Abstract
Protein kinases (PKs) control many aspects of plant physiology by regulating signaling networks through protein phosphorylation. Phototropins (phots) are plasma membrane–associated serine/threonine PKs that control a range of physiological processes that collectively serve to optimize photosynthetic efficiency in plants. These include phototropism, leaf positioning and flattening, chloroplast movement, and stomatal opening. Despite their identification over two decades ago, only a handful of substrates have been identified for these PKs. Progress in this area has been hampered by the lack of a convenient means to confirm the identity of potential substrate candidates. Here we demonstrate that the kinase domain of Arabidopsis phot1 and phot2 can be successfully engineered to accommodate non-natural ATP analogues by substituting the bulky gatekeeper residue threonine for glycine. This approach circumvents the need for radioactivity to track phot kinase activity and follow light-induced receptor autophosphorylation in vitro by incorporating thiophosphate from N6-benzyl-ATPγS. Consequently, thiophosphorylation of phot substrate candidates can be readily monitored when added or co-expressed with phots in vitro. Furthermore, gatekeeper-modified phot1 retained its functionality and its ability to accommodate N6-benzyl-ATPγS as a phosphodonor when expressed in Arabidopsis. We therefore anticipate that this chemical genetic approach will provide new opportunities for labeling and identifying substrates for phots and other related AGC kinases under in vitro and near-native in vivo conditions.
Keywords: Arabidopsis, ATP, protein engineering, protein kinase, photobiology, photoreceptor, chemical genetics, gatekeeper
Introduction
Phosphorylation of tyrosine, threonine, and serine residues is one of the most important posttranslational modifications controlling protein structure, activity, turnover, and subcellular localization (1). Protein phosphorylation is achieved by protein kinases (PKs),4 one of the largest protein families in eukaryotes. PKs are particularly prevalent in plants and regulate a wide range of biological processes, including innate immunity (2), responses to environmental signals (3), and many aspects of plant growth and development (4). Arabidopsis thaliana contains ∼1000 PKs (5), twice the number present in humans (6), and represents 4% of all protein-coding genes. Moreover, around 30% of the proteome in eukaryotes is phosphorylated (7). Therefore, dissecting the complexity of plant PK function and determining their substrate relationships represents a major challenge.
Recent methods have used small molecules to perturb the function of proteins in a way similar to genetic manipulation, hence the name chemical genetics. Such methods can be used to study the function of PKs, combining the specificity of genetics with the flexibility of chemistry, enabling the alteration of a PK in a conditional manner (8). These approaches rely on bio-orthogonal chemical reactions that are not found naturally to trace specific PK–substrate relationships. Specific substrate labeling can be performed by engineering the PK to accommodate bio-orthogonal ATP analogues that can facilitate visualization and identification of substrate targets by immunochemistry using antibodies recognizing the bio-orthogonal label (9). In practice, mutation of the so-called gatekeeper residue within the ATP-binding pocket of the PK can be used to accommodate ATPγS analogues containing a bulky adenine substitution. This leads to thiophosphorylation of proteins by the gatekeeper-engineered kinase and provides a selective means to distinguish specific PK substrates among a large and diverse phosphoprotein pool. We sought to implement this chemical genetic approach to label substrate targets of Arabidopsis phototropins (phots). Only a small number of substrate targets have been identified for these light-activated kinases despite the range of physiological responses they regulate. Hence, chemical genetic approaches such as the gatekeeper system could shed new light on this poorly characterized PK signaling network, which is ultimately important for optimizing photosynthetic productivity (10).
Phots are members of the AGCVIII kinase family (11) and bind to the intracellular side of the plasma membrane (12). Arabidopsis contains two phots (phot1 and phot2) that function as blue light receptors for controlling a variety of processes that serve to promote growth, particularly under low light conditions (12). These include stomatal opening, leaf blade flattening, leaf orientation, chloroplast movements, and phototropism (13). Light regulation of phot kinase activity is mediated by a photosensory region at the N terminus that comprises two light-, oxygen-, or voltage-sensing domains (LOV1 and LOV2) that bind flavin mononucleotide as a UV/blue light–absorbing cofactor (14). Phots can be viewed simply as molecular light switches, where the activity of the C-terminal kinase domain is repressed in darkness and activated upon illumination (12). Light sensing produces conformational changes associated with helical regions neighboring the LOV2 core that lead to an uncoupling of this repression, ATP binding, and, consequently, receptor autophosphorylation (12, 15). Autophosphorylation occurs on multiple residues throughout the protein (16–18) and can be readily detected in vitro by phosphate incorporation from radiolabeled ATP (19, 20). Alternatively, this process can be monitored in vivo by monitoring reduced electrophoretic mobility of the protein following immunoblotting (18, 21, 22). Phosphorylation of two serine residues within the activation loop of the kinase domain of phot1 (Ser-849 and Ser-851) and phot2 (Ser-761 or Ser-763) is essential for their function and signaling (17, 23). The majority of the remaining phosphorylation sites reside within either the N-terminal region upstream of LOV1 or in the linker sequence between LOV1 and LOV2 (12). Although the functional significance of these upstream phosphorylation sites is still largely unknown, phosphorylation at Ser-350, Ser-376, and Ser-410 within the LOV linker region of phot1 can facilitate binding of 14-3-3 regulatory proteins (17, 22). By contrast, 14-3-3 binding to phot2 is reported to involve Ser-747 within the kinase domain. Mutational analysis of Ser-747 suggests that 14-3-3-binding is required for phot2-mediated stomatal opening (24). However, the role of 14-3-3 binding to phot1 has yet to be determined because no impairment of stomatal opening was observed when Ser-350 and Ser-376 were mutated (17).
Although the photochemical and biochemical properties of phots have been well studied, less information is available regarding the signaling events that couple light perception to various biological outcomes. Indeed, the range of physiological responses controlled by phots is in stark contrast to the limited description of their substrates. So far, only four phosphorylation targets have been identified. ABCB19 is phosphorylated by phot1 and appears to contribute to controlling polar auxin transport during hypocotyl phototropism (25). Phytochrome kinase substrate 4 (PKS4) is also phosphorylated by phot1 (26) and, together with other PKS proteins, is involved in phototropism, leaf flattening, and positioning (27–29), whereas blue light signaling 1 (BLUS1) and convergence of blue light and CO2 1 (CBC1) are phosphorylated by phot1 and involved in blue light–induced stomatal opening (30, 31).
To date, a robust and convenient means to identify phosphorylation targets for phots is still lacking. We therefore implemented a chemical genetic method to facilitate specific labeling of phot kinase substrates. We show that the kinase domain of Arabidopsis phots can be engineered successfully to incorporate and process the unnatural ATP analogue N6-benzyl-ATPγS, which contains a bulky adenine modification. Mutation of the gatekeeper residue within the kinase domain does not significantly perturb the activity and biological function of phot1 in Arabidopsis but facilitates thiophosphorylation of substrate targets in vitro in the presence of N6-benzyl-ATPγS. Also, the activity of gatekeeper-engineered phot1 was rendered sensitive to a specific kinase inhibitor. Finally, we show that thiophosphorylation activity of gatekeeper-engineered phot1 can be readily detected in plant extracts close to in vivo native conditions. Consequently, this methodology provides a sensitive and non-radioactive means to monitor and specifically inhibit phot kinase activity in a conditional way and potentially identify new substrate targets for phots.
