Abstract
Autophagy is a highly conserved catabolic pathway that is vital for development, cell survival, and the degradation of dysfunctional organelles and potentially toxic aggregates. Dysregulation of autophagy is associated with cancer, neurodegeneration, and lysosomal storage diseases. Accordingly, autophagy is precisely regulated at multiple levels (transcriptional, post-transcriptional, translational, and post-translational) to prevent aberrant activity. Various model organisms are used to study autophagy, but the baker's yeast Saccharomyces cerevisiae continues to be advantageous for genetic and biochemical analysis of non-selective and selective autophagy. In this Minireview, we focus on the cellular mechanisms that regulate autophagy transcriptionally and post-transcriptionally in S. cerevisiae.
Keywords: autophagy, post-translational modification, RNA degradation, transcription, vacuole, yeast
Overview
In response to external environmental and internal homeostatic cues, cells must efficiently and successfully adapt to ensure survival during stress conditions. Macroautophagy/autophagy is a highly conserved (from yeast to human) catabolic mechanism of “self-eating” that is vital for homeostasis, development, and the clearance of damaged or superfluous organelles and protein aggregates, substrates that cannot be degraded by the proteasome, the other major degradative pathway in eukaryotic cells. Autophagy occurs in all eukaryotic cells (1), underlying its importance. The classical morphological feature of autophagy is the formation of the double-membrane structure termed the autophagosome. In most cells, basal autophagy generally occurs at a low level but is markedly induced in response to nutrient deprivation, pathogen infection, and other forms of stress. Autophagic flux results in the fusion of the autophagosome with the vacuole (in yeast or plants) or a lysosome (in mammalian cells). In yeast, vacuolar hydrolases degrade the autophagic cargo. This degradation is followed by efflux of the breakdown products for reuse, helping to safeguard cell survival particularly during starvation or low-energy conditions.
When induced by nutrient deprivation or pharmacological means, non-selective autophagy targets bulk cytoplasm for uptake into the phagophore, the autophagosome precursor. During selective types of autophagy, the phagophore sequesters specific cargo (such as organelles or invasive pathogens) through receptor-mediated interactions between selective autophagy receptors and Atg8 (which is located on both sides of the phagophore), and unique cargo-localized ligands, thereby generally excluding bulk cytoplasm. In addition to receptor targeting, specific cargos are further selected by proximity to the expanding phagophore membrane (2). In yeast, under nutrient-rich conditions, a biosynthetic form of selective autophagy, the cytoplasm-to-vacuole targeting (Cvt)2 pathway, delivers resident hydrolases to the vacuole (3–6). Multiple forms of selective autophagy in yeast have been characterized, including mitophagy (7, 8), pexophagy (9), reticulophagy (10, 11), ribophagy (12), granulophagy (13), aggrephagy (14), nucleophagy (11, 15), lipophagy (16, 17), and piecemeal microautophagy of the nucleus/micronucleophagy (18, 19). Although not considered to be a selective type of autophagy, the degradation of bulk RNA has also been described (20). At least in yeast, the receptors utilized to target cargos are unique to the form of selective autophagy that the cell is undergoing (5, 7, 8, 21). For further discussion on the topic of selective autophagy, see Refs. 22, 23.
Autophagy is a highly complex and rigorously coordinated process; at present, 41 unique autophagy-related (ATG) genes have been identified in fungi, and many have homologs in higher eukaryotes. Autophagy dysregulation is associated with multiple human pathologies such as cancer, neurodegeneration, microbial infection and lysosomal storage diseases. Thus, autophagy must be strictly modulated to maintain an appropriate level, as too much or too little can be deleterious to the cell. Accordingly, eukaryotic cells have evolved mechanisms to tightly control and coordinate autophagy at multiple levels (transcriptional, post-transcriptional, translational, and post-translational; see Fig. 1). In this Minireview, we focus on autophagy regulation, particularly at the transcriptional and post-transcriptional levels, in the yeast Saccharomyces cerevisiae. In addition to its high degree of conservation with other systems, S. cerevisiae continues to be particularly advantageous for genetic and biochemical analysis of non-selective and selective forms of autophagy.
Figure 1.
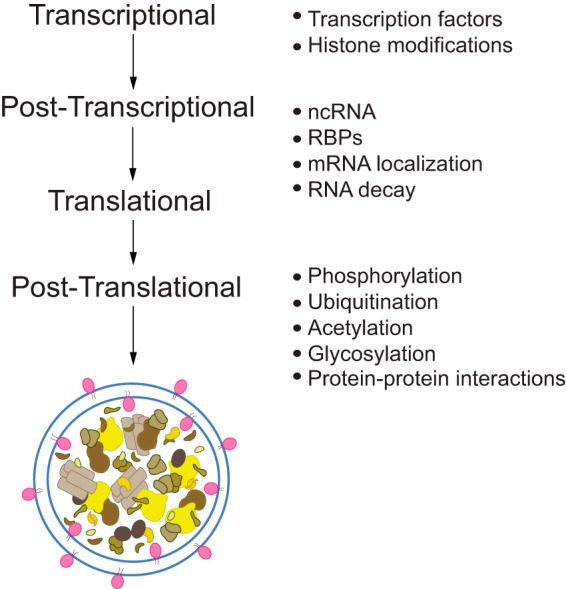
Autophagy regulation occurs at multiple levels. Because of the essential role that autophagy plays in maintaining homeostasis and the myriad of diseases that can result from perturbations to the pathway, cells must strictly modulate the entire process, beginning at transcription through post-translational modification. At the level of transcription, ATG gene expression can be regulated both positively and negatively through the action of specific transcription factors and epigenetic changes at histones. These transcripts can then be controlled further at the post-transcriptional and translational levels through the mechanisms of non-coding (nc) RNA, RNA-binding proteins (RBPs), mRNA localization, and RNA decay. At the protein level, post-translational modification such as phosphorylation, ubiquitination, acetylation, glycosylation, and protein–protein interactions can further regulate autophagy activity.
