Abstract
A novel protein-folding function of RNA has been recognized, which can outperform previously known molecular chaperone proteins. The RNA as a molecular chaperone (chaperna) activity is intrinsic to some ribozymes and is operational during viral infections. Our purpose was to test whether influenza hemagglutinin (HA) can be assembled in a soluble, trimeric, and immunologically activating conformation by means of an RNA molecular chaperone (chaperna) activity. An RNA-interacting domain (RID) from the host being immunized was selected as a docking tag for RNA binding, which served as a transducer for the chaperna function for de novo folding and trimeric assembly of RID-HA1. Mutations that affect tRNA binding greatly increased the soluble aggregation defective in trimer assembly, suggesting that RNA interaction critically controls the kinetic network in the folding/assembly pathway. Immunization of mice resulted in strong hemagglutination inhibition and high titers of a neutralizing antibody, providing sterile protection against a lethal challenge and confirming the immunologically relevant HA conformation. The results may be translated into a rapid response to a new influenza pandemic. The harnessing of the novel chaperna described herein with immunologically tailored antigen-folding functions should serve as a robust prophylactic and diagnostic tool for viral infections.—Yang, S. W., Jang, Y. H., Kwon, S. B., Lee, Y. J., Chae, W., Byun, Y. H., Kim, P., Park, C., Lee, Y. J., Kim, C. K., Kim, Y. S., Choi, S. I., Seong, B. L. Harnessing an RNA-mediated chaperone for the assembly of influenza hemagglutinin in an immunologically relevant conformation.
Keywords: protein folding, chaperna, neutralizing antibody, viral infection
Molecular chaperones are involved both in the facilitation of protein folding and prevention of protein aggregation, which would otherwise lead to cytotoxic consequences (1, 2). Conventionally, these pivotal functions have been considered unique properties of chaperones and chaperonins, which are proteins themselves (3). Nonetheless, recent reports suggest that RNA molecules can also function as chaperones and are extremely effective in executing the folding of a variety of proteins both in vivo and in vitro (4, 5). The potential role of RNAs in modulating aggregation and amyloid formation has been reported for the p53 tumor suppressor (6). Moreover, chaperone activity has been discovered as intrinsic to some ribozymes, which are RNA molecules that function as catalysts: both in the M1 RNA ribozyme that is responsible for the maturation of tRNAs and in the large rRNAs that function as peptidyl transferase during ribosome-dependent protein synthesis (7–9). Being distinct from the RNA chaperone, which is a protein that facilitates structural alteration of RNA molecules, the term for RNA as a molecular chaperone (chaperna) refers to an RNA molecule that serves as a chaperone for the folding of client proteins (10). Nevertheless, neither the extent of its involvement in the folding and homeostasis of normal cellular proteins nor its efficacy in the folding of difficult-to-express proteins has been explored. It is noteworthy that the proper use of molecular chaperones for the in vivo folding of recombinant proteins in Escherichia coli has been reported only in isolated cases, and no study of considerable size that has shown broad efficacy has been conducted (11). Therefore, the possibility that the moonlighting activity of RNAs as chaperones beyond the known canonical cellular functions could be used as an efficient vehicle for the folding and assembly of proteins requires dedicated exploration.
With respect to recombinant vaccines, the assembly of monomeric antigens into oligomeric structures is crucially important for the ligation of B cell receptors to enhance immunogenicity and to induce relevant neutralizing (NT) antibody responses toward protection (12, 13). In the present study, we show that a chaperna is remarkably effective in the folding and trimeric assembly of influenza hemagglutinin (HA) into an immunologically activating conformation. A convenient avenue for harnessing the chaperone function is to fuse genetically the target protein of interest with an RNA-interacting domain (RID) as a docking tag that enables interaction with cellular RNAs (14). A judicious choice of the RID is required for a biomolecule to function as a chaperone without physically interfering with the oligomeric structure of the target antigen. Therefore, the RID should preferably be small and sufficiently flexible not to interfere with the multimeric assembly of the target antigen, and moreover, it should be intrinsically nonimmunogenic (15). In this study, a small tRNA-binding domain of lysyl-tRNA synthetase (LysRS; aa positions 1–70), which was derived from the host being immunized, was selected as the transducer for the chaperna function of cellular RNAs. The N-terminal domain is intrinsically disordered but can switch from an unfolded structure into an α-helical conformation in vitro after tRNA binding (16, 17). This domain has been shown to interact in vitro with nonspecific tRNAs (18).
By exploiting the chaperna function, herein, we demonstrate, for the first time, to our knowledge, that influenza HA can be assembled into a soluble, trimeric, and immunologically relevant conformation. The influenza HA globular domain (HAgD), including the host receptor-binding domain, was produced predominantly in the trimeric form, and remarkably, mutations that affect the tRNA-binding domain rendered the trimer assembly defective.
Immunization of mice with this purified HA elicited a high degree of hemagglutination inhibition (HI) and high titers of NT antibodies without crossreaction with the selfRID and provided a sterile protection against a lethal challenge.
Pandemics and annual influenza epidemics have been the causes of considerable mortality and a heavy burden on human health (19, 20). The speedy and timely delivery of vaccines is crucial for effective responses to pandemics. Conventional systems that rely on virus culture, either from embryonated eggs or cell cultures, are not likely to meet the time requirements. Taking advantage of the novel concept of protein folding, as presented herein, should facilitate the development of pandemic vaccines. RNA viruses are responsible for most of the emerging and re-emerging viral infections (21), and the chaperna described herein may offer a novel prophylactic and diagnostic platform for the effective control and management of these infections.
MATERIALS AND METHODS
Cloning, expression, and protein purification
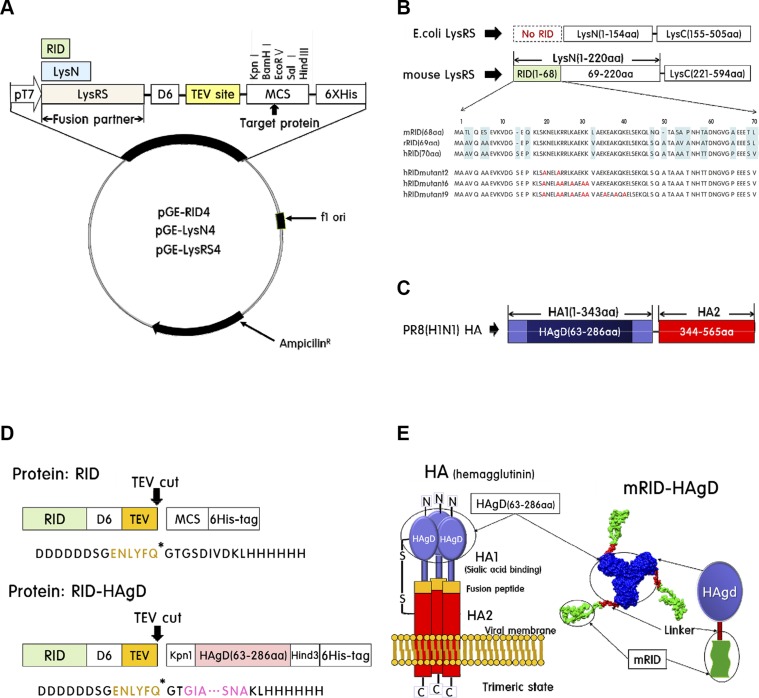
Basically, 3 different RNA-interaction domains—RID, N-terminal domain of LysRS (LysN), and LysRS—were used here for construction of expression vectors. N-Terminal appendage (70 aa long) of human LysRS, which is known to interact with nonspecific tRNA in vitro, served as RID of human origin (hRID). For comparison, LysRS from E. coli (eLysRS; 57.6 kDa) or LysN (24.8 kDa) were also used (14). Expression vectors of wild-type (WT) hRID and RNA-nonbinding mutant hRID (hRIDmu) were constructed in 3 versions. The first mutant has 2 aa substitutions (K23A and K27A), the second mutant carries 6 substitutions with alanine (K19A, K23A, R24A, K27A, K30A, and K31A), and the third mutant contains 9 substitutions with alanine (K19A, K23A, R24A, K27A, K30A, K31A, K35A, K38A, and K40A). Subsequently, the HAgD, originating from influenza A/Puerto Rico/8/34 (PR8; H1N1) virus, was inserted between the KpnI and HindIII restriction sites to construct the hRIDmu-HAgD.
