Abstract
Purpose
Ultrafast single‐shot T2‐weighted images are common practice in fetal MR exams. However, there is limited experience with fetal T1‐weighted acquisitions. This study aims at establishing a robust framework that allows fetal T1‐weighted scans to be routinely acquired in utero at 3T.
Methods
A 2D gradient echo sequence with an adiabatic inversion was optimized to be robust to fetal motion and maternal breathing optimizing grey/white matter contrast at the same time. This was combined with slice to volume registration and super resolution methods to produce volumetric reconstructions. The sequence was tested on 22 fetuses.
Results
Optimized grey/white matter contrast and robustness to fetal motion and maternal breathing were achieved. Signal from cerebrospinal fluid (CSF) and amniotic fluid was nulled and 0.75 mm isotropic anatomical reconstructions of the fetal brain were obtained using slice‐to‐volume registration and super resolution techniques. Total acquisition time for a single stack was 56 s, all acquired during free breathing. Enhanced sensitivity to normal anatomy and pathology with respect to established methods is demonstrated. A direct comparison with a 3D spoiled gradient echo sequence and a controlled motion experiment run on an adult volunteer are also shown.
Conclusion
This paper describes a robust framework to perform T1‐weighted acquisitions and reconstructions of the fetal brain in utero. Magn Reson Med 80:137–146, 2018. © 2017 The Authors Magnetic Resonance in Medicine published by Wiley Periodicals, Inc. on behalf of International Society for Magnetic Resonance in Medicine. This is an open access article under the terms of the Creative Commons Attribution NonCommercial License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited and is not used for commercial purposes.
Keywords: fetal MRI, T1 contrast, slice‐to‐volume reconstruction, hemorrhage, motion tolerance and efficiency
INTRODUCTION
MRI is an invaluable tool with which to assess brain development, and there has been a growing demand for in utero brain imaging for diagnosis and research. The workhorse sequence for fetal MRI is single‐shot‐fast‐spin‐echo (ssFSE), predominantly used for T2w (weighted) imaging 1, 2, but also applied with inversion recovery (IR) preparation for T1w imaging 3. In general, ssFSE acquisitions are characterized by both high contrast and signal‐to‐noise ratio (SNR) and they provide consistent image quality with robustness against artefacts caused by motion. To keep the specific absorption rate (SAR) within acceptable safety constraints 4, 5, it is common to reduce the refocusing angle of the radiofrequency (RF) pulses in the FSE echo train, and this results in mixed signal pathways with contributions from both stimulated and spin echoes, causing the signal evolution to depend both on T1 and T2 6. The addition of an inversion recovery preparation as applied in the snapshot inversion recovery (SNAPIR) approach 3 combined with a centric phase encode order shifts the balance toward T1w, but the inversion time was constrained to be short to preserve motion robustness by limiting the time for differential movement between inverting a slice and reading it out. Although a measure of T1 weighting was achieved by this sequence, it lacks sensitivity to pathology involving hemorrhage, which is generally better visualized by spoiled gradient‐echoes (GE) sequences, although as currently performed, the latter is very motion‐sensitive 1 (e.g., the use of 3D GE VIBE sequence targeting the fetal brain has been reported [7], but this scan often requires multiple repeats run under breath‐hold conditions). There remains an unfulfilled need for GE T1w methods that are robust to motion and that do not require a breath hold, so that reliable comprehensive T1w and T2w examinations can be achieved for the fetus as a direct parallel to routine practice in ex utero brain imaging. The approach reported in this paper seeks to address this need by exploring snapshot GE methods that can freeze fetal motion while still providing sufficient contrast‐to‐noise ratio (CNR) to be suitable for clinical diagnostic use as well as for scientific studies. As a first step, this work explores the optimal parameter space that maximizes T1 contrast between grey and white matter in the fetal brain while attenuating the signal from the CSF and amniotic fluid. The mechanism by which contrast is generated is similar to a 3D MP‐RAGE sequence 8, although achieving a snapshot capability results in a 2D acquisition and therefore we will refer to it as 2D MP‐RAGE. To render the sequence robust to anatomical displacements between inversion and readout caused by fetal motion and maternal breathing, a thicker inversion slab than the encoded slice is used. A fast single shot GE readout is then used to freeze fetal motion. Furthermore, the robustness of the sequence to motion is assessed by direct comparison to VIBE and via a controlled experiment on an adult moving volunteer. Finally, a simple rearrangement of the sequence building blocks as proposed in 9 was introduced allowing a much more efficient execution and hence a reduction of total scan time while preserving contrast. A clinical case is also presented and compared to established methods. Slice‐to‐volume (SVR) registration and reconstruction methods 10, 11 to achieve full high resolution 3D anatomical depiction of the fetal brain anatomy were also tested for the proposed acquisition and a sample result is presented in the Supporting Information (see Section 1.1 in the https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fmrm.27012&attachmentId=201902400 and https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fmrm.27012&attachmentId=201902400). A precursor approach that has since been improved on has been presented in abstract form 12.
METHODS
Our sequence is designed to:
provide the desired contrast with adequate SNR ratio
be robust to spontaneous fetal motion and to maternal breathing
be as efficient as possible thereby limiting total scan time
Each of these is discussed in turn.
