Similar to the observations above however, the GFP expression observed in the Synapsin I intron containing cells was stochastic. Some cells expressed high GFP fluorescence, significantly higher than that in the absence of the intron, but this was a very small fraction of the transduced cells (Fig. 5C). To explore if the screening system had intrinsic limitations, which resulted in false-negative results, we performed in vivo stereotactic injections of the vectors in the striatum of wild type mice. Four weeks post LV injection, the striatum was dissected out and total RNA extracted. Multiple rounds of RT-PCR failed to detect any trans-splicing, despite the fact that the RNA from the splice acceptor library was readily identified.
The consistent result from all experiments to this point is that nonintegrating splice acceptors from plasmid transfection result in robust trans-splicing events, while transduction of LV vectors shows dramatically lower efficacy. To mitigate this, we moved the splice acceptor constructs from the lentiviral backbone to a self-complementary, double-stranded adeno-associated viral (scAAV) vector backbone. Infection of replication-deficient recombinant AAV results in the formation of circular episomal plasmids, and thus this should be considerably similar to the plasmid transfection than to the LV transduction. To assess the relative efficacy of trans-splicing, we then performed a qPCR assay on HeLa cells stably expressing the TagBFP[+intron], which were transduced by the LV or scAAV P1 constructs (Fig. 5D). With this assay, we confirmed that the trans-splicing efficacy was not improved by the switch to the scAAV virus. On the contrary, this was reduced compared to the LV, as expected based on the relative transduction efficacy of immortalized cell lines, where the integrating LV is known to be superior. In vivo however, the scAAV is expected to provide significant increases in transduction efficacy over the LV. Therefore, we performed the transduction of the mouse striatum again, but now using either the scAAV-P1, or scAAV-Scr splice acceptor vectors expressing the full-length GFP. Similar to the LV in vivo transduction experiment, we failed to detect any trans-spliced mRNAs from the Synapsin I gene using PCR but significant amounts of nonspliced splice acceptor mRNA were detected. Finally, we performed immunohistochemical detection of the GFP proteins in striatal sections from remaining mouse brains injected with AAV vectors. While there were more GFP positive neurons in the striatum (Str) and Globus Pallidus (GP) in the animals receiving the active AAV-P1 construct (Fig. 5F-F′), compared to the scrambled control (Fig. 5E-E′), the expression was again stochastic and scattered. Furthermore, the total expression levels were significantly lower than what is achieved with a constitutive scAAV-GFP vector injected at the same titer (Fig. 5G-G′).
DISCUSSION
We have developed and validated an unbiased screening assay for the optimization of trans-splicing efficacy both in cell culture and in vivo. Generating a diverse and functional DNA library is a prerequisite for this type of study, and with novel cloning techniques, we have generated libraries with enough diversity to study a variety of biological functions. The use of molecular barcodes is a novel and highly efficient way of studying biological events, and they are crucial when studying events that precede expression of mRNA (as in trans-splicing but also when studying promoter activity). Using two types of screening assays, one fluorescence-based, and one sequencing-based, we were able to screen an entire intron, and map efficacy per base, as well as to identify specific fragments suitable for trans-splicing.
Our data set is not large and diverse enough to infer which acceptor sequence features promote efficient trans-splicing (i.e., optimal length, sequence composition, secondary structure, base-pairing stability, etc.). However, as observed with RNA-seq data, aggregation of data over time between genes, studies, and laboratories is very plausible using this approach. Thus, over time, such studies may be possible based on the provided analysis pipeline and the publicly available raw data.
Validations of selected fragments, recovered from this initial screening, showed reproducibility in transfected stable cell lines, but failed to show the expected efficacy following LV- or AAV-mediated transfer in cell culture and in vivo. Even though there are GFP expressing cells in the striatum and globus pallidus with the active trans-splicing constructs, the expression is far from constitutive levels.
An interesting observation throughout experiments performed using viral transduction was that although the number of cells that were positive for trans-splicing was low, the level of GFP was high, almost comparable to a constitutive GFP expression. Our findings are supported by several other studies (Tahara et al. 2004; Liu et al. 2005; Rindt et al. 2012; Shababi and Lorson 2012; Murauer et al. 2013). However, we believe that we in this study have excluded the possibility of silencing due to integration of the LV vector. We base this on two observations in the study. The first is that, through the use of a four-fluorophore approach we have been able to monitor the expression of an independent marker in both the donor and the splice acceptor (Supplemental Fig. S3). With this approach, we observe excellent correlation between the gene inserted in cis and in trans in the same vector as well as between the two control genes in the two vectors if delivered simultaneously. However, this correlation is not observed when quantifying the trans-splicing events (Fig. 4). This is regardless of the placement and orientation of the control gene in relation to the splice acceptor and choice of promoters. The second reason why we believe that insertional effects cannot solely explain the stochastic expression is that we observe the same effect using episomal scAAV vectors delivering the splice acceptor.