Autophagy is a dynamic process that requires constant fine-tuning at multiple levels to maintain the appropriate timing of induction and magnitude within the cell. Cells control the level of the autophagic response largely by regulating the size and frequency (i.e. number) of autophagosomes (24, 25). Here, we will briefly describe the main phases of the autophagic process and the machinery involved before further reviewing recently published work on its transcriptional and post-transcriptional regulation.
Autophagy in S. cerevisiae
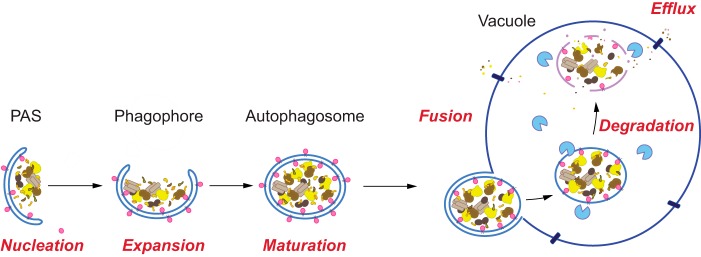
There are five main stages of autophagy, including: 1) induction and nucleation of the phagophore membrane; 2) expansion of the phagophore; 3) closure and maturation to form the autophagosome; 4) autophagosome–vacuole fusion; and 5) degradation/efflux of the breakdown products (see Fig. 2). Briefly, in yeast, autophagy is typically stimulated through nutrient deprivation (most commonly with nitrogen starvation) or with the use of a pharmacological agent such as rapamycin. Rapamycin treatment inhibits target of rapamycin (TOR), a serine/threonine kinase and major negative regulator of autophagy induction in yeast (26). In addition to TOR, upstream nutrient sensors such as protein kinase A and Snf1 (the homolog of mammalian AMP kinase) integrate signals for autophagy regulation (27). In yeast, the phagophore assembly site (PAS) is the intracellular location of autophagosome formation. At the PAS, Atg proteins assemble in a hierarchical order. The first complex recruited to the PAS includes Atg1, Atg13, and the Atg17–Atg31–Atg29 ternary subcomplex (28, 29). Next, Atg9 (along with Atg2 and Atg18) localizes to the PAS (30). The phagophore, a dynamic cup-shaped membrane structure, transiently envelops bulk cytoplasm or specific cargo. This sequestration event is followed by expansion and closure of the phagophore to form the mature autophagosome and requires two conserved ubiquitin-like (Ubl) conjugation systems, which involve Atg12 and Atg8.
Figure 2.
Non-selective autophagy in S. cerevisiae. Autophagy occurs through a sequential series of events in the yeast S. cerevisiae, including induction and nucleation of the phagophore at the PAS, expansion of the phagophore, closure and maturation to form the autophagosome, autophagosome–vacuole fusion, and cargo degradation followed by efflux of the breakdown products.
The Atg12 Ubl system includes Atg5, Atg7, Atg10, Atg12, and Atg16 and leads to the formation of the heterotrimeric complex Atg12—Atg5–Atg16, which may function as an E3-like enzyme for the Atg8 conjugation system, although this complex is not absolutely required for conjugation to occur (31–34). The conjugation of Atg12 to Atg5 at the phagophore occurs sequentially, requiring Atg7, an E1-like enzyme that activates Atg12, and Atg10, an E2-like enzyme (35, 36). The Atg8 Ubl system is necessary for membrane expansion and closure of the phagophore, and this second system includes Atg3, Atg4, Atg7, and Atg8 (30). Non-lipidated Atg8 is converted to its lipidated phosphatidylethanolamine-conjugated species following Atg4-mediated proteolytic processing of its C terminus, Atg7-dependent activation, and Atg3-facilitated conjugation at a conserved C-terminal glycine (37, 38).
The next major step in autophagy involves fusion between the outer membrane of the autophagosome and the vacuole. The resulting vesicle formed by this event is termed the autophagic body and consists of the remaining inner autophagosome membrane found within the vacuole lumen. The final events in autophagic flux include degradation of the cargo, followed by efflux of the resulting macromolecules. In yeast, the vacuole has enzymes for degrading the major macromolecules; however, an efflux mechanism has only been identified for amino acids (39), and catabolized RNA products appear to be secreted from the cell (20). For a more comprehensive review on the main stages of autophagy and the role of ubiquitin, please see Refs. 28, 40, 41.
Transcriptional regulation of autophagy
Background
As mentioned above, cells must successfully fine-tune and integrate signals at multiple regulatory levels to maintain appropriate control over autophagy. Here, we focus on transcriptional and post-transcriptional regulation of autophagy in S. cerevisiae. For further discussion on autophagy regulation, particularly at the epigenetic and post-translational levels, please see these recent reviews in Refs. 40, 42–45.
Major transcriptional regulators in yeast
Ume6
Ume6 is a DNA-binding protein that has dual functions both as a transcriptional activator and as a repressor depending on the growth conditions (46, 47). Ume6 is dependent on the corepressor Sin3 and histone deacetylase Rpd3, and all are components of the multisubunit Rpd3 large (Rpd3L) complex in yeast (48–50). Rpd3L is one of the 2 Rpd3 histone deacetylase complexes that regulate gene expression (50–52). A Ume6 consensus-binding site (URS1 region) is found within the ATG8 promoter (53); Ume6 directly binds (and thereby represses) the ATG8 promoter under nutrient-rich conditions (53). Ume6 undergoes phosphorylation by the kinase Rim15 (further detailed below) during nitrogen starvation, which leads to the derepression of ATG8, thus promoting ATG8 transcription (53). Deletion of UME6, SIN3, and/or RPD3 significantly up-regulates ATG8 mRNA (and consequently protein) under nutrient-rich conditions, and autophagy is more rapidly induced in ume6Δ cells during nitrogen starvation (53). Importantly, the amount of Atg8 directly correlates with the size of autophagosomes during starvation conditions (24). These findings support a mechanism whereby the cell is primed for a rapid autophagic response once it encounters starvation conditions. Although Ume6 is not conserved in more complex eukaryotes, the mammalian SIN3 proteins appear to play a similar role in regulating the expression of the Atg8 homolog MAP1LC3B (53).
