Abstract
Objectives To improve the accuracy of mining structured and unstructured components of the electronic medical record (EMR) by adding temporal features to automatically identify patients with rheumatoid arthritis (RA) with methotrexate-induced liver transaminase abnormalities.
Materials and methods Codified information and a string-matching algorithm were applied to a RA cohort of 5903 patients from Partners HealthCare to select 1130 patients with potential liver toxicity. Supervised machine learning was applied as our key method. For features, Apache clinical Text Analysis and Knowledge Extraction System (cTAKES) was used to extract standard vocabulary from relevant sections of the unstructured clinical narrative. Temporal features were further extracted to assess the temporal relevance of event mentions with regard to the date of transaminase abnormality. All features were encapsulated in a 3-month-long episode for classification. Results were summarized at patient level in a training set (N=480 patients) and evaluated against a test set (N=120 patients).
Results The system achieved positive predictive value (PPV) 0.756, sensitivity 0.919, F1 score 0.829 on the test set, which was significantly better than the best baseline system (PPV 0.590, sensitivity 0.703, F1 score 0.642). Our innovations, which included framing the phenotype problem as an episode-level classification task, and adding temporal information, all proved highly effective.
Conclusions Automated methotrexate-induced liver toxicity phenotype discovery for patients with RA based on structured and unstructured information in the EMR shows accurate results. Our work demonstrates that adding temporal features significantly improved classification results.
Keywords: natural language processing, electronic medical record, pharmacogenetic, rheumatoid arthritis, methotrexate, liver toxicity
Background and significance
Rheumatoid arthritis (RA) is one of the most common and serious forms of autoimmune arthritis costing the US economy nearly $128 billion per year in medical care and indirect expenses, including lost wages and productivity. Although there are several disease-modifying anti-rheumatic drugs (DMARDs) currently available, methotrexate (MTX) is currently the most widely used and has been the first-line therapy for RA since the 1980s.1 The drug is typically well tolerated in the doses used to treat RA; however, hepatic toxicity is a side effect of significant concern.1–3 Studies have reported cumulative elevations of liver transaminases associated with MTX use.2–4 Several guidelines recommend frequent checking of liver transaminases to monitor for evidence of liver injury,5,6 worsening the economic burden RA has on society.
Defining toxicity usually requires manual chart review because human expertise and reasoning ability are needed to recognize the nuances of relevant information scattered as free text throughout the electronic medical records (EMRs), as well as provide a temporal perspective for drug exposure preceding the adverse event. Yet, the manual reviewing process is labor intensive and inefficient for large-scale analysis.
Mining the EMR, both its structured and unstructured components, has increasingly become a substitute for traditional chart review. Success stories include the development of phenotyping algorithms within projects such as Electronic Medical Records and Genomics (eMERGE),7–10 Pharmacogenomic Research Network (PGRN),11–13 Informatics for Integrating Biology and the Bedside (i2b2),14–19 and Strategic Health Advanced Research Project: Area 4 (SHARP).20 Mining of structured information is executed through traditional database queries to include codified data for ICD-9 codes, lab results, and medication orders. For unstructured information, natural language processing (NLP) has been used to abstract the meaning from the surface textual representations. As an example, Liao et al.14 validated an algorithm to define RA cases in the Partners HealthCare EMR against a gold standard dataset. The algorithm combined variables from NLP and codified EMR data, achieved a high accuracy (AUC 93.7%, positive predictive value (PPV) 94%, and sensitivity 63%, when specificity was set at 97%), and was portable across three academic hospital EMRs.15 Lin et al.11,21,22 further explored multiple feature representations of EMR notes with feature selection methods to investigate algorithms for automatically discovering RA disease activity.
While the extraction of the codified data is fairly straightforward, information extraction from the clinical narrative requires sophisticated technologies grounded in linguistic, cognitive, and computational sciences to go beyond simple string matching and abstract the meaning in a normalized form. Harnessing the recent progress in NLP technologies, especially temporal relation discovery in the clinical domain,23–30 we aimed to develop an algorithm using codified EMR data and Apache clinical Text Analysis and Knowledge Extraction System (cTAKES)31,32 to identify patients with MTX-related liver toxicity within an RA cohort from Partners HealthCare and study whether the addition of temporal features extracted using NLP techniques improve the accuracy of the algorithm.
The challenge was to build an automatic classifier to eliminate patients with elevated transaminases who do not have relevant temporally positioned MTX mentions, or their liver abnormality was caused by factors other than MTX. This entailed higher level processing of the clinical narrative content, including detailed information about the medication, other potential toxicity factors, and temporal-causal indicators. This was the technical challenge and the main focus of the current study.
