Abstract
A variety of mechanisms deliver cytosolic materials to the lysosomal compartment for degradation through autophagy. Here, we focus on two autophagic pathways, the chaperone-mediated autophagy and the endosomal microautophagy that rely on the cytosolic chaperone hsc70 for substrate targeting. Although hsc70 participates in the triage of proteins for degradation by different proteolytic systems, the common characteristic shared by these two forms of autophagy is that hsc70 binds directly to a specific five-amino acid motif in the cargo protein for its autophagic targeting. We summarize the current understanding of the molecular machineries behind each of these types of autophagy.
Keywords: autophagy, chaperone, lysosome, membrane protein, protein targeting, selective degradation
Introduction
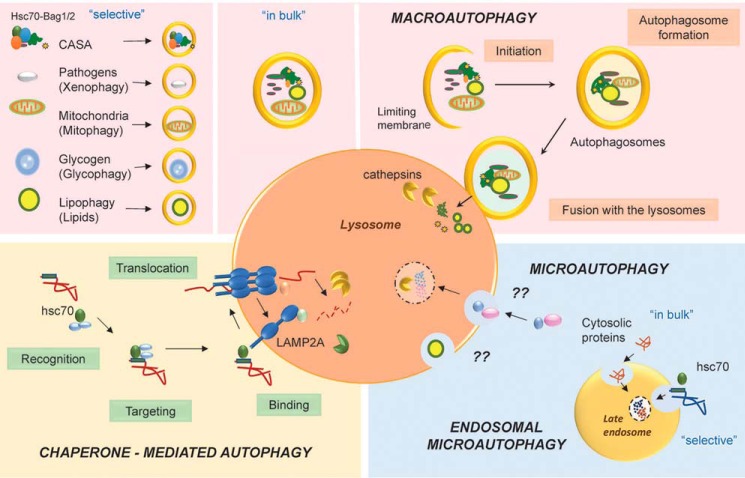
Autophagy refers to the degradation of a variety of cytoplasmic material in lysosomes (1). The delivery of autophagic cargo to lysosomes inside double membrane vesicles (autophagosomes) or macroautophagy has become the best characterized form of autophagy (Fig. 1) (2). However, cytosolic materials reach the lysosomal lumen through other mechanisms that also contribute to the overall intracellular autophagic activity (3). The lysosomal membrane can invaginate to internalize cargo in vesicles that pinch off from the invaginated membrane (4). This process known as microautophagy is conserved from yeast to mammals and contributes to degradation of proteins and organelles sequestered “in bulk” or in a selective manner (Fig. 1). Cytosolic proteins can also enter lysosomes for degradation through a protein translocation system at the lysosomal membrane, in a process known as chaperone-mediated autophagy (CMA)2 (Fig. 1) (5).
Figure 1.
CMA and eMI in the context of mammalian autophagic pathways. In macroautophagy, cargo (proteins and organelles) sequestered inside autophagosomes in bulk (non selective macroautophagy) or selectively (left) is then delivered to lysosomes through autophagosome/lysosome fusion. In CMA all cargo (proteins) are selectively delivered to lysosomes upon recognition by hsc70 and targeting and binding to the lysosomal membrane protein LAMP2A. Microautophagy requires invagination of the lysosomal membrane to degrade cytosolic material. Proteins and organelles can also be targeted to late endosomes for degradation in mammals through what is known as endosomal microautophagy. Whether mammalian cells are able to directly invaginate the lysosomal membrane to trap cytosolic cargo, as described in yeast, remains unknown (??). CASA, chaperone-assisted selective autophagy.
The first CMA studies saw the light (6) when autophagy was still considered a non-selective form of in bulk degradation. This made CMA the first evidence that autophagy can be selective, because only the subset of cytosolic proteins bearing in their amino acid sequence a pentapeptide recognized by hsc70 (heat-shock cognate protein of 70 kDa) was selected for degradation through CMA. The landscape of autophagy has changed, and selective forms of both macro- and microautophagy have been described (Fig. 1). Cargo recognition by chaperones has been described for these three autophagic processes, and even the same chaperone, hsc70, can triage cytosolic proteins to all of them. hsc70 binds exposed hydrophobic residues in misfolded or aggregated proteins in the macroautophagy variant known as chaperone-assisted selective autophagy (7). This is in clear contrast to the sequence-mediated targeting of proteins by hsc70 to CMA or a selective form of microautophagy, endosomal-microautophagy (eMI) (8). Here, we review these two forms of sequence-specific hsc70-mediated selective autophagy describing their substrates, molecular effectors and regulators, and the intracellular compartments where they occur.
Chaperone-mediated autophagy
General description
CMA is a selective form of autophagy with distinctive mechanisms for cargo recognition and internalization into the lysosomal lumen (5). Only proteins amenable to unfolding can be internalized in lysosomes by CMA through a mechanism with resemblance to protein transport systems into other organelles such as mitochondria or ER (5).
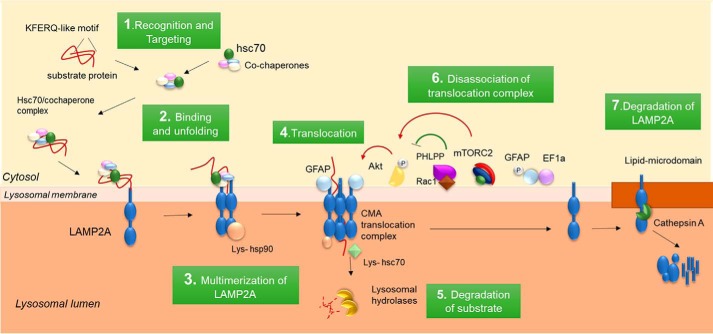
CMA starts with binding of hsc70 (9) to a consensus pentapeptide motif in the substrate protein (6). hsc70 targets these proteins to the lysosomal membrane, and after binding to the cytosolic tail of LAMP2A (lysosome-associated membrane protein type 2A) (10), the substrate proteins are unfolded (11) and translocated one-by-one into the lysosomal lumen (Fig. 2). Transport through the membrane requires multimerization of LAMP2A into the CMA translocation complex (12) and a form of the hsc70 resident in the lysosomal lumen required to complete substrate translocation (13, 14). The substrates are then rapidly degraded in the lysosomal lumen.
Figure 2.
Steps and lysosomal membrane components of CMA. Proteins degraded through CMA are recognized by hsc70 in the cytosol (step 1) and are targeted to the lysosomal membrane where they bind to LAMP2A (step 2). Substrate binding triggers multimerization of LAMP2A (step 3) to form the complex that mediates substrate translocation (step 4). hsp90 stabilizes LAMP2A through this transition, and luminal hsc70 assists with the internalization of the substrate that then is rapidly degraded by lysosomal proteases (step 5). The stability of LAMP2A in the translocation complex is regulated by the depicted subset of proteins. Once substrate translocate, LAMP2A, dissociates into monomers (step 6). Changes in the turnover of LAMP2A at the lysosomal membrane also contribute to modulate CMA activity (step 7).
Although effectors of other forms of autophagy are conserved from yeast to mammals, LAMP2A, the essential CMA component (10), appears late in evolution. LAMP2A is a spliced variant of the lamp2 gene, absent in yeast, fungi, and worms (15). A gene with homology to the mammalian lamp2 gene has been identified in Drosophila but with a C terminus homologous to LAMP2C, but there is no evidence of splicing. In zebrafish, the two lamp2 variants described show higher homology to the mammalian LAMP2B and -2C variants. The LAMP2A exon has so far been described only in birds and mammals (15).
CMA, as the other components of the cellular proteostasis networks (16), does not function in isolation. Blockage of CMA in vitro and in vivo is compensated for by up-regulation of macroautophagy and of the proteasome system in most cell types (17, 18). Conversely, cells respond to inhibition of macroautophagy or the proteasome by constitutively activating CMA (19–22). In most cases, compensation ensures the maintenance of cellular quality control and the energetic balance under basal conditions (17–19). However, these systems are not redundant, and upon persistent loss, compensation is no longer possible (17, 19). For example, CMA is up-regulated in cells of Huntington's disease patients to compensate for macroautophagy malfunctioning (23). However, this continuous overloading of CMA accelerates the normally occurring decline of CMA with age and contributes to accumulation of pathogenic huntingtin and other prone-to-aggregate proteins (23).
