Abstract
Cervical cancer is the leading cause of death with gynecological malignancies. We aimed to explore the molecular mechanism of carcinogenesis and biomarkers for cervical cancer by integrated bioinformatic analysis. We employed RNA-sequencing details of 254 cervical squamous cell carcinomas and 3 normal samples from The Cancer Genome Atlas. To explore the distinct pathways, messenger RNA expression was submitted to a Gene Set Enrichment Analysis. Kyoto Encyclopedia of Genes and Genomes and protein–protein interaction network analysis of differentially expressed genes were performed. Then, we conducted pathway enrichment analysis for modules acquired in protein–protein interaction analysis and obtained a list of pathways in every module. After intersecting the results from the 3 approaches, we evaluated the survival rates of both mutual pathways and genes in the pathway, and 5 survival-related genes were obtained. Finally, Cox hazards ratio analysis of these 5 genes was performed. DNA replication pathway (P < .001; 12 genes included) was suggested to have the strongest association with the prognosis of cervical squamous cancer. In total, 5 of the 12 genes, namely, minichromosome maintenance 2, minichromosome maintenance 4, minichromosome maintenance 5, proliferating cell nuclear antigen, and ribonuclease H2 subunit A were significantly correlated with survival. Minichromosome maintenance 5 was shown as an independent prognostic biomarker for patients with cervical cancer. This study identified a distinct pathway (DNA replication). Five genes which may be prognostic biomarkers and minichromosome maintenance 5 were identified as independent prognostic biomarkers for patients with cervical cancer.
Keywords: cervical cancer, bioinformatic analysis, DNA replication, survival analysis, biomarker, minichromosome maintenance 5
Introduction
Cervical cancer is the second most prevalent cancer and the fourth primary cause of cancer-related mortality in females around the world.1 Among all the patients with cervical cancer, more than 90% of the pathological type had been squamous cell cancer.2,3 With the development of a widespread program termed the Papanicolaou test for cervical cytologic screening, cervical cancer mortality has substantially been reduced. Infection of certain types of the human papillomavirus (HPV) has been proved to be the greatest risk factor for cancer,4,5 but whether viral infection can cause or promote the pathological process of cervical cancer alone is still controversial.4 Previous studies revealed that mutations of various genes like tumor protein p53 (TP53),6 PIK3CA, 7 phosphatase and tensin homolog (PTEN),8,9 as well as aberrant copy number alterations of many oncogenes and tumor suppressors may be involved in the development and progression of cervical carcinoma.10 Radical surgical treatment and radiotherapy are effective treatment methods; however, up to one-third of these patients will develop progressive or recurrent tumors.11,12 Although some clinicopathological parameters, such as grade and International Federation of Gynecology and Obstetrics (FIGO) stage, and several biomarkers, such as cancer antigen 125, have been proposed for recurrence prediction,13–16 insufficient sensitivity and specificity of those parameters limits their applications in clinic. Therefore, there is a pressing requirement to find out novel markers or models to predict the prognosis of cervical cancer, especially squamous cancer.
As a refined biological process, DNA replication exists in all kinds of living organisms and is the basis of biological inheritance. The role of DNA replication has been proved to be essential in the process of tumorigenesis and development in previous studies. Tomasetti et al proposed the hypothesis that random mistakes in DNA replication (R) were the third major contributors to cancer, which was verified by the investigation of the incidence of 17 different cancer types among 69 countries (the top 2 major contributors were identified to be environmental factors [E] and heredity [H] formerly).17 Because of the complicated clinical prognosis of cervical cancer, DNA replication may also have a decisive position in the cervical cancer pathological process. Furthermore, it is also indispensable to investigate the functional and clinical significance of genes in DNA replication pathway in cervical cancer.
Bioinformatic analysis is an effective and practical method to predict the possible oncogenes and gene set variation in tumorigenesis or other pathological process. In our study, we investigated the messenger RNA (mRNA) expression differences between cancer and normal tissues basing on the data sets from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/). Based on previous findings and the bioinformatic analysis performed, we evaluated the clinical significance of DNA replication pathway further. Our result highlighted 5 genes (minichromosome maintenance 2 [MCM2], MCM4, MCM5, proliferating cell nuclear antigen [PCNA], and ribonuclease H2 subunit A [RNASEH2A]) participating in DNA replication pathway to be promising prognosis markers for these patients. Finally, MCM5 was observed as an independent prognosis biomarker of overall survival (OS) in patients with cervical cancer.
Materials and Methods
Data Sets
Transcriptome profiling data and prognostic data of cervical squamous cancer were obtained from TCGA consortium. In total, 254 cervical squamous cell carcinoma samples and 3 normal cervical squamous samples details were obtained. Oncomine (version 4.5; www.oncomine.org) is an open database which contained 715 data sets and 86 733 samples. The Human Protein Atlas (THPA) program is a scientific research program exploring the whole human proteome, including protein expression profile of 44 different normal, 20 different cancer tissues, and 56 cell lines (www.proteinatlas.org).18 Four data sets in Oncomine: Scotto cervix (21 cervix squamous epithelium and 32 cervical squamous cell carcinoma), Pyeon Multi-cancer (8 cervix uteri and 20 cervical cancer), Biewenga cervix (5 cervix uteri and 40 cervical cancer), and Zhai cervix (10 cervix squamous epithelium and 21 cervical cancer) and protein expression level in clinical specimens from THPA were chosen to validate the results obtained from TCGA.
Identification of Differentially Expressed Genes
The transcriptome profiling data files for analysis were systemized and transferred into a .txt file which included expression and prognosis data using a Perl order line. Then, package “edgeR” of Bioconductor (version 3.4) was applied in RStudio (version 3.3.2) to screen out the differentially expressed genes (DEGs) with a fold change >2, and P value was defined as .05 to be statistically significant. Volcano plot was drafted in RStudio and genes whose fold-change >2 along with false discovery rate (FDR) <0.1 were marked with red (upregulated) and green (downregulated). Hierarchical clustering analysis was applied to categorize the data into 2 groups with similar expression patterns between cervical cancer and normal cervical epithelia.
Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis of DEGs
Kyoto Encyclopedia of Genes and Genomes (KEGG), which can link genomic information with higher level function information, is a knowledge base for systematic and comprehensive analysis of gene functions. For another, mapping of user’s gene to the related biological annotation in the Database for Annotation, Visualization and Integrated Discovery (DAVID) database (version 6.8; http://david.ncifcrf.gov) is an important foundation for any high-throughput gene functional analysis. As a plug-in app of Cytoscape, ClueGO (version 2.2.3) is also an excellent visualization tool to perform KEGG enrichment analysis basing on different database from DAVID. To analyze the DEGs at the functional level, KEGG pathway analysis was applied using both DAVID online tool and ClueGo. P < .05 was considered statistically significant; pathways including 4 more DEGs were showed in ClueGo-KEGG figures.