Pho23
Pho23 is another member of the Rpd3L complex (50, 54) and a negative regulator of autophagy activity in yeast (25). Deletion of PHO23 results in the up-regulation of multiple ATG transcripts, including ATG1, -7–9, -12, -14, and -29 and an increased frequency of autophagosome formation (i.e. number) (25). Pho23 represses ATG9 under nutrient-replete conditions, and levels of Atg9 directly affect the frequency of autophagosome formation (25). Atg9 is the only integral membrane protein component of the core autophagy machinery—those proteins that are essential for autophagosome formation (55). Atg9 cycles between the PAS and peripheral sites close to the mitochondria during autophagy; these sites are also known as tubulovesicular clusters and are thought to correspond to donor membranes (56–58). Phosphorylation of Atg9 at serine 122 (Ser-122) regulates anterograde movement between the peripheral sites and the PAS, thereby controlling the rate of autophagosome formation (59). These data support a model in which Atg9 functions to provide membrane or to direct membrane delivery for phagophore expansion; thus, increased levels of Atg9 (through Pho23 derepression) allow for a greater number of autophagosomes to form, which would have a direct effect on the magnitude of autophagy activity.
Rph1
Rph1 is a Jumonji C catalytic domain-containing histone demethylase (60). However, the role of Rph1 in autophagy is independent of its demethylase activity (61). Deletion of RPH1 enhances autophagy, and overexpression of Rph1 strongly inhibits autophagy and autophagosome formation (61). Rph1 functions as a negative transcriptional regulator of autophagy by repressing the expression of a subset of ATG genes under nutrient-replete conditions, particularly ATG7, but also including ATG1, -8, -9, -14, -29, and -32 (61). Furthermore, Rph1 directly regulates ATG7 by binding to its promoter; this binding does not occur when the DNA-binding domains of Rph1 are eliminated (61). As described above, Atg7 plays an essential role in the conjugation of phosphatidylethanolamine to Atg8, which is critical for autophagosome formation. Levels of Atg7 have an impact on the magnitude of the autophagic response (61, 62). When cells are starved for nitrogen, Rim15 phosphorylates Rph1, inhibiting its repression of ATG transcripts (61). KDM4A, a mammalian homolog of Rph1, has a conserved role in autophagy induction (61). Additionally, Rph1 may mediate transcriptional control over other as yet unidentified ATG genes (63).
Rim15
The Rim15 protein kinase integrates signals from the two major nutrient sensing pathways in yeast, TOR and protein kinase A, to positively regulate autophagy (64–65). Although not a direct transcriptional regulator, Rim15 phosphorylates the DNA-binding proteins Ume6 (53) and Rph1 (61) to influence ATG gene transcription (66); Rim15-dependent phosphorylation inhibits these transcription factors, leading to the derepression of ATG genes (61). Future studies will determine whether Rim15 mediates the phosphorylation of additional autophagy regulatory factors, particularly those involved in other aspects of transcriptional or post-transcriptional autophagy modulation.
Gcn4
Gcn4 is a basic leucine zipper (bZIP) transcriptional activator that primarily binds to the 5′-TGACTC-3′ consensus site in the promoter regions of target genes (67). Gcn4 was initially identified to function in the amino acid starvation response and has been described as a principal regulator of autophagy gene expression (68). During growing conditions, Gcn4 positively mediates delivery of precursor aminopeptidase I in the Cvt pathway (69). When cells undergo starvation, Gcn4 regulates non-selective autophagy activity through ATG gene transcription (69). Gcn4 activates ATG1, ATG13, and ATG14 gene expression during amino acid deprivation (68) and ATG1 during nitrogen starvation (69). Gcn4 controls ATG41 mRNA expression during nitrogen starvation-induced autophagy through transcriptional activation (70). Recently, Atg41 was identified to function in Atg9 cycling and the delivery of donor membrane to expand the phagophore at the PAS (70). The translation of GCN4 is stimulated by the phosphorylation of Sui2/eIF2α by the protein kinase Gcn2 (71). Gcn2 itself also positively regulates autophagy, presumably by its downstream effects on Gcn4-mediated ATG transcription (72, 73) and possibly through the inhibition of TOR (74).
Gln3
Gln3 is a GATA-like transcription factor that shares only 65% homology with other known GATA factors in yeast (75, 76). Gln3 binds to the consensus sequences 5′-GATAAG-3′ and 5′-GATTAG-3′, which have been previously identified as nitrogen-responsive upstream activation sequences within the promoter regions of target genes (77). During rich conditions, Gln3 mediates biosynthetic delivery of precursor aminopeptidase I to the vacuole (69). When cells are starved, Gln3 positively regulates non-selective autophagy by targeting ATG14 (78), -7–9, -29, and -32 (69). Unexpectedly, in nutrient-replete conditions, deletion of GLN3 results in the accumulation of ATG8 and ATG29 transcripts, demonstrating either direct or indirect negative control over basal autophagy (69).
Gat1
Gat1 is another GATA-type transcription factor that has a GATA1-type zinc finger DNA-binding motif and binds to 5′-GATAAG-3′ upstream activation sequence regions in the promoters of nitrogen-sensitive genes, similar to Gln3 (79). Gat1 functions as a positive factor for autophagy induction; deletion of GAT1 results in decreased autophagy activity (69). Also similar to Gln3, deletion of GAT1 significantly down-regulates ATG7–9, -29, and -32 mRNAs; however, no additive effect is observed on ATG transcript levels when both GAT1 and GLN3 are deleted in the same strain (69).
Other transcriptional regulators
Yap1 is a bZIP transcription factor with a preference for binding at promoter sites containing 5′-TTACTAA-3′ sequences (80). It has been recently reported that Yap1 positively regulates transcription of the lipase gene ATG15 during starvation-induced autophagy (81). Atg15 preferentially hydrolyzes phosphatidylserine and facilitates autophagic body lysis within the vacuole (82, 83). Sfl1 functions as a transcriptional repressor and activator (84, 85) and has also been recently identified to function as a positive regulator of the Cvt pathway, autophagy, and ATG8 expression (69). Fyv5 negatively regulates the expression of ATG1, -8, -9, and -14, but this effect may not be direct; in contrast, the Cvt pathway appears to be positively affected by this factor (69). Spt10, a histone H3 acetylase, represses ATG8 and ATG9 at both the RNA and protein levels (69). Although key transcriptional regulators have been identified to affect core ATG gene transcripts, the potential exists for novel factors to be discovered, particularly those that may target selective autophagy pathways.