This indicated that there may be unknown differences between the cells governing the trans-splicing efficacy where some cells are permissive, and others are not. This appears to be true both in cell culture and in vivo. What factors govern these differences are still unknown, but we have explored a number of potential candidates.
With the exception of microRNA biogenesis and RNA degradation, all double-stranded RNA is usually of viral origin and therefore it is possible that the trans-splicing is activating the anti-viral protein kinase R (PKR) pathway (Garcia et al. 2007). To assess whether this pathway does interfere with the trans-splicing, we have evaluated the addition of the two noncoding virus-associated RNAs from the adenovirus, which uses these sequences as a decoy for the PKR activation (O'Malley et al. 1986). However, this had no detectable positive effect. A second possibility is that the splice acceptor is not sufficiently maintained inside the nucleus for it to hybridize with the target intronic sequence. To assess this, we utilized multiple 3′ UTR sequences; the WPRE and pA sequences presented here and the U1 snRNA terminator and 3′ box (Shechner et al. 2015). However, none of them conferred any significant advantage over the other. A third possibility is that the 5′CAP formed on the splice acceptor by the Pol II promoter transcription is interfering with the RNA-RNA hybridization and may in itself also promote too rapid export from the nucleus. In the development of Cas9 mediated genome editing it was demonstrated that the 5′Cap is interfering with the synthetic guide RNA (sgRNA) function when it is expressed from a Pol II promoter. One potent way to solve this was to add a hammer head (HH) ribozyme sequence upstream of the sgRNA (Nissim et al. 2014). This ribozyme self-cleaves and leaves a “naked” 5′ end of the RNA, similar to that produced by a Pol III promoter (Gao and Zhao 2014). We have also generated constructs containing a HH sequence immediately upstream of the binding domain, but this again failed to improve the trans-splicing. Finally, it is possible that the RNA–RNA hybridization is not strong or selective enough to induce the splicing event. To evaluate this, we rebuilt the binding domain of the splice acceptor into an sgRNA for the spCas9 protein and supplied this in conjunction with the splice acceptor. The rationale for this is that it has been observed that numerous Cas9 proteins can selectively bind to mRNA as well as to RNA but not cleave the RNA (Price et al. 2015; Nelles et al. 2016). Again, this modification had no positive effect on the trans-splicing efficacy.
Despite the observed low level of trans-splicing efficacy, most likely below what is needed for therapeutic interventions in the CNS, there might be other cell-types and disease conditions, where the method could prove to be useful. This will depend on the levels of trans-spliced mRNA that are needed as well as the cell-type being targeted. This would however need to be further validated under those specific conditions. We show here that trans-splicing can work and that the design of the binding domain does have an important influence on the trans-splicing efficacy. Thus, in permissive systems this screening assay could prove to be very valuable for the further optimization of promising trans-splicing constructs.
In the present study, we demonstrate the generation of a number of useful tools and constructs. The novel G1564A LV backbone is useful for circumventing aberrant cis-splicing events without affecting titers and infectivity. The bidirectional LV construct enables the expression of two transgenes under separate Pol II promoters and can efficiently retain noncoding RNA sequences e.g., introns or lncRNA. However, the most broadly applicable development validated in this study is the novel screening assay using molecular barcodes. Using this approach to perform cloning, in vivo screening and NGS sequencing to map function, it is possible to study a wide range of biological functions where a functionally relevant measure can be stored in RNA. The robustness of this approach has been validated through the same results achieved under three different conditions both in cell culture and in vivo and the strength of the assay was confirmed by a fluorescence assay that showed very similar results.
MATERIALS AND METHODS
Experimental design
To be able to perform an unbiased screening assay of trans-splicing efficacy, a splice donor and a splice acceptor plasmid were generated with the splice donor plasmid expressing N-GFP and full-length Synapsin I intron 9–10, and the splice acceptor plasmid expressing intron fragment, molecular barcode and C-GFP. DNA was extracted from mouse tail biopsies and Synapsin I intron 9–10 was PCR amplified. The PCR product was further amplified using dUTPs and fragmented without sequence bias (Fig. 1A). Fragmented DNA was end-repaired, dA-tailed, and ligated into dT-tailed zero-background cloning vector. PCR and Gateway cloning was used to barcode and transfer the fragments from cloning vector into LV splice acceptor plasmid. Simultaneously, a negative control splice acceptor vector was created. A scrambled sequence was generated to have no or very little hybridization possibility to Synapsin I intron 9–10. PacBio sequencing was then used to characterize the LV splice acceptor plasmid library and to create a look-up table linking each molecular barcode to the corresponding intron fragment.