Promiscuous gene expression (pGE)-RID, derived from the pGE-LysRS4 vector, served as an expression vector in E. coli (22). First, the LysRS gene of the pGE-LysRS4 vector was deleted by means of restriction enzymes NdeI and KpnI. The genes of RIDs of human, murine, and rabbit origins were amplified by PCR, resulting in hRID, mouse RID (mRID), and rabbit RID (rRID); each was inserted into pGE-LysRS4 (separately). The RID4 expression cassette is composed of an RID, a D6 linker (hexa-aspartic acid sequence) to aid soluble expression, an alternating serine/glycine space linker, a tobacco etch virus (TEV) protease recognition cleavage site (ENLYFQ), a multiple cloning site (MCS; GTGSDIVDKL: KpnI/BamHI/EcoRV/SalI/HindIII), and a hexahistidine tag (6xHis) for nickel (Ni)-affinity purification, under control of the T7 promotor (23). HAgD cDNA of the PR8 (H1N1) strain was amplified by RT-PCR and was inserted between the KpnI and HindIII sites of the pGE-RID4 vector. All of the constructs were designed to yield 2 fragments—RID and HAgD—after cleavage with the TEV protease. hRID served as a template for mutagenesis of amino acid residues involved in RNA binding.
All of the recombinant proteins were expressed in an E. coli strain BL21 Star (DE3) pLysS (Invitrogen, Carlsbad, CA, USA) and were purified from the soluble fraction of cell lysates. The primary culture was grown overnight in 3 ml of the Luria-Bertani medium containing 50 μg/ml ampicillin and 30 μg/ml chloramphenicol at 37°C. Large-scale culture was implemented in 500 ml of the Luria-Bertani medium that was inoculated with 25 ml culture broth and incubated at 37°C until optical density at 600 nm (OD600) of 0.5–0.7 was reached. Protein expression was induced by the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside at various temperatures. Cells were harvested at 3000 g when an OD600 of 1.3–2.0 was reached. The cell lysates obtained by sonication were centrifuged at 12,000 g for 10 min, and this procedure separated them into supernatant and precipitate fractions.
Proteins were purified by Ni-affinity chromatography. Each soluble fraction equilibrated in Buffer A [50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 10% glycerol, 2 mM 2-ME, 0.05% Triton X-100, and 10 mM imidazole] was applied to an Ni-nitrilotriacetic acid resin (GE Healthcare, Little Chalfont, United Kingdom) in a column pre-equilibrated with Buffer A. After a wash with Buffer A, proteins were eluted with a linear gradient of imidazole 10–300 mM by means of Buffer B (Buffer A supplemented with 300 mM imidazole). Each fraction was analyzed by SDS-PAGE, and the fractions containing proteins of interest were pooled, concentrated by centrifugal filters (Centriprep; EMD Millipore, Billerica, MA, USA), and then dialyzed against Buffer C [50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 0.1 mM EDTA, and 0.1% Tween 20]. After SDS-PAGE, followed by Coomassie staining, the concentration of purified proteins was determined using bovine serum albumin (BSA; Amresco, Solon, OH, USA) of known concentrations as a standard.
The HAgD without RID fusion formed insoluble aggregates, which necessitated refolding in vitro. The E. coli lysate was centrifuged, and the inclusion bodies were washed with 1% Triton X-100. The pellets were denatured and solubilized with 6 M guanidine hydrochloride, supplemented with 1 mM DTT at a final protein concentration of 5 mg/ml. The solution containing the monomeric HAgD was dialyzed against 20 mM Tris-HCl (pH 8.0) to remove the denaturation agents. The extent of refolding into oligomeric assembly was next analyzed by size-exclusion chromatography (SEC).
Structural modeling
To predict the potential trimerization capacity of the mRID-HAgD recombinant protein, the crystal structures of influenza PR8 (H1N1) HA (Protein Data Bank ID: 1RU7) and the N-terminal domain of Brugia malayi asparaginyl-tRNA synthetase (Protein Data Bank ID: 2KQR) were retrieved from http://www.rcsb.org/. All heteroatoms and water molecules were eliminated from the structure. With the use of homology modeling by Modeler (https://salilab.org/), the RID-HAgD fusion version, including a 14-aa linker (DDDDDDSGENLYFQ), was built, and the asparaginyl-tRNA synthetase structure served as a template for mRID and rRID (24, 25). The trimerization of each mRID-HAgD and rRID-HAgD structure was confirmed by ClusPro 2.0 protein–protein docking (Boston University, Boston, MA, USA) (26, 27). The lowest energy of an mRID-HAgD trimer was −655.8 kcal/mol, and for the rRID-HAgD trimer, it was −755.9 kcal/mol. The final structures were visualized by means of a surface model in the University of California, San Francisco (San Francisco, CA, USA) Chimera software.
The effect of RNA on the solubility of proteins
To confirm the folding and stabilization effects of RNA on soluble proteins, cell lysates with a recombinant protein, mRID- or rRID-HAgD, were obtained by sonication or B-PER lysis (Bacterial Protein Extraction Reagent, 78248; Thermo Fisher Scientific, Waltham, MA, USA). Cell lysates (T) were centrifuged at 12,000 rpm for 10 min to obtain soluble (S) and pellet (P) fractions. RNase A (250 μg/ml; 27062; iNtRON Biotechnology, Seongnam, Korea) was added to the soluble fraction and incubated for 15 min at 37°C to deplete this fraction of RNA. The mixture (ST) was centrifuged at 14,000 rpm for 20 min and thus, separated into the supernatant of soluble fraction (SS) and the precipitate of soluble fraction (SP) fractions. The soluble fraction (S) without RNase A served as a control. All of the fractions obtained were identified by SDS-PAGE analysis in a 12% gel.
SEC
The oligomeric status of purified recombinant proteins was analyzed by SEC at 4°C on a Superdex-200 analytical gel-filtration column (GE Healthcare). The column was equilibrated with a buffer [50 mM Tris-HCl (pH 7.5), 300 mM NaCl, 2 mM 2-ME, and 0.05% Triton X-100] and was calibrated using broad-range MW markers: ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (44 kDa), and Blue Dextran 2000 (2000 kDa; GE Healthcare).
Fetuin-binding assay
For this assay, 200 µl fetuin (Sigma-Aldrich, St. Louis, MO, USA) per well at 200 µg/ml was used for coating 96-well Nunc plates (Thermo Fisher Scientific) and was incubated at 4°C overnight. The plates were washed with PBS containing 50 mM Tris-HCl (pH 7.4), with 0.05% Tween 20 (PBST), and were blocked with 1% BSA in PBST for 1 h at room temperature. Next, 100 μl of each recombinant HAgD protein at various concentrations was added into each well, and then the plates were incubated for 2 h at room temperature, followed by washing with PBST. Recombinant enhanced green fluorescent protein (eGFP) (or RID) served as a control protein. After that, 100 μl of an anti-6xHis tag antibody conjugated with horseradish peroxidase (HRP; Thermo Fisher Scientific) was added into each well at a dilution of 1:1000 and was incubated for 1 h at room temperature. After a wash with PBST, 100 μl of the HRP substrate solution (BD Biosciences, San Jose, CA, USA) was added into each well, and then the plate was developed in the dark for 30 min. The colorimetric reaction (blue to yellow) was stopped by addition of 50 μl/well 2 N H2SO4, and OD450 was measured on a microplate reader (FluoStar Optima; BMG Labtech, Ortenberg, Germany).
Immunization and a virus challenge
All of the animal experiments were carried out in strict accordance with the recommendations of the Korean Food and Drug Administration. Immunization of rabbits was conducted at animal facilities of Young-In Frontier (Gasandong, South Korea). Namely, 1.0 mg of an rRID fusion protein emulsified in Freund’s complete adjuvant was injected intradermally at multiple sites on the back of each rabbit. Starting from 4 wk after the first immunization, each animal was boosted twice with the same protein–adjuvant mixture every 2 wk. Rabbit blood was collected 1 wk after each boost and was centrifuged to prepare serum and stored at −70°C. The experimental protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Young-In Frontier.