Pathway Gene Signatures Analyzed Using Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) is a computational method for exploring whether a given gene set is significantly enriched in a group of gene markers ranked by their relevance with a phenotype of interest. The curated KEGG pathway V5.2 data set was used to compare the impaired pathways in normal and cervical cancer samples. In addition, the gene sets less than 15 genes or more than 500 genes were excluded. The phenotype label was set as cervical cancer versus control. The t-statistic mean of the genes was computed in each KEGG pathway using a permutation test with 1000 replications. The upregulated pathways were defined by a normalized enrichment score (NES) >0 and the downregulated pathways were defined by an NES <0. Pathways with an FDR P value ≤.1 were considered significantly enriched.
Integration of Protein–Protein Interaction Network and Module Analysis
Cytoscape is a software which can visualize and integrate complex networks. We mapped the DEGs to the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) to evaluate the protein–protein interaction (PPI) information. The experimentally validated interactions with a combined score >0.4 were selected. Another plug-in app Molecular Complex Detection (MCODE) was used to screen the modules of PPI network in Cytoscape. The criteria were set as follows: MCODE scores >6 and number of nodes >10. Moreover, the functional and pathway enrichment analyses were performed for DEGs in the modules. P < .05 was considered significantly different.
Survival Analysis of Distinct Gene Sets and Genes in Cervical Cancer
A single-sample (ssGSEA) score based on the expression of genes within each KEGG pathway was calculated for all cervical cancer samples in the TCGA cohort. Cancers with a ssGSEA score above the cohort median were considered to have high expression of a particular KEGG pathway gene set, and cancers with ssGSEA score below the cohort median were considered to have a low expression of the KEGG pathway gene set. For each KEGG pathway, a Kaplan-Meier curve was constructed to compare the OS of patients with cervical cancer with a high expression of the KEGG pathway against low expression of the KEGG pathway. A log-rank test was used to calculate the statistical significance of the difference in survival between the 2 groups. A Cox univariate hazard ratio (HR) was calculated as a measure of the magnitude of the difference in survival between the 2 groups.19,20 Survival analysis of single genes in survival-related gene sets was conducted to evaluate the clinical significance of each genes. “Survival” package was applied in RStudio, and a Kaplan-Meier curves were mapped based on the follow-up data from TCGA. A log-rank test was used to calculate the statistical significance of the difference in survival between the 2 groups, and the cutoff of P value defined as .05 was considered to be significant.
Statistical Analysis
The statistical analysis was applied using SPSS version 22.0 for Windows. Student t test was conducted to determine the mean of I-IIA versus IIB-IV for MCM5. Univariate and multivariate Cox regression models were performed to analyze the 5 survival-related genes from DNA replication pathway. The differences were considered significant at P value <.05.
Results
Workflow for the Identification of Key Pathways and Genes in Cervical Cancer
We compared cancer and normal tissues to identify significant signatures and prognostic markers for cervical cancer. The workflow of the bioinformatic process is outlined in Figure 1A. Clinical details of the patients with cervical squamous cancer from TCGA cohort including the age of diagnosis, vital status, tumor status, tumor size (T) stage, lymph node (N) stage, metastasis (M) stage, and tumor grade are summarized in Supplemental Table 1.
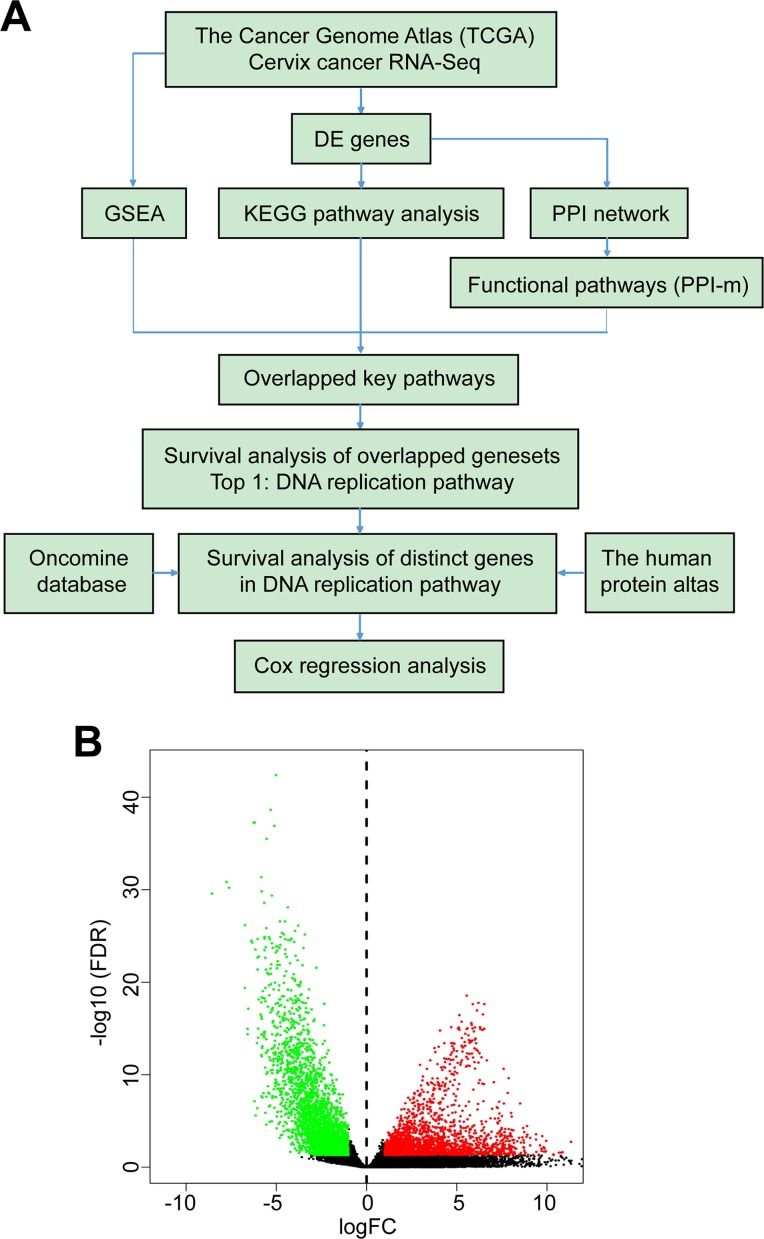
Figure 1.
Identification of DEGs. A, Flowchart of the bioinformatics work included in this research. Data were collected from TCGA database, Oncomine, and also THPA. B, Volcano plot of DEGs, each dot represents 1 gene, and colorized genes had statistical significance (log-fold change >1; red, upregulated genes; green, downregulated genes). DEG indicates differentially expressed genes; TCGA, The Cancer Genome Atlas; THPA, The Human Protein Atlas.
Identification of DEGs
A total of 3818 DEGs were identified, of which 1574 were upregulated and 2244 were downregulated. Genes whose fold-change >2 along with FDR <0.1 were marked with red (upregulated) and green (downregulated) in the volcano plot as shown in Figure 1B.
