Abstract
The post-transcriptional modification of tRNA at the wobble position is a universal process occurring in all domains of life. In eukaryotes, the wobble uridine of particular tRNAs is transformed to the 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) modification which is critical for proper mRNA decoding and protein translation. However, current methods to detect mcm5s2U are technically challenging and/or require specialized instrumental expertise. Here, we show that γ-toxin endonuclease from the yeast Kluyveromyces lactis can be used as a probe for assaying mcm5s2U status in the tRNA of diverse eukaryotic organisms ranging from protozoans to mammalian cells. The assay couples the mcm5s2U-dependent cleavage of tRNA by γ-toxin with standard molecular biology techniques such as northern blot analysis or quantitative PCR to monitor mcm5s2U levels in multiple tRNA isoacceptors. The results gained from the γ-toxin assay reveals the evolutionary conservation of the mcm5s2U modification across eukaryotic species. Moreover, we have used the γ-toxin assay to verify uncharacterized eukaryotic Trm9 and Trm112 homologs that catalyze the formation of mcm5s2U. These findings demonstrate the use of γ-toxin as a detection method to monitor mcm5s2U status in diverse eukaryotic cell types for cellular, genetic, and biochemical studies.
Keywords: endonuclease, γ-toxin, mcm5s2U, modification, tRNA
INTRODUCTION
The post-transcriptional modification of uridine at the wobble position of tRNA is ubiquitous in all organisms and plays a critical role in proper mRNA decoding during translation (for reviews, see Grosjean et al. 2010; Agris et al. 2017; Schaffrath and Leidel 2017). In eukaryotes, the wobble uridine of certain tRNAs can undergo a series of modifications to generate the modified nucleotide, methoxycarbonylmethyl-2-thiouridine (mcm5s2U) (Fig. 1A). While the specific order of biochemical intermediates leading to mcm5s2U remains to be clarified (Songe-Møller et al. 2010; Chen et al. 2011a; van den Born et al. 2011), several proteins have been identified in the yeast S. cerevisiae that are required for mcm5s2U formation in tRNA. The multisubunit Elongator complex is required for early steps of mcm5U formation through the generation of 5-carbonylmethyluridine (cm5U) and 5-carbamoylmethyluridine (ncm5U) via a proposed enzymatic reaction involving acetyl-CoA (for reviews, seed Karlsborn et al. 2014b; Dauden et al. 2017; Kolaj-Robin and Seraphin 2017). The cm5U and/or ncm5U modification is then further modified by a heterodimeric methyltransferase complex consisting of the Trm9 enzymatic subunit and the Trm112 structural protein (Kalhor and Clarke 2003; Mazauric et al. 2010; Liger et al. 2011; Letoquart et al. 2015; Bourgeois et al. 2017). In a subset of tRNAs, the mcm5U base can undergo even further modification by the Ncs2-Ncs6 thiolase to generate the final mcm5s2U modification (Dewez et al. 2008; Huang et al. 2008; Leidel et al. 2009; Noma et al. 2009; Nakai et al. 2017).
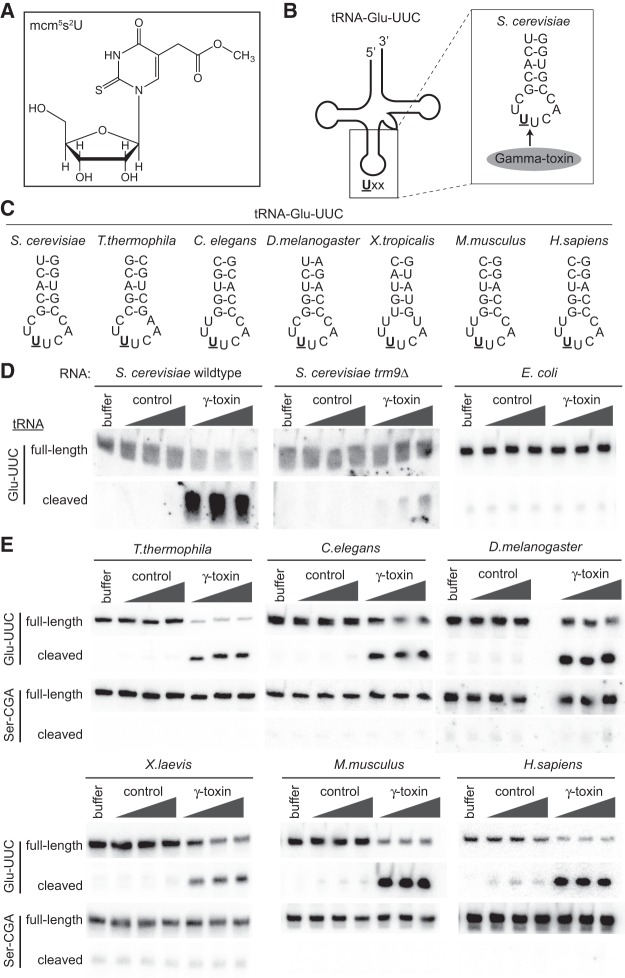
FIGURE 1.
Purified γ-toxin can cleave mcm5s2U containing tRNA species from a wide range of organisms. (A) Chemical structure of the mcm5s2U modification found on the wobble uridine of select tRNA species. (B) Schematic of S. cerevisiae tRNA-Glu-UUC highlighting the anticodon stem and position of γ-toxin cleavage 3′ of the mcm5s2U modification denoted by U. (C) Anticodon stems of tRNA-Glu-UUC from eukaryotic organisms tested in this study. The underlined U denotes the mcm5s2U modification. (D) Northern blots of S. cerevisiae or E. coli total RNA preincubated with buffer or increasing amounts of either a control purification eluate or γ-toxin. The blot was hybridized with a probe against tRNA-Glu-UUC to detect full-length or cleaved tRNA products. (E) Total RNA from the indicated eukaryotic organisms was incubated with either buffer, control purification eluate, or purified γ-toxin, followed by northern blot analysis, as in D. The blot was probed for tRNA-Ser-CGA as a specificity and loading control.
The mcm5s2U modification plays a key role in the efficiency and fidelity of mRNA translation to modulate protein levels in eukaryotes (Huang et al. 2005; Johansson et al. 2008; Bauer et al. 2012; Patil et al. 2012a; Fernández-Vázquez et al. 2013; Rezgui et al. 2013; Deng et al. 2015; Klassen et al. 2015, 2016a; Tükenmez et al. 2015). Due to its effects on protein synthesis, the presence of mcm5s2U at the wobble position is crucial for proper cellular growth, metabolism, and organismal development (Esberg et al. 2006; Björk et al. 2007; Lin et al. 2013; Leitner et al. 2015; Karlsborn et al. 2016; Klassen et al. 2016b; Schaffrath and Klassen 2017). Moreover, the mcm5U-class of tRNA modifications has been shown to play a critical role in the cellular stress response by regulating the translation of proteins involved in redox homeostasis, DNA repair, and telomere silencing (Begley et al. 2007, 2013; Chan et al. 2010; Fu et al. 2010a; Chen et al. 2011b; Patil et al. 2012b; Fernández-Vázquez et al. 2013; Zinshteyn and Gilbert 2013; Endres et al. 2015). Significantly, the mcm5s2U modification is also required for proper neurodevelopment in worms, mice, and humans since gene mutations that reduce mcm5s2U levels lead to a spectrum of neurological disorders including familial dysautonomia (Chen et al. 2009; Karlsborn et al. 2014a; Yoshida et al. 2015; Kojic and Wainwright 2016). Thus, the detection of mcm5s2U levels will be critical for understanding the role of tRNA modifications in diverse biological processes.
