Abstract
The Hippo pathway controls cell proliferation and differentiation through the precisely tuned activity of a core kinase cassette. The activity of Hippo kinase is modulated by interactions between its C-terminal coiled-coil, termed the SARAH domain, and the SARAH domains of either dRassF or Salvador. Here, we wanted to understand the molecular basis of SARAH domain–mediated interactions and their influence on Hippo kinase activity. We focused on Salvador, a positive effector of Hippo activity and the least well-characterized SARAH domain–containing protein. We determined the crystal structure of a complex between Salvador and Hippo SARAH domains from Drosophila. This structure provided insight into the organization of the Salvador SARAH domain including a folded N-terminal extension that expands the binding interface with Hippo SARAH domain. We also found that this extension improves the solubility of the Salvador SARAH domain, enhances binding to Hippo, and is unique to Salvador. We therefore suggest expanding the definition of the Salvador SARAH domain to include this extended region. The heterodimeric assembly observed in the crystal was confirmed by cross-linked MS and provided a structural basis for the mutually exclusive interactions of Hippo with either dRassF or Salvador. Of note, Salvador influenced the kinase activity of Mst2, the mammalian Hippo homolog. In co-transfected HEK293T cells, human Salvador increased the levels of Mst2 autophosphorylation and Mst2-mediated phosphorylation of select substrates, whereas Salvador SARAH domain inhibited Mst2 autophosphorylation in vitro. These results suggest Salvador enhances the effects of Hippo kinase activity at multiple points in the Hippo pathway.
Keywords: cell signaling, Hippo pathway, Salvador (sav), structure-function, crystal structure, protein phosphorylation, enzyme
Introduction
Genetic screens for overgrowth mutants in Drosophila melanogaster identified a set of genes that function together in a pathway that controls tissue growth during development and regeneration (1–6). The Hippo pathway controls the balance between cell proliferation and apoptosis by sequestering a transcriptional co-activator, Yorkie, in the cytoplasm (1, 3–8). Both the molecular function and organization of the Hippo pathway are largely conserved between flies and humans. Each component of the core kinase cassette in flies has a human homolog. Disruption of the pathway results in tumorigenesis in mouse models and is associated with numerous cancers in humans (8–11). Phenotypes arising from knockouts of two core components in flies can be rescued by complementation with the human gene encoding the homologous protein (5, 7, 10). Functionally what is learned about one protein can be applied to its homolog. The naming convention for Hippo pathway components is not conserved between flies and humans. Throughout this paper we will use the fly names when referring to general aspects of the pathway and the species appropriate name when highlighting specific results.
The outcomes of the Hippo pathway are a consequence of the activity of the core kinase cassette that include two kinases, Hippo and Warts, and two accessory proteins, Mats and Salvador. The coordinated activity of these proteins results in the phosphorylation of Yorkie by Warts resulting in the cytoplasmic sequestration of Yorkie (7, 8). To initiate these downstream events, Hippo kinase must be activated by phosphorylation of its activation loop, a consequence of homodimerization-induced autophosphorylation (12–14). An upstream kinase may also bypass the need for autophosphorylation by phosphorylating the activation loop of Hippo directly (1–6, 15, 16). Once activated, Hippo kinase phosphorylates other members of the pathway including Mats, Salvador, and Warts (1, 3–8). Hippo phosphorylates Warts kinase on its hydrophobic motif, which triggers the autophosphorylation and subsequent activation of Warts (5, 8–11, 17–19). The outcomes of the Hippo pathway are linked to the fidelity and activity of these kinases.
The activity of Hippo kinase is tuned by interactions with two other proteins, dRassF and Salvador. Hippo binds each protein through reciprocal interactions of the C-terminal SARAH domains, so named after each family member (Salvador-RassF-Hippo) and characterized by heptad repeats (5, 7, 10, 20). SARAH domains mediate complex formation between Hippo and either dRassF or Salvador as well as homodimerization of either Hippo and dRassF (5–8, 21–25). Each of the SARAH domain–mediated interactions with Hippo has distinct consequences for kinase activity. Autophosphorylation of Mst1/2, the human homolog of Hippo, is faster in proteins with the SARAH domain than without, presumably because the SARAH domain induced homodimerization (12–14, 26). Interactions of Hippo with either dRassF or Salvador are mutually exclusive and correlate with the phosphorylation state of Hippo, where phosphorylated Hippo binds Salvador and unphosphorylated Hippo binds dRassF (27). In insect cells, co-transfection of Salvador, Hippo, and Warts leads to higher levels of Warts phosphorylation than those lacking Salvador, suggesting that Salvador enhances Hippo kinase activity through an unidentified mechanism (5, 23). dRassF blocks autophosphorylation of Hippo resulting in reduced pathway activity (14, 26), while human homologs of dRassF (RassFs) stimulate apoptosis in cells suggesting an increase in Mst1/2 activity (14, 24, 27). The mechanism behind the differences in the activity of human versus Drosophila RassFs is not yet clear, but may involve human RassF homologs promoting phosphorylation of different substrates by Mst1/2 (28).
Understanding the physical basis of SARAH domain–mediated interactions may reveal the molecular basis of how Salvador stimulates the activity of Hippo kinase. Although SARAH domain structures have been determined for mammalian proteins Mst1, Mst2, and two dRassF homologs (Nore1 and RassF5) and complexes between either Mst1 or Mst2 and RassF5 (26, 29, 33, 34) (Protein Data Bank codes 4L0N, 4HKD, 4NR2, and 3WWS), there is no structural information for Salvador SARAH domain. In the available structures, the SARAH domains adopt an approximately 10 turn α-helix and are sometimes preceded by a single-turn 310 helix connected by a flexible linker. Two SARAH domains interact to form anti-parallel coiled-coils. Unlike most coiled-coils, the dimer interface is stabilized by hydrophobic interactions. SARAH domains were, initially, proposed to form trimers but subsequent studies failed to identify such a complex (20, 27, 29). The preponderance of evidence, including solution NMR studies, supports dimerization of SARAH domains (29). However, NMR studies also provide evidence for a tetrameric assembly of Mst1 and human Salvador (hSav1) SARAH domains. Upon addition of Salvador, residues of Mst1 located outside the canonical dimer interface undergo chemical shift perturbations, suggesting Salvador binding to Mst1 does not alter Mst1 homodimerization (29). Cross-linking studies revealed evidence for both homodimers of Mst1 and tetramers of RassF5 SARAH domains, suggesting SARAH domain-mediated complexes may not share a common stoichiometry (29). The lack of structural or biochemical information for the Salvador SARAH domain precludes a mechanistic understanding of how Salvador can influence Hippo kinase activity, perhaps because of difficulty in purifying recombinant SARAH domain protein from human Salvador (35).
To understand the molecular basis of how Salvador modulates Hippo function, we undertook structural and functional studies of the Salvador SARAH domain. First we identified a region N-terminal to the SARAH domain of D. melanogaster Salvador that increases the stability of recombinant SARAH domain protein and suggest the domain boundaries of Salvador SARAH be changed to include these residues. We report the crystal structure of a complex between the SARAH domains of Salvador and Hippo, hereafter referred to as Salvador–Hippo SARAH. This structure reveals a unique binding interaction between Salvador and Hippo SARAH domains mediated in part by the N-terminal extension of Salvador SARAH that also increases binding of Salvador to Hippo SARAH domain in solution over that of the canonical coiled-coil region alone. The heterodimeric assembly of the Salvador–Hippo SARAH in the crystal structure is validated by cross-linking and MS. This structure provides insight into the atomic level organization of any Salvador SARAH domain. We also investigated the effects of Salvador on Hippo kinase activity. In cells, co-transfection of hSav1 enhanced autophosphorylation of Mst2 and phosphorylation of select substrates, whereas in vitro Salvador SARAH domain inhibited autophosphorylation. Our work redefines the domain boundaries for the Salvador SARAH domain, identifies an unanticipated and extended binding surface between Salvador and Hippo SARAH domains, and reveals multiple points at which Salvador stimulates Hippo pathway activity.
