Abstract
Aim
This study assesses a clinical potential of immediate early responsive gene X‐1 (IEX‐1), also named IER3, in the diagnosis of epithelial ovarian carcinoma using blood and salivary specimens.
Methods
Immediate early responsive gene X‐1 was quantified in blood and saliva by real‐time quantitative reverse transcription polymerase chain reaction in 26 cases of epithelial ovarian carcinoma, 37 cases of benign ovarian tumor and 55 cases of healthy women. The receiver operating characteristic (ROC) curve was used to evaluate the diagnostic efficacy of IEX‐1.
Results
Immediate early responsive gene X‐1 was expressed in blood and saliva of the benign ovarian tumor group and the healthy women group, both at a level significantly higher than that of the ovarian carcinoma group (P < 0.017). There were no significant differences in IEX‐1 expression in blood and saliva (P = 0.376 or 0.621, respectively) between the benign ovarian tumor and the healthy women group. Comparison of IEX‐1 expression in blood between the ovarian carcinoma group and the benign ovarian tumor group or the healthy women group demonstrated the ROC‐area under curves (AUC) of 0.947 or 0.929, respectively. In discriminating the ovarian carcinoma group from the benign ovarian tumor group, IEX‐1 expression in blood demonstrated a sensitivity and specificity of 84.6% and 94.6%, respectively. Similarly, blood IEX‐1 expression conferred a sensitivity of 84.6% and specificity of 90.9% in distinguishing the ovarian carcinoma group from the healthy women group. Moreover, salivary IEX‐1 expression had ROC‐AUC of 0.851 when compared between the ovarian carcinoma group and the benign ovarian tumor group or 0.896 when compared between the ovarian cancer group and the healthy women group. IEX‐1 expression was able to discriminate the ovarian carcinoma group from the benign ovarian tumor group with a sensitivity and specificity of 65.4% and 94.6%, respectively, or the ovarian carcinoma from the healthy women with 92.3% sensitivity and 72.5% specificity.
Conclusion
These results suggest the clinical potential of IEX‐1 expression in blood and saliva as a sensitive and specific diagnosis for epithelial ovarian carcinoma.
Keywords: carcinoma, epithelial ovarian, IEX‐1, qRT‐PCR, ROC curve
Introduction
Ovarian cancer is the most common cause of mortality among gynecological cancers.1 Owing to atypical clinical symptoms and insufficient screening of available biomarkers, 70% patients have widespread metastasis at advanced stages when diagnosed. Ovarian cancer is an extremely invasive tumor, highly resistant to chemotherapy and 5 years’ survival rates only at 20–30% even if chemotherapy is delivered after surgical treatment.2 Although significant advancements have been made in ovarian cancer treatment, the survival of patients highly depends on early diagnosis. Serum tumor biomarkers offer important hints in the initial diagnosis of ovarian cancer, evaluation of curative effects and prognosis, but the specificity and sensitivity are relatively low.3, 4, 5 Therefore, it is essential to explore novel biomarkers with higher sensitivity and specificity for the diagnosis of ovarian cancer malignancy. Immediate early response gene X‐1 (IEX‐1) is a member of the immediate early responsive gene family. IEX‐1 is known to be involved in cell growth, differentiation and apoptosis under cellular stress. Our early study found a significant downregulation of IEX‐1 in epithelial ovarian tumors,6 suggesting that IEX‐1 may be involved in the suppression of epithelial ovarian carcinoma development. The finding raises a possibility that its level of expression may serve as a specific biomarker for early diagnosis of this disease. In clinical practice, blood‐based diagnostic assays are the most widely applied for tumor markers, with advantages of easy procedures, high sensitivity and cost‐effectiveness. Saliva‐based diagnostic assay is presently at a mature juncture of being evaluated as a means of detecting ovarian cancer.7 In this investigation, differential expressions and diagnostic values of IEX‐1 in human blood and saliva were validated among different groups using quantitative reverse transcription polymerase chain reaction (qRT‐PCR). Our results suggest the big clinical potential of using saliva as a specimen in the diagnosis of ovarian cancer.
