Ferrer et al. demonstrate that a homozygous inactivating mutation in the histone deacetylase SIRT6 results in severe congenital anomalies and perinatal lethality in four affected fetuses. Human induced pluripotent stem cells derived from D63H homozygous fetuses fail to differentiate into embryoid bodies, functional cardiomyocytes, and neural progenitor cells due to a failure to repress pluripotent genes.
Keywords: chromatin, embryonic development, epigenetics, human syndrome, SIRT6
Abstract
It has been well established that histone and DNA modifications are critical to maintaining the equilibrium between pluripotency and differentiation during early embryogenesis. Mutations in key regulators of DNA methylation have shown that the balance between gene regulation and function is critical during neural development in early years of life. However, there have been no identified cases linking epigenetic regulators to aberrant human development and fetal demise. Here, we demonstrate that a homozygous inactivating mutation in the histone deacetylase SIRT6 results in severe congenital anomalies and perinatal lethality in four affected fetuses. In vitro, the amino acid change at Asp63 to a histidine results in virtually complete loss of H3K9 deacetylase and demyristoylase functions. Functionally, SIRT6 D63H mouse embryonic stem cells (mESCs) fail to repress pluripotent gene expression, direct targets of SIRT6, and exhibit an even more severe phenotype than Sirt6-deficient ESCs when differentiated into embryoid bodies (EBs). When terminally differentiated toward cardiomyocyte lineage, D63H mutant mESCs maintain expression of pluripotent genes and fail to form functional cardiomyocyte foci. Last, human induced pluripotent stem cells (iPSCs) derived from D63H homozygous fetuses fail to differentiate into EBs, functional cardiomyocytes, and neural progenitor cells due to a failure to repress pluripotent genes. Altogether, our study described a germline mutation in SIRT6 as a cause for fetal demise, defining SIRT6 as a key factor in human development and identifying the first mutation in a chromatin factor behind a human syndrome of perinatal lethality.
Perinatal mortality is defined as intrauterine fetal demise or neonatal death until 1–4 wk after birth (Boyle et al. 2018). In this context, congenital cardiac and neural tube defects account for 21% of late stage fetal or neonatal loss (GBD 2013 Mortality and Causes of Death Collaborators). Chromosomal aberrations commonly contributing to fetal demise are routinely identified via karyotype or comparative genome hybridization (CGH) array analysis. However, these techniques do not identify single-nucleotide changes; thus, many affected pregnancies remain without a genetic diagnosis (Yang et al. 2013; Lamont et al. 2015).
Epigenetic modification patterns on DNA and histones play a critical role in altering global gene expression during early embryogenesis and fetal development (Nafee et al. 2008; Chen and Dent 2014). Inactivating mutations in DNA methyltransferases (DNMTs) such as DNMT3a and MECP2 (Rett syndrome) are associated with severe congenital immunological, craniofacial, and neurodevelopmental disorders (Nafee et al. 2008). Loss-of-function mutations in the chromatin regulator histone methyltransferase NSD1 result in Sotos syndrome, a disease characterized by physical overgrowth and macrocephaly (Douglas et al. 2003; Rayasam et al. 2003). Inherited mutations in the class I histone deacetylase HDAC8 contribute to the pathologies observed in Cornelia de Lange syndrome, with the majority of children living into adulthood (Kaiser et al. 2014; Feng et al. 2015). However, whether mutations in chromatin regulators contribute to embryonic or fetal demise in utero remains unknown.
The NAD+-dependent histone deacetylase SIRT6 is a chromatin-associated protein that targets H3K9ac and H3K56ac and is known to play roles in genomic stability, glucose homeostasis, and tumor suppression (Mostoslavsky et al. 2006; Michishita et al. 2009; Yang et al. 2009; Zhong et al. 2010; Sebastián et al. 2012; Toiber et al. 2013; Kugel et al. 2015, 2016). Sirt6 knockout mice from the 129SvJ background are viable and born at Mendelian ratios; however, they are much smaller than wild-type littermates (Mostoslavsky et al. 2006). At 2–3 wk of age, Sirt6 knockout mice demonstrate an overall failure to thrive, accompanied by loss of subcutaneous fat, osteopenia, and profound hypoglycemia, which eventually leads to their death at 4 wk of age (Mostoslavsky et al. 2006). Interestingly, Sirt6 knockout mice in the C57/B6 background display a strain-dependent embryonic-lethal phenotype (J-P Etchegaray and R Mostoslavsky, unpubl.), suggesting a role for SIRT6 during embryonic development. Indeed, our previous studies have shown that SIRT6 directly represses expression of core pluripotent genes, which in turn controls embryonic stem cell (ESC) differentiation through Tet-mediated production of 5-hydroxymethylcytosine (5hmC) specifically on neuroectoderm genes (Etchegaray et al. 2015). However, the role that SIRT6 plays in human embryonic and perinatal development is still unclear.
In this study, we identify a family presenting with four cases of perinatal loss consistent with a number of severe neurodevelopmental and cardiac anomalies. Through whole-exome sequencing (WES) and Sanger sequencing, we detected a single homozygous mutation in SIRT6 (c.187G > C [p.Asp63His]; NM_016539.3) in all of the affected fetuses. In vitro experiments indicate that this D63H mutation renders SIRT6 inactive on chromatin targets. Functional tests using induced pluripotent stem cells (iPSCs) derived from affected individuals and mouse-derived ESCs expressing the SIRT6 D63H mutation demonstrate a failure to suppress pluripotent gene expression and major defects in the ability of cells carrying the mutant SIRT6 to differentiate into embryoid bodies (EBs), cardiomyocytes, and neural progenitor cells (NPCs); altogether, these results identify a human congenital syndrome of SIRT6 loss-of-function mutation—the first such case for a chromatin factor involved in perinatal lethality.
Results
WES identifies a homozygous SIRT6 187G>C mutation in four affected fetuses
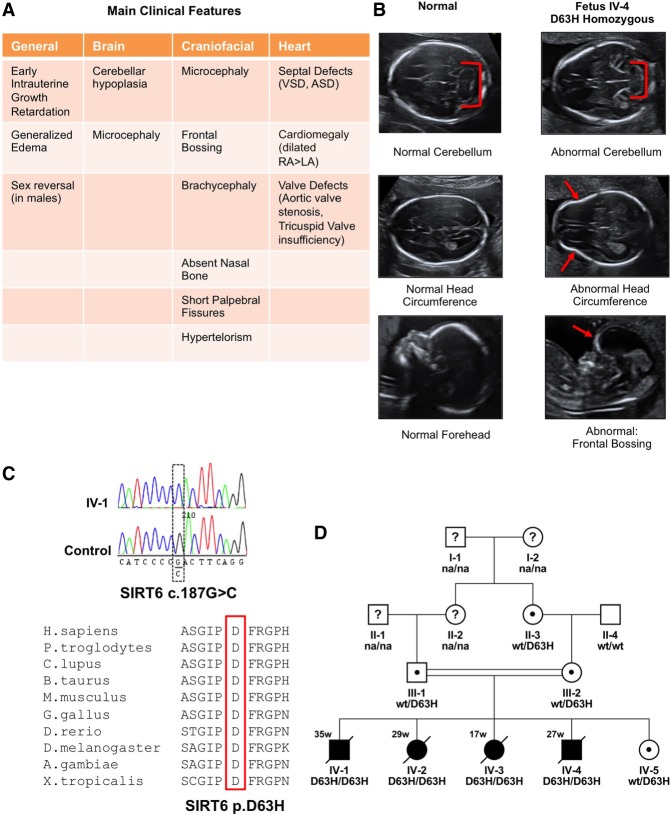
A consanguineous couple presented with four consecutive cases of late fetal loss with gestational ages ranging between 17 and 35 wk. Ultrasound imaging revealed a number of prenatal abnormalities, including intrauterine growth restriction (IUGR), microcephaly, craniofacial anomalies, sex reversal in male fetuses, and congenital heart defects (Fig. 1A,B). Cephalic and craniofacial fetal abnormalities included cerebellar hypoplasia, decreased head circumference, and frontal bossing. Moreover, all fetuses had an atypically shaped head (Fig. 1B). In the first trimester, an increased nuchal translucency was also noted. Cardiac insufficiencies included septal defects, cardiomegaly, and valve defects (Fig. 1A). All of these anomalies have been associated with chromosomal anomalies (Uchida et al. 1962; Gibson et al. 1963; Pandya et al. 1995; Srinivasan and Wright 2014).
Figure 1.
WES identifies the homozygous c.187G > C; p.(Asp63His) SIRT6 mutation in four fetuses with severe congenital defects and perinatal lethality. (A) Description of clinical findings of fetuses with SIRT6 mutation. (B) Representative ultrasound images of abnormal cerebella, head circumferences, and frontal bossing observed in four affected fetuses. (C) Sanger sequencing confirms the SIRT6 c.187G > C missense mutation in affected fetal amniocytes corresponding to SIRT6 p.D63H. Protein sequence alignment of various species for the area around the D63 amino acid showing strong evolutionary conservation at this position. (D) Pedigree showing consanguinity and cosegregation of the SIRT6 p.D63H mutation. (na) Genotype not available; (black symbols) affected; (white symbols) unaffected; (?) unknown phenotype; (dots) carriers of mutation.
Notably, CGH array analysis revealed no shared relevant copy number variants (data not shown) or major chromosomal aberrations; however, WES performed on amniocytes isolated from four fetuses identified a homozygous missense mutation (c.187G > C; p.[Asp63His]) in the SIRT6 gene (Supplemental Fig. S1A). This mutation affects a highly conserved residue (Fig. 1C) and is predicted to be damaging by multiple variant effect prediction tools (polyphen, SIFT, SNPdryad, and CADD). It is present at very low frequency in the general population (two out of 241,948 alleles in the Genome Aggregation Database; gnomAD; http://gnomad.broadinstitute.org) in a heterozygous form but has never been found before in an homozygous state. Sanger capillary sequencing confirmed the presence of this variant in the four fetuses (Fig. 1C,D). Both parents and one grandparent were tested and found to be heterozygous carriers (Fig. 1D). The fifth pregnancy presented with a healthy neonate identified as a heterozygous carrier for SIRT6 (c.187G > C; p.[Asp63His]) (Fig. 1D). The variant segregated among family members in a manner consistent with an autosomal recessive trait (Fig. 1D).