Results
N-terminal extension of Salvador SARAH domain
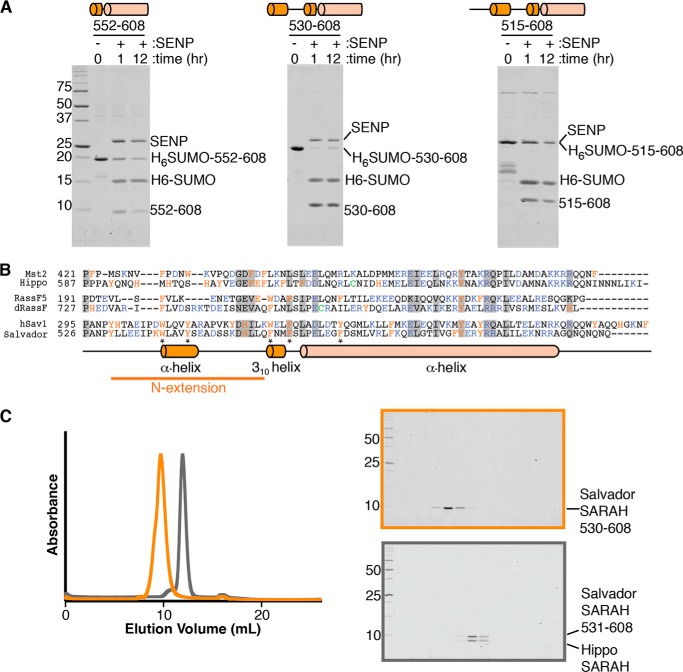
To investigate the role of the Salvador SARAH domain, we first determined a variant of the Salvador SARAH domain that was stable in solution. We started by purifying a D. melanogaster Salvador SARAH domain variant (residues 552–608) equivalent to the hSav1 variant used for NMR studies (29). We expressed the protein as a fusion to a His6 tag for purification and a SUMO domain for solubility in Escherichia coli and purified the protein by immobilized metal affinity chromatography (IMAC).2 The tags were removed by proteolysis with SENP but the free SARAH domain was not stable in solution overnight (Fig. 1A). We then further analyzed the sequence of Salvador for regions of conservation or with predicted secondary structure. We identified two regions of interest N-terminal to the coiled-coil. One region, roughly 20 residues preceding the coiled-coil, was predicted to form a short α-helix. The other comprised a series of conserved residues about 35 residues N-terminal to the SARAH domain (Fig. 1B). We expressed SARAH domain variants with both N-terminal extensions, residues 530–608 or 515–608, and tested their solubility following proteolytic removal of the tags (Fig. 1A). The SARAH domain variant that included the α-helix (residues 530–608) was stable overnight in solution, whereas the longest variant (residues 515–608) was not stable. The SARAH domain variant corresponding to residues 530–608 could be further purified to homogeneity and was monodisperse on gel-filtration chromatography (Fig. 1C).
Figure 1.
Additional residues are needed for the solubility of Salvador SARAH domain. A, three His6-SUMO tagged Salvador SARAH domain variants, corresponding to residues 552–608, 530–608, and 515–608, were purified by IMAC. Eluted proteins were incubated with SENP at 4 °C for 1 or 12 h and then separated by Coommassie-stained SDS-PAGE. SENP and tagged His6-SUMO-Salvador SARAH (residues 515–608) migrate at the same position on the gel. The experiment was performed three times and three panels cropped from the same representative gel are shown. A schematic corresponding to the variant used is shown above each panel. B, sequence alignments of the SARAH domains and N-terminal regions of human Mst2, RassF5, and Salvador (hSav1) as well as D. melanogaster Hippo, dRassF, and Salvador. Residues conserved among all six sequences are marked with a gray box. Aromatic residues are colored orange, charged residues purple, and cysteines green. Aligned below is a diagram of the secondary structure assignments derived from the crystal structure for Salvador SARAH domain. Residues that mediate sequence-specific interactions between Hippo and the N-terminal extension of Salvador are indicated by an asterisk. C, left panel are overlaid gel filtration chromatograms of either the Salvador SARAH domain (residues 530–608) (orange) or Salvador–Hippo SARAH (residues 531–608 and 606–662, respectively) (gray). Coomassie-stained SDS-PAGE for Salvador SARAH is highlighted in orange (top right) and Salvador–Hippo SARAH in gray (bottom right). Both gels contain fractions spanning from the void volume to the end of the column, and lanes with protein correspond to the peaks on the chromatograms.
Structure determination
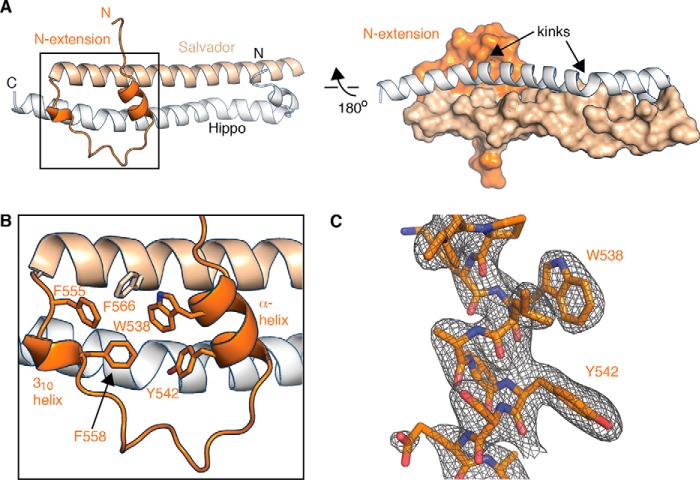
To isolate a homogenous and stoichiometric complex between Hippo and Salvador SARAH domains, we co-expressed Salvador SARAH domain (residues 531–608) and a Hippo SARAH domain with each SARAH domain bearing a different affinity tag. The complex was purified by tandem affinity purification followed by standard chromatographies. This approach resulted in a monodisperse and stoichiometric complex, as judged by gel-filtration chromatography and Coomassie-stained SDS-PAGE (Fig. 1C). Crystals with primitive tetragonal symmetry were grown by vapor diffusion against a mother liquor containing a mixture of polyethylene and hexylene glycols. These crystals diffracted X-rays to 2.6 Å and contained one copy of Salvador–Hippo SARAH in the asymmetric unit. Initial phases were determined by molecular replacement using an ensemble of SARAH domain structures as a search model. All but the last three C-terminal residues of Hippo SARAH and eight C-terminal residues of Salvador SARAH were sufficiently ordered in electron-density maps to be built and refined. The current model has an Rwork/Rfree of 20.2/24.6%, and all residues have favored or allowed Ramachandran geometry (Table 1) (Fig. 2).
Table 1.
Data collection and refinement statistics
| PDB 6BN1 | |
|---|---|
| Data collection | |
| Space group | P4322 |
| Unit cell | |
| a, b, c (Å) | 49.934 49.934 186.169 |
| α, β, γ (°) | 90.000 90.000 90.000 |
| Number of unique reflections | 7,921 |
| Number of observed reflections | 193,623 |
| Resolution (Å)a | 38.90–2.60 (2.72–2.60) |
| Rpim (%)a,b | 0.049 (0.719) |
| I/σa | 10.6 (1.4) |
| Completeness (%)a | 100.0 (100.0) |
| Redundancya | 24.4 (26.1) |
| Refinement | |
| Rcryst (%)c | 21.20 |
| Rfree (%)c | 24.59 |
| Ramachandran analysis | |
| Favored (%) | 99.17 |
| Allowed (%) | 0.83 |
| R.m.s. deviation bonds | 0.013 |
| R.m.s. deviation angles | 1.247 |
a The values in parentheses are for the highest resolution shell.
b Rpim = Σhkl√((1/(n-1)) Σi |Ii − 〈I〉|/ΣhklΣi Ii, where Ii is the intensity of an individual reflection and 〈I〉 is the mean intensity obtained from multiple observations of symmetry related reflections.
c Rcryst is Σ‖Fo − Fc‖/Fo, where Fo is an observed amplitude and Fc a calculated amplitude; Rfree is the same statistic calculated over a subset of the data that has not been used for refinement.
Figure 2.
Structure of Salvador–Hippo SARAH. A, two views of the Hippo SARAH domain (white schematic) bound to the Salvador SARAH domain (the N-terminal extension in darker orange and the coiled-coil in light orange) in either cartoon or surface. B, the aromatic residues of Salvador that mediate the arrangement of the N-terminal extension are shown as sticks. C, electron density from a 2Fo − Fc composite omit map contoured at +1.2σ (gray mesh) and the final refined model for the α-helix of the N-terminal extension is shown in a cartoon (dark orange).
Salvador–Hippo SARAH structure
The structure of Salvador–Hippo SARAH revealed anti-parallel, heterodimeric coiled-coils comprised of residues 606–659 of the Hippo SARAH domain and residues 531–600 of Salvador SARAH domain (Fig. 2). The interface between the coiled-coils is dominated by hydrophobic interactions and has a hydrogen bonding pattern that is both similar to other SARAH domain complexes and as predicted by the heptad repeats. α-Helices can adopt linear, kinked, or curved conformations based on the overall trajectory of the α-helix (36). Measurement of the two α-helices that form the coiled-coils revealed the Salvador SARAH domain is linear and Hippo SARAH domain is kinked (Fig. 2A). The α-helix of the Hippo SARAH domain has two kinks that correspond to two stutters in the heptad repeat (Fig. 2A). This pairing of one kinked and one unkinked α-helix is characteristic of the five other heterodimeric SARAH domain structures (Table 2). Salvador–Hippo SARAH superposes with an average root mean square deviation (r.m.s. deviation) of 1.3 Å over 39 Cα atoms to the three heterodimer structures that had full-length α-helices but superposes with an r.m.s. deviation of 1.8 Å over the same range to 15 homodimer structures (Table 2).
Table 2.
