Abstract
Cytochrome b (Cytb) is the only mitochondrial encoded subunit from the bc1 complex. Cbp3 and Cbp6 are chaperones necessary for translation of the COB mRNA and Cytb hemylation. Here we demonstrate that their role in translation is dispensable in some laboratory strains, whereas their role in Cytb hemylation seems to be universally conserved. BY4742 yeast requires Cbp3 and Cbp6 for efficient COB mRNA translation, whereas the D273-10b strain synthesizes Cytb at wildtype levels in the absence of Cbp3 and Cbp6. Steady-state levels of Cytb are close to wildtype in mutant D273-10b cells, and Cytb forms non-functional, supercomplex-like species with cytochrome c oxidase, in which at least core 1, cytochrome c1, and Rieske iron–sulfur subunits are present. We demonstrated that Cbp3 interacts with the mitochondrial ribosome and with the COB mRNA in both BY4742 and D273-10b strains. The polymorphism(s) causing the differential function of Cbp3, Cbp6, and the assembly feedback regulation of Cytb synthesis is of nuclear origin rather than mitochondrial, and Smt1, a COB mRNA-binding protein, does not seem to be involved in the observed differential phenotype. Our results indicate that the essential role of Cbp3 and Cbp6 is to assist Cytb hemylation and demonstrate that in the absence of heme b, Cytb can form non-functional supercomplexes with cytochrome c oxidase. Our observations support that an additional protein or proteins are involved in Cytb synthesis in some yeast strains.
Keywords: mitochondria, mitochondrial DNA (mtDNA), translation, respiratory chain, yeast, bc1 complex, Cbp3, Cbp6, Cytochrome b, Qcr7
Introduction
Mitochondrial respiratory complex bc1 is a dimeric enzyme composed of 10 subunits. Cytochrome b (Cytb)4 is the only subunit encoded in the mitochondrial genome by the COB gene. Assembly of the bc1 complex involves formation of different subcomplexes (1, 2): 1) early stage: subunits Cytb, Qcr7, and Qcr8 interact, forming the first subcomplex (Cytb–Qcr7–Qcr8); 2) intermediate stage: subunits Cytb, Qcr7, Qcr8, Qcr6, cytochrome c1 (Cytc1), Cor1, and Cor2 form a 500-kDa subcomplex; dimerization and interaction with the cytochrome c oxidase (CcO) occurs at this stage (2); and 3) late stage: Rip1, Qcr9, and Qcr10 assemble to form the complete bc1 complex. Although little is known about regulatory mechanisms in each step of the bc1 complex assembly, some studies show two important points of regulation, the formation of the subcomplex Cytb–Qcr7–Qcr8 and the assembly of the Rieske iron–sulfur protein Rip1 (3–5). Stability of each subunit in the subcomplex Cytb–Qcr7–Qcr8 depends on the presence of the other two subunits (6), and recently it was demonstrated that synthesis of Cytb is regulated at this early stage of assembly (7).
COB mRNA translation depends on a set of translational activators: Cbs1 and Cbs2, which act on the COB mRNA 5′-UTR (8, 9). Cbp1 is involved in maturation, stability (10), and translation of the COB mRNA by an unknown mechanism (11). These proteins interact with mitochondrial ribosomes (12, 13). Cbp3 and Cbp6 are chaperones of Cytb that in addition activate COB mRNA translation in a cooperative form. These proteins also interact with the tunnel exit of the mitochondrial ribosome, specifically with the ribosomal subunit MrpL4 (14). Cbp3 and Cbp6 interact with newly synthetized Cytb, probably immediately as the peptide exits the mitochondrial ribosome. This interaction is important for addition of heme bL to Cytb, and it is preserved until Qcr7, Qcr8, the chaperone Cbp4, and the heme bH assemble with Cytb (15). The model suggests that once Cbp3–Cbp6 dissociate from the newly synthesized Cytb, these chaperones are able to activate additional rounds of COB mRNA translation. This idea is supported by the observation that Cytb synthesis is reduced in mutants lacking some of the bc1 complex subunits, like Qcr7 and Qcr8. In this case, Cbp3 and Cbp6 remain sequestered in a complex with Cytb and are therefore proposed to be impaired for COB mRNA translational activation (7).
Feedback assembly regulation of mitochondrial mRNAs translation is a very well-known mechanism in mitochondria. In CcO, synthesis of the subunit Cox1 is highly repressed by assembly defects of the complex, and this process is mostly mediated by the Cox1 translational activator Mss51 (16–18). Synthesis of the mitochondrial-encoded ATP synthase subunits Atp6 and Atp8 decrease by assembly defects on the F1 portion (19). The negative effect on Atp6 and Atp8 synthesis is mediated by Smt1, an integral protein localized in the mitochondrial inner membrane facing the matrix. Smt1 physically interacts with the ATP8–ATP6 mRNA acting as a translational repressor. Surprisingly, it was shown that Smt1 also interacts with the COB mRNA. However, no effect on Cytb synthesis was observed in a Δsmt1-null mutant (20).
In this study, we report that Cbp3 and Cbp6 are dispensable for translational activation of Cytb synthesis in the D273-10b yeast strain, whereas in BY4742 cells these chaperones are necessary for Cytb synthesis as previously reported for W303 strain (7, 14). We characterized the role of Cbp3 and Cbp6 in Cytb biogenesis in both strains and studied whether Smt1 was involved in the differential regulation of Cytb synthesis by Cbp3/Cbp6 in the D273-10b and BY4742 strains. Our study demonstrated that the essential role of Cbp3 and Cbp6 is to regulate Cytb hemylation and that a regulatory function in COB mRNA translation prevails in some yeast strains. Moreover, non-hemylated Cytb can form non-functional supercomplexes with cytochrome c oxidase. Our results provide relevant information about the process of Cytb biogenesis and translational control of the COB mRNA.
Results
Cbp3 and Cbp6 are dispensable for Cytb synthesis, but not for bc1 complex activity in D273-10b lab strains
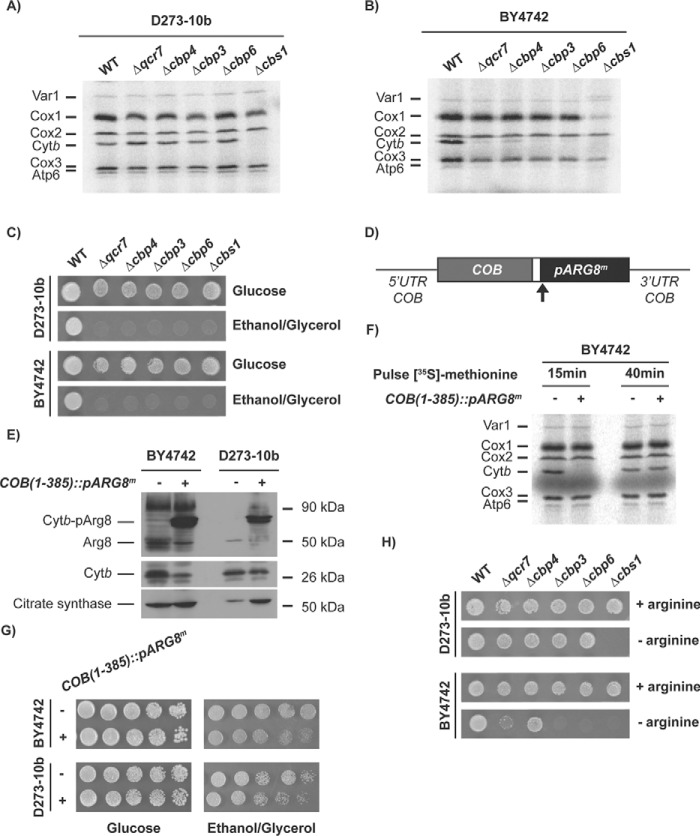
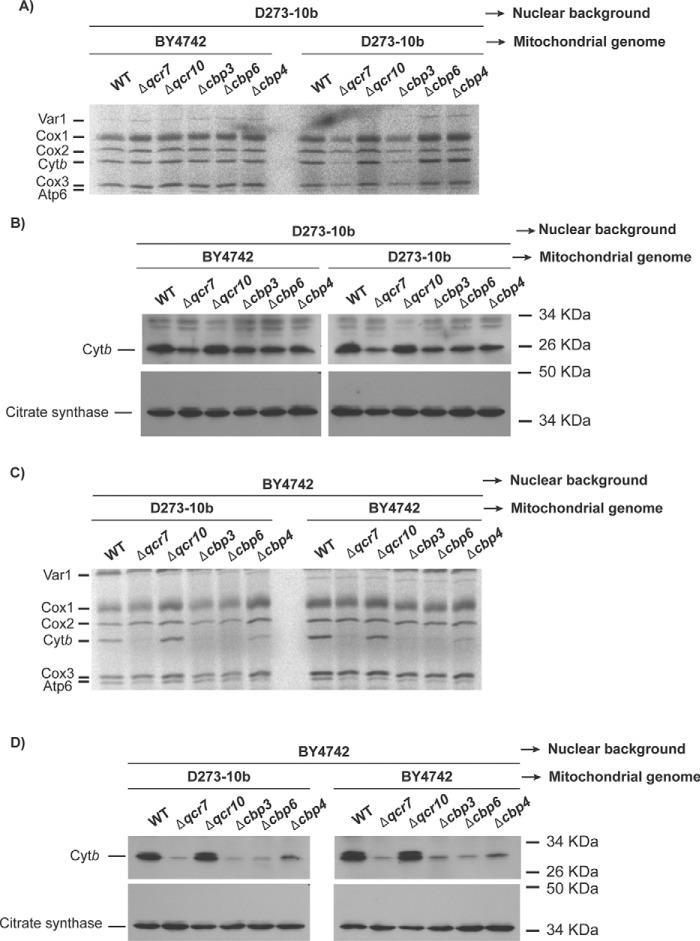
We were interested in the study of the mechanisms of Cytb synthesis. To see whether assembly defects in bc1 complex affect Cytb synthesis, we created Δqcr7, Δcbp4, Δcbp3, and Δcbp6 mutants in the D273-10b yeast strain. These proteins form the first intermediates at early stages of Cytb assembly and maturation (7, 15). To follow mitochondrial translation, the cells were incubated with [35S]methionine in the presence of cycloheximide, and mitochondrial products were analyzed by SDS-PAGE and autoradiography. Surprisingly, contrary to previous reports (7, 14), depletion of Cbp3 or Cbp6 did not negate Cytb labeling (Fig. 1A). Deletion of CBP4 also showed normal levels of Cytb labeling in agreement with previous reports supporting that this chaperone is not necessary for efficient Cytb synthesis (14). Likewise, the absence of subunit Qcr7 also permitted wildtype levels of Cytb labeling. As expected, cells lacking Cbs1, one of the translational activators of the COB mRNA (8), were completely impaired in Cytb synthesis. The majority of experiments regarding Cbp3 and Cbp6 function are based on the W303 strain, so we hypothesized that the difference in the observed phenotype could be due to the strain used. To test this, Cytb synthesis of the same mutants was analyzed in a third yeast strain, BY4742. In these BY4742 mutants, Cytb labeling dramatically decreased in the absence of Cbp3 and Cbp6, as previously reported for W303 nuclear background (7, 14, 15) (Fig. 1B). Interestingly, in BY4742 cells Δqcr7 and Δcbp4 mutations also decreased Cytb labeling, whereas absence of Cbs1 abolished labeling. In the Δcbs1 mutant, Cox1 labeling also decreased dramatically, probably because of the presence of introns in the COX1 gene and the dependence of a COB maturase for COX1 mRNA maturation (21). Even though Cytb [35S]methionine labeling was normal in Δcbp3 and Δcbp6 mutants in the D273-10b strain, respiratory growth was still compromised (Fig. 1C), indicating that in D273-10b cells, Cbp3 and Cbp6 are still necessary for a post-translational step of complex III biogenesis.