Materials and methods
Rheumatoid arthritis cohort
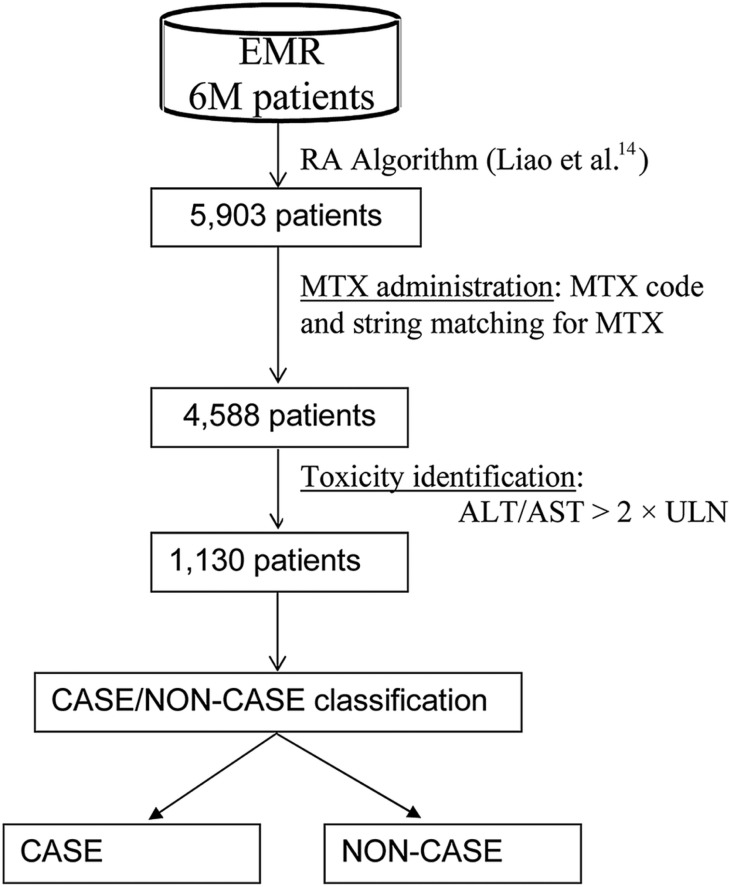
This study used the RA EMR cohort, which included 5903 patients with RA followed at Partners HealthCare since 1992.14 The data warehouse, Research Patient Data Repository, included detailed data with timestamps for diagnoses, medications, problem lists, laboratory tests, procedures, and clinical notes. Patients were followed for variable periods of time from 1992 to 2013, and may have received some of their care outside the Partners HealthCare network. To identify potential MTX-induced liver toxicity, we used rules based on codified and narrative EMR data:
Exposure: the patient had to be exposed to MTX before the transaminase date based on a medication code for MTX and string matching against the clinical narrative for text indicating MTX. The resulted set included 4588 patients (figure 1).
Outcome: among MTX exposed, we defined liver toxicity as any elevation of alanine transaminase (ALT) or aspartate transferase (AST) greater than two times the upper limit of normal (>2× ULN) based on studies that showed an association between elevation >2× ULN and change in hepatic architecture when liver biopsies were obtained annually in patients with RA on MTX.5,33 The number of RA cases was further reduced to 1130.
Figure 1:
Methotrexate (MTX)-induced liver toxicity cohort identification. >2 × ULN, greater than two times the upper limit of normal; ALT, alanine transaminase; AST, aspartate transferase; EMR, electronic medical record; RA, rheumatoid arthritis.
Chart reviews
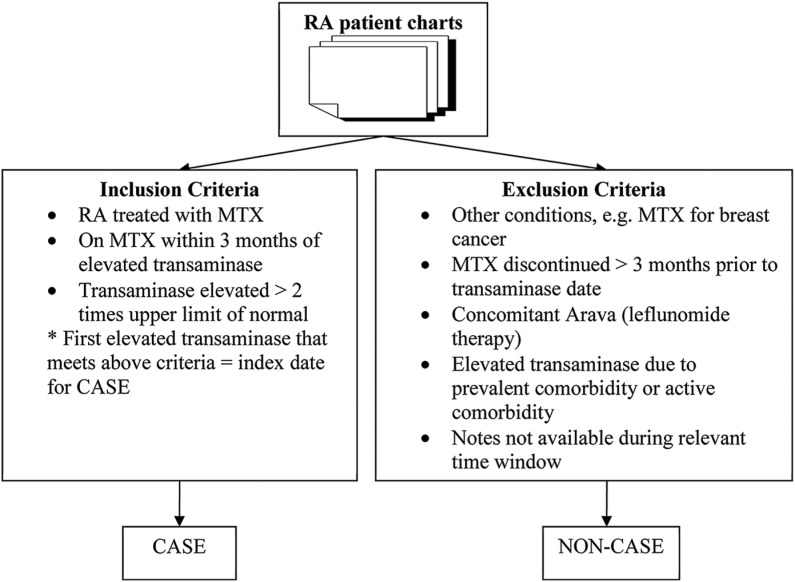
Chart annotation guidelines were developed by two domain experts to define CASE/NON-CASE groups (see online supplement). Two board certified rheumatologists (MPR and EWK) conducted chart reviews of EMR notes and laboratory data from randomly selected patients for the training set (N=480) and test set (N=120) from the 1130 potential cases. Clinical notes were reviewed for 3 months prior to each elevated transaminase timestamp for inclusion and exclusion criteria (figure 2). If there were sequential elevated transaminases, the review window was extended 3 months prior to each transaminase date. We limited the reviews to a clinically relevant window based on rheumatology practice guidelines for monitoring transaminases every 2–3 months. Inclusion criteria included exposure to MTX within 3 months of transaminase date, and reviewers attributed elevated transaminase to MTX exposure. Exclusion criteria were elevated transaminase being attributed to other hepatotoxic drugs such as leflunomide (Arava), and elevated transaminase being attributed to comorbidity, prevalent comorbidity occurring in the past such as hepatitis, or an active comorbidity occurring during the 3-month episode such as congestive heart failure, cholecystitis, sepsis, trauma, or surgery (see online supplement section 1.2 for complete list). If a reviewer was unable to confirm MTX exposure, they proceeded to the next transaminase episode and repeated the review until all instances of transaminase elevation had been reviewed. If any hepatitis C or hepatitis B diagnosis was found (prevalent comorbidity), the patient was classified as a NON-CASE. If an active comorbidity was present, the reviewer proceeded to the next episode and continued the review until all episodes were reviewed. If any single episode met the inclusion criteria without any exclusion criteria, the patient was coded as a CASE (box 1). If all episodes met exclusion criteria, the subject was coded as a NON-CASE. For algorithm development, we extracted notes from the Partner's Healthcare EMR within a time window of 3 months of each elevated transaminase date, defined as an ‘episode’ to mirror chart review methods. If there were multiple transaminase test dates, we used multiple windows and thus included multiple episodes.