In a first cell culture experiment, we screened trans-splicing efficacy based on fluorescence, where two stable cell lines were generated; one cell line with barcoded intron fragment library and one cell line with scrambled sequence. Cells were transiently transfected with splice donor and sorted individually into 96-well by FACS based on GFP expression using single cell accuracy, and the corresponding GFP expression was recorded. Cells were expanded and approximately 50% of the cells from each well were split out into new plates for a second round of transient transfection with splice donor. GFP expression from these cells was assessed by flow cytometry and then compared to the GFP expression from the first transfection (FACS). The remaining 50% of cells from each well were used for PCR followed by Sanger sequencing to obtain the sequence of intron fragment (Figs. 2, 3A,E).
In a second experiment, the mRNA-based screening assay utilizing the molecular barcodes was performed both in cell culture and in vivo. For the first screening, HEK293T cells were either transfected with splice acceptor plasmid library and splice donor mixed together or first transduced with a lentiviral vector containing the splice acceptor library and then transfected with splice donor. For the in vivo screening, C57BL/6 mice were injected in the striatum with lentivirus containing the barcoded splice acceptor library. RNA was extracted from HEK293T cells and brain tissue and molecular barcodes were sequenced and mapped back to corresponding intron fragments (PacBio sequencing) (Fig. 3B–D,F,G). Four fragments, P1, P2, N1, and N2, were identified using this approach and used individually in a transfection assay to validate trans-splicing efficacy (Fig. 3H).
Fragment P1 was further validated and compared to the complete splice acceptor library and the scrambled control through the generation of stable cell lines that were then transiently transfected with splice donor. Cells were analyzed by confocal microscopy and FACS (Fig. 4A–D).
Due to the strong splice site that was inserted to facilitate trans-splicing, an aberrant cis-splicing during LV production of LV 1.0 was observed removing the splice acceptor sequence. The discovered de novo 5′ splice site in the LV-backbone was disrupted by a mutation, G1564A, leading to LV 2.0. The 2.0 version of LV was validated in the same way as 1.0 (stable cell line transfected with splice donor), and the GFP expression was analyzed by FACS (Fig. 4F–G).
Subsequently, the constructs to be validated were moved into a bidirectional LV-vector containing a full-length GFP sequence. Several different terminators and combinations of such were validated in lentiviruses by FACS when sitting in trans. In the first assay, none of the terminators outperformed GFP without a terminator (Supplemental Fig. S3A), therefore a second validation was performed with new terminators (Supplemental Fig. S3B).
To facilitate trans-splicing efficacy, a new LV-donor was created containing TagBFP with Synapsin I intron 9–10 inserted, which was first validated in cell culture to ensure correct splicing of TagBFP, removing the inserted intron from the mature mRNA. A transfection assay was performed, followed by mRNA extraction, cDNA synthesis, and Sanger sequencing (Supplemental Fig. S3C). A second cell culture assay was then performed to validate a furin cleavage site together with a P2A ribosome skipping site or P2A followed by either one or four targets for miR15a (Fig. 5B,C; Supplemental Fig. S3D–G). Selected fragment P1 was validated using transfection of stable cell lines expressing TagBFP[+intron] and TagBFP[−intron], and cells were analyzed by confocal microscopy (Fig. 5B,C).
To validate continued functionality also in AAV and LV we performed a transduction of HEK293T cells. Cells expressing TagBFP[+intron] were transduced with AAV, and the experiment showed correct sequence and size of expressed fragment (Supplemental Fig. S3F,G). A comparison between LV and AAVs in HEK293 cells was also performed. Cells expressing TagBFP[+intron] and TagBFP[−intron] were transduced and trans-splicing efficacy was assessed by qPCR (Fig. 5D). Finally, an in vivo study of trans-splicing was conducted. AAV-P1 and AAV-Scr were injected bilaterally in striatum in C57Bl/6 mice and the mice were killed 3 wk later through trans-cardial perfusion and PFA fixation. The striatal region was then cut into coronal sections using a freezing sliding microtome, and sections were stained for GFP using immunohistochemistry (Fig. 5E–G).
Library generation
DNA was isolated from tail biopsies from mice and Synapsin I intron 9–10 was amplified by PCR. The PCR product was then further PCR amplified using Phusion U (Thermo Scientific), to incorporate dUTPs in the sequence. Intron 9–10 with dUTPs incorporated was then fragmented by using Uracil-DNA-Glycosylase (UDG) (Sigma-Aldrich) followed by NaOH (Speck et al. 2011). Size distribution and fragmentation efficacy was validated by PCR and gel electrophoresis. Fragmented DNA was end-repaired and dA-tailed using NEBNext end-repair and dA-tailing module (NEB). The zero-background cloning vector was digested with XcmI (NEB) restriction enzyme prior to the ligation (Chen et al. 2009). XcmI digestion separates the ccdB gene from the backbone and leaves T-overhangs on the backbone suitable for dA/dT ligation. Digestion products were separated by gel electrophoresis, and cleaved backbone without ccdB sequence was purified from gel. Digested vector and dA-tailed fragments were ligated using T4 DNA Ligase (NEB) in a 1:6 (vector:insert) ratio at 16°C overnight. To transfer insert from cloning vector to LV vector, Gateway cloning was used. AttB1 and barcode+AttB2 were added in one PCR reaction using a forward primer containing AttB1-site and a reverse primer containing barcode followed by AttB2-site. Fragments were inserted into LV-vector using Gateway BP Clonase II Enzyme Mix (Life Technologies).