Figure 1.
Cbp3 and Cbp6 are dispensable for Cytb synthesis in D273-10b lab strains. A and B, whole-cell mitochondrial translation products from WT and the indicated mutant strains in the D273-10b (A) and BY4742 (B) strains were labeled with [35S]methionine in the presence of cycloheximide. The proteins were analyzed by SDS-PAGE and autoradiography. Cox1, cytochrome c oxidase subunit 1; Cox2, cytochrome c oxidase subunit 2; Cox3, cytochrome c oxidase subunit 3; Atp6, ATP synthase subunit 6; Atp8, subunit 8; Atp9, subunit 9; Var1, ribosomal subunit Var1. C, WT and the indicated mutants from the BY4742 and D273-10b strains were spotted as serial dilutions in rich fermentative (glucose) and respiratory (ethanol/glycerol) media for 3 days at 30 °C. D, the pARG8m gene was fused in frame with the complete COB codons. The reporter gene contains the Arg8 mitochondrial targeting signal, and the processing site is indicated by an arrow. E, mitochondria from D273-10b and BY4742 lab strains carrying the COB(1–352)::pARG8m) construct were analyzed by SDS-PAGE and Western blotting using antibodies against Arg8, Cytb, and citrate synthase (as loading control). F, cells from the BY4742 lab strain carrying either the WT mitochondrial genome or the COB(1–352)::pARG8m construct were pulse-labeled with [35S]methionine for 15 min or 40 min as in A. G, respiratory growth of cells carrying the COB(1–352)::pARG8m construct were assessed by 10-fold serial dilutions spotted on YPD or YPEG media and were grown for 3 days at 30 °C. H, the indicated mutants from D273-10b and BY4742 lab strains bearing the COB(1–352)::pARG8m construct were spotted on synthetic media in the presence or absence of arginine. 10-fold serial dilutions were grown for 3 days at 30 °C.
Variations in the Cytb [35S]methionine labeling in D273-10b and BY4742 strains might be due to differences in the effect of Cbp3, Cbp6, Qcr7, and Cbp4 on COB mRNA translation. Alternatively, it could be due to differences in Cytb instability in the absence of these proteins. To differentiate between these two possibilities, we used the translation reporter gene ARG8m, which has been widely used to study mitochondrial translation. Arg8 is an acetylornithine aminotransferase involved in arginine biosynthesis. It is normally encoded by a nuclear gene and imported into mitochondria using a mitochondrial targeting signal. The mitochondrial encoded Arg8 protein activity is not linked to the respiratory chain function and therefore can be used to assess mitochondrial translation by monitoring growth on media lacking arginine (14, 16, 19, 22). In a strain with the endogenous ARG8 deleted, we inserted ARG8m in the COB locus, maintaining the COB untranslated regions and COB codons. The precursor pARG8m reporter was fused in frame to the last codon of the COB gene to create the construct COB(1–385)::pARG8m (Fig. 1D) and followed ARG8m expression by monitoring growth in media lacking arginine. After Western blotting analysis with antibodies against Arg8, we detected the presence of a Cytb-pArg8 fusion protein of the expected size, 90 kDa, as well as a faint band for processed Arg8 (Fig. 1E). Cytb was also observed albeit at lower levels as compared with mitochondria carrying wildtype mtDNA in both BY4742 and D273-10b strains. Mitochondrial translation products analysis indicated that processed Cytb is only observed after 40 min of [35S]methionine pulse labeling (Fig. 1F). Both strains carrying the COB(1–385)::pARG8m mitochondrial construct supported growth on media lacking arginine (Fig. 1H and Fig. S1A), as well as respiratory growth, albeit with less efficiency as compared with cells carrying wildtype mitochondrial DNA (Fig. 1G). We created Δcbp3, Δcbp6, Δqcr7, Δcbp4, and Δcbs1 mutants carrying the COB(1–385)::pARG8m construct in D273-10b and BY4742 strains. With the exception of Δcbs1 mutation, the rest of D273-10b mutants showed a robust growth on media lacking arginine (Fig. 1H). In contrast, Δcbp3 and Δcbp6 mutants completely lacked growth capacity in −Arg media in BY4742 cells. This is consistent with previous observations where absence of Cbp3 and Cbp6 reduced −Arg growth of cells carrying a complete COB codon replacement with the ARG8m reporter, cobΔ::pARG8m (14) (Fig. S1C).
Δqcr7 BY4742 mutants showed a very weak growth on media lacking arginine, whereas Δcbp4 showed a more robust growth on −Arg media (Fig. 1G). Growth of the Δqcr7 mutant in the absence of supplemental arginine was particularly impaired as compared with Δcbp4 and Δqcr8 mutants (Fig. S1B). However, growth in media lacking arginine was recovered after complete replacement of COB codons by ARG8m (Fig. S1C), indicating that translation of COB mRNA is regulated by the presence of Cytb. These phenotypes are consistent with previous observations where different feedback regulation levels of Cytb synthesis occurs in the absence of bc1 complex subunits and chaperones (7). These results strongly suggest that Cbp3 and Cbp6 are necessary for Cytb synthesis in BY4742 but not in D273-10b. In addition, the assembly feedback regulation of Cytb synthesis by complex III subunits is preserved in BY4742 cells but not in D273-10b cells.
This comparative analysis showed that Cbp3 and Cbp6 are dispensable as translational activators in the D273-10b strain but are still necessary for respiration, probably as chaperones, whereas in the BY4742 lab strain both proteins are necessary to achieve efficient Cytb synthesis. Moreover, our results indicate that assembly feedback regulation of COB mRNA translation is present in BY4742 strains, where some subunits and chaperones like Qcr7, Qcr8, and Cbp4 have an important participation in this process.
In D273-10b strain Cytb and Rip1 are stable in the absence of Cbp3 and Cbp6
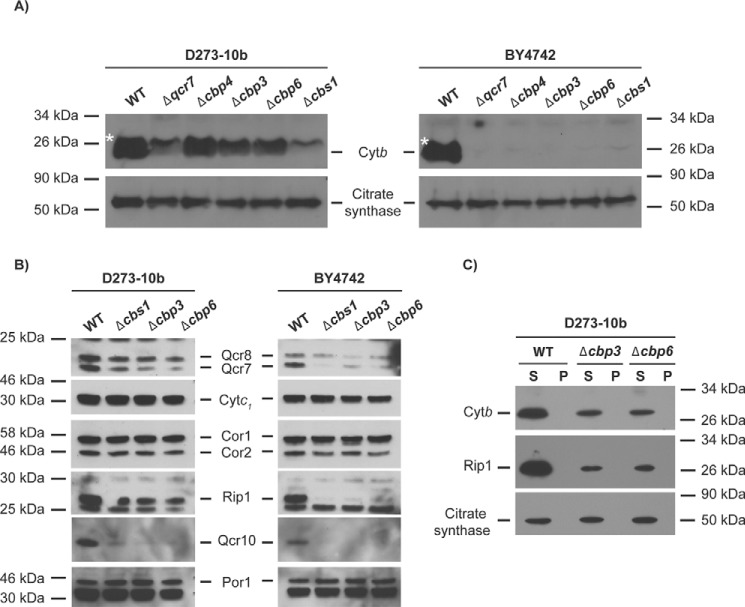
Previous reports indicate that cells lacking Cbp3, Cbp6, and Qcr7 exhibit high Cytb degradation (6, 23, 24). Because Cbp3, Cbp6, and Qcr7 were dispensable for COB mRNA translation in D273-10b strains, we analyzed steady-state levels of Cytb by Western blotting in mutants lacking these proteins. Cytb levels were slightly reduced in Δcbp3 and Δcbp6 as compared with wildtype in D273-10b cells; however, cells lacking Qcr7 showed strongly decreased Cytb levels. In contrast, in BY4742 cells, none of the mutants analyzed showed detectable levels of Cytb (Fig. 2A).
Figure 2.
Cytb and Rip1 accumulate in Δcbp3 and Δcbp6-null mutants from the D273-10b lab strain. A, mitochondrial proteins (20 μg) of the indicated mutants were resolved on a 16% SDS-PAGE in the presence of urea 6 m and transferred to PVDF membrane. Western blotting was carried out with anti-Cytb or anti-citrate synthase (as loading control). The asterisk shows an unspecific band. B, mitochondrial proteins (20 μg) of D273-10b and BY4742 mutants were analyzed by SDS-PAGE 12% and Western blotting with the indicated antibodies. C, isolated mitochondria from WT, Δcbp3, and Δcbp6 mutants were first treated with digitonin and sequentially with Triton X-100. After ultracentrifugation, supernatants from both solubilizations were combined. Supernatants (S) and pellet (P) were loaded on a 12% SDS-PAGE and analyzed by Western blotting using antibodies against Cytb, Rip1, and citrate synthase.
Because Cytb was stable in mutants lacking Cbp3 or Cbp6 in D273-10b cells, we next evaluated the steady-state levels of other bc1 complex subunits in these mutants and compared them to protein levels in the BY4742 strain. Subunits like Cytc1, Cor1, and Cor2 were stable in all mutants from both strains, probably because these three subunits form a stable subcomplex (25) (Fig. 2B). Whereas Qcr7, Qcr8, and Rip1 were practically undetectable in BY4742 mutants, these subunits were detected in D273-10b cells lacking Cbp3, Cbp6, and even Cbs1. Interestingly, the Qcr10 subunit was almost absent in all the mutants analyzed in both strains (Fig. 2B). This observation suggests that Rip1 is not correctly assembled into the bc1 complex in the mutants, and therefore Qcr10 stability is compromised, as previously reported (26).
The observed stability of Cytb, which is an early assembly subunit, and of Rip1, a late assembly subunit (25) in the absence of Cbp3 and Cbp6 in the D273-10b strain surprised us, because both mutants are non-respiratory. We asked whether Cytb and Rip1 aberrantly aggregated and became insensitive to mitochondrial protease activity. To test aggregation, we first solubilized mitochondria from Δcbp3 and Δcbp6 mutants in the D273-10b strain with digitonin and separated pellet from supernatant by ultracentrifugation. The pellet was next solubilized with Triton X-100, and pellet and supernatant fractions were again separated by ultracentrifugation. Aggregates are expected to migrate in the pellet portion of the digitonin–Triton X-100 sequential treatment (5). Western blotting analysis of both fractions showed that Cytb and Rip1 were present in the supernatant fractions, regardless of the presence of Cbp3 and Cbp6 (Fig. 2C). This result indicates that at least Cytb and Rip1 subunits are not forming protease-resistant aggregates in the absence of Cbp3 and Cbp6.