Figure 2:
Inclusion and exclusion criteria for chart reviews to define methotrexate (MTX)-induced liver toxicity in patients with rheumatoid arthritis (RA).
Box 1: Rules for labeling CASE or NON-CASE patients.
If all episodes of a patient were NON-CASES,– then patient was classified as a NON-CASE;
If at least one of the episodes was CASE positive,– then the patient was labeled as a CASE
Algorithm development
We aimed to develop an automatic CASE/NON-CASE classification algorithm using a combination of NLP and classification rules. The goal was to first build and test a series of machine learning baseline systems using several competing non-temporal feature sets (tested with a 10-fold cross-validation approach) in the training set. The preferred model was extended with combinations of temporal features to evaluate the contribution of each feature in the training set. The best feature rich model was then applied to a test set from Partners and an independent test set from the Vanderbilt.
We extracted Unified Medical Language System-based terms (UMLS)34 from highly relevant sections of the clinical notes based on Named Entity (NE) types, diseases/disorders, signs/symptoms, anatomical sites, procedures, and medications (eg, MTX, leflunomide) as defined by the UMLS.34 We used cTAKES to extract NE mentions with qualifying attributes such as negation and drug signatures from unstructured free-text clinical narrative. Each mention was mapped to a UMLS concept unique identifier (CUI), thus dealing with language variations. For example, the mentions RA and rheumatoid arthritis would be typed as disease/disorder and mapped to the same CUI (C0003873). The cTAKES drug name entity recognition module was used for extracting drug signatures (dosage, frequency, route, duration, status change, form, strength, and start date).
Two clinical experts developed a list of comorbidities that are associated with transaminase elevations as a customized dictionary based on domain knowledge, published data, and chart review. Concepts included acute events (eg, serious infection, congestive heart failure), trauma (eg, motor vehicle accident) and surgery (eg, cholecystectomy). This customized dictionary was used as input to the cTAKES dictionary look-up module to extract the listed terms and related attributes (see online supplement).
To incorporate temporality into our algorithm, we developed a novel module within cTAKES called DocTimeRel (Document Time Relation) which discovered the temporal relation between an event and the document creation time (DCT). The DocTimeRel values were before, after, overlap, and before/overlap (designed for events that started before DCT and continue to the present). DocTimeRel provided a coarse temporal framework for each event and enabled us to build temporally aware learning models. Events tagged as overlap or before/overlap were treated as temporally relevant for liver toxicity events. For example, in ‘Patient on MTX since 2009’, MTX has a DocTimeRel value of before/overlap; in ‘Patient was on MTX in 2009’, MTX has a value of before; in ‘Patient will start MTX next week’, MTX has a value of after; in ‘Patient is on MTX’, MTX has a value of overlap. The DocTimeRel module was developed and tested on the Temporal History of Your Medical Events (THYME) corpus35–37 which contained 78 clinical and pathology notes on colorectal cancer for 26 patients. The THYME corpus was richly annotated for events and their attributes as well as temporal relations between events. It contained 9730 DocTimeRel relations. The dataset was split 60/20/20 for training, development, and testing. A Support Vector Machine model was trained on the training data to classify the DocTimeRel attribute of each mention into one of the four categories. The most productive features were the part-of-speech (POS) pattern of nearby verbs aimed to capture tense, POS sequence between target event and its closest temporal expression to capture aspect, and domain-specific section headings. The DocTimeRel module performance was 0.814 F1 score. The DocTimeRel model was released as part of Apache cTAKES.
In addition, we restricted the information extraction to only highly relevant sections from the clinical notes. We utilized cTAKES’ sectionizer to parse the document into sections. We ignored information from the following sections as they tend to introduce noise: past medical history, past surgical history, surgical history, social history, family history, allergies, and adverse reactions.
We cast the MTX liver toxicity identification as a binary classification problem into CASE and NON-CASE groups with features as described below.
Features and learning algorithm
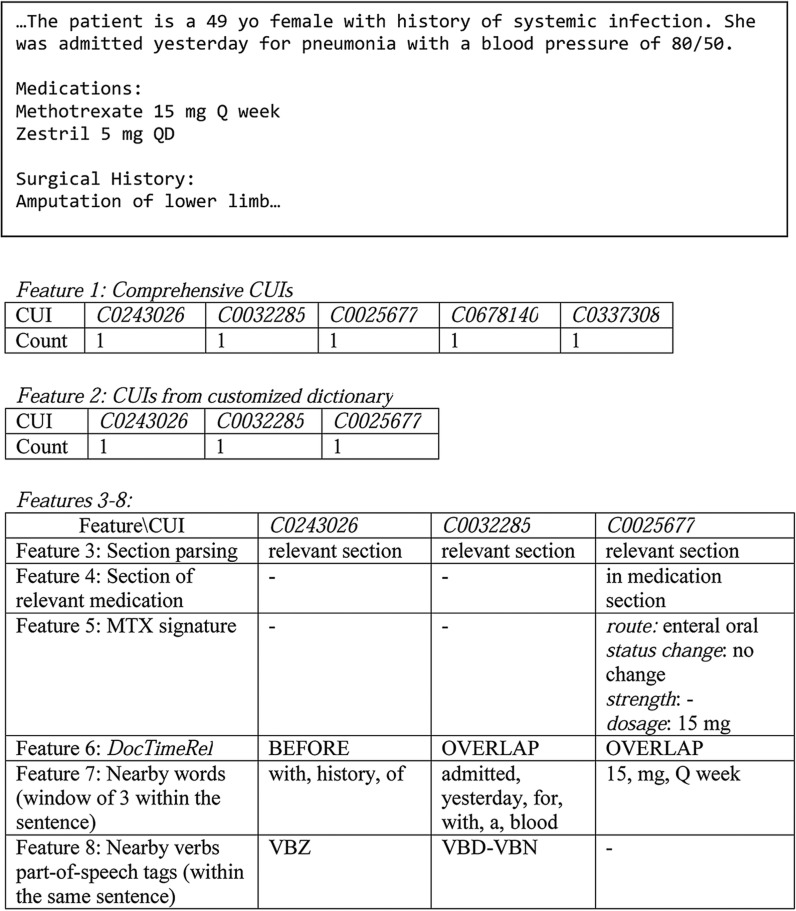
We represented a document by the following groups of features (see figure 3 for examples):
Comprehensive CUIs: we included mentions that map to CUIs from SNOMED-CT and RxNORM (filtered by the US Food and Drug Administration approved list of medications through the Orange book) belonging to UMLS semantic types as our baseline feature set.