Investigation of Significant Pathways in Cervical Cancer
Upregulated and downregulated DEGs were observed to excavate the most significantly enriched pathways by KEGG pathway analysis. The most significant pathways enriched in the upregulated DEGs were cell cycle, systemic lupus erythematosus (SLE), alcoholism, DNA replication, and viral carcinogenesis, while the downregulated DEGs were enriched in focal adhesion, vascular smooth muscle contraction, neuroactive ligand-receptor interaction, calcium signaling pathway, and cyclic guanosine monophosphate–protein kinase G signaling pathway (Table 1).
Table 1.
KEGG Pathway Analysis of Differentially Expression Genes Associated With Cervical Cancer.a
| Pathway ID | Name | Count | P Value | Gene ID |
|---|---|---|---|---|
| Upregulated DEGs | ||||
| hsa04110 | Cell cycle | 35 | 1.84E−17 | BUB1/BUB1B/CCNA2/CCNB1/CCNB2/CCNE1/CCNE2/CDC20/CDC25A/CDC25C/CDC45/CDC6/CDC7/CDK1/CDKN2A/CDKN2B/CHEK1/E2F1/E2F2/ESPL1/MAD2L1/MCM2/MCM4/MCM5/ORC1/ORC6/PCNA/PKMYT1/PLK1/PTTG1/RBL1/SFN/SKP2/SMC1B/TTK |
| hsa05322 | Systemic lupus erythematosus | 28 | 8.27E−11 | HIST1H2AD/HIST1H2AG/HIST1H2AI/HIST1H2AJ/HIST1H2AL/HIST1H2AM/HIST1H2BC/HIST1H2BD/HIST1H2BF/HIST1H2BG/HIST1H2BH/HIST1H2BI/HIST1H2BJ/HIST1H2BL/HIST1H2BO/HIST1H3B/HIST1H3C/HIST1H3D/HIST1H3F/HIST1H3G/HIST1H3H/HIST1H3J/HIST1H4D/HIST1H4E/HIST2H2AB/HIST2H2BF/HIST2H4A/HIST3H2BB |
| hsa05034 | Alcoholism | 33 | 8.29E−11 | CALML3/CALML5/GNGT1/GRIN1/GRIN2D/HIST1H2AD/HIST1H2AG/HIST1H2AI/HIST1H2AJ/HIST1H2AL/HIST1H2AM/HIST1H2BC/HIST1H2BD/HIST1H2BF/HIST1H2BG/HIST1H2BH/HIST1H2BI/HIST1H2BJ/HIST1H2BL/HIST1H2BO/HIST1H3B/HIST1H3C/HIST1H3D/HIST1H3F/HIST1H3G/HIST1H3H/HIST1H3J/HIST1H4D/HIST1H4E/HIST2H2AB/HIST2H2BF/HIST2H4A/HIST3H2BB |
| hsa03030 | DNA replication | 12 | 1.10E−07 | DNA2/FEN1/LIG1/MCM2/MCM4/MCM5/PCNA/POLD1/POLE/POLE2/RFC4/RNASEH2A |
| hsa05203 | Viral carcinogenesis | 30 | 1.12E−07 | ATP6V0D2/BAK1/CCNA2/CCNE1/CCNE2/CCR8/CDC20/CDK1/CDKN2A/CDKN2B/CHEK1/HIST1H2BC/HIST1H2BD/HIST1H2BF/HIST1H2BG/HIST1H2BH/HIST1H2BI/HIST1H2BJ/HIST1H2BL/HIST1H2BO/HIST1H4D/HIST1H4E/HIST2H2BF/HIST2H4A/HIST3H2BB/IRF7/PMAIP1/RBL1/SKP2/SYK |
| Downregulated pathways | ||||
| hsa04510 | Focal adhesion | 43 | 1.27E−12 | AKT3/BCL2/CAV1/CHAD/COL1A2/COL4A6/COL6A1/COL6A2/COL6A3/COL6A6/FLNA/FLNC/FLT1/FLT4/FYN/HGF/ILK/ITGA11/ITGA7/ITGA8/ITGA9/ITGB3/KDR/LAMA2/LAMA4/LAMB2/MAPK10/MYL9/MYLK/PARVA/PDGFC/PDGFD/PDGFRA/PDGFRB/PPP1R12B/PRKCA/RELN/THBS1/THBS4/TNXB/FIGF/VWF/ZYX |
| hsa04270 | Vascular smooth muscle contraction | 31 | 1.51E-11 | ACTA2/ACTG2/ADCY4/ADCY5/ADRA1A/ADRA1D/AGTR1/AVPR1A/CACNA1C/CALCRL/CALD1/EDNRA/GUCY1A2/ITPR1/KCNMA1/KCNMB1/KCNMB2/MRVI1/MYH11/MYL9/MYLK/NPR1/NPR2/PLA2G5/PPP1R12B/PPP1R14A/PRKCA/PRKG1/RAMP1/RAMP2/RAMP3 |
| hsa04080 | Neuroactive ligand-receptor interaction | 50 | 1.73E−11 | ADCYAP1R1/ADRA1A/ADRA1D/ADRA2A/ADRA2C/ADRB3/AGTR1/AGTR2/AVPR1A/AVPR2/BRS3/CALCRL/CHRM2/CTSG/EDNRA/EDNRB/GH1/GLP2R/GRIA2/GRIA3/GRID1/GRIK5/GRM7/HTR1E/HTR2A/HTR2B/HTR4/LEPR/LPAR4/MAS1/MCHR1/NPY1R/NPY5R/P2RX1/PRL/PRLHR/PRLR/PTGER2/PTGER3/PTGFR/PTH1R/S1PR1/S1PR2/S1PR3/SSTR3/TACR1/TACR2/TBXA2R/THRA/VIPR2 |
| hsa04020 | Calcium signaling pathway | 37 | 2.43E−10 | ADCY4/ADRA1A/ADRA1D/ADRB3/AGTR1/ATP2B4/AVPR1A/CACNA1C/CACNA1G/CACNA1H/CAMK2A/CHRM2/EDNRA/EDNRB/GNA14/HTR2A/HTR2B/HTR4/ITPKB/ITPR1/MYLK/NOS3/P2RX1/PDE1A/PDE1B/PDE1C/PDGFRA/PDGFRB/PLN/PRKCA/PTGER3/PTGFR/RYR3/SLC8A1/TACR1/TACR2/TBXA2R |
| hsa04022 | cGMP-PKG signaling pathway | 34 | 1.52E−09 | ADCY4/ADCY5/ADRA1A/ADRA1D/ADRA2A/ADRA2C/ADRB3/AGTR1/AKT3/ATP1A2/ATP1B2/ATP2B4/CACNA1C/EDNRA/EDNRB/GUCY1A2/ITPR1/KCNJ8/KCNMA1/KCNMB1/KCNMB2/MEF2C/MRVI1/MYL9/MYLK/NFATC4/NOS3/NPR1/NPR2/PDE2A/PLN/PRKG1/RGS2/SLC8A1 |
Abbreviations: cGMP-PKG, cyclic guanosine monophosphate–protein kinase G; DEG, differentially expressed genes; PCNA, proliferating cell nuclear antigen.
aTop 5 upregulated and downregulated KEGG pathways based on DEGs. Top 5 upregulated keg pathways concluded cell cycle, systemic lupus erythematosus, alcoholism, DNA replication, viral carcinogenesis, while top 5 downregulated DEGs were enriched in focal adhesion, vascular smooth muscle contraction, neuroactive ligand–receptor interaction, calcium signaling pathway, and cGMP-PKG signaling pathway.