Characteristics of the proteome amenable for CMA degradation
All proteins degraded by CMA contain a pentapeptide-targeting motif and can completely unfold (24). Cytosolic origin was considered a characteristic of all CMA substrates but, recent studies, support that proteins from other compartments (i.e. nucleus or mitochondria) can also be degraded by CMA if they access the cytosol (18, 25).
The need for substrate unfolding is imposed by the translocation complex. Studies using an artificial CMA substrate unable to unfold demonstrated that unfolding is not required for lysosomal binding, but it is absolutely necessary for lysosomal translocation (11). Protein aggregates, irreversible oligomers (26–28), and proteins part of multiprotein complexes can only be degraded by CMA after disassembly and complete unfolding (29).
The pentapeptide-targeting motif in CMA substrates is necessary for their identification by hsc70 and sufficient for their lysosomal targeting (6). In fact, insertion of the motif in fluorescent proteins leads to their lysosomal degradation via CMA and are used as reporters of CMA activity (21). The motif is defined as KFERQ-like and is based on specific biochemical and physical properties of its constituent amino acids (24). Early experimental studies demonstrated that hsc70 binds to targeting motifs that contain one or two of the positively charged amino acids lysine (K) or arginine (R), one or two of the hydrophobic amino acids, phenylalanine (F), valine (V), leucine (L) or isoleucine (I), and one of the two negatively charged amino acids, aspartic acid (D) or glutamic acid (E), flanked by a glutamine (Q) on either side of this pentapeptide (6, 24).
Approximately, 40% of proteins in mammalian proteomes contain this KFERQ-like motif making them amenable for CMA once it is exposed (i.e. by unfolding, dissociation from other proteins, or from membranes) and accessible to hsc70 (24). The number of potential CMA substrates is estimated to be even higher, as new motifs can be generated by post-translational modifications such as phosphorylation (that contributes the negative charge) or acetylation of a lysine (that behaves as a glutamine) (30–33).
The presence of a KFERQ-like motif is required to classify a protein as bona fide CMA substrate, but as described below, it is no longer sufficient because the same motif is also used by hsc70 to target cytosolic proteins to late endosomes via eMI (8). Hence, validation of proteins as CMA substrates requires experimental validation (5, 34).The list of experimentally validated CMA substrates continues to grow and includes a broad variety of proteins involved in diverse cellular processes such as glycolytic enzymes (18, 30, 35, 36), lipogenic enzymes (18), lipid droplet structural proteins (32), RNA-modifying enzymes (37), proteins involved in calcium biology (38, 39), transcription factors and their regulators (40–42), cell cycle regulators (25), ubiquitin–proteasome components (43), proteins involved in immune function (39, 44), and in cell survival/cell death decisions (45–48), as well as a subset of proteins that contribute to the pathogenesis of known neurodegenerative disorders (23, 26–28, 43, 49–51).
Specific lysosomes dedicated to CMA
Even though the LAMP2A receptor is present in all types of lysosomes, not all lysosomes can perform CMA. The presence of hsc70 in lysosomes defines their CMA capabilities (14). The abundance of CMA-competent lysosomes (enriched on hsc70) fluctuates depending on the CMA requirements. During high CMA activity demand (i.e. sustained periods of starvation (14) and mild oxidative stress (52)), the number of CMA-competent lysosomes increases at the expense of a reduction in the number of other types of lysosomes (53). A similar lysosomal switch occurs during aging, when the number of LAMP2A molecules per lysosome decreases and cells compensate for this loss by increasing the percentage of lysosomes containing hsc70 (54).
Experimental introduction of hsc70 in the lumen of CMA-incompetent lysosomes is sufficient to make them capable to perform CMA (14), suggesting that CMA-inactive lysosomes contain the rest of the CMA machinery. Despite their higher efficiency for CMA, these lysosomes can still engage in the other autophagic pathways. For example, CMA-active lysosomes can fuse with autophagosomes, albeit with lower efficiency (55).
Lysosomal and cytosolic hsc70 originate from the same gene, but they present different electrophoretic properties in support of compartment-specific post-translational modifications (13). The exact mechanism by which hsc70 reaches lysosomes is unknown. Neither blockage of macroautophagy (20) or CMA (56) reduced the content of hsc70 in lysosomes. The abundance of hsc70 in late endosomes (8) makes attractive the idea that endosome/lysosome fusion may contribute hsc70 to the lysosomal lumen. The resistance of hsc70 to degradation is only maintained at a very acidic pH and small increases in lysosomal pH render lysosomal hsc70 unstable (14). It is possible that fluctuations in lysosomal pH determine changes in hsc70 conformation and lead to its rapid degradation.
The relatively small fraction of total intracellular hsc70 present in lysosomes has made it difficult to study. The proposed contribution of luminal hsc70 to substrate translocation is based on the fact that lysosomes lacking luminal hsc70 can bind CMA substrates, but do not internalize them, and that blockage of luminal hsc70 with antibodies against hsc70 internalized by endocytosis abolished CMA degradation (13).
Key CMA components: lysosomal chaperones and LAMP2A
Both cytosolic and lysosomal hsc70 are indispensable for CMA. A subset of co-chaperones, including Hsp90, Hsp40, the Hsp70–Hsp90-organizing protein (Hop), the Hsp70-interacting protein (Hip), and the Bcl2-associated athanogene 1 protein (BAG-1) associate to the complex hsc70/KFERQ-containing proteins (57), although the specific contribution of each of them to CMA remains unknown. Their many other cellular functions make the phenotypes resulting from their genetic knockdown difficult to interpret and of little use for understanding their role in CMA. Incubation of isolated lysosomes with blocking antibodies against hsc70 co-chaperones present at the lysosomal surface reduces substrate translocation suggesting that they could play a dual function by assisting hsc70 in substrate targeting and in their unfolding at the lysosomal membrane (Fig. 2) (57). hsc70 also participates in a later CMA step when it actively mediates disassembly of LAMP2A from the CMA translocation complex (12). The coordination and switch between these distinctive functions of hsc70 in different steps of CMA remain poorly studied.
Another key player in CMA is the membrane protein LAMP2A, required for both substrate binding and translocation (10, 12). Binding to LAMP2A is the rate-limiting step of CMA, and therefore, changes in the levels of LAMP2A at the lysosomal membrane up-regulate or down-regulate CMA (58). The lysosomal content of LAMP2A depends on its rates of de novo synthesis, efficiency of lysosomal trafficking, and changes in its half-life once in lysosomes. For example, LAMP2A synthesis increases during CMA activation by oxidative stress (52) or during T cell activation (39). The increase in lysosomal levels of LAMP2A during starvation is attained through a decrease in its degradation at the lysosomal membrane (58, 59) and relocation of LAMP2A present in the lysosomal lumen toward the membrane (58). Sub-compartmentalization of LAMP2A at the lysosomal membrane is responsible for its dynamic regulation. Under resting conditions, LAMP2A is periodically sequestered into lipid microdomains for cleavage by cathepsin A that initiates its membrane release and rapid degradation in the lumen (60). Upon CMA activation, LAMP2A is actively excluded from these microdomains (60). Recent studies have shown that defective targeting of LAMP2A from the Golgi to lysosomes is behind the low efficiency of CMA in the lysosomal storage disease cystinosis (61, 62). These findings highlight LAMP2A trafficking as a possible additional mechanism for regulation of CMA activity.
Binding of CMA substrates to LAMP2A occurs through its short (12 amino acids) cytosolic tail. Blockage of this region with specific antibodies, addition of a 12-residue peptide with the same amino acid composition, or swapping of this cytosolic tail with the one present in LAMP2B or LAMP2C reduce CMA (10, 63). Four positively charged residues, only present in the LAMP2A tail but not in B or C, are necessary for substrate binding (63).