The use of thin layer chromatography (TLC) for separation of digested RNA nucleosides has been a classic technique for identifying and detecting modifications such as mcm5s2U (Grosjean et al. 2004). While TLC analysis remains a viable approach for analysis of mcm5s2U, the radiolabeling, digestion, spotting, and chromatographic separation of RNA on TLC plates is labor-intensive and time-consuming. In addition, the resolution of mcm5s2U by 2-dimensional TLC requires extensive optimization for clear interpretation (Grosjean et al. 2007).
High pressure-liquid chromatography (HPLC) provides an alternative method for detection of mcm5s2U through the separation of nucleosides generated by RNA digestion (Huang et al. 2005; Mazauric et al. 2010; Chen et al. 2011a). Furthermore, HPLC has been coupled with mass spectrometry (HPLC-MS) for sensitive identification and quantification of RNA modifications (Su et al. 2014; Cai et al. 2015; Cao and Limbach 2015; Basanta-Sanchez et al. 2016; Ross et al. 2016). However, HPLC-MS requires lengthy run times, extensive optimization, and the generation of mcm5s2U standards. Thus, the expertise and instrumentation costs associated with HPLC-MS instrumentation are beyond the capabilities of many laboratories wishing to study tRNA modifications such as mcm5s2U.
Many tRNA modifications can also be detected by reverse transcriptase (RT)-based primer extension methods, differential probe hybridization, or reactivity to certain chemical reagents (for reviews, see Schwartz and Motorin 2016; Helm and Motorin 2017). However, the mcm5s2U modification has no detectable effect on oligonucleotide hybridization, RT extension, or nucleotide misincorporation (Hiley et al. 2005; Motorin et al. 2007; Behm-Ansmant et al. 2011). Loss of the thiol group in mcm5s2U can be detected by differential tRNA migration in the presence of acryloylaminophenylmercuric chloride, but changes in the methylester group are undetectable using the same technique (Igloi 1992; Kaneko et al. 2003). While chemical treatment of tRNA with low concentrations of NaOH can remove the methyl group from mcm5s2U to generate cm5U, this treatment is incomplete, leads to RNA degradation, and does not lead to any change in RNA migration, or RT primer extension (Mazauric et al. 2010; Chen et al. 2011a; Leihne et al. 2011). Moreover, there are no known published antibodies that have been generated against mcm5s2U.
Due to the reasons noted above, we sought to develop an assay for detection of mcm5s2U that would utilize standard molecular biology assays and obviate the need for TLC or HPLC-MS. We focused our attention on γ-toxin, a ribonuclease secreted from the milk yeast Kluyveromyces lactis that catalyzes the endonucleolytic cleavage of foreign yeast tRNAs in a mcm5s2U-dependent manner (for reviews, see Lu et al. 2005; Jablonowski et al. 2006; Jablonowski and Schaffrath 2007; Lu et al. 2008). γ-Toxin cleaves tRNAs-Glu-UUC, Lys-UUU, and Gln-UUG between positions 34 and 35 of the anticodon loop with cleavage being highly dependent upon the presence of the mcm5s2U modification (Fig. 1B, tRNA-Glu-UUC shown). S. cerevisiae yeast cells that are deficient for any of the enzymes that form mcm5s2U in tRNA are resistant to γ-toxin–mediated tRNA cleavage and cell death (Frohloff et al. 2001; Fichtner and Schaffrath 2002; Jablonowski et al. 2006; Huang et al. 2008; Studte et al. 2008). γ-Toxin has been used in S. cerevisiae to uncover proteins and pathways involved in mcm5s2U modification (Huang et al. 2008; Studte et al. 2008; Mehlgarten et al. 2009, 2010, 2017; Jüdes et al. 2016). Based upon the sequence requirements and substrate specificity of γ-toxin, we predicted that γ-toxin could be used to cleave mcm5s2U-containing tRNAs isolated from other eukaryotic organisms. Here, we show that γ-toxin can be used as a molecular tool for monitoring mcm5s2U status across a variety of eukaryotic species, including model eukaryotic organisms. Moreover, we demonstrate that γ-toxin can be used for the discovery and in vitro validation of uncharacterized eukaryotic proteins involved in the formation of mcm5s2U.
RESULTS AND DISCUSSION
γ-Toxin cleaves mcm5s2U-containing tRNAs from diverse eukaryotic organisms
Due to the specificity of γ-toxin for yeast tRNAs containing mcm5s2U, we hypothesized that γ-toxin could be used to monitor mcm5s2U status in additional eukaryotic organisms besides S. cerevisiae (Fig. 1C). To assay for tRNA cleavage, we incubated recombinant γ-toxin purified from E. coli with total RNA isolated from different eukaryotic organisms followed by northern blot hybridization analysis. To monitor nonspecific RNA cleavage due to possible contaminating nucleases, we also incubated RNA with control purifications from E. coli cells containing vector alone. As a positive control, we tested the activity of γ-toxin against S. cerevisiae tRNA-Glu-UUC, the primary physiological target of γ-toxin endonuclease (Lu et al. 2005; Jablonowski et al. 2006). No tRNA cleavage products were detected when S. cerevisiae total RNA was incubated with buffer or increasing amounts of a control protein purification from E. coli cells (Fig. 1D, S. cerevisiae wild-type, buffer, and control lanes). However, incubation of S. cerevisiae RNA with purified γ-toxin led to the cleavage of tRNA-Glu-UUC and the accumulation of cleaved tRNA fragments (Fig. 1D, S. cerevisiae wild-type + γ-toxin). γ-Toxin cleavage of S. cerevisiae tRNA-Glu-UUC was greatly reduced with RNA extracted from trm9Δ cells, which are unable to form the mcm5s2U modification (Fig. 1D, trm9Δ). Moreover, γ-toxin was unable to cleave E. coli tRNA-Glu-UUC, which contains a 5-methylaminomethyluridine (mnm5U) modification instead of mcm5s2U (Fig. 1D, E. coli + γ-toxin). The inability of γ-toxin to cleave E. coli tRNA-Glu-UUC confirms the results of earlier studies (Lu et al. 2008) and validates the specificity of the γ-toxin preparation used in this study.
Upon confirming the activity of γ-toxin, we tested whether γ-toxin displays endonuclease activity against mcm5s2U-containing tRNAs of additional eukaryotic organisms. Remarkably, we find that γ-toxin can efficiently and specifically cleave tRNA-Glu-UUC isolated from a diversity of eukaryotes including Tetrahymena thermophila (ciliated protozoa), Caenorhabditis elegans (nematode), Drosophila melanogaster (fruit fly), Xenopus tropicalis (frog), Mus musculus (mouse embryonic fibroblasts) and Homo sapiens (human embryonic fibroblast cells) (Fig. 1E, Glu-UUC + γ-toxin). In addition to the organisms described here, we have also used γ-toxin to demonstrate the presence of mcm5s2U in tRNA-Glu-UUC of the parasitic protozoan, Toxoplasma gondii (Padgett et al. 2018). In all cases where total eukaryotic RNA was incubated with γ-toxin, we detected tRNA fragments indicative of only a single tRNA cleavage event within the anticodon loop of tRNA-Glu-UUC without any other products of nonspecific degradation (Fig. 1E, Glu-UUC, data not shown). In contrast to tRNA-Glu-UUC of each eukaryote, we detected no detectable cleavage of tRNA-Ser-CGA, which does not contain a wobble uridine and hence, lacks the mcm5s2U modification (Fig. 1E, Ser-CGA). Our results demonstrate that γ-toxin exhibits specific endonuclease activity for the mcm5s2U-containing tRNA-Glu(UUC) across a spectrum of eukaryotic organisms. Moreover, these studies provide the first experimental evidence that the mcm5s2U modification is conserved in the tRNA of single-celled protists as well as nonmammalian vertebrates such as frogs.