CUIs from customized dictionary: we pruned the CUI space through expert-guided feature selection done by domain experts. Similar terms were collapsed into one representation mapped to UMLS CUIs (see online supplement, section 2). Only positive mentions were retained, negated mentions were discovered, but filtered and not represented in the vectors.
Section parsing: we extracted customized dictionary terms only from relevant sections, excluding sections defined above. Section of relevant medication: indicated whether the medication occurred in the medication section of the clinical note which is likely to contain the richest data.
MTX signature: the occurrence of these four medication signature attributes—route, status change, strength, and dosage—of MTX mentions in relevant sections. If such attributes were mentioned with the drug, it was highly likely that it was in the context of a prescribed/administered drug rather than a discussion about a certain drug. Of note, the timestamp of the note was not assigned as the date of the medication.
DocTimeRel: we applied DocTimeRel to all MTX and leflunomide medication and comorbidity mentions from the customized dictionary to determine temporal relations.
Nearby words: three preceding and following words anchored around a mention from the customized dictionary (within the same sentence). The motivation for this feature was to capture temporally relevant signals. Of course, the nearby words might be an indicator of other types of information, not necessarily linked to temporality.
Nearby verbs’ POS tag: POS tags of same sentence verbs anchored around a mention from the customized dictionary (within the same sentence). POS tags could be indicative of the temporal positioning of an event. The tagset was based on Penn Treebank38: VB—verb, base form; VBD—verb, past tense; VBG—verb, gerund or present participle; VBN—verb, past participle; VBP—verb, non-third person singular present; VBZ—verb, third person singular present.
Figure 3:
A sample of features on dummy clinical text from a methotrexate (MTX)-induced liver-toxicity NON-CASE patient with rheumatoid arthritis (RA) (C0243026 for Systemic infection; C0032285 for Pneumonia; C0025677 for Methotrexate; C0678140 for Zestril; C0337308 for Amputation of lower limb). CUI, concept unique identifier; VBD, verb, past tense; VBN, verb, past participle; VBZ, verb, third person singular present.
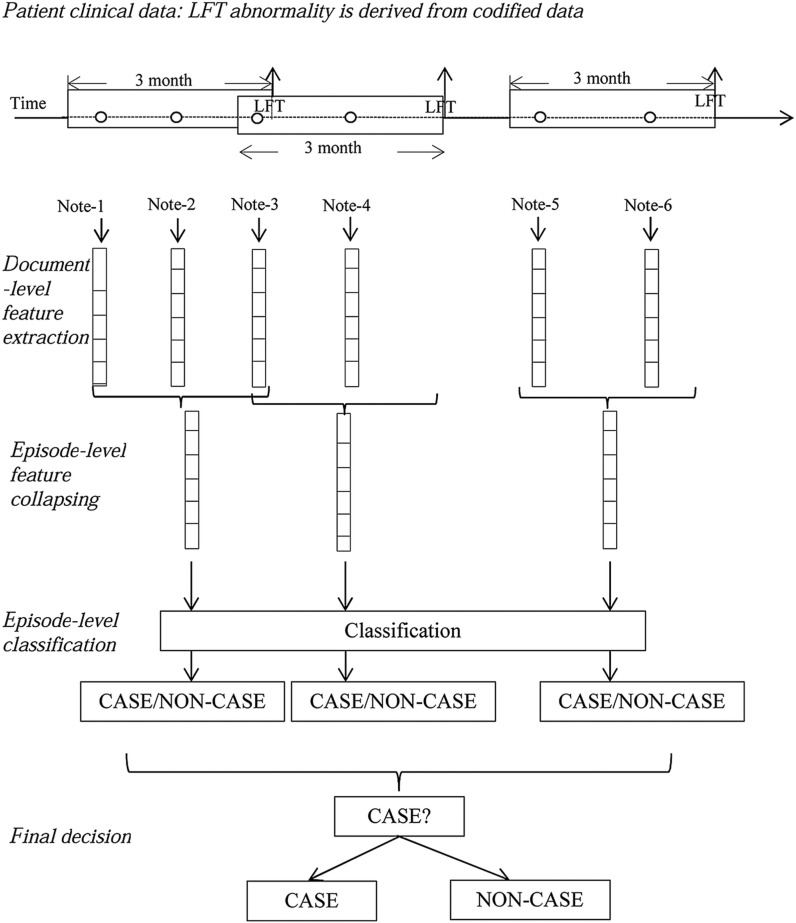
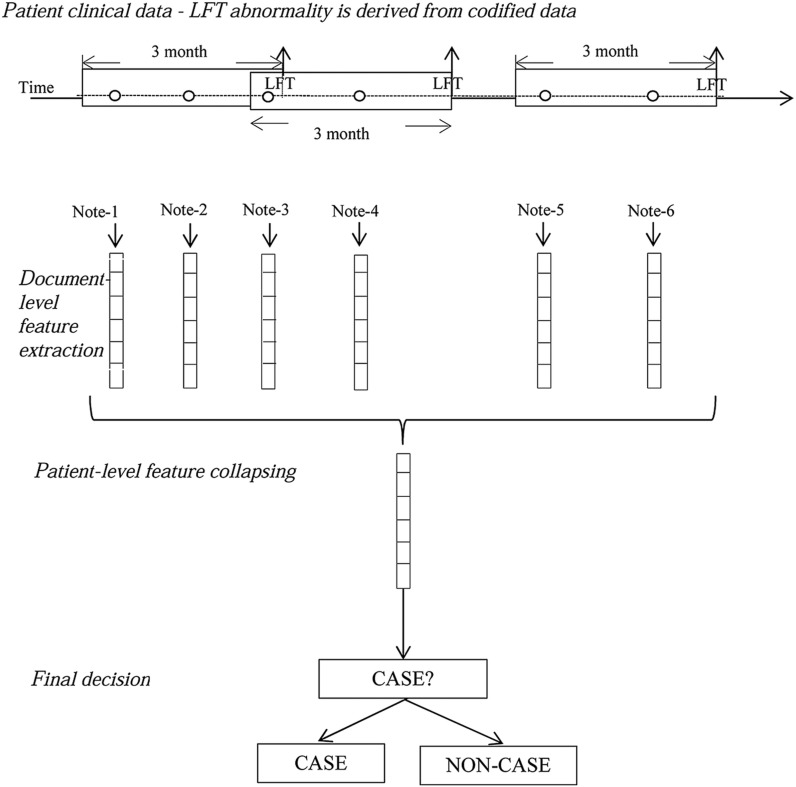
We used a learning strategy anchored around each transaminase episode (figure 4). For all documents that fell into an episode, their document-level features were collapsed into one episode-level feature vector. Classification algorithms (described below) were applied at the episode-level vectors for predicting MTX-induced liver toxicity. The final decision for patient-level classification was based on the same rule used in chart reviews (box 1).
Figure 4:
CASE/NON-CASE classification: episode-level classification (‘o’ signifies a clinical note). LFT, liver function test.
Two episode-level rules were implemented to filter out NON-CASE episodes (box 2).
Box 2: Episode-level NON-CASE filtering rules.
- If an episode-vector contained zero values for methotrexate (MTX), or there were zero drug signature attributes associated with MTX,