Substrate binding to LAMP2A triggers the multistep transition of monomeric forms of LAMP2A into multimers (64) and their final assembly with other proteins into a translocation complex of about 700 kDa (Fig. 2) (12). Formation of this complex is transient and dynamic, as it disassembles once the substrate is translocated into the lysosome (12, 65). Continuous cycles of assembly and disassembly of LAMP2A may occur because substrates can only bind monomeric LAMP2A and can only be transported into the lumen when LAMP2A is in the translocation complex (12).
A second chaperone implicated in CMA is the lysosomal hsp90, which localizes both at the cytosolic and luminal sides of the lysosomal membrane. The latter helps to stabilize LAMP2A as it transitions through the different stages of multimerization (12).
CMA regulation
CMA is under regulatory mechanisms self-contained in the lysosomal compartment (65). Cleavage by cathepsin A determines the stability of monomeric LAMP2A (60), whereas the stability of the multimeric LAMP2A complex is regulated in a GTP-dependent manner by two proteins: GFAP (glial fibrillary acidic protein) and EF1α (elongation factor 1α) (65). GFAP associates transiently with multimeric LAMP2A and prevents hsc70-mediated disassociation of this complex (Fig. 2). A phosphorylated variant of GFAP is also present at the lysosomal membrane but is bound to EF1α, which makes it inaccessible for binding to other proteins. In the presence of GTP, EF1α is released from the lysosomal membrane, and unmodified GFAP moves from the multimeric complex to bind the exposed phospho-GFAP (65). This results in disassembly of LAMP2A from the CMA translocation complex and its return to the monomeric state (65).
Phosphorylation of lysosomal GFAP is performed by Akt1 under the control of the mechanistic target of rapamycin complex 2 (mTORC2), both at the lysosomal membrane (31). mTORC1, shown to negatively regulate macroautophagy, is present in all type of lysosomes, including CMA-competent lysosomes, but modulation of mTORC1 with drugs such as rapamycin does not affect CMA activity (21). In contrast, mTORC2 and its effector kinase Akt1 are almost exclusively detected in CMA-competent lysosomes where they negatively regulate assembly of LAMP2A into the CMA translocation complex (31). When CMA activation is needed, the phosphatase PHLPP1 (pleckstrin homology domain and leucine-rich repeat protein phosphatase 1), responsible for dephosphorylating Akt1, is recruited to lysosomes and stabilized at the membrane by the GTPase Rac1 (31). Reduced Akt1 activity increases the pool of non-phosphorylated GFAP thus favoring formation of the CMA translocation complex (Fig. 2).
Besides the lysosomal regulation of CMA, signaling through the nuclear receptor retinoic acid receptor α (RARα) (66) and the calcineurin–nuclear factor of activated T cells (NFAT) pathway (39) also regulate CMA. Genetic knockdown of RARα activates CMA in a transcription-dependent manner, suggesting that this nuclear receptor is an endogenous inhibitor of the subset of genes required for CMA (66). In T cells, CMA is activated upon engagement of the T cell receptor (TCR) that through generation of mitochondrial ROS promotes nuclear translocation of NFAT1 and its binding to the lamp2 promoter (39). Inhibition of calcineurin (activator of NFAT) or blockage of ROS production prevents CMA activation in this context.
Physiological relevance of CMA
CMA activity is detectable at basal conditions in most cells, but maximal activation occurs during stress such as nutrient deprivation (53, 67), mild-oxidative stress (52, 68), exposure to genotoxic or proteotoxic stressors (25, 69), hypoxia (70, 71), and lipid overload (Fig. 3) (32, 72).
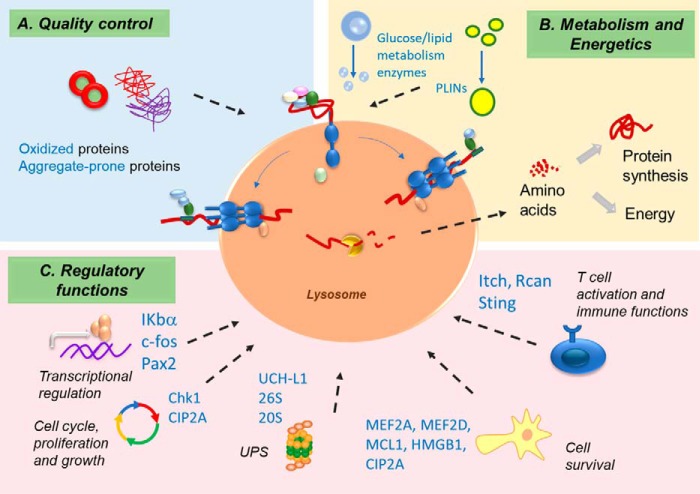
Figure 3.
Physiology of CMA. CMA participates in cellular quality control. CMA participates in (A) cellular quality control through the removal of damaged or abnormal proteins and in (B) cellular metabolism and energetics through recycling of the amino acids of the degraded proteins and by selectively degrading rate-limiting enzymes of lipid and glucose metabolism. C, timely degradation of specific proteins through CMA confers this pathway's regulatory function in multiple cellular processes. Some of these processes and the CMA substrates involved in these pathways are depicted.
In most cells, CMA is up-regulated after periods of nutrient deprivation of 10 h or longer (17, 53) following the earlier macroautophagy response to starvation. Although macroautophagy switches from proteins to lipid (lipophagy) after 4–6 h of starvation (73), CMA activation persists past 3 days of starvation (53) becoming a source of recycled amino acids for protein synthesis (74). Part of these amino acids may also be used as cellular fueling, because cells with compromised CMA have reduced ATP levels (17, 75) and restoration of normal CMA activity in the livers of old mice increases their ATP content (76). Recent studies support that CMA may modulate cellular energetics through mechanisms other than mere amino acid recycling. Mice with selective knockout of LAMP2A in hepatocytes display defective handling of glucose and lipids due to loss of their ability to regulate levels of key metabolic enzymes that are usually turned over by CMA when their activity needs to be suppressed (18).
CMA also participates in protein quality control through selective removal of altered or damaged proteins (Fig. 3). Up-regulation of CMA upon mild oxidative stress facilitates degradation of oxidized proteins (52) that otherwise will aggregate and persist as intracellular protein inclusions (17). This explains why CMA blockage associates with an increase in intracellular protein aggregates (17, 56) and why old mice with preserved CMA activity show reduction in the age-dependent increase in aggregate-oxidized proteins (76).
The ability of CMA to selectively degrade intracellular proteins confers it specialized functions such as regulation of transcription by degradation of several transcription factors or control of cell cycle progression through degradation of cell cycle arrest proteins. CMA contributes to regulation of neuronal survival by degrading inactive forms of the transcription factor MEF2D (myocyte enhancer factor D) (45), of NFκB-mediated transcription via IκBα degradation (41), and of kidney growth through regulation of transcription factor Pax2 (42). The role of CMA in T cell activation is a result of its ability to timely degrade the negative regulators of T cell activation Itch and RCAN1 (39). Contribution of CMA to the immune response through the presentation of antigens in macrophages has also been described (Fig. 3) (77).
Interestingly, the contribution of CMA to protein quality control is compensated for by activation of macroautophagy and proteasome (18), but CMA regulatory functions of CMA are not absorbed by these other systems (39). These findings further supporting that autophagic pathways are not redundant and that each participates in specific cellular functions depending on their timing of activation and on substrate selectivity.
Endosomal microautophagy
General description
The concept that the lysosomal membrane invaginates to trap cytosolic components for degradation was proposed in the very early days of the discovery of autophagy. This process, termed microautophagy (4), was first studied in liver (78). Later studies described that yeast use a similar process for the sequestration and degradation of peroxisomes when switched to glucose as a source of energy (79) and lead to the discovery that some of the genes required for peroxisome microautophagy (GSA genes) (80) were shared with macroautophagy (81). Reconstitution of yeast microautophagy in vitro with isolated vacuoles (82) has allowed us to further identify the molecular machinery involved in this process (83).