γ-Toxin is specific for tRNAs containing mcm5s2U but not similar derivatives
In addition to tRNA-Glu-UUC, the wobble position of eukaryotic tRNA-Arg-UCU, Gln-UUG, and Lys-UUU are also known to contain mcm5s2U (Kuntzel et al. 1975; Keith 1984; Chen et al. 2009; Fu et al. 2010a; Songe-Møller et al. 2010; van den Born et al. 2011; Fernández-Vázquez et al. 2013; Karlsborn et al. 2014a; Yoshida et al. 2015). γ-Toxin has been shown to cleave these mcm5s2U-containing tRNAs in S. cerevisiae, albeit at lower efficiency (Supplemental Fig. 1; Lu et al. 2005). Using RNA isolated from human cells, we find that γ-toxin can also cleave human tRNA-Arg-UCU, Gln-UUG, and Lys-UUU in the anticodon loop (Fig. 2, mcm5s2U). However, the cleavage of human tRNA-Arg-UCU, Gln-UUG, and Lys-UUU was much less efficient than tRNA-Glu-UUC, as predicted from previous studies with S. cerevisiae tRNA (Lu et al. 2005). These results indicate that γ-toxin can be used to monitor or verify the presence of mcm5s2U in multiple eukaryotic tRNAs.
FIGURE 2.
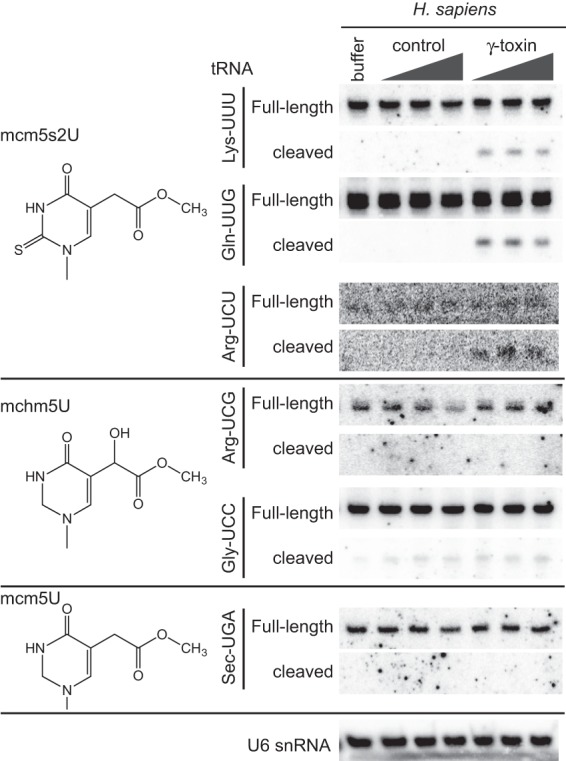
γ-Toxin is highly specific for tRNAs containing the mcm5s2U modification. Total RNA extracted from human HEK293T cells was incubated with either buffer or increasing amounts of a control purification, or γ-toxin followed by northern blot analysis with probes against the indicated tRNAs. The modification at the wobble uridine of each tRNA is denoted on the left. U6 snRNA serves as a loading and specificity control.
Besides mcm5s2U-containing tRNAs, we also tested whether γ-toxin could cleave human tRNAs known to contain modified wobble uridines that differ slightly in chemical composition from mcm5s2U. Previous studies have shown that the mcm5U side chain of mammalian tRNA-Arg-UCG and tRNA-Gly-UCC undergoes enzymatic oxidation to generate the hydroxylated modification, 5-methoxycarbonylhydroxymethyluridine (mchm5U) (Fu et al. 2010b; van den Born et al. 2011). However, γ-toxin did not exhibit any detectable cleavage activity on either tRNA-Arg-UCG or Gly-UCC (Fig. 2, mchm5U). γ-Toxin was also inactive for cleaving tRNA-Sec-UGA, which contains the 5-methoxycarbonylmethyl-2′-O-methyluridine (mcm5Um) modification at the wobble uridine position (Fig. 2, mcm5U). These results demonstrate that γ-toxin is highly specific for the mcm5s2U modification with lack of the thiol, addition of a hydroxyl group, or presence of 2′O-methylation possibly interfering with recognition and cleavage.
Using γ-toxin cleavage to test the role of eukaryotic proteins in mcm5s2U formation
In S. cerevisiae, the heterodimeric Trm9–Trm112 complex catalyzes the final formation of the methoxycarbonylmethyl side chain in mcm5s2U (Mazauric et al. 2010; Chen et al. 2011a). Trm9 is the catalytic methyltransferase subunit while Trm112 is required for enzymatic activity by serving as a platform for tRNA interaction and cofactor binding (Letoquart et al. 2015; Bourgeois et al. 2017). Numerous homologs of S. cerevisiae Trm9 and Trm112 have been identified in eukaryotic and archaeal species (Grosjean et al. 2010; Leihne et al. 2011; Phillips and de Crecy-Lagard 2011; Pastore et al. 2012; Towns and Begley 2012; Zdżalik et al. 2014). However, the activities of the predicted enzymes remain unverified in most cases. Thus, we investigated whether the γ-toxin assay could be used to validate the methyltransferase activities of Trm9 and Trm112 homologs. For this assay, we used total RNA extracted from S. cerevisiae trm9Δ cells as a substrate since it contains the available cm5s2U modification in tRNA as the final methyl acceptor (Mazauric et al. 2010; Chen et al. 2011a). The trm9Δ cellular RNA is first incubated with a putative tRNA methyltransferase in the presence of S-adenosyl-methionine followed by incubation with γ-toxin. The level of γ-toxin cleavage of tRNA is then used as an indicator of mcm5s2U formation catalyzed by the Trm9–Trm112 complex (Fig. 3A).
FIGURE 3.
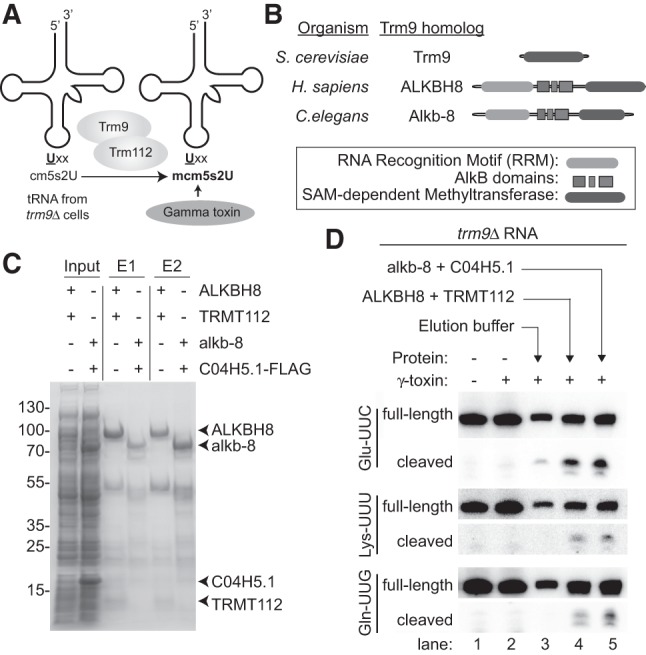
Using the γ-toxin assay to validate the biochemical activity of putative Trm9–Trm112 homologs. (A) Schematic of γ-toxin assay to characterize Trm9–Trm112 enzymes for methyltransferase activity. The tRNA of S. cerevisiae trm9Δ strains lack the mcm5s2U modification and instead harbors the cm5U modification that can be methylated in vitro by purified Trm9–Trm112 enzyme complexes. Formation of mcm5s2U can be subsequently detected by the γ-toxin assay. (B) Schematic of S. cerevisiae Trm9p along with the H. sapiens and C. elegans homologs, ALKBH8 and Alkb-8, respectively. Both human and C. elegans homologs contain a SAM-dependent methyltransferase domain in addition to a RNA recognition motif (RRM) and AlkB dioxygenase domain. (C) Purified protein expressed in E. coli cells along with cellular inputs were fractionated by SDS-PAGE and visualized by Coomassie stain. Arrows denote respective proteins. (D) Total RNA extracted from the yeast trm9Δ line was incubated with either elution buffer, human ALKBH8–TRMT112, or C. elegans alkb-8-C04H5.1 and then subjected to the γ-toxin assay.