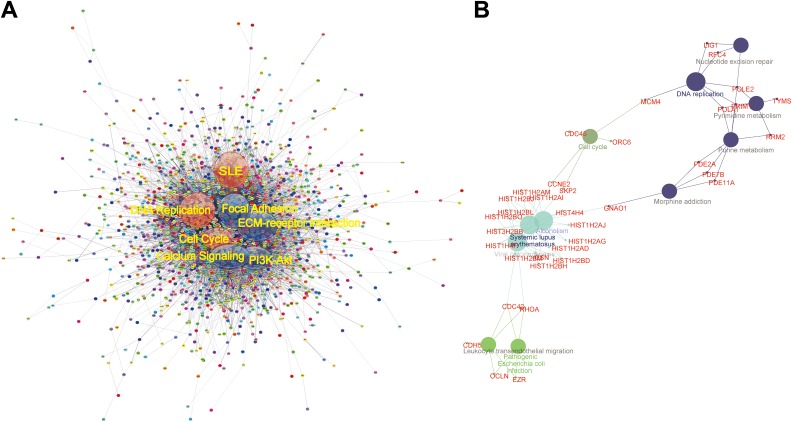
In order to understand the system-level functional interactions of the DEGs we obtained, PPI network analysis was performed based on the information in the STRING database. After mapping the whole DEGs into STRING, the top 5 hub nodes with higher degrees were screened. These hub genes included somatostatin receptor 1 (SSTR1), NPBWR1, S1PR3, NPY1R, and CXCR3. Among these genes, SSTR1 showed the highest node degree, which was 31. Figure 2A illustrates the whole DEG PPI network. Moreover, a total of 3818 nodes and 17 128 edges were analyzed using plug-in MCODE. The top 3 significant modules were selected, and KEGG enrichment analysis (functional annotation) of the genes involved in the modules were analyzed. Enrichment analysis obtaining the pathways involved in the PPI modules (PPI-m) showed that the genes in submodules were mainly associated with neuroactive ligand-receptor interaction, calcium signaling pathway, chemokine signaling pathway, extracellular matrix-receptor interaction, SLE, DNA replication, and also cell cycle. Figure 2B displays the significantly enriched pathways and DEGs in top-ranked submodules based on the PPI network, including DNA replication, SLE, cell cycle, viral carcinogenesis, and so on.
Figure 2.
The PPI network analysis. A, PPI network of the whole DEGs based on the STRING database (edges: 17128; nodes: 2458), dots refer proteins encoded by DEGs and lines represent the correlation between proteins. The 6 identified subnetworks indicated by circles were named after pathway enrichment of the genes encoding the proteins in the circles. They represent as cell cycle, DNA replication, SLE, focal adhesion, ECM-receptor interaction, P13K-Akt pathway, and calcium signaling pathway. B, Representative KEGG pathway of submodule based on the PPI network. Bigger circles indicate pathways and smaller circles are genes in the pathways. If the genes belonged to more than 2 pathways, it will be indicated as a fusion of the 2 colors from the 2 pathways. DEG indicates differentially expressed genes; ECM, extracellular matrix; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein–protein interaction; SLE, systemic lupus erythematosus; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins.
Gene Set Enrichment Analysis was conducted using the RNA-sequencing (RNA-seq) data. In total, 24 pathways were upregulated in cancer group and 16 pathways were upregulated in normal group (data not shown).
Identification of Key Gene Signatures in Cervical Cancer
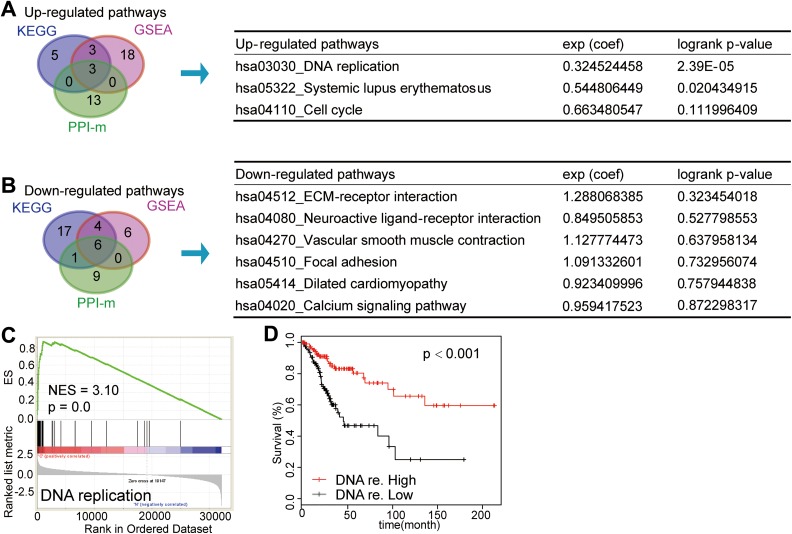
To comprehend the pathways we investigated above further, we intersected the results of 3 enriched approaches (KEGG, GSEA, and PPI-m) to get mutual pathways. Three upregulated pathways, namely, cell cycle, SLE, and DNA replication, were obtained and 6 pathways of downregulated were obtained. To further investigate the clinical signature of 9 mutual pathways we found above, we conducted the gene set survival analysis. The results as shown in Figure 3A and B demonstrated that 2 (DNA replication and SLE pathway) of 9 pathways were significantly related to survival. Survival rate of patients with high expression score of DNA replication was significantly superior to those patients with low expression score (P < .001). The SLE signature was related to survival (P = .02104) as well while other pathways showed no significant association with survival. Corresponding GSEA results of DNA replication is shown in Figure 3C with NES = 3.10. The peak of the curve was inclined left, which means the expression of most genes in DNA replication pathway was increased when compared between cancer and normal samples. The survival curve of DNA replication is illustrated in Figure 3D.
Figure 3.
Overlap of up- and downregulated pathways among KEGG, GSEA, and PPI-m. A and B, Venn diagram illustrated that the overlapped pathways among KEGG, GSEA, and PPI-m were on left and the survival analysis of gene set in mutual pathways were described in the right table. A, Three mutual upregulated pathways. B, Six mutual downregulated pathways. C, The GSEA result of DNA replication pathway was shown with NES equaling 3.10 when comparing between cervical squamous cancer and normal patients. Peak of curve was inclined left which means the expression of most genes in DNA replication pathway in cancer was increased when comparing with normal samples. D, Survival curve of DNA replication was shown. Survival rate of patients with high-expression score of DNA replication was significantly superior to those patients with low-expression score (P < .001). GSEA indicates Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; PPI, protein–protein interaction; PPI-m, PPI modules.