Cytb, Rip1, and Cor1 associate into supercomplexes in the absence of Cbp3 and Cbp6 in the D273-10b strain
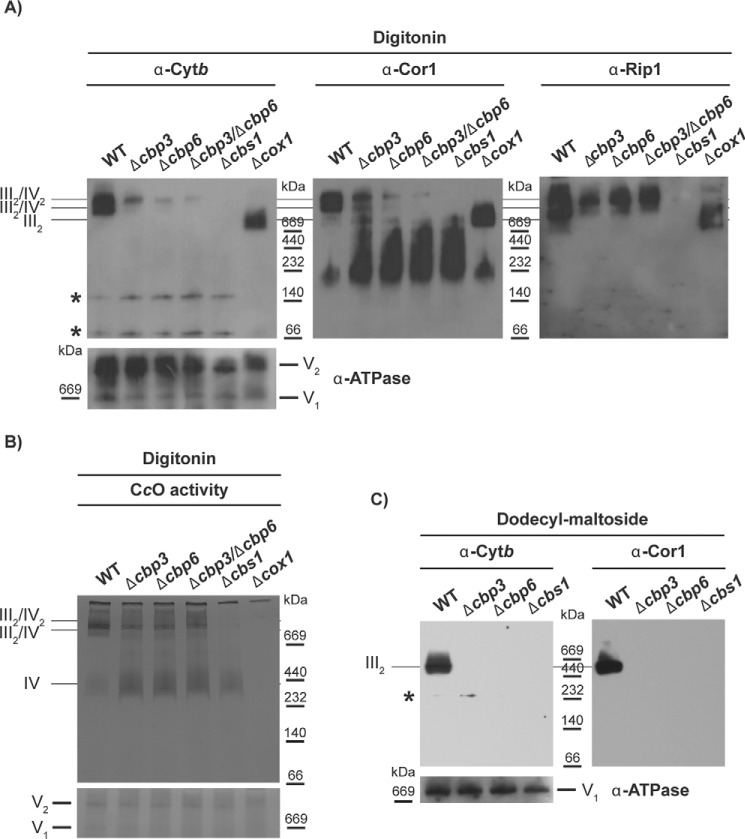
Because Cytb and Rip1 are stable in D273-10b strains in the absence of Cbp3 and Cbp6, we asked whether these subunits could form high molecular weight complexes related to supercomplexes III2/IV2 and III2/IV. Digitonin-solubilized mitochondria from D273-10b containing the mutations Δcbp3, Δcbp6, Δcbp3/Δcbp6, and Δcbs1 were separated by blue native (BN)-PAGE. Western blotting analysis indicated that Cytb, Cor1, and Rip1 co-migrated into III2/IV2 and III2/IV supercomplexes in the wildtype strain (Fig. 3A). The Cytb, Cor1, and Rip1 high molecular weight bands were not present after deletion of the cytochrome c oxidase subunit COX1, where the main species was the dimeric III2 complex. Cor1 and Rip1 were unable to form the high molecular weight complex after deletion of Cbs1, where Cytb synthesis was abolished. Surprisingly, in the Δcbp3 and Δcbp6 mutants, and even in the Δcbp3/Δcbp6 double mutant, a fraction of Cytb, Cor1, and Rip1 co-migrated in high molecular weight complexes with sizes corresponding to supercomplexes III2/IV2 and III2/IV and enriched in III2/IV2 complex-like size. In the Δcbp3, Δcbp6, Δcbp3/Δcbp6, and Δcbs1-null mutants, Cor1 migrates in complexes of 232–669 kDa that are absent in the wildtype strain and in the Δcox1 strain. These bands probably correspond to the assembly intermediate subcomplex Cor1–Cor2–Cytc1 (25) (Fig. 3A). It calls our attention that most of Rip1 is observed in high mass complexes, suggesting association with supercomplexes, but very little Cor1 fractionates in these high mass bands. To explore the possibility that Rip1 might form another high mass complex perhaps associated with the Bcs1 hexamer (27), we combined a Δcpb6 mutation with a deletion of Cox1 (by elimination of Pet309, translational activator of the COX1 mRNA (28)). In the double mutant, association of Rieske protein, Cor1, and also Cytb with supercomplex-like bands was lost, and only co-migration with a III2 dimer-like size band was detected (Fig. S2A). These observations support that the high-molecular-weight complexes observed in the absence of Cbp3/Cbp6 are related to complex IV and to III/IV supercomplexes. The supercomplex-like band contained active cytochrome c oxidase, as revealed by an in-gel activity assay (Fig. 3B and Fig. S2B). Mitochondria from the Δcbs1 mutant presented a ∼450-kDa band corresponding to the monomeric CcO enzyme, not present in a Δcox1-null mutant. Taken together, these data support the idea that the observed complex in Δcbp3, Δcbp6, and Δcbp3/Δcbp6 mutants is related to respiratory supercomplexes, even if the bc1 complex lacks activity.
Figure 3.
Cytb forms supercomplex-like species in the absence of Cbp3 and Cbp6 in D273-10b lab strains. 200 μg of mitochondrial protein from the indicated mutants were solubilized with digitonin, divided, and then loaded in two different BN-PAGE systems. A, one BN-PAGE was analyzed by Western blotting using antibodies against Cytb, Cor1, Rip1, and ATP synthase (as loading control). Asterisks indicate unspecific bands not related to Cytb. B, the second BN-PAGE was treated with 3,3′-diaminobenzidine and horse Cytc to observe the CcO in-gel activity. Coomassie stain is shown as a loading control. C, 100 μg of mitochondrial protein were solubilized with dodecyl-maltoside and analyzed by BN-PAGE and Western blotting using antibodies against Cytb, Cor1, and ATP synthase (the monomeric complex, V1, was used as loading control).
Solubilization of mitochondria with dodecyl-maltoside usually separates the III–IV associations and maintains individual IV and III2 complexes (29). Thus, mitochondria from D273-10b strains carrying the Δcbp3, Δcbp6, Δcbp3/Δcbp6, or Δcbs1 mutations were solubilized with dodecyl-maltoside and analyzed by BN-PAGE. Only wildtype mitochondria maintained Cytb and Cor1 in the ∼670-kDa band corresponding to dimeric complex III (Fig. 3C), whereas Δcbp3, Δcbp6, Δcbp3/Δcbp6, or Δcbs1 mitochondria showed undetectable levels of this band. These data indicate that in the absence of Cbp3 and Cbp6, the bc1 complex subunits Cytb and Cor1 are not forming a stable complex III. Moreover, these data suggest that association with the CcO is necessary to maintain the interaction of bc1 complex subunits.
Cbp3 and Cbp6 are still necessary for Cytb hemylation in the D273-10b strain
Even though Cytb synthesis proceeds and aberrant supercomplexes form in the absence of Cbp3 and Cbp6, these chaperones are still necessary for respiration. In W303 lab strain, it was observed that Cbp3 and Cbp6 are necessary for heme b assembly into the Cytb subunit (15). We asked whether Cbp3 and Cbp6 were still necessary for Cytb hemylation in the D273-10b lab strain. Hemes were extracted from purified mitochondria by incubation with acidic acetone (1% HCl), and the extract was analyzed by reverse phase HPLC. We used hemin as a control to identify heme b, with a retention time of 12 min (data not shown). HPLC analysis showed a dramatic decrease of the total heme b present in Δcbp3 and Δcbp6 mutants (Fig. 4). This level was similar to the one observed in the absence of Cbs1, used as negative control because Cytb is completely absent in this strain (8). Our results indicate that even when Cbp3 and Cbp6 are not necessary for COB mRNA translation in D273-10b strains, they are still necessary for Cytb hemylation.
Figure 4.
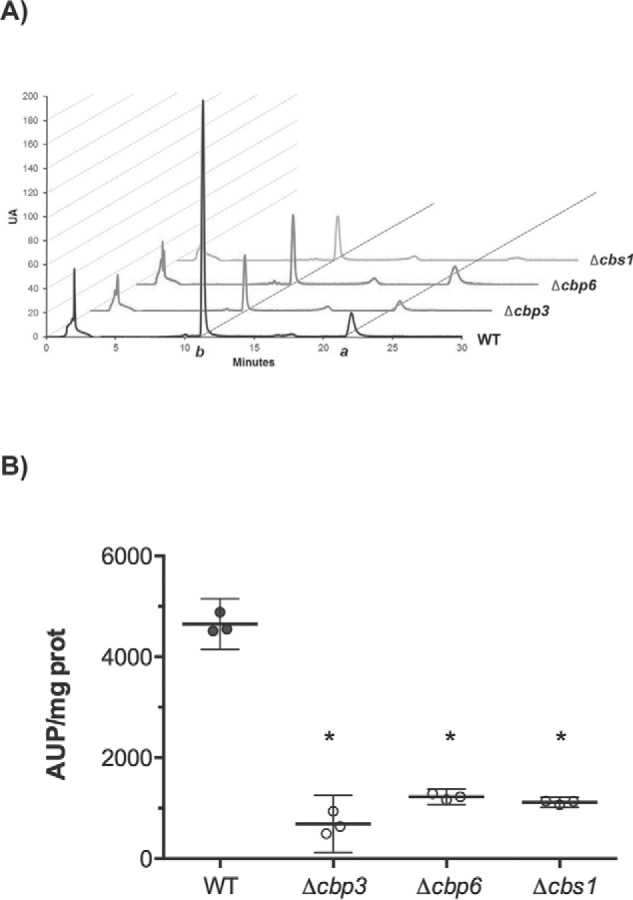
Cbp3 and Cbp6 are still necessary for Cytb hemylation in the D273-10b lab strain. Mitochondrial proteins (700 μg) from the indicated mutants were treated with acidic acetone (1% HCl), clarified, and then separated by reverse-phase HPLC. A, retention time peaks of heme b (peak b) and heme a (peak a) are represented by arbitrary units (UA). B, the area under the peak (AUP)/mg protein was calculated by the mean of two biological and one technical replicate (n = 3). Confidence interval (95%) is represented by error bars. *, p < 0.0001 versus WT.