Optimizing CNR
Given the performance issues noted above for the SNAPIR sequence 3, this work focused on gradient echo methods and requirement 2 drives toward snapshot approaches based on slice‐selective short pulse repetition time (TR) spoiled sequences. Spoiled gradient echo (SPGR) and inversion recovery magnetization prepared rapid acquisition gradient echo (MP‐RAGE) methods were compared in a simulation. The sequence structure of the latter is represented in Figure 1. To achieve robust inversions, an adiabatic pulse (arrow) is applied and then the longitudinal magnetization recovers back toward equilibrium during an inversion time (TI). Following this, a gradient echo readout (represented in Fig. 1 with a box labelled RAGE), which requires application of a train of RF pulses of flip angle α each separated by a TR period, disturbs the recovery for the entire readout duration (TL). Finally, a post readout delay (TD) can be introduced for magnetization recovery. We define the time between inversion (TS) as TS = TI + TL + TD.
Figure 1.
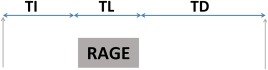
Basic inversion recovery gradient echo structure: inversions and readout are represented with arrows and boxes respectively. Timings of the sequence are also indicated.
To investigate an optimal operating point for all contrast parameters, 2 simulations were carried out: the first quantifies contrast in the SPGR, and the second in the MP‐RAGE sequence. All simulations used literature T1 values 13 for CSF and neonatal white and grey matter at 3T (4000, 2800, and 2000 ms) to explore optimal settings. For the SPGR sequence, a steady‐state value was obtained using the Ernst angle formalism 14 for different α‐TR combinations. Contrast between grey and white matter was then calculated as the difference of the transverse magnetization expressed in % and referenced to 1.
For the MP‐RAGE sequence, 3 subcases were considered. The first sequence achieves full motion tolerance using a non‐selective inversion, whereas the second is slice‐selective. A slab‐selective sequence, whose structure lies in between, is characterized by an inversion thickness considerably larger than the encoded slice.
In the non‐selective case, when the number of acquired slices is sufficient, the application of a train of global inversions at intervals TS sets up a steady state. This steady state is sampled 1 slice at a time, and then it is reasserted during subsequent TS periods. The situation differs substantially with a slice‐selective inversion, as the inversion pulses only act on their designated slice so do not contribute in determining an initial steady‐state value for the magnetization of other slices. Furthermore, because many slices are needed to cover the fetal brain with margin for motion, we can reasonably assume that fully recovered magnetization before any inversion is achieved for a wide range of relevant readout durations and TI (or, equivalently, that TS → ∞). Finally, the achieved inversion to inversion time in the slab‐selective sequence will lie in between these 2 extremes.
To investigate contrast, the forward simulation was divided into 3 steps: first, an initial steady‐state value of the longitudinal magnetization was obtained using the Ernst angle formula for a wide range of TS values. The magnetization is then allowed to evolve by T1 recovery for increasing TI to determine the longitudinal magnetization immediately before the RAGE loop. Finally, the signal evolution through the RAGE module is obtained by alternating excitations and signal recoveries for different combinations of the repetition time TR and the flip angle α. In here, is the magnetization at equilibrium and and are, respectively, the longitudinal magnetization before and after each process. Contrast between grey and white matter is calculated as in the SPGR simulation from the difference in predicted tissue signals when the center of k‐space is acquired. In all simulations and in vivo, a center‐out phase encoding order is adopted.
Motion Robustness
Motion can disrupt the sequence performance in 2 distinct ways. First, movement during the RAGE module causes motion artefacts and blurring and therefore it is advantageous to minimize TL. More generally, the use of short readouts in the context of fetal imaging has always been recommended 1, 2, 7, 15. However, reducing this time limits the achievable combinations of in plane resolution and field of view although this can be mitigated to some extent using SENSE 16 or Grappa 17. In past work, we have found that a time per full RAGE module <1250 ms is sufficient to freeze fetal motion for most slices acquired during a fetal examination. Second, movement during the inversion time TI can cause sampled slices to contain non‐inverted magnetization. In 3, this issue was addressed by limiting the time delay TI to 400 ms. However, optimal performance in terms of contrast and SNR may require longer TI periods, so in this work other approaches have been explored. Complete immunity from movement during TI can be achieved by employing a non‐spatially selective inversion. As described in the previous section, the application of a train of global inversions at intervals TS sets up a dynamic steady state. If TS is short, then there can be significant saturation of longitudinal magnetization throughout, reducing both SNR and CNR, so that the time between inversions can become a critically limiting parameter. An intermediate approach is to simply increase the thickness of the inverted slab so that its profile is considerably thicker than the one used for the slice‐selective gradient echo readout. This approach is more flexible allowing adjustment of the amount of motion that can be accommodated during TI, but it must be combined with a suitable slice interleaving strategy as it is then a requirement that successively inverted slabs do not overlap in space. As many slices are needed to cover the fetal head, there is substantial scope in this regard and the approach inherently increases TS and hence potentially ameliorates problems of saturation without sacrificing total scanning time. In this study, to create reasonable room for the inversions, the selected slice acquisition order was 1–7–14…2–8–15…42, so that it was possible to set the inversion slab thickness to 18 mm (i.e., 4.5 times larger than the slice thickness of the gradient echo readout).