Identification of the Prognostic Biomarkers and Verification of Differential Expression of DNA Replication Signature for the Patients With Cervical Squamous Cancer
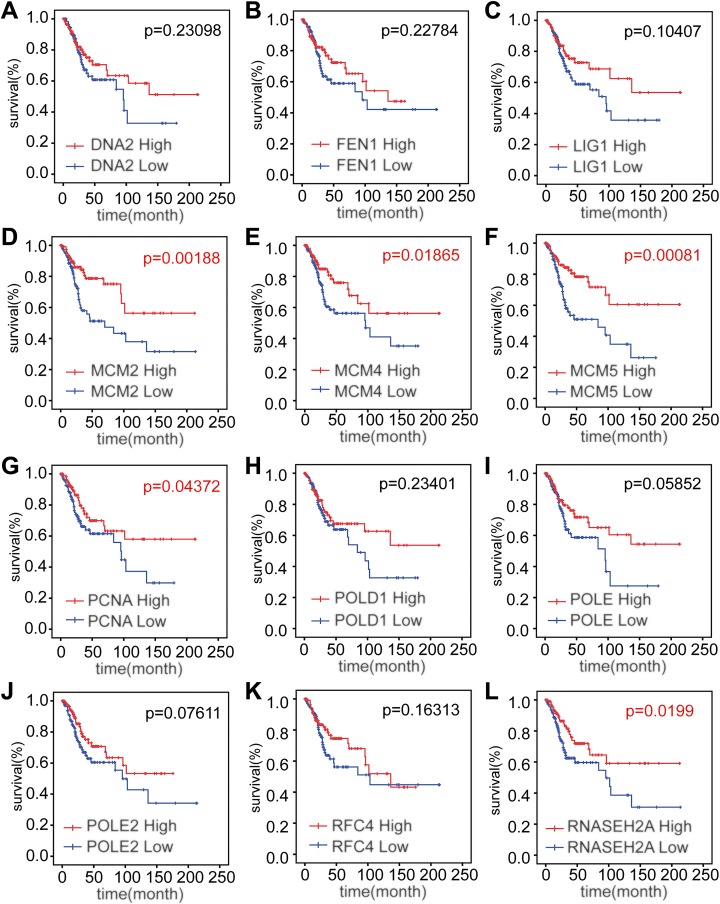
Expression levels of MCM2, MCM4, MCM5, PCNA, and RNASEH2A were significantly related to the OS of patients with cervical squamous cancer (P = .00188, .01865, .00081, .04372, and .01991, respectively), while the expression levels of the other 7 genes were uncorrelated with the survival rate of patients with cervical squamous cancer (P = .23098, .22784, .10407, .23401, .05852, .07611, and .16313, respectively; Figure 4A-L). On the other hand, high expression of MCM2, MCM4, MCM5, PCNA, and RNASEH2A could bring high rate of survival, which was consistent with the result of gene set survival curves we presented before. Together, high level of these 5 genes may be the promising prognostic factors to predict the better survival of cervical cancer.
Figure 4.
Survival curves of 12 genes in DNA replication pathway. A-C and H-K, The expression of other 7 genes was uncorrelated with the survival rate of patients with cervical squamous cancer (P = .23098, .22784, .10407, .23401, .05852, .07611, and .16313, respectively). D-G and L, Expression level of MCM2, MCM4, MCM5, PCNA, and RNASEH2A was significantly related to the overall survival of patients with cervical squamous cancer (P = .00188, .01865, .00081, .04372, and .0199, respectively). MCM indicates minichromosome maintenance; PCNA, proliferating cell nuclear antigen; RNASEH, ribonuclease H2 subunit A.
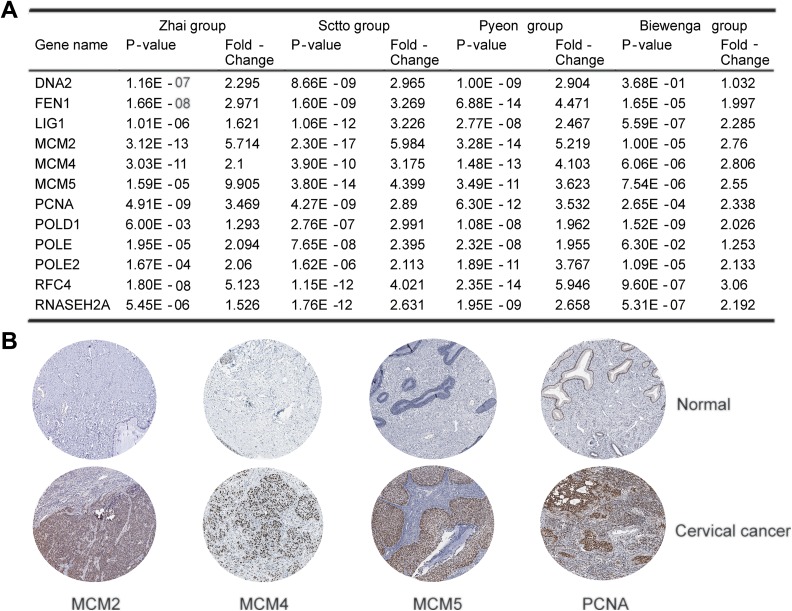
We further investigated whether these genes in DNA replication signature were also upregulated in 4 other cervical cancer cohorts: Scotto cervix group, Pyeon Multi-cancer group, Biewenga cervix group, and also Zhai cervix group. The fold-change and P value of each gene in DNA replication pathway are shown in Figure 5A, normal versus cervical cancer. The genes were consistent with RNA-seq result as they were upregulated in patients with cervical cancer at mRNA level except DNA2. Then, we analyzed the protein expression level of 12 genes in clinical specimens from THPA. Protein expression levels of survival-related genes MCM2, 4, 5, and PCNA are shown in Figure 5B. All of them had positive strong or moderate expression in cervical squamous cancer tissues and negative weak expression in normal cervical squamous tissues. The protein expression level of RNASEH2A was not available on the website.
Figure 5.
Validation of genes in the selected distinct DNA replication pathway. A, Four data sets concluding Zhai, Sctto, Pyeon, and Biewenga cervical cancer groups from Oncomine data were analyzed and all the 12 genes were significantly upregulated in cervical squamous cancer compared with normal, except DNA2 of Biewenga group. B, The expression of MCM2, MCM4, MCM5, and also PCNA in normal cervix and cervical squamous cancer tissues. Images were taken from the Human Protein Atlas (http://www.proteinatlas.org) online database. MCM indicates minichromosome maintenance; PCNA, proliferating cell nuclear antigen.