Mouse experiments were carried out on 6-wk-old female BALB/c mice (Orient Bio, Seongnam, South Korea). Every group of mice (3–6/group) was immunized via the intraperitoneal route with 4–20 μg of a purified RID-fused recombinant protein or HAgD without RID, along with 50 μg Imject Alum adjuvant (Thermo Fisher Scientific) on d 0 (primary), 14 (first boost), and 28 (second boost). After the first immunization, each mouse was boosted twice, at 2 wk apart. PBS-immunized mice served as a control. Blood samples were collected via transorbital bleeding, 14 d after each boost, and serum was prepared, aliquoted, and stored at −70°C. The protocol of the experiment was evaluated and approved by the IACUC of the Yonsei Laboratory Animal Research Center (Permits: IACUC-A-201503-202-02, IACUC-A-201512-558-02, and IACUC-A-20160331-061).
Two weeks after the secondary immunization, all of the mice were challenged intranasally with 1 time LD50 [i.e., 103 plaque-forming units (PFUs), of the PR8 (H1N1) virus under anesthesia with avertin or mock infected with PBS as a control]. The survival rate and changes in weight of the challenged mice were measured every day for 2 wk. After the virus challenge, mouse blood samples were collected via transorbital bleeding and were centrifuged at 4°C for 30 min to prepare serum samples.
Determination of serum antibody titers by an ELISA
To this end, 96-well Nunc plates (Thermo Fisher Scientific) were coated with 100 ng/well RID, an RID-fused protein, or 106 PFU/well PR8 (H1N1) virus and were incubated at 4°C overnight. The plates were washed with PBST and were blocked with 1% BSA in PBST for 1 h at room temperature. Next, 100 μl serum samples, at various dilutions, was added into each well, and the plates were incubated for 1 h at room temperature, followed by washing with PBST. One hundred microliters of a secondary goat anti-rabbit IgG antibody or goat anti-mouse IgG antibody conjugated with HRP (Sigma-Aldrich) was added into each well at a dilution of 1:10,000 and was incubated for 1 h. After a wash with PBST, 100 μl of the substrate 3,3′,5,5′-tetramethylbenzidine solution (BD Biosciences) was added into each well, and then the plate was developed in the dark for 30 min. The colorimetric reaction (blue to yellow) was stopped by addition of 2 N H2SO4 50 μl/well, and OD450 was measured on a microplate reader (FLUOstar Optima; BMG Labtech).
Western blot analysis
This analysis was carried out by means of the final serum samples from immunized rabbits and mice. Various dilutions of each serum were used for the analysis of the antibodies in the serum samples. As secondary antibodies, a goat anti-rabbit IgG mAb or goat anti-mouse IgG antibody conjugated with HRP (Sigma-Aldrich) was applied. For the detection of antibodies against HA in serum samples of PR8 (H1N1) virus-infected mice, 10 μg of an RID-HAgD protein was loaded onto a gel for SDS-PAGE, followed by Western blot (WB), including mouse serum (dilution of 1:1000). The protein gels were blotted in the Trans-Blot Turbo transfer apparatus with PVDF Midi transfer packs (Bio-Rad Laboratories, Hercules, CA, USA). The PVDF membranes were immediately transferred to a blocking solution—5% skim milk in Tris-buffered saline/Tween 20 (TBST)—and were incubated with gentle agitation for 1 h at room temperature. After a wash with TBST, the membranes were incubated for 2 h or overnight with gentle agitation at 4°C in 15 ml of the primary-antibody solution with rabbit or mouse serum (1:1000 dilution) and 3% BSA in TBST. The blots were rinsed 3 times with TBST and were incubated in 15 ml of a secondary-antibody solution containing an HRP-conjugated goat anti-mouse IgG antibody (Bio-Rad Laboratories; 1:10,000 dilution) or an HRP-conjugated goat anti-rabbit IgG antibody (Bio-Rad Laboratories; 1:10,000 dilution) in PBST for 40 min with gentle agitation at room temperature. The membranes were washed with TBST, followed by development by means of an ECL detection reagent (Bio-Rad Laboratories) for 1 min. After that, images were captured on X-ray film using a developing and fixing solution in a darkroom.
A virus NT assay
Blood samples were collected from mice by transorbital bleeding and were allowed to clot at room temperature. The clots were removed, and the samples were centrifuged in a microcentrifuge at 4°C for 10 min at 5500 rpm. The clarified serum samples were transferred to sterile microcentrifuge tubes and were heat inactivated at 56°C for 30 min. The heat-inactivated serum samples were subjected to 2-fold serial dilutions in 96-well plates with minimum essential medium (Thermo Fisher Scientific). For the analysis of titers of NT antibodies against the influenza virus, equal volumes (50 μl) of the influenza virus (100 PFUs) were added to the diluted serum samples and mixed. The 96-well plates containing virus and serum samples were incubated at 37°C for 2 h. These mixtures were then adsorbed to 24-well plates containing confluent Madin-Darby canine kidney cells. The agar-overlaid plates were incubated at 32°C in a 5% CO2 incubator. The cell monolayers were stained with crystal violet at 4 d postinfection. In each assay, the serum samples were diluted and analyzed in duplicate, and each assay was performed at least twice. NT titers were calculated from dilutions that correspond to a 50% plaque reduction compared with the control.
The HI assay
Serum samples were treated with a receptor-destroying enzyme to remove nonspecific agglutination inhibitors and heated at 56°C for 1 h. The serum samples were subjected to serial 2-fold dilutions with PBS in 96-well plates. Equal volumes (50 μl) of an influenza virus solution (4 hemagglutination units) were then added to the diluted serum samples and mixed; next, the plate was incubated at 37°C for 1 h. After that, an equal volume of a 1% chicken erythrocyte suspension was added with incubation at 4°C for 1 h. The HI antibody titer of each serum was expressed as the reciprocal of the highest dilution of the sample that completely inhibited hemagglutination.
Mouse infection and preparation of lung-tissue samples
Six-week-old female BALB/c mice (Orient Bio) were anesthetized before the intranasal infection with 50 μl of a virus suspension and euthanized by cervical dislocation to minimize suffering. The viral replication in the respiratory tract was measured in the lungs. Whole lungs were excised for viral titration. The whole lungs were homogenized with a homogenizer in PBS and centrifuged to remove cell debris. These supernatants were transferred to a new tube and stored at −80°C until analysis.
RESULTS
Cloning and soluble expression of RID-fused recombinant antigens
Schematic presentation of expression vectors is given in Fig. 1. As initial screening for soluble expression, a variety of fusion vectors—pGE-LysRS (4), pGE-RID4, and pGE-LysN4—was tested (Fig. 1A). LysRS, which is composed of LysN and a C-terminal domain, was derived from E. coli (Fig. 1B). LysRS of eukaryotic origin carries an extra N-terminal domain (RID) that is absent in the E. coli counterpart (Fig. 1B). As a target antigen, the HAgD of strain PR8 (H1N1), containing the receptor-binding site, was used (Fig. 1C, D). As an RID, the most N-terminal tRNA-binding domain of ∼70 aa from LysRS was derived from each of 3 species—humans, mice, and rabbits—to obtain hRID, mRID, and rRID, respectively, and hRIDmu (Fig. 1B). In addition to RIDs, the LysN (amino acid positions 1–220) and the whole LysRS from mice and rabbits were cloned as fusion partners for the soluble expression of HAgD and other test proteins. The fusion linker contains the ENLYFQ sequence for the site-specific cleavage by TEV protease, if needed (Fig. 1D). The schematic diagrams of HA trimers in the mature virion and of the recombinant RID-HAgD fusion protein are shown in Fig. 1E. According to some reports, the solubility of fusion proteins is somewhat influenced by the location of the fusion tag (28, 29). The N-terminal fusion of mRID increased water-soluble expression by ∼30% compared with the C-terminal fusion (Supplemental Fig. 1B). In addition, the N-terminal fusion protein mRID-HAgD was eluted mainly in a trimeric form, whereas the C-terminal fusion protein HAgD-mRID was eluted at ∼70% in a dimeric state (Supplemental Fig. 1D). Therefore, all fusion tags in this study were placed in the N-terminal part of target proteins.