- – then this episode of liver transaminase abnormality was not due to MTX, thus this episode was a NON-CASE (Rule 1)
- If leflunomide occurred in the medication section of the patient's clinical notes for a given episode,
- – then this episode was a NON-CASE (Rule 2)
For classification, L2-regularized logistic regression as implemented by LIBLINEAR39 was used for all models. We used logistic regression for classification purposes and did not analyze feature weights explicitly. Because of LIBLINEAR's fast convergence we could efficiently train and classify thousands of episodes and then summarize the result into one final patient-level label.
Evaluation
Performance metrics included PPV (or precision), sensitivity (or recall), and F1 score. To compare model performance, six machine-learning baselines were used. Baselines 1, 2, and 3 were patient-level classifications (figure 5), in which all features were collapsed into a patient-level vector therefore ignoring episode groupings. Bag-of-words (BOW) was used as the feature representation for Baseline 1, and all CUIs found (comprehensive CUIs) for Baseline 2, and customized CUIs for Baseline 3. Baselines 4, 5, and 6 were episode-level classifications (figure 4), in which all features were collapsed into an episode-level vector for classification. BOW features were used for Baseline 4, CUIs (comprehensive CUIs) for Baseline 5, and customized CUIs for Baseline 6.
Figure 5:
CASE/NON-CASE classification: patient-level classification (‘o’ signifies a clinical note). LFT, liver function test.
Tenfold cross validation was performed on the training set. Models were additionally evaluated on the test sets for portability. Our final algorithm was further ported to a different EMR system, the Vanderbilt University RA set, for cross-site evaluation.
Results
From the pool of 1130 patients with RA with MTX exposure and elevated ALT/AST outcome, chart reviews of 480 patients confirmed 132 MTX liver toxicity CASES (27.5%) in the training set and 38 of 120 CASES for the test set (31.7%) (table 1). Cohen's κ measurement for inter-annotator agreement on the test set was 0.828.
Table 1:
Training and test set characteristics
| Set | CASE | NON-CASE | Total | Inter-annotator agreement (κ) |
|---|---|---|---|---|
| Training | 132 | 348 | 480 | Single annotated |
| Test | 38 | 82 | 120 | 0.828 |
Table 2 shows the 10-fold cross-validation performance of the six baseline systems. Table 3 shows the 10-fold cross-validation results of feature addition experiments. The best performing episode-level baseline was the simple BOW (Baseline 4). However, in that baseline not all words equated to events. For instance, words like ‘patient’ and ‘liver’ cannot be temporally anchored. At the same time, temporality (through DocTimeRel) is essential for this phenotype. Therefore, we decided not to build upon the BOW baseline. A much better strategy is to build upon CUIs indicating potentially clinically relevant events. Taking the entire CUI space is highly likely to lead to overfitting. Therefore, we pruned the CUI space through expert-guided feature selection (rather than automatic feature selection), using the customized dictionary, and we based our further system development on Baseline 6.