Evaluation of MCM5 as a Prognostic Marker of Patients With Cervical Cancer
In a Cox regression analysis, 5 survival-related genes expression, node (N) stage and FIGO stage were found to be significantly associated with OS of patients with cervical cancer by a univariate analysis. Multivariate analysis after reducing the nonsignificant variables demonstrated that FIGO stage (HR = 2.245, 95% confidence interval [CI] 1.866-2.904, P = .035) and MCM5 (HR = 0.461, 95% CI 0.259-0.821, P = .009) were independent predictors of OS (Table 2). The clinical relevance of MCM5 was also investigated. As shown in Figure S1a, MCM5 expression decreases when stage increased (P = .0434).The sensitivity, specificity, positive predictive value, and negative predictive value of MCM2, 4, 5, PCNA, and RNASEH2A, for discriminating between grade 1 versus grade 2 + grade 3, FIGO stage I-IIA versus stage IIB-IV, and N0 versus N1, were also estimated. Both of sensitivity and specificity of stage I-IIA versus stage IIB-IV for MCM5 are more than 60% (60.00% and 60.20%, respectively). All the ROC (receiver operating characteristic) curves of the 5 genes are shown in supplemental Figure S1b.
Table 2.
Univariate and Multivariate Cox Proportional Hazards Analysis of MCM2, 4, 5, PCNA, RNASEH2A Expression, and Overall Survival of Patients With Cervical Cancer in the TCGA Cohort.
| Factor | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age | 0.555 (0.251-1.225) | .139 | ||
| Grade | 1.277 (0.171-9.555) | .805 | ||
| T, (T1+T2 vs T3+T4) | 1.520 (0.521-4.431) | .443 | ||
| N, (N0 vs N1) | 1.341 (0.893-2.013) | .004a | 1.411 (1.182-1.930) | .133 |
| STAGE, (I-IIA vs IIB-IV) | 2.395 (1.191-4.842) | .012a | 2.245 (1.866-2.904) | .035a |
| MCM2 | 0.729 (0.574-0.968) | .014a | 1.180 (0.795-1.751) | .412 |
| MCM4 | 0.820 (0.489-1.025) | .038a | 1.305 (0.868-1.961) | .201 |
| MCM5 | 0.270 (0.180-0.404) | <.001a | 0.461 (0.259-0.821) | .009a |
| PCNA | 0.706 (0.438-0.951) | .036a | 1.115 (0.743-1.673) | .598 |
| RNASEH2A | 0.952 (0.537-1.117) | .042a | 0.984 (0.657-1.474) | .938 |
Abbreviations: CI, confidence interval; HR, hazard ratio; MCM, minichromosome maintenance; PCNA, proliferating cell nuclear antigen; RNASEH2A, ribonuclease H2 subunit A; vs, versus; TCGA, The Cancer Genome Atlas.
a Statistical significance (P < .05).
Discussion
In this study, we comprehensively demonstrated 10 significant pathways in cervical cancer using different bioinformatic methods based on TCGA data. Among these pathways, DNA replication (P < .001) has the strongest positive correlation with survival. We also validated the expression level of genes in the DNA replication pathway in other different cohorts which also supported our results. Of the 12 DEGs identified in the DNA replication pathway, MCM2, 4, 5, PCNA, and RNASEH2A were upregulated in cervical squamous cancer samples compared to normal samples, and the survival rate had a positive correlation with the high expression of these genes in patients with cancer. These genes may act as the independent prognostic biomarkers to predict the survival of cervical cancer. According to the results of Cox multivariate analysis, MCM5 was indicated to be an independent prognostic factor for OS of patients with cervical cancer.
As we all know, the univariate Cox regression is the most traditional and simplest method to select prognostic genes.21 Therefore, we also performed the univariate Cox regression analysis of all genes sequenced by RNA-seq. Finally, a total of 1930 genes which were significant in univariate Cox regression analysis (P < .05, the genes list is attached as Supplemental Table 3) were obtained and may be related to the prognosis of cervical cancer. After aligning these genes with the DEGs, we found that a total of 397 genes were common to both, including many genes in DNA replication pathway. After performing KEGG pathway and PPI analysis of these 1930 and 397 genes, we found that the DNA replication pathway obtained by our integrative bioinformatic methods was also screened out; these results are shown in Supplemental Figure 2. These results suggested that our methods could achieve similar results when compared with traditional methods, and both could find key pathways and genes in the process of tumorigenesis.
Eukaryotic MCM is made up of 6 protein MCM2-7 complex (also known as MCM2-MCM7), which is an essential replicative helicase.22–24 Many studies have proved the roles of MCM2 and MCM4 in cancers such as human non-small cell lung carcinomas25 and oral squamous cell carcinomas.26 Amaro Filho et al reported an increasing expression of MCM2 in invasive cervical cancers compared to controls, which was due to the correlation between the MCM2-positive cells and the presence of HPV DNA detected by in situ hybridization.27 Another study indicates that cervical cancer cells may use excess MCMs as a backup for replicative stress.28 The level of MCM4 and PCNA was reported to be upregulated by mutant p53, a primary determinant in variety cancers.29 Tatsumi and Ishim also reported the detection of an MCM4 mutation in HeLa cells, and those cells where the mutant MCM4 was expressed had abnormal nuclear morphology, suggesting that DNA replication was perturbed in the presence of the mutant MCM4.30 Since the mutant MCM4 may affect human MCM4/6/7 complex formation, the complex containing the mutant MCM4 protein is unstable and tend to be degraded, which would affect the proliferation of cancer further.30,31
Pruitt et al have developed a transgenic Mcm2IRES-CreERT2/IRES-CreERT2 mouse in which the expression of MCM2 was two-third reduced compared to wide-type mouse in homozygous embryos or mouse embryonic fibroblasts (MEFs); the life span of the transgenic mice was greatly reduced due to cancer such as lymphoma.32 Shima et al reported that the instable mutation Chaos3 (chromosome aberrations occurring spontaneously 3) in mouse chromosome is an allele of MCM4. After finding the slightly reduced amount of protein level of MCM4 in Mcm4Chaos3/Chaos3 MEFs, they observed the development of breast cancer in Mcm4Chaos3 homozygous females further and proved the disposition to breast cancer and worse survival latency in homozygous females compared to wild-type mice.33 These findings suggest that hypomorphic alleles of the genes encoding the subunits of the MCM2-7 complex may increase breast cancer risk. In our research, MCM2, MCM4, and PCNA were upregulated in cervical squamous cancer samples compared to normal, and for those patients with cervical squamous cancer, their survival rate had a positive correlation with the high expression of these genes. Mutation of these genes may be a possible hypothesis of the result.
Minichromosome maintenance 5 is expressed in a wide variety of cells as a main protein to control replication origins and is involved in the cell cycle of both normal and cancer cells. Several studies had linked MCM5 to the progression of oral squamous cell carcinoma,34 urothelial cancer,35 breast cancer,36 ovarian cancer,37 and so on. Qing et al conducted a proteomic identification of potential biomarkers for cervical squamous cell carcinoma and found that MCM5 might be important in cervical cancer.38 In this study, we observed that MCM5 was an independent prognostic biomarker in cervical squamous cell carcinoma, and the results showed that MCM5 expression decreases as stage increases. A possible reason might be that MCM5 was present throughout the cell cycle of proliferating cells but not in nonproliferating quiescent cells.39 With the progress of the tumor, tumor proliferation in situ is slow,40,41 leading to the decreasing of MCM5 expression. According to the NCCN (National Comprehensive Cancer Network) guidelines of cervical cancer (https://www.nccn.org), surgical indications would become scarce for those patients with advanced stage IIB. Thus, the relationship between MCM5 and stage made MCM5 possible to serve as a biomarker in predicting disease progression.