Cbp3 interacts with the COB mRNA and with the mitochondrial ribosome in both D273-10b and BY4742 strains
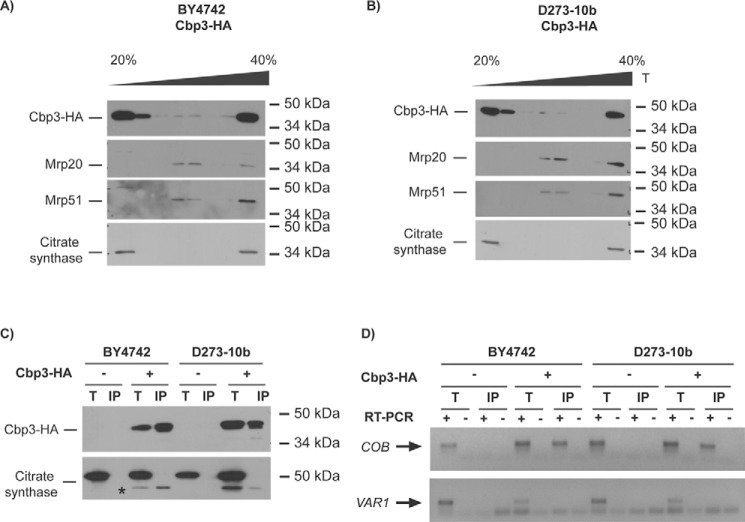
Cbp3 and Cbp6 interact with the mitochondrial ribosome tunnel exit in W303 lab strains (14). Thus, we investigated whether Cbp3 associates with the mitoribosome in BY4742 strains, in which this protein is necessary for Cytb synthesis, and in D273-10b strains, in which Cbp3 is dispensable for Cytb synthesis. To detect Cbp3, we fused a hemagglutinin (HA) epitope to the C-terminal end of Cbp3. The construction expressing this protein was expressed from a centromeric plasmid and transformed into Δcbp3 cells from either BY4742 or D273-10b strains. The presence of the HA epitope on Cbp3 did not disrupt respiratory growth of otherwise wildtype cells (Fig. S3). Mitochondria was digitonin-solubilized in the presence of 10 mm Mg+, a condition in which both ribosomal subunits are associated. The extracts were loaded on a discontinuous sucrose gradient (20–40%) and ultracentrifuged. Six fractions were collected and analyzed by SDS-PAGE and Western blotting. In both strains, BY4742 and D273-10b, Cbp3–HA co-migrated with Mrp20 (protein from the large ribosomal subunit) and with Mrp51 (protein from the small ribosomal subunit) (Fig. 5, A and B), indicating that in both strains Cbp3–HA interacts with the mitoribosome.
Figure 5.
Cbp3 and Cbp6 interact with the mitochondrial ribosome and with the COB mRNA in both D273-10b and BY4742 lab strains. A and B, 500 μg of isolated mitochondria from BY4742 (A) and D273-10b (B) cells expressing CBP3-HA from a low-copy plasmid were solubilized with digitonin, clarified, and loaded into a discontinuous sucrose gradient (20–40%) for ultracentrifugation. 1/10 of the clarified portion was taken as a total control (T). Proteins from seven fractions were TCA-precipitated and analyzed by SDS-PAGE (12%) and Western blotting. Cbp3–HA was detected with an antibody against the HA epitope. Antibodies against Mrp20 and Mrp51 were used as markers for large and small ribosomal subunits, respectively. Citrate synthase was used as a control for a non-ribosomal mitochondrial protein. C, 500 μg from isolated mitochondria were solubilized with dodecyl maltoside, and Cbp3–HA or untagged Cbp3 were subjected to immunoprecipitation with an antibody against HA. Immunoprecipitate (IP, one-third) and total (T, 5% of the mitochondrial extract used for immunoprecipitation) fractions were separated by SDS-PAGE and analyzed by Western blotting with antibodies against HA and citrate synthase (as a negative control for interaction). The asterisk corresponds to the previous HA immunoblot analysis of Cbp3–HA. D, RNA was extracted from the total (T) and immunoprecipitate (IP) fractions. Each fraction was divided in two, and cDNA was prepared in the presence (+) or absence (−) of reverse transcriptase (RT) using primers for the COB and VAR1 5′-UTRs. The (−) RT lanes represent a negative control for DNA contamination. The PCR products were run on agarose gel, and the gel pictures were color-inverted.
Considering that Cbp3 is necessary for efficient Cytb synthesis in W303 (14) and BY4742 lab strains (this study), we asked whether this protein physically interacts with the COB mRNA. Mitochondria from BY4742 and D273-10b strains expressing the CBP3-HA or CBP3 gene were solubilized with dodecyl-maltoside. The mitochondrial extract was immunoprecipitated using antibodies against the HA epitope, and RNA was isolated and analyzed by reverse transcription-PCR (30). For cDNA synthesis and PCR amplification, we used primers specific for COB and also for VAR1 (used as a negative control because translation of this mRNA is independent of Cbp3). After HA immunoprecipitation, Western blotting analysis showed that Cbp3 was efficiently immunoprecipitated in both BY4742 and D273-10b mitochondria (Fig. 5C). First strand cDNA and PCR amplification indicated that in both strains Cbp3 physically interacts with the COB mRNA (Fig. 5D).
The polymorphism(s) between D273-10b and BY4742 producing differential function of Cbp3, Cbp6, and Qcr7 in Cytb synthesis has a nuclear origin and is not present on Smt1
The observed difference on the role of Cbp3, Cbp6, Qcr7, and Cbp4 in Cytb synthesis regulation between the two lab strains suggests the existence of polymorphism(s) producing these distinct phenotypes. This polymorphism(s) could locate either on nuclear or in mitochondrial genes. To investigate this, we exchanged the mitochondrial genomes between the two lab strains, creating two different cybrid strains: D273-10b with the BY4742 mtDNA and BY4742 with the D273-10b mtDNA. Next, by whole-cell [35S]methionine labeling in the presence of cycloheximide, we analyzed the effect of Δqcr7, Δcbp3, Δcbp6, and Δcbp4 mutations on Cytb synthesis. We included a Δqcr10 mutant as control of a subunit whose absence has no effect on Cytb synthesis (7, 31). D273-10b mutants with BY4742 mtDNA showed Cytb labeling pattern as a D273-10b strain, where Δqcr7, Δcbp3, and Δcbp6 mutants exhibited wildtype Cytb labeling (Fig. 6A). Δcbp4 and Δqcr10 cells also labeled Cytb as wildtype. In concordance, steady-state levels of Cytb in the mutants are close to wildtype levels (Fig. 6B). BY4742 mutants with D273-10b mtDNA behaved as BY4742 cells: the absence of Cbp3, Cbp6, and Qcr7 dramatically reduced Cytb labeling, whereas the absence of Cbp4 reduced only mildly Cytb labeling (Fig. 6C). In agreement, steady-state levels of Cytb in these mutants were also reduced (Fig. 6D). These results suggest that the polymorphism(s) between D273-10b and BY4742 is located in a nuclear gene or genes.
Figure 6.
The polymorphism(s) associated with the differential phenotype of Δcbp3 and Δcbp6 mutants between D273-10b and BY4742 lab strains is located in the nuclear genome. A, mitochondrial DNA from BY4742 was introduced in rho0, D273-10b cells by cytoduction. These cells were labeled with [35S]methionine in the presence of cycloheximide, and the proteins were analyzed by SDS-PAGE and autoradiography. B, mitochondria (20 μg) from the same strains as in A were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. C, mitochondrial DNA from D273-10b was introduced in rho0, BY4742 cells by cytoduction. The cells were [35S]methionine-labeled in the presence of cycloheximide, and proteins were analyzed as in A. D, mitochondria (20 μg) from the same strains in C were analyzed by SDS-PAGE and Western blotting with antibodies against Cytb and citrate synthase.
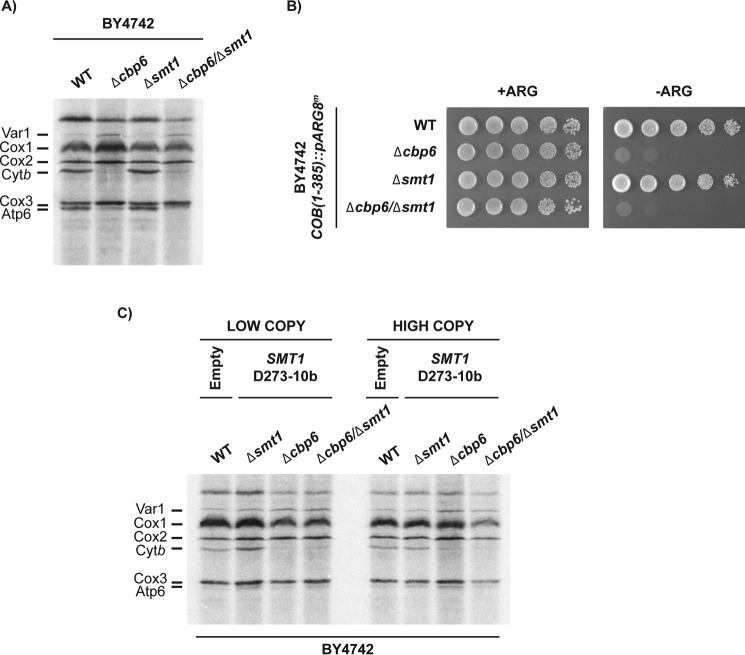
Unfortunately, many attempts to identify the polymorphism(s) associated with the differential phenotype have failed. One candidate was the nuclear gene SMT1. This gene has three polymorphisms: A → G, T → C, and A → G at positions 400, 644, and 668 of the orf, respectively. These polymorphisms change three amino acids in D273-10b with respect to BY4742 and W303: methionine 134 to valine, phenylalanine 206 to serine, and histidine 223 to arginine (data obtained from the Saccharomyces genome database). Smt1 was found to interact with the COB mRNA and to act as a translational repressor of the ATP8–ATP6 mRNA (20). To study whether Smt1 was involved in the differential regulation of Cytb synthesis by Cbp3 and Cbp6, we first tested whether in the BY4742 strain Smt1 was acting as a translational repressor of the COB mRNA as was demonstrated for the ATP8–ATP6 mRNA (20). In this case, we expected that double mutants Δcbp3 or Δcbp6 combined with Δsmt1 resulted in wildtype levels of Cytb synthesis. Whole cell [35S]methionine labeling in the presence of cycloheximide showed that Cytb synthesis had normal levels in the Δsmt1 mutant. However, in the Δcbp6/Δsmt1 double mutant, Cytb synthesis was not recovered in comparison to labeling of a Δcbp6 mutant (Fig. 7A). These data were confirmed by analyzing growth on media lacking arginine of the same strains in Fig. 7A but carrying the COB(1–385)::ARG8m construct in the mitochondrial genome. Neither the single mutant Δcbp6 nor the double mutant Δcbp6/Δsmt1 was able to grow on media lacking arginine (Fig. 7B). Because these experiments tested the effect of the absence of Smt1, we next asked whether the presence of Smt1 expressing the D273-10b gene would compensate Cytb synthesis in the BY4742 Δcbp6 mutant. Thus, we cloned SMT1 from D273-10b lab strain in low-copy and high-copy yeast expression plasmids and transformed them into BY4742 cells carrying the Δcbp6 mutation. In vivo translation assays indicated that Cytb synthesis was not recovered after overexpression of SMT1 carrying the D273-10b lab strain polymorphisms (Fig. 7C). Together, these results suggest that Smt1 is not related to the role of Cbp3 and Cbp6 in Cytb synthesis. Because Smt1 physically interacts, directly or indirectly, with the COB mRNA, it must have a different role in Cytb biogenesis.
Figure 7.
SMT1 is not involved in the differential regulation of Cytb synthesis between BY4742 and D273-10b lab strains. A, mitochondrial translation products from the WT, Δcbp6, Δsmt1, and Δcbp6/Δsmt1-null mutants were labeled with [35S]methionine in the presence of cycloheximide. Proteins were analyzed by SDS-PAGE and autoradiography. B, D273-10b cells carrying the COB(1–352)::pARG8m mitochondrial construct were grown on 10-fold serial dilutions on complete media (+ARG) or media lacking arginine (−ARG) for 3 days at 30 °C. C, BY4742 cells carrying the D273-10b SMT1 gene expressed form low-copy or high-copy plasmids were [35S]methionine pulse-labeled, and mitochondrial translation products were analyzed as in A. Wildtype cells were transformed with empty plasmid as indicated.