The term microautophagy has been reserved for degradation of intracellular proteins and organelles directly engulfed by lysosomes or the vacuole (in yeast) (3). This degradation has now proven able to discriminate cargo, giving rise to terms such as micropexophagy (for peroxisomes) (79, 80), micromitophagy (for mitochondria) (84), or microlipophagy (for lipid droplets) (85). Even portions of the nucleus can undergo degradation through this invagination-mediated process (piecemeal microautophagy) (86). Selectivity of yeast microautophagy has been further supported by the discovery of specific cargo receptors, such as Nvj1p in piecemeal microautophagy (87).
The study of mammalian microautophagy has been slower because of the inability to detect an invagination-like process in secondary lysosomes and the fact that essential genes for yeast microautophagy have no conserved function in mammals. Relatively recent studies demonstrated that a degradative process of similar characteristics to yeast microautophagy occurs in mammals and in late endosomes/multivesicular bodies (LE/MVB) instead of lysosomes (8). This process, termed eMI, contributes to in bulk degradation of proteins present in cytosol trapped in vesicles forming at the LE membrane. However, some cytosolic proteins can also be selectively degraded by eMI after hsc70 binds in their sequence to the same pentapeptide motif previously described for CMA (Fig. 4) (8). hsc70 is not necessary for cargo targeting to microautophagy in yeast, but recent studies support the occurrence of hsc70-mediated eMI in Drosophila (88, 89). Below, we describe this type of hsc70-dependent eMI in the context of other types of microautophagy and in comparison with CMA (Fig. 5).
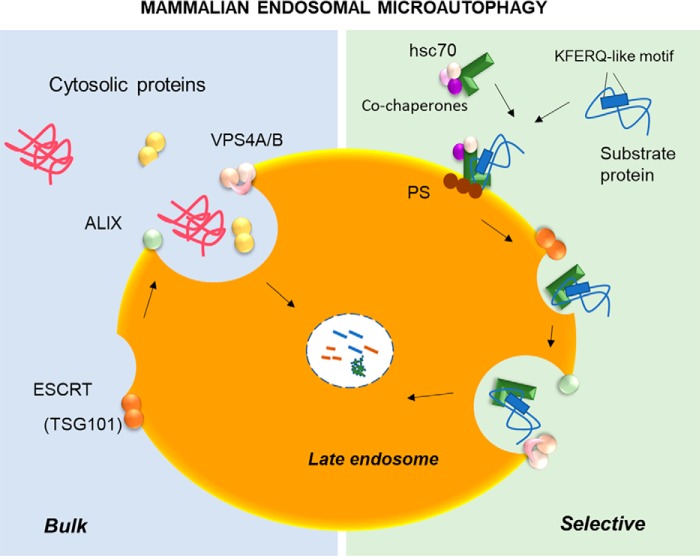
Figure 4.
Mammalian endosomal microautophagy. Left, cytosolic proteins can be sequestered along with other cytosolic components by the invaginations that form in the surface of the endosomal membrane through the coordinated function of ESCRT (VPS4A/B and TSG101) and accessory proteins (Alix). Right, selective targeting to late endosomes of proteins bearing a KFERQ-like motif is mediated by hsc70. Upon cargo binding, hsc70 interacts directly with phosphatidylserine (PS) moieties of the endosomal membrane and is internalized along with the substrate in ESCRT-mediated microvesicles. Part of the internalized vesicles undergoes degradation in the endosomal lumen.
Figure 5.
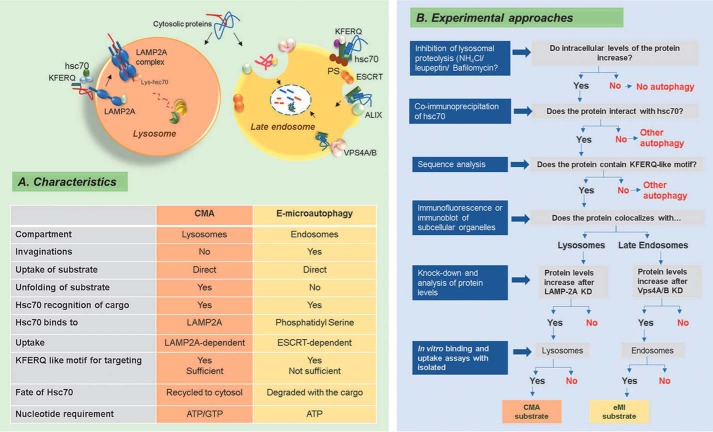
Similarities and differences between CMA and eMI and assays to monitor protein degradation through them. A, summary of common and distinctive characteristics between CMA and eMI identified to date. B, recommended experimental approach to follow to discriminate involvement of CMA or eMI in the degradation of a protein. Blue boxes, methods; gray boxes, observed results.
eMI substrates
Mammalian eMI originated from studies attempting to characterize the contribution of autophagic pathways to antigen presentation in dendritic cells. Analysis of LE/MVB, where cytosolic antigens are processed for presentation, revealed that arrival of some cytosolic proteins persisted even upon genetic blockage of macroautophagy or CMA (8). However, blockage of components of the ESCRT complex I, required for MVB formation, abrogated trafficking of these cytosolic proteins into endosomes. Interestingly, most of the cytosolic proteins internalized through this ESCRT-dependent process underwent full degradation instead of limited cleavage for antigen presentation, thus fulfilling the criteria of an autophagic pathway (8). In fact, studies in other cell types and organs (i.e. liver and brain) confirmed that non-immunological cells also display this form of autophagy (8, 90) that has been named eMI to highlight its cellular localization.
Comparative proteomic analysis of MVB in cells with functional or disrupted ESCRT identified the pool of proteins usually degraded by eMI and confirmed the cytosolic origin of most of them (8). Some of these proteins were present in the vesicles at a similar ratio as in the cytosol, suggesting in bulk internalization. However, hsc70 and proteins bearing the KFERQ-like motif, previously associated only with CMA, were highly enriched in the vesicles (8). In vitro studies reconstituting eMI with isolated LE confirmed that proteins such as GAPDH or RNase A, classic examples of CMA substrates, can also be internalized in an hsc70- and ESCRT-dependent manner by eMI and that mutations in the KFERQ-like motif disrupt their targeting (8). In fact, an early proposed reporter for CMA activity based on fluorescent tagging of GAPDH has now been shown to undergo degradation by both pathways, thus limiting its usability to differentiate between them (91). Endogenous proteins such as Tau, a cytoskeletal protein associated with neurodegeneration, also undergo simultaneous degradation by both eMI and CMA (90). The fact that pathogenic mutations in Tau switch the percentage of the protein degraded by each of these pathways (90) suggests that intrinsic properties of the protein (i.e. mutations, post-translational modifications, oligomeric state, etc.) may be responsible for the autophagic switch of the substrate.
Despite sharing the KFERQ-like motif, CMA and eMI substrates are not fully overlapping. KFERQ-like motif-bearing proteins in a state of semi-aggregation, organized into higher molecular weight complexes or unable to unfold, cannot by degraded by CMA but are still amenable for eMI degradation. Thus, similar experiments to the ones that showed that proteins unable to unfold cannot undergo degradation by CMA (11) support that unfolding of cytosolic KFERQ-like bearing proteins is not a requirement for their association with late endosomes (8). Furthermore, contrary to CMA where the presence of the KFERQ-like targeting motif is necessary and sufficient for hsc70-mediated lysosomal targeting, adding a KFERQ-like motif is not sufficient for targeting of proteins through mammalian eMI. For example, the CMA reporter with a KFERQ-like sequence added to photo-switchable proteins is only targeted to CMA-competent lysosomes but not to endosomes (21), suggesting that the motif is necessary but not sufficient for mammalian eMI. As more eMI substrates become validated, identification of protein sequence or structure requirements for eMI targeting should become possible. In this respect, recent studies in Drosophila have demonstrated that a subset of synaptic proteins is turned over selectively by eMI (88). Interestingly, the requirements for Drosophila eMI are different, and addition of the KFERQ motif to a fluorescent protein is sufficient for its targeting through this pathway (89). Co-existence of CMA and eMI in mammals but not in Drosophila could have forced the need of a second requirement in mammals. Fig. 5 summarizes the main differences described so far between CMA and eMI and the experimental steps currently recommended to differentiate substrates for each of these pathways.