Results
Gatekeeper identification in Arabidopsis phot1
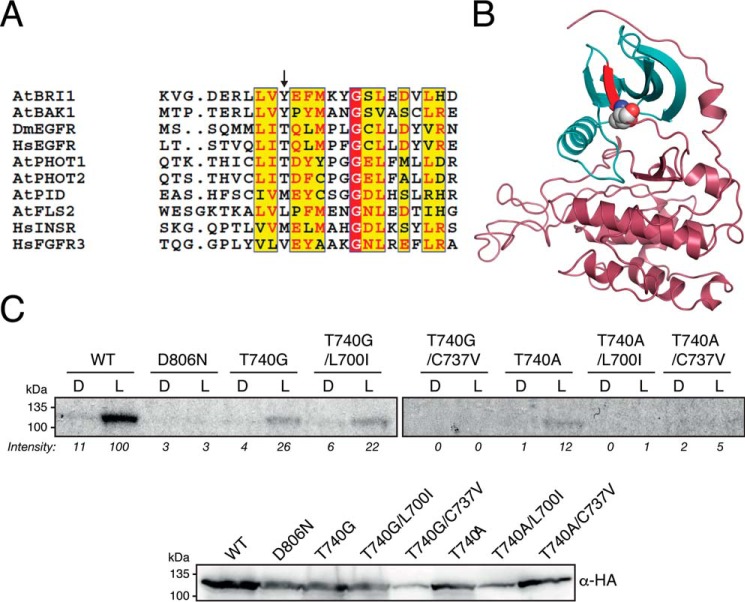
The gatekeeper residue generally bears a large side chain that, when mutated to a smaller amino acid, creates an enlarged substrate binding pocket. This residue lines the bottom of the kinase active site and controls the sensitivity of the kinase to ATP-competitive inhibitors (32). Mutation of the gatekeeper residue within the kinase active site has emerged as a promising approach to incorporate unnatural ATP analogues as well as alter kinase selectivity toward binding specific inhibitors (33). By contrast, the native kinase is resistant to binding such analogues because of the presence of the larger gatekeeper residue. To identify the gatekeeper residue in the kinase domain of Arabidopsis phot1, we aligned its amino acid sequence with PK sequences from other organisms in which the identity of the gatekeeper residue had been determined previously. The gatekeeper is typically a methionine or threonine residue (34) that belongs to the ΦΦX+Φ motif (35, 36) where Φ represents a hydrophobic side chain residue, X represents the gatekeeper, and + represents a positively charged residue. Sequence homology to other PKs indicated that Thr-740 was the candidate gatekeeper in the kinase domain of Arabidopsis phot1 (Fig. 1A). From a structural perspective, the gatekeeper is localized at the end of the β5 strand in the N lobe of the kinase domain (34) and interacts with a group of hydrophobic residues that form a cluster called the R spine, which regulates the kinase (37). Both of these properties were observed for Thr-740 of phot1 after homology modeling (Fig. 1B).
Figure 1.
Threonine 740 is the gatekeeper residue of Arabidopsis phot1. A, amino acid sequence alignment of the kinase domain of A. thaliana (At) phototropin 1 with other protein kinases for which the gatekeeper residue has been identified (AtBRI1 and AtFLS2 (35)) or for which the gatekeeper has been engineered to accommodate bulky ATP analogues (Homo sapiens (Hs) AMPKa2 (51) and Saccharomyces cerevisiae (Sc) CDK1 (63)). B, structural model of the kinase domain of Arabidopsis phot1 generated by homology using SwissModel and displayed using PyMOL. Secondary structures are displayed as a schematic. The N-terminal lobe of the kinase domain is colored blue, whereas the C-terminal lobe is colored pink. The β5 strand in the N-terminal lobe is shown in red, and threonine 740 is shown in spheres colored by atom (carbon is gray, oxygen is red, nitrogen is blue). C, cell-free expression and autophosphorylation analysis of wildtype phot1 and different gatekeeper mutants in the presence of [γ-32P]ATP. Reactions were carried out in the absence (D) or presence of 10 s of white light (L). Samples were separated between two SDS-PAGE gels but exposed to autoradiography simultaneously (top panel). The extent of autophosphorylation in the autoradiogram was quantified by ImageJ, and the band intensity (percent) relative to the phot1 light-treated sample is shown below each lane. An immunoblot of phot1 protein levels using anti-HA antibody is shown below.
Engineering phot1 to process bulky ATP analogues
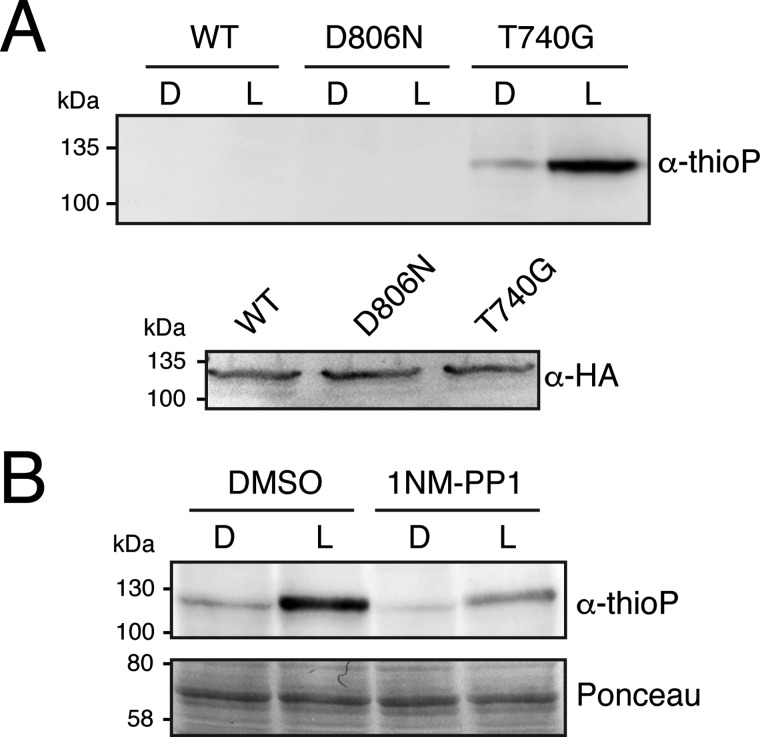
Mutation of the gatekeeper to alanine or glycine in over 80 PKs has been successful in accommodating N6-benzyl-ATPγS (9, 36). We therefore mutated Thr-740 to alanine or glycine to determine whether this variant of phot1 could accept this ATP analogue. A cell-free in vitro transcription/translation system was used to express phot1 in the presence of flavin mononucleotide. Autophosphorylation activity was initially examined in the presence of radiolabeled ATP to assess the effect of different gatekeeper mutations. Light-induced autophosphorylation was detected for phot1 in this system but not for the kinase inactive mutant D806N (Fig. 1C). Both the T740A and T740G variants of phot1 displayed light-induced receptor kinase activity, but at a reduced level relative to the wildtype (Fig. 1C). Additional active site mutations were therefore introduced (L700I and C737V) in an attempt to compensate for this decrease in activity, as reported previously for the gatekeeper engineering of other PKs (38). However, no activity enhancement was observed for phot1-T740A or phot1-T740G in the presence of these mutations (Fig. 1C). Our subsequent efforts therefore focused on phot1-T740G because it appeared to display higher activity than the T740A mutant (Fig. 1C). We rationalized that mutation of the gatekeeper should enable phot1 to accommodate the bulky ATP analogue N6-benzyl-ATPγS and promote receptor thiophosphorylation, which can be detected by immunoblotting with anti-thiophosphoester antibody following chemical alkylation of the incorporated thiophosphates (9). Indeed, light-induced autophosphorylation was readily detected for phot1-T740G when N6-benzyl-ATPγS was used as a phosphate donor (Fig. 2A). In contrast, no signal was evident in reactions containing wildtype phot1.
Figure 2.
Phot1 containing a modified gatekeeper residue (T740G) can accommodate N6-benzyl-ATPγS and undergo thiophosphorylation in vitro. A, immunoblot of a kinase assay containing cell-free expressed wildtype phot1, phot1-D806N, or phot1-T740G in the presence of N6-benzyl-ATPγS. Reactions were carried out in the absence (D) or presence of 20 s of white light (L), and thiophosphorylation was detected using anti-thiophosphoester antibody (α-thioP). An immunoblot analysis of phot1 protein levels using anti-HA antibody is shown below. B, phot1-T740G thiophosphorylation in the presence of the kinase inhibitor 1-NM-PP1 or DMSO as a control. Ponceau staining of cell-free expression reactions is shown below to indicate equal protein loading.
In addition to accommodating unnatural ATP analogues, the enlarged ATP-binding site created by the gatekeeper mutation also permits small-molecule inhibitors to bind selectively to the kinase active site. The cell-permeable kinase inhibitor 4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine (1-NM-PP1) has been reported to effectively block the activity of gatekeeper-mutated kinases, including members of the Src family of tyrosine kinases (39), the tomato resistance protein serine/threonine kinase Pto (40), and MAPK4 in Arabidopsis (41). Similarly, we found that thiophosphorylation of phot1 was reduced when treated with 1-NM-PP1 prior to blue light stimulation, demonstrating that this approach can also be used to reduce the activity of gatekeeper-modified phot1 (Fig. 2B).