Methods
Patients and controls
All the blood and saliva samples were obtained from patients presenting with pelvic mass at the First Affiliated Hospital of Zhengzhou University between December 2014 and June 2015. According to the postoperative pathological report, the patients were divided into epithelial ovarian carcinoma and benign ovarian tumor groups. The control individuals comprised 55 cases of healthy women seeking routine health checkup during the corresponding period. The epithelial ovarian carcinoma stage was categorized according to FIGO criteria (2014), based on which nine were in stage I/II and 17 were in stage III/IV. The tumors were classified after surgery as serous carcinoma (n = 13), mucinous carcinoma (n = 4) and other carcinoma (n = 9). Eight cases were histologically graded as G1/G2 and 18 cases as G3. Exclusion criteria were the presence or suspicion of any infectious disease, radiotherapy or chemotherapy. The benign ovarian tumor group included 13 mature cystic teratomas, 15 simple ovarian cysts, six serous cystadenomas and three mucinous cystadenomas. Hypertension, diabetes, hepatitis B virus (HBV) infection and AIDS were excluded in all the control subjects. In addition, liver and kidney function as well as chest X‐rays were normal. Above all, thyroid and breast mass must be ruled out by ultrasound examination. Mean ages of the epithelial ovarian carcinoma group, the benign ovarian tumor group and the healthy women group were 44.1 ± 6.2, 39 ± 7.4 and 43 ± 8.7 years, respectively. There were no significant differences in the age among the three groups. Each participant provided a written consent form and an ethics permission was obtained before sample collection.
Sample collection and storage
Peripheral blood of 1–3 mL was drawn from each patient or control into a tube containing ethylenediaminetetraacetic acid. Blood samples were stored at 4°C from which RNA was extracted within 12 h. For saliva collection, spontaneous 3–5 mL unstimulated saliva specimens were expectorated into clean tubes after rinsing the mouth with distilled water.7 Saliva samples were centrifuged at 3000 g for 15 min at 4°C, then at 12 000 g for 10 min at 4°C and precipitated to obtain the supernatant. The supernatant samples were stored at −80°C until use.
RNA extraction
Total RNA of blood samples was extracted using Trizol BD Reagent (MRC) by mixing 200 μL whole blood with 750 μL Trizol BD Reagent and incubated for 5 min at room temperature, after which 200 μL chloroform was added to extraction. The mixture was then centrifuged at 12 000 g for 15 min at 4°C, the upper aqueous layer was collected, to which 600 μL of isopropyl alcohol was added, followed by centrifugation at 12 000 g for 10 min at 4°C to precipitate the RNA. The resultant RNA was washed once by 1 mL 75% ethanol and dried at room temperature. RNA was dissolved in the final volume of 40 μL RNase‐free water.
Salivary RNA was isolated from the cell‐free saliva supernatant according to the modified protocol from the manufacturer8 (QIAamp Viral RNA Kit). Saliva of 140 μL was mixed well with 560 μL AVL buffer for 10 min at room temperature. Absolute ethanol (560 μL) was added to the solution, followed by loading the solution onto a silica column and centrifugation at 6000 g for 1 min. The column was then washed twice, centrifuged at 6000 g for 2 min, after which 500 μL of Buffer AW1 first and then 500 μL of Buffer AW2 were added to the column. Finally, 60 μL of Buffer AVE was used to elute the RNA. Aliquots of RNA were then treated with RNase‐free DNase (DNase I‐DNA‐free; Ambion Inc.) according to the manufacturer’s instructions. The RNA quality and quantity were measured by spectrophotometer analysis (NanoDrop ND‐1000 Spectrophotometers; Thermo Scientific).