SIRT6 D63H mutation results in loss of deacetylase and demyristoylase activity
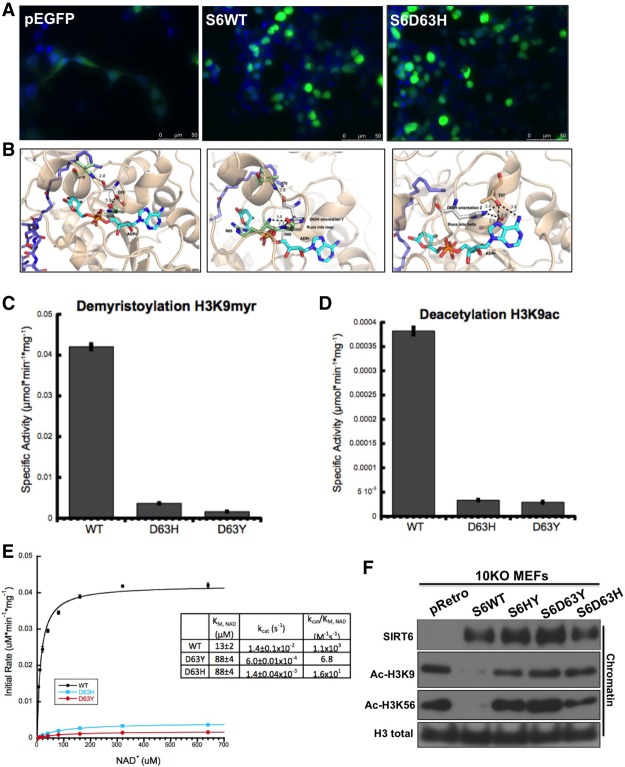
We first investigated the consequences of the Asp63His (D63H) mutation on SIRT6 protein localization and function. We generated the D63H SIRT6 mutation using site-directed mutagenesis in a GFP-tagged construct. Upon fluorescence imaging, D63H SIRT6 demonstrated nuclear localization similar to wild-type SIRT6 (Fig. 2A). To gain further insight into the functional significance of the D63H mutation, we analyzed in silico the cocrystal structure of SIRT6 bound to ADP-ribose and the H3K9 myristoylated peptide (Protein Data Bank [PDB] 3ZG6) (Fig. 2B; Jiang et al. 2013; Kugel et al. 2015). Asp63 is located in the NAD+-binding pocket, forming hydrogen bonds with neighboring amino acids and thus providing structure to the NAD+-binding loop (Jiang et al. 2013). Similar to the cancer-associated Asp63 mutation to tyrosine (D63Y) (Kugel et al. 2015), the D63H mutation on SIRT6 should hinder NAD+ binding and catalysis (Fig. 2B). In vitro deacetylase assays using purified recombinant wild-type, D63Y, and D63H SIRT6 demonstrated that the mutations almost completely abolished the enzymatic activity of SIRT6 toward a H3K9ac peptide (Fig. 2C). Next, we wanted to determine whether SIRT6 D63H retains the previously reported demyristoylase activity (Jiang et al. 2013). Consistently, D63Y and D63H recombinant SIRT6 mutants displayed decreased demyristoylase activity when compared with wild-type SIRT6 (Fig. 2D). To test whether D63H mutation decreases SIRT6 affinity for NAD+, we measured steady-state rates of demyristoylation with increasing concentrations of NAD+. Steady-state kinetics revealed that the kcat/Km value for D63H mutant is 688 times less than wild-type SIRT6 (Fig. 2E). The Km for NAD+ for both D63Y and D63H is increased 6.8-fold, suggesting a decreased affinity for NAD+ (Fig. 2E). Last, we performed site-directed mutagenesis and overexpressed doxycycline-inducible SIRT6 constructs in SIRT6 knockout mouse embryonic fibroblasts (MEFs) to determine whether the SIRT6 D63H mutant is able to deacetylate H3K9 and H3K56 in vivo. Notably, the D63H mutant was unable to deacetylate its targets, as clearly seen even on bulk chromatin (Fig. 2F). Together, these results indicate that a mutation of Asp63 to histidine inhibits the affinity of SIRT6 for NAD+ and therefore significantly reduces its catalytic activity both in vitro and on bulk chromatin in vivo.
Figure 2.
The SIRT6 D63H mutant localizes to the nucleus but is catalytically inactive both in vitro and in vivo. (A) Subcellular localization of wild-type and mutant SIRT6 proteins. Fluorescent images of fixed 293T cells transiently transfected with the indicated eGFP-tagged SIRT6 deletion and point mutants are shown. Nuclei were costained with 4′,6′-diamidino-2-phenylindole (DAPI; 20×). (B) Locations of mutated residues mapped on the crystal structure of SIRT6 (PDB 3GZ6). (C) SIRT6 demyristoylase activity measured in vitro. Wild-type and SIRT6 mutants (0.5 mM) were incubated with 50 mM H3K9Myr peptide and 0.5 mM NAD+. (D) SIRT6 deacetylase activity measured in vitro. Recombinant wild-type and SIRT6 mutants (2 mM) were incubated with 50 mM H3K9ac peptide and 0.5 mM NAD+. (E) Steady-state rates of demyristoylation were measured by varying NAD+ (5–640 µM) in the presence of 0.5 mM wild-type and SIRT6 mutants and 50 mM H3K9Myr peptide. Calculated values determined from nonlinear regression fits to Michaelis-Menten are shown below for wild-type, D63Y, and D63H (n = 3; ±SD). (F) Western blot of chromatin fractions in SIRT6 knockout MEFs with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 mutants.
SIRT6 D63H mutant mESCs fail to form EBs and retain pluripotent gene expression
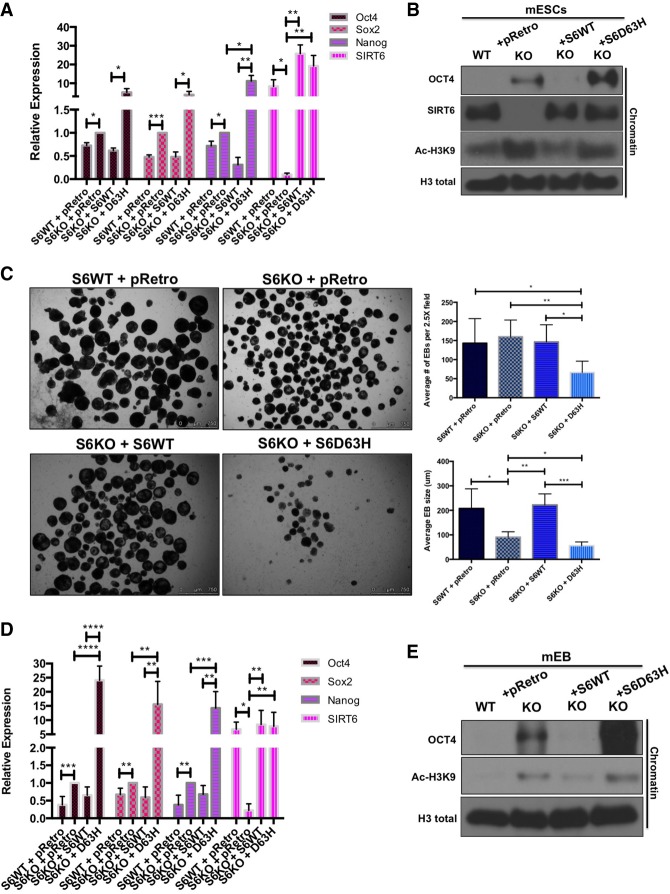
An overall down-regulation of the core pluripotent genes is essential for proper ESC differentiation (Young et al. 2011). We found previously that SIRT6 acts as a histone deacetylase to directly silence pluripotent genes, and SIRT6-deficient EB cells fail to repress pluripotent genes, significantly altering normal development of the three germ layers during differentiation (Etchegaray et al. 2015). To functionally test whether the SIRT6 D63H mutation is causing an aberrant developmental phenotype, we first measured the expression of pluripotent genes in SIRT6 knockout mouse ESCs (mESCs) stably transduced with doxycycline-inducible SIRT6 wild-type and D63H constructs. Interestingly, quantitative RT–PCR (qRT–PCR) results revealed that D63H mESCs express significantly higher levels of the core pluripotent genes Oct4, Sox2, and Nanog compared with SIRT6 knockout and wild-type reconstituted mESCs (Fig. 3A). Additionally, we observed an increased presence of the pluripotent factor Oct4 on bulk chromatin in D63H mutant mESCs when compared with SIRT6 knockout mESCs (Fig. 3B). This result was accompanied by a failure to deacetylate H3K9, a SIRT6 target (Fig. 3B).
Figure 3.
SIRT6 D63H-expressing mESCs fail to differentiate into EBs and suppress pluripotent genes. (A) Core pluripotent gene expression in wild-type versus SIRT6 knockout (S6KO) ESCs with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 D63H assessed by qRT–PCR analysis. Data are expressed relative to knockout mESCs. (B) Western blot analysis on bulk chromatin for Oct4, SIRT6, and Ac-H3K9 in wild-type versus SIRT6 knockout ESCs with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 D63H. (C, left panel) EBs derived from wild-type versus SIRT6 knockout ESCs with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 D63H (2.5×). (Right panel) Quantified average EB size and number. (D) Core pluripotent gene expression in wild-type versus SIRT6 knockout EBs with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 D63H assessed by qRT–PCR analysis. Data are expressed relative to knockout mouse EBs. (E) Western blot analysis on bulk chromatin for Oct4 and Ac-H3K9 in wild-type versus SIRT6 knockout EBs with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 D63H.
SIRT6 knockout ESCs cultured to form EBs are significantly smaller than their wild-type counterparts (Etchegaray et al. 2015). Therefore, we tested the ability of D63H reconstituted SIRT6 knockout mESCs to differentiate in vitro into EBs. As demonstrated previously, expression of wild-type SIRT6 in SIRT6 knockout mESCs is sufficient to rescue the SIRT6 knockout EB phenotype (Etchegaray et al. 2015, Fig. 3C). Interestingly, we observed that stable expression of SIRT6 D63H leads to an even more severe differentiation defect with a significantly smaller and decreased number of EBs compared with SIRT6 knockout ESCs (Fig. 3C). This phenotype was associated with a significant derepression of pluripotent genes in the context of the mutant SIRT6 (Fig. 3D), increased binding of pluripotent factors to chromatin, and a failure to deacetylate H3K9 (Fig. 3E). Such changes were significantly worse than in SIRT6 knockout cells, suggesting that the presence of the mutant protein could be biologically worse than the absence of the enzyme (see the Discussion below). Interestingly, shRNA-mediated knockdown of Oct4 and SOX2 in SIRT6 knockout mESCs reconstituted with D63H SIRT6 (Supplemental Fig. S2B,C) resulted in a partial rescue of the EB defect observed in control shRNA-treated cells (Supplemental Fig. S2A). Together, the data suggest that the D63H mutant SIRT6 severely impairs mESC differentiation due to a failure to suppress pluripotent genes.