Post-transcriptional regulation of autophagy
Background
Although novel transcriptional mediators of autophagy have recently been identified (25, 53, 61, 69), post-transcriptional regulation of autophagy in yeast is largely uncharacterized. In mammals, non-coding RNAs such as microRNAs (miRNAs) and RNA-binding proteins can modulate autophagy at the post-transcriptional level (42, 45, 86–89). However, the RNA interference system (which is required for miRNA processing (90, 91)) is not present in S. cerevisiae (92). Alternatively, another mechanism whereby cells exert post-transcriptional control over gene expression is through RNA decay pathways (which can degrade transcripts in either the 5′ to 3′ or the 3′ to 5′ direction). During canonical 5′ to 3′ degradation, transcripts undergo a reversible process known as deadenylation (which removes the 3′ poly(A) tail), followed by decapping. The decapping enzyme Dcp2 removes the 5′-methylguanosine cap of the mRNA, resulting in an exposed 5′-monophosphate. Finally, the decapped cytoplasmic mRNAs undergo 5′- to 3′-mediated degradation by the cytoplasmic exoribonuclease Xrn1.
Dcp2 and RCK family RNA helicases Dhh1/Vad1/DDX6
A role has recently been described for Dcp2 and RCK family RNA-binding proteins as post-transcriptional regulators of ATG mRNAs, autophagy, and autophagy-dependent innate immune responses in yeast and mammalian cells (93). RCK family members–Dhh1 in S. cerevisiae, Vad1 in Cryptococcus neoformans, and DDX6/p54 in mammals–are RNA helicases acting in part as decapping accessory factors that interact with target mRNAs through recruitment to the decapping complex by binding the 5′- and/or 3′-untranslated region of selected transcripts (40, 94). Dhh1 also physically interacts with the decapping enzyme Dcp2 (95).
In the report by Hu et al. (96), deletion of DHH1 or a temperature-sensitive (ts) mutation of DCP2 caused a significant up-regulation of ATG transcripts. In particular, of the ATG transcripts that were examined in this study, dhh1Δ cells demonstrated significantly increased levels of ATG3, -7, -8, -19, -20, -22, and -24 mRNA under nutrient-replete conditions (96). In the dcp2-7Δ ts strain, transcript levels for ATG1 through ATG9, ATG11, ATG13 through ATG24, and ATG29, -31, -32, and -34 were significantly up-regulated in rich conditions (96). Both the dhh1Δ and dcp2-7Δ ts strains demonstrated higher levels of autophagy activity through multiple assays when starved for nitrogen (96). This mechanism is highly conserved across the two fungal species examined (S. cerevisiae and the pathogen C. neoformans) and up through mammalian cells (96). In C. neoformans, the Dhh1 homolog Vad1 mediates decapping of ATG mRNAs, especially ATG8 (96). Furthermore, Hu et al. (96) demonstrated that Dcp2 is phosphorylated by TOR under nutrient-rich conditions in C. neoformans, which drives its association with (at least) ATG8 mRNA, stimulating recruitment of the transcript to the decapping machinery, followed by subsequent decapping. Thus, TOR acts as a negative regulator of the translation of ATG genes under conditions where it promotes the translation of the vast majority of cellular transcripts. In mammalian cells, mechanistic TOR phosphorylates DDX6, the homolog of Dhh1/Vad1, resulting in decapping of the MAP1LC3B transcript under nutrient-rich conditions to repress autophagy (96). Transcripts are then presumably degraded to maintain autophagy at a basal level. In contrast, starvation conditions inactivate TORC1, arresting degradation of the targeted transcripts and promoting sustained autophagy (96).
Xrn1
The data on Dhh1/Vad1 and Dcp2 imply a role for 5′ to 3′ mRNA degradation in autophagy regulation. Recent work shows that the RNase Xrn1/XRN1, which functions downstream of the decapping complex in the canonical 5′ to 3′ RNA decay pathway, is also a negative post-transcriptional regulator of autophagy in both yeast and mammalian cells (97). Chromosomal deletion of XRN1 induces a more rapid and robust autophagy response as determined through multiple assays in yeast (97). We also found that the frequency of autophagosome formation increases in starved xrn1Δ cells compared with wild-type cells based on transmission electron microscopy (97). Furthermore, when xrn1Δ cells are assessed by quantitative PCR under nutrient-rich conditions, select ATG transcripts are found to be up-regulated, including ATG1, -4, -5, -7, -8, -12, -14, -16, -29, and -31. Regulation of (at least) ATG8, ATG12, and ATG29 is dependent upon the RNase activity of Xrn1 (97).
In mammalian cells, there is enhanced autophagy activity in a starvation-independent manner when XRN1 is depleted by small interfering RNA (97). The impact on autophagy is blocked in BECN1 or ATG5 CRISPR knockout cells, supporting the role of the canonical autophagy pathway (97); BECN1 is a component of the phosphatidylinositol 3-kinase complex required for autophagy induction (98). In addition, reduction of XRN1 levels is associated with an up-regulation of poliovirus infection in an autophagy-dependent manner (97), underlying the role of Xrn1/XRN1 as a conserved autophagy regulator. Poliovirus, similar to other picornaviruses, utilizes host membranes that are proposed to be derived from autophagosomes to support viral genome replication (97, 99, 100).