Phot-mediated thiophosphorylation of BLUS1
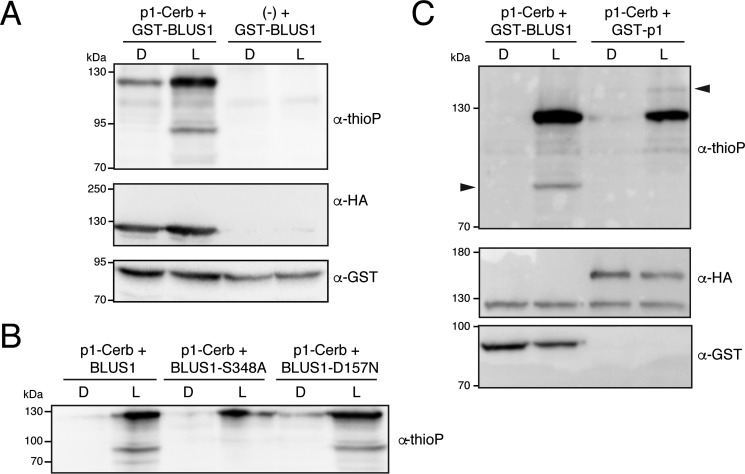
The above results confirmed Thr-740 as the gatekeeper residue in phot1 and showed how mutation of this residue can be used to produce a low-background, non-radioactive means to monitor phot1 kinase activity in the presence of the bio-orthogonal ATP analogue N6-benzyl-ATPγS. We therefore named this phot1 variant Cerberus, after the “keeper” of the underworld in Greek mythology. We next determined whether phot1-Cerberus could be used to phosphorylate known substrate targets in addition to tracking receptor autophosphorylation. BLUS1 is a guard cell–specific PK that is phosphorylated by phot1 on serine 348, a signaling event that is required to regulate stomatal opening (31). BLUS1 was therefore expressed in Escherichia coli as a glutathione S-transferase (GST) fusion, purified by affinity chromatography, and used as a substrate target for in vitro kinase assays. In addition to phot1 autophosphorylation, thiophosphorylation of BLUS1 was clearly detected following blue light irradiation (Fig. 3A). Mutation of Ser-348 in BLUS1 abolished its thiophosphorylation by phot1-Cerberus, whereas thiophosphorylation of the kinase inactive variant of BLUS1 (D157N) was still clearly evident (Fig. 3B). Together, these results are consistent with previous findings (31) and demonstrate that the engineered phot1-Cerberus retains native substrate specificity.
Figure 3.
Phot1-Cerberus (p1-T740G) directly phosphorylates BLUS1 in vitro. A, thiophosphorylation analysis of in vitro kinase assays containing phot1-Cerberus (p1-Cerb) and GST-BLUS1 or the cell-free expression extract alone (−) with GST-BLUS1. Reactions were carried out in the absence (D) or presence of 20 s of white light (L), and thiophosphorylation was detected using anti-thiophosphoester antibody (α-thioP). Phot1-Cerberus thiophosphorylation is evident, ∼130 kDa, whereas GST-BLUS1 is shown above 70 kDa. Blots were probed with anti-HA antibody to detect phot1 or anti-GST antibody to detect BLUS1 (shown below). B, thiophosphorylation analysis of in vitro kinase assays containing phot1-Cerberus together with GST-BLUS1-S348A or GST-BLUS1-D157N. C, thiophosphorylation analysis of in vitro kinase assays containing phot1-Cerberus co-expressed with either GST-BLUS1 or GST-phot1 (GST-p1). Reactions were carried out in the absence or presence of 20 s of white light, and thiophosphorylation was detected using anti-thiophosphoester antibody. Phot1-Cerberus thiophosphorylation is evident, ∼130 kDa, whereas thiophosphorylation of GST-phot1 and GST-BLUS1 is indicated above 130 and 70 kDa, respectively, by arrowheads. Blots were probed with anti-HA antibody to detect phot1 and GST-phot1 or with anti-GST antibody to detect BLUS1 (shown below).
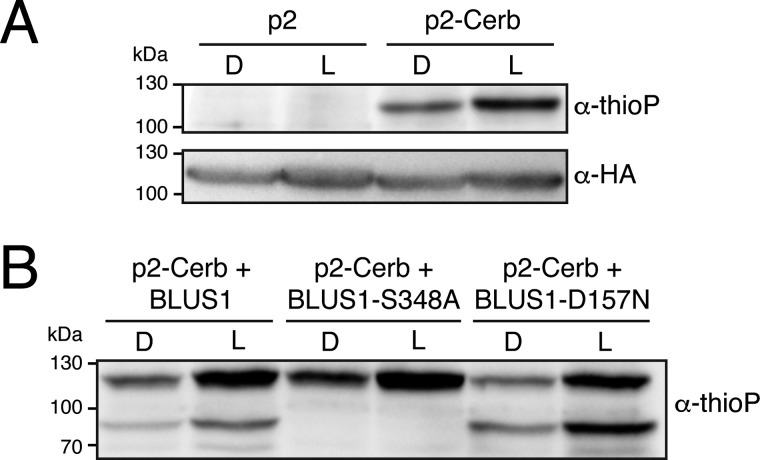
BLUS1 has also been shown to act as a substrate for phot2 kinase activity (42). To determine whether this substrate commonality between phot1 and phot2 could be detected using the gatekeeper system, we generated a Cerberus variant for phot2 (T654G). Light-induced autophosphorylation of phot2-Cerberus was observed using this approach, although the level of kinase activity in the dark was evidently higher compared with that observed for phot1 (Fig. 4A). Blue light–induced thiophosphorylation of GST-BLUS1 and its D157N variant were clearly evident in the presence of phot2-Cerberus (Fig. 4B). By contrast, no substrate phosphorylation was observed for BLUS1 S348A (Fig. 4B). Together, these results confirm that BLUS1 is a direct substrate of phot2 (42) and indicate that phot1 and phot2 share the same molecular mechanism to regulate BLUS1 activity through serine 348 phosphorylation.
Figure 4.
Phot2-Cerberus phosphorylates BLUS1 at Ser-348. A, thiophosphorylation analysis of in vitro kinase assays containing wildtype phot2 (p2) or phot2-Cerberus (p2-Cerb). Reactions were carried out in the absence (D) or presence of 20 s of white light (L), and thiophosphorylation was detected using anti-thiophosphoester antibody (α-thioP). Phot2-Cerberus thiophosphorylation is evident above 100 kDa. Blots were probed with anti-HA antibody to detect phot2 protein levels (shown below). B, thiophosphorylation analysis of in vitro kinase assays containing phot2-Cerberus together with GST-BLUS1-S348A or GST-BLUS1-D157N. Reactions were carried out as in A. Thiophosphorylation of GST-BLUS1 is shown above 70 kDa, whereas phot2 thiophosphorylation is evident above 100 kDa.
Detection of phot substrate phosphorylation by co-expression
Our results so far demonstrate the utility of the gatekeeper system combined with in vitro transcription/translation to monitor phot substrate phosphorylation. However, a potential limitation of this approach is the requirement to produce and purify the substrate from E. coli. To circumvent the need for recombinant protein expression, we investigated whether substrate targets could be co-expressed along with phot1 in the cell-free expression system. We found that GST-BLUS1 was thiophosphorylated in a light-dependent manner when co-expressed with phot1-Cerberus (Fig. 3C).
We have also shown previously that phot1 exhibits light-dependent dimerization (43) and that receptor autophosphorylation can occur intermolecularly between two distinct phot molecules (43, 44). Consistent with these findings, light-dependent thiophosphorylation of GST-phot1 was detected when co-expressed with phot1-Cerberus (Fig. 3C). Similar results were also found when a kinase-inactive version of GST-phot1 (D806N) was used as substrate (Fig. S1). Taken together, these findings demonstrate the utility of co-expression in the cell-free expression system combined with gatekeeper engineering to rapidly confirm the identity of phot substrate targets.