Figure 1.
Construction of the pGE-RID4 vector and HAgD viral protein. A) Schematic illustration of the pGE-RID (4) vector used for the expression of fusion proteins in E. coli. B) Sequence homology of RIDs of LysRS from humans, mice, and rabbits. E. coli LysRS has no RID sequence. C) The globular head domain of HA consists of aa 63–286 in the HA1 subunit. D) Modular representation of the fusion protein. By the TEV protease, the fusion protein is cleaved into an RID and HAgD carrying a 6xHis tag. E) Left: schematic illustration of trimeric HA composed of the HA1 globular domain (containing HAgD) and HA2 of mostly α-helical conformation. Right: structure modeling of mRID-HAgD.
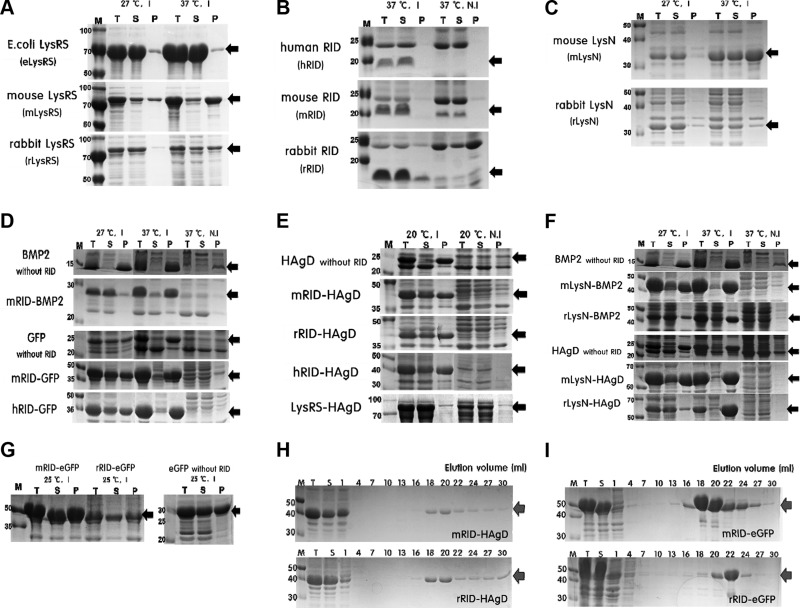
Expression at lower temperature increased the solubility of the proteins (Fig. 2B–F), in agreement with findings that the slowing down of translation enhances the proper folding and solubility of proteins (30). As a control, direct expression of GFP or bone morphogenetic protein 2 (BMP2) without fusion to an RID yielded predominantly a precipitate of insoluble aggregates under all of the temperature conditions tested (Fig. 2D). Initial screening of LysRS proteins for solubility revealed that the eLysRS had superb solubility compared with the counterparts of mouse or rabbit origin (Fig. 2A). Parallel analyses showed that the RIDs of all 3 species are produced predominantly in the soluble form (>95% for hRID and mRID and ∼80% for rRID, respectively; Fig. 2B). Of note, the RID equivalent is absent in E. coli LysRS (Fig. 1B) and therefore, could not be tested. Thus, the RIDs of human and animal origins were further explored as a docking tag for potential soluble expression of target proteins: BMP2, GFP, and HAgD (Fig. 2D, E). As shown in Fig. 2E, unlike the HAgD without RID, HAgDs as a fusion to an RID of varied origin were expressed as soluble proteins with a good yield (20–40% of total protein). The solubility of all of the recombinant proteins was greatly enhanced (by ∼5–40%) compared with direct expression without fusion (Fig. 2D, E). Overall, the expression of fusion partners of mouse origin (mRID, mLysN, mLysRS) was higher than the expression of fusion partners of rabbit origin, but their solubility levels were similar (Fig. 2A, C, E, F). Despite minor differences, each RID fusion showed a strongly increased total soluble yield of the respective proteins. Because there were no significant differences between LysN and an RID in promotion of the solubility of a reporter protein, RIDs were chosen as a fusion tag for the rest of the experiments: an RID of small size (∼8 kDa) is not expected to interfere physically with trimeric assembly of HAgD and as a docking tag, is less immunogenic than LysN (∼28 kDa) or LysRS (∼57.6 kDa; Fig. 2A–C). Thus, mRID-HAgD and rRID-HAgD were chosen as immunogens for inoculation of mice or rabbits, respectively. In parallel, eGFP was used as a reporter for solubility as a fusion to RID, where its solubility ratio was ∼50% at 25°C (Fig. 2G).
Figure 2.
Soluble expression and purification of recombinant proteins in E. coli. A) A comparison of LysRS proteins derived from E. coli, mice, and rabbits in terms of their solubility and expression levels. I, induction; N.I, noninduction; T, total extract; S, soluble fraction; P, pellet fraction. Black arrows, target proteins. B) A comparison of RIDs derived from humans, mice, and rabbits in terms of their solubility and expression levels. All of the RID proteins were expressed solubly at 37°C. C) Soluble expression data on each LysN (mouse and rabbit origin). D) Soluble expression data on RID-fused proteins. E) Expression profiles of recombinant proteins RID-HAgD and eLysRS-HAgD and HAgD without RID. F) Soluble expression of LysN-fused proteins. G) Soluble expression of RID-eGFP and eGFP without RID, a control fusion protein. H) SDS-PAGE data on purification of RID-HAgD proteins that were expressed at 20°C. I) Purification data on mRID-eGFP and rRID-eGFP proteins expressed at 25°C as a control protein. Red arrows, purified target proteins. All of the data were obtained in duplicate.
Purification and biochemical characterization of HA proteins
The proteins were purified from 500 ml culture by 1-step Ni chromatography. After cell lysis and centrifugation, the soluble fraction containing the bulk of recombinant proteins was loaded onto the Ni column, and proteins were eluted with a linear gradient of imidazole, ranging from 10 to 300 mM. RID-HAgD proteins with the 6xHis tag were purified effectively from the soluble fraction. The SDS-PAGE analysis revealed that the major protein band corresponds to the expected molecular mass of ∼40 kDa (Fig. 2E, G). RID-fused eGFP with the 6xHis tag was also purified in abundance from the soluble fraction as a control. Fractions enriched in RID-HAgD (Fig. 2H) or RID-eGFP (Fig. 2I) were pooled and concentrated by centrifugal filters and then quantified. The final concentrations of purified proteins were ∼2 and 1.8 mg/ml (mRID-HAgD and rRID-HAgD, respectively). The final concentrations of purified RID-eGFPs were ∼9 and 3.7 mg/ml (mRID-eGFP and rRID-eGFP, respectively). Likewise, the final concentrations of proteins without fusion to mRID after chemical refolding were ∼1.4 and 7.3 mg/ml for the HAgD and eGFP, respectively. All of the recombinant proteins were dialyzed against PBS.
The oligomeric state of RID-fused proteins and of the HAgD without RID was determined by SEC at 4°C on a Superdex-200 Increase column (GE Healthcare), and the MW of the eluted fractions was estimated by calibration with marker proteins of known size (Fig. 3). Based on the elution volume and partition coefficient (Kav), the MW of mRID-HAgD was estimated to be 140.54 kDa. Likewise, rRID-HAgD showed an elution volume of 17.60 ml and the estimated size of 143.87 kDa (Fig. 3A). Of note, most of the protein was eluted as a single peak, without any peaks in void volume, suggesting that the protein is predominantly in a defined conformation without any insoluble aggregates of ill-defined conformations. According to the elution pattern, the RID-fused HAgD proteins were predominantly in the trimeric state, resembling the assembly status of the whole HA protein in an influenza virus. The estimated size of trimeric RID-HAgD (140–144 kDa) is slightly bigger than the calculated value (40 kDa × 3 = 120 kDa). The flexible nature of the fusion junction and the elongated nature of the RID are expected to cause slight overestimation of the overall size of a fusion protein (17, 18) (Fig. 1E). SDS-PAGE analysis also confirmed that the bulk of RID-HAgD proteins was present in the trimer fraction. Apparently, the bound tRNAs dissociated and failed to be copurified with the HA proteins during the purification process, probably owing to relatively low affinity for RID (31). Serving as a control, mRID-eGFP was eluted at elution volumes 12.80 and 22.50 ml, indicating a mixture of oligomeric (∼30%) and monomeric (∼70%) conformations, with the estimated size of the monomer 34.2 kDa, close to calculated 37.8 kDa (Fig. 3A). eLysRS-HAgD (85.8 kDa) was eluted as a mixture of oligomeric (60%) and dimeric (40%; estimated as 181.97 kDa) conformations (Fig. 3B). According to the elution pattern, eLysRS-HAgD protein was a mixture of oligomers and the dimer in solution. The HAgD without RID, refolded from inclusion bodies (25.4 kDa), was eluted mainly as a monomeric conformation (Fig. 3C). The results showed the efficacy of the RID in promoting the folding and assembly of its reporter HA protein into the native trimeric conformation.