The pluripotent factor Sox2 is responsible for promoting neuroectoderm differentiation (Wang et al. 2012). Previously, our laboratory had demonstrated that SIRT6 knockout mESCs display a differentiation phenotype skewed toward the neuroectoderm lineage compared with wild-type counterparts (Etchegaray et al. 2015). D63H EBs showed significantly elevated expression of SOX2 consistent with increases in neuroectoderm markers as measured by increased Nestin, Pax6, and Hoxa7 compared with SIRT6 knockout EBs (Supplemental Fig. S1B), without compromising mesoderm and endoderm markers (Supplemental Fig. S1C). Interestingly, combined knockdown of Oct4 and Sox2 was able to reverse the increase in neuroectoderm markers in SIRT6 D63H EBs (Supplemental Fig. S2D), together indicating that sustained SOX2 expression skews D63H EBs further toward the neuroectoderm lineage.
SIRT6 D63H mutant mESCs fail to differentiate into functional cardiomyocyte foci
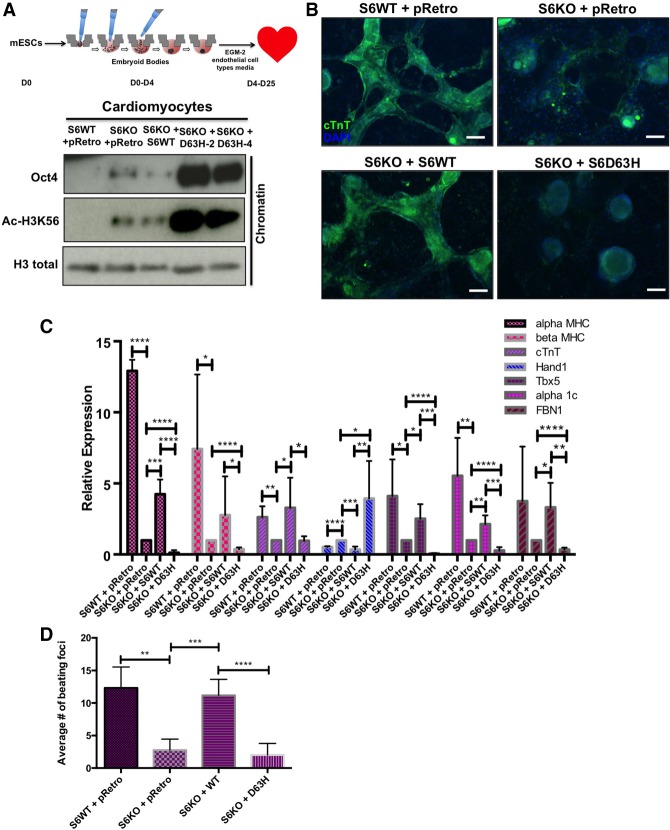
SIRT6 in the adult heart has been shown to protect against concentric cardiac hypertrophy (Sundaresan et al. 2012). However, the role of SIRT6 in cardiac development and morphogenesis still remains to be investigated. Analysis of SIRT6 knockout mESC-derived cardiomyocytes revealed decreased expression of cardiomyogenesis markers, a phenotype associated with irregular beating patterns and a decreased number of beating foci when compared with wild-type SIRT6 cardiomyocytes (data not shown). Since a number of cardiac anomalies were identified in D63H homozygote fetuses (Fig. 1A), we tested the capacity of D63H mutant mESCs to differentiate into functional cardiomyocytes. To address this question, we used an EB-directed differentiation protocol in mESCs to aid in mesoderm layer specification (Lee et al. 2011). From day 0 to day 4, mESCs were cultured under hanging drop conditions as EBs. Subsequently, from day 4 to day 25, cells were placed in EGM-2 (endothelial cell type) medium containing growth factors essential for normal cardiac development (Fig. 4A, top panel; Lee et al. 2011). On day 25, wild-type SIRT6 differentiated cardiomyocytes morphologically resembled cardiomyocyte foci and exhibited regular spontaneous contraction patterns (Supplemental Fig. S3A; Supplemental Movies S1–S4). SIRT6 knockout ESCs, on the other hand, formed less differentiated cardiomyocyte foci, a phenotype rescued with the re-expression of wild-type SIRT6 but not with expression of the mutant D63H protein (Supplemental Fig. S3A; Supplemental Movies S1–S4). This severe phenotype was accompanied by a dramatic derepression of Oct4 and increased acetylation of the SIRT6 target H3K56 when the mutant protein was expressed (Fig. 4A, bottom panel). Immunofluorescence staining for the cardiac differentiation marker cardiac troponin T (cTnT) demonstrated intense staining in SIRT6 wild-type cardiomyocytes when compared with SIRT6 knockout, which exhibited weak staining and aberrant morphology (Fig. 4B). cTnT staining was rescued when wild-type SIRT6 was reconstituted in SIRT6 knockout cardiomyocytes (Fig. 4B). However, D63H-expressing mouse cardiomyocytes failed to stain for cTnT, morphologically remaining as EBs (Fig. 4B). These data indicate that expression of SIRT6 D63H suppresses cardiomyocyte differentiation, likely through failure to suppress Oct4 expression.
Figure 4.
D63H mutant mESCs fail to differentiate into functional cardiomyocyte foci and retain pluripotent gene expression. (A, top panel) Schematic showing the differentiation from mESCs into cardiomyocytes. (Bottom panel) Western blot analysis on bulk chromatin for Oct4 and Ac-H3K56 in wild-type versus SIRT6 knockout ESC-derived cardiomyocytes with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 D63H. (B) Immunofluorescence staining for cTnT in mESC-derived cardiomyocytes (20×). (C) Cardiac marker expression in wild-type versus SIRT6 knockout differentiated cardiomyocytes with doxycycline-inducible overexpression of wild-type SIRT6 and SIRT6 D63H assessed by qRT–PCR analysis. Data are expressed relative to knockout cardiomyocytes. (D) Spontaneous contractile foci were quantified on day 21 as the average number of foci from five clones.
Next, we sought to characterize molecular signatures of SIRT6 D63H mESC-derived cardiomyocytes. SIRT6 wild-type and SIRT6 knockout cardiomyocytes reconstituted with wild-type SIRT6 exhibit expression of cardiac sarcomeric genes αMHC, βMHC, and cTnT, indicating fully differentiated foci (Fig. 4C; Lian et al. 2012). Interestingly, SIRT6 D63H-expressing cardiomyocytes expressed significantly lower levels of αMHC and βMHC when compared with SIRT6 knockout cardiomyocytes (Fig. 4C), consistent with a failure to differentiate. The transcription factor TBX5 is an important regulator of cardiomyocyte development, and decreased levels are associated with ventral septal defects (VSDs) and atrial septal defects (ASDs) (Horb and Thomsen 1999; Posch et al. 2010). Tbx5 expression was decreased in SIRT6 knockout cardiomyocytes compared with wild-type counterparts (Fig. 4C). However, TBX5 expression was further compromised in D63H-expressing cardiomyocytes compared with knockout alone (Fig. 4C), consistent with septal defects observed in all four affected fetuses. Sustained HAND1 expression and decreased FBN1 expression are associated with delayed ventricle morphogenesis, aortic valve thickening (Vincentz et al. 2011), and tricuspid valve prolapse (Lima et al. 2010), respectively. Gene expression analysis indicated that SIRT6 D63H mutant cardiomyocytes failed to suppress HAND1 expression while exhibiting significantly reduced FBN1 levels when compared with SIRT6 knockout cells (Fig. 4C), in line with the ventricular and cardiac valve defects observed in the D63H homozygous fetuses (Fig. 4C). Thus, molecular characterization of SIRT6 D63H mESCs indicates a failure to differentiate and express cardiac morphogenesis markers, likely due to a failure to repress pluripotent gene expression.
To test the functional significance of the D63H mutation on cardiomyocyte differentiation, we quantified the number of spontaneously contractile foci in SIRT6 D63H cardiomyocytes. SIRT6 D63H cardiomyocytes formed significantly less spontaneous contractile foci, exhibiting irregular beating patterns (Supplemental Fig. S2B) compared with SIRT6 wild-type and wild-type reconstituted SIRT6 knockout cardiomyocytes (Fig. 4D). This was consistent with gene expression data demonstrating significantly decreased levels of the α1-adrenergic receptor α1c (Fig. 4C; Jensen et al. 2014). Collectively, SIRT6 D63H mESCs exhibit a robust failure to differentiate into cardiomyocyte foci and display irregular beating patterns.