Quantification of IEX‐1 RNA by qRT‐PCR
Briefly, 1 μL of total RNA was reverse transcribed to cDNA according to the manufacturer’s instructions of Super Quick RT cDNA Kit (Kangweishiji) and the final product was diluted 20 times for qRT‐PCR. The cDNA was amplified by qPCR with specific primers in a reaction (20 μL) including cDNA template 2 μL, distilled water 4.4 μL, SYBR Real‐time PCR Master Mix 10 μL (TOYOBO), Plus solution 2 μL (TOYOBO), forward and reverse primer 0.8 μL, separately. qRT‐PCR was performed on the Applied Biosystems 7500 Sequence Detection System (Applied Biosystems). The qPCR reactions were carried out using the following conditions: 95°C for 1 min, 95°C for 15 s, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, then hold at 4°C. IEX‐1 primers were 5′‐GCCGCCTTCTAACTGTGACTC‐3′(forward) and 5′‐GTCTCCGCTGTAGTGTTCTGAG‐3′(reverse) and the primer pair of β‐actin were 5′‐TGACGTGGACATCCGCAAAG‐3′ and 5′‐CTGGAAGGTGGACAGCGAGG‐3′ for endogenous control. The raw data were analyzed using the automatic cycle threshold (Ct) setting for assigning the baseline and the threshold for Ct determination. IEX‐1 expression of epithelial ovarian carcinoma group was used as the exogenous control. ΔCt = IEX‐1 mean Ct − β‐actin mean Ct, 2−ΔΔCt was used to represent the expression level of IEX‐1. The relative expression of IEX‐1 was calculated by considering IEX‐1 expression of epithelial ovarian carcinoma group as 1 (expressed in multiple).
Statistical analysis
All statistical analyses were performed using statistical software spss 17.0. Quantitative variables were reported as mean ± standard deviation and processed with one‐way anova accounting and Bonferroni test. Correct bilateral P = 0.017 was considered as a test standard. ROC curve and AUC can be used to evaluate the diagnostic capacity of IEX‐1. The best cutoff values were determined by ROC curves and gold standards were determined by pathological reports after operation. Four standard tables were applied to calculate the sensitivity, specificity, positive predictive value, negative predictive value and the total coincidence rate.
Results
IEX‐1 expression in blood and saliva in the three groups
Relative IEX‐1 expression in blood in the benign ovarian tumor group and the healthy women group were 1.81 ± 0.31 and 1.72 ± 0.32 times higher than that of the ovarian carcinoma group, respectively. Also, IEX‐1 expression in saliva in the benign ovarian tumor group and the healthy women group were 1.67 ± 0.37 and 1.80 ± 0.40 times higher than that of the ovarian carcinoma group, respectively. The differences in IEX‐1 expression in blood and saliva were both statistically significant (P < 0.017) between the ovarian carcinoma and benign ovarian tumor groups and also between the ovarian carcinoma and healthy women groups. However, there was no significant difference in IEX‐1 expression between the benign ovarian tumor group and the healthy women groups regardless of whether the samples were collected from blood or saliva (P = 0.376 and P = 0.621, respectively; Fig. 1).
Figure 1.
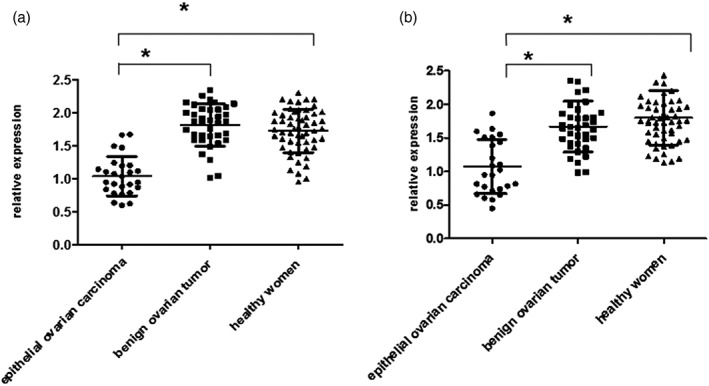
(a) Relative immediate early responsive gene X‐1 (IEX‐1) expression in blood of the three groups of women. (b) Relative IEX‐1 expression in saliva of the three groups of women (*P < 0.017).