Figure 3.
Determination of the oligomeric status of fusion antigens by gel-filtration chromatography and a fetuin-binding assay. A) mRID-HAgD and rRID-HAgD were eluted in trimeric form on the basis of Kav, followed by analysis of RID-HAgD by SDS-PAGE. mRID-eGFP, as a control, was eluted as a mixture of oligomeric (30%) and monomeric (70%; estimated at 34.2 kDa) forms. B) LysRS-HAgD, as a control, was eluted as a mixture of oligomeric (60%) and dimeric (40%; estimated as 182 kDa) forms. C) The refolded HAgD without RID was eluted predominantly in monomeric form (80%; estimated at 25.6 kDa). D) The glycoprotein (fetuin), which contains terminal sialic acid residues as a receptor for HA, was adsorbed to a microplate and analyzed for binding to mRID-HAgD, HAgD without RID, and eLysRS-HAgD. The mRID-eGFP protein served as a negative control. All of the data were obtained in duplicate.
The recombinant HAgD proteins were evaluated regarding their capacity for binding to terminal sialic acid residues by an ELISA. Trimeric HA glycoproteins are responsible for binding of the influenza virus to sialic acid on the host cell membrane for initiation of the infection cycle. Fetuin is a glycoprotein that contains sialic acid and can be used in a convenient assay for evaluation of binding of a functional trimeric assembly of an HA protein (32). In this assay, the quality of HA folding could be assessed regarding receptor binding at the initial stage of viral infection. Our results showed that the E. coli-derived recombinant HA, which is not glycosylated, binds to fetuin (Fig. 3D), suggesting that glycosylation is not important for the folding of HA (33). Of note, the mRID-HAgD trimer manifested stronger binding than did monomeric HAgD (without RID) obtained by refolding (Fig. 3D). eLysRS-HAgD, which is predominantly in a dimeric form, failed to bind, as was the case for mRID-eGFP, which was thus selected for negative controls (Fig. 3D). Consistent with other reports (34), the receptor-binding activity of mRID-HAgD reflects its proper folding and assembly into the biologically relevant trimer conformation.
The effect of RNA on the solubility of antigens
Depletion of RNA by RNase A was performed to test the potential effect of RNAs on the solubility of proteins. The total E. coli lysate (T) was centrifuged to prepare two fractions: the soluble (S) and pellet (P). Subsequently, the total of soluble fraction (ST) was treated with RNase A and centrifuged again to obtain the supernatant of soluble fraction (SS) and the precipitate of soluble fraction (SP). Solubility of the protein was greatly reduced by RNase A treatment for both mRID-HAgD and rRID-HAgD, according to Coomassie staining (Fig. 4A). The results were further substantiated by the semiquantitation of protein bands in WB by densitometric scanning (S/P < 95%; SS/SP without RNase A < 90%; SS/SP with RNase A < 30%; Fig. 4B). This finding showed that RNA plays an important role in the maintenance of the solubility of RID-fused HA proteins.
Figure 4.
The chaperoning role of RNA facilitates the folding of RID-fused proteins. A, B) SDS-PAGE analysis of mRID-HAgD in cell lysates after RNAse A treatment; Coomassie staining (A) and WB (B). The total extract (T) was centrifuged to obtain soluble (S) and pellet (P) fractions. The total of soluble fraction (ST) was then treated with RNAse A and centrifuged to obtain supernatant of soluble (SS) and precipitate of soluble (SP) fractions. The elimination of RNA by RNase A digestion reduced the solubility of RID-HAgD. Black arrows, target proteins; +, treatment with RNase A; −, nontreatment with RNase A. C, D) SDS-PAGE data on WT hRID-HAgD and 3 types of hRIDmu-HAgD, which were expressed and purified. Red arrows, purified target proteins. E) Determination of oligomeric status of WT hRID-HAgD and various hRIDmu-HAgD by gel-filtration chromatography. WT hRID-HAgD and hRIDmu2-HAgD were eluted as a mixture of trimeric and oligomeric forms on the basis of Kav, followed by analysis of RID-HAgD by SDS-PAGE. hRIDmu6-HAgD and hRIDmu9-HAgD were eluted as a mixture of dimeric and oligomeric forms. All data were obtained in duplicate.
The effect of RNA binding on HA trimer assembly
To address further the RNA dependence of folding and assembly, mutations were introduced into key amino acid positions in hRID that are crucially involved in tRNA binding. The choice of mutations was guided by detailed reports on mutagenesis (31). Cumulative mutations (lysine or arginine to alanine) were introduced into each site to generate three mutants—hRIDmu2, hRIDmu6, and hRIDmu9—carrying mutations at 2, 6, and all 9 sites involved in RNA binding, respectively (Fig. 1B). All of these proteins were expressed in soluble form, purified by Ni-affinity chromatography, and subjected to SEC for the analysis of trimeric assembly. All 3 mutants showed enhanced solubility compared with the WT hRID-HAgD protein at 37°C (Fig. 4C), although the solubility levels were not noticeably different at 20°C. All of these proteins were purified by 1-step Ni-affinity chromatography (Fig. 4D). Remarkably, SEC data revealed that as the number of mutations increased, the proportion of trimeric assembly decreased, with a corresponding increase in the amount of the oligomer in the void volume (Fig. 4E). These results strongly indicated that the interaction with RNA is indeed required for the assembly of the HA antigen into the immunologically relevant conformation.