Phot1-Cerberus is functional in Arabidopsis
To determine whether phot1-Cerberus retains biological activity in vivo, we stably expressed this variant as a translational fusion to GFP in the phot1 phot2 double mutant of Arabidopsis under the control of the native PHOT1 promoter. Five independent homozygous lines were isolated for analysis, and transgenic Arabidopsis expressing wildtype phot1-GFP was used as a control (21, 45). The abundance of phot1 in three of the phot1-Cerberus lines (10, 14 and 21) was comparable with phot1 protein levels detected both in wildtype seedlings and seedlings expressing phot1-GFP (Fig. S2). Comparable levels of GFP fluorescence at the cell periphery of etiolated hypocotyls were also detected in lines expressing phot1-Cerberus (Fig. S3), consistent with the association of phot1 with the plasma membrane (46).
Examination of phot1 protein levels in extracts isolated from dark-grown (etiolated) seedlings showed that in vivo blue light treatment was sufficient to induce a reduced electrophoretic mobility shift for phot1-Cerberus (Fig. 5A), which is characteristic of receptor autophosphorylation (47). We therefore concluded that, despite the gatekeeper mutation, phot1-Cerberus is still capable of processing endogenous ATP as a phosphate donor. These findings are consistent with our in vitro analysis (Fig. 1C).
Figure 5.
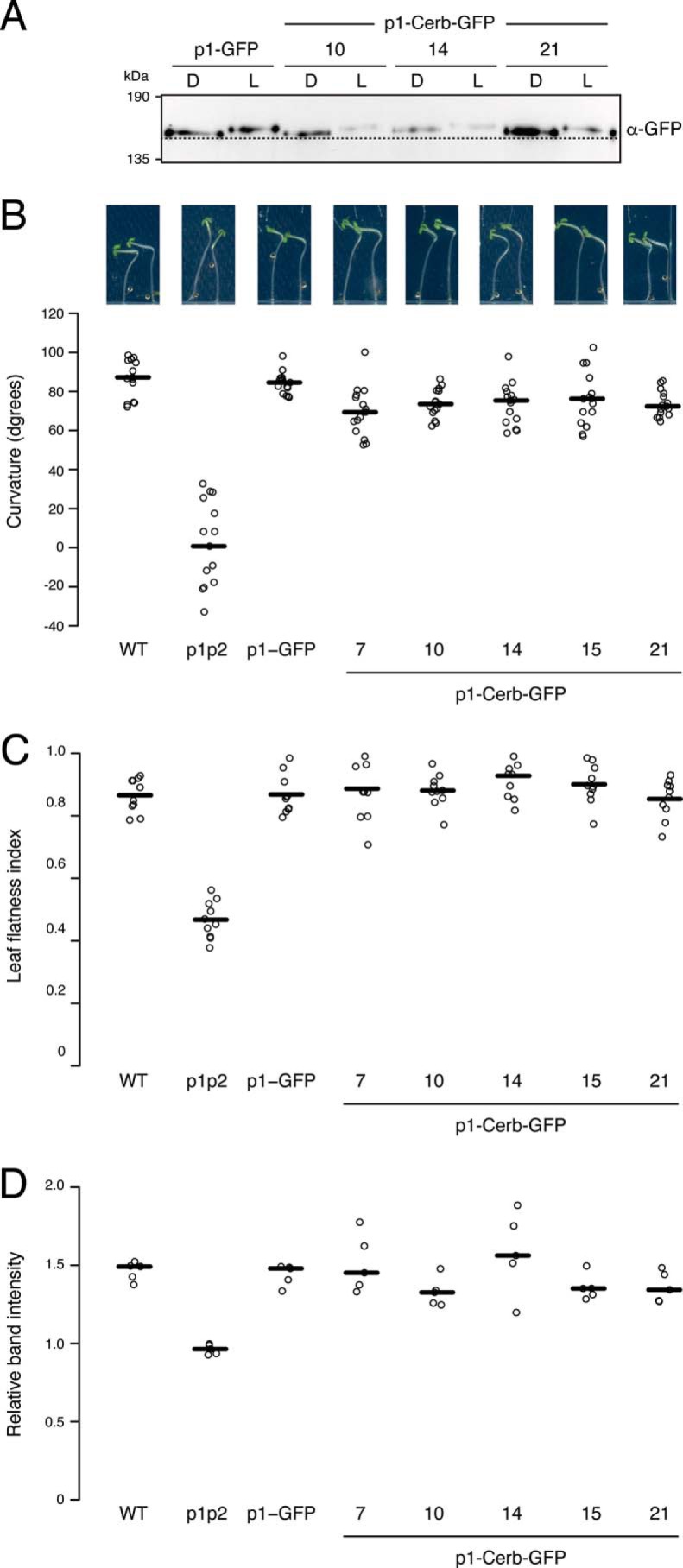
Phot1-Cerberus is functional when expressed in the phot1 phot2 double mutant. A, immunoblot analysis of phot1 protein abundance in transgenic Arabidopsis expressing phot1-GFP (p1-GFP) and three independent lines expressing phot1-Cerberus fused to GFP (p1-Cerb-GFP). Protein extracts were isolated from 3-day-old etiolated seedlings either maintained in darkness (D) or irradiated with 20 μmol m−2 s−1 of blue light for 15 min (L) and probed with anti-GFP antibody. The dashed line indicates the lowest mobility edge. B, phototropic responses of 3-day-old etiolated wildtype (gl-1) seedlings, the phot1 phot2 double mutant (p1p2), plants expressing p1-GFP, and five independent lines expressing p1-Cerb-GFP. Measurements represent the angle made by the tip of the hypocotyl with the vertical. Representative seedling images are shown above. C, leaf flatness index (ratio of the leaf area before and after leaf uncurling) for the wildtype (gl-1), the phot1 phot2 double mutant (p1p2), plants expressing p1-GFP, and five independent lines expressing p1-Cerb-GFP. D, chloroplast accumulation measurements in the same genotypes as in A. Detached leaves were illuminated with 1.5 μmol m−2 s−1 of blue light through a 1-mm slit for 90 min, and the slit band intensity was quantified. The relative band intensity shown is expressed as the ratio of the irradiated to the non-irradiated areas. Ratios above 1 indicate accumulation. For B–D, quantifications of plant responses are represented as the median (horizontal line) and individual data points (open circles).
To further verify the functionality of phot1-Cerberus in vivo, we examined whether it could restore several different phot1-mediated responses in Arabidopsis. All lines expressing phot1-Cerberus displayed a robust hypocotyl phototropism response when irradiate at 10 μmol m−2 s−1 for 24 h (Fig. 5B), indicating that phot1-Cerberus retains functionality for this response. In addition to its phototropic impairment, the rosette leaves of the phot1 phot2 double mutant display a downward-curled or epinastic phenotype (46). Each of the phot1-Cerberus lines was able to complement the epinastic leaf phenotype of the phot1 phot2 double mutant (Fig. 5C), again demonstrating functionality. Phot1 and phot2 are also known to control chloroplast accumulation movement, which allows plants to maximize light capture for photosynthesis (48). This response can be visualized by the slit band assay, where a dark band appears on the exposed section of the leaf when irradiated with low fluence rates of blue light (1.5 μmol m−2 s−1) through a 1-mm slit (49). Although chloroplasts failed to accumulate on the upper cell surface in leaves from the phot1 phot2 mutant, a dark band was readily observed in lines expressing phot1-Cerberus (Fig. 5D). Hence, these findings indicate that, as found for phototropism and leaf expansion, phot1-Cerberus retains an ability to utilize endogenous ATP to mediate this blue light response.