ROC curves were constructed to evaluate the diagnostic ability of IEX‐1 in blood and saliva
ROC‐AUC of IEX‐1 expression in blood and saliva were 0.947 (95% confidence interval [CI], 0.896~0.998) and 0.851 (95% CI, 0.757~0.946), respectively, when compared between the ovarian carcinoma group and the benign ovarian tumor group, or 0.929 (95% CI, 0.874~0.984) and 0.896 (95% CI, 0.825~0.967), respectively, when compared between the ovarian cancer group and healthy women group (Fig. 2). Because there are currently no known cutoff values of IEX‐1 in ovarian cancer diagnosis, the largest Youden’s indexes were used to identify the appropriate cutoff points.9 Calculation using the formula (sensitivity +specificity − 1) yielded the cut off values in the ovarian carcinoma group versus the benign ovarian tumor group or the healthy women group to be 1.263 and 1.244 in blood or 1.118 and 1.554 in saliva, respectively.
Figure 2.
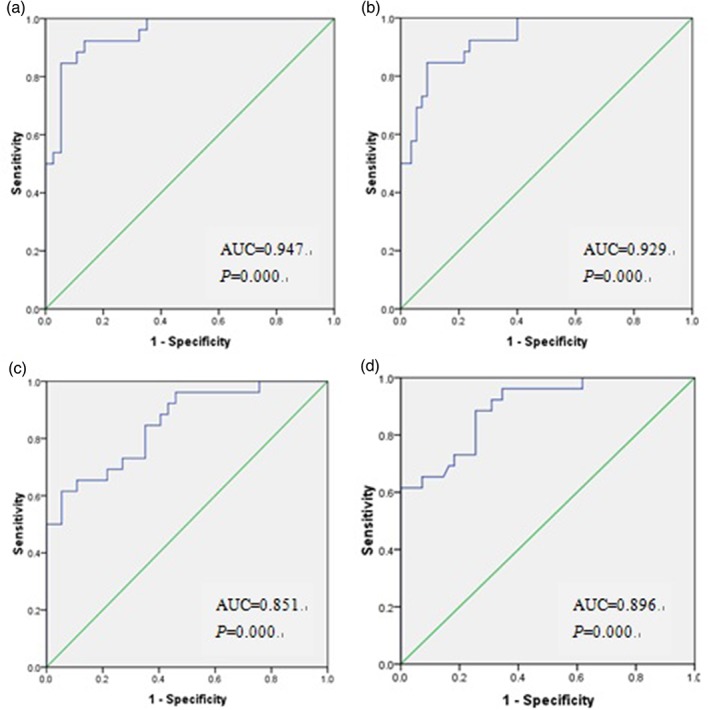
Areas under the receiver operating curves (AUC) were calculated evaluating a diagnostic ability of immediate early responsive gene X‐1 (IEX‐1) in blood and saliva. IEX‐1 expression in blood in the ovarian carcinoma group versus the benign ovarian tumor group (a) or the healthy women group (b). IEX‐1 expression in saliva in the ovarian carcinoma group versus the benign ovarian tumor group (c) or the healthy women group (d).
Diagnostic sensitivity and specificity of IEX‐1 expression in ovarian cancer
Four standard tables were next applied to calculate a positive rate among the three groups based on the blood and saliva assays (Table 1). Using cutoff values established above, the sensitivity, specificity, positive and negative predictive values and the total coincidence rate were calculated. IEX‐1 expression in blood could discriminate the ovarian carcinoma group from the benign ovarian tumor group with a sensitivity and specificity of 84.6% and 94.6%, respectively. The panel also provided an 84.6% sensitivity and 90.9% specificity in distinguishing the ovarian carcinoma group from the healthy women group. Likewise, IEX‐1 expression in saliva discriminated the ovarian carcinoma group from the benign ovarian tumor group with sensitivity and specificity of 65.4% and 94.6%, respectively, while demonstrating 92.3% sensitivity and 72.5% specificity in distinguishing the ovarian carcinoma group from the healthy women group (Table 2).
Table 1.