To explore the use of the γ-toxin assay for identifying eukaryotic proteins involved in mcm5s2U formation, we expressed and purified putative Trm9 and Trm112 homologs encoded by the C. elegans genome. Sequence alignment has revealed potential C. elegans homologs of yeast Trm9 and Trm112 that are encoded by the Alkb-8 and C04H5.1 genes, respectively (Vilella et al. 2009; Pastore et al. 2012). The Alkb-8 gene product encodes a 591-amino acid protein that displays 32% identity and 47% similarity to the methyltransferase domain of S. cerevisiae Trm9p (Fig. 3B). In addition to the SAM-dependent methyltransferase domain, C. elegans Alkb-8 contains an AlkB dioxygenase domain and RNA recognition motif (Pastore et al. 2012). The C. elegans C04H5.1 gene contains an open reading frame encoding a polypeptide of 125 amino acid residues with 23% identity and 55% similarity to S. cerevisiae Trm112. However, neither the gene product of Alkb-8 nor C04H5.1 have been characterized in terms of biochemical activity so their status as a functional Trm9–Trm112 complex remains to be shown.
We coexpressed the C. elegans homologs of Trm9 and Trm112 in bacteria followed by metal affinity purification. Alkb-8 was tagged with a hexa-histidine tag for purification purposes while the C. elegans Trm112 homolog was epitope-tagged with a 3×FLAG tag for detection. As a positive control, we compared the C. elegans proteins to purified human ALKBH8–TRMT112, which forms a heterodimeric complex that has previously been shown to exhibit methyltransferase activity to form mcm5s2U (Fu et al. 2010a). As expected, eluted fractions of affinity purified human ALKBH8 led to the copurification of TRMT112 (Fig. 3C, E1 and E2, +ALKBH8+TRMT112, Supplemental Fig. 2). We also detected the coprecipitation of the C. elegans C04H5.1 protein with Alkb-8, thereby demonstrating the formation of a heterodimeric complex between the predicted C. elegans Trm9 and Trm112 homologs (Fig. 3C; Supplemental Fig. 2).
The purified Trm9–Trm112 complexes were then tested for methyltransferase activity on tRNA isolated from Trm9Δ yeast strains. As noted previously, we detected only slight cleavage of either tRNA-Glu-UUC, Lys-UUU, or Gln-UUG when preincubated with γ-toxin due to the lack of mcm5s2U in trm9Δ tRNA that is critical for recognition by γ-toxin (Fig. 3D, lanes 1 and 2). In contrast, we could detect γ-toxin cleavage of tRNA-Glu-UUC, Lys-UUU, or Gln-UUG if the trm9Δ RNA was first preincubated with human ALKBH8–TRMT112 (Fig. 3D, ALKBH8+TRMT112, lane 4). This result suggests that recombinant human ALKBH8–TRMT112 has generated the final mcm5s2U modification in tRNA isolated from trm9Δ cells that is now a substrate for γ-toxin cleavage in vitro. While γ-toxin treatment of RNA isolated from wild-type S. cerevisiae cells led to a single tRNA cleavage event (Fig. 1D), we detected the presence of two closely migrating 5′-half cleavage products for tRNA-Glu-UUC and Gln-UUG isolated from trm9Δ cells pretreated with ALKBH8–TRMT112. We hypothesize that the different 5′-half products represent tRNA species that are differentially hypomodified for other modifications besides mcm5s2U in trm9Δ cells (Chan et al. 2010; Begley et al. 2013; Deng et al. 2015). To confirm that the tRNA products were due to formation of mcm5s2U that allowed for γ-toxin cleavage, we also treated tRNA samples with ALKBH8 alone, which is inactive without the TRMT112 subunit (Fu et al. 2010a; Songe-Møller et al. 2010). In this case, we detected only background cleavage for tRNA-Glu-UUC after preincubation with ALKBH8 alone (Supplemental Fig. 3A). Notably, we could also detect robust cleavage of tRNA-Glu-UUC, Lys-UUU, or Gln-UUG by γ-toxin after preincubation with the reconstituted C. elegans Trm9–Trm112 complex (Fig. 3D, alkb-8+C04H5.1, lane 5). While a discrete cleavage product was detected for tRNA-Glu-UUC, Lys-UUU, and Gln-UUG, we detected only background levels of cleavage for tRNA-Ser-CGA, which does not contain the mcm5s2U modification (Supplemental Fig. 3B). Moreover, no γ-toxin cleavage products were detected when tRNA was preincubated with human ALKBH8–TRMT112 or C. elegans Trm9–Trm112 that had previously been inactivated by thermal denaturation (Supplemental Fig. 3C). Thus, the γ-toxin assay provides the first demonstration that C. elegans Alkb-8 and C04H5.1 (Trm112) form an enzymatically-active tRNA methyltransferase complex that can catalyze the formation of mcm5s2U. Altogether, these results highlight the use of γ-toxin as a tool to discover and probe the methyltransferase activity of uncharacterized Trm9 and Trm112 homologs from diverse eukaryotic organisms.
Interestingly, γ-toxin cleavage of tRNA-Lys-UUU and Gln-UUG isolated from trm9Δ cells occurred with similar efficiency as tRNA-Glu-UUC after treatment with either human or C. elegans Trm9–Trm112. This result contrasts with the much less efficient γ-toxin cleavage of tRNA-Lys-UUU and Gln-UUG relative to tRNA-Glu-UUC when isolated from wild-type S. cerevisiae cells (Supplemental Fig. 1; Lu et al. 2005). We speculate that a much greater fraction of tRNA-Glu-UUC contains the mcm5s2U modification compared to that of tRNA-Lys-UUU and Gln-UUG in wild-type S. cerevisiae cells, accounting for the difference in γ-toxin cleavage efficiency. In contrast, trm9Δ cells lack mcm5s2U in all tRNAs and exhibit increased levels of both cm5s2U and ncm5s2U (Chen et al. 2011a). Thus, trm9Δ cells could have similar amounts of incompletely modified tRNA-Glu-UUC, Lys-UUU, and Gln-UUG that are now substrates for methylation by Trm9–Trm112 in vitro leading to the comparable levels of cleavage by γ-toxin.