D63H homozygous iPSCs fail to differentiate into EBs and retain pluripotent gene expression
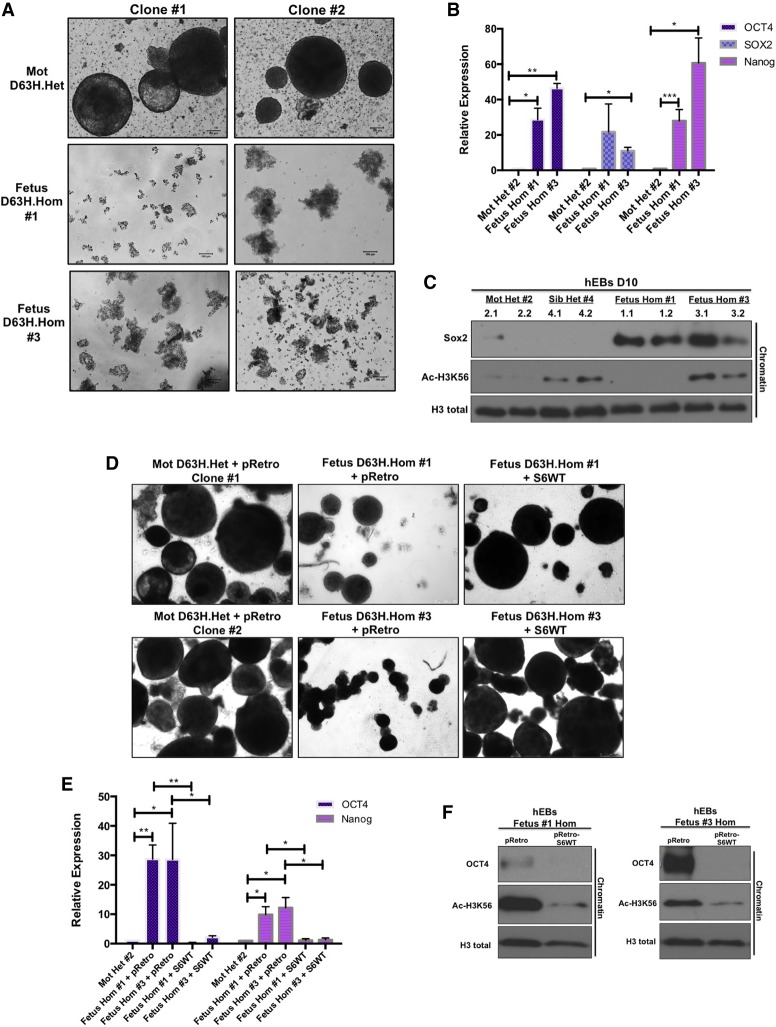
Mouse-derived ESCs expressing SIRT6 D63H displayed aberrant pluripotent gene expression and failure to differentiate into EBs and functional cardiomyocytes (Figs. 3, 4). To determine whether the endogenous expression of the human homozygous D63H mutation is sufficient to cause differentiation defects, we generated iPSCs from amniocytes isolated from two D63H homozygous fetuses (one female gestational age 20 wk and one male gestational age 27 wk) as well as peripheral blood mononuclear cells (PBMCs) from the heterozygous mother and viable sibling (Supplemental Fig. S4A). Immunofluorescence staining for pluripotency surface markers SSEA-4 and TRA-1-81 confirmed the undifferentiated state of multiple generated iPSC clones (Supplemental Fig. S4A). Interestingly, undifferentiated iPSCs derived from homozygous amniocytes did not exclusively retain pluripotent factors on bulk chromatin (Supplemental Fig. S4B) or display increased pluripotent gene expression (Supplemental Fig. S4C). To test whether the presence of the homozygous D63H mutation prevents these cells from undergoing proper differentiation, we enabled spontaneous differentiation by generating human EBs. Strikingly, both fetal homozygous iPSCs collapsed and failed to form EBs compared with two iPSC clones generated from the unaffected heterozygous mother (Fig. 5A). This severe in vitro differentiation and morphogenic phenotype observed in homozygous D63H EBs was associated with a failure to suppress pluripotent gene expression (Oct4, SOX2, and Nanog) (Fig. 5B) and the sustained presence of Sox2 on chromatin (Fig. 5C). Consistent with the microcephalic and hypoplastic cerebellar phenotype observed in SIRT6 D63H homozygous fetuses (Fig. 1A), EBs expressed significantly lower levels of the neuronal precursor Nestin and atypically high levels of the dosage-dependent transcription factor GBX2 (Supplemental Fig. S4D). To confirm that the phenotype observed in these patient-derived cells was driven by the mutant SIRT6 protein (as opposed to some unidentified mutation in another gene), we re-expressed wild-type SIRT6 in these cells. Notably, expression of wild-type SIRT6 in D63H homozygous iPSCs resulted in full rescue of the severe EB differentiation phenotype (Fig. 5D) accompanied by suppression of pluripotent gene expression (Fig. 5E) and removal of Oct4 from chromatin (Fig. 5F), clearly indicating that the phenotype observed in these cells was caused by the lack of a functional SIRT6 protein.
Figure 5.
Homozygous D63H human iPSCs fail to form EBs in vitro and retain pluripotent gene expression. (A) EBs derived from heterozygous (mother) and homozygous (fetuses #1 and #3) SIRT6 D63H iPSCs (two clones each; 10×). (B) iPSC-derived EB core pluripotent gene expression assessed by qRT–PCR analysis (average of two clones). Data are expressed relative to iPSC-derived EBs from a heterozygous mother. (C) Western blot analysis on bulk chromatin for Sox2 and Ac-H3K56 in iPSC-derived EBs. (D) EBs derived from iPSCs with doxycycline-inducible overexpression of pRetro empty vector and wild-type SIRT6 (4×). (Right panel) Quantified average EB size and number. (E) Core pluripotent gene expression in EBs with doxycycline-inducible overexpression of pRetro empty vector and wild-type SIRT6 assessed by qRT–PCR analysis. Data are expressed relative to iPSC-derived EBs from a heterozygous mother infected with pRetro empty vector. (F) Western blot analysis on bulk chromatin for Oct4 and Ac-H3K56 in EBs with doxycycline-inducible overexpression of pRetro empty vector and wild-type SIRT6.
D63H homozygous iPSCs fail to form cardiomyocyte foci and retain pluripotent gene expression
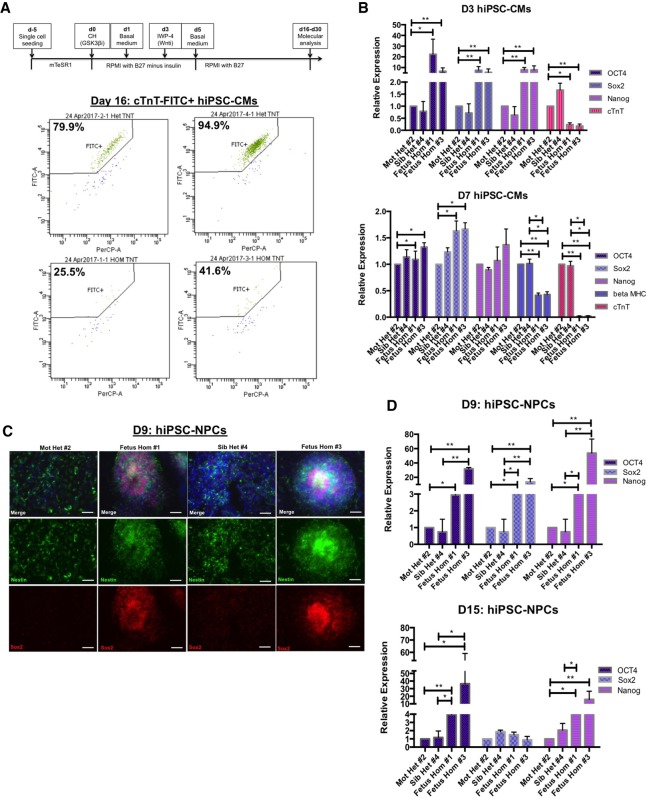
As mentioned previously, a number of congenital cardiac defects were identified in the SIRT6 D63H homozygous fetuses (Fig. 1A), and mESCs expressing the D63H mutation failed to differentiate into functional cardiomyocytes (Fig. 4). Human EBs derived from SIRT6 D63H homozygous iPSCs failed to express typical cardiac markers such as cTnT (Supplemental Fig. S4D). In order to test whether reprogrammed SIRT6 D63H homozygous iPSCs have the ability to undergo lineage-specific differentiation into functional cardiomyocytes, we performed directed monolayer differentiation by modulating Wnt signaling (Fig. 6A, top panel; Lian et al. 2012, 2013; Aguilar et al. 2015; Rajasingh et al. 2015). Day 3 after Wnt activation resulted in retained expression of pluripotent genes and significantly lower levels of cTnT in the D63H homozygous cells (Fig. 6B, top panel). Further differentiation (day 7) exacerbated the phenotype, where both βMHC and cTnT were severely reduced in the mutant iPSC cardiomyocytes (Fig. 6B, bottom panel), consistent with delayed differentiation of the overall population as measured by cTnT-FITC+ staining (Supplemental Fig. S5A). Notably, Wnt-mediated differentiation of D63H homozygous iPSC cardiomyocytes resulted in a failure to fully differentiate, and the cells crashed prior to completion on day 16 with only a few cells remaining, and, from those, <50% of cells stained positive for cTnT-FITC+ (Fig. 6A, bottom panel). Genome-wide RNA sequencing (RNA-seq) analysis confirmed the lack of suppression of pluripotent genes (Supplemental Fig. S4C). In addition, gene ontology analysis identified an altered gene expression signature with defects in HOX genes, Wnt signaling, aorta and cardiac morphogenesis, and cardiomyopathy (Supplemental Fig. S7C). Interestingly, many of these aberrantly expressed genes are critical for cardiac development (TBX20, NKX2-3, MYOCD, NKX2-5, GATA4, FBN1, Wnt1, and Wnt3A), with deregulation of these genes contributing to cardiomyopathy, proper valve formation, and cardiac septation (Supplemental Fig. S7D; Brand et al. 2003; Perrino and Rockman 2006; Pepe et al. 2014).
Figure 6.
Homozygous D63H human iPSCs fail to differentiate into functional cardiomyocytes and NPCs. (A, top panel) Schematic showing the differentiation from iPSCs into cardiomyocytes. (Bottom panels) Flow cytometry analysis of day 16 cardiomyocytes stained for cTnT-FITC. (B) Day 3 (top panel) and day 7 (bottom panel) iPSC-derived cardiomyocyte expression for core pluripotent genes and cardiac sarcomeric genes assessed by qRT–PCR analysis. Data are expressed relative to iPSC-derived cardiomyocytes from a heterozygous mother. (C) Immunofluorescence staining of day 9 iPSC-derived NPCs for Nestin, Sox2, and DAPI (10×). Images are representative of two clones for each genotype. (D) Day 9 (top panel) and day 15 (bottom panel) iPSC-derived NPC expression for core pluripotent genes assessed by qRT–PCR analysis. Data are expressed relative to iPSC-derived NPCs from a heterozygous mother.
D63H homozygous iPSCs display delayed NPC differentiation
As described above, EBs derived from SIRT6 D63H homozygous iPSCs failed to express the early CNS intermediate filament protein Nestin (Supplemental Fig. S4D). In order to directly test whether reprogrammed SIRT6 D63H homozygous iPSCs properly differentiate into CNS precursors, we performed directed monolayer differentiation into NPCs (Fig. 6C). By day 9 of differentiation, heterozygous iPSCs readily expressed Nestin while suppressing Oct4, SOX2, and Nanog expression (Fig. 6C,D, top panel). However, iPSC-derived NPCs generated from SIRT6 D63H homozygous fetuses, while expressing Nestin, retained pluripotent gene expression (Fig. 6C,D, top panel). Furthermore, even late stage (day 15) D63H homozygous iPSCs–NPCs retained Oct4 and Nanog expression (Fig. 6D, bottom panel). This was confirmed by RNA-seq analysis (Supplemental Fig. S4C), and, consistent with the severe neural phenotypes observed in the fetuses, we also found aberrant expression of HOX and POU genes, limb morphogenesis, Wnt signaling, and neural differentiation pathways (Supplemental Fig. S7A). Interestingly, many of these aberrantly expressed genes are critical for CNS development (POU4F1, FOXG1, SHH, NEUROD1, GBX2, MSX1, and SOX10), and deregulation of these genes has been associated with microcephaly and cerebellar hypoplasia (Supplemental Fig. S7B; Nanni et al. 1999; Ramos et al. 2004; Kortüm et al. 2011; Szulc et al. 2013). Together, directed differentiation of D63H homozygous iPSCs resulted in delayed NPC generation characterized by sustained pluripotent gene expression, which could explain the neurodevelopmental defects observed in the fetuses.