Motion Experiments
The robustness of the sequence to motion is assessed in 2 distinct experiments. The first compares the proposed slab‐selective 2D sequence with 2 variants of VIBE (7), and the second is a motion‐controlled brain experiment on an adult volunteer comparing 2D versus 3D MP‐RAGE.
VIBE experiment
Two subjects of 31 + 6 and 34 + 5 weeks were scanned with/without spectral attenuated inversion recovery (SPAIR) pulses 18 for efficient fat suppression using VIBE (7).
Sequence parameters were: field of view 300 × 330 × 100 mm3 with in plane resolution matched to 2D MP‐RAGE (see below) and 4 mm voxel size along the second phase encode, foot/head (FH). Similarly to what was used in 7, TR, TE, and flip angle were 3.7 ms, 1.98 ms, and 9°, respectively. To further shorten the echo train length allowing breath holds scans in pregnant women, a SENSE factor of 2 was used along the anterior/posterior (AP) direction (first phase encode) in conjunction to partial Fourier factors of 0.7 and 0.85 along AP and FH. With these settings, the achieved acquisition time per volume was 9.4 and 11.7 s, respectively, and the shot duration was slightly different because of the absence/presence of SPAIR pulses in between contiguous kz planes. Finally, in the free‐breathing scans, 20 volumes were acquired in total for an acquisition time of 188 and 234 s.
Adult volunteer
In the adult experiment, we compare a (modified) 2D MP‐RAGE scan with a conventional 3D MP‐RAGE sequence on an adult moving volunteer.
Because it is not feasible to match contrast for these 2 scans without compromising performance of either or both, we opted to run each in a way that is individually tuned. Therefore, the 3D sequence was adapted directly from a standard high contrast vendor adult protocol, and the 2D sequence was kept close to the fetal protocol (see below). This meant that there was little internal brain tissue contrast in the latter, but because it has a high CSF–brain contrast, any motion‐related issues would be clearly seen.
Both scans had 1 mm in plane resolution and a field of view (FOV) providing coverage of a cubic region measuring 240 mm along AP, right/left (RL), and FH directions. Furthermore, they had identical shot durations of 1126 ms, which was kept close to the one used in the fetal sequence. The 3D scan had 1‐mm section thickness and the 2D scans had 2.2‐mm slice thickness. Both would later be reconstructed to 1 mm isotropic voxels. The 2D scan used an inversion slab thickness of 17.6 mm, and a shift of 2 to reduce scanning time (see Efficiency), for a TS of 12 s and TI of 2500 ms (i.e., the same inversion time as in the fetus) (see below). In total, 110 slices were acquired with slice order: 1–11–…101–2–12–102–10–20…–110. Each 2D scan took 2 min and 16 s and there were 4 repeats (1 × transverse, 2 × sagittal, and 1 × coronal), taking a total of 9 min and 4 s.
Other parameters for the 3D case were: SENSE factor 1.84 along the primary (RL) phase encode direction. TI, TR, TE, and flip angle were 900, 8.4, 3.8 ms, and 8°, respectively. Total scan time was 7 min and 33 s. Finally, in the 2D scan, SENSE factor, TR, TE. and flip angle were matched to 3D MP‐RAGE.
The full protocol was run twice in the same session—once with the volunteer (26‐year‐old male) staying still and once with the subject instructed to move once every 10 s and then to rest. This motion pattern was chosen to represent typical fetal head motion, where the fetus often alternates brief periods of activity, particularly rotations of the head, interspersed with longer quiet periods 19. To ensure reproducibility between different scans, motion triggering was achieved by a tone played through the subject's MR‐compatible headset. SVR was finally used to reconstruct the 2D MP‐RAGE data into 3D volumes.
Efficiency
Long inversion times and, in the case of global inversions, long durations for TS can result in excessive acquisition time with very low efficiency. Slab‐selectivity speeds up the acquisition compared to the global inversion for a fixed level of contrast, but simple‐slice interleaving approaches (Fig. 2a) in which inversion and readout are treated as a single block result in massive inefficiency with the scanner being idle for as much as 70% of the time for some relevant TI and TL combinations. We therefore implemented a sequence modification that allows full interleaving of inversion pulses and RAGE modules to minimize dead time 9. More specifically, Figure 2a shows the conventional 2D MP‐RAGE sequence as implemented in our scanner, whereas Figure 2b shows the fully interleaved structure for a case with 5 acquired slices. Different colors signify different slice locations. The efficiency of the fully interleaved approach depends on the relationship between TI and TL, with the optimum being achieved when the former is an exact multiple of the latter. In the example of Figure 2b, the applied shift is equal to 1 slice location. In the case of the in vivo sequence, given that 2 readouts could fit within the inversion time TI that maximizes contrast (see below), the number of shifts is 2. Note that to ensure that initially acquired slices have the correct contrast, it is necessary to introduce dummy inversions equal to the applied shifts (indicated by the asterisk in Fig. 2b).