γ-Toxin-coupled real-time qRT-PCR to quantify mcm5s2U levels
Thus far, we have detected γ-toxin cleavage using northern blotting with radiolabeled probes. While northern blotting provides a sensitive method for RNA detection, the protocol requires expertise in RNA electrophoresis, blotting equipment and radioactive probe labeling. As an alternative to northern blotting, previous studies have used conventional RT-PCR to monitor tRNA cleavage by γ-toxin (Bär et al. 2008; Nandakumar et al. 2008; Studte et al. 2008). Inspired by these approaches, we developed a quantitative real-time qRT-PCR assay to measure γ-toxin cleavage of tRNA as a rapid, nonradioactive alternative to northern blot analysis for monitoring the levels of mcm5s2U. For this method, we treated sample RNAs with γ-toxin followed by reverse transcription using a primer downstream from the tRNA anticodon sequence to generate a mixture of cDNA products representing full-length or truncated cDNA products (Fig. 4A). Next, we PCR amplified the mixture of cDNA products using primers spanning the γ-toxin cleavage site. Since the presence of mcm5s2U would lead to γ-toxin cleavage of tRNA, the expectation would be to detect less cDNA product by quantitative PCR. Conversely, a decrease in mcm5s2U modification would lead to less γ-toxin cleavage and more full-length cDNA product detectable by PCR. We then normalized the signal to rRNA to control for the amount of RNA within the same sample.
FIGURE 4.
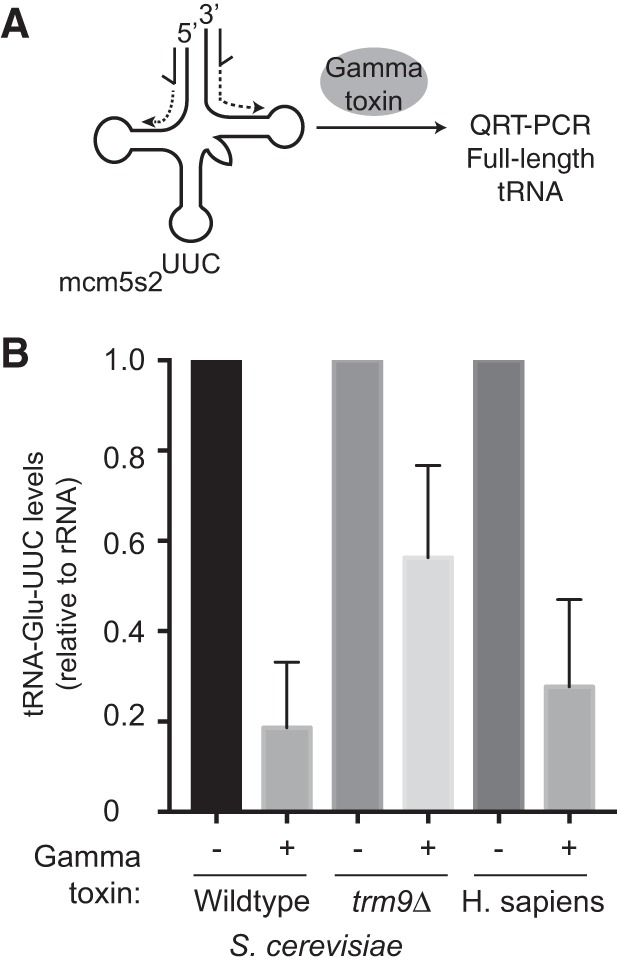
Quantitative detection of γ-toxin cleavage of mcm5s2u-containing tRNAs using qRT-PCR. (A) Schematic of qRT-PCR assay for detecting tRNA cleavage by γ-toxin. The location of PCR primers for amplification of full-length tRNA-Glu-UUC is denoted by half-arrows. (B) Real-time PCR analysis of cDNA generated from reverse transcription of total RNA isolated from the indicated yeast strain or human cells that were either untreated or pretreated with γ-toxin. The total amount of RNA was normalized using 25S rRNA (yeast) or 5.8S (human) and expressed relative to the untreated total RNA sample of each organism. All assays were performed in triplicate on multiple independent samples and repeated greater than 3× to ensure reproducibility.
As a benchmark comparison to northern blotting, we tested the detection sensitivity of qRT-PCR for monitoring mcm5s2U in S. cerevisiae tRNA after γ-toxin cleavage. We found that qRT-PCR could detect an ∼80% decrease in full-length tRNA-Glu-UUC after treatment with γ-toxin, comparable to the detection sensitivity by northern blot analysis (Fig. 4B). We also used qRT-PCR to monitor γ-toxin cleavage of tRNA-Glu-UUC isolated from trm9Δ strains, which are known to lack mcm5s2U and cannot be cleaved by γ-toxin. Indeed, we found that the levels of full-length tRNA-Glu-UUC after γ-toxin treatment was higher for trm9Δ strains versus WT yeast cells (Fig. 4B, compare WT versus trm9Δ + γ-toxin). To demonstrate the utility of the assay beyond S. cerevisiae, we also monitored mcm5s2U levels in human tRNA-Glu-UUC with our integrated γ-toxin qRT-PCR approach. We found that qRT-PCR could detect a ∼70% decrease in tRNA-Glu-UUC levels after γ-toxin treatment (Fig. 4B). Thus, the combination of γ-toxin cleavage with qRT-PCR provides a rapid and sensitive method to probe the levels of mcm5s2U levels in different genetic backgrounds or conditions.
Future applications
Here, we have shown that recombinant γ-toxin displays robust and specific endonuclease activity to cleave mcm5s2U-containing tRNAs in diverse eukaryotic organisms. These results demonstrate that γ-toxin can be applied as a tool for investigating the levels of mcm5s2U modification in a variety of model eukaryotic systems including D. melanogaster, C. elegans, and mammalian cells. The use of γ-toxin technique is straightforward by using short incubation, northern blotting, and detection of cleaved products. The γ-toxin protocol can also be combined with real-time qRT-PCR, thus providing a rapid and sensitive alternative to northern blotting for quantifying the presence of mcm5s2U in the tRNA of eukaryotes. Thus, γ-toxin represents an efficient and specific detection method for assaying the mcm5s2U tRNA modification in all known tRNAs containing the mcm5s2U modification.
In addition to monitoring the levels of mcm5s2U in tRNA, the γ-toxin assay can be used to identify and validate putative eukaryotic proteins that play a role in mcm5s2U formation. Indeed, we have used the γ assay to demonstrate the methyltransferase activity of human and worm homologs of Trm9–Trm112. We envision that the γ-toxin assay can be extended to discover putative homologs of the Elongator and thiolase complexes as well as novel subunits involved in mcm5s2U formation. Moreover, the γ-toxin assay can be used as an assay to distinguish Trm9 paralogs with different targets such as the case of ALKBH8 and KIAA1456 in humans (Fu et al. 2010a; Songe-Møller et al. 2010; Begley et al. 2013). Furthermore, the γ-toxin assay could be applied to the numerous predicted Archaeal homologs of Elongator and Trm9 that have not been characterized in terms of their final biochemical activities (Grosjean et al. 2008; Phillips and de Crecy-Lagard 2011; Naor et al. 2012).
Finally, our findings suggest that γ-toxin could be expressed in heterologous organisms as a way to test translation dependent upon tRNAs containing the mcm5s2U modification. For example, γ-toxin could be transiently expressed in mouse or human cells to cleave endogenous mcm5s2U-containing tRNAs followed by proteomic analysis as a tunable method to analyze the role of mcm5s2U modifications in translation. Altogether, these studies will pave the way for understanding the biological role of mcm5s2U modification and their ubiquitous nature in eukaryotes.