Discussion
As many as one-third of cases of late stage fetal loss remain unexplained, and idiopathic stillbirth is now the most common contributor to human perinatal mortality (Robson and Leader 2010; Bukowski et al. 2014; Lamont et al. 2015). Here we identify, for the first time, a family with four cases of perinatal lethality carrying a naturally occurring homozygous loss-of-function mutation in the histone deacetylase SIRT6. This mutation resulted in severe brain and heart developmental defects. When tested in culture, we observed a failure of mESCs and iPSCs carrying the homozygous mutant D63H SIRT6 to undergo proper differentiation (when challenged to differentiate into EBs, cardiomyocytes, and NPCs) due to sustained pluripotent gene expression. Importantly, ectopic expression of the human SIRT6 D63H mutant in SIRT6 knockout mESCs recapitulates the severe differentiation phenotype observed in D63H homozygous human iPSCs, thereby confirming a conserved role for SIRT6 in mouse and human pluripotency and development. In previous studies, we found that SIRT6 knockout mESCs fail to properly differentiate due to an inability to repress pluripotent genes (Etchegaray et al. 2015). Interestingly, the presence of the mutant D63H SIRT6 presents a more severe ESC differentiation phenotype than the one observed in the absence of SIRT6. It is plausible that the presence of the enzymatically inactive SIRT6 D63H on chromatin may impede compensatory mechanisms from other histone deacetylases from removing H3K9 and H3K56 marks. Alternatively, the SIRT6 D63H protein may exhibit a slower turnover than the wild-type protein, altering the balance between protein synthesis and degradation and disrupting histone acetylation in a more severe manner than when SIRT6 is completely absent. Of note, we observed a severe defect in the ability of the mutant iPSCs to differentiate into human EBs in vitro, yet D63H homozygous fetuses are miscarried relatively late in gestation. It is possible that in vivo, there is some redundancy with other chromatin factors that manage to keep early embryos differentiating and viable, and the phenotype manifests only in later developmental stages. Such a mechanism may be lost in the EB in vitro setting, where the differentiation to the three germ layers is driven at a much faster pace. Another explanation is that maternal contribution from the wild-type copy of SIRT6 may be weakening the developmental consequences of the D63H homozygous mutant fetuses in vivo. In this case, the embryos exhibit a milder defect when compared with the in vitro EBs due to compensation from the maternal wild-type SIRT6. For instance, Dnmt1o (Howell et al. 2001) and Dnmt3l (Bourc'his et al. 2001) are other epigenetic modifiers that exhibit similar maternal contribution in vertebrates, demonstrating late developmental phenotypes in vivo.
A number of mutations in DNA methylation and chromatin factors have been associated with neurodevelopmental syndromes with affected individuals living into adulthood (Douglas et al. 2003; Nafee et al. 2008; Jakovcevski and Akbarian 2012; Kaiser et al. 2014; Feng et al. 2015; Vallianatos and Iwase 2015). Although severe neurological phenotypes are observed in patients with chromatin-associated mutations in NSD1 (Sotos syndrome), EHMT1 (Kleefstra syndrome), and KDM5C (X-linked mental retardation), these mutations rarely result in perinatal lethality, thus pointing to possible compensation for the enzymatic activities of these histone modifiers during development (Douglas et al. 2003; Iwase et al. 2007; Nafee et al. 2008). For example, it has been suggested that the mild phenotype observed in KDM5C (JARID1C)-deficient mice is due to compensation by other Jumonji domain-containing histone demethylases (Iwase et al. 2007; Klose et al. 2007; Vallianatos and Iwase 2015). In contrast, our data indicate a key role for SIRT6 as a critical enzyme required for proper cell lineage differentiation and organogenesis throughout fetal development. A loss-of-function mutation results in a completely penetrant perinatal lethal phenotype, likely due to an inability to repress early embryonic genes. The main features observed in these fetuses were severe heart and neurological defects, a phenotype that we mimicked in vitro when forcing mutant iPSCs derived from amniocytes to differentiate into neural progenitors and cardiomyocytes. Previous studies have shown critical roles for SIRT6 in modulating glucose homeostasis, with SIRT6-deficient mice in certain strains dying early after birth due to severe hypoglycemia (Mostoslavsky et al. 2006; Zhong et al. 2010). Unfortunately, we were unable to measure glucose in these fetuses before their demise; therefore, it remains to be determined whether a glucose imbalance also contributed to the lethal phenotype. Although we detected an increase in LDHA and PDK1 at the protein level in the context of the homozygous SIRT6 mutation (iPSCs), one fetus (fetus #3) failed to derepress these genes to a lesser extent than the other fetus (fetus #1) (Supplemental Fig. S6A,B). Notably, the glucose transporter GLUT1 (SLC2A1) was actually down-regulated in D63H homozygous fetus-derived EBs (Supplemental Fig. S6B), suggesting some compensatory mechanism. Despite this increase in LDHA and PDK1 expression, treatment with the glycolytic inhibitor dichloroacetate (DCA) did not rescue the severe EB phenotype (Supplemental Fig. S6C), as we observed for previous SIRT6-related phenotypes (Zhong et al. 2010; Sebastián et al. 2012). Additionally, genome-wide RNA-seq expression analysis demonstrated that iPSCs, NPCs, and cardiomyocytes derived from D63H homozygous fetuses do not contain elevated levels of glycolytic genes and ribosome biogenesis genes (Supplemental Fig. S6D). These results confirm that SIRT6-dependent metabolic changes cannot account for the developmental defects seen in the D63H homozygous iPSCs, but rather the inability of SIRT6 D63H fetuses to suppress pluripotent gene expression is likely the dominant molecular mechanism contributing to perinatal loss.
In separate studies, SIRT6 has been shown to prevent genomic instability and maintain pericentromeric silencing, inhibiting cellular senescence (Michishita et al. 2009; Mao et al. 2011; Toiber et al. 2013; Tasselli et al. 2016; Van Meter et al. 2016). However, cytogenetic analysis of fetal amniocytes indicated no evidence of copy number variations or structural aneuploidy (data not shown). Although D63H-overexpressing mESC-derived EBs fail to reduce DNA damage signaling in the context of the SIRT6 knockout (Supplemental Fig. S6E), the D63H homozygous patient-derived iPSCs and EBs do not exhibit increased cleaved caspase and γ-H2AX (both signs of DNA damage) compared with the phenotypically normal heterozygous iPSCs (Supplemental Fig. S6F). This suggests that these cells somehow are able to compensate for the absence of SIRT6 function in order to sustain genomic stability, further confirming that DNA damage is not causal in the developmental defects observed in D63H homozygous fetuses.
Although SIRT6 has been shown to protect against cardiac hypertrophy in the adult (Sundaresan et al. 2012), our studies identify a novel role for this chromatin modifier in regulating the balance between pluripotency and cardiac lineage specification during development. Contrary to results seen in adult hearts, SIRT6 D63H-expressing cells exhibit inconsistent IGF signaling, with increased p-AKT and decreased p-ERK levels (data not shown). Additionally, iPSC-derived cardiomyocytes from D63H homozygous fetuses fail to exhibit elevated IGF signaling compared with cardiomyocytes derived from the heterozygous mother, as measured by RNA-seq (Supplemental Fig. S5B), suggesting that, unlike the adult, IGF signaling may not drive the phenotypes that we observed during development. However, RNA-seq analysis demonstrates aberrant expression of important downstream mediators of cardiac specification in the D63H homozygous fetuses (for example, GATA4 and Wnt3a) (Supplemental Fig. S7D). Future studies will investigate the functional role of these genes in cardiac differentiation in the context of the homozygous SIRT6 D63H mutation. Whether they are direct targets of SIRT6 or function downstream from the pluripotent genes or the Tet enzymes (Etchegaray et al. 2015) remains to be determined.
Patients with de novo mutations in H3K4me regulators (MLL2, KDM6A, and CHD7) display diverse congenital heart defects commonly accompanied by neurodevelopmental and growth abnormalities (Zaidi et al. 2013). H3K4me2 is an activating epigenetic mark associated with both promoters and enhancers involved in transcriptional activation of key developmental genes (Iwase et al. 2007; Klose et al. 2007; Vallianatos and Iwase 2015). Previously, our laboratory demonstrated that SIRT6 knockout mESCs contain elevated Tet1 levels and enriched 5hmC within exons encoding neuroectoderm genes (Etchegaray et al. 2015). Importantly, these regions were also enriched for H3K4me2, and the up-regulation of coregulated genes was rescued upon Tet inhibition (Etchegaray et al. 2015). Interestingly, D63H-expressing mouse cardiomyocytes retain Tet2 on chromatin (data not shown). In the future, it will be interesting to explore the link between SIRT6 deacetylation and H3K4me during cardiac differentiation. Whether such a link represents an example of coordination of two related marks (histone code) or whether SIRT6 may directly deacetylate nonchromatin targets, including lysine methyltransferases and demethylases, remains to be explored.
In summary, our work identified a novel human syndrome defined by a naturally occurring loss-of-function mutation in the chromatin enzyme SIRT6, associated with a fully penetrant human perinatal lethal phenotype. We demonstrated a failure to suppress pluripotent gene expression in homozygous SIRT6 D63H iPSCs, resulting in an inability of the cells to undergo proper differentiation into EBs, cardiomyocytes, and NPCs. Together, our data implicate SIRT6 chromatin remodeling activity as a critical modulator of human development, regulating the transition from pluripotency to differentiated states.