Late endosomes: Hosts for eMI
LEs are the point of entry of cytosolic proteins for eMI as their membrane contains a dedicated machinery (the ESCRT proteins) for invagination, formation of MVB, and their excision for release into the endosomal lumen. This process of LE membrane microvesiculation has been well-characterized in the context of degradation of membrane proteins internalized by endocytosis and in the extracellular release of cytosolic material in the form of exosomes. Components of the ESCRT complex I (i.e. TSG101), II (VPS25), and III (VPS32) and two of the accessory proteins, VPS4 and Alix, have been proven necessary for eMI (Fig. 4) (8, 89). Whether the full ESCRT machinery is required for eMI or whether biogenesis and properties of MVB for eMI differ from those utilized in membrane protein recycling is still unknown. Similarly, it is not clear whether degradation of plasma membrane proteins and eMI occur in the same type of late endosomes, or whether, as in the case of CMA, there is a specific subpopulation of LE dedicated to eMI.
Studies in vitro support that some eMI substrates undergo degradation in LE (i.e. Tau (90)), whereas other proteins (i.e. GAPDH) are internalized in LE, but their degradation is markedly less efficient than in lysosomes (8, 90). It is possible that in these cases most of the degradation occurs by endosomal/lysosomal fusion. Studies in Drosophila support this final degradation of eMI cargo in lysosomes, because an artificial eMI fluorescent substrate can be detected in LAMP1-positive compartments that lack endosome markers (89).
It is important to clarify that the process of eMI is different from the process of loading MVB for release as exosomes. Although both processes share ESCRT components for the loading of cytosolic proteins in MVB, the fate of the cargo is different. Thus, the term eMI should be limited for cytosolic proteins loaded in MVB that undergo degradation in this compartment or upon lysosomal fusion, whereas MVB loading for extracellular release of cytosolic proteins inside exosomes is a type of exocytosis and not a type of autophagy.
Molecular machinery of eMI
Although not required for all forms of microautophagy, hsc70 is the component that defines the type of selective eMI described in this Minireview. Upon binding the eMI substrate proteins through their KFERQ-like motif, hsc70 targets them to LE (8). The co-chaperone Sgt modulates the switch between the chaperone and eMI functions of hsc70 in Drosophila (88). However, the determinants of cargo triage between CMA and eMI remain unknown. During eMI, hsc70 binds directly to LE membranes, but despite their abundance on LAMP2A, hsc70 binds directly instead to phosphatidylserine (PS) at the LE membrane through a stretch of 4–5 lysine residues in the C terminus of the hsc70 LID domain (Fig. 4) (8). Mutations in this region have also revealed that PS binding is required to trigger cargo internalization (92). The role of hsc70 in eMI substrate internalization may be mediated by its ability to deform membranes through oligomerization (88). Whether hsc70 piggybacks in forming MVB or whether it actively triggers their formation is still unknown. Different from CMA, where after transferring the substrate to LAMP2A hsc70 is released back to the cytosol, in the case of eMI, hsc70 undergoes internalization and degradation with the cargo protein (8).
Several protein complexes have been implicated in other forms of microautophagy. For example, clathrin and a family of ER proteins (class E VPS) are necessary for yeast microlipophagy (93, 94), the family of proteins Niemann-Pick type C and the phosphoinositide-binding protein Ivy1 for the formation of membrane invaginations in yeast microautophagy (95, 96), and specific cargo-recognizing proteins contribute to micropexophagy, micromitophagy, and piecemeal microautophagy of the nucleus (84, 85, 87). However, the lack of systematic studies makes it difficult to sort out which molecular players are common to all microautophagy processes and which ones are process-specific. Similarly, the involvement of macroautophagy proteins (Atg) in microautophagy seems to depend on the type of microautophagy. Thus, yeast microlipophagy occurs independent of Atgs, whereas Drosophila eMI requires Atg1 and Atg13 (89).
eMI regulation
Activation of mammalian eMI has not been observed late upon starvation (8), in clear contrast with Drosophila eMI, which is maximally activated after starvation exceeding 24 h (89). This responsiveness of eMI to starvation in flies has led us to propose that eMI functions could have split late in evolution between eMI and CMA. The fact that Atg1 and Atg13, required for Drosophila eMI, act downstream of TOR suggests that this major nutrient sensor may be behind the starvation-induced activation of eMI. TOR and EGO also regulate yeast microautophagy (97) and microlipophagy (96), and 5′-AMP-activated protein kinase and Atg14 have also been implicated in the activation of microlipophagy (85).
Cytosolic hsc70 is a very abundant protein whose levels remain rather constant making it unlikely that changes in hsc70 levels are physiologically used to regulate eMI. Vesicle formation is the limiting step of microautophagy in yeast (82) and that also seems to be the case in eMI (92). It is thus more likely that changes in levels and dynamics of the assembly of ESCRT proteins may contribute to eMI regulation. It is also possible that availability of specific nutrients may contribute to modulate eMI through direct changes in the lipid composition of the late endosomal membrane, for example by expanding the raft-like membrane regions as recently described in yeast (95).
Physiological functions of eMI
Cellular functions of eMI remain for the most part unknown. A role for eMI in protein quality control has been proposed in light of its constitutive nature and the accumulation of oxidized proteins (most of them bearing KFERQ-like motifs) in MVB from old animals (98). Failure to timely eliminate these oxidized products in the dendritic cells of old mice negatively impacts LE and antigen processing and presentation with age. Yeast microautophagy contributes to quality control of intracellular membranes (99).
Selective targeting of proteins through eMI makes possible a regulatory effect of eMI on specific cellular processes by controlling intracellular levels of their limiting proteins. In this respect, blockage of eMI in Drosophila slows down neurotransmission by altering degradation of specific synaptic proteins (88). Active search for eMI substrates will help in gaining an understanding of the physiological relevance of this pathway.
Concluding remarks and pending questions
The landscape of selective autophagy has undergone major changes in recent years. Although CMA initially pioneered the concept of selectivity in lysosomal degradation, nowadays some level of selectivity in the cargo degraded has been described for almost every type of autophagy. As the molecular determinants of each pathway become known, it should become easier to understand the differences among these autophagic processes and their specific physiological relevance. The fact that the two different pathways described in this Minireview share not only the same chaperone but even the specific way in which hsc70 binds to the cargo highlights the level of cross-talk among different autophagic pathways and the existence of the mechanism in place, whereby substrates of a pathway can be easily rerouted to another pathway. However, the different dynamics and timing of activation of CMA and eMI make it impossible to compensate for specific regulatory functions of each of these pathways and result in functional phenotypes. Many open questions remain about the molecular mechanisms behind CMA and more so about eMI, their regulation and bases of their cross-communication with other pathways, and how hsc70 triages substrates between CMA and eMI. Further understanding of which intrinsic properties of the substrate proteins contribute to the hsc70-mediated triage will also help to complete this picture.
This work was supported by National Institutes of Health Grants AG021904, AG031782, AG038072, and AG054108 from NIA, Grant NS100717 from NINDS, Grant DK098408 from NIDDK, Leducq Network Award RA15CVD04, the Rainwaters Foundation, Glen Foundation, and Backus Foundation, and the generous support of Robert and Renee Belfer. This is the fourth article in the Thematic Minireview series “Autophagy.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CMA
- chaperone-mediated autophagy
- eMI
- endosomal-microautophagy
- ROS
- reactive oxygen species
- GFAP
- glial fibrillary acidic protein
- PS
- phosphatidylserine
- RARα
- retinoic acid receptor
- NFAT
- nuclear factor of activated T cell
- LE
- late endosome
- MVB
- multivesicular body
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TOR
- target of rapamycin.