Figure 2.
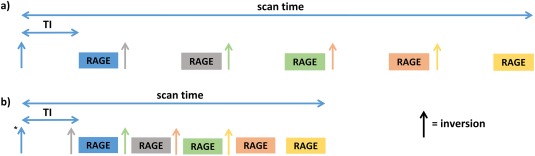
Conversional (a) versus proposed approach leading to maximal efficiency (b). In both figures, inversions and GE modules are represented with arrow and boxes and different colors encode different slice locations.
Experimental Factors
All slices data was acquired with a FOV of 320 × 320 mm2, encoded at 1.5 mm isotropic in plane resolution. To accommodate a typical fetal head up to late gestational age, 120 mm coverage is required and this was assumed to be acquired using at least 42 slices. To explore SNR issues, tests included thicker slices with overlap as needed to preserve constant slice spacing. A range of interleaved slice orders were available to allow flexibility needed to avoid slice cross talk and to achieve motion robustness. Whenever possible, the phase encoding direction was set to be AP, which requires less encoding. Scans were all performed with the mother in the supine position 20.
The approach was tested on 21 fetuses ranging from 26 to 38 weeks gestational age (wGA) and a clinical case of 30 weeks. Written informed consent was obtained from all the women. All data in this study was acquired on a Philips 3T Achieva scanner using a 32‐channel cardiac array coil wrapped around the abdomen.
Stacks of T1‐weighted data were acquired transverse, sagittal, and coronal to the fetal head or to the maternal abdomen. Other parameters were TE of 4.8 ms, readout with minimum bandwidth to maximize SNR, and a SENSE factor of 2. Taken together, the above settings imply 113 phase encode steps with a total RAGE readout time of 1130 ms. Finally, all data acquired in a single examination was reconstructed jointly at 0.75 mm isotropic resolution using SVR 11, and 1 example is reported in https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fmrm.27012&attachmentId=201902400 in Supporting Information Section 1.1.
RESULTS
Figure 3a shows grey–white matter contrast of the steady‐state SPGR sequence plotted as a function of repetition time TR and flip angle α. The dotted line follows the optimal settings for SNR, obtained using the Ernst angle formalism for a T1 value of 2000 ms. The blue line in plot Figure 3b shows the same quantity as in Figure 3a plotted as a function of the inversion recovery time TI for the slice selective 2D MP‐RAGE sequence (hence TS → ∞).
Figure 3.
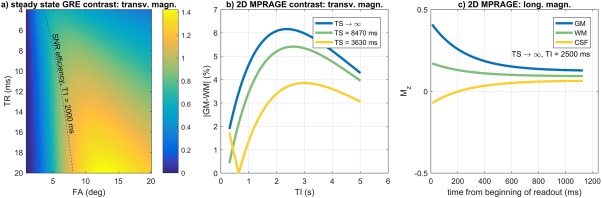
(a) Contrast between grey and white matter in steady‐state gradient echo as a function of TR and flip angle α. The dash line represents the optimal settings for SNR using the Ernst angle formula. (b) MP‐RAGE contrast simulations. In blue, contrast between grey and white matter as a function of the inversion time TI (quantity evaluated at k = 0). In green and yellow: same quantity using TS values of 8470 and 3630 ms. (c) Evolution of the longitudinal magnetization for grey (blue), white (green) matter, and CSF (yellow) for 2D MP‐RAGE against a readout formed by 113 phase encodes, TR of 10 ms, and flip angle of 15 °. TI = 2500 ms and TS is approaching infinity.
In Figure 3c, the behavior of the longitudinal magnetization as a function of time during the RAGE module for CSF, white, and grey matter is plotted for the TS → ∞ and TI of 2500 ms, which is close to a global optimum. The readout is formed by 113 phase encodes, TR of 10 ms, and a flip angle of 15°.
The comparison between Figures 3a and 3b (blue line) suggests 2 things: first, a higher contrast can be achieved with an inversion pre‐pulse (maximum of ∼6.1% with a TI of ∼2.5 s) than the steady‐state gradient echo sequence (maximum of ∼1.4% for the best TR‐α combination); second, whereas in Figure 3a the choices for optimal contrast and SNR do not necessarily coincide and hence a trade‐off has to be made, in Figure 3b, given that contrast is set by TI using a centre‐out k space order, there is more operational room for SNR optimization. In practice, increasing TR increases SNR but rapidly makes the shot duration excessive causing vulnerability to motion, and a high flip angle saturates the signal quickly. In this study, the final values for TR and α were chosen as the result of these trade‐offs during in vivo tests for a fixed encoding matrix. Note also that in Figure 3c, this combination for TI, TR, and α suppresses the signal from CSF (and amniotic fluid) throughout the echo train. Figure 4 shows the comparison between different data sets acquired at increasing TI values on a subject of 32 + 3 weeks (1500, 2000, 2500, and 3000 ms). Whereas at TI = 1500 ms the brain tissue appears close to being nulled and the CSF is inverted (see arrows), the image at TI = 2000 ms shows inconsistencies at the CSF‐brain tissue boundaries (arrows) because of destructive signal interaction (negative longitudinal magnetization for CSF versus positive in brain tissue within the same voxel). The image at 2500 ms has the CSF nulled and grey–white matter boundaries that are clearly distinguishable (arrow). At 3000 ms, the SNR is greater because of a higher degree of recovery, but contrast is decreased.