Specificity of antiserum against the recombinant viral proteins
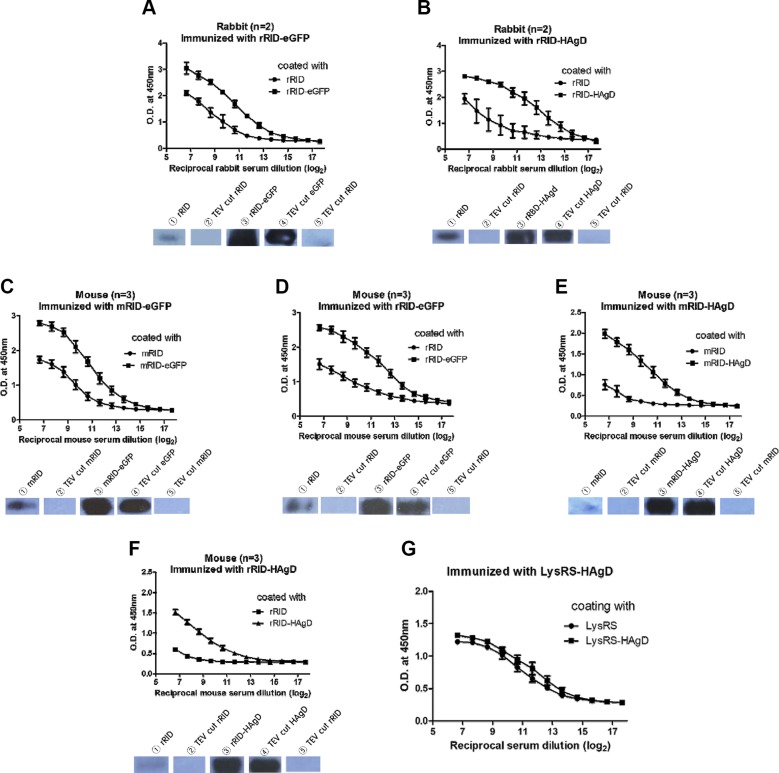
The purified proteins were applied to antibody production in mice and rabbits. After immunization with an RID-HAgD or RID-eGFP fusion protein, antiserum was obtained, and an ELISA was performed to estimate the amount of a specific antibody against the docking tag and the reporter proteins, respectively (Fig. 5A–F). ELISA data confirmed that immunization elicited robust antibody responses against target reporter antigen HAgD or eGFP but much less toward the RID docking tag. First, irrespective of the reporter protein, the ELISA signal was much stronger against fusion proteins than against the RID itself (for rRID, Fig. 5A, B for eGFP and HAgD, respectively; for mRID, Fig. 5C, E for the respective proteins). Second, for the same reporter antigen, the ELISA signal was much stronger for the reporter antigen regardless of the source of the docking protein (compare Fig. 5E with 5F for the HAgD and Fig. 5C with 5D for eGFP). Third, the same conclusions were drawn, regardless of the animal species to be immunized (compare Fig. 5A with 5B for rabbits and Fig. 5C–F for mice). It should be noted that specificity of the antibody response to a reporter protein relative to the RID docking protein was not appreciably different regardless of the origin of an RID and the animal species to be immunized (compare Fig. 5A with 5D for eGFP and Fig. 5B with 5F for the HAgD), probably as a result of high homology (∼80%) in an amino acid sequence between the murine and rabbit counterparts. In contrast, data from the ELISA involving eLysRS-HAgD as an immunogen showed a similar magnitude of the responses to fusion proteins eLysRS and eLysRS-HAgD, suggesting that the immune response was directed predominantly against the eLysRS docking tag with a negligible response to the desired HAgD region (Fig. 5G). High immunogenicity of eLysRS may be because of low sequence homology of LysRS between the mouse and E. coli (∼36%). In addition, WB analysis was carried out to examine the binding specificity of the antibodies produced against recombinant proteins. After treatment with the TEV protease, the 6xHis tag and the peptides corresponding to the MCS were expected to be removed from an RID and RID-HAgD (Fig. 1D). Accordingly, the antibody against RID-HAgD bound specifically to the HAgD (cleaved) and RID-HAgD (remained uncleaved) but not to the RID docking tag alone (cleaved; Fig. 5A–F and Supplemental Fig. 2A–D).
Figure 5.
Analysis of differences in antibody production levels by an ELISA and WB. A) The difference in antibody levels (according to an ELISA) between groups rRID and rRID-eGFP relative to the serum samples from rabbits (n = 2) immunized with rRID-eGFP; (lower) WB analysis for the detection of antibodies in the rabbit serum samples. B) Differences in antibody levels between groups rRID and rRID-HAgD relative to the serum samples from rabbits (n = 2) immunized with rRID-HAgD; (lower) WB analysis for the detection of antibodies in the rabbit serum samples. C–F) Differences in antibody levels between groups RID and RID-fused protein relative to the serum samples from mice (n = 3) immunized with an RID-fused immunogen; (lower) WB analysis for the detection of antibodies in the mouse serum samples. G) Differences in antibody levels between groups eLysRS and eLysRS-HAgD relative to the serum samples from mice (n = 5) immunized with eLysRS-HAgD. Error bars indicate the sd of each cohort. The x-axis indicates reciprocal serum dilution (log2). The initial serum dilution value was 1/200 and 1/3200 for mice and rabbits, respectively.
The full-spectrum analysis of the antibody specificity is shown in WB data in Supplemental Fig. 2A–E. The RID fusion tags, RID-eGFP and RID-HAgD, were digested with TEV, resulting in cleavage products of the expected size (Supplemental Fig. 2A). For convenience, protein fragments, with or without the His tag + MCS peptide moiety, are indicated with black and white arrows, respectively. rRID was detected by rabbit antiserum raised against rRID fusion proteins but failed to be detected after TEV treatment (Supplemental Fig. 2B). These results suggested that the positive detection, albeit a weak one, by WB was a result of the immune response to the His tag + MCS moiety, rather than to the self-rRID docking tag. To determine whether anti-histidine (His) tag antibodies raised against 1 protein could bind to the His tag attached to another (irrelevant) protein, we performed an additional WB analysis. Serum samples from rRID-HAgD-immunized rabbits showed an immunoreaction with the eGFP protein, and conversely, serum samples from mRID-eGFP-immunized mice showed an immunoreaction with the HAgD protein (Supplemental Fig. 2E). Given that only the His tag was a common region between rRID-HAgD and eGFP and between mRID-eGFP and HAgD, it is likely that the bands indicated by black arrows represent a specific interaction between the anti-His tag antibody and the His tag. Likewise, in rRID-eGFP or rRID-HAgD, the predominant response was directed against the eGFP or HAgD reporter proteins, rather than rRID. Likewise, mRID was weakly reactive with the murine antiserum against mRID-eGFP (Supplemental Fig. 2C), but the reactivity disappeared after TEV cleavage (white arrow), suggesting that the self-mRID was not immunogenic in mice. The lack of crossreactivity between mouse antiserum and rRID of rabbit origin was probably a result of high homology of RIDs between the 2 host species (Supplemental Fig. 2C, D). Therefore, the weak but distinct ELISA response to the RID docking tag (Fig. 5) is not actually a result of immunogenicity of the RID but of the peptides artificially introduced for affinity purification and cloning purposes. This finding indicates that the RID docking tag itself is nonimmunogenic, and the immune response was aimed predominantly at the desired viral antigen after immunization. The WB analyses, along with the ELISA, confirmed that RID can serve as an excellent enhancer of folding and assembly of a target antigen without compromising the specific immune responses.
Specificity of anti-influenza virus antiserum against recombinant antigens
Mice were infected intranasally with a sublethal dose (102 PFUs) of the PR8 (H1N1) virus to obtain antiserum against the PR8 (H1N1) virus. Four weeks later, blood was drawn from each mouse via transorbital collection, and serum was prepared. All of the mice survived with a maximal weight loss of 23% by 7 d postchallenge (dpc) and recovered slowly. Furthermore, 2 wk later, blood was collected from the recovered mice, and the resulting serum was subjected to ELISA and WB analyses (Fig. 6A, B and Supplemental Fig. 3A). The results confirmed that RID-HAgD (trimeric form; Fig. 3A), as a coating antigen, was detected by highly diluted antisera from infected mice (n = 5), whereas the refolded HAgD without RID (monomeric form; Fig. 3C) was detected at a much lower dilution of the antisera (∼50-fold lower) than RID-HAgD was. The RID docking tag, as a negative control, failed to show any immunoreactivity (Fig. 6A, B). WB then confirmed that the antibody response was directed against HAgD, whereas RID, either before or after TEV cleavage, failed to react with the antiserum (Supplemental Fig. 3A). These results indicated that recombinant RID-HAgD may serve as a diagnostic antigen with high specificity and sensitivity for influenza infection. In addition, we compared ELISA reactivity among various antisera from mice infected with different subtypes of influenza strains. As expected, the highest reactivity was seen with the homologous PR8 (H1N1) antiserum (Fig. 6C). Potential crossreactivity with different HA antigens was examined using cold-adapted X-31-based reassortant strains carrying 6 internal genes from the cold-adapted X-31 virus and the genes encoding surface proteins HA and neuraminidase from WT viruses (35, 36). Lower reactivity was observed with heterologous virus New Caledonia (NC; H1N1) or heterosubtypes, including Panama (PA; H3N2), Indonesia (ID; H5N1), or Korea (KR; H5N2; Fig. 6C and Supplemental Fig. 3B). Alternatively, to test whether the antibody elicited by the RID-HAgD vaccine binds to the homologous PR8 (H1N1) virus, we conducted an ELISA with the antisera from mice immunized with mRID-HAgD or HAgD without RID (as a control; n = 5), by means of the PR8 (H1N1) virus as a coating antigen (Fig. 6D, E). The results confirmed that mRID-HAgD elicited specific antibodies against the homologous virus and that repeated boosts with the fusion protein further amplified the antibody response, suggesting that the quality of the recombinant antigen is immunologically relevant. Despite 3 immunizations, the antisera to the HAgD without RID rarely bound to PR8 (H1N1) viruses, indicating that the HAgD without RID could not induce antibody responses specific to the virus in mice (Fig. 6E). These results suggested that recombinant RID-HAgD may serve as a good antigen for accurate diagnosis of infection or quantification of vaccine antigens.