Materials and methods
Patient amniocyte/PBMC collection and sequencing
Peripheral blood was collected from the mother, father, and grandmother. Amniotic fluid was collected from amniocentesis during all five pregnancies. Written consent was obtained in accordance with guidelines of the Academic Medical Centre (AMC), Netherlands Institutional Review Board. DNA was extracted from blood or amniotic fluid cells using standard methods. WES was performed on child #4 and both parents. Exonic targets were enriched with the SeqCap EZ human exome library version 3·0 kit (NimbleGen). The resulting libraries were sequenced on a HiSeq 2500 version 4 chemistry (Illumina) according to the manufacturer's recommendations for paired-end 125-base-pair (bp) reads. Alignment of sequence reads to the human reference genome (hg19) was done using BWAMEM 0.7.5 (http://www.bio-bwa.sourceforge.net), and variants were called using the GATK3.3 software package (http://www.broadinstitute.org/gatk). Annotation and filtering of variants was done using Cartagenia Bench Lab NGS (Agilent). Variants with a frequency of >1% or homozygously present in public (ESP, ExAC, dbSNP, and 1KG) and/or in-house databases were excluded. Evaluation of the homozygous variants revealed possible causal variants in five genes (ARHGEF17, EGLAM, RMB4N, RAF1, and SIRT6). Subsequent analysis of the three affected siblings using Sanger sequencing showed that only the SIRT6 variant (c.187G > C p.[Asp63His]) segregated with the disorder.
Expression and purification of recombinant human SIRT6
His-tagged wild-type and mutant SIRT6 proteins were overexpressed in the BL21(DE3) Escherichia coli strain as described previously (Feldman et al. 2015). Cells were harvested by centrifugation and stored at −80°C. SIRT6 wild-type and mutant proteins were purified by nickel affinity resin chromatography as reported previously (Feldman et al. 2015). Protein concentrations were determined by the Bradford reagent assay.
HPLC deacylation assay
Peptides corresponding to residues 4–17 of histone H3 (acetyl: AcKQTARKacSTGGKAPR-WW-NH2, and myristoyl: Ac-KQTARKmyrSTGGKAPRWW-NH2) were synthesized as described previously (Feldman et al. 2013). Deacylation reactions were analyzed by reversed-phase high-performance liquid chromatography on a Kinetex C18 column (100 Å, 100 mm × 4.6 mm, 2.6 µm; Phenomenex) by monitoring the formation of the deacylated product at 214 nm (Feldman et al. 2013).
Steady-state kinetic analyses
Steady-state rates were measured by varying NAD+ (5–640 µM) in the presence of 0.5 µM wild-type, D63Y, and D63H SIRT6 in 20 mM sodium phosphate (pH 7.5) at 37°C. Initial velocities were determined, and data were fitted to the Michaelis-Menten equation. Deacetylase and demyristoylase activities were measured in the presence of 50 µM corresponding substrate, 0.5 mM NAD+, and 2 µM wild-type or mutant SIRT6. Reactions were quenched at <15% substrate depletion by addition of 2% TFA final concentration.
Generation of human SIRT6 constructs and virus production
Generation of human eGFP and pRetroX-Tight-Pur SIRT6 constructs have been described previously (Kugel et al. 2015). Site-directed mutagenesis of wild-type SIRT6 was performed using the QuikChange Lightning site-directed mutagenesis kit (Stratagene). pLVX-Tet-On was obtained from Clontech. Viral particles containing the above-mentioned plasmids were synthesized using retroviral packaging plasmids pCL-ECO (Addgene) and pCMV-VSV-G (Addgene).
Subcellular localization of SIRT6 constructs
293T cells were transfected using Trans-IT 293 (Mirus) with empty vector, pEGFP-SIRT6, or the SIRT6 mutant constructs. Twenty-four hours after transfection, cells were trypsinized and seeded onto eight-well chamber slides and allowed to adhere overnight. Cells were then fixed using 2% paraformaldehyde in a 1% phosphate-buffered saline (PBS) solution and permeabilized with 0.1% sodium citrate and 0.1% Triton X-100, and nuclei were stained using 4′,6′-diamidino-2-phenylindole (DAPI). Images were taken using a fluorescent microscope.
Mouse-derived cell lines and tissue culture conditions
SIRT6 knockout primary MEFs were generated from 13.5-d-old embryos as described (Mostoslavsky et al. 2006). Cells were cultured at 37°C under 5% CO2 in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% 100 U/mL penicillin/streptomycin (Invitrogen), 2 mM L-glutamine, 0.1 mM NEAA, 1 mM sodium pyruvate, and 20 mM HEPES. 293T cells were cultured in high-glucose DMEM supplemented with 10% FBS and 1% 100 U/mL penicillin/streptomycin (Invitrogen). SIRT6 knockout MEFs were infected by incubating with virus and 10 µg/mL polybrene. Forty-eight hours later, cells were selected in 2.5 µg/mL puromycin, and the pooled populations were used for various experiments. For all experiments involving the doxycycline-inducible SIRT6 knockout MEFs, cells were treated with 1 µg/mL doxycycline for 48 h.
mESC cultures, formation, and immunostaining of EBs
mESCs derived from Sirt6 knockout and wild-type 129 mouse strain were maintained on γ-irradiated MEFs in knockout DMEM (Gibco) containing 15% ES-qualified FBS, 0.1 mM (each) nonessential amino acids, 2 mM L-glutamine, 0.1 mM β-mercaptoethanol, and 50 U/mL penicillin/streptomycin (Invitrogen) and supplemented with leukemia-inhibiting factor. SIRT6 knockout and wild-type mESCs were infected by incubating with virus and 10 µg/mL polybrene. Forty-eight hours later, cells were selected in 2.5 µg/mL puromycin, and five clones were selected for use in various experiments. For all experiments involving doxycycline-inducible SIRT6 knockout ESCs, cells were treated with 1 µg/mL doxycycline for 7 d prior to collection and differentiation. For all experiments described, cells were trypsinized and plated for 30 min on standard tissue culture dishes to remove feeder cells before floating ESCs were collected and replated on gelatin-coated dishes or wells before differentiation toward EBs. ESCs were dissociated with trypsin (day 0) and cultured at a density of 5000 cells per milliliter in Iscove's modified Dulbecco's medium (IMDM) with 15% fetal calf serum (Atlanta Biologicals), 2 mM L-glutamine (Gibco), 200 µg/mL transferrin (Roche), 0.5 mM ascorbic acid (Sigma), and 4.5° 10−4 M monothioglycerol (Sigma). Differentiation was carried out using the standard hanging drop method, and EBs were collected from days 6 to 12. EB size was quantified using ImageJ 64 software as an average of five independent clones in a 2.5× field. EB number was quantified as the number of EBs in a 2.5× field and was an average of five independent clones.
Generation of Oct4 and Sox2 knockdown ESC lines
Oct4 and Sox2 shRNA constructs were used as described previously (Etchegaray et al. 2015). Briefly, ESCs were infected by incubating with virus and 10 µg/mL polybrene. Forty-eight hours later, cells were selected in 2.5 µg/mL puromycin on puromycin-resistant γ-irradiated feeders. Clones were picked and screened by qRT–PCR for knockdown of Oct4 and Sox2 expression. Prior to experiments, clones were removed from feeders and grown on gelatin-coated plates.
Differentiation of mESCs into cardiomyocytes
SIRT6 wild-type and knockout ESCs were infected and treated with doxycycline as described above. Differentiation toward the cardiomyocyte lineage was carried out using the standard hanging drop method with EBs collected on day 4 and plated onto adherent plates in EGM-2 bullet kit medium (Lonza). Spontaneous contractile foci within differentiating EBs were detected as early as day 15 and quantified as the average number of foci per five clones. The medium was subsequently changed until day 21, and cells were collected for bulk chromatin and RNA isolation and fixed for immunofluorescence staining for cTnT.
Human iPSC generation and immunostaining
Frozen amniocytes were thawed and maintained on 0.1% gelatin-coated six-well plates using amniocyte medium. The medium contained Alpha-MEM (Invitrogen) supplemented with 10% FBS (Hyclone), 1× GlutaMax (Invitrogen), 1× nonessential amino acids (Invitrogen), and 0.1 mg/mL Primocin (InvivoGen). Two days before transduction, amniocytes were plated onto 0.1% gelatin-coated six-well plates to obtain 2 × 105 and 5 × 105 cells on the day of transduction, which represents 50%–80% confluency. To transduce the cells, CytoTune 2.0 Sendai reprogramming kit (Invitrogen) was thawed and prepared according to the manufacturer's protocol. Amniocytes were infected with each virus at multiplicity of infection (KOS:hc-Myc:hKlf4 = 5:5:3) using amniocyte medium with 5 µg/mL polybrene (MilliporeSigma). The viruses were washed the following day, and the infected cells were cultured for six more days by changing the amniocyte medium every other day. At day 7 after transduction, the cells were passaged and plated onto human ESC (hESC)-qualified Matrigel-coated (Corning) six-well plates at ∼1.0 × 104 to 8.5 × 104 cells per well. From day 8 after transduction, the cells were fed with complete ReproTeSR (Stem Cell Technologies). The medium was changed daily. Once human iPSC colonies appeared, clonal human iPSCs were picked and expanded using mTeSR (Stem Cell Technologies).
Frozen PBMCs were thawed and expanded for 9 d using Expansion medium containing QBSF-60 stem cell medium (Quality Biological) supplemented with 50 µg/mL ascorbic acid (MilliporeSigma), 50 ng/mL SCF (R&D Systems), 10 ng/mL IL-3 (R&D Systems), 2 U/mL EPO (R&D Systems), 40 ng/mL IGF-1 (R&D Systems), 1 µM dexamethasone (MilliporeSigma), and 0.1 mg/mL Primocin (InvivoGen). Expanded PBMCs were transduced with CytoTune 2.0 Sendai reprogramming kit (Invitrogen) by spinoculation at 2250 rpm for 90 min at room temperature. After 1 d of incubation, the viruses were washed, and the cells were plated onto hESC-qualified Matrigel-coated (Corning) six-well plates at ∼8.0 × 104 to 2 × 105 cells per well using Expansion medium. At day 3 after transduction, the infected cells started being fed with ReproTeSR (Stem Cell Technologies). Once human iPSC colonies appeared, clonal human iPSCs were picked and expanded using mTeSR (Stem Cell Technologies).
All generated human iPSCs were fixed with 4% paraformaldehyde/PBS and immunofluorescence-stained with mouse anti-human SSEA-4, mouse anti-human TRA-1-81, and mouse anti-human TRA-1-60 (EMD Millipore) followed by secondary antibody, Alexa fluor 488-conjugated goat anti-mouse immunoglobulin M (ThermoFisher Scientific), and Alexa fluor 488-conjugated goat anti-mouse immunoglobulin G (ThermoFisher Scientific). All human iPSC differentiation experiments were performed using two iPSC clones from each genotype.