Monitoring phot1-Cerberus activity ex vivo
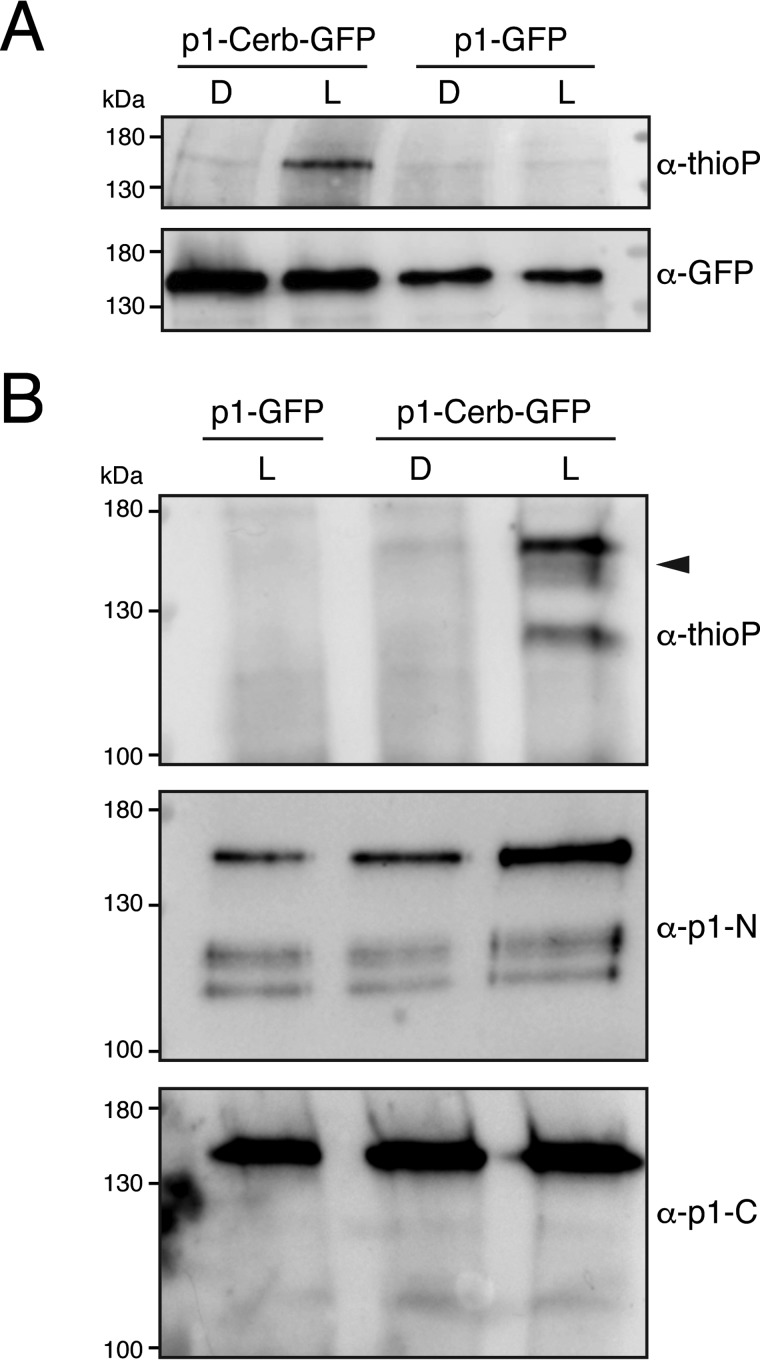
Given the functionality observed for phot1-Cerberus in planta, we rationalized that it was still capable of phosphorylating endogenous substrate targets in vivo. Phot1-Cerberus–expressing lines could therefore offer a new means to monitor substrate phosphorylation. To explore this possibility, we performed ex vivo thiophosphorylation screening in protein extracts isolated from etiolated seedlings expressing phot1-Cerberus. Light-dependent autophosphorylation of phot1-Cerberus was readily detected in microsomal membrane fractions in the presence of N6-benzyl-ATPγS but was absent in microsomes isolated from phot1-GFP–expressing seedlings (Fig. 6A). Thus, these findings demonstrate that phot1-Cerberus retains its ability to utilize the bulky ATP analogue as a phosphodonor when expressed in Arabidopsis.
Figure 6.
Phot1-Cerberus thiophosphorylation in plant protein extracts. A, thiophosphorylation analysis of in vitro kinase assays on microsomal extracts isolated from transgenic Arabidopsis expressing phot1-Cerberus fused to GFP (p1-Cerb-GFP) or phot1-GFP (p1-GFP). Reactions were carried out in the absence (D) or presence of 20 s of white light (L), and thiophosphorylation was detected using anti-thiophosphoester antibody (α-thioP). Phot1-Cerberus thiophosphorylation is evident above 130 kDa. Blots were probed with anti-GFP antibody to detect phot1 protein levels (shown below). B, thiophosphorylation analysis of in vitro kinase assays on total protein extracts isolated from the p1-Cerb-GFP or p1-GFP lines. Reactions were carried out as described in A, except that only the light-treated p1-GFP sample was used as a negative control. Phot1-Cerberus thiophosphorylation is evident above 130 kDa. Additional thiophosphorylation products are indicated by the arrowhead. Blots were also probed with anti-phot1 N-terminal (p1-N) or C-terminal (p1-C) antibodies to detect phot1 and its proteolytic cleavage products.
We found that light-dependent thiophosphorylation of phot1-Cerberus could also be detected in total protein extracts isolated from etiolated seedlings (Fig. 6B). Additional light-dependent thiophosphorylation products were also observed in these reactions that ran immediately below phot1-Cerberus. Moreover, probing with antibody raised against the N (19) or C terminus of phot1 (18, 50) suggested that these signals did not result from phot1 proteolysis (Fig. 6B), although we cannot exclude that these signals could correspond to internal fragments that lack both native termini. These findings therefore suggest that the chemical genetic approach described here has the potential to identify phot1 phosphorylation targets ex vivo in addition to tracking receptor phosphorylation.
Discussion
Characterization of signaling networks controlled by plant PKs is a major challenge because of their large number and identical catalytic function, which obscures identifying specific kinase–substrate relationships. Our research focused on identifying the underlying signaling processes associated with a small family of AGC kinases in plants known as phots (phot1 and phot2). Despite regulating a range of responses that collectively optimize photosynthetic productivity, minimal substrate targets have been identified for these light-activated kinases (12). Indeed, genetic screens in Arabidopsis have only been successful in identifying one substrate candidate for these kinases, which is designated BLUS1 (31). Biochemical attempts to isolate further substrate candidates based on yeast two-hybrid screening has met with minimal success (22, 25). Identifying new substrates remains a major challenge not just for phots but also for other AGC-related kinases.
In this study, we adopted a chemical genetic approach to facilitate tracking phot autophosphorylation as well as substrate phosphorylation. This approach relies on engineering the kinase domain of phots to accommodate the bio-orthogonal ATP analogue N6-benzyl-ATPγS, which enables thiophosphorylation of substrate candidates. This was achieved by replacing the threonine gatekeeper residue within the ATP-binding site with an amino acid bearing a smaller side chain (Fig. 1). This synthetic approach has been successful in identifying kinase substrate relationships in several different organisms. For example, Banko et al. (51) have exploited this methodology to thiophosphorylate and label substrates of AMPKα2 in cultured mammalian cells. These thiophosphorylation targets were subsequently immunopurified and identified by mass spectrometry, uncovering 28 unknown substrates. More recently, Leissing et al. (52) have used this gatekeeper approach to identify substrate targets for Arabidopsis mitogen-activated protein kinases.
Here we demonstrate that engineering of the kinase domain of Arabidopsis phots can be successfully employed to create the analogue-sensitive variants designated phot1- and phot2-Cerberus. These variants are capable of light-induced receptor thiophosphorylation in the presence of N6-benzyl-ATPγS (Figs. 2–4). Moreover, our analysis in transgenic Arabidopsis has shown that phot1-Cerberus retains its functionality when expressed in the phot-deficient double mutant (Fig. 5), likely because of its ability to still process natural ATP as a phosphodonor (Fig. 1C). Functionality was still observed in transgenic lines expressing low levels of phot1-Cerberus (Fig. S1 and Fig. 5) These findings are consistent with previous reports showing that very low levels of phot1 are capable of restoring receptor responsiveness in the phot1 phot2 double mutant (21, 50, 53, 54). Further detailed physiological analysis, however, would be required to determine whether lines expressing phot1-Cerberus exhibit quantitative differences in receptor activity compared with wildtype phot1.
Although phot1-Cerberus is capable of using endogenous ATP to facilitate light-induced receptor autophosphorylation in vitro (Fig. 1C) and in vivo (Fig. 5A), it is also capable of processing N6-benzyl-ATPγS as a thiophosphodonor ex vivo (Fig. 6). This chemical genetic approach should aid further probing of phot kinase–substrate relationships without the need for radioactivity. Furthermore, we found that the engineered kinase of phot1-Cerberus is able to accommodate the small-molecule inhibitor 1-NM-PP1 and reduce its activity in vitro (Fig. 2B). These findings demonstrate the potential for using a chemical genetic strategy to inhibit phot1 function. Such inhibitors could be used in a conditional manner to facilitate spatial dissection of phot1 signaling in planta through localized application. Such an approach has been shown to be particularly useful when kinase loss-of-function strategies are not possible because of embryo lethality (55).