Figure 4.
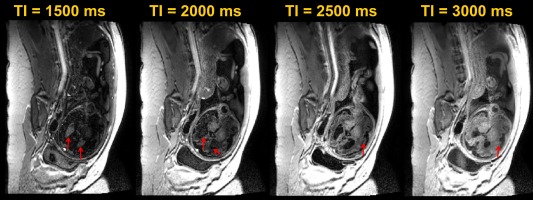
Transverse slice of a 32‐ + 3‐week‐old fetus acquired at increasing TI values (1500, 2000, 2500, and 3000 ms).
The yellow and green lines in Figure 3b report the simulated grey‐white matter contrast using the global inversion and the slab‐selective inversion for values of TS of 3630 and 8470 ms, respectively. These values correspond to the times between successive inversion pulses operating on the same spatial locations for sequence parameters required to achieve typical resolution and coverage suitable for fetal brains up to term age. Note that in each case there is an optimal contrast, but this decreases and shifts to larger TI values as TS declines. Although a global inversion would ensure robustness to motion at all TIs, a longer TS is generally preferable—and therefore, in this study, the slab‐selective sequence was chosen as the best compromise between complete motion robustness and enhanced contrast.
Figures 5a and 5b show coronal and sagittal T1 data acquired on a fetus of 26 weeks of gestation. In both images, grey and white matter are easily distinguishable, whereas the signal from CSF and amniotic fluid is uniformly nulled. Fat, muscles, and soft maternal tissue appear bright because of their short T1. Despite the substantial amount of motion present in the second scan, grey and white matter are still distinguishable in the native sagittal plane.
Figure 5.
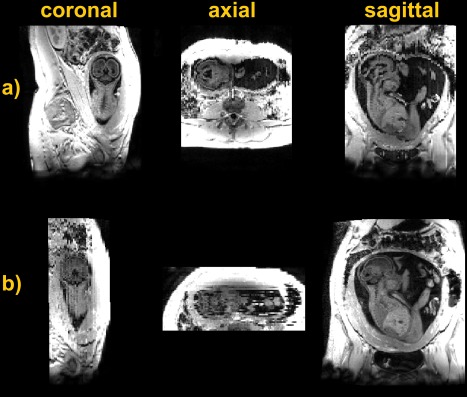
Data acquired (a) coronal and (b) sagittal to a fetus of 26 weeks with 2D slab‐selective MPRAGE.
In what follows, we compare the proposed approach to 3D VIBE 7. Figure 6 shows 2D MP‐RAGE data (Fig. 6a) and a 3D single shot VIBE sequence run during free breathing (Fig. 6b) and during a breath hold (Fig. 6c) with and without fat suppression using SPAIR pulses (1st and 2nd rows). By comparing the data in Figure 6, the following observations can be made.
Figure 6.
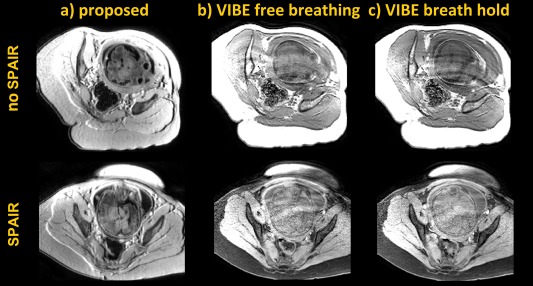
: 2D MP‐RAGE (a) and 3D VIBE (b and c) run on two subjects of 31 + 6 and 34 + 5 weeks (first and second rows). b and c) Free breathing and breath hold VIBE scans. In the second experiment, SPAIR pulses for efficient fat suppression were employed.
First, artefacts deriving from maternal respiration significantly degrade the quality of the acquired data in both VIBE acquisitions, especially in the regions close to the fetal head. This is because of the long echo train length required for VIBE where, in 9.4 and 11.7 s the mothers often perform multiple breaths. Furthermore, all of the 20 volumes in the non‐breath hold scans were damaged. The breath hold scans (Fig. 6c) show a lower level of damage, but are not as artefact free as would be hoped. We attribute this to challenge of the mothers to achieve even the limited duration of the required breath hold.
Second, and in accordance with the simulation of Figure 3, increased contrast between grey and white matter can generally be observed with 2D MP‐RAGE as opposed to 3D VIBE, which is a steady‐state gradient echo sequence. This disparity is made worse by the inherent requirements of VIBE where, for motion and respiration robustness, the shortest TR must be used, whereas longer TRs and higher flip angles are required for increased contrast (Fig. 3a).