Summary
The studies by Hu et al. (96) and Delorme-Axford et al. (97) provide a paradigm for post-transcriptional autophagy regulation, but they also represent the limit of what is known about this type of regulation of autophagy in yeast. Although these findings are exciting and novel, it is still not clear why there appears to be differential targeting of transcripts in the strains examined (dcp2–7Δ ts, dhh1Δ, and xrn1Δ) even though they encode components of a related pathway of RNA degradation (96, 97). Thus, despite these insights, much work remains to further understand how autophagy is controlled post-transcriptionally. For example, how are transcripts selected for degradation under nutrient-replete conditions? Although Dhh1 and Xrn1 are likely to drive some degree of specificity, many more ATG transcripts are affected in the dcp2-7Δ ts cells compared with the dhh1Δ and xrn1Δ strains (96, 97). Based on these data, Dhh1 and Xrn1 are likely not the only factors mediating selective transcript targeting. Furthermore, although S. cerevisiae does not post-transcriptionally regulate gene expression through miRNA, additional non-coding RNA mechanisms such as long non-coding RNA could potentially play a role in autophagy regulation. Future studies should be aimed toward uncovering additional components moderating ATG gene expression through RNA decay.
Conclusions
Here, we present a concise review of what is currently known regarding the transcriptional and post-transcriptional regulation of autophagy in the yeast S. cerevisiae (see Fig. 3 for a summary). Recent advances in the field have provided additional clues as to the myriad of regulatory mechanisms that cells maintain to tightly control and coordinate autophagy. However, many questions remain, particularly concerning how autophagy is regulated post-transcriptionally, not only in yeast but in other eukaryotic systems as well. Additional novel post-transcriptional regulatory factors have yet to be identified and characterized. Further investigation into the mechanisms by which cells control the major stages of autophagy—especially induction and magnitude—at multiple regulatory levels is critical to enhance our understanding of how this essential process is maintained in the cell to promote normal physiological processes and how its dysregulation contributes to disease pathogenesis.
Figure 3.
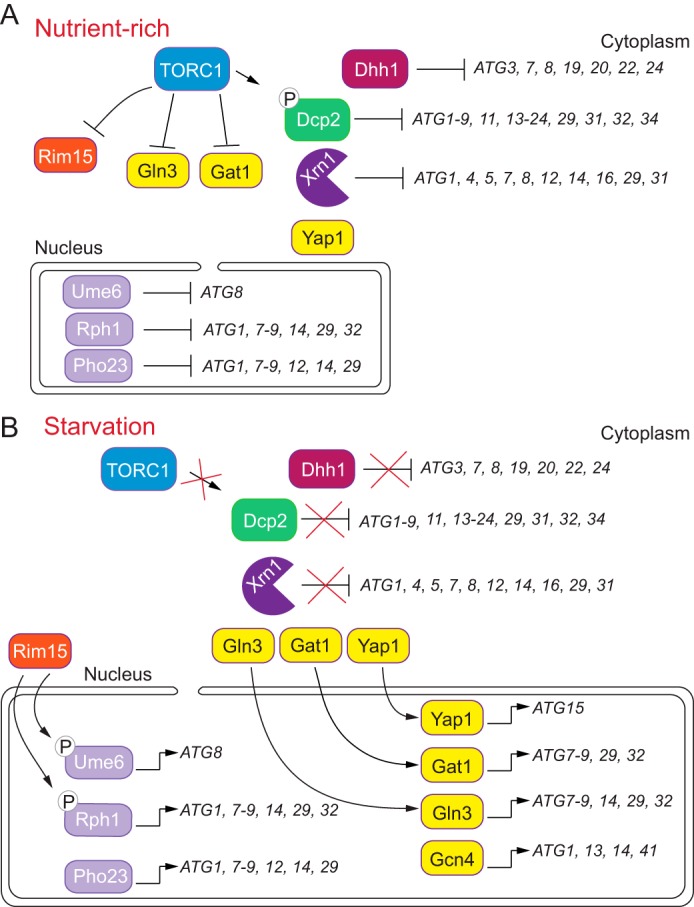
Summary of transcriptional and post-transcriptional regulatory factors in S. cerevisiae. A, during nutrient-replete conditions, TORC1 is active and negatively regulates key autophagy-promoting factors, such as Rim15, while activating negative regulatory elements, including the decapping enzyme Dcp2. Dcp2 mediates decapping of target transcripts for subsequent RNA degradation. Repressors such as Rph1, Ume6, and Pho23 bind the promoters of ATG7, ATG8, and ATG9, respectively, to maintain autophagy at a basal level. B, when autophagy is stimulated by an external stress, such as nitrogen or amino acid starvation, TORC1 becomes inactivated. This inhibition of TORC1 allows for the downstream activation of positive autophagy regulators. Activators such as Gcn4, Gln3, Gat1, and Yap1 bind to target genes for transcription as indicated. RNA decay mediators Dhh1, Dcp2, and Xrn1 no longer target ATG mRNAs for down-regulation; instead, these ATG transcripts are presumably translated to sustain autophagy.
Acknowledgment
We apologize to those whose work was not included here due to space limitations.
This is the second article in the Thematic Minireview series “Autophagy.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- Cvt
- cytoplasm-to-vacuole targeting
- TOR
- target of rapamycin
- miRNA
- microRNA
- PAS
- phagophore assembly site
- Ubl
- ubiquitin-like
- bZIP
- basic leucine zipper.