Barcode design
Barcodes were ordered as High-Purity Salt-Free purified oligos (Eurofins Genomics) where the barcode length was 20 nt, defined as ambiguity nucleotides by using the sequence V-H-D-B (IUPAC ambiguity code) repeated five times, and flanked by static sequences containing AttB2 sequence.
Cloning
Throughout the paper several different cloning techniques were used. In brief, for generating the barcoded intron fragment library, Gateway cloning was used, for generating LV constructs with fluorescent marker proteins, restriction enzyme digestion (FastDigest, Thermo Fisher Scientific) and ligation were used, and for generating LV constructs with different terminators, Gibson Assembly (NEB) was used. To utilize the bidirectional LV vector together with the bicistronic splice acceptor vector, and to enable the studies on off-target expression, we had to expand the fluorescence color palette from two to four nonoverlapping fluorophores. Besides the eGFP (ex 488 nm|em 509 nm) and dsRed2 (ex 556 nm|em 586 nm), we added TagBFP (ex 405 nm|em 457 nm) and iRFP713 (ex 690 nm|em 713 nm). This allowed for selective identification of all four native fluorophores both in the FACS and in confocal microscopy.
PCR free sequencing using PacBio RSII
Plasmids from the library were digested by unique restriction enzyme to result in products with the length averaging around 800 bp (min 696 bp, max 1726 bp). The fragments were then end-repaired using NEBNext End Repair Kit (NEB). SMRTbell adaptor sequences (Pacific Biosciences) were ligated on to the sequences according to the supplier's protocol and sequenced using two SMRT-cells in the PacBio RSII sequencer.
Sequencing using Ion Torrent
Plasmid from the library was digested using SalI enzyme, and the digestion products were separated by gel electrophoresis, and cleaved backbone without 3′domain sequence was purified from gel. Seventy-five nanograms of the linear product containing the fragment and barcode was circularized using T4 DNA Ligase (NEB) 16°C overnight. The ligation product was treated with Lambda Exonuclease (NEB) and RecJF (NEB) for 16 h at 37°C (Balagurumoorthy et al. 2008). The remaining, circularized product was PCR amplified with primers containing Ion Torrent sites P1 & A. Concentration was determined by Bioanalyzer (Agilent) and sequenced on Ion Torrent using a 316 V2 chip (Thermo Fisher). Sequencing was conducted as 500-bp-long reads using an extended number of reagent flows in the 400 bp kit.
Illumina sequencing
cDNA was subjected to two PCR reactions to add Illumina compatible ends to the fragments. A P5/P7 Illumina adapter PCR was performed followed by a Nextera XT index PCR where each sample was labeled with a unique Nextera index. The DNA was then sequenced on Illumina MiSeq or NextSeq500.
Cell culture
HEK293T and HeLa cells were grown in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS and P/S. Cells were kept in 37°C with 5% CO2 and passaged every third day.
Transfection assay
HEK293T cells stably expressing splice acceptor (Library or Scrambled sequence) were transfected using splice donor DNA. The first round of transfection was carried out in six-well plates using 2500 ng DNA and 5.0 µL Lipofectamine 2000 (Thermo Fisher Scientific) per well. Cells were single sorted by FACS into 96-well plates and GFP expression was analyzed. Cells were expanded over a few weeks, transferred to 24-well plates and then subjected to a second round of transfection with splice donor DNA. The second round of transfection was carried out using 500 ng DNA and 1.0 µL Lipofectamine 2000 per well. Approximately 50% of the cells from each well were used for GFP expression analysis by flow cytometry, and the remaining cells were used for DNA extraction followed by PCR amplification of the intron fragment and Sanger Sequencing to obtain the sequence of the integrated intron fragment.
LV production
Lentivirus was produced using standard HEK293T PEI transfection with either second or third packaging vectors (see previous section). Forty-eight hours post-transfection, virus was harvested, filtered through a 0.45 µm filter and ultra-centrifuged at 77,000g for 90 min. LV was resuspended in PBS and stored in −80°C. Lentiviral titers were determined by qPCR on DNA extracted from transduced HEK293T cells.