Table 2:
Tenfold cross-validation results of machine learning baseline models in the training set
| No. of features | PPV | Recall | F1 score | |
|---|---|---|---|---|
| Baseline 1 (patient-level BOW) | 48 078 | 0.711 | 0.727 | 0.719 |
| Baseline 2 (patient-level comprehensive CUI) | 14 265 | 0.738 | 0.727 | 0.733 |
| Baseline 3 (patient-level customized CUI) | 107 | 0.616 | 0.682 | 0.647 |
| Baseline 4 (episode-level BOW) | 48 078 | 0.813 | 0.758 | 0.784 |
| Baseline 5 (episode-level comprehensive CUI) | 14 265 | 0.797 | 0.742 | 0.769 |
| Baseline 6 (episode-level customized CUI) | 107 | 0.742 | 0.523 | 0.613 |
BOW, bag-of-words; CUI, concept unique identifier; PPV, positive predictive value.
Table 3:
Tenfold cross-validation results of customized dictionary with added features in the training set (feature contribution)
| Features | No. of features | PPV | Recall | F1 score |
|---|---|---|---|---|
| Setting 1: Baseline 6 + section parsing to discover mentions in relevant sections + section of relevant medications | 109 | 0.759 | 0.806 | 0.782 |
| Setting 2: Setting 1 + MTX signature | 169 | 0.738 | 0.829 | 0.781 |
| Setting 3: Setting 2 + DocTimeRel | 409 | 0.740 | 0.868 | 0.798 |
| Setting 4: Setting 2 + nearby words | 4806 | 0.781 | 0.884 | 0.829 |
| Setting 5: Setting 2 + nearby verbs’ part-of-speech tags | 875 | 0.762 | 0.891 | 0.821 |
| Setting 6: Setting 2 + nearby words + nearby verbs part-of-speech tags | 5512 | 0.780 | 0.907 | 0.839 |
| Setting 7: Setting 2 + nearby words + nearby verbs’ part-of-speech tags + DocTimeRel | 5752 | 0.800 | 0.899 | 0.847 |
| Setting 8: same feature settings as Setting 7 but patient-level classification | 5752 | 0.814 | 0.727 | 0.768 |
For examples with the features, see figure 3
MTX, methotrexate; PPV, positive predictive value.
To evaluate the best-performing baseline (table 2, Baseline 4) and the feature-rich model (table 3, Setting 7) based on highest F1 score, we further tested them on the test set (table 4).The best performing baseline (Baseline 4) achieved an F1 score of 0.642. The best performing feature-rich model from table 3 (Setting 7) achieved an F1 score of 0.829, which was comparable to its F1 score of 0.847 on the 10-fold cross validation in the training set. A rule-based baseline including only codified information from the structured EMR (see online supplement) was added and tested on the test set (table 4).
Table 4:
Results on the test set
| Models | PPV (95% CI) | Recall (95% CI) | F1 score (95% CI) |
|---|---|---|---|
| Best machine learning baseline from table 2—Baseline 4 | 0.590 (0.433 to 0.737) | 0.703 (0.530 to 0.841) | 0.642 (0.476 to 0.785) |
| Rule-based baseline (codified data only) | 0.750 (0.551 to 0.893) | 0.568 (0.345 to 0.729) | 0.646 (0.460 to 0.803) |
| Setting 7, table 3 | 0.756 (0.605 to 0.871) | 0.919 (0.781 to 0.983) | 0.829 (0.682 to 0.924) |
PPV, positive predictive value.
In addition, to further validate the PPV, we ran the best performing feature-rich model (table 3, Setting 7) on the remaining patients with RA and possible liver toxicity not selected for the training and test sets. From the patients labeled as CASE by the model, 40 charts were randomly pulled and evaluated by one of our domain experts (EWK). The resulting PPV was 0.75 (algorithm produced 30 correct labels out of 40 total labels). To test the portability of our algorithm in a different EMR system, a rheumatologist at Vanderbilt (NB) performed chart reviews on patients with RA with any MTX use, elevated transaminases >2× ULN, and available notes from Vanderbilt University for 103 patients and identified 41 CASES (39.8%) and 62 NON-CASES. The algorithm produced a PPV of 0.66 (95% CI 0.517 to 0.785), a sensitivity of 0.853 (95% CI 0.708 to 0.944), and F1 score of 0.745 (95% CI 0.598 to 0.857).
Discussion
We validated an EMR algorithm for automatic classification of RA cases with MTX-related liver toxicity that includes novel temporal relation discovery techniques applied to the clinical domain. We demonstrate the improvement in performance of episode-level classification (Baselines 4 and 5 from table 2, and Setting 7 from table 3) compared with patient-level classification (Baselines 1 and 2 from table 2, and Setting 8 from table 3). To avoid over-fitting, we chose to build on the customized dictionary with a rich set of features (table 2, Baseline 6). Our best system (table 3, Setting 7) that included temporal relations outperformed any of the machine-learning baselines in 10-fold validation in the training set. In addition, it maintained its performance when tested on the test set from Partners. The PPV was lower in a test set from Vanderbilt, possibly due to sparse temporal cues. However, sensitivity was maintained, which is arguably more important for a rare phenotype.40,41 To our knowledge this study is the first to address a highly temporally sensitive phenotype: liver toxicity secondary to recent RA-related MTX treatment using NLP. Our approach provides compelling evidence for using informatics approaches and state-of-the-art temporality NLP that might be relevant for developing EMR mining algorithms for other classes of pharmaceutical agents.
Section parsing alone increased the system performance from F1 score 0.613 (table 2, Baseline 6) to 0.782 (table 3, Setting 1). Through section parsing, the information extraction focused on only highly informative and relevant sections, thus reducing noise in the data. For example, the medication section of the clinical notes contains the most accurate source of medication information compared with past medical history that could include information on past use of medications.