MATERIAL AND METHODS
Protein expression constructs
The protein expression construct for γ-toxin was provided by the Shuman Laboratory (pET28-His10Smt3-γ-toxin-C13A-C177A-C231A) (Keppetipola et al. 2009). The open reading for human TRMT112 was PCR amplified from cDNA plasmid HsCD00323319 (PlasmID Repository, Harvard Medical School) and cloned into the BglII-KpnI sites of pET-Duet1 (EMD Millipore) to generate pET-Duet1-TRMT112. The open reading frame for human ALKBH8 was PCR amplified from pcDNA3.1-FLAG-ALKBH8 (Fu et al. 2010a) and cloned into pET-Duet-TRMT112 using SacI-SalI. The dual ALKBH8–TRMT112 construct was generated by cloning NotI-XhoI fragments from pDuet-TRM112 into the identical sites of pET-Duet1-ALKBH8. The open reading frames of C. elegans Alkb-8 and C04H5.1 were PCR amplified from the cDNA clones, C14B1.10 (OCE1182-202123704) and C04H5.1 (OCE1182-202123244), respectively (Dharmacon, GE Life Sciences). The Alkb-8 and C04H5.1 inserts were cloned into pET-Duet1 using BglII-KpnI and SacI-HindIII, respectively. All inserts were sequenced and verified for the absence of mutations.
Protein expression and purification
The pET28a bacterial expression vector containing either N-terminal His-tagged γ-toxin or control vector (empty pET28a) was transformed into BL21 (DE3)-RIPL E.coli cells (Agilent Technologies) and grown in 50 mL of Luria-Bertani media containing kanamycin and chloramphenicol to an OD600 of 0.6–0.8. Protein expression was induced by the addition of Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 18 h at 16°C at a final concentration of 0.4 M. Cells were then pelleted at 4000g for 10 min and resuspended in 4 mL of bacterial lysis buffer (20 mM Tris, 5% glycerol, 0.3%Triton, 1 mM DTT, 0.1 mM PMSF, 150 mM NaCl, 25 mM imidazole). Cells were lysed via sonication and cellular debris was pelleted at 20,000g for 30 min at 4°C. Cellular extracts were incubated with HisPur Ni-NTA Resin (Thermo Fisher Scientific) and rotated at 4°C for 2 h before being washed three times in the aforementioned lysis buffer. Protein was eluted using a buffer containing 300 mM imidazole, 200 mM NaCl, 20 mM Tris, 5% glycerol, 0.3% Triton, and PMSF. Purified protein was visualized on a 15% SDS-PAGE gel stained with Coomassie protein stain.
BL21 (DE3)-RIPL E. coli cells harboring the pETDuet1 bacterial expression vectors containing either Homo sapiens or Caenorhabditis elegans Trm9–Trm112 homologs were grown as mentioned above. Protein expression of Human ALKBH8–TRMT112 by IPTG was induced for 15 h at 20°C. C. elegans Alkb-8 and TRM-112 bacterial expression vector was induced for 15 h at 16°C. Cells were then pelleted at 4000g for 10 min and resuspended in 4 mL of bacterial lysis buffer (20 mM Tris, 5% glycerol, 0.3%Triton, 1 mM DTT, 0.1 mM PMSF, 150 mM NaCl [200 mM for Human homologs], and 25 mM imidazole). Cells were lysed and proteins were purified as described above for γ-toxin.
RNA isolation
Wild-type and trm9Δ S. cerevisiae strains were obtained from the Sia Laboratory (University of Rochester). Colony PCR was performed to ensure the correct genotype. S. cerevisiae strains were grown until OD600 of 1.0 and total RNA was purified using the hot acid phenol technique (Collart and Oliviero 2001). E. coli RNA was extracted following the RNAsnap method (Stead et al. 2012). For Tetrahymena thermophila, human, and mouse samples, RNA was purified directly from cell pellets using TRIzol LS RNA extraction (Invitrogen). Tetrahymena strain SB210 were grown in standard proteose peptone growth media to mid-log phase before harvesting. Human embryonic kidney (HEK) 293T cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS), 1× antibiotics and 1× Glutamax at 37°C with 5% CO2. Mouse embryonic fibroblast (MEF) were grown in DMEM supplemented with 15% FBS at 37°C with 5% CO2.
Whole organism RNA extraction was performed on C. elegans and D. melanogaster. Vials containing adult D. melanogaster were frozen at −80°C for 10 min. Twenty-five milligrams of frozen adult flies were homogenized with a plastic pestle in a 1.5 mL microfuge tube while adding 250 µL of TRIzol reagent at a time until 1 mL was added. RNA was then extracted via an adapted TRIzol extraction protocol (Bogart and Andrews 2006). For extraction of C. elegans RNA, a confluent 60 mm plate of adult worms was scraped into a 1.5 mL microfuge tube and resuspended in 1 mL of RNase-free water. Tubes were centrifuged at 14,000g for 10 min and 750 µL water was removed. The worm pellet was resuspended in the remaining 250 µL water and processed using TRIzol LS.
For X. laevis samples, the liver from one adult X. laevis was surgically extracted, flash frozen in liquid nitrogen and stored immediately at −80°C. One quarter of an adult liver was used to extract RNA at a given time. While still frozen, one fourth of the liver was cut into multiple slices, transferred to a 1.5 mL microfuge tube and homogenized with 750 µL of TRIzol using a plastic pestle. Following this step, TRIzol purification was followed according to standard protocol.
γ-Toxin assay
For all species tested except for C. elegans, 5 µg of total RNA was incubated for 10 min at 30°C with increasing amounts of purified γ-toxin noted below or control eluate in 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 mM NaCl, and 1 mM dithiothreitol (DTT, pH 7.5) in a total volume of 15 µL. For C. elegans, 1 µg of total RNA was used. For assays with RNA from S. cerevisiae or C. elegans, the titration of γ-toxin ranged from 45, 112, and 224 nM. For assays with RNA from E. coli, D. melanogaster, M. musculus, or H. sapiens, the titration of γ-toxin ranged from 560, 1400, and 2800 nM. For RNA from T. thermophila or X. tropicalis, γ-toxin was used at a concentration of 293, 733, and 1466 nM. Each sample was run on a 10% polyacrylamide, 7 M urea gel and transferred to an Amersham Hybond-XL membrane (GE Healthcare). Oligonucleotides used to detect tRNAs are listed in Supplemental Table 1. The oligos were radiolabeled by T4 polynucleotide kinase (NEB) with adenosine [γ32P]-triphosphate (6000 Ci/mmol, Amersham Biosciences) following standard protocols. Northern blots were visualized by Phosphor-Imager analysis. Blots were subsequently stripped via two incubations at 80°C for 20 min in a buffer containing 0.15 M NaCl, 0.015 M Na-citrate, and 0.1% SDS.
Reconstitution and activity assay of Trm9–Trm112 homologs
Purified human and C. elegans Trm9–Trm112 homologs were mixed with 5 µg total RNA harvested from trm9Δ yeast deletion strain along with RNase inhibitor, 160 µM S-adenosylmethionine, 250 mM Tris (pH 7.5), 25 mM CaCl2, 400 µM NH4Ac, 500 µM MgCl2, and 100 µM EDTA. In place of purified protein, water was used as a control to ensure nonspecific cleavage was not occurring throughout the process. Reactions were incubated at 37°C for 2 h before being RNA purified through Zymo-Spin RNA Clean and Concentrator IC columns (Zymo Research). The γ-toxin assay was then performed on each purified RNA sample as described above using 45 nM of γ-toxin.