Figure 6.
Detection of anti-influenza virus antibodies in mouse serum samples by means of the recombinant viral proteins and the PR8 (H1N1) virus. A, B) ELISA and WB data revealed that the recombinant proteins, RID-HAgD, were detected by highly diluted antibodies against the PR8 (H1N1) virus in serum samples from infected mice (n = 5), whereas the refolded HAgD without RID as a coating protein (compared with RID-HAgD) was detected only at much lower dilution (50-fold or ∼26-fold lower) of the antisera, and the RID docking tag failed to show any immunoreactivity. C) ELISA data for detection of antibodies in serum samples from mice infected with various viruses (n = 3). As a coating antigen, the recombinant mRID-HAgD protein and cold-adapted X-31-based genetic reassortants (carrying 6 internal genes from cold-adapted X-31 virus and the surface HA and neuraminidase genes from WT viruses) were used as the source of the HA antigen. NC, New Caledonia; PA, Panama; ID, Indonesia; KR, Korea; B/Y, Influenza B virus Yamagata. D) ELISA data showing that according to the number of boosts, high-titer antibodies in serum samples from mice (n = 5) immunized with mRID-HAgD (20 μg/mouse) bound to the PR8 (H1N1) virus (104 PFU/well). Error bars indicate the sd of each cohort. The x-axis indicates reciprocal serum dilution (log2). The initial serum dilution value was 1/50. E) ELISA data showing that regardless of the number of boosts, antibodies in serum samples from mice (n = 5) immunized with the HAgD without RID (15 μg/mouse) hardly bound to the PR8 (H1N1) virus (104 PFU/well). Error bars indicate the sd of each group. The x-axis indicates reciprocal serum dilution (log2). The initial serum dilution was 1/50.
Immunogenicity of the RID-HAgD recombinant protein vaccine
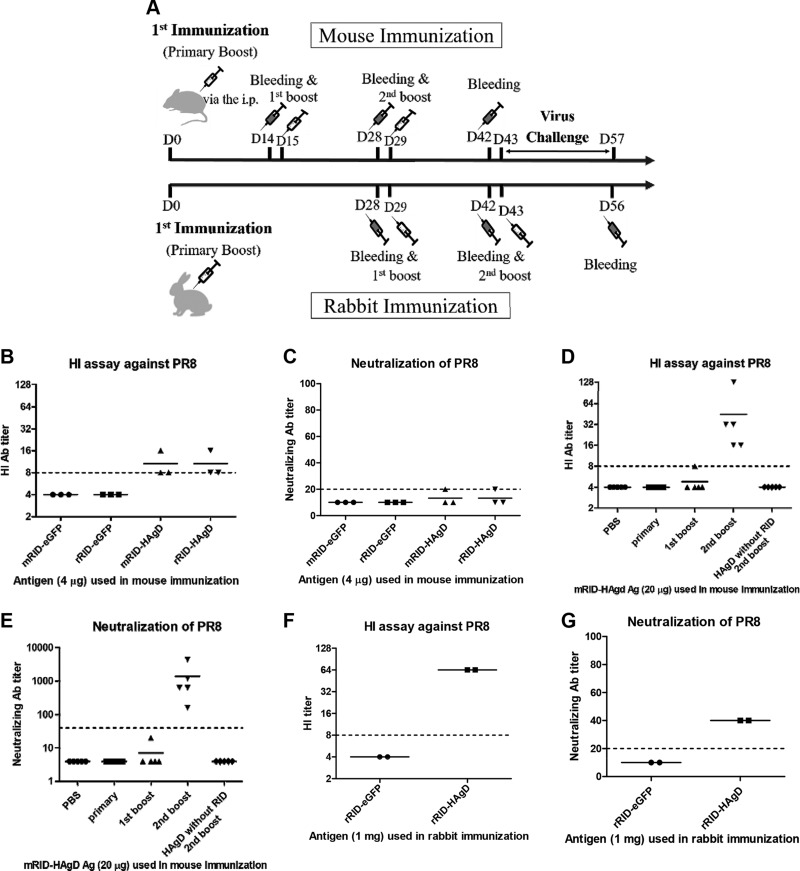
To determine whether the RID-HAgD recombinant proteins were suitable as vaccine antigens and elicited protective antibodies, we conducted an HI assay and an NT assay using the antisera derived from the immunized animals. The HI assay against the PR8 (H1N1) virus was carried out by means of antisera collected from the mice (n = 3) immunized with RID-HAgD (4 μg/mouse) or RID-eGFP (as a control, 4 μg/mouse), along with an Alum adjuvant (Thermo Fisher Scientific) via the intraperitoneal route. The mouse antisera against RID-HAgD showed detectable levels of HI titers and NT antibody titers, whereas those from the RID-eGFP group failed to show such an activity (Fig. 7B, C). The HI activity appeared to be similar, regardless of the origin of the RID docking protein. Therefore, with the use of mRID-HAgD (20 μg/mouse), HAgD without RID (15 μg/mouse), and PBS, detailed analyses of immune responses were conducted after prime-boost immunization (n = 5) via the intraperitoneal route (Fig. 7D, E). As shown in Fig. 7D, E, boost immunization with mRID-HAgD (20 μg/mouse) augmented the immune responses, and high HI titers (>32) were achieved by the second boost immunization, whereas the NT antibody levels (>103) correlated with the HI antibody levels. The multiple immunizations with the HAgD without RID did not generate HI or NT antibodies to the PR8 (H1N1) virus (Fig. 7D, E). Antisera from rabbits (n = 2) immunized with 1 mg of the rRID-HAgD recombinant vaccine (Fig. 7F, G) showed higher HI titers (>64) and higher NT antibody levels (>40) than those shown by mouse antisera (Fig. 7B, C). The control rabbit antisera against the RID-eGFP proteins or PBS showed neither the HI ability nor NT titers against the homologous PR8 (H1N1) virus (Fig. 7F, G). These results clearly showed that the RID-HAgD fusion proteins could be further optimized into effective vaccine antigens that can produce protective NT antibody responses against influenza viruses.
Figure 7.
Immunogenicity of the RID-HAgD recombinant vaccine. A) Schematic illustration of the immunization and challenge schedule. B) An HI assay against the PR8 (H1N1) virus was conducted with the antisera collected from the mice (n = 3) immunized (intraperitoneally) with RID-HAgD (4 μg/mouse) or RID-eGFP (as a control, 4 μg/mouse), along with the Alum adjuvant. C) An assay of NT of the PR8 (H1N1) virus was performed on the serum samples from mice (n = 3) immunized with RID-HAgD (4 μg/mouse) or RID-eGFP (as a control, 4 μg/mouse). Detection limits are 8 and 20 for the HI assay and NT assay, respectively. D) An HI assay toward the PR8 (H1N1) virus was performed on serum samples from the mice (n = 5) immunized (intraperitoneally) with mRID-HAgD (20 μg/mouse), PBS, or the HAgD without RID (15 μg/mouse). The HI antibody titers increased ∼32-fold, depending on the number of immunization boosts with mRID-HAgD (20 μg/mouse). However, immunization boosting with PBS or HAgD without RID did not yield immune responses. E) An assay of NT of the PR8 (H1N1) virus was performed on the serum samples from mice (n = 5) immunized with mRID-HAgD (20 μg/mouse), PBS, or HAgD without RID (15 μg/mouse), respectively. The NT antibody titers increased by the factor of ∼103, depending on the number of boosts. However, immunization boosting with PBS or HAgD without RID did not yield immune responses. Detection limits are 8 and 40 for the HI assay and NT assay, respectively. F) Antisera from rabbits (n = 2) immunized with 1 mg of the rRID-HAgD recombinant vaccine showed higher HI titers than mouse antisera did (B). G) The rabbit antisera (n = 2) obtained by immunization with 1 mg rRID-HAgD showed higher titers of NT antibodies than mouse antisera did (C). The control rabbit antisera (n = 2) after immunization with 1 mg of an RID-eGFP protein showed neither an HI ability nor NT antibodies against the homologous PR8 (H1N1) virus.