AAV production
AAV8 was produced using standard PEI transfection of HEK293T cells using pDP8 (PlasmidFactory) and transfer vector. AAVs were harvested 72 h post-transfection using polyethylene glycol 8000 (PEG8000) precipitation and chloroform extraction followed by PBS exchange in Amicon Ultra-0.5 Centrifugal filters (Merck Millipore) (Wu et al. 2001). Purified AAVs were titered using qPCR with primers specific for either promoter or transgene.
Generating stable cell lines
All stable cell lines were generated using lentiviral supernatant transduction. In brief, Lentivirus was produced using standard HEK293T PEI transfection with a second generation packaging system for LVs named 1.0 and a third generation packaging system for LVs named 2.0. Forty-eight hours post-transfection, medium containing virus was collected and filtered through a 0.45 µm filter. The crude viral supernatant was then used to transduce cells in a 1:1 ratio with new medium. Polybrene (Sigma-Aldrich) was added to a final concentration of 8 µg/mL to improve transduction efficacy. One week post-transduction, transduced cells were enriched by FACS based on fluorescence (e.g., mRFP, dsRed2, iRFP, and TagBFP).
mRNA extraction and cDNA synthesis
In vivo brain samples for mRNA analysis were swiftly dissected, flash frozen and collected in Lysing Matrix D tubes (MP Bio). For all mRNA extractions, Quick-RNA MiniPrep Plus (Zymo Research) was used. In vivo samples were homogenized in lysing buffer using a FastPrep-24 (MP Biomedicals). Cell culture samples were collected in lysing buffer; both cell and tissue samples were then processed according to manufacturer's protocol. cDNA synthesis for all samples was carried out using a qScript Flex cDNA Synthesis kit (QuantaBio) with either a mix of oligo dTs and random hexamers or with a gene specific primer.
RT-qPCR
HEK293T cells with stable integration of TagBFP[+intron] or TagBFP[−intron] were transduced with LV-P1 2.0 or AAV8-P1, both containing full GFP sequences. cDNA from transduced cells were used in a qPCR assay to validate trans-splicing efficacy. Forward primer was designed to be specific for TagBFP and reverse primer for GFP. A PCR product therefore had to be a successful trans-splicing event. For normalizing between variations in transduction efficacy, primers specific for iRFP and TagBFP were used. The qPCR reaction mix was made up of 5 µL Sso Advanced (Bio-Rad), 0.25 µL forward and reverse primer, 0.5 µL Cdna, and 4 µL H2O and ran on CFX96 Real Time System (Bio-Rad).
Flow cytometry
HEK293T cells were double transfected with splice donor (N-GFP) and splice acceptor P1, P2, N1, N2, Lib, or Scr (C-GFP). In 24-well format, 500 ng DNA and 1.0 µL Lipofectamine were used for transfection, which was performed in triplicate. Seventy-two hours post-transfection, cells were analyzed in a flow cytometer (BD Accuri C6). The MESF ratio of GFP was normalized to iRFP (splice donor) and mRFP (splice acceptor). CytoCal beads (Thermo Fisher Scientific) were used for MESF normalization.
Confocal microscopy
All LSM (laser-scanning confocal microscopy) was conducted using a Leica SP8 setup where images were captured using a HyD detector and always with the lasers activated in sequential mode using solid-state lasers at wavelengths of 405, 488, 552, and 650 nm (a pinhole of 1 AU).
Animal experiments
Adult, female, wild-type C57BL/6 mice were housed in standard laboratory cages with ad libitum access to food and water, under a 12:12 h dark–light cycle in temperature-controlled rooms. All experimental procedures performed in this study were approved by the local ethics committee in the Malmo/Lund region “Malmö/Lunds regional djurförsöksetiska nämnd” in accordance with national and EU regulations.
Stereotactic injections
Mice were anesthetized using isoflurane prior to surgeries and placed in a stereotactic frame with the tooth bar individually adjusted for flat skull. Coordinates for all injections were performed in relation to bregma. The animals received small burr hole through the skull and were then infused with viral vectors into the brain using a pulled glass capillary (60–80 µm i.d. and 120–160 µm o.d.) attached to a 25 µL Hamilton syringe connected to an automated infusion pump. Lentiviral vectors were injected bilaterally into the striatum at two infusion sites with two deposits/site at the following coordinates and volumes: Rostral injection site (1.5 + 1 µL): AP = +0.8; ML = −1.8/+1.8; DV = −3.0/−2.7. Caudal injection site (1.5 + 1 µL): AP = +0.4; ML = −2.2/+2.2; DV = −3.1/−2.7. LVs were injected with a titer in the range of 4–6 × 108 TU/mL. AAVs were injected bilaterally into the striatum with two deposits/site at the following coordinates and volumes: Rostral injection site (1.25 µL): AP = +0.8; ML = −1.8/+1.8; DV = −2.8. Caudal injection site (1.25 µL): AP = +0.4; ML = −2.2/+2.2; DV = −2.9. AAVs were injected with a titer of 1.5 × 1012 gc/mL. All viral vector solutions were injected with an infusion rate of 0.4 µL/min.