References
- 1. Deter R. L., Baudhuin P., and De Duve C. (1967) Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J. Cell Biol. 35, C11–C16 10.1083/jcb.35.2.C11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Levine B., and Klionsky D. J. (2017) Autophagy wins the 2016 Nobel Prize in physiology or medicine: breakthroughs in baker's yeast fuel advances in biomedical research. Proc. Natl. Acad. Sci. U.S.A. 114, 201–205 10.1073/pnas.1619876114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Galluzzi L., Baehrecke E. H., Ballabio A., Boya P., Bravo-San Pedro J. M., Cecconi F., Choi A. M., Chu C. T., Codogno P., Colombo M. I., Cuervo A. M., Debnath J., Deretic V., Dikic I., Eskelinen E. L., et al. (2017) Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marzella L., Ahlberg J., and Glaumann H. (1981) Autophagy, heterophagy, microautophagy and crinophagy as the means for intracellular degradation. Virchows Arch B. Cell Pathol. Incl. Mol. Pathol. 36, 219–234 10.1007/BF02912068 [DOI] [PubMed] [Google Scholar]
- 5. Kaushik S., and Cuervo A. M. (2018) The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chiang H. L., and Dice J. F. (1988) Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J. Biol. Chem. 263, 6797–6805 [PubMed] [Google Scholar]
- 7. Arndt V., Dick N., Tawo R., Dreiseidler M., Wenzel D., Hesse M., Fürst D. O., Saftig P., Saint R., Fleischmann B. K., Hoch M., and Höhfeld J. (2010) Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr. Biol. 20, 143–148 10.1016/j.cub.2009.11.022 [DOI] [PubMed] [Google Scholar]
- 8. Sahu R., Kaushik S., Clement C. C., Cannizzo E. S., Scharf B., Follenzi A., Potolicchio I., Nieves E., Cuervo A. M., and Santambrogio L. (2011) Microautophagy of cytosolic proteins by late endosomes. Dev. Cell 20, 131–139 10.1016/j.devcel.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chiang H. L., Terlecky S. R., Plant C. P., and Dice J. F. (1989) A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382–385 10.1126/science.2799391 [DOI] [PubMed] [Google Scholar]
- 10. Cuervo A. M., and Dice J. F. (1996) A receptor for the selective uptake and degradation of proteins by lysosomes. Science 273, 501–503 10.1126/science.273.5274.501 [DOI] [PubMed] [Google Scholar]
- 11. Salvador N., Aguado C., Horst M., and Knecht E. (2000) Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J. Biol. Chem. 275, 27447–27456 [DOI] [PubMed] [Google Scholar]
- 12. Bandyopadhyay U., Kaushik S., Varticovski L., and Cuervo A. M. (2008) The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol. Cell. Biol. 28, 5747–5763 10.1128/MCB.02070-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Agarraberes F. A., Terlecky S. R., and Dice J. F. (1997) An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J. Cell Biol. 137, 825–834 10.1083/jcb.137.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cuervo A. M., Dice J. F., and Knecht E. (1997) A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 272, 5606–5615 10.1074/jbc.272.9.5606 [DOI] [PubMed] [Google Scholar]
- 15. Gough N. R., and Fambrough D. M. (1997) Different steady state subcellular distributions of the three splice variants of lysosome-associated membrane protein LAMP-2 are determined largely by the COOH-terminal amino acid residue. J. Cell Biol. 137, 1161–1169 10.1083/jcb.137.5.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaushik S., and Cuervo A. M. (2015) Proteostasis and aging. Nat. Med. 21, 1406–1415 10.1038/nm.4001 [DOI] [PubMed] [Google Scholar]
- 17. Massey A. C., Kaushik S., Sovak G., Kiffin R., and Cuervo A. M. (2006) Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U.S.A. 103, 5805–5810 10.1073/pnas.0507436103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schneider J. L., Suh Y., and Cuervo A. M. (2014) Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 20, 417–432 10.1016/j.cmet.2014.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodríguez-Muela N., Koga H., García-Ledo L., de la Villa P., de la Rosa E. J., Cuervo A. M., and Boya P. (2013) Balance between autophagic pathways preserves retinal homeostasis. Aging Cell 12, 478–488 10.1111/acel.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaushik S., Massey A. C., Mizushima N., and Cuervo A. M. (2008) Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell 19, 2179–2192 10.1091/mbc.E07-11-1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koga H., Martinez-Vicente M., Macian F., Verkhusha V. V., and Cuervo A. M. (2011) A photoconvertible fluorescent reporter to track chaperone-mediated autophagy. Nat. Commun. 2, 386 10.1038/ncomms1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chava S., Lee C., Aydin Y., Chandra P. K., Dash A., Chedid M., Thung S. N., Moroz K., Wu T., Nayak N. C., and Dash S. (2017) Chaperone-mediated autophagy compensates for impaired macroautophagy in the cirrhotic liver to promote hepatocellular carcinoma. Oncotarget 8, 40019–40036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koga H., Martinez-Vicente M., Arias E., Kaushik S., Sulzer D., and Cuervo A. M. (2011) Constitutive upregulation of chaperone-mediated autophagy in Huntington's disease. J. Neurosci. 31, 18492–18505 10.1523/JNEUROSCI.3219-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dice J. F. (1990) Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 15, 305–309 10.1016/0968-0004(90)90019-8 [DOI] [PubMed] [Google Scholar]
- 25. Park C., Suh Y., and Cuervo A. M. (2015) Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat. Commun. 6, 6823 10.1038/ncomms7823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cuervo A. M., Stefanis L., Fredenburg R., Lansbury P. T., and Sulzer D. (2004) Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science 305, 1292–1295 10.1126/science.1101738 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y., Martinez-Vicente M., Krüger U., Kaushik S., Wong E., Mandelkow E. M., Cuervo A. M., and Mandelkow E. (2009) τ fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 18, 4153–4170 10.1093/hmg/ddp367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orenstein S. J., Kuo S. H., Tasset I., Arias E., Koga H., Fernandez-Carasa I., Cortes E., Honig L. S., Dauer W., Consiglio A., Raya A., Sulzer D., and Cuervo A. M. (2013) Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 16, 394–406 10.1038/nn.3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cuervo A. M., Palmer A., Rivett A. J., and Knecht E. (1995) Degradation of proteasomes by lysosomes in rat liver. Eur. J. Biochem. 227, 792–800 10.1111/j.1432-1033.1995.tb20203.x [DOI] [PubMed] [Google Scholar]
- 30. Lv L., Li D., Zhao D., Lin R., Chu Y., Zhang H., Zha Z., Liu Y., Li Z., Xu Y., Wang G., Huang Y., Xiong Y., Guan K. L., and Lei Q. Y. (2011) Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol. Cell 42, 719–730 10.1016/j.molcel.2011.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arias E., Koga H., Diaz A., Mocholi E., Patel B., and Cuervo A. M. (2015) Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol. Cell 59, 270–284 10.1016/j.molcel.2015.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaushik S., and Cuervo A. M. (2015) Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17, 759–770 10.1038/ncb3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bonhoure A., Vallentin A., Martin M., Senff-Ribeiro A., Amson R., Telerman A., and Vidal M. (2017) Acetylation of translationally controlled tumor protein promotes its degradation through chaperone-mediated autophagy. Eur. J. Cell Biol. 96, 83–98 10.1016/j.ejcb.2016.12.002 [DOI] [PubMed] [Google Scholar]
- 34. Arias E. (2017) Methods to study chaperone-mediated autophagy. Methods Enzymol. 588, 283–305 10.1016/bs.mie.2016.10.009 [DOI] [PubMed] [Google Scholar]
- 35. Aniento F., Roche E., Cuervo A. M., and Knecht E. (1993) Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 268, 10463–10470 [PubMed] [Google Scholar]