Discussion
An essential step on bc1 complex biogenesis is assembly of the only mitochondrial encoded subunit, Cytb, where chaperones and translational activators of the COB mRNA coordinate to form the first Cytb assembly intermediaries (7). Cbp3 and Cbp6 were described as specific translational activators of the COB mRNA (14). Moreover, Cbp3 and Cbp6 act as chaperones necessary for respiratory growth (14, 23, 32), and it is proposed that they sense the assembly state of Cytb on early stages and coordinate synthesis and assembly (7, 14). In the course of our studies to understand the function of Cbp3 and Cbp6, we observed that these proteins have a different role in Cytb biogenesis depending on the yeast lab strain we used. Cbp3 and Cbp6 are necessary for Cytb synthesis in the BY4742 strain, whereas in D273-10b lab strain, these proteins are dispensable for translational regulation. However, in both strains Cbp3 and Cbp6 are necessary for respiratory growth and Cytb hemylation.
The dual function of Cbp3 and Cbp6 in Cytb synthesis/assembly was first observed in W303 yeast lab strains (14). It was not surprising that BY4742 lab strains displayed a similar phenotype as W303 cells, because both strains share a more recent common ancestor than with the D273-10b strain (33, 34), which arose from a different lineage (35). Previous reports also observed that in D273-10b lab strains Cbp3 and Cbp6 were only involved in bc1 complex assembly rather than in Cytb synthesis regulation (23, 32). Moreover, the homologs of Cbp3 and Cbp6 in Schizosaccharomyces pombe are only involved in the bc1 complex assembly, but not in Cytb synthesis (24). These observations suggest that the role of Cbp3 and Cbp6 in COB mRNA translational regulation was acquired at a very recent event, after the divergence of Saccharomyces cerevisiae D273-10b strain from W303 and BY4742 strains.
Even though COB mRNA translation occurs normally in D273-10b strain in the absence of Cbp3 and Cbp6, cells are incapable of supporting respiratory growth. This could be explained because these chaperones are still required for Cytb hemylation in the D273-10b lab strain. Accordingly, Hildenbeutel et al. (15) reported that Cbp3–Cbp6 coordinate apocytochrome b hemylation. These chaperones interact with a heme-free Cytb intermediate, as well as with a Cytb protein carrying heme bL. This result shows that the function of Cbp3 and Cbp6 as chaperones for Cytb hemylation is well-conserved in different S. cerevisiae laboratory strains. The exact mechanism by which heme b assembles into Cytb is still unknown, the heme b could be assembled from the mitochondrial matrix or from the inner membrane (36), and interaction of Cbp3 and Cbp6 with newly synthesized Cytb may keep it in an “accessible” conformation to be hemylated. Moreover, association of Cbp3 and Cbp6 with the ribosomal tunnel exit might be important for this process (15). Accordingly, we observed that even when Cbp3 is dispensable for translation in D273-10b cells, this chaperone interacts with the mitochondrial ribosome in both the BY4742 and in the D273-10b strains, as previously reported for W303 cells (14). This interaction could be important for early interaction with Cytb or for efficient communication between the translational machinery and assembly of Cytb. In this translation/assembly coordination process, Rrf1 and Mif3 might also be involved as previously proposed (37).
We also observed that in D273-10b cells, Cytb interacts with cytochrome c oxidase to form non-functional supercomplexes in the absence of Cbp3 and Cbp6. These supercomplexes contain at least Cytb, Core 1, and the Rieske iron–sulfur subunits. Several reports show that supercomplexes formation can occur at early steps of CcO and bc1 complex assembly (2, 38, 39). Moreover, CcO subunit Cox2 can form a 230-kDa subcomplex with the early assembly intermediate composed of Cytb, Qcr7, and Qcr8 (2). These data support a model in which bc1 complex interacts with CcO at early assembly stages and suggest that this interaction is independent from Cytb hemylation and Cbp3/Cbp6. Alternatively, Cbp3/Cbp6 might participate on regulation of Cytb incorporation into supercomplexes, explaining why in the absence of these chaperones, non-functional supercomplexes form.
How is Cytb synthesis differentially regulated by Cbp3/Cbp6 in D273-10b versus BY4742/W303 strains? We hypothesize that there are additional proteins participating in the Cbp3/Cbp6-dependent translational regulation observed in BY4742 and W303 lab strains. This function may arise from polymorphism(s) affecting the protein(s) sequence and therefore activity. Here we showed that the differential role of Cbp3/Cbp6 comes from polymorphisms in the nuclear genome rather than in the mitochondrial DNA. This polymorphism(s) confers a dominant phenotype to D273-10b cells over BY4742 cells, meaning that diploids resulting from the cross of D273-10b and BY4741 have normal levels of Cytb synthesis in the absence of Cbp3 and Cbp6 (Fig. S4). Moreover, tetrad analysis of diploid, CBP3 heterozygous cells carrying the COB(1–385)::ARG8m suggested that there are two nuclear genes involved in the observed differential phenotype of Δcbp3 mutants (data not shown).
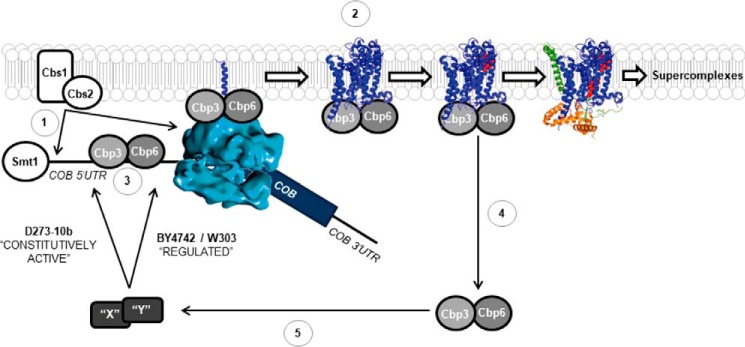
We propose some modifications to the current model for Cytb synthesis and assembly (40). Translational activation of the COB mRNA depends on Cbs1 and Cbs2, which act on the COB mRNA 5′-UTR and also interact with the mitochondrial ribosome (8, 12) (Fig. 8, part 1). Cbp3 and Cbp6 interact with the ribosomal tunnel exit (7) and with the Cytb peptide probably to facilitate Cytb hemylation (15) and/or assembly (Fig. 8, part 2). We also demonstrated that Cbp3 physically interacts with the COB mRNA. This interaction could be direct or mediated by another protein or by the ribosome (Fig. 8, part 3). Interestingly, the Cbp3-COB mRNA association was observed in both lab strains, suggesting that this interaction alone is not sufficient for COB mRNA translational regulation. Once Cytb assembly proceeds to form the first intermediaries containing Qcr7 and Qcr8 subunits (and Cbp4; not shown), then Cbp3/Cbp6 release from the Cytb complex (7) (Fig. 8, part 4). Protein “X” (or perhaps proteins X and Y) becomes active only upon release of Cbp3/Cbp6 from the Cytb complex in BY4742 and W303 lab strains to allow efficient COB mRNA translation (Fig. 8, part 5). In these lab strains, absence of Cbp3/Cbp6 prevents the factors X and Y from becoming active, and therefore Cytb synthesis is reduced. In contrast, in D273-10b lab strain factors X and Y are constitutively active, even in the absence of Cbp3 and Cbp6, possibly because of a loss of communication with these chaperones. Thus, Cytb synthesis is normal even in the absence of Cbp3/Cbp6.
Figure 8.
Model showing a mechanism for the differential regulation of Cytb synthesis by Cbp3 and Cbp6 in distinct laboratory yeast strains. Translational activation of the COB mRNA depends on Cbs1 and Cbs2 (8, 9) (1). Cbp3 and Cbp6 interact with the ribosomal tunnel exit (14) and with newly synthesized Cytb to promote hemylation/assembly (hemes in red) (7, 15) (2). Cbp3 interacts with the COB 5′-UTR mRNA (this work), probably to promote efficient translation (3). These interactions are present in both D273-10b and BY4742 lab strains. Cytb assembly proceeds to form the first intermediary, containing Qcr7 (orange) and Qcr8 (green) subunits, and then Cbp3/6 release from the Cytb complex (7). We propose the existence of additional factors X and Y, which in turn become active in BY4742 and W303 lab strains, allowing an efficient COB mRNA translation (5). Absence of Cbp3/6 prevents factors X and Y from becoming active, and thereby Cytb synthesis is reduced. In contrast, in D273-10b strain factors X and Y are constitutively active, even in the absence of Cbp3 and Cbp6. In addition, Smt1 physically interacts with the COB mRNA to carry over an unknown function that does not seem to be related to Cbp3/Cbp6 (20). Tertiary and quaternary structures were taken from the Protein Data Bank (code 1EZV).
The identity of proteins X and Y remains to be elucidated. Laboratory S. cerevisiae strains contain polymorphisms, introduced mutations, and genetic markers that confer differential phenotypes on mitochondrial function (41, 42). Chaperones that are currently known to be involved in Cytb biogenesis were discarded (Cbs1, Cbs2, Cbp1, and Cbp4), because sequence comparisons did not show any changes between D273-10b and BY4742/W303 strains. A candidate was Smt1, which physically interacts with the COB mRNA (20) and has three amino acid changes in the D273-10b protein as compared with BY4742 and W303. However, our results did not support that Smt1 has a regulatory role of Cytb synthesis that is related to Cbp3 and Cbp6. Other previously identified alleles affecting mitochondrial function that might be candidates are HAP1 and MRM1(42). However, none of these genes are likely to be involved in the differential function of Cbp3/Cbp6 that we observe. Hap1, a heme-responsive transcriptional factor of genes involved in electron transfer reactions (43) has a C-terminal end mutation in some S288c-derived strains that modifies respiratory and oxygen metabolism (44). However, although BY4742 has the hap1 mutation, W303 and D273-10b have wildtype HAP1, ruling out the possibility that HAP1 could be involved in the Cbp3/Cbp6 differential function we observed between D273-10b and BY4742/W303 strains. Mutations on the promoter of MRM1, encoding a mitochondrial methyl-transferase, are associated with respiratory defects in some lab strains (42, 45). However, this gene is wildtype in W303, D273-10b, and BY4742 strains.
Experimental procedures
Yeast strains and genetic methods
S. cerevisiae D273-10b (ATCC24657), BY4742 (ATCC4040004), and BY4741 (ATCC4040002) strains used in this study are listed on Table 1. Genetic methods and media were as previously described (46, 47). Strains were cultured in complete fermentable media containing 1% yeast extract, 2% Bacto-peptone, and 2% glucose (YPD) or 2% galactose (YPGal), or in synthetic complete media containing 0.67% yeast nitrogen base and 2% glucose and lacking uracil or the indicated amino acids. Nuclear deletion constructs with LEU2, URA3, HIS3MX6, or KanMX4 cassettes were made by PCR. Correct integration of the different constructs into the nuclear genome was confirmed by PCR. The CBP3-HA construct, including 320 and 352 bp of the CBP3 5′- and 3′-UTR, respectively, was amplified by fusion PCR (48). This product was ligated into NotI sites of the yeast expression plasmid pRS416.