QRT-PCR assay
Five micrograms of total RNA underwent γ-toxin treatment as previously described with 45 nM γ toxin. RNA was extracted via the TRIzol method and resuspended in 15 µL of RNase-free water. One microgram of purified RNA was used to create cDNA using SuperScript IV reverse transcriptase (Thermo Fisher Scientific). cDNA was created for both tRNA-Glu for both yeast and human as well as an rRNA normalizing control (25S for yeast and 5.8S for human). RNA was first incubated with 10 mM dNTPs and 10 µM reverse primer (Listed in Supplemental Table 1). Reactions were incubated at 65°C for 5 min and then left on ice for 2 min before the addition of RNase Inhibitor (Thermo Fisher Scientific), 100 mM DTT, SuperScript IV reverse transcriptase and its accompanying buffer at 50°C for an hour followed by 80°C heat inactivation for 10 min. Each reaction was then cleaned up using the Qiagen PCR clean-up kit. 2 µL of cDNA was added to a reaction with SYBR Green Real-time PCR Master Mixes (Thermo Fisher) and 10 µM of forward and reverse primers (Supplemental Table 1). Quantitative PCR analysis was conducted with a Biorad PCR thermocycler. Each reaction was performed in triplicate. Relative tRNA levels were calculated using the ΔΔCt method and normalized to either yeast 25S rRNA or human 5.8S rRNA.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stewart Shuman (Memorial Sloan Kettering) for the γ-toxin expression plasmid, Elaine Sia and Christopher Prevost (University of Rochester) for yeast strains, Jacques Robert (University of Rochester Medical Center) for X. laevis liver samples, Douglas Portman (University of Rochester Medical Center) for C. elegans, Danna Eickbush and Amanda Larracuente (University of Rochester) for D. melanogaster, Suzanne Lee (Western Washington University) for Tetrahymena thermophila cells, and Yan Li for cloning of C. elegans Trm9–Trm112. This work was supported by the National Science Foundation, USA (NSF CAREER Award 1552126 to D.F.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065581.118.
REFERENCES
- Agris PF, Eruysal ER, Narendran A, Vare VYP, Vangaveti S, Ranganathan SV. 2017. Celebrating wobble decoding: half a century and still much is new. RNA Biol: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bär C, Zabel R, Liu S, Stark MJ, Schaffrath R. 2008. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol Microbiol 69: 1221–1233. [DOI] [PubMed] [Google Scholar]
- Basanta-Sanchez M, Temple S, Ansari SA, D'Amico A, Agris PF. 2016. Attomole quantification and global profile of RNA modifications: epitranscriptome of human neural stem cells. Nucleic Acids Res 44: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F, Matsuyama A, Candiracci J, Dieu M, Scheliga J, Wolf DA, Yoshida M, Hermand D. 2012. Translational control of cell division by Elongator. Cell Rep 1: 424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. 2007. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 28: 860–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begley U, Sosa M, Avivar-Valderas A, Patil A, Endres L, Estrada Y, Chan C, Su D, Dedon P, Aguirre-Ghiso J, et al. 2013. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-α. EMBO Mol Med 5: 366–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Helm M, Motorin Y. 2011. Use of specific chemical reagents for detection of modified nucleotides in RNA. J Nucleic Acids 2011: 408053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Huang B, Persson OP, Byström AS. 2007. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 13: 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogart K, Andrews J. 2006. Extraction of total RNA from Drosophila. CGB Technical Report 10: 1–4. [Google Scholar]
- Bourgeois G, Létoquart J, van Tran N, Graille M. 2017. Trm112, a protein activator of methyltransferases modifying actors of the eukaryotic translational apparatus. Biomolecules 7: E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WM, Chionh YH, Hia F, Gu C, Kellner S, McBee ME, Ng CS, Pang YL, Prestwich EG, Lim KS, et al. 2015. A platform for discovery and quantification of modified ribonucleosides in RNA: application to stress-induced reprogramming of tRNA modifications. Methods Enzymol 560: 29–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Limbach PA. 2015. Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LC-MS/MS. Anal Chem 87: 8433–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. 2010. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 6: e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tuck S, Byström AS. 2009. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 5: e1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Anderson JT, Byström AS. 2011a. Unexpected accumulation of ncm5U and ncm5S2U in a trm9 mutant suggests an additional step in the synthesis of mcm5U and mcm5S2U. PLoS One 6: e20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Eliasson M, Ryden P, Byström AS. 2011b. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet 7: e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S. 2001. Preparation of yeast RNA. Curr Protoc Mol Biol Chapter 13: Unit 13.12. [DOI] [PubMed] [Google Scholar]
- Dauden MI, Jaciuk M, Muller CW, Glatt S. 2017. Structural asymmetry in the eukaryotic Elongator complex. FEBS Lett. 10.1002/1873-3468.12865. [DOI] [PubMed] [Google Scholar]
- Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. 2015. Trm9-catalyzed tRNA modifications regulate global protein expression by codon-biased translation. PLoS Genet 11: e1005706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. 2008. The conserved Wobble uridine tRNA thiolase Ctu1–Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci 105: 5459–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres L, Begley U, Clark R, Gu C, Dziergowska A, Malkiewicz A, Melendez JA, Dedon PC, Begley TJ. 2015. Alkbh8 regulates selenocysteine-protein expression to protect against reactive oxygen species damage. PLoS One 10: e0131335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Byström AS. 2006. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 24: 139–148. [DOI] [PubMed] [Google Scholar]
- Fernández-Vázquez J, Vargas-Pérez I, Sansó M, Buhne K, Carmona M, Paulo E, Hermand D, Rodríguez-Gabriel M, Ayté J, Leidel S, et al. 2013. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 9: e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner L, Schaffrath R. 2002. KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol Microbiol 44: 865–875. [DOI] [PubMed] [Google Scholar]
- Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. 2001. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J 20: 1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. 2010a. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 30: 2449–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dai Q, Zhang W, Ren J, Pan T, He C. 2010b. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew Chem Int Ed Engl 49: 8885–8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, Keith G, Droogmans L. 2004. Detection and quantification of modified nucleotides in RNA using thin-layer chromatography. Methods Mol Biol 265: 357–391. [DOI] [PubMed] [Google Scholar]
- Grosjean H, Droogmans L, Roovers M, Keith G. 2007. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol 425: 55–101. [DOI] [PubMed] [Google Scholar]
- Grosjean H, Gaspin C, Marck C, Decatur WA, de Crécy-Lagard V. 2008. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics 9: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H, de Crécy-Lagard V, Marck C. 2010. Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett 584: 252–264. [DOI] [PubMed] [Google Scholar]
- Helm M, Motorin Y. 2017. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet 18: 275–291. [DOI] [PubMed] [Google Scholar]
- Hiley SL, Jackman J, Babak T, Trochesset M, Morris QD, Phizicky E, Hughes TR. 2005. Detection and discovery of RNA modifications using microarrays. Nucleic Acids Res 33: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Byström AS. 2005. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 11: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Lu J, Byström AS. 2008. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 14: 2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL. 1992. Affinity electrophoretic detection of primary amino groups in nucleic acids: application to modified bases of tRNA and to aminoacylation. Anal Biochem 206: 363–368. [DOI] [PubMed] [Google Scholar]
- Jablonowski D, Schaffrath R. 2007. Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem Soc Trans 35: 1533–1537. [DOI] [PubMed] [Google Scholar]
- Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. 2006. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 59: 677–688. [DOI] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Björk GR, Byström AS. 2008. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 28: 3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jüdes A, Bruch A, Klassen R, Helm M, Schaffrath R. 2016. Sulfur transfer and activation by ubiquitin-like modifier system Uba4*Urm1 link protein urmylation and tRNA thiolation in yeast. Microb Cell 3: 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor HR, Clarke S. 2003. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 23: 9283–9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Suzuki T, Kapushoc ST, Rubio MA, Ghazvini J, Watanabe K, Simpson L, Suzuki T. 2003. Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J 22: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsborn T, Tükenmez H, Chen C, Byström AS. 2014a. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem Biophys Res Commun 454: 441–445. [DOI] [PubMed] [Google Scholar]