Structural analysis of SARAH domain dimers
| General |
Superpositions |
Helical analysis |
Buried surface area (BSA) (Å2) | |||||
|---|---|---|---|---|---|---|---|---|
| SARAH domain dimers | PDB code | Chain IDs of dimers | R.m.s. deviation (Å) | Residue range chain 1 | Residue range chain 2 | Chain 1 | Chain 2 | |
| Hippo/Salvadora | 6BN1 | A, B | 0 | 615–654 | 581–620 | Kinked | NK-Lb | 3358 |
| Mst2/RassF5 (26) | 4LGD | A, E | 1.9 | 445–484 | 374–412 | Kinked | NK-L | 2755 |
| Mst2/RassF5 (26) | 4LGD | B, F | NAc | NA | NA | Kinked | NK-L | 1483 |
| Mst2/RassF5 (26) | 4LGD | C, G | 0.9 | 445–484 | 374–412 | Kinked | NK-Cd | 2863 |
| Mst2/RassF5 (26) | 4LGD | D, H | NA | NA | NA | Kinked | NK-L | 2106 |
| Mst1/RassF5 (33) | 4OH8 | A, B | 1.1 | 12–51 | 9–47 | Kinked | NK-L | 2402 |
| Mst2 | 3WWS | A, B | 1.8 | 445–484 | 445–484 | Kinked | Kinked | 2912 |
| Mst2 | 3WWS | C, D | 1.8 | 445–484 | 445–484 | Kinked | Kinked | 2815 |
| Mst2 (33) | 4OH9 | A, B | 1.3 | 12–51 | 12–51 | Kinked | NK-L | 3069 |
| Mst1 | 4NR2 | A, B | 1.8 | 441–480 | 441–480 | Kinked | Kinked | 2759 |
| Mst1 | 4NR2 | C, D | 1.8 | 441–480 | 441–480 | Kinked | Kinked | 2830 |
| Mst1 | 4NR2 | E, F | 1.6 | 441–480 | 441–480 | Kinked | Kinked | 2749 |
| Mst1 | 4NR2 | G, H | 1.9 | 441–480 | 441–480 | Kinked | Kinked | 2846 |
| Mst2 | 4HKD | A, B | 1.3 | 445–484 | 445–484 | Kinked | Kinked | 2901 |
| Mst2 | 4HKD | C, D | 1.3 | 445–484 | 445–484 | Kinked | Kinked | 2992 |
| Mst2 | 4L0N | A, B | 2.1 | 445–484 | 445–484 | Kinked | Kinked | 2861 |
| Mst2 | 4L0N | C, D | 1.9 | 445–484 | 445–484 | NK-L | Kinked | 2934 |
| Mst2 | 4L0N | E, F | 2.3 | 445–484 | 445–484 | NK-C | Kinked | 2799 |
| Mst2 | 4L0N | G, H | 2.0 | 445–484 | 445–484 | NK-C | Kinked | 2850 |
| Mst2 | 4L0N | I, I*e | 2.5 | 445–484 | 445–484 | Kinked | Kinked | 1911 |
| Mst2 | 4L0N | J, J*e | 2.0 | 445–484 | 445–484 | NK-L | NK-L | 1894 |
| Mst1 (29) | 2JO8 | A, B | 2.4 | 369–408 | 369–408 | Kinked | Kinked | 2795 |
| Nore1 (34) | 2YMY | A, B | 2.2 | 369–408 | 369–408 | NK-L | NK-L | 1768 |
a Current work.
b Not kinked, curved (NK-C).
c α-Helices were of insufficient length to compare.
d Not kinked, linear (NK-L).
e *, represents a second chain generated by crystal symmetry.
Additionally the structure revealed the additional N-terminal residues of Salvador (residues 531–559) adopted a fold separate from the main α-helix (Fig. 2, A and B). This extension is a continuation of the 310 helix and is preceded by a poorly ordered loop, as evidenced by high B-factors, a two-turn α-helix, and another loop. A series of hydrophobic interactions between aromatic residues in both the extension (Trp-538, Tyr-542, Phe-555, Phe-558) and the coiled-coil (Phe-566) of Salvador SARAH domain stabilize the arrangement of this extension (Fig. 2B). The N-terminal extension of the Salvador SARAH domain forms part of the binding interface with the Hippo SARAH domain (Fig. 2A). Such an extended binding interface has not been observed in other SARAH domain structures. The Hippo SARAH domain interacts with this extension using residues with short, hydrophobic side chains (Lys-644, Pro-647, Ile-648, Ala-651). In addition, the interaction is stabilized by a salt bridge between Lys-640 of Hippo and Glu-544 in the α-helix in the N-terminal extension of Salvador and by hydrogen bonds between the ϵ-amino group of Lys-644 of Hippo and the backbone carbonyls of Asp-546, Ala-545, and Tyr-542 of the extension (Fig. 2B). The complex between Hippo and Salvador SARAH domains buries a total of 3358 Å2 surface area, 506 Å2 of which arises from the interface mediated by the N-terminal extension.
The N-terminal extension of Salvador SARAH domain mediates binding to Hippo
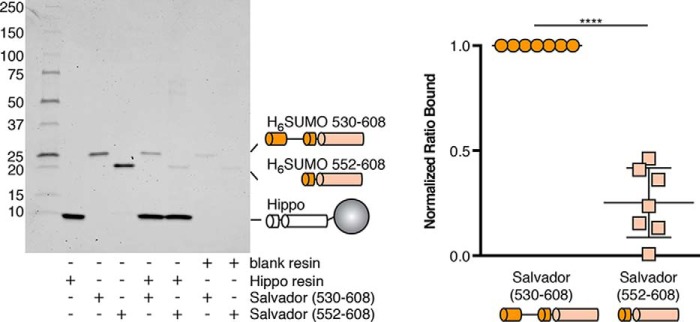
To validate the role of the N-terminal extension in binding Hippo, we performed pulldown experiments comparing the ability of immobilized Hippo SARAH domain to bind Salvador SARAH variants either with or without the N-terminal extension (residues 530–608 or 552–608, respectively). Although both Salvador variants bound the Hippo SARAH domain, more binding was observed for the variant containing the N-terminal extension (Fig. 3).
Figure 3.
The N-terminal extension of Salvador SARAH enhances binding to Hippo. A pulldown experiment was performed to analyze the amount of binding of His6-SUMO-tagged Salvador SARAH domain variants with (residues 530–608) or without (residues 552–608) the N-terminal extension to either blank resin or resin coupled to the Hippo SARAH domain and analyzed by Coomassie-stained SDS-PAGE. The binding experiment was performed seven times and a representative gel is shown (left). The ratio of bound Salvador to Hippo SARAH domains was calculated from band intensities and normalized between replicates (right). The p value between Salvador SARAH domain variants either with (530–608) or without the N-terminal extension (552–608) is ≤0.0001.
Hippo and Salvador form a heterodimer
To determine the stoichiometry of Salvador–Hippo SARAH, we performed native MS to measure the mass of the complex in solution. The buffer conditions required for the assay, however, did not stabilize the complex sufficiently to maintain its structure during transition to the gas phase. To increase the stability of the complex prior to ionization, the sample was cross-linked with a homobifunctional cross-linker. The cross-linked complex was then analyzed by SDS-PAGE and MALDI-TOF MS. Following cross-linking, the predominate species on the gel was a dimer and the molecular weight of the cross-link complex as measured by MS corresponded to a heterodimer between Salvador and Hippo SARAH domains (Fig. 4, A and B).
Figure 4.
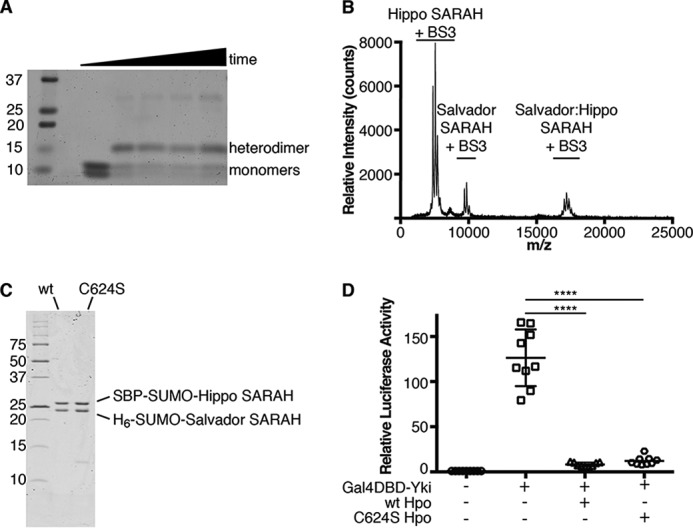
Salvador–Hippo SARAH is a heterodimer. A, SDS-PAGE gel of cross-linking reactions containing Salvador and Hippo SARAH domains and BS3 at 0, 30, 60, 120, or 180 min. Initially the two SARAH domains run as monomers, and a second band appears at 13 kDa corresponding to the heterodimer during incubation with BS3 and remains present throughout the time course. B, MALDI-MS spectrum of the cross-linking reaction at the 30-min time point. This spectrum shows that the 17-kDa species matches a Salvador–Hippo SARAH heterodimer rather than either possible homodimer. The samples taken for analysis using SDS-PAGE and MALDI-MS came from the same reaction. The difference in apparent abundance between the gel image and the mass spectrum is caused by the difference in ionizability between the different protein species. C, SDS-PAGE following IMAC purification of tagged Salvador–Hippo SARAH complexes containing either WT (wt) or C624S Hippo SARAH domains (C624S). D, S2 cells were transfected with plasmids encoding Renilla, a Gal4 responsive luciferase reporter (white circle), Yorkie fused to the Gal4 DNA-binding domain (Gal4DBD-Yki) (white square), and either wt (white triangle) or C624S Hippo (white hexagon). After 3 days, the levels of luciferase were measured. Each measurement was normalized to the signal from Renilla luciferase. The data were plotted as the relative firefly luciferase signal compared with that from Gal4DBD-Yki and are from nine measurements from four independent experiments, the mean is indicated by a horizontal bar with error bars corresponding to the S.D. among measurements. p value ≤ 0.0001, compared with control. The p value between wt and C624S Hippo is 0.8906.