References
- 1. Klionsky D. J., and Emr S. D. (2000) Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sawa-Makarska J., Abert C., Romanov J., Zens B., Ibiricu I., and Martens S. (2014) Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat. Cell Biol. 16, 425–433 10.1038/ncb2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harding T. M., Morano K. A., Scott S. V., and Klionsky D. J. (1995) Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602 10.1083/jcb.131.3.591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hutchins M. U., and Klionsky D. J. (2001) Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 276, 20491–20498 10.1074/jbc.M101150200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scott S. V., Guan J., Hutchins M. U., Kim J., and Klionsky D. J. (2001) Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell 7, 1131–1141 10.1016/S1097-2765(01)00263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yorimitsu T., and Klionsky D. J. (2005) Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell 16, 1593–1605 10.1091/mbc.E04-11-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanki T., Wang K., Cao Y., Baba M., and Klionsky D. J. (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Okamoto K., Kondo-Okamoto N., and Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- 9. Hutchins M. U., Veenhuis M., and Klionsky D. J. (1999) Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112, 4079–4087 [DOI] [PubMed] [Google Scholar]
- 10. Bernales S., McDonald K. L., and Walter P. (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 4, e423 10.1371/journal.pbio.0040423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mochida K., Oikawa Y., Kimura Y., Kirisako H., Hirano H., Ohsumi Y., and Nakatogawa H. (2015) Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362 10.1038/nature14506 [DOI] [PubMed] [Google Scholar]
- 12. Kraft C., Deplazes A., Sohrmann M., and Peter M. (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- 13. Buchan J. R., Kolaitis R. M., Taylor J. P., and Parker R. (2013) Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474 10.1016/j.cell.2013.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu K., Psakhye I., and Jentsch S. (2014) Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158, 549–563 10.1016/j.cell.2014.05.048 [DOI] [PubMed] [Google Scholar]
- 15. Mijaljica D., Prescott M., and Devenish R. J. (2012) A late form of nucleophagy in Saccharomyces cerevisiae. PloS one 7, e40013 10.1371/journal.pone.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Zutphen T., Todde V., de Boer R., Kreim M., Hofbauer H. F., Wolinski H., Veenhuis M., van der Klei I. J., and Kohlwein S. D. (2014) Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 25, 290–301 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vevea J. D., Garcia E. J., Chan R. B., Zhou B., Schultz M., Di Paolo G., McCaffery J. M., and Pon L. A. (2015) Role for lipid droplet biogenesis and microlipophagy in adaptation to lipid imbalance in yeast. Dev. Cell 35, 584–599 10.1016/j.devcel.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roberts P., Moshitch-Moshkovitz S., Kvam E., O'Toole E., Winey M., and Goldfarb D. S. (2003) Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 10.1091/mbc.E02-08-0483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E. L., Millen J., Goldfarb D. S., and Thumm M. (2008) Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 19, 4492–4505 10.1091/mbc.E08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H., Kawamata T., Horie T., Tsugawa H., Nakayama Y., Ohsumi Y., and Fukusaki E. (2015) Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 34, 154–168 10.15252/embj.201489083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Motley A. M., Nuttall J. M., and Hettema E. H. (2012) Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31, 2852–2868 10.1038/emboj.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Farré J. C., and Subramani S. (2016) Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat. Rev. Mol. Cell Biol. 17, 537–552 10.1038/nrm.2016.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Anding A. L., and Baehrecke E. H. (2017) Cleaning house: selective autophagy of organelles. Dev. Cell 41, 10–22 10.1016/j.devcel.2017.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xie Z., Nair U., and Klionsky D. J. (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin M., He D., Backues S. K., Freeberg M. A., Liu X., Kim J. K., and Klionsky D. J. (2014) Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr. Biol. 24, 1314–1322 10.1016/j.cub.2014.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Noda T., and Ohsumi Y. (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 273, 3963–3966 10.1074/jbc.273.7.3963 [DOI] [PubMed] [Google Scholar]
- 27. Cebollero E., and Reggiori F. (2009) Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793, 1413–1421 10.1016/j.bbamcr.2009.01.008 [DOI] [PubMed] [Google Scholar]
- 28. Parzych K. R., and Klionsky D. J. (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid. Redox Signal. 20, 460–473 10.1089/ars.2013.5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mao K., Chew L. H., Inoue-Aono Y., Cheong H., Nair U., Popelka H., Yip C. K., and Klionsky D. J. (2013) Atg29 phosphorylation regulates coordination of the Atg17–Atg31–Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc. Natl. Acad. Sci. U.S.A. 110, E2875–E2884 10.1073/pnas.1300064110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng Y., He D., Yao Z., and Klionsky D. J. (2014) The machinery of macroautophagy. Cell Res. 24, 24–41 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mizushima N., Noda T., and Ohsumi Y. (1999) Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 18, 3888–3896 10.1093/emboj/18.14.3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuma A., Mizushima N., Ishihara N., and Ohsumi Y. (2002) Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 277, 18619–18625 10.1074/jbc.M111889200 [DOI] [PubMed] [Google Scholar]
- 33. Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., and Ohsumi Y. (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 10.1074/jbc.C700195200 [DOI] [PubMed] [Google Scholar]
- 34. Sakoh-Nakatogawa M., Matoba K., Asai E., Kirisako H., Ishii J., Noda N. N., Inagaki F., Nakatogawa H., and Ohsumi Y. (2013) Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat. Struct. Mol. Biol. 20, 433–439 10.1038/nsmb.2527 [DOI] [PubMed] [Google Scholar]