Our analysis demonstrates that the gatekeeper approach provides a means to conveniently monitor phot1 kinase activity in protein extracts from Arabidopsis (Fig. 6). These initial findings also suggest that thiophosphorylation of proteins other than phot1-Cerberus can be detected under these near native in vivo conditions. Future studies will now be focused on attempting to isolate and identity these thiophosphorylated products using immunoprecipitation and mass spectrometry approaches. Two purification strategies have been successfully employed for such purposes. The first involves immunoprecipitation using anti-thiophosphoester antibodies following thiophosphate alkylation (9). The second uses covalent capture of thiol-containing peptides with iodoacetyl resin (56). Although this approach can lead to the binding of cysteine-containing peptides in addition to thiophosphorylated peptides, the latter can be released preferentially through oxidation of the phosphate diester bond.
The power of the gatekeeper approach used here relies on the ability to directly and specifically label substrates of a single protein kinase in a complex protein mixture, an important step toward large-scale elucidation of PK–substrate relationships. Although our studies on transgenic Arabidopsis were performed ex vivo, in vivo labeling strategies may be possible by employing non-ionic detergents to permeabilize the plasma membrane and facilitate the uptake of ATP analogues while maintaining cell viability (51, 57). We anticipate that this approach will complement recent large-scale phosphoproteomic methods (30) to tackle the challenge of characterizing phot kinase signaling in Arabidopsis. Notwithstanding, we envisage that the co-expression strategy adopted here will provide a rapid and robust means to confirm the identity of newly isolated substrate targets for phot and related AGC kinases.
Experimental procedures
Plant material and growth conditions
Wildtype (gl-1 ecotype Columbia) and phot1–5 phot2–1 mutants have been described previously (58, 59), as have transgenic Arabidopsis expressing phot1-GFP (21). Seeds were planted on soil or on Murashige and Skoog salts with 0.8% agar (w/v). After 4 °C treatment for 2 days, seeds were grown in a controlled environment room (Fitotron, Weiss-Gallenkamp) under 16/8 h, 22/18 °C light/dark cycles.
Plasmid construction
Plasmids for cell-free expression were constructed using the plasmid pSP64 (Promega). A cDNA fragment encoding full-length Arabidopsis PHOT1 was PCR-amplified using a plasmid containing PHOT1 or PHOT1-D806N cDNA as a template. PCR fragments were inserted into the pSP64 vector using HindIII and PstI restriction sites, and the resulting constructs were confirmed by DNA sequencing. The hemagglutinin (HA) tag was encoded and included in the reverse oligonucleotide. Similarly, PHOT2 cDNA fused to HA was PCR-amplified and inserted into the pSP64 poly(A) vector via HindIII and PstI restriction sites. cDNAs encoding GST-BLUS1 and GST-phot1 were PCR-amplified from cDNA templates (31, 43) and cloned into the pSP64 vector by Gibson assembly (New England Biolabs) via HindIII and PstI restriction sites. Mutation of the gatekeeper codon was performed by sited-directed mutagenesis (Agilent). The plant transformation vector encoding PHOT1-T740G-GFP under control of the PHOT1 promoter was generated by replacing the coding sequence of wildtype PHOT1 in the binary vector pEZR(K)-LN via EcoRI and BamHI sites (21).
Transformation of Arabidopsis
Vectors for plant expression were transformed into the phot1–5 phot2–1 double mutant using a modified version of the floral dip method (60). Homozygous plants lines containing a single transgene locus were selected for analysis based on segregation of kanamycin resistance.
Cell-free expression
Reactions were performed using the TnT® SP6 High-Yield Wheat Germ Protein Expression System (Promega) with 4–8 μg of vector for a 20-μl reaction. For co-expression, 2–4 μg of each plasmid was used. Flavin mononucleotide (Sigma-Aldrich) was added to a final concentration of 10 μm. Protein expression was carried out in the dark at room temperature (20–25 °C) for 2 h.
Expression and purification of GST-BLUS1
GST-BLUS1, GST-BLUS1-D157N, and GST-BLUS1-S348A were expressed as described previously (31) and purified from E. coli using the Glutathione HiCap Kit (Qiagen) following the manufacturer's instructions. Purified proteins were dialized overnight at 4 °C in a buffer containing 37.5 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 1 mm DTT. Dialysis was performed using Slide-A-Lyzer dialysis cassettes (Thermo Fisher Scientific).
Protein extraction from Arabidopsis
Proteins were extracted from 3-day-old etiolated seedlings under dim red light on ice in extraction buffer (50 mm Tris-HCl (pH 7.5), 300 mm sucrose, 150 mm NaCl, 10 mm potassium acetate, 5 mm EGTA, 1 mm DTT, 1 mm phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor mixture (Roche)). Microsomal membranes were isolated by centrifugation at 45,000 rpm for 45 min and resuspended in extraction buffer.
In vitro phosphorylation assays
Radiolabeled kinase assays were performed as described previously in Ref. 61. Reactions were prepared under dim red light and performed in a final volume of 20 μl containing 10 μl of cell-free expression extract or 10 μg of protein extract from Arabidopsis. N6-benzyl-ATPγS (Jena Bioscience) was used at a final concentration of 100 μm for phot1 autophosphorylation or 500 μm for substrate phosphorylation and for phosphorylation screening with plant protein extracts. Light-treated samples were illuminated for 20 s with white light at a total fluence of 60,000 μmol m−2. Reactions were incubated at room temperature for 5 min and stopped by adding EDTA (pH 8.0) to a final concentration of 20 mm. Thiophosphorylated molecules were alkylated for 1–2 h at room temperature by adding p-nitrobenzyl mesylate (Abcam) to a final concentration of 2.5 mm. For inhibitor studies, 1-NM-PP1 (CAS 221244-14-0, Merck Millipore) at a final concentration of 100 μm, or solvent (DMSO) was added to the reactions before light exposure.
Western blot analysis
Proteins were detected by Western blot analysis on a nitrocellulose membrane with rabbit anti-thiophosphoester monoclonal antibody (clone 51-8, Abcam), anti-phot1 C-terminal antibody (50), anti-phot1 N-terminal antibody (19), rabbit anti-GFP-HRP monoclonal antibody (Miltenyi Biotech), rat anti-hemagglutinin monoclonal antibody (Roche), or mouse anti-GST monoclonal antibody (Novagen). Blots were developed with anti-rabbit or anti-mouse horseradish peroxidase (HRP)–conjugated secondary antibody (Promega) or anti-rat HRP–conjugated secondary antibody (Dako Denmark A/S) and Pierce ECL Plus Western blotting substrate (Thermo Fisher Scientific).
Measurement of phototropic curvature
Three-day-old etiolated seedlings grown vertically on Petri dishes containing Murashige and Skoog agar were exposed to unilateral light provided by a fluorescent lamp filtered through one layer of blue Plexiglas for 24 h. Images of the seedlings were captured using a scanner, and hypocotyl curvature was measured using ImageJ.
Chloroplast accumulation
Measurements of chloroplast positioning were performed as described previously (20). Rosette leaves detached from 3-week-old plants grown on soil were placed on agar plates and irradiated with 1.5 μmol m−2 s−1 blue light through a 1-mm slit or placed into darkness for 1 h. The plates were placed on a white light transilluminator and photographed. Band intensities were quantified using ImageJ, and the relative band intensities were expressed as the ratio of the irradiated to the non-irradiated areas.
Leaf expansion
Measurement of leaf expansion was carried out as described previously (20). Plants were grown on soil under white light at 70 μmol m−2 s−1 for 4 weeks. The fifth rosette leaves were detached and photographed. The leaves were then uncurled manually and photographed again. Leaf areas were measured before and after uncurling, and the ratio of the curled to uncurled area was designated the leaf expansion index. The leaf area was measured using ImageJ.
Confocal microscopy
Imaging of GFP-tagged proteins in Arabidopsis seedlings was visualized using a laser-scanning confocal microscope (Zeiss LSM 510) as described previously (20).
Homology modeling of the phot1 kinase domain
Modeling was performed using the automated online SwissModel server (62). Based on coverage and percentage of sequence identity, several templates were selected to build independent structural models. The template giving the best score for model quality was chosen (PDB code 4GV1 for PKB α serine/threonine kinase).