- 36. Xia H. G., Najafov A., Geng J., Galan-Acosta L., Han X., Guo Y., Shan B., Zhang Y., Norberg E., Zhang T., Pan L., Liu J., Coloff J. L., Ofengeim D., Zhu H., et al. (2015) Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J. Cell Biol. 210, 705–716 10.1083/jcb.201503044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dice J. F., Chiang H.-L., Spencer E. P., and Backer J. M. (1986) Regulation of catabolism of microinjected ribonuclease A: identification of residues 7–11 as the essential pentapeptide. J. Biol. Chem. 261, 6853–6859 [PubMed] [Google Scholar]
- 38. Cuervo A. M., Gomes A. V., Barnes J. A., and Dice J. F. (2000) Selective degradation of annexins by chaperone-mediated autophagy. J. Biol. Chem. 275, 33329–33335 10.1074/jbc.M005655200 [DOI] [PubMed] [Google Scholar]
- 39. Valdor R., Mocholi E., Botbol Y., Guerrero-Ros I., Chandra D., Koga H., Gravekamp C., Cuervo A. M., and Macian F. (2014) Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat. Immunol. 15, 1046–1054 10.1038/ni.3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Aniento F., Papavassiliou A. G., Knecht E., and Roche E. (1996) Selective uptake and degradation of c-Fos and v-Fos by rat liver lysosomes. FEBS Lett. 390, 47–52 10.1016/0014-5793(96)00625-4 [DOI] [PubMed] [Google Scholar]
- 41. Cuervo A. M., Hu W., Lim B., and Dice J. F. (1998) IκB is a substrate for a selective pathway of lysosomal proteolysis. Mol. Biol. Cell 9, 1995–2010 10.1091/mbc.9.8.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sooparb S., Price S. R., Shaoguang J., and Franch H. A. (2004) Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 65, 2135–2144 10.1111/j.1523-1755.2004.00639.x [DOI] [PubMed] [Google Scholar]
- 43. Kabuta T., Furuta A., Aoki S., Furuta K., and Wada K. (2008) Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J. Biol. Chem. 283, 23731–23738 10.1074/jbc.M801918200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu M. M., Yang Q., Xie X. Q., Liao C. Y., Lin H., Liu T. T., Yin L., and Shu H. B. (2016) Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45, 555–569 10.1016/j.immuni.2016.08.014 [DOI] [PubMed] [Google Scholar]
- 45. Yang Q., She H., Gearing M., Colla E., Lee M., Shacka J. J., and Mao Z. (2009) Regulation of neuronal survival factor MEF2D by chaperone-mediated autophagy. Science 323, 124–127 10.1126/science.1166088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Suzuki J., Nakajima W., Suzuki H., Asano Y., and Tanaka N. (2017) Chaperone-mediated autophagy promotes lung cancer cell survival through selective stabilization of the pro-survival protein, MCL1. Biochem. Biophys. Res. Commun. 482, 1334–1340 10.1016/j.bbrc.2016.12.037 [DOI] [PubMed] [Google Scholar]
- 47. Wu J. H., Guo J. P., Shi J., Wang H., Li L. L., Guo B., Liu D. X., Cao Q., and Yuan Z. Y. (2017) CMA down-regulates p53 expression through degradation of HMGB1 protein to inhibit irradiation-triggered apoptosis in hepatocellular carcinoma. World J. Gastroenterol. 23, 2308–2317 10.3748/wjg.v23.i13.2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gomes L. R., Menck C. F. M., and Cuervo A. M. (2017) Chaperone-mediated autophagy prevents cellular transformation by regulating MYC proteasomal degradation. Autophagy 13, 928–940 10.1080/15548627.2017.1293767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bauer P. O., Goswami A., Wong H. K., Okuno M., Kurosawa M., Yamada M., Miyazaki H., Matsumoto G., Kino Y., Nagai Y., and Nukina N. (2010) Harnessing chaperone-mediated autophagy for the selective degradation of mutant huntingtin protein. Nat. Biotechnol. 28, 256–263 10.1038/nbt.1608 [DOI] [PubMed] [Google Scholar]
- 50. Qi L., Zhang X. D., Wu J. C., Lin F., Wang J., DiFiglia M., and Qin Z. H. (2012) The role of chaperone-mediated autophagy in huntingtin degradation. PLoS ONE 7, e46834 10.1371/journal.pone.0046834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xilouri M., Vogiatzi T., Vekrellis K., Park D., and Stefanis L. (2009) Abberant alpha-synuclein confers toxicity to neurons in part through inhibition of chaperone-mediated autophagy. PLoS ONE 4, e5515 10.1371/journal.pone.0005515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kiffin R., Christian C., Knecht E., and Cuervo A. M. (2004) Activation of chaperone-mediated autophagy during oxidative stress. Mol. Biol. Cell 15, 4829–4840 10.1091/mbc.E04-06-0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cuervo A. M., Knecht E., Terlecky S. R., and Dice J. F. (1995) Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 269, C1200–C1208 10.1152/ajpcell.1995.269.5.C1200 [DOI] [PubMed] [Google Scholar]
- 54. Cuervo A. M., and Dice J. F. (2000) Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275, 31505–31513 10.1074/jbc.M002102200 [DOI] [PubMed] [Google Scholar]
- 55. Koga H., Kaushik S., and Cuervo A. M. (2010) Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 24, 3052–3065 10.1096/fj.09-144519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schneider J. L., Villarroya J., Diaz-Carretero A., Patel B., Urbanska A. M., Thi M. M., Villarroya F., Santambrogio L., and Cuervo A. M. (2015) Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 14, 249–264 10.1111/acel.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Agarraberes F. A., and Dice J. F. (2001) A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J. Cell Sci. 114, 2491–2499 [DOI] [PubMed] [Google Scholar]
- 58. Cuervo A. M., and Dice J. F. (2000) Regulation of lamp2a levels in the lysosomal membrane. Traffic 1, 570–583 10.1034/j.1600-0854.2000.010707.x [DOI] [PubMed] [Google Scholar]
- 59. Cuervo A. M., Mann L., Bonten E. J., d'Azzo A., and Dice J. F. (2003) Cathepsin A regulates chaperone-mediated autophagy through cleavage of the lysosomal receptor. EMBO J. 22, 47–59 10.1093/emboj/cdg002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kaushik S., Massey A. C., and Cuervo A. M. (2006) Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 25, 3921–3933 10.1038/sj.emboj.7601283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Napolitano G., Johnson J. L., He J., Rocca C. J., Monfregola J., Pestonjamasp K., Cherqui S., and Catz S. D. (2015) Impairment of chaperone-mediated autophagy leads to selective lysosomal degradation defects in the lysosomal storage disease cystinosis. EMBO Mol. Med. 7, 158–174 10.15252/emmm.201404223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang J., Johnson J. L., He J., Napolitano G., Ramadass M., Rocca C., Kiosses W. B., Bucci C., Xin Q., Gavathiotis E., Cuervo A. M., Cherqui S., and Catz S. D. (2017) Cystinosin, the small GTPase Rab11, and the Rab7 effector RILP regulate intracellular trafficking of the chaperone-mediated autophagy receptor LAMP2A. J. Biol. Chem. 292, 10328–10346 10.1074/jbc.M116.764076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cuervo A. M., and Dice J. F. (2000) Unique properties of lamp2a compared to other lamp2 isoforms. J. Cell Sci. 113, 4441–4450 [DOI] [PubMed] [Google Scholar]
- 64. Rout A. K., Strub M. P., Piszczek G., and Tjandra N. (2014) Structure of transmembrane domain of lysosome-associated membrane protein type 2a (LAMP-2A) reveals key features for substrate specificity in chaperone-mediated autophagy. J. Biol. Chem. 289, 35111–35123 10.1074/jbc.M114.609446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bandyopadhyay U., Sridhar S., Kaushik S., Kiffin R., and Cuervo A. M. (2010) Identification of regulators of chaperone-mediated autophagy. Mol. Cell 39, 535–547 10.1016/j.molcel.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anguiano J., Garner T. P., Mahalingam M., Das B. C., Gavathiotis E., and Cuervo A. M. (2013) Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat. Chem. Biol. 9, 374–382 10.1038/nchembio.1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Berger J. J., and Dice J. F. (1985) Effect of serum deprivation and replacement on proteolysis in cultured human fibroblasts. Prog. Clin. Biol. Res. 180, 479–481 [PubMed] [Google Scholar]