In the crystal, the asymmetric unit contains one heterodimer with the same molecular weight as the heterodimer detected by MS. A dimer of dimers, however, is generated by crystal symmetry and stabilized by a disulfide bond between a nonconserved cysteine, Cys-624, of the Hippo SARAH domain and the same residue of a symmetry mate. This interaction is on the solvent-exposed surface of the coiled-coils and buries only 620 Å2 of surface area, compared with the 3358 Å2 of surface area buried in the heterodimer. To determine whether the disulfide bond arises from simple crystal packing and not a biologically relevant interaction, we substituted a serine for the cysteine in the Hippo SARAH domain and tested the ability of this variant to bind Salvador SARAH domain. Hippo C624S SARAH domain can co-purify as a complex with Salvador SARAH domain (Fig. 4C). To assay if Cys-624 is required for Hippo activity, we monitored the pathway function in insect cells using a reporter assay that uses the levels of luciferase expression as a proxy for Yorkie localization. There was no discernable difference in the ability of either WT or Hippo C624S to suppress transcription of the luciferase reporter gene (Fig. 4D). Cys-624 of Hippo is not required for either binding to the Salvador SARAH domain or proper pathway activity.
The effects of Salvador on Mst2 activity
We next wanted to investigate the consequences of complex formation with Salvador on Hippo kinase activity. We started our investigation with in vitro assays monitoring autophosphorylation of the purified components by Western blotting using an antibody that recognizes Mst2 phosphorylated on the activation loop (pThr-180). Full-length unphosphorylated Mst2 was incubated with ATP and Mg2+ in the presence or absence of an excess of Salvador SARAH domain (100 μm). This heterologous system comprised of human and fly proteins should replicate native interactions and activity, as Mst2 rescues the phenotype of a hpo knockout (5) and the sequence identity between human and fly SARAH domains is 41% for Salvador and 52% for Hippo. Reports of the disassociation constants for SARAH domain–mediated interactions range from nanomolar to tens of micromolar (28, 29, 34, 37, 38), so the conditions of the assay should favor complex formation. Addition of Salvador SARAH domain lowered the amount of Mst2 autophosphorylation compared with Mst2 alone (Fig. 5A). We then wanted to know if these results would be recapitulated using full-length Salvador in cells. HEK293T cells were transfected with Mst2 alone or with full-length hSav1 and the amount of Mst2 autophosphorylation monitored by Western blotting. Cells transiently expressing Mst2 and hSav1 had higher levels of Mst2 phosphorylated on the activation loop than cells transfected with Mst2 alone with no change in total Mst2 protein levels (Fig. 5B).
Figure 5.
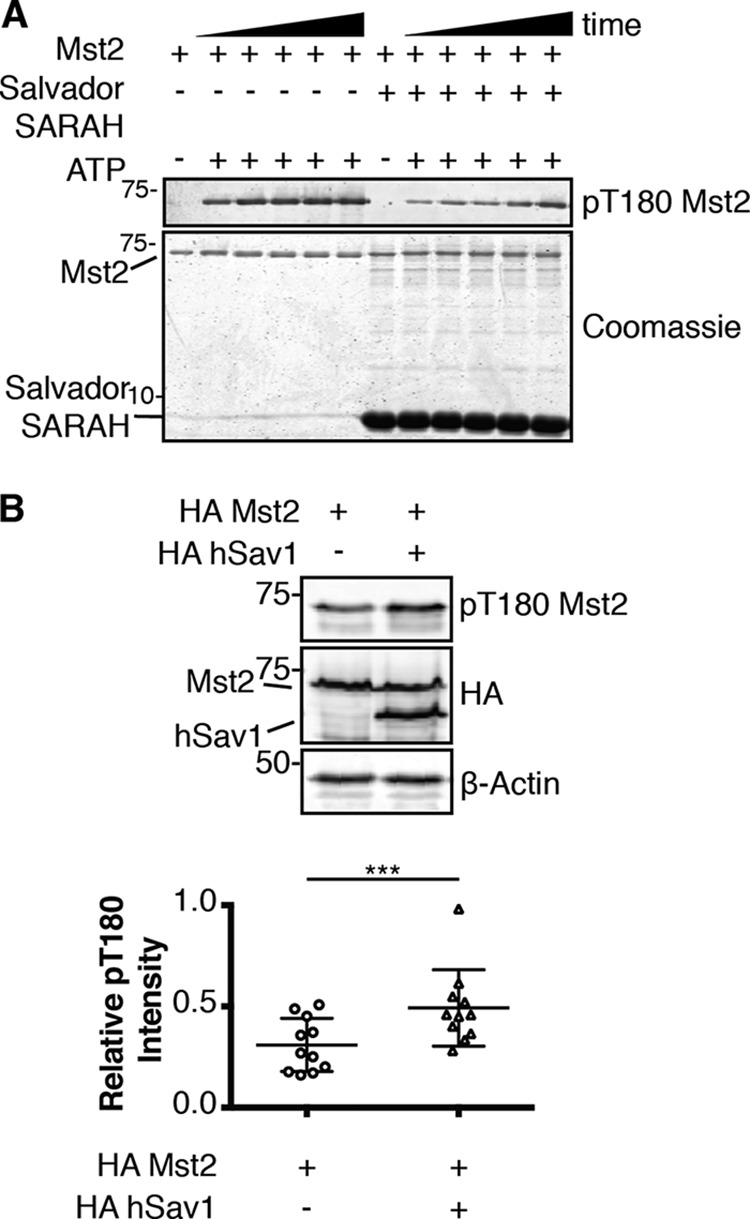
The differential effects of Salvador on autophosphorylation of Mst2 in vitro or in cells. A, purified Mst2 was incubated with ATP and Mg2+ with or without 100 μm Salvador SARAH. Samples were taken at different times (0, 5, 10, 15, 30, 60, and 120 min), quenched with EDTA, and analyzed by Coomassie-stained SDS-PAGE and the level of autophosphorylation was determined by Western blotting with an antibody that recognized phosphorylated Thr-180 (pT180 Mst2). The assay was run three times and representative blots are shown. B, HEK293T cells were transiently transfected with the indicated plasmids (HA-Mst2 and HA-hSav1). Normalized cell lysates were analyzed by Western blotting for protein expression with antibodies that recognize the epitope tag (HA), the phosphorylated activation loop of Mst2 (pT180 Mst2), or β-Actin as a loading control. Representative blots from one experiment (top) and a scatter plot of 11 replicates (bottom) are shown. Data are plotted as pThr-180 Mst2 intensity divided by total HA-Mst2 intensity (p value ≤ 0.001).
We next monitored the effect of full-length Salvador on substrate phosphorylation in cells. HEK293T cells were co-transfected with Mst2 and Mob1 (the mammalian homolog of Mats) with or without hSav1, and the amount of Mob1 phosphorylation at two known phosphorylation site sites, Thr-12 and Thr-35, detected by Western blotting using phospho-specific antibodies (Fig. 6A). Phosphorylation of Mob1 at either site was not significantly altered by co-transfection of hSav1. We next asked if this result was consistent for different substrates, so we performed an analogous experiment and monitored the levels of hydrophobic motif phosphorylation of Lats1 (the mammalian homolog of Warts) by Mst2 (Fig. 6B). In contrast to Mob1, co-transfection of hSav1 in HEK293T cells resulted in an increase in the fraction of Lats1 phosphorylated on the hydrophobic motif (Thr-1079).
Figure 6.