Author contributions
J. S. and J. M. C. conceptualization; J. S., P. H., T. W., G. G., J. P., and J. M. C. formal analysis; J. S. and J. M. C. supervision; J. S., P. H., T. W., G. G., and J. P. investigation; J. S. and J. M. C. writing-original draft; J. S., P. H., T. W., G. G., J. P., and J. M. C. writing-review and editing; J. M. C. funding acquisition; J. M. C. project administration.
Supplementary Material
Acknowledgment
We thank Atsushi Takemiya for providing GST-BLUS1 expression constructs.
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (BB/J016047/1 and BB/M002128/1 to J. M. C.). This work was also supported by a Dobbie Smith Scholarship (to G. G.), an Erasmus+ Scholarship (to P. H.), and a Doctoral Training Programme Studentship from the College of Medical, Veterinary, and Life Sciences, University of Glasgow (to T. W.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- PK
- protein kinase
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- phot
- phototropin
- GST
- glutathione S-transferase
- cDNA
- complementary DNA
- HA
- hemagglutinin
- HRP
- horseradish peroxidase
- 1-NM-PP1
- 4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d]pyrimidine.
References
- 1. Jensen O. N. (2006) Interpreting the protein language using proteomics. Nat. Rev. Mol. Cell Biol. 7, 391–403 10.1038/nrm1939 [DOI] [PubMed] [Google Scholar]
- 2. Schwessinger B., and Ronald P. C. (2012) Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482 10.1146/annurev-arplant-042811-105518 [DOI] [PubMed] [Google Scholar]
- 3. Mishra N. S., Tuteja R., and Tuteja N. (2006) Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452, 55–68 10.1016/j.abb.2006.05.001 [DOI] [PubMed] [Google Scholar]
- 4. Katsir L., Davies K. A., Bergmann D. C., and Laux T. (2011) Peptide signaling in plant development. Curr. Biol. 21, R356–R364 10.1016/j.cub.2011.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lehti-Shiu M. D., and Shiu S. H. (2012) Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R Soc. Lond. B Biol. Sci. 367, 2619–2639 10.1098/rstb.2012.0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Manning G., Whyte D. B., Martinez R., Hunter T., and Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 10.1126/science.1075762 [DOI] [PubMed] [Google Scholar]
- 7. Ahn N. G., and Resing K. A. (2001) Toward the phosphoproteome. Nat. Biotechnol. 19, 317–318 10.1038/86687 [DOI] [PubMed] [Google Scholar]
- 8. Alaimo P. J., Shogren-Knaak M. A., and Shokat K. M. (2001) Chemical genetic approaches for the elucidation of signaling pathways. Curr. Opin. Chem. Biol. 5, 360–367 10.1016/S1367-5931(00)00215-5 [DOI] [PubMed] [Google Scholar]
- 9. Allen J. J., Li M., Brinkworth C. S., Paulson J. L., Wang D., Hübner A., Chou W. H., Davis R. J., Burlingame A. L., Messing R. O., Katayama C. D., Hedrick S. M., and Shokat K. M. (2007) A semisynthetic epitope for kinase substrates. Nat. Methods 4, 511–516 10.1038/nmeth1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takemiya A., Inoue S., Doi M., Kinoshita T., and Shimazaki K. (2005) Phototropins promote plant growth in response to blue light in low light environments. Plant Cell 17, 1120–1127 10.1105/tpc.104.030049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rademacher E. H., and Offringa R. (2012) Evolutionary adaptations of plant AGC kinases: from light signaling to cell polarity regulation. Front. Plant Sci. 3, 250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Christie J. M., Blackwood L., Petersen J., and Sullivan S. (2015) Plant flavoprotein photoreceptors. Plant Cell Physiol. 56, 401–413 10.1093/pcp/pcu196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Christie J. M. (2007) Phototropin blue-light receptors. Annu. Rev. Plant Biol. 58, 21–45 10.1146/annurev.arplant.58.032806.103951 [DOI] [PubMed] [Google Scholar]
- 14. Christie J. M., Salomon M., Nozue K., Wada M., and Briggs W. R. (1999) LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc. Natl. Acad. Sci. U.S.A. 96, 8779–8783 10.1073/pnas.96.15.8779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christie J. M., Gawthorne J., Young G., Fraser N. J., and Roe A. J. (2012) LOV to BLUF: flavoprotein contributions to the optogenetic toolkit. Mol. Plant 5, 533–544 10.1093/mp/sss020 [DOI] [PubMed] [Google Scholar]
- 16. Deng Z., Oses-Prieto J. A., Kutschera U., Tseng T. S., Hao L., Burlingame A. L., Wang Z. Y., and Briggs W. R. (2014) Blue light-induced proteomic changes in etiolated Arabidopsis seedlings. J. Proteome Res. 13, 2524–2533 10.1021/pr500010z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inoue S., Kinoshita T., Matsumoto M., Nakayama K. I., Doi M., and Shimazaki K. (2008) Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 5626–5631 10.1073/pnas.0709189105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sullivan S., Thomson C. E., Lamont D. J., Jones M. A., and Christie J. M. (2008) In vivo phosphorylation site mapping and functional characterization of Arabidopsis phototropin 1. Mol. Plant 1, 178–194 10.1093/mp/ssm017 [DOI] [PubMed] [Google Scholar]
- 19. Christie J. M., Reymond P., Powell G. K., Bernasconi P., Raibekas A. A., Liscum E., and Briggs W. R. (1998) Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701 10.1126/science.282.5394.1698 [DOI] [PubMed] [Google Scholar]
- 20. Sullivan S., Petersen J., Blackwood L., Papanatsiou M., and Christie J. M. (2016) Functional characterization of Ostreococcus tauri phototropin. New Phytol. 209, 612–623 10.1111/nph.13640 [DOI] [PubMed] [Google Scholar]
- 21. Sullivan S., Takemiya A., Kharshiing E., Cloix C., Shimazaki K. I., and Christie J. M. (2016) Functional characterization of Arabidopsis phototropin 1 in the hypocotyl apex. Plant J. 88, 907–920 10.1111/tpj.13313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sullivan S., Thomson C. E., Kaiserli E., and Christie J. M. (2009) Interaction specificity of Arabidopsis 14-3-3 proteins with phototropin receptor kinases. FEBS Lett. 583, 2187–2193 10.1016/j.febslet.2009.06.011 [DOI] [PubMed] [Google Scholar]
- 23. Inoue S., Matsushita T., Tomokiyo Y., Matsumoto M., Nakayama K. I., Kinoshita T., and Shimazaki K. (2011) Functional analyses of the activation loop of phototropin2 in Arabidopsis. Plant Physiol. 156, 117–128 10.1104/pp.111.175943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tseng T. S., Whippo C., Hangarter R. P., and Briggs W. R. (2012) The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell 24, 1114–1126 10.1105/tpc.111.092130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Christie J. M., Yang H., Richter G. L., Sullivan S., Thomson C. E., Lin J., Titapiwatanakun B., Ennis M., Kaiserli E., Lee O. R., Adamec J., Peer W. A., and Murphy A. S. (2011) phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 9, e1001076 10.1371/journal.pbio.1001076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Demarsy E., Schepens I., Okajima K., Hersch M., Bergmann S., Christie J., Shimazaki K., Tokutomi S., and Fankhauser C. (2012) Phytochrome kinase substrate 4 is phosphorylated by the phototropin 1 photoreceptor. EMBO J. 31, 3457–3467 10.1038/emboj.2012.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boccalandro H. E., De Simone S. N., Bergmann-Honsberger A., Schepens I., Fankhauser C., and Casal J. J. (2008) PHYTOCHROME KINASE SUBSTRATE1 regulates root phototropism and gravitropism. Plant Physiol. 146, 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Carbonnel M., Davis P., Roelfsema M. R., Inoue S., Schepens I., Lariguet P., Geisler M., Shimazaki K., Hangarter R., and Fankhauser C. (2010) The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol. 152, 1391–1405 10.1104/pp.109.150441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lariguet P., Schepens I., Hodgson D., Pedmale U. V., Trevisan M., Kami C., de Carbonnel M., Alonso J. M., Ecker J. R., Liscum E., and Fankhauser C. (2006) PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc. Natl. Acad. Sci. U.S.A. 103, 10134–10139 10.1073/pnas.0603799103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hiyama A., Takemiya A., Munemasa S., Okuma E., Sugiyama N., Tada Y., Murata Y., and Shimazaki K. I. (2017) Blue light and CO2 signals converge to regulate light-induced stomatal opening. Nat. Commun. 8, 1284 10.1038/s41467-017-01237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Takemiya A., Sugiyama N., Fujimoto H., Tsutsumi T., Yamauchi S., Hiyama A., Tada Y., Christie J. M., and Shimazaki K. (2013) Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat. Commun. 4, 2094 [DOI] [PubMed] [Google Scholar]