Tissue preparation and immunohistochemistry
Animals were sacrificed 4 wk post-viral infusion by sodium pentobarbital overdose (Apoteksbolaget) and transcardially perfused with physiological saline solution followed by fresh, ice-cold, 4% paraformaldehyde (PFA) prepared in 0.1 M phosphate buffer (pH = 7.4). The brains were removed and post-fixed for 2 h in ice-cold PFA before storing in 25% buffered sucrose in order to ensure cryoprotection for at least 24 h until further post-mortem analysis. The brains were then cut into 35 µm thick coronal sections, using a sliding microtome (HM 450, Thermo Scientific) and stored in anti-freeze solution (0.5 M sodium phosphate buffer, 30% glycerol, and 30% ethylene glycol) at −20°C until further processing. For 3,30-di-aminobenzidine (DAB) immunohistochemical analysis of GFP expression, tissue sections were first washed (3×) with TBS (pH 7.4) and incubated for 1 h in 3% H2O2 in 0.5% TBS Triton solution in order to quench endogenous peroxidase activity. Following another washing step, the sections were blocked in 5% bovine serum and incubated for 1 h. The tissue sections were subsequently incubated with primary antibodies overnight in 2.5% bovine serum. GFP positive cells were identified using a polyclonal antibody (chicken anti-GFP, Abcam Cat# ab13970 RRID:AB_300798, 1:20000). Following overnight incubation, the primary antibody was first washed away with TBS (×3) and then incubated with secondary antibodies for 2 h using the anti-chicken (Vector Laboratories Cat# BA-9010 RRID:AB_2336114, 1:250) secondary antibody. Following incubation and washing (TBS ×3) of the tissue sections, the ABC-kit (Vectorlabs) was used to amplify the staining intensity through streptavidin–peroxidase conjugation and followed by a DAB in 0.005% H2O2 color reaction.
Data assessment workflow
A complete interaction free workflow was implemented using the R statistical package together with a number of packages from the Bioconductor repository. From these scripts, a number of broad-utility external applications (bbmap, Starcode and Bowtie2, and SAMtools) were called and output returned to R for further analysis. This is publically available as a Git repository at https://bitbucket.org/MNM-LU/rna-editing and as a self-sustained Docker image Bjorklund/RNA-edit.
In brief: Barcode and sequence identification, trimming and quality filtration, were conducted using the bbmap software package (Bushnell 2016), which allows for kmere matching of known backbone sequences against the reads. As a vast majority of barcode reads were sequenced to the length of 20 with most barcodes of a deviating length ending up being 19 bp long, for all analysis in this study, length filtration of 18 ≤ BC ≤ 22 was applied.
The genomic sequence fragments were similarly isolated using the bbmap software package, but this time without any application of length restrictions.
The key component of the R-based analysis framework is a parallelized implementation of the MapReduce programming philosophy (Dean and Ghemawat 2008; McKenna et al. 2010). For more details on this process, please refer to Davidsson et al. (2016). In this process, Bowtie2 was utilized to align the sequences to the genomic reference sequence, and a purpose-built R workflow was implemented to select the pure sequencing results filtering out erroneous reads generated through template switching in the PCR-based sample preparation.
Availability of data and materials
The data sets supporting the conclusions of this article are available in the NCBI Sequence Read Archive (SRA) with accession number PRJNA403798. The R-based workflow is publically available as a Git repository at https://bitbucket.org/MNM-LU/rna-editing and as a Docker image: Bjorklund/RNA-edit. All plasmid sequences can be found on http://rna2018.neuromodulation.se and plasmids can be requested directly from the authors.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the staff at the National Genomics Infrastructure (NGI) of SciLifeLab, Sweden and UCLA Clinical Microarray Core, USA for expert assistance in the sequencing performed in this paper using Ion Torrent, PacBio RSII, and Illumina MiSeq technologies. We would also like to thank Anna Hammarberg for excellent assistance and much appreciated help with cell sorting and flow cytometri. FR_DsRed2 was a gift from Gerhart Ryffel (Addgene plasmid # 31444), pgLAP2 was a gift from Peter Jackson (Addgene plasmid # 19703), piRFP was a gift from Vladislav Verkhusha (Addgene plasmid # 31857), and pscAAV-GFP was a gift from John T. Gray (Addgene plasmid # 32396). This work was supported by grants from Parkinson's Disease Foundation International Research (PDF-IRG-1303); Swedish Research Council (K2014-79X-22510-01-1 and ÄR-MH-2016-01997 Starting grant); Swedish Parkinson Foundation; Swedish Alzheimer Foundation; Crafoord Foundation; The Bagadilico Linnaeus consortium; Schyberg Foundation; Thuring Foundation; Kocks Foundation; Åke Wiberg Foundation; Åhlén Foundation; Magnus Bergvall Foundation; Tore Nilsson Foundation; The Swedish Neuro Foundation; OE and Edla Johanssons Foundation, and the Lars Hierta foundation. T.B. is supported by an Associate Senior lectureship from the Bente Rexed Foundation.