Table 1.
Yeast strains used in this study
The mitochondrial genome is indicated in parentheses.
| Strain | Nuclear (mitochondrial) genotype | Reference/source |
|---|---|---|
| NB40-36a | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, D273-10B (ρ+) | Ref. 16 |
| DAU1 | Matα,ade2, ura3Δ, D273-10b (ρ+) | Ref. 58 |
| XPM201 | Matα, arg8::hisG, leu2-3,112, lys2, ura3-52, D273-10b (ρ+, ΔΣai)b | Ref. 50 |
| NAB69 | Mata, ade2-101, arg8-delta::hisG, ura3-52, kar1-1 (ρ°) | Ref. 49 |
| NB71 | Matα, ade2-101, ura3-52, leu2-delta, arg8-delta::URA3, kar1-1(ρ+, cox3Δ::ARG8m-1) | Ref. 49 |
| BY4742 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, BY4742 (ρ+) | YKO Matx Strain Collection–Glycerol Stocks (Open Biosystems |
| BY4741 | Mata, his3-delta1, leu2-delta0, met15-delta0, ura3-delta0, BY4741 (ρ+) | YKO Matx Strain Collection–Glycerol Stocks (Open Biosystems |
| Δqcr7 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, qcr7::KANMX4, BY4742 (ρ+) | YKO Matx Strain Collection–Glycerol Stocks (Open Biosystems) |
| Δqcr10 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, qcr10::KANMX4, BY4742 (ρ+) | YKO Matx Strain Collection–Glycerol Stocks (Open Biosystems) |
| Δcbp4 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, cbp4::KANMX4, BY4742 (ρ+) | YKO Matx Strain Collection–Glycerol Stocks (Open Biosystems) |
| Δsmt1 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, smt1::KANMX4, BY4742 (ρ+) | YKO Matx Strain Collection–Glycerol Stocks (Open Biosystems) |
| Δcbp3 | Mata, his3-delta1, leu2-delta0, met15-delta0, ura3-delta0, cbp3::KANMX4,BY4741 (ρ+) | Collection–Glycerol Stocks (Open Biosystems) |
| AGG24 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, D273-10B (ρ+, BY4742 mtDNA)a | This study |
| AGG25 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp3::KANMX4, D273-10B (ρ+, BY4742 mtDNA) | This study |
| AGG28 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp4::KANMX4, D273-10B (ρ+, BY4742 mtDNA) | This study |
| AGG29 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, qcr7::KANMX4, D273-10B (ρ+, BY4742 mtDNA) | This study |
| AGG30 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, qcr10::KANMX4, D273-10B (ρ+, BY4742 mtDNA) | This study |
| AGG33 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, BY4742 (ρ+, ΔΣaib, COB(1-352)::pARG8m) | This study |
| AGG34 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, qcr7::KANMX4, BY4742 (ρ+, ΔΣaib, COB(1-352)::pARG8m) | This study |
| AGG35 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, qcr8::KANMX4, BY4742 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG36 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp3::KANMX4, BY4742 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG37 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp4::KANMX4, BY4742 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG38 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, BY4742 (ρ+, NB40-36a mtDNA) | This study |
| AGG39 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, qcr7::KANMX4, BY4742 (ρ+, NB40-36 mtDNA) | This study |
| AGG40 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, qcr10::KANMX4, BY4742 (ρ+, NB40-36 mtDNA) | This study |
| AGG41 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp3::KANX4, BY4742 (ρ+, NB40-36 mtDNA) | This study |
| AGG42 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp6::LEU2, BY4742 (ρ+, NB40-36 mtDNA) | This study |
| AGG43 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, qcr7::KANMX4, D273-10B (ρ+) | This study |
| AGG46 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp3::KANMX4, D273-10B (ρ+) | This study |
| AGG47 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp4::KANMX4, D273-10B (ρ+) | This study |
| AGG48 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, D273-10B (ρ+, ΔΣai, COB(--352)::pARG8m) | This study |
| AGG56 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp6::LEU2, D273-10B (ρ+) | This study |
| AGG57 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp6::LEU2, D273-10B (ρ+, BY4742 mtDNA) | This study |
| AGG58 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, cbp3::KANMX4, BY4742 (ρ+) | This study |
| AGG59 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, cbp6::LEU2, BY4742 (ρ+) | This study |
| AGG60 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp4::KANMX4, BY4742 (ρ+, NB40-36a mtDNA) | This study |
| AGG61 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp3::KANMX4, cbp6::LEU2, D273-10B (ρ+) | This study |
| AGG62 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp6::LEU2, BY4742 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG63 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, qcr7::KANMX4, D273-10B (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG65 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp3::KANMX4, D273-10B (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG66 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp6::LEU2, D273-10B (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG67 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp4::KANMX4, D273-10B (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG68 | Mata, his3-delta1, leu2-delta0, met15-delta0, ura3-delta0 qcr7::KANMX4, BY4741 (ρ+) | This study |
| AGG69 | Mata, his3-delta1, leu2-delta0, met15-delta0, ura3-delta0 cbp6::LEU2, BY4741 (ρ+) | This study |
| AGG70 | Mata, his3-delta1, leu2-delta0, met15-delta0, ura3-delta0 cbp4::KANMX4, BY4741 (ρ+) | This study |
| AGG71 | Matα/a, lys2+/−, arg8::hisG+/−, ura3-52/ura3-delta0, leu2-3,112/leu2-delta0, met15-delta0−/+, his-delta1−/+, D273-10b/BY4741 (ρ+)c | This study |
| AGG72 | Matα/a, lys2+/−, arg8::hisG+/−, ura3-52/ura3-delta0, leu2-3,112/leu2-delta0, met15-delta0−/+, his-delta1−/+, qcr7::KANMX4+/+, D273-10b/BY4741 (ρ+) | This study |
| AGG73 | Matα/a, lys2+/−, arg8::hisG+/−, ura3-52/ura3-delta0, leu2-3,112/leu2-delta0, met15-delta0−/+, his-delta1−/+,cbp6::LEU2+/+, D273-10b/BY4741 (ρ+) | This study |
| AGG74 | Matα/a, lys2+/−, arg8::hisG+/−, ura3-52/ura3-delta0, leu2-3,112/leu2-delta0, met15-delta0−/+, his-delta1−/+, cbp4::KANMX4+/+, D273-10b/BY4741 (ρ+) | This study |
| AGG75 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbs1::URA3, D273-10B (ρ+, ΔΣaib, COB(1-352)::pARG8m) | This study |
| AGG76 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbs1::HIS3MX6, BY4742 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG77 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, cbs1::URA3, BY4742 (ρ+) | This study |
| AGG78 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbs1::URA3, D273-10B (ρ+) | This study |
| AGG87 | Matα/a, lys2+/−, arg8::hisG+/−, ura3-52/ura3-delta0, leu2-3,112/leu2-delta0, met15-delta0−/+, his-delta1−/+, cbp3::KANMX4+/+, D273-10b/BY4741 (ρ+) | This study |
| AGG89 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, smt1::KANMX4, cbp6::LEU2, BY4742 (ρ+) | This study |
| AGG91 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, smt1::KANMX4, BY4742 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG92 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, smt1::KANMX4, cbp6::LEU2, BY4742 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| AGG93 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3,qcr7::KANMX4, BY4742 (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| AGG95 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3,cbp4::KANMX4 BY4742 (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| AGG96 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp3::KANMX4, BY4742 (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| AGG97 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, qcr7::KANMX4, D273-10B (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| AGG99 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp4::KANMX4, D273-10B (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| AGG100 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp3::KANMX4 D273-10B (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| AGG101 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, pet309::URA3, cpb6::LEU2, D273-10Ba (ρ+) | This study |
| AGG102 | Matα/Mata, lys2+/−, arg8::hisG+/−, ura3-52+/[minus], leu2-3,112+/+, ade2 −/+, his3-11,15−/+, trp1-1−/+, ura3-1−/+, cbp3::KANMX4+/−, D273-10B/W303 (ρ+, ΔΣai, COB(1-352)::pARG8m) | This study |
| DFM2 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, BY4742 (ρ+, ΔΣai, cobΔ::ARG8m) | Daniel Flores-Mireles |
| DFM5 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, D273-10B (ρ+, ΔΣai, cobΔ::ARG8m) | Daniel Flores-Mireles |
| YC140 | Matα, ade2, ura3-delta, MSS51-3XHA, pet309::URA3, D273-10b (ρ+, cox1-delta) | This study |
| YC166 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbs1::URA3, D273-10B (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| YC173 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbp6::LEU2, BY4742 (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| YC174 | Matα, lys2, arg8::hisG, ura3-52, leu2-3,112, cbp6::LEU2, D273-10B (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
| YC175 | Matα, his3-delta1, leu2-delta0, lys2-delta0, ura3-delta0, arg8::URA3, cbs1::HIS3MX6, BY4742 (ρ+, ΔΣai, cobΔ::ARG8m) | This study |
a mtDNA refers to mitochondrial genome.
b ΔΣai refers to the intronless COX1 gene.
c +/− corresponds presence (+) or absence (−) of the allele in both chromosomes in the diploid strain.
Mitochondrial transformation
The construct COB(1–385)::pARG8m was made by PCR amplification of three fragments: one included the last 351 bp of the COB orf and another covered the first 470 bp of the ARG8m orf encoding the Arg8 mitochondrial targeting signal (22). These two products were used as templates for fusion PCR (48). The fusion product was cut with XhoI and EcoRI and cloned into equally digested pBluescript SK(+) vector to obtain plasmid pYCV55. The third PCR product included the complete ARG8m orf and 625pb of COB 3′-UTR; this product was cloned in pYCV55 digested with NcoI and EcoRI. The resulting plasmid, pYCV56, was transformed by high-velocity microprojectile bombardment into the rho0 strain NAB69 (49). Transformants were selected by their ability to rescue growth in media lacking arginine when mated with the strain NB71 (49). Transformants with the COB(1–385)::ARG8m plasmid were mated with XPM201 (derived from D273-10b lab strain) (50) and AGG26 (derived from BY4742 lab strain), and haploid cytoductants were selected for their ability to grow in media lacking arginine. Correct integration of the COB(1–385)::ARG8m constructs into mtDNA was confirmed by PCR and DNA sequencing.
Analysis of mitochondrial proteins
Mitochondria were isolated from cells grown in YPGal media until late log phase. The cells were disrupted with glass beads or by zymolyase 20T treatment as described (51). The proteins were resolved by SDS-PAGE on 12% gels (52) or 16% in the presence of 6 m urea (53), transferred to a polyvinylidene fluoride (PVDF) membrane and detected by immunoblotting with horseradish peroxidase–conjugated antibodies to HA (Roche) or the indicated rabbit polyclonal antibodies: anti-Qcr7, anti-Qcr8, anti-Cor1, anti-Cor2, anti-Rip1, and anti-Qcr10 (Vicenzo Zara); anti-Rip1, anti-Cor1, and anti-Mrp20 (Rosemary Stuart); anti-citrate synthase and anti-Mrp51 (Thomas D. Fox); and anti-Cytb. Secondary goat IgG anti-mouse or anti-rabbit (Santa Cruz Biotechnology) conjugated to horseradish peroxidase was detected with the Pierce ECL (Thermo Scientific) or ImmobilonTM Western Chemiluminescent HRP Substrate (Millipore). All loaded proteins were normalized by protein quantification using the Lowry method (54).