Positive rate of immediate early responsive gene X‐1 in different groups in blood and saliva
| Group | Number | Blood | Saliva | ||
|---|---|---|---|---|---|
| + | − | + | − | ||
| Ovarian carcinoma group | 26 | 22 (84.6%) | 4 (15.4%) | 17 (65.4%) | 9 (34.6%) |
| Benign ovarian tumor group | 37 | 2 (5.4%) | 35 (94.6%) | 2 (5.4%) | 35 (94.6%) |
| Group | Number | Blood | Saliva | ||
|---|---|---|---|---|---|
| + | − | + | − | ||
| Ovarian carcinoma group | 26 | 22 (84.6%) | 4 (15.4%) | 24 (92.3%) | 2 (7.7%) |
| Healthy women group | 55 | 5 (9.1%) | 50 (90.9%) | 14 (25.5%) | 41 (74.5%) |
Table 2.
Diagnostic sensitivity and specificity of immediate early responsive gene X‐1 expression in blood and saliva
| Sample | Sensitivity (%) | Specificity (%) | Total coincidence rate (%) | Positive predictive value (%) | Negative predictive value (%) | |
|---|---|---|---|---|---|---|
| Ovarian carcinoma group versus benign ovarian tumor group | Blood | 84.6 | 94.6 | 90.5 | 91.7 | 89.7 |
| Saliva | 65.4 | 94.6 | 82.5 | 89.5 | 79.5 | |
| Ovarian carcinoma group versus healthy women group | Blood | 84.6 | 90.9 | 95.1 | 81.5 | 92.6 |
| Saliva | 92.3 | 72.5 | 80.2 | 63.2 | 95.3 |
Discussion
Feasibility of tumor‐associated genes as biomarkers
Tumor biomarkers generally include three categories: cell surface proteins, serum markers and intracellular genes. Serum biomarkers such as CA125 have great practical value in tumor screening, diagnosis, prognosis, outcome and assessment of curative effect.10 However, these markers also confer false‐positive outcomes, because these diagnostic approaches can readily be affected by other gynecological benign diseases such as adenomyosis, endometriosis and pelvic inflammatory diseases.11 Moreover, CA125 concentrations may fluctuate throughout the menstrual cycle and pregnancy.12 On the contrary, specific origin tumors have more direct relationship with certain cancer genes. In recent years, many studies have shown that tumor‐associated genes possess a high prediction accuracy and can be used as molecular markers to help the diagnosis. For instance, HE4 showed a higher specificity than CA125 for differentiating benign from malignant cancers and in the differential diagnosis of gynecologic diseases.13 In this regard, HE4 biomarker could reach 79.6% sensitivity and 95% specificity with the AUC of 0.933 when identifying ovarian cancer and endometriosis.12 Other biomarkers in this category are P53, GADD45, Cyclin E, MDM2 and Bcl‐2, which can be used to distinguish among ovarian cancer, benign ovarian tumor and borderline ovarian tumors.14 A panel of five serum protein markers (MSP‐alpha, TIMP‐4, PDGF‐R alpha, OPG and CA125) was identified using antibody array technology and enzyme‐linked immuno sorbent assay (ELISA) assay, which could effectively detect ovarian cancer with a high specificity (95%) and a high sensitivity (100%).15 These studies strongly suggest that tumor‐associated genes are expected to be one of the valuable indexes in differentiating benign from malignant tumors.
IEX‐1 expression and prognosis of cancers
IEX‐1 is also known as IER3, p22/PRG1, Dif‐2 and gly96.16 It frequently comes out as an outliner in global gene expression studies in cancers, although the underlying mechanism is not completely understood. Induction of IEX‐1 expression confers survival advantages for some cells, but promotes apoptosis in others.17, 18 Positive IEX‐1 expression was a significant favorable factor for the survival of pancreatic cancer19 and myelodysplastic syndromes.20 Mice lacking IEX‐1 developed secondary brain injury following a mild brain injury21 and myelodysplastic syndromes‐like disease after a sublethal γ irradiation or bone marrow transplantation.20 And these mice were prone to lipopolysaccharide‐induced endotoxemia.22 On the contrary, a high level of IEX‐1 expression appears to link poor survival of patients with breast cancer,23 AML,24 myeloma25 and colon cancer.26 IEX‐1 deficiency also induced browning and activated thermogenic genes program.27 Our previous studies suggest that positive IEX‐1 expression is associated with a good prognosis in ovarian cancer.6 These gene expression profiles not only offer unbiased recognition about an importance of IEX‐1 in the prognosis but also possible diagnostic value of these cancers.