Human EB formation
Human iPSC lines were mechanically dissociated using StemPro EZPassage (Invitrogen) to make clumps of a consistent size. Human EBs were generated in suspension culture over a low-speed shaker for 10 d. Culturing conditions included medium containing DMEM/F12, 20% knockout serum replacement, 1 mM L-glutamine, 100 µM M MEM nonessential amino acids, and 0.1 mM β-mercaptoethanol in the absence of FGF-2.
For rescue experiments, human iPSCs were infected by incubating three separate clones with either pRetro empty vector or pRetro-S6 wild-type virus, 10 µg/mL polybrene, and 10 µM Rock inhibitor. Forty-eight hours later, cells were selected with 2.5 µg/mL puromycin. After puromycin selection, cells were treated with 1 µg/mL doxycycline for 6 d prior to differentiation into EBs. For glycolysis inhibition, iPSCs were treated with either dH2O or 5 mM dichloroacetate (DCA), and, 24 h later, cells were mechanically dissociated into EBs as described above.
Gel electrophoresis and Western blotting
Chromatin extraction and Western blot analysis were performed as described previously (Sebastián et al. 2012). Briefly, the cell pellet was resuspended in lysis buffer containing 10 mM HEPES (pH 7.4), 10 mM KCl, 0.05% NP-40 supplemented with a protease inhibitor cocktail (Complete EDTA-free, Roche Applied Science), 5 µM TSA, 5 mM sodium butyrate, 1 mM DTT, 1 mM PMSF, 50 mM NaF, 0.2 mM sodium orthovanadate, and phosphatase inhibitors (phosphatase inhibitor cocktail sets I and II, Calbiochem) and incubated for 20 min on ice. The lysate was then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was removed (cytosolic fraction), and the pellet (nuclei) was acid-extracted using 0.2 N HCl and incubated for 20 min on ice. The lysate was then centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant (containing acid-soluble proteins) was neutralized using 1 M Tris-HCl (pH 8). Protein concentration was quantified by Bio-Rad protein assay. Ten micrograms of protein was electrophoresed on a 10%–20% gradient polyacrylamide gel with SDS (Bio-Rad) and electroblotted onto polyvinylidene difluoride membranes (PVDF) (Millipore). Membranes were blocked in TBS with 5% nonfat milk and 0.1% Tween and probed with antibodies. Bound proteins were detected with horseradish peroxidase-conjugated secondary antibodies (Vector Biolaboratories) and SuperSignal West Pico luminol/enhancer solution (Thermo Scientific). The antibodies used were as follows: anti-Sirt6 (Cell Signaling, D8D12), anti-Oct4 (Santa Cruz Biotechnology, sc5279), anti-Sox2 (Abcam, ab92494), anti-H3K56ac (Abcam, ab76307), anti-H3K9ac (Millipore, 07-352) anti-H3 (Abcam, ab1791), anti-LDHA (Cell Signaling, 2012), anti-PDK1 (Cell Signaling, 3820), anti-p-AKT (S473) (Cell Signaling, 4060), anti-p-ERK (Sigma, M8159), anti-cleaved caspase-3 (Cell Signaling, 9664), and anti-γH2AX (Abcam, ab2893).
RNA extraction and qRT–PCR
Total RNA was extracted with the TriPure isolation reagent (Roche) as described by the manufacturer. For cDNA synthesis, 1 mg of total RNA was retrotranscribed by using the QuantiTect reverse transcription kit (Qiagen). Real-time PCR was performed using SYBR Green master mix (Roche), following the manufacturer's instructions, with the exception that the final volume was 12.5 mL of SYBR Green reaction mix. Real-time monitoring of PCR amplification was performed using the LightCycler 480 detection system (Roche). Data were expressed as relative mRNA levels normalized to the β-actin or GAPDH expression level in each sample. The primer sequences can be obtained on request.
Directed monolayer iPSC differentiation into cardiomyocytes
Human directed cardiomyocyte differentiation from monolayer iPSCs has been described previously (Lian et al. 2012, 2013; Aguilar et al. 2015; Rajasingh et al. 2015). Briefly, human iPSCs derived from patients (see above) were grown and maintained on Matrigel-coated plates in mTESR1 medium until ∼95% confluence. On day 0, cells were treated with RPMI-1640 containing fresh 50× B27-insulin, 12 µM CHiR0991 (GSK3B inhibitor), 50 µg/mL sodium ascorbate, and 1:100 Geltrex (reduced growth factor matrix) overnight. Early on day 1, morphological epithelial–mesenchymal transition (EMT) changes were observed, and the medium was changed to RPMI-1640 containing fresh 50× B27-insulin and 50 µg/mL sodium ascorbate. On day 3, cells were treated with RPMI-1640 containing fresh 50× B27-insulin, 10 µM IWP-4 (Wnt inhibitor), and 50 µg/mL sodium ascorbate. On day 5, the cell medium was changed to RPMI-1640 containing fresh 50× B27-insulin. On day 6, the medium was changed to RPMI-1640 containing fresh 50× B27 complete. The medium was changed until day 16, when spontaneous contractile foci were seen. For experiments, time points were collected on days 3, 7, and 16 for RNA expression and cTnT-FITC flow cytometry analysis.
Analysis of cTnT-FITC+ using flow cytometry
Cells were fixed in 1 mL of 1.0% formaldehyde for 30 min at 37°C in the well and then harvested using 0.25% trypsin-EDTA. Cells were then resuspended in 1 mL of 90% cold methanol and incubated for 15 min at 4°C using an end-over-end rotator. Pellets were washed twice with FACS buffer (0.5% BSA in 1× PBS) and then resuspended in 1 mL of Tween-20 buffer (0.5% Tween-20 in 1× PBS). Pellets were then spun down and incubated in primary antibody (anti-cTnT-FITC [Abcam, ab105439] or IgG isotype control) in 100 µL of Tween-20 buffer at a 1:50 dilution for 30 min at room temperature using an end-over-end rotator. Pellets were washed twice with Tween-20 buffer and then resuspended in FACS buffer for flow cytometry analysis.
Directed monolayer iPSC differentiation into NPCs
Human NPC differentiation from monolayer iPSCs was accomplished using the STEMdiff neural system according to the manufacturer's instructions (Stem Cell Technologies). Briefly, on day 0, human iPSCs maintained in mTESR1 medium on Matrigel-coated plates were dissociated and plated in STEMdiff neural induction medium + 10 µM Y-27632. The medium was changed, and cells were split (as required per the manufacturer's instructions) until day 6. On day 6, the medium was changed to STEMdiff neural progenitor medium until the completion of differentiation. Time points were collected every 3 d for RNA and immunofluorescence staining.
Immunofluorescence staining of mouse and human differentiated cells
In brief, cells were fixed with 4% formalin for 20 min at room temperature. Fixed cells were washed with PBS–glycine (130 mM NaCl, 7 mM Na2HPO4, 100 mM glycine) three times for 10 min. Wells were then blocked using immunofluorescence buffer (130 mM NaCl, 7 mM Na2PO4, 7.7 mM NaN3, 0.1% bovine serum, 0.2% Triton X-100, 0.05% Tween 20) plus 2% goat serum for 1 h. Cells were incubated with primary antibodies diluted in blocking buffer overnight at 4°C on a shaker. After three 20-min washes with immunofluorescence buffer, cells were incubated with anti-mouse or anti-rabbit secondary antibodies coupled with Alexa fluor dyes (Invitrogen) diluted in immunofluorescence buffer supplemented with 10% goat serum for 1 h at room temperature. After incubation with secondary antibodies, cells were washed three times with immunofluorescence buffer and then incubated with 0.5 ng/mL DAPI (Sigma-Aldrich) for 10 min at room temperature. Cells were washed again for 5 min with immunofluorescence buffer prior to imaging using a Nikon Eclipse Ti-E microscope. Ten images were taken per genotype. The antibodies used were as follows: anti-Sox2 (Abcam, ab92494), anti-Nestin (Abcam, ab18102), and anti-cTnT (Abcam, ab8295).
RNA isolation and library construction
Total RNA was isolated using Trizol-LS from three biological replicates for each condition. Library construction was performed using NEBNext poly(A) mRNA magnetic isolation module (New England Biolabs, E7490) and NEBNext Ultra Directional RNA library preparation kit for Illumina (New England Biolabs, E7420). The total RNA input amount for the poly(A) kit was 150 ng total. After poly(A) selection, all material was taken through library construction. Fifteen cycles of PCR were performed for PCR amplification. After library constructions, the samples were validated using a 2200 Tapestation system and high-sensitivity D1000 ScreenTape kit. Libraries were quantified using the Kapa Biosystems library quantification kit (catalog no. KK4828) and the Bio-Rad CFX96 instrument. Each lane of sequencing was pooled into a 21-plex (21 samples per lane) with unique barcodes over four total lanes. Pooled libraries are also quantified using the Kapa Biosystems library quantification kit (catalog no. KK4828) and the Bio-Rad CFX96 instrument. These pools are then denatured to 16 pM with 1% phix and sequenced on an Illumina HiSeq 2000 instrument. The sequencing length read was paired-end 50 bp.
Methods used for data analysis
STAR aligner (Dobin et al. 2013) was used to map sequencing reads to the mouse reference transcriptome (mm9 assembly). Read counts over transcripts were calculated using HTSeq version 0.6.0 (Anders et al. 2015) based on a current Ensembl annotation file for NCBI37/mm9 assembly. Differential expression analysis was performed using EdgeR (Robinson et al. 2010); genes were classified as differentially expressed based on the cutoffs of at least twofold change and false discovery rate <0.05. Analysis of enriched functional categories among detected genes was performed using DAVID (Subramanian et al. 2005).
Statistics
For steady-state deacetylation/demyristoylation assays, real-time RT–PCR analysis, and EB size and number and spontaneous contractile focus quantification, significance was analyzed using two-tailed Student's t-test. A P-value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank A. Ilgun and J. Stap for technical assistance, and the Mostoslavsky laboratory for helpful discussions and critical reading of the manuscript. This work was supported in part by National Institutes of Health (NIH) grants 5R01CA175727 (to R.M., G.M., and C.M.F.) and P30 DK040561 (to R.I.S. and M.C.). R.M. is the Kristine and Bob Higgins Massachusetts General Hospital Research Scholar and The Laurel Schwartz Associate Professor in Oncology, a Warshaw Institute Fellow, and a Howard Goodman Awardee. A.V.P. was supported by Cardiovasculair Onderzoek Nederland (CVON) project CONCOR-genes (CVON 2014-18).