Figure 7 shows the results obtained in the adult experiment. The first and second rows report the 3D MP‐RAGE data acquired without and with motion. Motion artefacts spread along all 3 orthogonal directions when the subject moves because of the individual shots being inconsistent with each other 21. The third row shows 2D MP‐RAGE coronal data acquired during the motion condition. Here, the disruption caused by motion is mostly evident in the through slice direction, whereas individual slices are less damaged in the native plane (Fig. 7, center). The fourth and last row reports the result of SVR reconstruction, which produces 3D volumetric image data that is free of significant motion corruption. As expected, grey–white matter contrast was poorer in all 2D scans. This is because of the shorter T1 of the adult brain 22 compared to fetal brain, in combination with the long inversion time that was used. The latter parameter had to be kept long to null the signal contribution from the CSF, which otherwise would have been negative. Luckily, in the fetal case, the TI that maximizes gray–white contrast roughly corresponds to the point when the CSF nulls (Fig. 3c).
Figure 7.
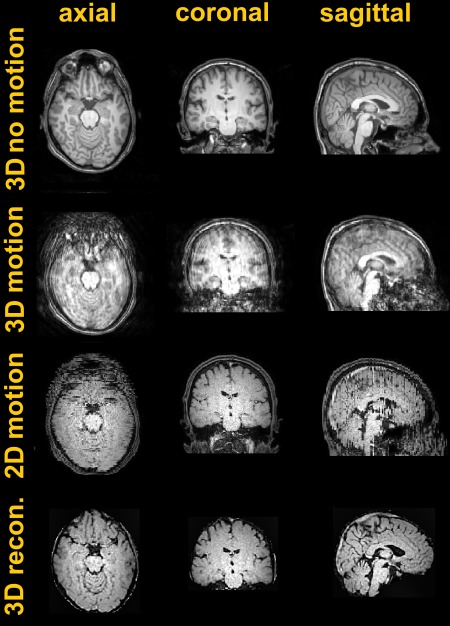
Motion experiment run on an adult volunteer. First and second rows: 3D MP‐RAGE data without and with motion. Third row: 2D MP‐RAGE data acquired coronal to the brain during motion. Fourth row: SVR reconstruction obtained by combining 4 orthogonal views.
Although the sequence was designed to be robust to motion, there can be instances in which 2D MP‐RAGE fails, and examples from examinations on 4 babies are shown in Figure 8. In these examples, rotations and translations during the inversion time TI were so large that the gradient echo readout excited a slice outside the inverted slab, and hence it read fully recovered magnetization that appears brighter than the surrounding tissue. This type of artefact requires large motion excursions and, in most of the cases, the problem does not arise. Out of the total 4536 slices that were collected from all subjects in this study, only 39 (0.8%) were affected. Furthermore, and as mentioned above, a global inversion could cope with this problem at the expenses of efficiency—and a sequence with equivalent contrast properties would indeed require ∼6 min to run, as opposed to 56 s for the proposed slab‐selective approach (or 2 min 30 s without optimal inversion and readout arrangement).
Figure 8.
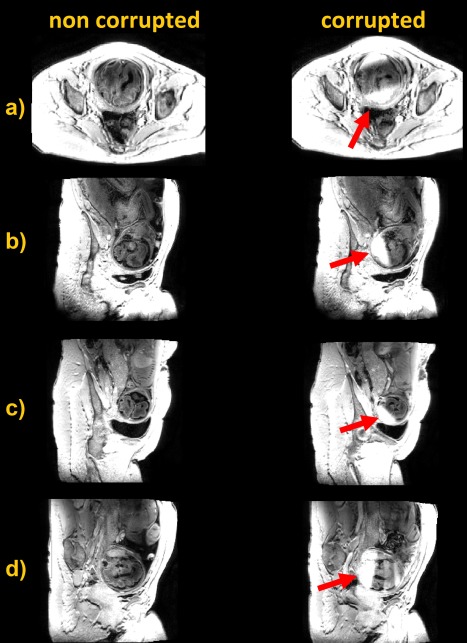
Artefact free data (left) and slab‐inversion motion artefact (right) deriving from motion during the inversion recovery time TI in 4 different subjects.
Finally, a scan on a clinical subject is presented comparing our approach with SNAPIR. The SNAPIR sequence 3 gives excellent tissue contrast but key features of the brain may not be well visualized because of loss of short T1 signal (e.g., the fetal pituitary gland). Our optimized T1 sequence retains all short T1 elements within the brain (Fig. 9a) with excellent detection of the fetal pituitary on a subject of 30 weeks, identification of which is important when evaluating the fetus with a midline defect such as agenesis of the corpus callosum. Fetal brain pathology includes a variety of short T1 lesions such as hemorrhage or sub‐ependymal nodules, the depiction of which is more readily seen with the T1 sequence than SNAPIR (Figs. 9b and 9c). Single‐shot T2 FSE are also reported as reference and its acquisition parameters together with the SNAPIR sequence are described in the figure caption.
Figure 9.