Overall, the inclusion of medication attributes as features did not contribute to improved overall performance (table 3, Setting 2). However, it increased the sensitivity from 0.806 to 0.829 (at the expense of the PPV). This could be due to text describing discontinuing MTX which would be coded as a feature (status change) resulting in more false positives and reducing the PPV. It is possible that by adding MTX signature attributes, the MTX usage signal was strengthened. More decision power would then be shifted towards the MTX usage. As a balance, the decision weights assigned to other features, such as comorbidities, would be reduced accordingly. As a result, many CASE instances with weak but correct MTX usage signal would now be picked up, increasing the true positives; many NON-CASE instances with weak comorbidity signals would be identified as CASE as well, adding to the false positives and reducing PPV. The overall F1 score thus stagnates.
There are three groups of features aimed at capturing the temporally relevant information of a target term—DocTimeRel, the nearby words (admittedly, capture more than temporality), and the nearby verbs’ POS tag (table 3, Settings 3–7). Each of them brought a performance increase in F1 scores. The DocTimeRel model made use of nearby words and nearby verb tense, and linguistic cues such as prepositions. There may be a functional overlap among these three groups of features. The difference is that the DocTimeRel model also takes the POS sequence between the target event and its closest temporal expression to capture the temporal aspect for the final prediction. The Apache cTAKES DocTimeRel model was trained on a different data set, the THYME corpus, which comprised colon and brain cancer pathology, radiology, and oncology notes. By explicitly modeling the other two temporally relevant feature sets and by adding episode-level rules (MTX absent, leflunomide mention) to filter out NON-CASE episodes, we enhanced performance of DocTimeRel in the RA dataset. DocTimeRel anchored each term to the DCT, providing a coarse timeframe. Nearby words captured some important lexical features like ‘weekly’, ‘tomorrow’, ‘stopped’, while nearby verbs’ POS could help identify useful tense patterns like past tense for ‘stopped’. Combining the three temporality feature sets gave the best performance, which demonstrates that they captured comprehensive temporal information associated with the liver-toxicity phenotype.
We analyzed the results on the Vanderbilt dataset and found that the counts of the overlap value for DocTimeRel in Vanderbilt's data are much higher than those in Partners data despite the relatively similar dataset sizes (2516 vs 1432 for overlap; 786 vs 1104 for before; 9 vs 82 for before/overlap; 40 vs 300 for after). If there is little or no context discovered information (ie, not enough temporality cues) around a key term, then the model assigns overlap as the DocTimeRel. However, the model requires a strong signal to assign the values of before, after, before/overlap. This suggests that the Vanderbilt data are less diverse in terms of temporal cues, making DocTimeRel less informative.
In summary, we achieved our best performance results by adding the temporal features. These temporal features aligned the temporal perspective of the relevant events; thus, drug exposure and comorbidity mentions could be differentiated by their temporal relevance to the transaminase date. Comparing the settings with or without temporal features (table 3, Settings 2 and 7), PPV increased by 6.2 points, sensitivity by 7 points, and F1 by 6.6 points. Such improvements demonstrate the usefulness of temporal features for this time-sensitive task.
Conclusion
In this paper we present a methodology for mining the wealth of clinical data in EMRs to automatically identify a temporally sensitive phenotype—MTX-related liver toxicity among patients with RA using a novel cTAKES module, DocTimeRel. We innovatively cast this task as an episode-level classification problem where knowledge is represented through a CUI-coded customized dictionary, temporal signals, drug attributes, and section parsing. In addition to enabling classification of adverse drug events among large cohorts of patients with RA, our work contributes to the general trend of methodology development for phenotyping using the EMR data, including its free text.
Contributors
All authors contributed to the design, experiments, analysis, and writing the manuscript.
Funding
The project described is supported by Grant Number R01LM010090 (THYME) and U54LM008748 (i2b2) from the National Library of Medicine and NIH grants U01 GM092691 (PGRN), AR049880, AR052403, AR047782, and 1R01GM103859-01A1.
Competing interests
GKS is on the Advisory Board of Wired Informatics, LLC which provides services and products for clinical NLP applications.
Ethics approval
The study was approved by the Partners HealthCare Institutional Review Board.
Provenance and peer review
Not commissioned; externally peer reviewed.
SUPPLEMENTARY MATERIAL
Supplementary material is available online at http://jamia.oxfordjournals.org/.