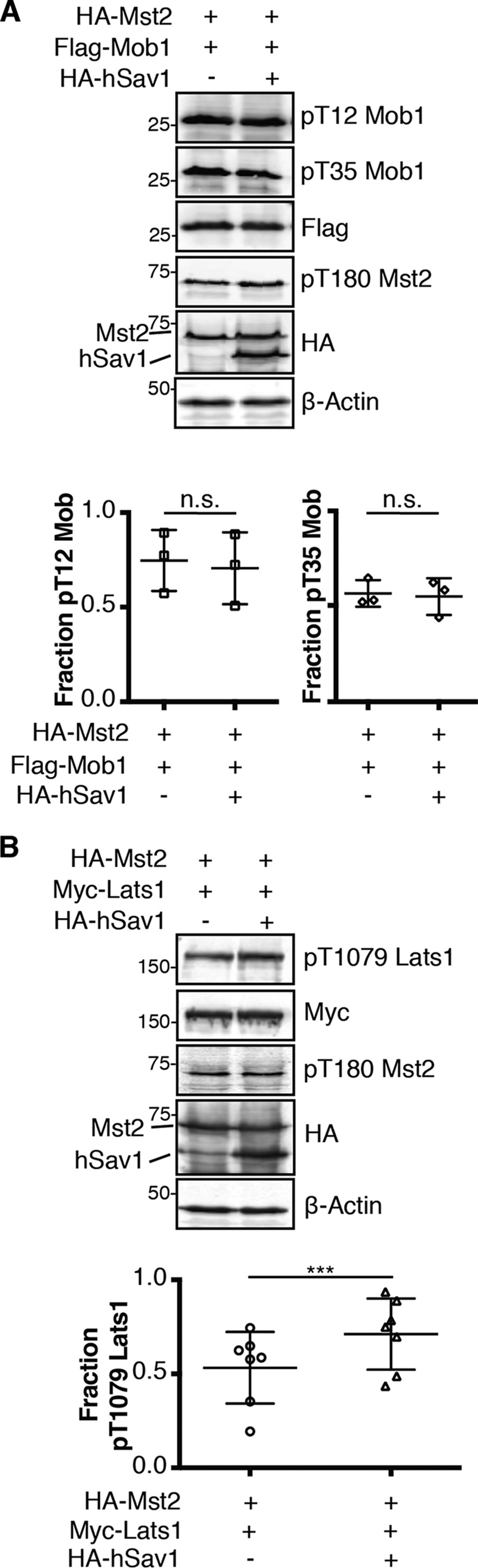
hSav1 changes the levels of phosphorylation of specific substrates by Mst2. A, plasmids encoding a HA-Mst2 or FLAG-Mob1 were transiently co-transfected with or without HA-hSav1 in HEK293T cells. Normalized cell lysates were analyzed by Western blotting for protein expression with antibodies that recognize each epitope tag (HA or Flag), the two phosphorylation sites of Mob1 (pT12 or pT35), or β-Actin as a loading control. Representative blots from one experiment (top) and a scatter plot of three replicates (bottom) are shown. Data are plotted as pThr-12 or pThr-35 Mob1 intensity divided by total FLAG-Mob1 intensity (p values for both > 0.05). B, the same experiment as in panel A but with a plasmid encoding HA-Lats1 instead of FLAG-Mob1 and antibodies specific to the hydrophobic motif phosphorylation of Lats1 (pT1079) instead of those specific to phosphorylated Mob1. Representative blots from one experiment (top) and a scatter plot of seven replicates (bottom) are shown. Data are plotted as pThr-1079 Lats1 intensity divided by total Myc-Lats1 intensity (p values ≤ 0.001).
Discussion
Our results suggest the canonical definition of the SARAH domain of Salvador be expanded to include approximately 20 residues N-terminal to the canonical coiled-coil that are characterized by the presence of a short, hydrophobic α-helix. This extension increases the solubility of the isolated SARAH domain (Fig. 1A). Partial domains of protein are less stable than complete domains (39) supporting the notion the N-terminal extension is an integral part of the Salvador SARAH domain-fold. The domain does not extend further as even longer N-terminal variants had reduced solubility (Fig. 1A). Upon realizing the significance of the N-terminal extension, we asked if this region was conserved among Salvador SARAH domains and, indeed, both the aromatic residues (Trp-538, Tyr-542, Phe-555, Phe-558) as well as those residues that comprise the short α-helix (residues 537–544) are conserved in both the fly and human sequences (Fig. 1B). In fact, sequence comparisons of 100 Salvador SARAH domain sequences reveal that Trp-538, Tyr-542, and Phe-558 are absolutely conserved, and the residue at position 555 (phenylalanine or tryptophan) could maintain the hydrophobic packing. Next, we wondered if the SARAH domains of Hippo, dRassF, or their homologs also contain similar N-terminal extensions. Sequence alignments failed to reveal a similar pattern of aromatic residues upstream of the coiled-coil, and secondary structure predications did not identify N-terminal regions likely to form an α-helix (Fig. 1B). Therefore, this extension is a unique feature of the Salvador SARAH domain, and we argue the Salvador SARAH domain be redefined to include these residues.
The N-terminal extension of the Salvador SARAH domain facilitates binding to Hippo. The X-ray structure revealed an unique interaction between Hippo and Salvador SARAH domains due to the novel interactions arising from the N-terminal extension (Fig. 2, A and B). These interactions expand the surface area buried by complex formation by nearly 18% (506 Å2) over that of just the canonical coiled-coils as well as enhance binding as monitored by solution binding studies (Fig. 3). Previous studies on Salvador SARAH domains lacked this extension, and this may explain the poor stability or weak binding of these variants to Hippo. There is precedent for residues beyond those included in the canonical domain definition contributing to complex formation. For example, the binding interface between the proline-rich region of β-dystroglycan and the WW domain of dystrophin is partially mediated by residues from the neighboring EF domain of dystrophin (40, 41).
Hippo and Salvador SARAH domains form a heterodimeric complex. In our crystal the complex with the largest buried surface area, 3358 Å2, corresponds to the heterodimer mediated by the coiled-coils. Biologically relevant interfaces have larger buried surface areas than simple crystal lattice contacts, often larger than 800 Å2, and are mediated by conserved residues (42–44). The disulfide-mediated dimer of dimers, generated by crystal symmetry, meets neither criteria as it buries only 620 Å2 of the surface area and relies on a nonconserved cysteine that is neither required for binding to Salvador or Hippo activity (Fig. 4, C and D). The interaction between Mst1 and hSav1 SARAH domains was proposed to be tetrameric because residues on the Mst1 SARAH domain that mediated complex formation with the human Salvador SARAH domain mapped to the solvent-exposed surface of the coiled-coils (29). If we map the equivalent residues onto Hippo in our Salvador–Hippo SARAH structure, those residues are either at or near the binding interface with the N-terminal extension of Salvador. The chemical shift perturbations, therefore, are in agreement with the heterodimeric assembly of Salvador–Hippo SARAH presented here.
Identification of the stoichiometry of the Salvador–Hippo SARAH domain interaction provides a structural basis for the long-standing observation that interactions between Hippo and dRassF or Salvador are mutually exclusive (25). Both complexes form heterodimers with interfaces mediated by largely the same residues of Hippo or Mst1/2 SARAH domains. These complexes are also mutually exclusive with Hippo homodimerization. The 23 previous independent observations of SARAH domain homodimers or heterodimers have buried surface areas ranging from 1483 to 3069 Å2 (Table 2). Salvador–Hippo SARAH has the largest buried surface, 3358 Å2, arising from the N-terminal extension. This suggests the affinity of Hippo for Salvador may be stronger than other SARAH domains and may explain why Salvador can outcompete dRassF for binding to Hippo (27).
With this new insight into the organization of Salvador–Hippo SARAH, we can analyze the structure of the entire complement of SARAH domains. SARAH domains have relatively simple secondary structures making superpositions of isolated α-helices or even two α-helices not very informative. Our analysis instead focused on the overall shape of the α-helices that form the coiled-coils. α-Helices can be characterized as kinked, linear, or curved (36), and individual SARAH domain coils can be grouped into either kinked or not-kinked, which encompasses both linear or curved (Table 2). Of the 36 independent observations of Hippo or Mst1/2 SARAH domain coils, 83% of the time the main α-helix is kinked. This arrangement is a consequence of two bends that occur at each of the stutters in the heptad repeat of Hippo and Mst1/2. The larger bend is caused by Pro-647 in Hippo and is absolutely conserved among 250 Hippo or Mst1/2 SARAH domain sequences (Fig. 1B). The degree of these bends varies from 18° to 82° suggesting Hippo and Mst1/2 SARAH domains can sample a large region of conformational space. In contrast, none of the eight independent observations of either a RassF or Salvador SARAH domain are kinked. These SARAH domains sample a smaller area of conformational space. So far, all heterodimeric SARAH domain structures contain a kinked Hippo or Mst1/2 SARAH domain bound to a nonkinked binding partner such as a RassF5 or Salvador SARAH domain (Table 2). The significance of this arrangement is unclear as homodimers can be formed between all possible combinations of kinked and unkinked SARAH domains. These patterns may provide the basis for the different binding patterns or varying affinities among SARAH domain complexes.
Salvador has long been thought to be positive effector of Hippo activity as transfection of Salvador in cells resulted in higher levels of Warts phosphorylation but the molecular mechanism underlying this outcome was unknown (5). We compared autophosphorylation of Mst2 in the presence of either Salvador SARAH domain in vitro or full-length hSav1 in cells and found the outcomes different. Recombinant, Salvador SARAH domain inhibited autophosphorylation of Mst2 by presumably forming a SARAH domain–mediated heterodimer thereby blocking Mst2 homodimerization and, thus, autophosphorylation (Fig. 5A). RassF5 SARAH domain also blocks Mst2 autophosphorylation via heterodimerization (26). We were, therefore, surprised to discover hSav1 had the opposite effect on autophosphorylation of Mst2 in cells (Fig. 5B). These differences could arise from changes in protein stability, localization, or factors absent from the in vitro studies. The total amount of Mst2 protein remained constant so the only change was the fraction of autophosphorylated Mst2 (Fig. 5B), suggesting other regions of Salvador may be responsible for the changes. In addition to a SARAH domain, both hSav1 and Salvador contain two WW domains, one mediates binding to proline-rich motifs and the other mediates homodimerization (45), and a large region containing polyglutamine tracts with little predicted secondary structure and no assigned function. Because none of these domains are obvious candidates to stimulate Mst2 autophosphorylation we conclude the stimulation of autophosphorylation comes from another cellular component. In both flies and in human cells, STRIPAK, a phosphatase containing complex, associates with SARAH domain–containing components of the Hippo pathway (46, 47). Knockdown of dRassF reduces association between Hippo and dSTRIPAK, whereas knockdown of Salvador promotes association, a pattern that is consistent with our cell-based results. We hypothesize Salvador competes with dRassF for binding to Hippo and thereby blocking formation of a dRassF–dSTRIPAK complex and subsequent dephosphorylation of Hippo. This model also provides a potential explanation for the phosphospecific nature of the interactions between Hippo and either dRassF or Salvador, which cannot be rationalized simply by stoichiometry. The specificity is not a consequence of either dRassF or Salvador altering the activity of Hippo or selectively binding to a phosphorylation induced conformation of Hippo, but rather a result of regulating the activity of a phosphatase. We next compared if full-length hSav1 affected substrate phosphorylation by Mst2 and found a substrate-specific effect. Although the fraction of phosphorylated Mob was unaffected by transient transfection of hSav1, the fraction of Lats1 phosphorylated on the hydrophobic motif was increased (Fig. 6). Because the stimulatory effects of hSav1 were not universal, they most likely stem from specific interactions between Salvador and the substrate. The first WW domain of Salvador binds the proline-rich motifs of Warts (3). We hypothesize that hSav1 may act as a scaffold simultaneously binding both Mst2 and Lats1 that would increase the effective concentration of the substrate resulting in higher levels of Lats1 phosphorylation.