RNASEH2A gene can encode an enzyme in humans named Ribonuclease H2 subunit A42; mutations in this gene cause a rare genetic disorder, Aicardi-Goutieres syndrome, which affects the brain and the skin.43,44 Flanagan et al have validated RNASEH2A as an putative anticancer drug target45 and other studies identified RNASEH2A may be the susceptible gene in aggressive prostate cancer46 or played roles in lung adenocarcinoma,47 but the mechanism is still unclear.
The Cancer Genome Atlas is a public funded project that aims to catalogue and discover major cancer-causing genomic alterations to create a comprehensive “atlas” of cancer genomic profiles. It is possible and challenging to identify molecular biomarkers using genomics data. Recently, many researchers are also using the sequencing data to predict and analyze tumor markers. Methods vary according to the integrity of the available data, but each has its own advantages.21,48 We have comprehensively analyzed the cervical cancer RNA-seq data set from TCGA, finally obtained 2 significant pathways, 12 genes in one of the most significant pathway and 5 prognostic-related genes and MCM5 which may be an independent prognostic biomarker in cervical cancer. It should be noted that there are some certain limitations in the present study. Due to some missing clinical data in the database, 5 genes except MCM5 were not validated as independent prognostic factors of prognosis, which may also play important roles in cervical cancer, and need further investigation. Moreover, the validation of these biomarkers in an independent clinical cohort should be conducted in our future research to achieve more support for our results.
In conclusion, we identified a distinct pathway (DNA replication) and also 5 genes in it (MCM2, 4, 5, PCNA, and RNASEH2A) which were likely associated with prognosis of patients with cervical cancer. Of the 5 genes, MCM5 was an independent prognostic marker. The findings could be helpful for us to understand the pathological process of cervical carcinogenesis, predict outcomes, and develop therapeutic targets for patients with cervical cancer. As for linking genotype to phenotype, further in vitro and in vivo studies are necessary to improve our understanding of cervical cancer.
Supplemental Material
Supplementary_material for Identification of Significant Gene Signatures and Prognostic Biomarkers for Patients With Cervical Cancer by Integrated Bioinformatic Methods by Xiaofang Li, Run Tian, Hugh Gao, Feng Yan, Le Ying, Yongkang Yang, Pei Yang and Yan’e Gao in Technology in Cancer Research & Treatment
Acknowledgments
The authors thank the favor Dakang Xu provided in this article and Jasmine for polishing the manuscript. Y.E.G., P.Y., and X.F.L. made a substantial contribution to the concept and design of the work. R.T. and H.G. did acquisition, analysis, and interpretation of data. X.F.L. and R.T. drafted the article. L.Y., F.Y., and Y.K.Y. revised it critically for important intellectual content.
Abbreviations
- CI
confidence interval
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DEGs
differentially expressed genes
- FDR
false discovery rate
- FIGO
International Federation of Gynecology and Obstetrics
- GSEA
Gene Set Enrichment Analysis
- HPV
human papillomavirus
- HR
hazard ratio
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MCODE
Molecular Complex Detection
- MCM
minichromosome maintenance
- MEF
mouse embryonic fibroblast
- mRNA
messenger RNA
- NES
normalized enrichment score
- OS
overall survival
- PCNA
proliferating cell nuclear antigen
- PPI
protein–protein interaction
- PPI-m
PPI modules
- RNA-seq
RNA-sequencing
- RNASEH
ribonuclease H2 subunit A
- TCGA
The Cancer Genome Atlas
- THPA
The Human Protein Atlas
- SLE
systemic lupus erythematosus
- SSTR1
somatostatin receptor 1
- ssGSEA
single-sample GSEA
- STRING
Search Tool for the Retrieval of Interacting Genes/Proteins.
Authors’ Note: This article does not contain any studies with human participants or animals performed by any of the authors.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by the Sci-tech Program Foundation of Shannxi Province, China (Award number: 2017SF-013).
ORCID iD: Yan’e Gao, MD  http://orcid.org/0000-0002-7894-7312
http://orcid.org/0000-0002-7894-7312
Supplemental Material: Supplementary material for this article is available online.
References
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA A Cancer Jr Clin. 2015;65(2):87–108. [DOI] [PubMed] [Google Scholar]
- 2. Wang SS, Sherman ME, Hildesheim A, Lacey JV, Devesa S. Cervical adenocarcinoma and squamous cell carcinoma incidence trends among white women and black women in the United States for 1976-2000. Cancer. 2004;100(1):1035–1044. [DOI] [PubMed] [Google Scholar]
- 3. Galic V, Herzog TJ, Lewin SN, et al. Prognostic significance of adenocarcinoma histology in women with cervical cancer. Gynecologic Oncol. 2012;125(2):287–291. [DOI] [PubMed] [Google Scholar]
- 4. Burk RD, Chen Z, Saller C, et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382(9285):889–899. [DOI] [PubMed] [Google Scholar]
- 6. Crook T, Wrede D, Tidy JA, Mason WP, Evans DJ, Vousden KH. Clonal p53 mutation in primary cervical cancer: association with human-papillomavirus-negative tumours. Lancet. 1992;339(8801):1070–1073. [DOI] [PubMed] [Google Scholar]
- 7. McIntyre JB, Wu JS, Craighead PS, et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecologic Oncol. 2013;128(3):409–414. [DOI] [PubMed] [Google Scholar]
- 8. Lee M-S, Jeong M-H, Lee H-W, et al. ARTICLE PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nature Commun. 2015;6:7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Analysis identifies mutations linked to cervical cancer. Cancer Discov. 2014;4(3):OF2. [DOI] [PubMed] [Google Scholar]
- 10. Narayan G, Murty VV. Integrative genomic approaches in cervical cancer: implications for molecular pathogenesis. Future Oncol. 2010;6(10):1643–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munro A, Codde J, Spilsbury K, et al. Risk of persistent and recurrent cervical neoplasia following incidentally detected adenocarcinoma in situ. Am J Obstet Gynecol. 2017;216(3):272.e271–e272.e277. [DOI] [PubMed] [Google Scholar]
- 12. Kim S-W, Chun M, Ryu H-S, et al. Salvage radiotherapy with or without concurrent chemotherapy for pelvic recurrence after hysterectomy alone for early-stage uterine cervical cancer. Strahlenther Onkol. 2017;193(7):534–542. [DOI] [PubMed] [Google Scholar]
- 13. Feng Y, He F, Yan S, et al. The role of GOLPH3L in the prognosis and NACT response in cervical cancer. J Cancer. 2017;8(3):443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Higgins GD, Davy M, Roder D, Uzelin DM, Phillips GE, Burrell CJ. Increased age and mortality associated with cervical carcinomas negative for human papillomavirus RNA. Lancet. 1991;338(8772):910–913. [DOI] [PubMed] [Google Scholar]