The feasibility of saliva specimen as a test specimen
Saliva is one of the important fluids in the human body. As a mirror of the body’s physiological conditions, saliva is readily accessible and harbors diverse components, including proteins,28 RNA29 and exosomes.30 There are active and nonspecific exchanges of substances between blood and saliva upon systemic disease development, and thus, significant changes can occur in the salivary biomarker profile.31 Using oral fluids for diagnosis has been developed beyond oral diseases.29 In the past decade, a number of findings have evoked interests in the use of saliva as a source of biomarkers. Mouse models have demonstrated that Runx1, Mlxipl, Trim30 and IEX‐1 were significantly altered between melanoma‐bearing mice and control mice in salivary gland tissue.31 Salivary genomic biomarkers have been successfully developed for the detection of lung cancer,32 pancreatic cancer,29 oral cancer33 and breast cancer.34 Zhang et al.35 used cDNA microarray to detect seven significant differences in genes between cancer and healthy samples. Logistic regression approach was used to assess the ability of discriminating ovarian cancer from different biomarker combinations. As a diagnostic fluid, saliva offers distinct advantages over serum because it can be collected noninvasively at home by individuals with modest training, making it assessable to measure biomarkers at multiple time points.
This article extends the diagnostic application of saliva to ovarian cancers and evaluates its feasibility. About 5.6 ~ 8.4 ng of mRNA molecules can be obtained from 140 μL of cell‐free saliva supernatant samples using QIAamp Viral RNA Mini Kit. Two clear bands of 28S and 18S rRNA were observed by electrophoresis; 28S band fluorescence intensity is about two times more than 18S band. These findings demonstrate high quantity and quality of isolated salivary mRNA to ensure the sufficiency and accuracy for qRT‐PCR. In the past decades, focus has been on finding tumor markers in biological fluids such as saliva that can be used in association with therapy for noninvasive detection of epithelial ovarian carcinoma and our current study extends those findings.
qRT‐PCR assays and ROC curve analysis
Quantitative reverse transcription polymerase chain reaction was used to measure IEX‐1 expression. IEX‐1 expression in blood and saliva was lower in the epithelial ovarian carcinoma group than in the benign ovarian tumor group or in the healthy women group. ROC curves were generated to analyze the clinical significance of IEX‐1 expression in distinguishing among the three groups. According to an objective standard, AZ less than or equal to 0.7 indicates low diagnostic value; AZ less than 0.7 and greater than 0.9 indicates medium diagnostic value; and AZ greater than 0.9 indicates high diagnostic value. Accordingly, the diagnostic value of IEX‐1 in ovarian cancer is high in blood and medium in saliva, and IEX‐1 diagnosis efficiency is higher in blood than in saliva. These results indicate that blood and saliva IEX‐1 expression represents a suitable marker able to differentiate between the epithelial ovarian carcinoma group and the benign ovarian tumor group or the healthy women group. In general, IEX‐1 expression in blood has a relatively higher degree of sensitivity and specificity when it comes to distinguishing between benign and epithelial ovarian carcinoma. We found that IEX‐1 expression in saliva may have a higher specificity and slightly lower sensitivity in the differentiation of ovarian carcinoma from benign ovarian tumor. In view of the less sensitivity of IEX‐1 expression alone, it may be applied clinically in combination with other cancer‐specific biomarkers such as CA125 and CA19–9. Our findings enhance the prospect of an important role for salivary diagnostics in the detection of systemic diseases.