We set out to understand the nature of the interaction between the Hippo and Salvador SARAH domains and the significance of this complex formation on Hippo activity. We identified an additional region of Salvador that behaves as part of the Salvador SARAH domain and both mediates and enhances binding to the Hippo SARAH domain. The domain boundaries of Salvador SARAH should be expanded to include this region and future studies of Salvador SARAH domains reflect this change. In cells hSav1 enhances the effects of Hippo activity at both the level of autophosphorylation and phosphorylation of select substrates but the molecular basis for these changes most likely differs. These results demonstrate Salvador can enhance Hippo signaling at multiple nodes and further work is needed to fully identify the molecular basis behind these changes.
During preparation of this manuscript, a similar structure was reported for a complex of the hSav1 SARAH domain bound to an Mst2 variant (48). That structure reveals a similar N-terminal extension of hSav1 and a heterodimeric assembly of the SARAH domains. The authors do not comment on the contribution of the N-terminal extension of Salvador SARAH domain to complex formation or on the stability of the Salvador SARAH domain. Their functional studies also highlight a positive role for Salvador in Mst2 autophosphorylation and show this effect arises from the inhibition of STRIPAK by Salvador.
Experimental procedures
Expression and purification of SARAH domains
Genes encoding D. melanogaster Sav (residues 552–608, 530–608, or 515–608) were cloned into a pBAD4 vector derivative downstream from His6 and SUMO tags (49). Proteins were expressed in T7 Express cells (New England BioLabs) grown at 37 °C, induced with 0.5 mm IPTG at 0.5 A600, and grown at 37 °C. Cells were lysed in 40 mm Tris, pH 8, 200 mm NaCl, 5 mm ATP, 5 mm MgCl2, and supplemented with protease inhibitor mixture (Sigma). To improve the solubility of some variants, lysis buffer was further supplemented with 10% glycerol and 0.1% Nonidet P-40. Salvador SARAH domain was isolated by a IMAC using Profinity-IMAC resin (Bio-Rad) and eluted with 125 mm imidazole. For Salvador SARAH-(530–608), the affinity tag was removed by incubation with a SUMO-specific protease (SENP) at 4 °C overnight, and the protein was further purified by a combination of anion-exchange and gel-filtration chromatographies. Protein was concentrated in 10 mm Tris, pH 8, 200 mm NaCl and flash frozen in liquid nitrogen. Hippo SARAH domain (residues 606–662) was expressed and purified in the same manner as the Salvador SARAH domain except 1 mm tris(2-carboxyethyl)phosphine (TCEP) was included in all buffers after the IMAC purification step.
Expression and purification of Salvador–Hippo SARAH
Genes encoding either D. melanogaster Sav (residues 531–608) or Hpo (residues 606–662) were cloned into pRSF-Duet Vector (EMD-Millipore) as SUMO-tagged fusion with either a SBP or His6 tag, respectively. The variant of the Salvador SARAH domain that crystallized is one residue shorter on the N terminus than the variant used in biochemical assays. T7 Express cells transformed with the plasmid were grown at 37 °C and protein expression was induced at 0.5 A600 with 0.5 mm IPTG at 20 °C for 16 h. Cells were lysed in 40 mm Tris, pH 8, and 200 mm NaCl, and supplemented with protease inhibitor mixture and Salvador–Hippo SARAH purified by sequential affinity purification, IMAC followed by Streptactin (GE Healthcare). Eluted protein was cleaved with SENP to remove both tags and further purified by a combination of anion exchange and gel-filtration chromatographies. The final protein was stored in 10 mm Tris, pH 8, 200 mm NaCl, 1 mm TCEP and flash frozen in liquid nitrogen at 10 mg/ml. Expression and purification of the Salvador–Hippo SARAH variant with C624S substitution in Hippo was expressed and purified in a similar fashion but proceeded only to the tandem affinity steps.
Expression and purification of full-length Mst2 and SENP
T7 Express cells were transformed with two plasmids, pRSF-Duet encoding a maltose-binding protein fused to λ-phosphatase and a pBAD4 derivative encoding full-length Mst2 fused to an N-terminal His6 and SUMO tags, and grown to 0.8 A600. Protein expression was induced with 0.5 mm IPTG and grown for 16 h at 20 °C. Cells were lysed in 50 mm Tris, pH 8.0, and 400 mm NaCl supplemented with a protease inhibitor mixture (Sigma), and clarified lysate was incubated with IMAC resin. The affinity tag was removed from the eluted protein by incubation with SENP, and untagged protein was further purified using anion exchange and gel-filtration chromatographies. To make phosphorylated Mst2, the protein was incubated with 5 mm ATP and 10 mm MgCl2 for 30 min at room temperature prior to gel-filtration. Final concentrated proteins were stored in 10 mm Tris, pH 8.0, 150 mm NaCl, and 1 mm TCEP. SENP was purified as previously described (50).
SARAH domain solubility assays
3 μg of each Salvador SARAH domain variant was incubated with 2.5 μg of SENP in 10 mm Tris, pH 8, 200 mm NaCl, 150 mm imidazole in 60 μl for 1 or 12 h at 4 °C, and the reaction was quenched by boiling for 5 min in SDS loading buffer. Proteins were analyzed by Coomassie-stained SDS-PAGE.
Crystallization and structure determination
Crystals were grown by sitting-drop vapor diffusion at 20 °C. Purified Salvador–Hippo SARAH at 3–6 mg/ml was mixed in a 2:1 ratio with well solution (30–40%) 2-methyl-2,4-pentanediol (Hampton Research), 0–4% polyethylene glycol 20,000 (Hampton Research), and 100 mm sodium cacodylate, pH 6. Rectangular-shaped crystals appeared after 2 days, and continued to grow for up to 1 week to dimensions of 120 × 30 × 10 μm. Crystals were harvested directly from the drop and frozen in liquid nitrogen.
Diffraction data were collected at the 12-2 beamlines at the Stanford Synchrotron Radiation Lightsource (SSRL) and processed with XDS/Aimless(51). Phases were determined by molecular replacement with PHASER (52) with an ensemble search model comprised of two copies each of SARAH domain dimers from both Protein Data Bank codes 3WWS and 4HKD. Initial electron-density maps were improved by prime-and-switch density modification (54). The final model is the final, converged product of iterative rounds of model building in COOT (55) and refinement in PHENIX (56).
Sequence and structure analysis
The measurement of the bend angle of α-helices and characterization of shape were performed with HELANAL-Plus (57, 58). PSI-Pred was used for the secondary structure assignments (59). Sequence alignments were performed using Clustal Omega (30, 31). Buried surface area calculations were performed using PISA (32). All structure figures were drawn with PyMOL (Schrödinger, LLC).
Binding experiments
Profinity Epoxide resin (Bio-Rad) was coupled to either Hippo SARAH domain in 100 mm NaHCO3, pH 11.3, or incubated with just buffer according to the manufacturer's directions. 20 μl of either Hippo SARAH resin or blank resin were mixed with 6.25 μm of the indicated H6-SUMO-tagged Salvador SARAH domain variants in 50 μl of buffer containing 200 mm NaCl and 10 mm Tris, pH 8, and incubated for 1 h at 4 °C. The resin was collected and washed five times with 10 mm Tris, pH 8, and 800 mm NaCl. The resin was boiled in SDS-loading buffer. Samples were analyzed by SDS-PAGE stained with Coomassie Brilliant Blue. Gels were imaged using the Odyssey IR Imaging System (LI-COR), and bands were quantified using Image Studio Software. The ratio of bound Salvador was calculated by subtracting the amount of Salvador bound to a blank resin from the amount bound to Hippo resin, and then dividing that value by the amount of Hippo bound to the resin. To normalize the signal between the seven independent replicates, this ratio was divided by a ratio of bound Salvador with the N-terminal extension. Ratios were analyzed by a paired t test using GraphPad Prism to determine a p value.