Author contributions: C.M.F. performed and designed most of the experiments, analyzed the data, and wrote the manuscript. M.C. and R.I.S. performed bioinformatic analysis. M.A., A.V.P., E.P., A.G., S.v.K., V.M.C., M.M.A.M.M., L.K., and M.C.v.M. identified the family, collected materials from the patients, and performed the clinical analysis and sequencing of the individuals. M.C.v.M also helped design the study and provided conceptual input and discussions to write the manuscript. S.P. and G.M. derived human iPSCs from the patients and helped with some of the iPSC experiments. M.A.K. and J.M.D. performed the in vitro assays with recombinant SIRT6. J-P.E. assisted with ESC experiments and provided advice with experimental design. R.M. supervised and designed the study, interpreted the results, analyzed the data, and wrote the manuscript with C.M.F.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.307330.117.
References
- Aguilar JS, Begum AN, Alvarez J, Zhang X-b, Hong Y, Hao J. 2015. Directed cardiomyogenesis of human pluripotent stem cells by modulating Wnt/β-catenin and BMP signalling with small molecules. Biochem J 469: 235–241. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539. [DOI] [PubMed] [Google Scholar]
- Boyle B, Addor MC, Arriola L, Barisic I, Bianchi F, Csáky-Szunyogh M, de Walle HEK, Dias CM, Draper E, Gatt M, et al. 2018. Estimating global burden of disease due to congenital anomaly: an analysis of European data. Arch Dis Child Fetal Neonatal Ed 103: F22–F28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand T. 2003. Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol 258: 1–19. [DOI] [PubMed] [Google Scholar]
- Bukowski R, Hansen NI, Willinger M, Reddy UM, Parker CB, Pinar H, Silver RM, Dudley DJ, Stoll BJ, Saade GR, et al. 2014. Fetal growth and risk of stillbirth: a population-based case-control study. PLoS Med 11: e1001633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Dent SY. 2014. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 15: 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas J, Hanks S, Temple IK, Davies S, Murray A, Upadhyaya M, Tomkins S, Hughes HE, Trevor Cole RP, Rahman N. 2003. NSD1 mutations are the major cause of Sotos syndrome and occur in some cases of Weaver syndrome but are rare in other overgrowth phenotypes. Am J Hum Genet 72: 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Chavez L, Huang Y, Ross KN, Choi J, Martinez-Pastor B, Walsh RM, Sommer CA, Lienhard M, Gladden A, et al. 2015. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat Cell Biol 17: 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, Denu JM. 2013. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem 288: 31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Dittenhafer-Reed KE, Kudo N, Thelen JN, Ito A, Yoshida M, Denu JM. 2015. Kinetic and structural basis for acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry 54: 3037–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Zhou D, Zhang Z, Liu Y, Yang Y. 2015. Exome sequencing identifies a de novo mutation in HDAC8 associated with Cornelia de Lange syndrome. J Hum Genet 60: 165. [DOI] [PubMed] [Google Scholar]
- GBD 2013 Mortality and Causes of Death Collaborators. 2015. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 385: 117–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DA, Uchida IA, Lewis AJ. 1963. A review of the 18 trisomy syndrome. Med Biol Illus 13: 80–88. [PubMed] [Google Scholar]
- Horb ME, Thomsen GH. 1999. Tbx5 is essential for heart development. Development 126: 1739–1751. [DOI] [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104: 829–838. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Jakovcevski M, Akbarian S. 2012. Epigenetic mechanisms in neurological disease. Nat Med 18: 1194–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BC, O'Connell TD, Simpson PC. 2014. α-1-adrenergic receptors in heart failure: the adaptive arm of the cardiac response to chronic catecholamine stimulation. J Cardiovasc Pharmacol 63: 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. 2013. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 496: 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser FJ, Ansari M, Braunholz D, Concepción Gil-Rodríguez M, Decroos C, Wilde JJ, Fincher CT, Kaur M, Bando M, Amor DJ, et al. 2014. Loss-of-function HDAC8 mutations cause a phenotypic spectrum of Cornelia de Lange syndrome-like features, ocular hypertelorism, large fontanelle and X-linked inheritance. Hum Mol Genet 23: 2888–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG. 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128: 889–900. [DOI] [PubMed] [Google Scholar]
- Kortüm F, Das S, Flindt M, Morris-Rosendahl DJ, Stefanova I, Goldstein A, Horn D, Klopocki E, Kluger G, Martin P, et al. 2011. The core FOXG1 syndrome phenotype consists of postnatal microcephaly, severe mental retardation, absent language, dyskinesia, and corpus callosum hypogenesis. J Med Genet 48: 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel S, Feldman JL, Klein MA, Silberman DM, Sebastián C, Mermel C, Dobersch S, Clark AR, Getz G, Denu JM, et al. 2015. Identification of and molecular basis for SIRT6 loss-of-function point mutations in cancer. Cell Rep 13: 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel S, Sebastián C, Fitamant J, Ross KN, Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al. 2016. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell 165: 1401–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont K, Scott NW, Jones GT, Bhattacharya S. 2015. Risk of recurrent stillbirth: systematic review and meta-analysis. BMJ 350: h3080. [DOI] [PubMed] [Google Scholar]
- Lee MY, Cagavi Bozkulak E, Schliffke S, Amos PJ, Ren Y, Ge X, Ehrlich BE, Qyang Y. 2011. High density cultures of embryoid bodies enhanced cardiac differentiation of murine embryonic stem cells. Biochem Biophys Res Commun 416: 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. 2012. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci 109: E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. 2013. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 8: 162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima BL, Santos EJC, Fernandes GR, Merkel C, Mello MRB, Gomes JPA, Soukoyan M, Kerkis A, Massironi SMG, Visintin JA, et al. 2010. A new mouse model for marfan syndrome presents phenotypic variability associated with the genetic background and overall levels of Fbn1 expression. PLoS One 5: e14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. 2011. SIRT6 promotes DNA repair under stress by activating PARP1. Science 332: 1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. 2009. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 8: 2664–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. 2006. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329. [DOI] [PubMed] [Google Scholar]
- Nafee TM, Farrell WE, Carroll WD, Fryer AA, Ismail KM. 2008. Epigenetic control of fetal gene expression. BJOG 115: 158–168. [DOI] [PubMed] [Google Scholar]
- Nanni L, Ming JE, Bocian M, Steinhaus K, Bianchi DW, de Die-Smulders C, Giannotti A, Imaizumi K, Jones KL, Del Campo M, et al. 1999. The mutational spectrum of the sonic hedgehog gene in holoprosencephaly: SHH mutations cause a significant proportion of autosomal dominant holoprosencephaly. Hum Mol Genet 8: 2479–2488. [DOI] [PubMed] [Google Scholar]
- Pandya PP, Kondylios A, Hilbert L, Snijders RJ, Nicolaides KH. 1995. Chromosomal defects and outcome in 1015 fetuses with increased nuchal translucency. Ultrasound Obstet Gynecol 5: 15–19. [DOI] [PubMed] [Google Scholar]
- Pepe G, Nistri S, Giusti B, Sticchi E, Attanasio M, Porciani C, Abbate R, Bonow RO, Yacoub M, Gensini GF. 2014. Identification of fibrillin 1 gene mutations in patients with bicuspid aortic valve (BAV) without Marfan syndrome. BMC Med Genet 15: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrino C, Rockman HA. 2006. GATA4 and the two sides of gene expression reprogramming. Circ Res 98: 715–716. [DOI] [PubMed] [Google Scholar]
- Posch MG, Perrot A, Berger F, Ozcelik C. 2010. Molecular genetics of congenital atrial septal defects. Clin Res Cardiol 99: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasingh S, Thangavel J, Czirok A, Samanta S, Roby KF, Dawn B, Rajasingh J. 2015. Generation of functional cardiomyocytes from efficiently generated human iPSCs and a novel method of measuring contractility. PLoS One 10: e0134093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C, Fernández-Llebrez P, Bach A, Robert B, Soriano E. 2004. Msx1 disruption leads to diencephalon defects and hydrocephalus. Dev Dyn 230: 446–460. [DOI] [PubMed] [Google Scholar]
- Rayasam GV, Wendling O, Angrand PO, Mark M, Niederreither K, Song L, Lerouge T, Hager GL, Chambon P, Losson R. 2003. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J 22: 3153–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson SJ, Leader LR. 2010. Management of subsequent pregnancy after an unexplained stillbirth. J Perinatol 30: 305–310. [DOI] [PubMed] [Google Scholar]
- Sebastián C, Zwaans BMM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. 2012. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151: 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Wright D. 2014. Pallister-Killian syndrome. Am J Case Rep 15: 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam V, et al. 2012. The sirtuin SIRT6 blocks IGF–Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med 18: 1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc KU, Nieman BJ, Houston EJ, Bartelle BB, Lerch JP, Joyner AL, Turnbull DH. 2013. MRI analysis of cerebellar and vestibular developmental phenotypes in Gbx2 conditional knockout mice. Magn Reson Med 70: 1707–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, Chua KF. 2016. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol 23: 434–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Erdel F, Bouazoune K, Silberman DM, Zhong L, Mulligan P, Sebastian C, Cosentino C, Martinez-Pastor B, Giacosa S, et al. 2013. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol Cell 51: 454–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida IA, Bowman JM, Wang HC. 1962. The 18-trisomy syndrome. N Engl J Med 266: 1198–1201. [DOI] [PubMed] [Google Scholar]
- Vallianatos CN, Iwase S. 2015. Disrupted intricacy of histone H3K4 methylation in neurodevelopmental disorders. Epigenomics 7: 503–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Meter M, Simon M, Tombline G, May A, Morello TD, Hubbard BP, Bredbenner K, Park R, Sinclair DA, Bohr VA, et al. 2016. JNK phosphorylates SIRT6 to stimulate DNA double-strand break repair in response to oxidative stress by recruiting PARP1 to DNA breaks. Cell Rep 16: 2641–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz JW, Barnes RM, Firulli AB. 2011. Hand factors as regulators of cardiac morphogenesis and implications for congenital heart defects. Birth Defects Res A Clin Mol Teratol 91: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Oron E, Nelson B, Razis S, Ivanova N. 2012. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 10: 440–454. [DOI] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. 2009. The sirtuin SIRT6 deacetylates H3 K56ac in vivo to promote genomic stability. Cell Cycle 8: 2662–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, et al. 2013. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369: 1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. 2011. Control of the embryonic stem cell state. Cell 144: 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. 2013. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498: 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. 2010. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell 140: 280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.