Our study suggests that IEX‐1 has the potential to serve as an additional biomarker for diagnosing ovarian cancer. Similar to many other novel biomarkers at their early stages of research, extensive investigation to validate its clinical potential and to establish its standard cutoff range is key to the success of its clinical application in the future. Moreover, a better understanding of the mechanism and regulation of the gene may enhance the potential of this novel diagnostic approach for ovarian tumors.
Discloure
None declared.
Acknowledgments
The authors thank the Department of Obstetrics and Gynecology, the First Affiliated Hospital of Zhengzhou University for providing samples and clinical data. This study was supported by the National Natural Science Foundation of China (U1604172).
References
- 1. Gentry‐Maharaj A, Menon U. Screening for ovarian cancer in the general population. Best Pract Res Clin Obstet Gynaecol 2012; 26: 243–256. [DOI] [PubMed] [Google Scholar]
- 2. Jacobs IJ, Menon U. Progress and challenges in screening for early detection of ovarian cancer. Mol Cell Proteomics 2004; 3: 355–366. [DOI] [PubMed] [Google Scholar]
- 3. Moore RG, McMeekin DS, Brown AK et al A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol 2009; 112: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Molina R, Auge JM, Bosch X et al Usefulness of serum tumor markers, including progastrin‐releasing peptide, in patients with lung cancer: Correlation with histology. Tumour Biol 2009; 30: 121–129. [DOI] [PubMed] [Google Scholar]
- 5. Anastasi E, Granato T, Marchei GG et al Ovarian tumor marker HE4 is differently expressed during the phases of the menstrual cycle in healthy young women. Tumour Biol 2010; 31: 411–415. [DOI] [PubMed] [Google Scholar]
- 6. Han L, Geng L, Liu X, Shi H, He W, Wu MX. Clinical significance of IEX‐1 expression in ovarian carcinoma. Ultrastruct Pathol 2011; 35: 260–266. [DOI] [PubMed] [Google Scholar]
- 7. Lee Y‐H, Wong DT. Saliva: An emerging biofluid for early detection of diseases . Am J Dent 2009; 22: 241–248. [PMC free article] [PubMed] [Google Scholar]
- 8. Ballantyne J. Validity of messenger RNA expression analyses of human saliva. Clin Cancer Res 2007; 13: 1350–1351. [DOI] [PubMed] [Google Scholar]
- 9. Li Y, Zhou X, St John MA, Wong DT. RNA profiling of cell‐free saliva using microarray technology. J Dent Res 2004; 83: 199–203. [DOI] [PubMed] [Google Scholar]
- 10. Swets JA. Measuring the accuracy of diagnostic system. Science 1988; 240: 1285–1293. [DOI] [PubMed] [Google Scholar]
- 11. Van Dalen A, Favier J, Hallensleben E et al Significance of serum CA125 and TPS antigen levels for determination of overall survival after three chemotherapy courses in ovarian cancer patients during long‐term follow‐up. Eur J Gynaecol Oncol 2009; 30: 609–615. [PubMed] [Google Scholar]
- 12. Escudero JM, Auge JM, Filella X, Torne A, Pahisa J, Molina R. Comparison of serum human epididymis protein 4 with cancer antigen 125 as a tumor marker in patients with malignant and nonmalignant diseases. Clin Chem 2011; 57: 1534–1544. [DOI] [PubMed] [Google Scholar]
- 13. Nolen B, Velikokhatnaya L, Marrangoni A et al Serum biomarker panels for the discrimination of benign from malignant cases in patients with an adnexal mass. Gynecol Oncol 2010; 117: 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu YN, Ye X, Cheng HY et al Measurement of serum human epididymis secretory protein 4 combined with CA125 assay in differential diagnosis of endometriosis cyst and ovarian benign and malignant tumors. Chin J Obstet Gynecol 2010, 45: 363–366. [PubMed] [Google Scholar]
- 15. Lee H, Park G, Jung JH et al Diagnostic approach using the expression profiling of the P53 tumor suppressor gene and its related proteins in ovarian epithelial tumors. Int J Gynecol Cancer 2005; 15: 453–461. [DOI] [PubMed] [Google Scholar]