- 35. Klionsky D. J., and Codogno P. (2013) The mechanism and physiological function of macroautophagy. J. Innate Immun. 5, 427–433 10.1159/000351979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakatogawa H., Suzuki K., Kamada Y., and Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- 37. Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., Noda T., and Ohsumi Y. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 38. Kirisako T., Ichimura Y., Okada H., Kabeya Y., Mizushima N., Yoshimori T., Ohsumi M., Takao T., Noda T., and Ohsumi Y. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Z., Huang J., Geng J., Nair U., and Klionsky D. J. (2006) Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 17, 5094–5104 10.1091/mbc.E06-06-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wen X., and Klionsky D. J. (2016) An overview of macroautophagy in yeast. J. Mol. Biol. 428, 1681–1699 10.1016/j.jmb.2016.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Delorme-Axford E., Guimaraes R. S., Reggiori F., and Klionsky D. J. (2015) The yeast Saccharomyces cerevisiae: an overview of methods to study autophagy progression. Methods 75, 3–12 10.1016/j.ymeth.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feng Y., Yao Z., and Klionsky D. J. (2015) How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 25, 354–363 10.1016/j.tcb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Popelka H., and Klionsky D. J. (2015) Post-translationally-modified structures in the autophagy machinery: an integrative perspective. FEBS J. 282, 3474–3488 10.1111/febs.13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie Y., Kang R., Sun X., Zhong M., Huang J., Klionsky D. J., and Tang D. (2015) Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 11, 28–45 10.4161/15548627.2014.984267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Füllgrabe J., Klionsky D. J., and Joseph B. (2014) The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 15, 65–74 10.1038/nrm3716 [DOI] [PubMed] [Google Scholar]
- 46. Strich R., Surosky R. T., Steber C., Dubois E., Messenguy F., and Esposito R. E. (1994) UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 8, 796–810 10.1101/gad.8.7.796 [DOI] [PubMed] [Google Scholar]
- 47. Steber C. M., and Esposito R. E. (1995) UME6 is a central component of a developmental regulatory switch controlling meiosis-specific gene expression. Proc. Natl. Acad. Sci. U.S.A. 92, 12490–12494 10.1073/pnas.92.26.12490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kadosh D., and Struhl K. (1997) Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell 89, 365–371 10.1016/S0092-8674(00)80217-2 [DOI] [PubMed] [Google Scholar]
- 49. Washburn B. K., and Esposito R. E. (2001) Identification of the Sin3-binding site in Ume6 defines a two-step process for conversion of Ume6 from a transcriptional repressor to an activator in yeast. Mol. Cell. Biol. 21, 2057–2069 10.1128/MCB.21.6.2057-2069.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carrozza M. J., Florens L., Swanson S. K., Shia W. J., Anderson S., Yates J., Washburn M. P., and Workman J. L. (2005) Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim. Biophys. Acta 1731, 77–87 10.1016/j.bbaexp.2005.09.005 [DOI] [PubMed] [Google Scholar]
- 51. Kadosh D., and Struhl K. (1998) Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12, 797–805 10.1101/gad.12.6.797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vidal M., and Gaber R. F. (1991) RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 11, 6317–6327 10.1128/MCB.11.12.6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bartholomew C. R., Suzuki T., Du Z., Backues S. K., Jin M., Lynch-Day M. A., Umekawa M., Kamath A., Zhao M., Xie Z., Inoki K., and Klionsky D. J. (2012) Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc. Natl. Acad. Sci. U.S.A. 109, 11206–11210 10.1073/pnas.1200313109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Loewith R., Smith J. S., Meijer M., Williams T. J., Bachman N., Boeke J. D., and Young D. (2001) Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 276, 24068–24074 10.1074/jbc.M102176200 [DOI] [PubMed] [Google Scholar]
- 55. Noda T., Kim J., Huang W.-P., Baba M., Tokunaga C., Ohsumi Y., and Klionsky D. J. (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465–480 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Reggiori F., Tucker K. A., Stromhaug P. E., and Klionsky D. J. (2004) The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79–90 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- 57. Reggiori F., Shintani T., Nair U., and Klionsky D. J. (2005) Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 1, 101–109 10.4161/auto.1.2.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mari M., Griffith J., Rieter E., Krishnappa L., Klionsky D. J., and Reggiori F. (2010) An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J. Cell Biol. 190, 1005–1022 10.1083/jcb.200912089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Feng Y., Backues S. K., Baba M., Heo J. M., Harper J. W., and Klionsky D. J. (2016) Phosphorylation of Atg9 regulates movement to the phagophore assembly site and the rate of autophagosome formation. Autophagy 12, 648–658 10.1080/15548627.2016.1157237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tu S., Bulloch E. M., Yang L., Ren C., Huang W. C., Hsu P. H., Chen C. H., Liao C. L., Yu H. M., Lo W. S., Freitas M. A., and Tsai M. D. (2007) Identification of histone demethylases in Saccharomyces cerevisiae. J. Biol. Chem. 282, 14262–14271 10.1074/jbc.M609900200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bernard A., Jin M., González-Rodriguez P., Füllgrabe J., Delorme-Axford E., Backues S. K., Joseph B., and Klionsky D. J. (2015) Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr. Biol. 25, 546–555 10.1016/j.cub.2014.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bernard A., and Klionsky D. J. (2015) Rph1 mediates the nutrient-limitation signaling pathway leading to transcriptional activation of autophagy. Autophagy 11, 718–719 10.1080/15548627.2015.1018503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liang C. Y., Wang L. C., and Lo W. S. (2013) Dissociation of the H3K36 demethylase Rph1 from chromatin mediates derepression of environmental stress-response genes under genotoxic stress in Saccharomyces cerevisiae. Mol. Biol. Cell 24, 3251–3262 10.1091/mbc.E12-11-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yorimitsu T., Zaman S., Broach J. R., and Klionsky D. J. (2007) Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 4180–4189 10.1091/mbc.E07-05-0485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reggiori F., and Klionsky D. J. (2013) Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194, 341–361 10.1534/genetics.112.149013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Devenish R. J., and Prescott M. (2015) Autophagy: starvation relieves transcriptional repression of ATG genes. Curr. Biol. 25, R238–R240 10.1016/j.cub.2015.01.045 [DOI] [PubMed] [Google Scholar]