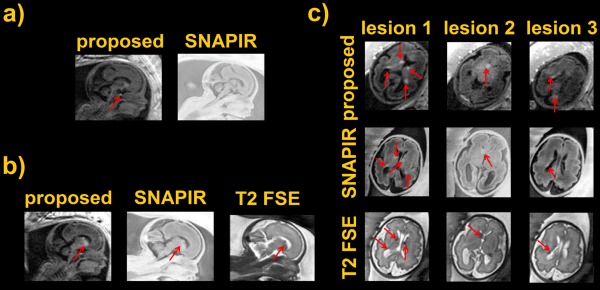
(a) Proposed sequence versus standard SNAPIR. Fetal pituitary is indicated by the red arrow. (b,c) Sagittal scans comparing 2D MP‐RAGE, SNAPIR, and a T2 single‐shot acquisition. Arrows indicate the location of pathology consistent with a diagnosis of tuberous sclerosis. The SNAPIR sequence was acquired at a resolution of 1 × 1 × 4 mm³ with TI of 400 ms. The T2 single shot scan at 1.25 × 1.25 × 2.5 mm³ with a TE of 180 ms and a TR of 30.9 s.
DISCUSSION
Assessing the health of the fetus is standard clinical practice to exclude both genetic and acquired abnormalities and to ensure appropriate growth and development. This is routinely performed with ultrasound (US), however, numerous studies confirm the additional importance of MR imaging. In current practice, the clinical assessment of the health of the fetus with MR is mainly performed using single‐shot T2w FSE. Other clinical sequences include diffusion‐weighted images 23, rapid gradient echo bSSFP images 19, spectroscopy 24, and rapid gradient echoes with either T1 or T2 contrast 1. However, T1w imaging is both less used and poorly optimized then T2w imaging.
There are formidable challenges to overcome during a fetal MRI exam, as the fetus lies within the mother and it performs its own sporadic and unpredictable movements. Therefore, motion is a core issue for any fetal MR scan. In this study, a T1‐weighted acquisition that maximizes contrast between grey and white matter in the fetal brain is explored. The approach, with the primary aim to decrease the effects of motion, uses a combined approach that integrates the classical single shot readout paradigm to freeze motion and a thick slab inversion. Because the long inversion time needed for contrast preparation creates an inefficient sequence, a simple modification 9 was introduced and a 2.67‐fold reduction in total scan time was achieved without loss in data quality. Finally, slice to volume registration and super resolution reconstruction 11 was used to achieve full 3D representation of the fetal head at 0.75 mm isotropic resolution (see https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fmrm.27012&attachmentId=201902400 and https://onlinelibrary.wiley.com/action/downloadSupplement?doi=10.1002%2Fmrm.27012&attachmentId=201902400). The sensitivity of the sequence to pathology is compared to standard methods in an example clinical case. Robustness against motion is also assessed by direct comparison to other techniques.
As shown in Figure 8, the slab‐selective approach can occasionally fail especially in the presence of very marked fetal motion. An alternative strategy is represented by the use of a non‐selective inversion, at the cost of either saturating the signal and hence ruining contrast, or increasing total scan time considerably by inserting a post readout delay. The use of multiband imaging, where several slices are excited simultaneously and the sensitivity information from all coils is used to unfold the signal, represents an attractive way to speed up acquisition 25. However, a multiband factor of 6 would be needed to match contrast and total scan time to the described slab‐selective sequence. It is clear that such a high acceleration would incur in large SNR penalty and be prone to unfolding artefacts, suggesting that the slab‐selective approach is likely to remain the more efficient of the two.
CONCLUSION
The present work proposes an efficient high contrast motion tolerant approach to perform T1 acquisitions and reconstructions of the fetal brain in utero. It shows promise for both detection of pathology and may be valuable for quantifying fetal brain development.
Supporting information
Fig. S1. SVR reconstruction. First row: stack of data acquired on a fetus of 32 + 2 weeks. Second row: slice to volume and super resolution reconstruction.
ACKNOWLEDGMENTS
This work has been performed entirely at the Centre for the Developing Brain, King's College London. This work received funding from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007‐2013)/ERC grant agreement 319456 and was supported by the Wellcome EPSRC Centre for Medical Engineering at King's College London (WT 203148/Z/16/Z), and by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St. Thomas' NHS Foundation Trust and King's College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
REFERENCES
- 1. Prayer D, Brugger PC, Prayer L. Fetal MRI: techniques and protocols. Pediatr Radiol 2004;34:685–693. [DOI] [PubMed] [Google Scholar]
- 2. Yamashita Y, Tomohiro N, Abe Y, Takahashi M, Iwamasa J, Miyazaki K, Okamura H. MR imaging of the fetus by a HASTE sequence. AJR Am J Roentgenol 1997;168:513–519. [DOI] [PubMed] [Google Scholar]
- 3. Malamateniou C, McGuinness A, Allsop J, O'Regan D, Rutherford M, Hajnal JV. Snapshot inversion recovery: an optimized single‐shot T1‐weighted inversion‐recovery sequence for improved fetal brain anatomic delineation. Radiology 2011;258:229–235. [DOI] [PubMed] [Google Scholar]