Cross-linking and mass spectrometry
Purified Salvador–Hippo SARAH domain was buffer exchanged into 50 mm HEPES, pH 8, 150 mm NaCl, 1 mm TCEP using a 3,000 molecular weight cutoff filter. The sample was reacted with the homobifunctional cross-linker bis(sulfosuccinimidyl)suberate (BS3) (ThermoFisher Scientific) in a 1:20 molar ratio of protein to cross-linker. Reactions were quenched with an excess of ammonium acetate at 30 min, 1, 2, and 4 h. For the analysis of the cross-linked samples using MS, each sample was diluted 1:1 (v/v) into a supersaturated solution of α-cyano-4-hydroxycinnamic acid dissolved in 50% acetonitrile, 49% water, 1% formic acid. 2 μl of each sample was spotted onto a Bruker MTP 384 polished steel plate and analyzed using a Bruker Autoflex and the data were analyzed using Bruker flexAnalysis version 3.3.
Luciferase assay
Schneider's Drosophila Line 2 cells (S2) (ATCC) were cultured in Schneider's medium (Gibco) supplemented with 10% fetal bovine serum (Corning) at 24 °C. S2 cells were transiently transfected with the indicated plasmids using FuGENE 6 (Promega) as described by the manufacturer. Plasmids were transiently transfected into S2 cells, plated 48 h prior, with FuGENE 6 (Promega) following the manufacturer's suggestions. For luciferase assays, a total of 0.5 μg of DNA was transfected per well. Seventy-two h following transfection, cells were harvested and lysed in Passive Lysis Buffer (Promega) and the luciferase signal was measured with the Dual-Glo Luciferase Assay (Promega) according to the manufacturer's directions using a Synergy H1 hybrid reader (Biotek). Scatter plots and statistical analysis of Luciferase Assay results were performed using Prism (GraphPad Software). To control for differences such as cell number and transfection efficiency between experiments, the total luciferase signal was expressed as a ratio of the firefly luciferase signal divided by the Renilla luciferase signal. The total luciferase signal was then normalized by division by the total luciferase signal from transfections without. A one-way analysis of variance test was performed to determine p values. The following plasmids were used in this assay Gal4DBD-YkiV5-pAc, Luciferase-pUAST, and Renilla luciferase-pRL all of which were a gift from the Irvine Lab as well as FLAG-Hpo-pAc5.1 and FLAG-Hpo-C624S-pAc5.1, which were cloned in the Kavran Lab.
In vitro kinase assays
To monitor autophosphorylation of Mst2, 0.5 μm unphosphorylated Mst2 was incubated in 10 mm Tris, pH 8.0, 150 mm NaCl, 5 mm MgCl2, 0.5 mm MnCl2, 1 mm NaF, 1 mm Na3VO4, 5% glycerol, 1 mm TCEP on ice for 5 min alone or with 100 μm Salvador SARAH. The reactions were then supplemented with 1 mm ATP and incubated for up to 2 h at room temperature. Samples were taken at 0, 5, 10, 15, 30, 60, and 120 min and quenched by mixing with SDS loading buffer supplemented with 17 mm EDTA and analyzed by a combination of Coomassie-stained SDS-PAGE or Western blotting using a primary antibody that recognized pThr-180 of Mst2 (Cell Signaling, number 3681) followed by goat α-rabbit 800 RD (LI-COR, number 925-32211) secondary antibody. Blots were scanned on an Odyssey IR Imaging System (LI-COR).
Cell-based activity assays
HEK293T cells (ATCC) were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 5% FBEssence (VWR) and 2 mm glutamine at 37 °C and 5% CO2. Cells were transfected with pCDNA3.1 plasmids (ThermoFisher Scientific) encoding the indicated proteins using X-tremeGENE HP DNA Transfection Reagent (Sigma) according to the manufacturer's protocol. Cells were harvested at 48 h following transfection in ice-cold RIPA buffer supplemented with 1 mm NaF, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, and Universal Nuclease (ThermoFisher Scientific). Cell lysates were clarified, and total protein in the lysate of each sample was quantified by BCA (ThermoFisher Scientific). Normalized cell lysates were analyzed by Western blotting as described above. For these experiments, the following antibodies were used pT12 Mob1 (Cell Signaling, number 8843) followed by goat α-rabbit 800RD, pT35 Mob1 (Cell Signaling, number 8699) followed by goat α-rabbit 800RD, pT180 Mst2 followed by goat α-rabbit 800RD, or pT1079 Lats1 (Cell Signaling, number 8654) followed by goat α-rabbit 800RD, Myc (Santa Cruz, number sc40) followed by goat α-mouse 800RD (LICOR, number 925-32210), HA (Roche, number 11867423001) followed by goat α-rat 800RD (LICOR, number 925-32219), β-actin (Abcam, number 8227) followed by goat α-rabbit 800RD, FLAG (Sigma, number F1804) followed by goat α-mouse 800RD. Band intensities were quantified using ImageJ (53). Scatter plots and statistical analysis were performed using Prism.
Author contributions
L. C., T. T., B. B., and J. M. K. conceptualization; L. C. and J. M. K. resources; L. C., T. T., and J. M. K. data curation; L. C., T. T., B. H. F., A. P., Y. J. K., B. B., and J. M. K. formal analysis; L. C. and J. M. K. supervision; L. C., B. B., and J. M. K. funding acquisition; L. C., T. T., B. H. F., A. P., Y. J. K., B. B., and J. M. K. investigation; L. C. and J. M. K. visualization; L. C., T. T., B. H. F., A. P., Y. J. K., B. B., and J. M. K. methodology; L. C. and J. M. K. writing-original draft; L. C. and J. M. K. project administration; L. C., T. T., B. H. F., A. P., Y. J. K., B. B., and J. M. K. writing-review and editing.
Acknowledgments
The Montana State University Proteomics, Metabolomics, and Mass Spectrometry Facility is supported in part by funding from the Murdock Charitable Trust and National Institutes of Health Grant P20GM103474 of the IDEA program. Plasmids encoding Renilla and firefly luciferase and Gal4DBD-Yki used in the reporter assay were provided by K. Irvine.
This work was supported by National Institutes of Health Grants T32GM007445 (to L. C.) and T32CA009110 (to T. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The atomic coordinates and structure factors (code 6BN1) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- IMAC
- immobilized metal affinity chromatography
- r.m.s. deviation
- root mean square deviation
- BS3
- bis(sulfosuccinimidyl)suberate
- TCEP
- tris(2-carboxyethyl)phosphin
- SUMO
- small ubiquitin-like modifier
- SENP
- SUMO-specific protease
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
References
- 1. Justice R. W., Zilian O., Woods D. F., Noll M., and Bryant P. J. (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 10.1101/gad.9.5.534 [DOI] [PubMed] [Google Scholar]
- 2. Xu T., Wang W., Zhang S., Stewart R. A., and Yu W. (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 3. Tapon N., Harvey K. F., Bell D. W., Wahrer D. C., Schiripo T. A., Haber D. A., and Hariharan I. K. (2002) salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 10.1016/S0092-8674(02)00824-3 [DOI] [PubMed] [Google Scholar]
- 4. Kango-Singh M. (2002) Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719–5730 10.1242/dev.00168 [DOI] [PubMed] [Google Scholar]
- 5. Wu S., Huang J., Dong J., and Pan D. (2003) Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 10.1016/S0092-8674(03)00549-X [DOI] [PubMed] [Google Scholar]
- 6. Harvey K. F., Pfleger C. M., and Hariharan I. K. (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 10.1016/S0092-8674(03)00557-9 [DOI] [PubMed] [Google Scholar]
- 7. Huang J., Wu S., Barrera J., Matthews K., and Pan D. (2005) The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- 8. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., and Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. St. John M. A., Tao W., Fei X., Fukumoto R., Carcangiu M. L., Brownstein D. G., Parlow A. F., McGrath J., and Xu T. (1999) Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21, 182–186 10.1038/5965 [DOI] [PubMed] [Google Scholar]
- 10. Lai Z.-C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L.-L., and Li Y. (2005) Control of cell proliferation and apoptosis by mob as tumor suppressor, Mats. Cell 120, 675–685 10.1016/j.cell.2004.12.036 [DOI] [PubMed] [Google Scholar]
- 11. Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., and Brummelkamp T. R. (2007) YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 10.1016/j.cub.2007.10.039 [DOI] [PubMed] [Google Scholar]
- 12. Creasy C. L., Ambrose D. M., and Chernoff J. (1996) The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J. Biol. Chem. 271, 21049–21053 10.1074/jbc.271.35.21049 [DOI] [PubMed] [Google Scholar]
- 13. Deng Y., Pang A., and Wang J. H. (2003) Regulation of mammalian STE20-like kinase 2 (MST2) by protein phosphorylation/dephosphorylation and proteolysis. J. Biol. Chem. 278, 11760–11767 10.1074/jbc.M211085200 [DOI] [PubMed] [Google Scholar]
- 14. Praskova M., Khoklatchev A., Ortiz-Vega S., and Avruch J. (2004) Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem. J. 381, 453–462 10.1042/BJ20040025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poon C. L., Lin J. I., Zhang X., and Harvey K. F. (2011) The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell 21, 896–906 10.1016/j.devcel.2011.09.012 [DOI] [PubMed] [Google Scholar]