- 67. Arndt K., and Fink G. R. (1986) GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc. Natl. Acad. Sci. U.S.A. 83, 8516–8520 10.1073/pnas.83.22.8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Natarajan K., Meyer M. R., Jackson B. M., Slade D., Roberts C., Hinnebusch A. G., and Marton M. J. (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21, 4347–4368 10.1128/MCB.21.13.4347-4368.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bernard A., Jin M., Xu Z., and Klionsky D. J. (2015) A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy 11, 2114–2122 10.1080/15548627.2015.1099796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yao Z., Delorme-Axford E., Backues S. K., and Klionsky D. J. (2015) Atg41/Icy2 regulates autophagosome formation. Autophagy 11, 2288–2299 10.1080/15548627.2015.1107692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wek S. A., Zhu S., and Wek R. C. (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol. 15, 4497–4506 10.1128/MCB.15.8.4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tallóczy Z., Jiang W., Virgin H. W. 4th, Leib D. A., Scheuner D., Kaufman R. J., Eskelinen E.-L., and Levine B. (2002) Regulation of starvation- and virus-induced autophagy by the eIF2α kinase signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 190–195 10.1073/pnas.012485299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hinnebusch A. G., and Fink G. R. (1983) Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 80, 5374–5378 10.1073/pnas.80.17.5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yuan W., Guo S., Gao J., Zhong M., Yan G., Wu W., Chao Y., and Jiang Y. (2017) General Control Nonderepressible 2 (GCN2) Kinase inhibits target of rapamycin complex 1 in response to amino acid starvation in Saccharomyces cerevisiae. J. Biol. Chem. 292, 2660–2669 10.1074/jbc.M116.772194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Magasanik B., and Kaiser C. A. (2002) Nitrogen regulation in Saccharomyces cerevisiae. Gene 290, 1–18 10.1016/S0378-1119(02)00558-9 [DOI] [PubMed] [Google Scholar]
- 76. Stanbrough M., Rowen D. W., and Magasanik B. (1995) Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. U.S.A. 92, 9450–9454 10.1073/pnas.92.21.9450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Blinder D., and Magasanik B. (1995) Recognition of nitrogen-responsive upstream activation sequences of Saccharomyces cerevisiae by the product of the GLN3 gene. J. Bacteriol. 177, 4190–4193 10.1128/jb.177.14.4190-4193.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chan T. F., Bertram P. G., Ai W., and Zheng X. F. (2001) Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J. Biol. Chem. 276, 6463–6467 10.1074/jbc.M008162200 [DOI] [PubMed] [Google Scholar]
- 79. Coffman J. A., Rai R., Cunningham T., Svetlov V., and Cooper T. G. (1996) Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 847–858 10.1128/MCB.16.3.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fernandes L., Rodrigues-Pousada C., and Struhl K. (1997) Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol. Cell. Biol. 17, 6982–6993 10.1128/MCB.17.12.6982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ramya V., and Rajasekharan R. (2016) ATG15 encodes a phospholipase and is transcriptionally regulated by YAP1 in Saccharomyces cerevisiae. FEBS Lett. 590, 3155–3167 10.1002/1873-3468.12369 [DOI] [PubMed] [Google Scholar]
- 82. Epple U. D., Suriapranata I., Eskelinen E.-L., and Thumm M. (2001) Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 183, 5942–5955 10.1128/JB.183.20.5942-5955.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Teter S. A., Eggerton K. P., Scott S. V., Kim J., Fischer A. M., and Klionsky D. J. (2001) Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 276, 2083–2087 10.1074/jbc.C000739200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Conlan R. S., and Tzamarias D. (2001) Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. J. Mol. Biol. 309, 1007–1015 10.1006/jmbi.2001.4742 [DOI] [PubMed] [Google Scholar]
- 85. Galeote V. A., Alexandre H., Bach B., Delobel P., Dequin S., and Blondin B. (2007) Sfl1p acts as an activator of the HSP30 gene in Saccharomyces cerevisiae. Curr. Genet. 52, 55–63 10.1007/s00294-007-0136-z [DOI] [PubMed] [Google Scholar]
- 86. Delorme-Axford E., Donker R. B., Mouillet J. F., Chu T., Bayer A., Ouyang Y., Wang T., Stolz D. B., Sarkar S. N., Morelli A. E., Sadovsky Y., and Coyne C. B. (2013) Human placental trophoblasts confer viral resistance to recipient cells. Proc. Natl. Acad. Sci. U.S.A. 110, 12048–12053 10.1073/pnas.1304718110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhu H., Wu H., Liu X., Li B., Chen Y., Ren X., Liu C. G., and Yang J. M. (2009) Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy 5, 816–823 10.4161/auto.9064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Korkmaz G., le Sage C., Tekirdag K. A., Agami R., and Gozuacik D. (2012) miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy 8, 165–176 10.4161/auto.8.2.18351 [DOI] [PubMed] [Google Scholar]
- 89. Frankel L. B., Lubas M., and Lund A. H. (2017) Emerging connections between RNA and autophagy. Autophagy 13, 3–23 10.1080/15548627.2016.1222992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., and Tuschl T. (2004) Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197 10.1016/j.molcel.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 91. Liu J., Carmell M. A., Rivas F. V., Marsden C. G., Thomson J. M., Song J. J., Hammond S. M., Joshua-Tor L., and Hannon G. J. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- 92. Drinnenberg I. A., Weinberg D. E., Xie K. T., Mower J. P., Wolfe K. H., Fink G. R., and Bartel D. P. (2009) RNAi in budding yeast. Science 326, 544–550 10.1126/science.1176945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hu G., McQuiston T., Bernard A., Park Y. D., Qiu J., Vural A., Zhang N., Waterman S. R., Blewett N. H., Myers T. G., Maraia R. J., Kehrl J. H., Uzel G., Klionsky D. J., and Williamson P. R. (2015) A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 17, 930–942 10.1038/ncb3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Presnyak V., and Coller J. (2013) The DHH1/RCKp54 family of helicases: an ancient family of proteins that promote translational silencing. Biochim. Biophys. Acta 1829, 817–823 10.1016/j.bbagrm.2013.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Decker C. J., Teixeira D., and Parker R. (2007) Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179, 437–449 10.1083/jcb.200704147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hu G., McQuiston T., Bernard A., Park Y. D., Qiu J., Vural A., Zhang N., Waterman S. R., Blewett N. H., Myers T. G., Kehrl J. H., Uzel G., Klionsky D. J., and Williamson P. R. (2016) Tor-dependent post-transcriptional regulation of autophagy: implications for cancer therapeutics. Mol. Cell. Oncol. 3, e1078923 10.1080/23723556.2015.1078923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Delorme-Axford E., Abernathy E., Lennemann N. J., Bernard A., Ariosa A., Coyne C. B., Kirkegaard K., and Klionsky D. J. (2018) The exoribonuclease Xrn1 is a post-transcriptional negative regulator of autophagy. Autophagy 14, 10.1080/15548627.2018.1441648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., and Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- 99. Jackson W. T., Giddings T. H. Jr., Taylor M. P., Mulinyawe S., Rabinovitch M., Kopito R. R., and Kirkegaard K. (2005) Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3, e156 10.1371/journal.pbio.0030156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wong J., Zhang J., Si X., Gao G., Mao I., McManus B. M., and Luo H. (2008) Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82, 9143–9153 10.1128/JVI.00641-08 [DOI] [PMC free article] [PubMed] [Google Scholar]