Protein aggregation analysis
Protein aggregation analysis was performed as described previously (5). Samples (100 μg) of mitochondrial protein were washed with 250 mm sorbitol, 50 mm Bis-Tris and lysed with 1% digitonin, 30 mm Tris, pH 7.4, 200 mm KCl, 5 mm EDTA, and 0.5 mm PMSF for 30 min on ice. After ultracentrifugation at 4 °C for 30 min at 100,000 × g in a TLA 120.2 rotor, supernatant was recovered into a new tube (S1). The resultant pellet was lysed with 1% Triton, 30 mm Tris, pH 7.4, 200 mm KCl, 5 mm EDTA, and 0.5 mm PMSF for 5 min on ice. Pellet (P) and supernatant (S2) were separated by ultracentrifugation at 4 °C for 30 min at 100,000 × g in a TLA 120.2 rotor. Supernatants S1 and S2 were mixed (S) and treated with TCA for protein precipitation.
Heme b analysis
Total heme extraction was performed as previously described (55). Samples (700 μg) of Histodenz (Sigma) purified mitochondrial protein were treated with 300 μl acidic acetone (3% HCl) and incubated for 5 min at room temperature. Supernatant was recovered after centrifugation for 5 min at 16,200 × g. Supernatant was mixed with 1% trifluoroacetic acid in a proportion 1:1. HPLC analysis was performed as described (56) in a Beckman HPLC unit with System Gold. Supernatant was injected onto a 4.6- by 250-μm SunFire C18 5-mm column (Waters). Hemes were eluted from the column at a flow rate of 1 ml/min using a 0–100% gradient of acetonitrile containing 1% of trifluoroacetic acid. Elution of heme compounds was monitored at 400 nm. Purified heme b (Sigma) was used as standard to determine elution time. For heme b analysis, two different mitochondria purifications were used as biological replicates with one technical replicate. For each biological replicate, heme b extractions and HPLC analysis were made with one technical replicate (n = 3). Statistical analysis was made by one-way analysis of variance followed by Bonferroni post hoc (GraphPad Prism software, version 6.0). Two-sided adjusted p values for multiple comparisons are presented.
Synthesis of mitochondrial proteins
In vivo labeling of cells in the presence of [35S]methionine and cycloheximide was performed as previously described (16). After 15 or 40 min of pulse labeling, the cells were chilled on ice and disrupted by vortexing with glass beads to obtain mitochondria by centrifugation. Mitochondrial proteins were resolved on a 16% polyacrylamide gel, transferred to a PVDF membrane, and analyzed with a Typhoon 8600 phosphorimaging device (GE Healthcare).
Blue native electrophoresis
BN-PAGE was performed as described previously (29). Samples (100 μg) of mitochondrial protein were washed with 250 mm sorbitol, 50 mm Bis-Tris and lysed with 750 mm aminocaproic acid, 50 mm Bis-Tris, and either digitonin or n-dodecyl β-d-maltoside on a protein detergent relation of 2:1 and 1:2, respectively, for 15 min (n-dodecyl β-d-maltoside) or 30 min (digitonin) on ice. Mitochondrial extracts were cleared at 16,200 × g for 12 min, and the supernatants were mixed with 2.5 μl of 5% Coomassie solution (750 mm aminocaproic acid, 50 mm Bis-Tris). Extracts were loaded on a 5% to 12% polyacrylamide gel and transferred to a PVDF membrane. The proteins were detected by immunoblotting with the indicated antibodies. In-gel CcO activity was performed after BN-PAGE using 0.04% diaminobenzidine (Sigma–Aldrich) and 0.02% of horse heart cytochrome c (Sigma–Aldrich) in phosphate buffer, pH 7.4 (57).
RNA immunoprecipitation assay
This technique was performed as previously described (30). Mitochondria (500 μg) were lysed with 0.7% n-dodecyl β-d-maltoside, 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, RNaseOUT (Invitrogen), and Minicomplete protease inhibitors (Roche). The solubilized fractions were incubated with an anti-HA antibody coupled to protein A-Sepharose (GE Healthcare). Immunoprecipitates were washed twice with 500 μl of lysis buffer and twice with 1 ml of 20 mm HEPES-KOH, pH 7.4, and then resuspended in 150 μl of the same buffer. One-third of the precipitate fractions were saved for Western blotting analysis, and the remainder was used for RNA extraction. RNA from total and immunoprecipitated fractions was extracted by incubation with TRIzol® reagent (Invitrogen). 20 ng of RNA were treated with 1 unit of DNase I (Invitrogen). The first strand of cDNAs were prepared by the addition of primers for COB or VAR1 in the presence of SuperScript III reverse transcriptase (Invitrogen). The resulting cDNAs were used as PCR templates to amplify COB or VAR1 5′-UTRs. Note that under these conditions, RT-PCRs are not quantitative.
Sucrose fractionation of mitochondrial lysates
Samples (500 μg) of mitochondrial protein were lysed with 1% digitonin, 10 mm MgOAc, 50 mm NaCl, 20 mm HEPES-KOH, pH 7.4, and 1 mm PMSF for 30 min on ice and clarified by centrifugation at 16,200 × g for 10 min. Supernatants were loaded into a discontinuous sucrose gradient of 40, 30, and 20% containing 0.1% digitonin, 10 mm MgOAc, 20 mm DTT, 10 mm Tris-HCl, pH 7.4, and 0.5 mm PMSF. Sucrose gradients were ultracentrifuged at 145,000 × g in a SW-55Ti rotor for 2 h at 4 °C. Fractions of 600 μl were taken and TCA-treated for protein precipitation. Proteins were resolved by SDS-PAGE, transferred to PVDF membranes, and detected by immunoblotting with the indicated antibodies.
Imaging and figure edition
For better visualization, all figures from pictures, scanning, or phosphorimaging were adjusted in contrast and brightness using the Adobe Photoshop software. Pictures of serial dilutions, agarose gels, in gel activity, and Coomassie Blue gels from Figs. 1 (G and H), 3B, 6D, and 7B and Figs. S1 (A–C), S2 (A and B), and S3 were taken by a camera. In vivo labeling from Figs. 1 (A, B, and F), 6 (A and C), and 7 (A and C) and Fig. S4 (A and B) was visualized by exposure to a storage phosphor screen and then scanned in a Typhoon FLA700 or Typhoon 9400 (GE Healthcare). Western blotting images from Figs. 1E, 2 (A and B), 2C, 3 (A and C), 5 (A–C), and 6 (B and D) and Fig. S2A were visualized by exposure to a film and then scanned in a scanner Color LaserJet Pro MFP M477fmw (HP).
Author contributions
A. E. G.-G. data curation; A. E. G.-G. and A. Z.-O. formal analysis; A. E. G.-G., Y. C.-V., A. Z.-O., D. R. W., and X. P.-M. investigation; A. E. G.-G. and X. P.-M. writing-original draft; Y. C.-V. project administration; Y. C.-V., A. Z.-O., and D. R. W. writing-review and editing; D. R. W. and X. P.-M. supervision; X. P.-M. conceptualization; X. P.-M. funding acquisition.
Supplementary Material
Acknowledgments
We thank Thomas D. Fox, Rosemary Stuart, Martin Ott, and Vicenzo Zara for the gift of antisera; Daniel Flores-Mireles, Gabriel del Río-Guerra, and Teresa Lara-Ortiz for the gift of yeast deletion strains; Thomas D. Fox for helpful insights of tetrad segregation analysis; Emma Bertha Gutiérrez-Cirlos, Tecilli Cabellos-Avelar, Ana Paula García-García, Mi-Young Jeong, Hyoung J. Kim, Miriam Vázquez-Acevedo, Guadalupe Códiz-Huerta, Minerva Mora-Cabrera, and Laura Ongay-Larios for technical assistance; and Claudia Rivera-Cerecedo and Héctor Malagón-Rivero for technical assistance obtaining antisera.
This work was supported by Consejo Nacional de Ciencia y Tecnología Grant 47514 (to X. P.-M.) and Fellowship 255917 (to A. E. G.-G.) and Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Universidad Nacional Autónoma de México Grants IN204414 and IN209217 (to X. P.-M.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S4.
- Cytb
- cytochrome b
- CcO
- cytochrome c oxidase
- Cytc1
- cytochrome c1
- BN
- blue native
- HA
- hemagglutinin
- PVDF
- polyvinylidene fluoride.
