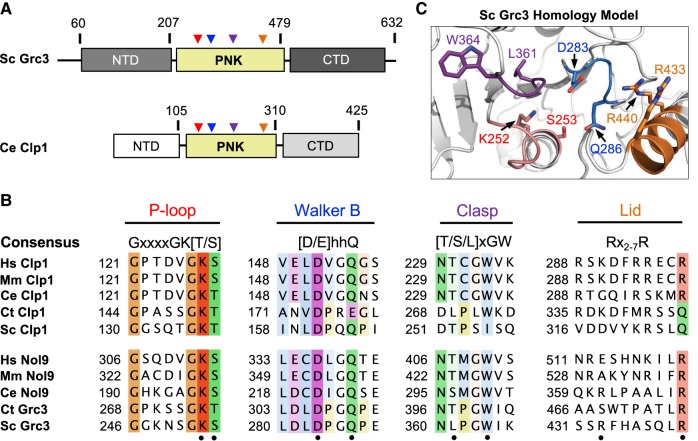
FIGURE 4.
Conserved Grc3 PNK active site motifs. (A) Cartoon diagram of S. cerevisiae (Sc) Grc3 and C. elegans (Ce) Clp1. The homologous central polynucleotide kinase (PNK) domain is colored in yellow, and the variable N- and C-terminal domains (NTD and CTD) are shaded in light and dark gray, respectively. Numbering above the bar diagrams marks the amino acid residue domain boundaries. Arrowheads identify the relative position of the P-loop (red), Walker B (blue), Clasp (purple), and Lid (orange) motifs. (B) Conserved sequence motifs of the Clp1/Grc3 PNK family from Homo sapiens (Hs), Mus musculus (Mm), C. elegans (Ce), Chaetomium thermophilum (Ct), and S. cerevisiae (Sc). Fully conserved and semi-conserved residues are highlighted in dark and light colors, respectively. Sc Grc3 residues targeted for mutagenesis in this study are marked by a black dot. (C) S. cerevisiae Grc3 PNK active site modeled using the SWISS-MODEL server (Arnold et al. 2006) and derived from the crystal structure of S. cerevisiae Clp1 (PDB ID: 2NPI) (Noble et al. 2007). Grc3 P-loop (red), Walker B (blue), Clasp (purple), and Lid (orange) motifs are colored and residues targeted for mutagenesis are labeled.