- Karlsborn T, Tükenmez H, Mahmud AK, Xu F, Xu H, Byström AS. 2014b. Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol 11: 1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsborn T, Mahmud A, Tükenmez H, Byström AS. 2016. Loss of ncm5 and mcm5 wobble uridine side chains results in an altered metabolic profile. Metabolomics 12: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith G. 1984. The primary structures of two arginine tRNAs (anticodons C-C-U and mcm5a2U-C-ψ) and of glutamine tRNA (anticodon C-U-G) from bovine liver. Nucleic Acids Res 12: 2543–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Jain R, Meineke B, Diver M, Shuman S. 2009. Structure-activity relationships in Kluyveromyces lactis γ-toxin, a eukaryal tRNA anticodon nuclease. RNA 15: 1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Grunewald P, Thuring KL, Eichler C, Helm M, Schaffrath R. 2015. Loss of anticodon wobble uridine modifications affects tRNALys function and protein levels in Saccharomyces cerevisiae. PLoS One 10: e0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Bruch A, Schaffrath R. 2016a. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol 14: 1252–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen R, Ciftci A, Funk J, Bruch A, Butter F, Schaffrath R. 2016b. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res 44: 10946–10959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic M, Wainwright B. 2016. The many faces of elongator in neurodevelopment and disease. Front Mol Neurosci 9: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaj-Robin O, Seraphin B. 2017. Structures and activities of the elongator complex and its cofactors. Enzymes 41: 117–149. [DOI] [PubMed] [Google Scholar]
- Kuntzel B, Weissenbach J, Wolff RE, Tumaitis-Kennedy TD, Lane BG, Dirheimer G. 1975. Presence of the methylester of 5-carboxymethyl uridine in the wobble position of the anticodon of tRNAIII Arg from brewer's yeast. Biochimie 57: 61–70. [DOI] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. 2009. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458: 228–232. [DOI] [PubMed] [Google Scholar]
- Leihne V, Kirpekar F, Vågbø CB, van den Born E, Krokan HE, Grini PE, Meza TJ, Falnes PO. 2011. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res 39: 7688–7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J, Retzer K, Malenica N, Bartkeviciute R, Lucyshyn D, Jager G, Korbei B, Byström A, Luschnig C. 2015. Meta-regulation of Arabidopsis auxin responses depends on tRNA maturation. Cell Rep 11: 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letoquart J, van Tran N, Caroline V, Aleksandrov A, Lazar N, van Tilbeurgh H, Liger D, Graille M. 2015. Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res 43: 10989–11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liger D, Mora L, Lazar N, Figaro S, Henri J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgue-Hamard V, Graille M. 2011. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res 39: 6249–6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Shen L, Jang CW, Falnes PO, Zhang Y. 2013. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet 9: e1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johansson MJ, Byström AS. 2005. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 11: 1648–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Esberg A, Huang B, Byström AS. 2008. Kluyveromyces lactis γ-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 36: 1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazauric MH, Dirick L, Purushothaman SK, Björk GR, Lapeyre B. 2010. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem 285: 18505–18515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Breunig KD, Stark MJ, Schaffrath R. 2009. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol 73: 869–881. [DOI] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jager G, Gong Z, Byström AS, Schaffrath R, Breunig KD. 2010. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 76: 1082–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C, Prochaska H, Hammermeister A, Abdel-Fattah W, Wagner M, Krutyholowa R, Jun SE, Kim GT, Glatt S, Breunig KD, et al. 2017. Use of a yeast tRNase killer toxin to diagnose Kti12 motifs required for tRNA modification by elongator. Toxins (Basel) 9: E272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Muller S, Behm-Ansmant I, Branlant C. 2007. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol 425: 21–53. [DOI] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Yano T. 2017. Sulfur modifications of the wobble U34 in tRNAs and their intracellular localization in eukaryotic cells. Biomolecules 7: E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. 2008. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol Cell 31: 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naor A, Thiaville PC, Altman-Price N, Cohen-Or I, Allers T, de Crecy-Lagard V, Gophna U. 2012. A genetic investigation of the KEOPS complex in halophilic Archaea. PLoS One 7: e43013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, Suzuki T. 2009. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 37: 1335–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett LR, Lentini JM, Holmes MJ, Stilger KL, Fu D, Sullivan WJ Jr. 2018. Elp3 and RlmN: a tale of two mitochondrial tail-anchored radical SAM enzymes in Toxoplasma gondii. PLoS One 13: e0189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore C, Topalidou I, Forouhar F, Yan AC, Levy M, Hunt JF. 2012. Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification. J Biol Chem 287: 2130–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Chan CT, Dyavaiah M, Rooney JP, Dedon PC, Begley TJ. 2012a. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol 9: 990–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A, Dyavaiah M, Joseph F, Rooney JP, Chan CT, Dedon PC, Begley TJ. 2012b. Increased tRNA modification and gene-specific codon usage regulate cell cycle progression during the DNA damage response. Cell cycle 11: 3656–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G, de Crecy-Lagard V. 2011. Biosynthesis and function of tRNA modifications in Archaea. Curr Opin Microbiol 14: 335–341. [DOI] [PubMed] [Google Scholar]
- Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG. 2013. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci 110: 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R, Cao X, Yu N, Limbach PA. 2016. Sequence mapping of transfer RNA chemical modifications by liquid chromatography tandem mass spectrometry. Methods 107: 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffrath R, Klassen R. 2017. Combined tRNA modification defects impair protein homeostasis and synthesis of the yeast prion protein Rnq1. Prion 11: 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffrath R, Leidel SA. 2017. Wobble uridine modifications—a reason to live, a reason to die? RNA Biol 14: 1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Motorin Y. 2016. Next-generation sequencing technologies for detection of modified nucleotides in RNAs. RNA Biol 14: 1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songe-Møller L, van den Born E, Leihne V, Vågbø CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PØ, Klungland A. 2010. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Molecular and cellular biology 30: 1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead MB, Agrawal A, Bowden KE, Nasir R, Mohanty BK, Meagher RB, Kushner SR. 2012. RNA snap™: a rapid, quantitative and inexpensive, method for isolating total RNA from bacteria. Nucleic Acids Res 40: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studte P, Zink S, Jablonowski D, Bär C, von der Haar T, Tuite MF, Schaffrath R. 2008. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol 69: 1266–1277. [DOI] [PubMed] [Google Scholar]
- Su D, Chan CT, Gu C, Lim KS, Chionh YH, McBee ME, Russell BS, Babu IR, Begley TJ, Dedon PC. 2014. Quantitative analysis of ribonucleoside modifications in tRNA by HPLC-coupled mass spectrometry. Nat Protoc 9: 828–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towns W, Begley T. 2012. Transfer RNA methytransferases and their corresponding modifications in budding yeast and humans: activities, predications, and potential roles in human health. DNA Cell Biol 31: 434–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tükenmez H, Xu H, Esberg A, Byström AS. 2015. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res 43: 9489–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born E, Vågbø CB, Songe-Møller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, et al. 2011. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun 2: 172. [DOI] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Kataoka N, Miyauchi K, Ohe K, Iida K, Yoshida S, Nojima T, Okuno Y, Onogi H, Usui T, et al. 2015. Rectifier of aberrant mRNA splicing recovers tRNA modification in familial dysautonomia. Proc Natl Acad Sci 112: 2764–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdżalik D, Vågbø CB, Kirpekar F, Davydova E, Puścian A, Maciejewska AM, Krokan HE, Klungland A, Tudek B, van den Born E, et al. 2014. Protozoan ALKBH8 oxygenases display both DNA repair and tRNA modification activities. PLoS One 9: e98729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinshteyn B, Gilbert WV. 2013. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 9: e1003675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.