References
- 1. Zara V., Conte L., and Trumpower B. L. (2009) Biogenesis of the yeast cytochrome bc1 complex. Biochim. Biophys. Acta 1793, 89–96 10.1016/j.bbamcr.2008.04.011 [DOI] [PubMed] [Google Scholar]
- 2. Conte A., Papa B., Ferramosca A., and Zara V. (2015) The dimerization of the yeast cytochrome bc1 complex is an early event and is independent of Rip1. Biochim. Biophys. Acta 1853, 987–995 10.1016/j.bbamcr.2015.02.006 [DOI] [PubMed] [Google Scholar]
- 3. Atkinson A., Khalimonchuk O., Smith P., Sabic H., Eide D., and Winge D. R. (2010) Mzm1 influences a labile pool of mitochondrial zinc important for respiratory function. J. Biol. Chem. 285, 19450–19459 10.1074/jbc.M110.109793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Atkinson A., Smith P., Fox J. L., Cui T. Z., Khalimonchuk O., and Winge D. R. (2011) The LYR protein Mzm1 functions in the insertion of the Rieske Fe/S protein in yeast mitochondria. Mol. Cell. Biol. 31, 3988–3996 10.1128/MCB.05673-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui T. Z., Smith P. M., Fox J. L., Khalimonchuk O., and Winge D. R. (2012) Late-stage maturation of the Rieske Fe/S protein: Mzm1 stabilizes Rip1 but does not facilitate its translocation by the AAA ATPase Bcs1. Mol. Cell. Biol. 32, 4400–4409 10.1128/MCB.00441-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crivellone M. D., Wu M. A., and Tzagoloff A. (1988) Assembly of the mitochondrial membrane system. Analysis of structural mutants of the yeast coenzyme QH2-cytochrome c reductase complex. J. Biol. Chem. 263, 14323–14333 [PubMed] [Google Scholar]
- 7. Gruschke S., Römpler K., Hildenbeutel M., Kehrein K., Kühl I., Bonnefoy N., and Ott M. (2012) The Cbp3–Cbp6 complex coordinates cytochrome b synthesis with bc1 complex assembly in yeast mitochondria. J. Cell Biol. 199, 137–150 10.1083/jcb.201206040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rödel G. (1986) Two yeast nuclear genes, CBS1 and CBS2, are required for translation of mitochondrial transcripts bearing the 5′-untranslated COB leader. Curr. Genet. 11, 41–45 10.1007/BF00389424 [DOI] [PubMed] [Google Scholar]
- 9. Rödel G., Michaelis U., Forsbach V., Kreike J., and Kaudewitz F. (1986) Molecular cloning of the yeast nuclear genes CBS1 and CBS2. Curr. Genet. 11, 47–53 10.1007/BF00389425 [DOI] [PubMed] [Google Scholar]
- 10. Dieckmann C. L., Koerner T. J., and Tzagoloff A. (1984) Assembly of the mitochondrial membrane system. CBP1, a yeast nuclear gene involved in 5′ end processing of cytochrome b pre-mRNA. J. Biol. Chem. 259, 4722–4731 [PubMed] [Google Scholar]
- 11. Islas-Osuna M. A., Ellis T. P., Marnell L. L., Mittelmeier T. M., and Dieckmann C. L. (2002) Cbp1 is required for translation of the mitochondrial cytochrome b mRNA of Saccharomyces cerevisiae. J. Biol. Chem. 277, 37987–37990 10.1074/jbc.M206132200 [DOI] [PubMed] [Google Scholar]
- 12. Krause-Buchholz U., Barth K., Dombrowski C., and Rödel G. (2004) Saccharomyces cerevisiae translational activator Cbs2p is associated with mitochondrial ribosomes. Curr. Genet. 46, 20–28 [DOI] [PubMed] [Google Scholar]
- 13. Kehrein K., Möller-Hergt B. V., and Ott M. (2015) The MIOREX complex: lean management of mitochondrial gene expression. Oncotarget 6, 16806–16807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gruschke S., Kehrein K., Römpler K., Gröne K., Israel L., Imhof A., Herrmann J. M., and Ott M. (2011) Cbp3–Cbp6 interacts with the yeast mitochondrial ribosomal tunnel exit and promotes cytochrome b synthesis and assembly. J. Cell Biol. 193, 1101–1114 10.1083/jcb.201103132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hildenbeutel M., Hegg E. L., Stephan K., Gruschke S., Meunier B., and Ott M. (2014) Assembly factors monitor sequential hemylation of cytochrome b to regulate mitochondrial translation. J. Cell Biol. 205, 511–524 10.1083/jcb.201401009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perez-Martinez X., Broadley S. A., and Fox T. D. (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 10.1093/emboj/cdg566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perez-Martinez X., Butler C. A., Shingu-Vazquez M., and Fox T. D. (2009) Dual functions of Mss51 couple synthesis of Cox1 to assembly of cytochrome c oxidase in Saccharomyces cerevisiae mitochondria. Mol. Biol. Cell 20, 4371–4380 10.1091/mbc.E09-06-0522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barrientos A., Zambrano A., and Tzagoloff A. (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23, 3472–3482 10.1038/sj.emboj.7600358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rak M., and Tzagoloff A. (2009) F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 106, 18509–18514 10.1073/pnas.0910351106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rak M., Su C. H., Xu J. T., Azpiroz R., Singh A. M., and Tzagoloff A. (2016) Regulation of mitochondrial translation of the ATP8/ATP6 mRNA by Smt1p. Mol. Biol. Cell 27, 919–929 10.1091/mbc.E15-09-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Labouesse M., Netter P., and Schroeder R. (1984) Molecular basis of the “box effect,” A maturase deficiency leading to the absence of splicing of two introns located in two split genes of yeast mitochondrial DNA. Eur. J. Biochem. 144, 85–93 10.1111/j.1432-1033.1984.tb08434.x [DOI] [PubMed] [Google Scholar]
- 22. Steele D. F., Butler C. A., and Fox T. D. (1996) Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc. Natl. Acad. Sci. U.S.A. 93, 5253–5257 10.1073/pnas.93.11.5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu M., and Tzagoloff A. (1989) Identification and characterization of a new gene (CBP3) required for the expression of yeast coenzyme QH2-cytochrome c reductase. J. Biol. Chem. 264, 11122–11130 [PubMed] [Google Scholar]
- 24. Kühl I., Fox T. D., and Bonnefoy N. (2012) Schizosaccharomyces pombe homologs of the Saccharomyces cerevisiae mitochondrial proteins Cbp6 and Mss51 function at a post-translational step of respiratory complex biogenesis. Mitochondrion 12, 381–390 10.1016/j.mito.2012.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zara V., Palmisano I., Conte L., and Trumpower B. L. (2004) Further insights into the assembly of the yeast cytochrome bc1 complex based on analysis of single and double deletion mutants lacking supernumerary subunits and cytochrome b. Eur. J. Biochem. 271, 1209–1218 10.1111/j.1432-1033.2004.04024.x [DOI] [PubMed] [Google Scholar]
- 26. Cui T. Z., Conte A., Fox J. L., Zara V., and Winge D. R. (2014) Modulation of the respiratory supercomplexes in yeast: enhanced formation of cytochrome oxidase increases the stability and abundance of respiratory supercomplexes. J. Biol. Chem. 289, 6133–6141 10.1074/jbc.M113.523688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagener N., Ackermann M., Funes S., and Neupert W. (2011) A pathway of protein translocation in mitochondria mediated by the AAA-ATPase Bcs1. Mol. Cell 44, 191–202 10.1016/j.molcel.2011.07.036 [DOI] [PubMed] [Google Scholar]
- 28. Manthey G. M., and McEwen J. E. (1995) The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 14, 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wittig I., Braun H. P., and Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 10.1038/nprot.2006.62 [DOI] [PubMed] [Google Scholar]
- 30. Zamudio-Ochoa A., Camacho-Villasana Y., García-Guerrero A. E., and Pérez-Martínez X. (2014) The Pet309 pentatricopeptide repeat motifs mediate efficient binding to the mitochondrial COX1 transcript in yeast. RNA Biol. 11, 953–967 10.4161/rna.29780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brandt U., Uribe S., Schägger H., and Trumpower B. L. (1994) Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex. J. Biol. Chem. 269, 12947–12953 [PubMed] [Google Scholar]
- 32. Dieckmann C. L., and Tzagoloff A. (1985) Assembly of the mitochondrial membrane system. CBP6, a yeast nuclear gene necessary for synthesis of cytochrome b. J. Biol. Chem. 260, 1513–1520 [PubMed] [Google Scholar]
- 33. Ralser M., Kuhl H., Werber M., Lehrach H., Breitenbach M., and Timmermann B. (2012) The Saccharomyces cerevisiae W303-K6001 cross-platform genome sequence: insights into ancestry and physiology of a laboratory mutt. Open Biol. 2, 120093 10.1098/rsob.120093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., and Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 10.1002/(SICI)1097-0061(19980130)14:2%3C115::AID-YEA204%3E3.0.CO%3B2-2 [DOI] [PubMed] [Google Scholar]
- 35. Sherman F. (1963) Respiration-deficient mutants of yeast: I. Genetics. Genetics 48, 375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim H. J., Khalimonchuk O., Smith P. M., and Winge D. R. (2012) Structure, function, and assembly of heme centers in mitochondrial respiratory complexes. Biochim. Biophys. Acta 1823, 1604–1616 10.1016/j.bbamcr.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ostojić J., Panozzo C., Bourand-Plantefol A., Herbert C. J., Dujardin G., and Bonnefoy N. (2016) Ribosome recycling defects modify the balance between the synthesis and assembly of specific subunits of the oxidative phosphorylation complexes in yeast mitochondria. Nucleic Acids Res. 44, 5785–5797 10.1093/nar/gkw490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Su C. H., McStay G. P., and Tzagoloff A. (2014) The Cox3p assembly module of yeast cytochrome oxidase. Mol. Biol. Cell 25, 965–976 10.1091/mbc.E13-10-0575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. García-Villegas R., Camacho-Villasana Y., Shingú-Vázquez M. A., Cabrera-Orefice A., Uribe-Carvajal S., Fox T. D., and Pérez-Martínez X. (2017) The Cox1 C-terminal domain is a central regulator of cytochrome c oxidase biogenesis in yeast mitochondria. J. Biol. Chem. 292, 10912–10925 10.1074/jbc.M116.773077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ott M., Amunts A., and Brown A. (2016) Organization and regulation of mitochondrial protein synthesis. Annu. Rev. Biochem. 85, 77–101 10.1146/annurev-biochem-060815-014334 [DOI] [PubMed] [Google Scholar]
- 41. Sherman F. (2002) Getting started with yeast. Methods Enzymol. 350, 3–41 10.1016/S0076-6879(02)50954-X [DOI] [PubMed] [Google Scholar]
- 42. Young M. J., and Court D. A. (2008) Effects of the S288c genetic background and common auxotrophic markers on mitochondrial DNA function in Saccharomyces cerevisiae. Yeast 25, 903–912 10.1002/yea.1644 [DOI] [PubMed] [Google Scholar]
- 43. Zitomer R. S., and Lowry C. V. (1992) Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 56, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gaisne M., Bécam A. M., Verdière J., and Herbert C. J. (1999) A “natural” mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Curr. Genet. 36, 195–200 10.1007/s002940050490 [DOI] [PubMed] [Google Scholar]
- 45. Struhl K. (1985) Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl. Acad. Sci. U.S.A. 82, 8419–8423 10.1073/pnas.82.24.8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burke D., Dawson D., and Stearns T. (2000) Methods in Yeast Genetics, pp. 39–72, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 47. Guthrie C., and Fink G. R. (eds). (2002) Guide to Yeast Genetics and Molecular and Cell Biology, p. 23, Academic Press, San Diego [Google Scholar]
- 48. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., and Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 10.1016/0378-1119(89)90358-2 [DOI] [PubMed] [Google Scholar]
- 49. Bonnefoy N., and Fox T. D. (2001) Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol. 65, 381–396 10.1016/S0091-679X(01)65022-2 [DOI] [PubMed] [Google Scholar]
- 50. Shingú-Vázquez M., Camacho-Villasana Y., Sandoval-Romero L., Butler C. A., Fox T. D., and Pérez-Martínez X. (2010) The carboxyl-terminal end of Cox1 is required for feedback assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J. Biol. Chem. 285, 34382–34389 10.1074/jbc.M110.161976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Diekert K., de Kroon A. I., Kispal G., and Lill R. (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 65, 37–51 10.1016/S0091-679X(01)65003-9 [DOI] [PubMed] [Google Scholar]
- 52. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 53. Schägger H., Aquila H., and Von Jagow G. (1988) Coomassie blue-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for direct visualization of polypeptides during electrophoresis. Anal. Biochem. 173, 201–205 10.1016/0003-2697(88)90179-0 [DOI] [PubMed] [Google Scholar]
- 54. Markwell M. A., Haas S. M., Bieber L. L., and Tolbert N. E. (1978) A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87, 206–210 10.1016/0003-2697(78)90586-9 [DOI] [PubMed] [Google Scholar]
- 55. Barros M. H., and Tzagoloff A. (2002) Regulation of the heme A biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett. 516, 119–123 10.1016/S0014-5793(02)02514-0 [DOI] [PubMed] [Google Scholar]
- 56. Bestwick M., Khalimonchuk O., Pierrel F., and Winge D. R. (2010) The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wittig I., Karas M., and Schägger H. (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics 6, 1215–1225 10.1074/mcp.M700076-MCP200 [DOI] [PubMed] [Google Scholar]
- 58. Costanzo M. C., and Fox T. D. (1988) Specific translational activation by nuclear gene products occurs in the 5′ untranslated leader of a yeast mitochondrial mRNA. Proc. Natl. Acad. Sci. U.S.A. 85, 2677–2681 10.1073/pnas.85.8.2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.