REFERENCES
- 1.Weinblatt ME, Trentham DE, Fraser PA, et al. Long-term prospective trial of low-dose methotrexate in rheumatoid arthritis. Arthritis Rheum. 1988;31:167–75. [DOI] [PubMed] [Google Scholar]
- 2.Kent PD, Luthra HS, Michet C, Jr, et al. Risk factors for methotrexate-induced abnormal laboratory monitoring results in patients with rheumatoid arthritis. J Rheumatol. 2004;31:1727–31. [PubMed] [Google Scholar]
- 3.Curtis JR, Beukelman T, Onofrei A, et al. Elevated liver enzyme tests among patients with rheumatoid arthritis or psoriatic arthritis treated with methotrexate and/or leflunomide. Ann Rheum Dis. 2010;69:43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kremer JM, Furst DE, Weinblatt ME, et al. Significant changes in serum AST across hepatic histological biopsy grades: prospective analysis of 3 cohorts receiving methotrexate therapy for rheumatoid arthritis. J Rheumatol. 1996;23:459–61. [PubMed] [Google Scholar]
- 5.Kremer JM, Alarcon GS, Lightfoot RW, Jr, et al. Methotrexate for rheumatoid arthritis. Suggested guidelines for monitoring liver toxicity. American College of Rheumatology. Arthritis Rheum. 1994;37:316–28. [DOI] [PubMed] [Google Scholar]
- 6.Visser K, Katchamart W, Loza E, et al. Multinational evidence-based recommendations for the use of methotrexate in rheumatic disorders with a focus on rheumatoid arthritis: integrating systematic literature research and expert opinion of a broad international panel of rheumatologists in the 3E Initiative. Ann Rheum Dis. 2009;68:1086–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annual Symp Proc. 2009;2009:497–501. [PMC free article] [PubMed] [Google Scholar]
- 8.Waudby CJ, Berg RL, Linneman JG, et al. Cataract research using electronic health records. BMC Ophthalmol. 2011;11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kho AN, Pacheco JA, Peissig PL. et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kullo IJ, Fan J, Pathak J, et al. Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J Am Med Inform Assoc. 2010;17:568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C, Karlson EW, Canhao H, et al. Automatic prediction of rheumatoid arthritis disease activity from the electronic medical records. PLoS One. 2013;8:e69932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Jiang M, Oetjens M, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc. 2011;18:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther. 2011;89:379–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao K, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res. 2010;62:1120–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll R, Thompson W, Eyler A. et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc. 2012;19:e162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ananthakrishnan AN, Cai T, Savova G, et al. Improving case definition of Crohn's disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis. 2013;19:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ananthakrishnan AN, Cagan A, Gainer VS, et al. Normalization of plasma 25-hydroxy vitamin D is associated with reduced risk of surgery in Crohn's disease. Inflamm Bowel Dis. 2013;19:1921–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ananthakrishnan AN, Gainer VS, Cai T, et al. Similar risk of depression and anxiety following surgery or hospitalization for Crohn's disease and ulcerative colitis. Am J Gastroenterol. 2013;108:594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ananthakrishnan AN, Gainer VS, Perez RG, et al. Psychiatric co-morbidity is associated with increased risk of surgery in Crohn's disease. Aliment Pharmacol Ther. 2013;37:445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathak J, Bailey KR, Beebe CE. et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. J Am Med Inform Assoc. 2013;20:e341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin C, Miller T, Dligach D, et al. Feature engineering and selection for rheumatoid arthritis disease activity classification using electronic medical records. ICML Workshop on Machine Learning for Clinical Data analysis Edinburgh, UK, 2012. [Google Scholar]
- 22.Lin C, Miller T, Dligach D, et al. Maximal information coefficient for feature selection for clinical document classification (extended abstract). ICML Workshop on Machine Learning for Clinical Data Edinburgh, UK, 2012. [Google Scholar]
- 23.Tang B, Wu Y, Jiang M, et al. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc. 2013;20:828–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grouin C, Grabar N, Hamon T, et al. Eventual situations for timeline extraction from clinical reports. J Am Med Inform Assoc. 2013;20:820–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sohn S, Wagholikar KB, Li D, et al. Comprehensive temporal information detection from clinical text: medical events, time, and TLINK identification. J Am Med Inform Assoc. 2013;20:836–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Wang Y, Liu T, et al. An end-to-end system to identify temporal relation in discharge summaries: 2012 i2b2 challenge. J Am Med Inform Assoc. 2013;20:849–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovacevic A, Dehghan A, Filannino M, et al. Combining rules and machine learning for extraction of temporal expressions and events from clinical narratives. J Am Med Inform Assoc. 2013;20:859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts K, Rink B, Harabagiu SM. A flexible framework for recognizing events, temporal expressions, and temporal relations in clinical text. J Am Med Inform Assoc. 2013;20:867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irvine AK, Haas SW, Sullivan T. TN-TIES: a system for extracting temporal information from emergency department triage notes. AMIA Annual Symposium Proceedings 2008;328.–. [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan T, Irvine A, Haas SW. It's All Relative: Usage of Relative Temporal Expressions in Triage Notes. Silver Spring, Maryland: American Society for Information Science and Technology, 2008. [Google Scholar]
- 31.Apache Clinical Text Analysis and Knowledge Extraction System (cTAKES). 2013. http://ctakes.apache.org(accessed 9 Oct 2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savova G, Masanz J, Ogren P, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kremer JM, Lee RG, Tolman KG. Liver histology in rheumatoid arthritis patients receiving long-term methotrexate therapy. A prospective study with baseline and sequential biopsy samples. Arthritis Rheum. 1989;32:121–7. [DOI] [PubMed] [Google Scholar]
- 34.Unified Medical Language System (UMLS). 2013. http://www.nlm.nih.gov/research/umls/ (accessed 9 Oct 2014). [Google Scholar]
- 35.Miller T, Bethard S, Dligach D, et al. Discovering Temporal Narrative Containers in Clinical Text. Proceedings of the 2013 Workshop on Biomedical Natural Language Processing Sofia, Bulgaria: Association for Computational Linguistics; , 2013. [Google Scholar]
- 36.THYME. 2013. http://thyme.healthnlp.org(accessed 9 Oct 2014).
- 37.Styler W, Bethard S, Finan S, et al. Temporal annotations in the clinical domain. Trans Assoc Comput Linguist, 2014. [PMC free article] [PubMed] [Google Scholar]
- 38.Marcus M, Santorini B, Marcinkiewicz M. Building a large annotated corpus of English: the Penn Treebank. Comput Linguistic. 1993;19:313–30. [Google Scholar]
- 39.Fan R, Chang K, Hsieh C, et al. LIBLINEAR: a library for large linear classification. J Mach Learn Res. 2008;9:4. [Google Scholar]
- 40.Overby CL, Pathak J, Gottesman O, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc. 2013;20:e243–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denny JC. Chapter 13: mining electronic health records in the genomics era. PLoS Comput Biol. 2012;8:e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]