- 15. Buamah PK, Cornell C, Skillen AW, Cantwell BM, Harris AL. Initial assessment of tumor-associated antigen CA-125 in patients with ovarian, cervical, and testicular tumors. Clin Chem. 1987;33(7):1124–1125. [PubMed] [Google Scholar]
- 16. Li M, Feng YM, Fang SQ. Overexpression of ezrin and galectin-3 as predictors of poor prognosis of cervical cancer. Braz J Med Biol Res. 2017;50(4):e5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355(6331):1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uhlén M, Fagerberg L, Hallström BM, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 19. Subramanian A, Tamayo P, Mootha VK, et al. Gene Set Enrichment Analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barbie DA, Tamayo P, Boehm JS, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou X, Liu J. A computational model to predict bone metastasis in breast cancer by integrating the dysregulated pathways. BMC cancer. 2014;14:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brewster AS, Chen XS. Insights into the MCM functional mechanism: lessons learned from the archaeal MCM complex. Crit Rev Biochem Mol Biol. 2010;45(3):243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Forsburg SL. Eukaryotic MCM proteins: beyond replication initiation. Microbiol Mol Biol Rev. 2004;68(1):109–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31(2):287–293. [DOI] [PubMed] [Google Scholar]
- 25. Zhang X, Teng Y, Yang F, et al. MCM2 is a therapeutic target of lovastatin in human non-small cell lung carcinomas. Oncol Rep. 2015;33(5):2599–2605. [DOI] [PubMed] [Google Scholar]
- 26. Razavi SM, Jafari M, Heidarpoor M, Khalesi S. Minichromosome maintenance-2 (MCM2) expression differentiates oral squamous cell carcinoma from pre-cancerous lesions. Malays J Pathol. 2015;37(3):253–258. [PubMed] [Google Scholar]
- 27. Amaro Filho SM, Nuovo GJ, Cunha CB, et al. Correlation of MCM2 detection with stage and virology of cervical cancer. Int J Biol Markers. 2014;29(4):363–371. [DOI] [PubMed] [Google Scholar]
- 28. Alvarez S, Díaz M, Flach J, et al. Replication stress caused by low MCM expression limits fetal erythropoiesis and hematopoietic stem cell functionality. Nature Commun. 2015;6:8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Polotskaia A, Xiao G, Reynoso K, et al. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc Natl Acad Sci U S A. 2015;112(11):E1220–E1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tatsumi R, Ishimi Y. An MCM4 mutation detected in cancer cells affects MCM4/6/7 complex formation. J Biochem. 2017;161(3):259–268. [DOI] [PubMed] [Google Scholar]
- 31. Hughes CR, Guasti L, Meimaridou E, et al. MCM4 mutation causes adrenal failure, short stature, and natural killer cell deficiency in humans. J Clin Invest. 2012;122(3):814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25(12):3121–3132. [DOI] [PubMed] [Google Scholar]
- 33. Shima N, Alcaraz A, Liachko I, et al. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nature Genetics. 2007;39(1):93–98. [DOI] [PubMed] [Google Scholar]
- 34. Yu SY, Wang YP, Chang JY, Shen WR, Chen HM, Chiang CP. Increased expression of MCM5 is significantly associated with aggressive progression and poor prognosis of oral squamous cell carcinoma. J Oral Pathol Med. 2014;43(5):344–349. [DOI] [PubMed] [Google Scholar]
- 35. Korkolopoulou P, Givalos N, Saetta A, et al. Minichromosome maintenance proteins 2 and 5 expression in muscle-invasive urothelial cancer: a multivariate survival study including proliferation markers and cell cycle regulators. Human Pathol. 2005;36(8):899–907. [DOI] [PubMed] [Google Scholar]
- 36. Eissa S, Matboli M, Shehata HH, Essawy NO. MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer. Tumour Biol. 2015;36(6):4487–4494. [DOI] [PubMed] [Google Scholar]
- 37. Levidou G, Ventouri K, Nonni A, et al. Replication protein A in nonearly ovarian adenocarcinomas: correlation with MCM-2, MCM-5, Ki-67 index and prognostic significance. Int J Gynecol Pathol. 2012;31(4):319–327. [DOI] [PubMed] [Google Scholar]
- 38. Qing S, Tulake W, Ru M, et al. Proteomic identification of potential biomarkers for cervical squamous cell carcinoma and human papillomavirus infection. Tumour Biol. 2017;39(4):1010428317697547. [DOI] [PubMed] [Google Scholar]
- 39. Williams GH, Romanowski P, Morris L, et al. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci U S A. 1998;95(25):14932–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Szymanska Z, Cytowski M, Mitchell E, Macnamara CK, Chaplain MAJ. Computational modelling of cancer development and growth: modelling at multiple scales and multiscale modelling [published online ahead of print June 20, 2017]. Bull Math Biol. 2017. [DOI] [PubMed] [Google Scholar]
- 41. Chaplain MA, Sleeman BD. Modelling the growth of solid tumours and incorporating a method for their classification using nonlinear elasticity theory. J Math Biol. 1993;31(5):431–473. [DOI] [PubMed] [Google Scholar]
- 42. Chon H, Vassilev A, DePamphilis ML, et al. Contributions of the two accessory subunits, RNASEH2B and RNASEH2C, to the activity and properties of the human RNase H2 complex. Nucleic Acids Res. 2009;37(1):96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aicardi J, Goutières F. A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol. 1984;15(1):49–54. [DOI] [PubMed] [Google Scholar]
- 44. Rice GI, Hill Building A, Vanderver A, Orcesi S. Characterization of human disease phenotypes associated with mutations in HHS public access. Am J Med Genet A. 2015;167A(2):296–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flanagan JM, Funes JM, Henderson S, Wild L, Carey N, Boshoff C. Genomics screen in transformed stem cells reveals RNASEH2A, PPAP2C, and ADARB1 as putative anticancer drug targets. Mol Cancer Ther. 2009;8(1):249–260. [DOI] [PubMed] [Google Scholar]
- 46. Williams KA, Lee M, Hu Y, et al. A systems genetics approach identifies CXCL14, ITGAX, and LPCAT2 as novel aggressive prostate cancer susceptibility genes. PLoS Genet. 2014;10(11):e1004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu H, Ma J, Wu J, et al. Gene expression profiling analysis of lung adenocarcinoma. Braz J Med Biol Res. 2016;49(3):e4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_material for Identification of Significant Gene Signatures and Prognostic Biomarkers for Patients With Cervical Cancer by Integrated Bioinformatic Methods by Xiaofang Li, Run Tian, Hugh Gao, Feng Yan, Le Ying, Yongkang Yang, Pei Yang and Yan’e Gao in Technology in Cancer Research & Treatment