- 4. Hand JW, Li Y, Thomas EL, Rutherford MA, Hajnal JV. Prediction of specific absorption rate in mother and fetus associated with MRI examinations during pregnancy. Magn Reson Med 2006;55:883–893. [DOI] [PubMed] [Google Scholar]
- 5. Murbach M, Neufeld E, Samaras T, Córcoles J, Robb FJ, Kainz W, Kuster N. Pregnant women models analyzed for RF exposure and temperature increase in 3T RF shimmed birdcages. Magn Reson Med 2017;77:2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weigel M. Extended phase graphs: dephasing, RF pulses, and echoes ‐ pure and simple. J Magn Reson Imaging 2015;41:266–295. [DOI] [PubMed] [Google Scholar]
- 7. Gholipour A, Estroff J, Barnewolt C, et al. Fetal MRI: a technical update with educational aspirations. Concepts Magn Reson Part A Bridg Educ Res 2014;43:237–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mugler JP, Brookeman JR. Three‐dimensional magnetization‐prepared rapid gradient‐echo imaging (3D MP RAGE). Magn Reson Med 1990;15:152–157. [DOI] [PubMed] [Google Scholar]
- 9. Oh CH, Hilal SK, Mun IK, Cho ZH. An optimized multislice acquisition sequence for the inversion‐recovery MR imaging. Magn Reson Imaging 1991;9:903–908. [DOI] [PubMed] [Google Scholar]
- 10. Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. IEEE Trans Med Imaging 2007;26:967–980. [DOI] [PubMed] [Google Scholar]
- 11. Kuklisova‐Murgasova M, Quaghebeur G, Rutherford MA, Hajnal JV, Schnabel JA. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Med Image Anal 2012;16:1550–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ferrazzi G, Price AN, Teixeira RP, Padormo F, Cordero‐Grande L, Hughes E, McCabe L, Rutherford M, Kuklisova Murgasova M, Hajnal JV. An optimised 2D MPRAGE sequence for T1 contrast in the fetal brain: application to slice to volume reconstruction and multiband acceleration. In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, Hawaii, USA, 2017. Abstract 1942.
- 13. Williams LA, Gelman N, Picot PA, Lee DS, Ewing JR, Han VK, Thompson RT. Neonatal brain: regional variability of in vivo MR imaging relaxation rates at 3.0 T ‐ initial experience. Radiology 2005;235:595–603. [DOI] [PubMed] [Google Scholar]
- 14. Haacke EM, Brown RW, Thompson MR, Venkatesan R. Magnetic resonance imaging: physical principles and sequence design. New York: Wiley‐Liss; 1999. 944 p. [Google Scholar]
- 15. Stehling MK, Mansfield P, Ordidge RJ, Coxon R, Chapman B, Blamire A, Gibbs P, Johnson IR, Symonds EM, Worthington BS. Echo‐planar imaging of the human fetus in utero. Magn Reson Med 1990;13:314–318. [DOI] [PubMed] [Google Scholar]
- 16. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med 1999;42:952–962. [PubMed] [Google Scholar]
- 17. Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002;47:1202–1210. [DOI] [PubMed] [Google Scholar]
- 18. Lauenstein TC, Sharma P, Hughes T, Heberlein K, Tudorascu D, Martin DR. Evaluation of optimized inversion‐recovery fat‐suppression techniques for T2‐weighted abdominal MR imaging. J Magn Reson Imaging 2008;27:1448–1454. [DOI] [PubMed] [Google Scholar]
- 19. Hayat TT, Nihat A, Martinez‐Biarge M, McGuinness A, Allsop JM, Hajnal JV, Rutherford M. Optimization and initial experience of a multisection balanced steady‐state free precession cine sequence for the assessment of fetal behavior in utero. AJNR Am J Neuroradiol 2011;32:331–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hughes E, Price AN, McCabe L, Pegoretti Baruteau K, Hutter J, Carney O, Gaspar AO, Hajnal JV, Rutherford M. Magnetic resonance imaging quantification of venous return in pregnant women: a comparison between supine and left lateral tilt position. In Proceedings of the 24th Annual Meeting of ISMRM, Singapore, 2016. Abstract 0969.
- 21. Cordero‐Grande L, Hughes EJ, Hutter J, Price AN, Hajnal JV. Three‐dimensional motion corrected sensitivity encoding reconstruction for multi‐shot multi‐slice MRI: application to neonatal brain imaging. Magn Reson Med 2018;79:1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wansapura JP, Holland SK, Dunn RS, Ball WS Jr. NMR relaxation times in the human brain at 3.0 tesla. J Magn Reson Imaging 1999;9:531–538. [DOI] [PubMed] [Google Scholar]
- 23. Kasprian G, Del Río M, Prayer D. Fetal diffusion imaging: pearls and solutions. Top Magn Reson Imaging 2010;21:387–394. [DOI] [PubMed] [Google Scholar]
- 24. Kok RD, Van Den Berg PP, Van Den Bergh AJ, Nijland R, Heerschap A. Maturation of the human fetal brain as observed by 1H MR spectroscopy. Magn Reson Med 2002;48:611–616. [DOI] [PubMed] [Google Scholar]
- 25. Larkman DJ, Hajnal JV, Herlihy AH, Coutts GA, Young IR, Ehnholm G. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J Magn Reson Imaging 2001;13:313–317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. SVR reconstruction. First row: stack of data acquired on a fetus of 32 + 2 weeks. Second row: slice to volume and super resolution reconstruction.