Author contributions: T.B. and M.D. designed the experiment; M.D., P.D.F., O.D.S., M.T., P.A., A.H., L.Q., and G.W. performed the wet experiments; T.B. analyzed the sequencing data; M.D. and T.B. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063925.117.
REFERENCES
- Adachi K, Enoki T, Kawano Y, Veraz M, Nakai H. 2014. Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nat Commun 5: 3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagurumoorthy P, Adelstein SJ, Kassis AI. 2008. Method to eliminate linear DNA from mixture containing nicked circular, supercoiled, and linear plasmid DNA. Anal Biochem 381: 172–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A, Lorain S, Joséphine C, Desrosiers M, Peccate C, Voit T, Garcia L, Sahel JA, Bemelmans AP. 2015. Repair of rhodopsin mRNA by spliceosome-mediated RNA trans-splicing: a new approach for autosomal dominant retinitis pigmentosa. Mol Ther 23: 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulaire J, Balani P, Wang S. 2009. Transcriptional targeting to brain cells: engineering cell type-specific promoter containing cassettes for enhanced transgene expression. Adv Drug Deliv Rev 61: 589–602. [DOI] [PubMed] [Google Scholar]
- Bushnell B. 2016. BBMap short read aligner. University of California; Berkeley, California. [Google Scholar]
- Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt FG. 1998. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci 95: 12185–12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. 2009. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150: 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BR, Hale DC, Ciolek PJ, Runge KW. 2012. Generation and analysis of a barcode-tagged insertion mutant library in the fission yeast Schizosaccharomyces pombe. BMC Genomics 13: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsson M, Diaz-Fernandez P, Schwich OD, Torroba M, Wang G, Björklund T. 2016. A novel process of viral vector barcoding and library preparation enables high-diversity library generation and recombination-free paired-end sequencing. Sci Rep 6: 37563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean J, Ghemawat S. 2008. MapReduce: simplified data processing on large clusters. Commun ACM 51: 107–113. [Google Scholar]
- Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu WL, Yang B, Huber N, Pasca SP, et al. 2016. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol 34: 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer SE, Butler MD, Pan Q, Ruvkun G. 2008. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature 455: 491–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G, Brand H, Seraphin B, Gannon F. 2002. Natural trans-spliced mRNAs are generated from the human estrogen receptor-α (hER alpha) gene. J Biol Chem 277: 26244–26251. [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. 2014. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol 56: 343–349. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89: 799–811. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA. 2003. Messenger RNA reprogramming by spliceosome-mediated RNA trans-splicing. J Clin Invest 112: 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor B, Bailey RM, Wimberly K, Kalburgi SN, Gray SJ. 2014. Methods for gene transfer to the central nervous system. Adv Genet 87: 125–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller U, Wally V, Mitchell LG, Klausegger A, Murauer EM, Mayr E, Gruber C, Hainzl S, Hintner H, Bauer JW. 2011. A novel screening system improves genetic correction by internal exon replacement. Nucleic Acids Res 39: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Luo M, Zhang LN, Yan Z, Zak R, Ding W, Mansfield SG, Mitchell LG, Engelhardt JF. 2005. Spliceosome-mediated RNA trans-splicing with recombinant adeno-associated virus partially restores cystic fibrosis transmembrane conductance regulator function to polarized human cystic fibrosis airway epithelial cells. Hum Gene Ther 16: 1116–1123. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Kole J, Puttaraju M, Yang CC, Garcia-Blanco MA, Cohn JA, Mitchell LG. 2000. Repair of CFTR mRNA by spliceosome-mediated RNA trans-splicing. Gene Ther 7: 1885–1895. [DOI] [PubMed] [Google Scholar]
- Mansfield SG, Chao H, Walsh CE. 2004. RNA repair using spliceosome-mediated RNA trans-splicing. Trends Mol Med 10: 263–268. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller KM, Stebel SC, Knall S, Zipf G, Bernauer HS, Arndt KM. 2005. Nucleotide exchange and excision technology (NExT) DNA shuffling: a robust method for DNA fragmentation and directed evolution. Nucleic Acids Res 33: e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murauer EM, Koller U, Hainzl S, Wally V, Bauer JW. 2013. A reporter-based screen to identify potent 3′ trans-splicing molecules for endogenous RNA repair. Hum Gene Ther Methods 24: 19–27. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Pergolizzi RG, Crystal RG. 2005. Gene transfer-mediated pre-mRNA segmental trans-splicing as a strategy to deliver intracellular toxins for cancer therapy. Cancer Res 65: 254–263. [PubMed] [Google Scholar]