- 68. Finn P. F., and Dice J. F. (2005) Ketone bodies stimulate chaperone-mediated autophagy. J. Biol. Chem. 280, 25864–25870 10.1074/jbc.M502456200 [DOI] [PubMed] [Google Scholar]
- 69. Cuervo A. M., Hildebrand H., Bomhard E. M., and Dice J. F. (1999) Direct lysosomal uptake of α2-microglobulin contributes to chemically induced nephropathy. Kidney Int. 55, 529–545 10.1046/j.1523-1755.1999.00268.x [DOI] [PubMed] [Google Scholar]
- 70. Ferreira J. V., Soares A. R., Ramalho J. S., Pereira P., and Girao H. (2015) K63 linked ubiquitin chain formation is a signal for HIF1A degradation by chaperone-mediated autophagy. Sci. Rep. 5, 10210 10.1038/srep10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hubbi M. E., Hu H., Kshitiz, Ahmed I., Levchenko A., and Semenza G. L. (2013) Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J. Biol. Chem. 288, 10703–10714 10.1074/jbc.M112.414771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rodriguez-Navarro J. A., Kaushik S., Koga H., Dall'Armi C., Shui G., Wenk M. R., Di Paolo G., and Cuervo A. M. (2012) Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc. Natl. Acad. Sci. U.S.A. 109, E705–E714 10.1073/pnas.1113036109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., and Czaja M. J. (2009) Autophagy regulates lipid metabolism. Nature 458, 1131–1135 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Backer J. M., and Dice J. F. (1986) Covalent linkage of ribonuclease S-peptide to microinjected proteins causes their intracellular degradation to be enhanced by serum withdrawal. Proc. Natl. Acad. Sci. U.S.A. 83, 5830–5834 10.1073/pnas.83.16.5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kon M., Kiffin R., Koga H., Chapochnick J., Macian F., Varticovski L., and Cuervo A. M. (2011) Chaperone-mediated autophagy is required for tumor growth. Sci. Transl. Med. 3, 109ra117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang C., and Cuervo A. M. (2008) Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 14, 959–965 10.1038/nm.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhou D., Li P., Lin Y., Lott J. M., Hislop A. D., Canaday D. H., Brutkiewicz R. R., and Blum J. S. (2005) Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity 22, 571–581 10.1016/j.immuni.2005.03.009 [DOI] [PubMed] [Google Scholar]
- 78. Mortimore G. E., Lardeux B. R., and Adams C. E. (1988) Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J. Biol. Chem. 263, 2506–2512 [PubMed] [Google Scholar]
- 79. Yuan W., Tuttle D. L., Shi Y. J., Ralph G. S., and Dunn W. A. Jr. (1997) Glucose-induced microautophagy in Pichia pastoris requires the α-subunit of phosphofructokinase. J. Cell Sci. 110, 1935–1945 [DOI] [PubMed] [Google Scholar]
- 80. Sakai Y., Koller A., Rangell L. K., Keller G. A., and Subramani S. (1998) Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J. Cell Biol. 141, 625–636 10.1083/jcb.141.3.625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Leão-Helder A. N., Krikken A. M., Gellissen G., van der Klei I. J., Veenhuis M., and Kiel J. A. (2004) Atg21p is essential for macropexophagy and microautophagy in the yeast Hansenula polymorpha. FEBS Lett. 577, 491–495 10.1016/j.febslet.2004.10.055 [DOI] [PubMed] [Google Scholar]
- 82. Kunz J. B., Schwarz H., and Mayer A. (2004) Determination of four sequential stages during microautophagy in vitro. J. Biol. Chem. 279, 9987–9996 10.1074/jbc.M307905200 [DOI] [PubMed] [Google Scholar]
- 83. Uttenweiler A., Schwarz H., Neumann H., and Mayer A. (2007) The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell 18, 166–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lemasters J. J. (2014) Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox Biol. 2, 749–754 10.1016/j.redox.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Seo A. Y., Lau P. W., Feliciano D., Sengupta P., Gros M. A. L., Cinquin B., Larabell C. A., and Lippincott-Schwartz J. (2017) AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. Elife 6, e21690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Roberts P., Moshitch-Moshkovitz S., Kvam E., O'Toole E., Winey M., and Goldfarb D. (2003) Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kvam E., and Goldfarb D. S. (2004) Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 117, 4959–4968 10.1242/jcs.01372 [DOI] [PubMed] [Google Scholar]
- 88. Uytterhoeven V., Lauwers E., Maes I., Miskiewicz K., Melo M. N., Swerts J., Kuenen S., Wittocx R., Corthout N., Marrink S. J., Munck S., and Verstreken P. (2015) Hsc70–4 deforms membranes to promote synaptic protein turnover by endosomal microautophagy. Neuron 88, 735–748 10.1016/j.neuron.2015.10.012 [DOI] [PubMed] [Google Scholar]
- 89. Mukherjee A., Patel B., Koga H., Cuervo A. M., and Jenny A. (2016) Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy 12, 1984–1999 10.1080/15548627.2016.1208887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Caballero B., Wang Y., Diaz A., Tasset I., Juste Y. R., Mandelkow E., Mandelkow E., and Cuervo A. M. (2018) Interplay of pathogenic forms of human τ with different autophagic pathways. Aging Cell 17, 10.1111/acel.12692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sato M., Seki T., Konno A., Hirai H., Kurauchi Y., Hisatsune A., and Katsuki H. (2016) Fluorescent-based evaluation of chaperone-mediated autophagy and microautophagy activities in cultured cells. Genes Cells 21, 861–873 10.1111/gtc.12390 [DOI] [PubMed] [Google Scholar]
- 92. Morozova K., Clement C. C., Kaushik S., Stiller B., Arias E., Ahmad A., Rauch J. N., Chatterjee V., Melis C., Scharf B., Gestwicki J. E., Cuervo A. M., Zuiderweg E. R., and Santambrogio L. (2016) Structural and biological interaction of hsc-70 protein with phosphatidylserine in endosomal microautophagy. J. Biol. Chem. 291, 18096–18106 10.1074/jbc.M116.736744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Oku M., Maeda Y., Kagohashi Y., Kondo T., Yamada M., Fujimoto T., and Sakai Y. (2017) Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 216, 3263–3274 10.1083/jcb.201611029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Vevea J. D., Garcia E. J., Chan R. B., Zhou B., Schultz M., Di Paolo G., McCaffery J. M., and Pon L. A. (2015) Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599 10.1016/j.devcel.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tsuji T., Fujimoto M., Tatematsu T., Cheng J., Orii M., Takatori S., and Fujimoto T. (2017) Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. Elife 6, e25960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Numrich J., Péli-Gulli M. P., Arlt H., Sardu A., Griffith J., Levine T., Engelbrecht-Vandré S., Reggiori F., De Virgilio C., and Ungermann C. (2015) The I-BAR protein Ivy1 is an effector of the Rab7 GTPase Ypt7 involved in vacuole membrane homeostasis. J. Cell Sci. 128, 2278–2292 10.1242/jcs.164905 [DOI] [PubMed] [Google Scholar]
- 97. Dubouloz F., Deloche O., Wanke V., Cameroni E., and De Virgilio C. (2005) The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 19, 15–26 10.1016/j.molcel.2005.05.020 [DOI] [PubMed] [Google Scholar]
- 98. Cannizzo E. S., Clement C. C., Morozova K., Valdor R., Kaushik S., Almeida L. N., Follo C., Sahu R., Cuervo A. M., Macian F., and Santambrogio L. (2012) Age-related oxidative stress compromises endosomal proteostasis. Cell Rep. 2, 136–149 10.1016/j.celrep.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kalachev A. V., and Yurchenko O. V. (2017) Microautophagy in nutritive phagocytes of sea urchins. Protoplasma 254, 609–614 10.1007/s00709-016-0963-1 [DOI] [PubMed] [Google Scholar]