- 16. Boggiano J. C., Vanderzalm P. J., and Fehon R. G. (2011) Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell 21, 888–895 10.1016/j.devcel.2011.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan E. H., Nousiainen M., Chalamalasetty R. B., Schäfer A., Nigg E. A., and Silljé H. H. (2005) The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24, 2076–2086 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- 18. Millward T. A., Hess D., and Hemmings B. A. (1999) Ndr protein kinase is regulated by phosphorylation on two conserved sequence motifs. J. Biol. Chem. 274, 33847–33850 10.1074/jbc.274.48.33847 [DOI] [PubMed] [Google Scholar]
- 19. Stegert M. R., Hergovich A., Tamaskovic R., Bichsel S. J., and Hemmings B. A. (2005) Regulation of NDR protein kinase by hydrophobic motif phosphorylation mediated by the mammalian Ste20-like kinase MST3. Mol. Cell. Biol. 25, 11019–11029 10.1128/MCB.25.24.11019-11029.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scheel H., and Hofmann K. (2003) A novel interaction motif, SARAH, connects three classes of tumor suppressor. Curr. Biol. 13, R899–900 10.1016/j.cub.2003.11.007 [DOI] [PubMed] [Google Scholar]
- 21. Udan R. S., Kango-Singh M., Nolo R., Tao C., and Halder G. (2003) Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 5, 914–920 10.1038/ncb1050 [DOI] [PubMed] [Google Scholar]
- 22. Pantalacci S., Tapon N., and Léopold P. (2003) The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat. Cell Biol. 5, 921–927 10.1038/ncb1051 [DOI] [PubMed] [Google Scholar]
- 23. Callus B. A., Verhagen A. M., and Vaux D. L. (2006) Association of mammalian sterile twenty kinases, Mst1 and Mst2, with hSalvador via C-terminal coiled-coil domains, leads to its stabilization and phosphorylation. FEBS J. 273, 4264–4276 10.1111/j.1742-4658.2006.05427.x [DOI] [PubMed] [Google Scholar]
- 24. Khokhlatchev A., Rabizadeh S., Xavier R., Nedwidek M., Chen T., Zhang X.-F., Seed B., and Avruch J. (2002) Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 12, 253–265 10.1016/S0960-9822(02)00683-8 [DOI] [PubMed] [Google Scholar]
- 25. Polesello C., and Tapon N. (2007) Salvador-warts-hippo signaling promotes Drosophila posterior follicle cell maturation downstream of notch. Curr. Biol. 17, 1864–1870 10.1016/j.cub.2007.09.049 [DOI] [PubMed] [Google Scholar]
- 26. Ni L., Li S., Yu J., Min J., Brautigam C. A., Tomchick D. R., Pan D., and Luo X. (2013) Structural basis for autoactivation of human Mst2 kinase and its regulation by RASSF5. Struct. Fold. Des. 21, 1757–1768 10.1016/j.str.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Polesello C., Huelsmann S., Brown N. H., and Tapon N. (2006) The Drosophila RASSF homolog antagonizes the hippo pathway. Curr. Biol. 16, 2459–2465 10.1016/j.cub.2006.10.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bitra A., Sistla S., Mariam J., Malvi H., and Anand R. (2017) Rassf proteins as modulators of Mst1 kinase activity. Sci. Rep. 7, 45020 10.1038/srep45020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hwang E., Ryu K.-S., Pääkkönen K., Güntert P., Cheong H.-K., Lim D.-S., Lee J.-O., Jeon Y. H., and Cheong C. (2007) Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc. Natl. Acad. Sci. U.S.A. 104, 9236–9241 10.1073/pnas.0610716104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., and Lopez R. (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state: ScienceDirect. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 33. Hwang E., Cheong H.-K., Il Mushtaq A. U., Kim H.-Y., Yeo K. J., Kim E., Lee W. C., Hwang K. Y., Cheong C., and Jeon Y. H. (2014) Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway. Acta Crystallogr. D Biol. Crystallogr. 70, 1944–1953 10.1107/S139900471400947X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Makbul C., Constantinescu Aruxandei D., Hofmann E., Schwarz D., Wolf E., and Herrmann C. (2013) Structural and thermodynamic characterization of Nore1-SARAH: a small, helical module important in signal transduction networks. Biochemistry. 52, 1045–1054 10.1021/bi3014642 [DOI] [PubMed] [Google Scholar]
- 35. Song J., Hong H. R., Yamashita E., Park I. Y., and Lee S. J. (2015) Low pH-driven folding of WW45-SARAH domain leads to stabilization of the WW45-Mst2 complex. J. Biochem. 158, 181–188 10.1093/jb/mvv031 [DOI] [PubMed] [Google Scholar]
- 36. Barlow D. J., and Thornton J. M. (1988) Helix geometry in proteins. J. Mol. Biol. 201, 601–619 10.1016/0022-2836(88)90641-9 [DOI] [PubMed] [Google Scholar]
- 37. Constantinescu Aruxandei D., Makbul C., Koturenkiene A., Lüdemann M.-B., and Herrmann C. (2011) Dimerization-induced folding of MST1 SARAH and the influence of the intrinsically unstructured inhibitory domain: low thermodynamic stability of monomer. Biochemistry 50, 10990–11000 10.1021/bi201110h [DOI] [PubMed] [Google Scholar]
- 38. Liu G., Shi Z., Jiao S., Zhang Z., Wang W., Chen C., Hao Q., Zhang M., Feng M., Xu L., Zhang Z., Zhou Z., and Zhang M. (2014) Structure of MST2 SARAH domain provides insights into its interaction with RAPL. J. Struct. Biol. 185, 366–374 10.1016/j.jsb.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 39. Street T. O., Courtemanche N., and Barrick D. (2008) Protein folding and stability using denaturants. Methods Cell Biol. 84, 295–325 10.1016/S0091-679X(07)84011-8 [DOI] [PubMed] [Google Scholar]
- 40. Huang X., Poy F., Zhang R., Joachimiak A., Sudol M., and Eck M. J. (2000) Structure of a WW domain containing fragment of dystrophin in complex with β-dystroglycan. Nat. Struct. Biol. 7, 634–638 10.1038/77923 [DOI] [PubMed] [Google Scholar]
- 41. Rentschler S., Linn H., Deininger K., Bedford M. T., Espanel X., and Sudol M. (1999) The WW domain of dystrophin requires EF-hands region to interact with β-dystroglycan. Biol. Chem. 380, 431–442 [DOI] [PubMed] [Google Scholar]
- 42. Chothia C., and Janin J. (1975) Principles of protein-protein recognition. Nature 256, 705–708 10.1038/256705a0 [DOI] [PubMed] [Google Scholar]
- 43. Valdar W. S., and Thornton J. M. (2001) Conservation helps to identify biologically relevant crystal contacts. J. Mol. Biol. 313, 399–416 10.1006/jmbi.2001.5034 [DOI] [PubMed] [Google Scholar]
- 44. Bahadur R. P., Chakrabarti P., Rodier F., and Janin J. (2004) A dissection of specific and nonspecific protein-protein interfaces. J. Mol. Biol. 336, 943–955 10.1016/j.jmb.2003.12.073 [DOI] [PubMed] [Google Scholar]
- 45. Ohnishi S., Güntert P., Koshiba S., Tomizawa T., Akasaka R., Tochio N., Sato M., Inoue M., Harada T., Watanabe S., Tanaka A., Shirouzu M., Kigawa T., and Yokoyama S. (2007) Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form. FEBS Lett. 581, 462–468 10.1016/j.febslet.2007.01.008 [DOI] [PubMed] [Google Scholar]
- 46. Ribeiro P. S., Josué F., Wepf A., Wehr M. C., Rinner O., Kelly G., Tapon N., and Gstaiger M. (2010) Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol. Cell. 39, 521–534 10.1016/j.molcel.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 47. Couzens A. L., Knight J. D., Kean M. J., Teo G., Weiss A., Dunham W. H., Lin Z.-Y., Bagshaw R. D., Sicheri F., Pawson T., Wrana J. L., Choi H., and Gingras A.-C. (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal. 6, rs15 10.1126/scisignal.2004712 [DOI] [PubMed] [Google Scholar]
- 48. Bae S. J., Ni L., Osinski A., Tomchick D. R., Brautigam C. A., and Luo X. (2017) SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK. eLife. 10.7554/eLife.30278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Peränen J., Rikkonen M., Hyvönen M., and Kääriäinen L. (1996) T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373 10.1006/abio.1996.0187 [DOI] [PubMed] [Google Scholar]
- 50. Malakhov M. P., Mattern M. R., Malakhova O. A., Drinker M., Weeks S. D., and Butt T. R. (2004) SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genomics 5, 75–86 [DOI] [PubMed] [Google Scholar]
- 51. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schneider C. A., Rasband W. S., and Eliceiri K. W. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Terwilliger T. C. (2004) Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 60, 2144–2149 10.1107/S0907444904019535 [DOI] [PubMed] [Google Scholar]
- 55. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 56. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bansal M., Kumar S., and Velavan R. (2000) HELANAL: a program to characterize helix geometry in proteins. J. Biomol. Struct. Dyn. 17, 811–819 10.1080/07391102.2000.10506570 [DOI] [PubMed] [Google Scholar]
- 58. Kumar P., and Bansal M. (2012) HELANAL-Plus: a web server for analysis of helix geometry in protein structures. J. Biomol. Struct. Dyn. 30, 773–783 10.1080/07391102.2012.689705 [DOI] [PubMed] [Google Scholar]
- 59. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]