- 32. Liu Y., Bishop A., Witucki L., Kraybill B., Shimizu E., Tsien J., Ubersax J., Blethrow J., Morgan D. O., and Shokat K. M. (1999) Structural basis for selective inhibition of Src family kinases by PP1. Chem. Biol. 6, 671–678 10.1016/S1074-5521(99)80118-5 [DOI] [PubMed] [Google Scholar]
- 33. Salomon D., Zhang C., Shokat K. M., and Sessa G. (2011) Sensitizing plant protein kinases to specific inhibition by ATP-competitive molecules. Methods Mol. Biol. 779, 185–197 10.1007/978-1-61779-264-9_10 [DOI] [PubMed] [Google Scholar]
- 34. Kornev A. P., and Taylor S. S. (2015) Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 40, 628–647 10.1016/j.tibs.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geldner N., and Robatzek S. (2008) Plant receptors go endosomal: a moving view on signal transduction. Plant Physiol. 147, 1565–1574 10.1104/pp.108.120287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lopez M. S., Kliegman J. I., and Shokat K. M. (2014) The logic and design of analog-sensitive kinases and their small molecule inhibitors. Methods Enzymol. 548, 189–213 10.1016/B978-0-12-397918-6.00008-2 [DOI] [PubMed] [Google Scholar]
- 37. Taylor S. S., and Kornev A. P. (2011) Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36, 65–77 10.1016/j.tibs.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang C., Kenski D. M., Paulson J. L., Bonshtien A., Sessa G., Cross J. V., Templeton D. J., and Shokat K. M. (2005) A second-site suppressor strategy for chemical genetic analysis of diverse protein kinases. Nat. Methods 2, 435–441 10.1038/nmeth764 [DOI] [PubMed] [Google Scholar]
- 39. Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D., Wood J. L., Morgan D. O., and Shokat K. M. (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 10.1038/35030148 [DOI] [PubMed] [Google Scholar]
- 40. Salomon D., Bonshtien A., Mayrose M., Zhang C., Shokat K. M., and Sessa G. (2009) Bypassing kinase activity of the tomato Pto resistance protein with small molecule ligands. J. Biol. Chem. 284, 15289–15298 10.1074/jbc.M809724200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brodersen P., Petersen M., Bjørn Nielsen H., Zhu S., Newman M. A., Shokat K. M., Rietz S., Parker J., and Mundy J. (2006) Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47, 532–546 10.1111/j.1365-313X.2006.02806.x [DOI] [PubMed] [Google Scholar]
- 42. Takemiya A., and Shimazaki K. (2016) Arabidopsis phot1 and phot2 phosphorylate BLUS1 kinase with different efficiencies in stomatal opening. J. Plant Res. 129, 167–174 10.1007/s10265-015-0780-1 [DOI] [PubMed] [Google Scholar]
- 43. Kaiserli E., Sullivan S., Jones M. A., Feeney K. A., and Christie J. M. (2009) Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation and endocytosis by blue light. Plant Cell 21, 3226–3244 10.1105/tpc.109.067876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Petersen J., Inoue S. I., Kelly S. M., Sullivan S., Kinoshita T., and Christie J. M. (2017) Functional characterization of a constitutively active kinase variant of Arabidopsis phototropin 1. J. Biol. Chem. 292, 13843–13852 10.1074/jbc.M117.799643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sullivan S., Hart J. E., Rasch P., Walker C. H., and Christie J. M. (2016) Phytochrome A mediates blue-light enhancement of second-positive phototropism in Arabidopsis. Front Plant Sci. 7, 290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sakamoto K., and Briggs W. R. (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14, 1723–1735 10.1105/tpc.003293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Christie J. M., and Murphy A. S. (2013) Shoot phototropism in higher plants: new light through old concepts. Am J. Bot. 100, 35–46 10.3732/ajb.1200340 [DOI] [PubMed] [Google Scholar]
- 48. Kong S. G., and Wada M. (2014) Recent advances in understanding the molecular mechanism of chloroplast photorelocation movement. Biochim. Biophys. Acta 1837, 522–530 10.1016/j.bbabio.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 49. Suetsugu N., Kagawa T., and Wada M. (2005) An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol. 139, 151–162 10.1104/pp.105.067371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cho H. Y., Tseng T. S., Kaiserli E., Sullivan S., Christie J. M., and Briggs W. R. (2007) Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol. 143, 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Banko M. R., Allen J. J., Schaffer B. E., Wilker E. W., Tsou P., White J. L., Villén J., Wang B., Kim S. R., Sakamoto K., Gygi S. P., Cantley L. C., Yaffe M. B., Shokat K. M., and Brunet A. (2011) Chemical genetic screen for AMPKα2 substrates uncovers a network of proteins involved in mitosis. Mol. Cell 44, 878–892 10.1016/j.molcel.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Leissing F., Nomoto M., Bocola M., Schwaneberg U., Tada Y., Conrath U., and Beckers G. J. (2016) Substrate thiophosphorylation by Arabidopsis mitogen-activated protein kinases. BMC Plant Biol. 16, 48 10.1186/s12870-016-0731-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doi M., Shigenaga A., Emi T., Kinoshita T., and Shimazaki K. (2004) A transgene encoding a blue-light receptor, phot1, restores blue-light responses in the Arabidopsis phot1 phot2 double mutant. J. Exp. Bot 55, 517–523 10.1093/jxb/erh044 [DOI] [PubMed] [Google Scholar]
- 54. Preuten T., Blackwood L., Christie J. M., and Fankhauser C. (2015) Lipid anchoring of Arabidopsis phototropin 1 to assess the functional significance of receptor internalization: should I stay or should I go? New Phytol. 206, 1038–1050 10.1111/nph.13299 [DOI] [PubMed] [Google Scholar]
- 55. Xu J., and Zhang S. (2014) RNA interference of plant MAPK cascades for functional studies. Methods Mol. Biol. 1171, 91–103 10.1007/978-1-4939-0922-3_8 [DOI] [PubMed] [Google Scholar]
- 56. Blethrow J. D., Glavy J. S., Morgan D. O., and Shokat K. M. (2008) Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc. Natl. Acad. Sci. U.S.A. 105, 1442–1447 10.1073/pnas.0708966105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trejo-Tapia G., Cuevas-Celis J., Salcedo-Morales G., Trejo-Espino J. L., Arenas-Ocampo M. L., and Jiménez-Aparicio A. (2007) Beta vulgaris L. suspension cultures permeabilized with Triton X-100 retain cell viability and betacyanines production ability: a digital image analysis study. Biotechnol. Prog. 23, 359–363 10.1021/bp0602597 [DOI] [PubMed] [Google Scholar]
- 58. Kagawa T., Sakai T., Suetsugu N., Oikawa K., Ishiguro S., Kato T., Tabata S., Okada K., and Wada M. (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141 10.1126/science.291.5511.2138 [DOI] [PubMed] [Google Scholar]
- 59. Liscum E., and Briggs W. R. (1995) Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7, 473–485 10.1105/tpc.7.4.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Logemann E., Birkenbihl R. P., Ülker B., and Somssich I. E. (2006) An improved method for preparing Agrobacterium cells that simplifies the Arabidopsis transformation protocol. Plant Methods 2, 16 10.1186/1746-4811-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sakai T., Kagawa T., Kasahara M., Swartz T. E., Christie J. M., Briggs W. R., Wada M., and Okada K. (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. U.S.A. 98, 6969–6974 10.1073/pnas.101137598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Allen J. J., Lazerwith S. E., and Shokat K. M. (2005) Bio-orthogonal affinity purification of direct kinase substrates. J. Am. Chem. Soc. 127, 5288–5289 10.1021/ja050727t [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.