- Naldini L. 2015. Gene therapy returns to centre stage. Nature 526: 351–360. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O'Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. 2016. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell 165: 488–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. 2014. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell 54: 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RP, Mariano TM, Siekierka J, Mathews MB. 1986. A mechanism for the control of protein synthesis by adenovirus VA RNAI. Cell 44: 391–400. [DOI] [PubMed] [Google Scholar]
- Pan H, Mostoslavsky G, Eruslanov E, Kotton DN, Kramnik I. 2008. Dual-promoter lentiviral system allows inducible expression of noxious proteins in macrophages. J Immunol Methods 329: 31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran P, Jalili R, Tao L, Shokralla S, Gharizadeh B, Ronaghi M, Fire AZ. 2007. A pyrosequencing-tailored nucleotide barcode design unveils opportunities for large-scale sample multiplexing. Nucleic Acids Res 35: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan RP, Lee C, Litvin O, Young DL, Pe'er D, Shendure J. 2009. High-resolution analysis of DNA regulatory elements by synthetic saturation mutagenesis. Nat Biotechnol 27: 1173–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. 2015. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci 112: 6164–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA, Mitchell LG. 1999. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat Biotechnol 17: 246–252. [DOI] [PubMed] [Google Scholar]
- Puttaraju M, DiPasquale J, Baker CC, Mitchell LG, Garcia-Blanco MA. 2001. Messenger RNA repair and restoration of protein function by spliceosome-mediated RNA trans-splicing. Mol Ther 4: 105–114. [DOI] [PubMed] [Google Scholar]
- Rindt H, Yen PF, Thebeau CN, Peterson TS, Weisman GA, Lorson CL. 2012. Replacement of huntingtin exon 1 by trans-splicing. Cell Mol Life Sci 69: 4191–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martin T, Garcia-Blanco MA, Mansfield SG, Grover AC, Hutton M, Yu Q, Zhou J, Anderton BH, Gallo JM. 2005. Reprogramming of tau alternative splicing by spliceosome-mediated RNA trans-splicing: implications for tauopathies. Proc Natl Acad Sci 102: 15659–15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shababi M, Lorson CL. 2012. Optimization of SMN trans-splicing through the analysis of SMN introns. J Mol Neurosci 46: 459–469. [DOI] [PubMed] [Google Scholar]
- Sharp PA, Burge CB. 1997. Classification of introns: U2-type or U12-type. Cell 91: 875–879. [DOI] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. 2015. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods 12: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck J, Stebel SC, Arndt KM, Müller KM. 2011. Nucleotide exchange and excision technology DNA shuffling and directed evolution. Methods Mol Biol 687: 333–344. [DOI] [PubMed] [Google Scholar]
- Tahara M, Pergolizzi RG, Kobayashi H, Krause A, Luettich K, Lesser ML, Crystal RG. 2004. Trans-splicing repair of CD40 ligand deficiency results in naturally regulated correction of a mouse model of hyper-IgM X-linked immunodeficiency. Nat Med 10: 835–841. [DOI] [PubMed] [Google Scholar]
- Wang JL, Chang WT, Tong CW, Kohno K, Huang AM. 2009. Human synapsin I mediates the function of nuclear respiratory factor 1 in neurite outgrowth in neuroblastoma IMR-32 cells. J Neurosci Res 87: 2255–2263. [DOI] [PubMed] [Google Scholar]
- Weeratna RD, Wu T, Efler SM, Zhang L, Davis HL. 2001. Designing gene therapy vectors: avoiding immune responses by using tissue-specific promoters. Gene Ther 8: 1872–1878. [DOI] [PubMed] [Google Scholar]
- Wu X, Dong X, Wu Z, Cao H, Niu D, Qu J, Wang H, Hou Y. 2001. A novel method for purification of recombinant adenoassociated virus vectors on a large scale. Chin Sci Bull 46: 485–488. [Google Scholar]
- Yang Y, Walsh CE. 2005. Spliceosome-mediated RNA trans-splicing. Mol Ther 12: 1006–1012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets supporting the conclusions of this article are available in the NCBI Sequence Read Archive (SRA) with accession number PRJNA403798. The R-based workflow is publically available as a Git repository at https://bitbucket.org/MNM-LU/rna-editing and as a Docker image: Bjorklund/RNA-edit. All plasmid sequences can be found on http://rna2018.neuromodulation.se and plasmids can be requested directly from the authors.