- 16. Jiang W, Huang R, Duan C et al Identification of five serum protein markers for detection of ovarian cancer by antibody arrays. PLoS One 2013; 8: e76795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arlt A, Schafer H. Role of the immediate early response 3 (IER3) gene in cellular stress response, inflammation and tumorigenesis. Eur J Cell Biol 2011; 90: 545–552. [DOI] [PubMed] [Google Scholar]
- 18. Wu MX, Ustyugova IV, Han L, Akilov OE. Immediate early response gene X‐1, a potential prognostic biomarker in cancers. Expert Opin Ther Targets 2013; 17: 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brahmbhatt A, NievesTorres E, Yang B et al The role of Iex‐1 in the pathogenesis of venous neointimal hyperplasia associated with hemodialysis arteriovenous fistula. PLoS One 2014; 9: e102542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sasada T, Azuma K, Hirai T et al Prognostic significance of the immediate early response gene X‐1 (IEX‐1) expression in pancreatic cancer. Ann Surg Oncol 2008; 15: 609–617. [DOI] [PubMed] [Google Scholar]
- 21. Ramsey H, Zhang Q, WMX. Mitoquinone restores platelet production in irradiation‐induced thrombocytopenia. Platelets 2015; 26: 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Q, Zhou C, Hamblin MR, Wu MX. Low‐level laser therapy effectively prevents secondary brain injury induced by immediate early responsive gene X‐1 deficiency. J Cereb Blood Flow Metab 2014; 34: 1391–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramsey H, Wu MX. Mitochondrial anti‐oxidant protects IEX‐1 deficient mice from organ damage during endotoxemia. Int Immunopharmacol 2014; 23: 658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang C, Trent S, Ionescu‐Tiba V et al Identification of cyclin D1‐ and estrogen‐regulated genes contributing to breast carcinogenesis and progression. Cancer Res 2006; 66: 11649–11658. [DOI] [PubMed] [Google Scholar]
- 25. Steensma DP, Neiger JD, Porcher JC et al Rearrangements and amplificationof IER3 (IEX‐1) represent a novel and recurrent molecular abnormality in myelodysplastic syndromes. Cancer Res 2009; 69: 7518–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ria R, Todoerti K, Berardi S et al Gene expression profiling of bone marrow endothelial cells in patients with multiple myeloma. Clin Cancer Res 2009; 15: 5369–5378. [DOI] [PubMed] [Google Scholar]
- 27. Nambiar PR, Nakanishi M, Gupta R et al Genetic signatures of high‐ and low‐risk aberrant crypt foci in a mouse model of sporadic colon cancer. Cancer Res 2004; 64: 6394–6401. [DOI] [PubMed] [Google Scholar]
- 28. Shahid M, Javed AA, Chandra D et al IEX‐1 deficiency induces browning of white adipose tissue and resists diet‐induced obesity. Sci Rep 2016; 11: 24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiao H, Wong DT. Proteomics and its applications for biomarker discovery in human saliva. Bioinformation 2011; 5: 294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang L, Farrell JJ, Zhou H et al Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology 2010; 138: 949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gonzalez‐Begne M, Lu BW, Han XM et al Proteomic analysis of human parotid gland exosomes by multidimensional protein identification technology (MudPIT). J Proteome Res 2009; 8: 1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gao K, Zhou H, Zhang L et al Systemic disease‐induced salivary biomarker profiles in mouse models of melanoma and non‐small cell lung cancer. PLoS One 2009; 4: e5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang L, Xiao H, Zhou H et al Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci 2012; 69: 3341–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Elashoff D, Zhou H, Reiss J et al Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev 2012; 21: 664–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L, Xiao H, Karlan S et al Discovery and preclinical validation of salivary transcriptomic and proteomic. PLoS One 2010; 5: e15573. [DOI] [PMC free article] [PubMed] [Google Scholar]