Protection from a lethal challenge
All of the immunized mice were challenged intranasally with one time LD50 of the PR8 (H1N1) virus (103 PFUs) under anesthesia. The survival rate and changes in weight of the challenged mice were monitored daily for 14 d. Mice immunized intraperitoneally with 4 μg RID-HAgD antigens (mRID-HAgD: n = 3; rRID-HAgD: n = 3) lost ∼13% of body weight by 8 dpc (Fig. 8A, B). Alternatively, immunization with 20 μg mRID-HAgD (n = 5) via the intraperitoneal route resulted in ∼10% loss of body weight by 7 dpc, but the weight fully recovered thereafter (Fig. 8C, D). The complete protection was achieved by vaccination with the RID-HAgD recombinant antigens, whereas the mice (n = 5) immunized with PBS as a control died within 7 dpc. Of note, immunization with eLysRS-HAgD even at a high dose (75 μg), even at high reactivity in ELISA (Fig. 5G), failed to provide protection, and all of these mice (n = 5) succumbed within 7 dpc (Fig. 8C, D). The immunization with the HAgD without RID (15 μg) resulted in less protective efficacy compared with the mRID-HAgD, judging by weight changes and survival rates of the mice (n = 5) after the challenge (Fig. 8C, D). The lung viral titers from the mRID-HAgD–immunized group were below the detection limit at 7 dpc, whereas persistent viral infection was detected in the PBS control group (Fig. 8C). To evaluate potential crossreactivity with heterosubtypes, the mice were challenged with the H5N2 virus (2 times LD50) after vaccination. All of the immunized mice died within 8 dpc, and the viral titer remained high at 7 dpc (Fig. 8F), indicating that RID-HAgD provided strain-specific protection (with rapid weight loss) from a homologous infection.
Figure 8.
Protective efficacy of the recombinant vaccine against a PR8 (H1N1) virus challenge. A, B) The mice immunized (intraperitoneally) with 4 μg RID-HAgD (n = 6) or RID-eGFP control (n = 6), along with the Alum adjuvant, were challenged with 1 times LD50 of the PR8 (H1N1) virus (103 PFUs), and the body weight changes (A) and survival rates (B) of the infected mice were monitored daily. Immunization with the RID-eGFP control (n = 6) failed to provide protection, and all of these mice (n = 6) succumbed within 7 dpc. C, D) The mice vaccinated intraperitoneally with 20 μg of the mRID-HAgD immunogen (n = 5) or mock infected (PBS control; n = 5) were challenged with 1 times LD50 of the PR8 (H1N1) virus (103 PFUs), and the body weight changes (C) and survival rates (D) of the infected mice were monitored daily. The mice immunized with 75 μg LysRS-HAgD (n = 5) or 15 μg of the HAgD without RID (n = 5) were also challenged with 1 times LD50 of the PR8 (H1N1) virus (103 PFUs), and body weight changes of the infected mice were monitored daily (C). Immunization with eLysRS-HAgD or PBS failed to provide protection, and all of these mice (n = 5) succumbed within 7 dpc. Immunization with the HAgD without RID provided partial protection, and only one mouse succumbed within 7 dpc (D). E, F) Replication of the challenge virus in the lungs of the immunized mice. The titers of challenge virus in the lungs were measured by a viral plaque assay. Dashed lines denote a detection limit, 1.0. Error bars indicate the sd of each cohort.
DISCUSSION
A novel protein-folding function of RNA molecules has been recognized (5, 37, 38), and some of these RNAs even outperform previously known molecular chaperone proteins in specific cases (4, 14). Initially recognized in HIV infections (10), the chaperna activity is intrinsic to some ribozymes (7, 8) and probably operational during the folding and homeostasis of normal cellular proteins (37, 39). Whether a chaperna operates in combination with molecular chaperone proteins during the de novo folding of proteins remains an exciting possibility. Nonetheless, the variety of guest proteins that are amenable to RNA-dependent folding is not yet known, and the identity of RNA molecules among the RNA-interacting proteins is not well documented. A convenient avenue for exploiting the chaperone activity of RNA for the folding of difficult-to-express proteins is to use RIDs that interact with known, identified RNA molecules for fusion with target proteins (14, 15).
By taking advantage of the chaperna function herein, for the first time, we show that influenza HA can be assembled in a soluble, trimeric, and immunologically activating conformation. The influenza HA antigen could be produced in bacteria as a soluble trimeric structure that can induce the production of NT antibodies for effective protection against a lethal challenge. As revealed during the H1N1 pandemic in 2009, the supply of sufficient doses of vaccines within a short period is a prerequisite for mitigating the economic effect and addressing public health concerns during a pandemic. Previously, all efforts toward the bacterial production of recombinant HA vaccines have been dependent on the chemical refolding of misfolded insoluble aggregates (34, 40–43). All of these processes have depended on the initial production of HA as insoluble aggregates and required subsequent chemical refolding procedures, involving solubilization with chaotropic agents, dilution, and concentration before an immunologically relevant conformation was obtained. Therefore, the production of immunologically relevant antigens in mammalian or insect cells in baculovirus vector systems has been favored. Nevertheless, these systems invariably require cell culture methods that are highly expensive and difficult to scale up. The production of HA in a soluble, trimeric conformation should greatly simplify the production process, ensuring the quality of the resultant vaccines and making them amenable to implementation of pandemic preparedness (43).
In the proposed system, an RID serves as a docking tag for transducing the chaperone function of RNA for the folding and assembly of the downstream reporter protein (HA) via interaction with resident cellular tRNAs. From the functional perspective, the coupled action of this chaperone during protein folding and assembly is similar to that of chaperone proteins (44, 45). Our results also indicate that the RIDs that were derived from various animals can be used as immunologically tailored, antigen-folding vehicles for antigens, which could serve as diagnostic and prophylactic tools for viral infections. In addition to the de novo folding of monomers, the subsequent assembly of monomeric antigens into multimeric-ordered structures is crucially important, especially for augmentation of the immune response and proper presentation of epitopes by immune cells toward protection (12, 13). Therefore, a judicious choice of an RID is required; while mediating the chaperna function, the docking tag should not physically interfere with the formation of oligomeric structures. In the present study, an N-terminal-appended tRNA-interaction domain (18) of a eukaryotic LysRS was chosen as the RID. This domain is small (∼70 aa long) and belongs to the family of intrinsically unfolded domains; it is expected to be sufficiently flexible so that it does not physically interfere with the trimer assembly of HA located downstream (46). Intrinsically, the influenza HAgD proteins were expressed and purified predominantly as trimers (Fig. 3A). Although the expression levels and solubility of eLysRS-HAgD containing eLysRS as an alternative tRNA-binding domain were substantially higher, the fusion protein existed primarily as a dimer, as confirmed by gel-filtration chromatography under native conditions (Fig. 3B). Possibly, the stabilization energy of dimerization of the relatively large LysRS (∼57.6 kDa) may physically interfere with the trimerization of HAgD (∼25.4 kDa) (47). Therefore, despite high immunogenicity, as demonstrated by an ELISA (Fig. 5G), immunization failed to provide protection against a lethal challenge (Fig. 8C, D). However, the HAgD, without an attached RID protein, yielded interesting results when administered to mice. Even without eliciting antibody responses, three immunizations with the HAgD without RID partially protected the mice from the lethal challenge. We assume that this partial protection may be because of the T cell-mediated immunity against HA. There are several reports indicating the existence of T cell epitopes in the HA of influenza viruses (48–50). It is likely that multiple immunizations with the HAgD without RID induced specific T cell responses, contributing to the protection. The results emphasize the importance of the assembly of antigens in a native conformation for induction of protective NT antibody responses. The process has distinctive advantages over the chemical refolding of insoluble aggregates (42, 43, 51), which require solubilization with chaotropic agents, dilution, and concentration before adopting an immunologically relevant conformation. The production of the HAgD with a receptor-binding domain in a soluble, trimeric, immunologically activating conformation should ensure the quality of viral antigens and make them amenable to implementation of pandemic preparedness (52–54).
It is worth comparing the mechanistic differences of the process described herein from other approaches to the oligomeric assembly of HA. The existing approaches involve fusion of HA with ferritin into nanoparticles or with a foldon that is derived from the natural trimerization domain of isoleucine zippers or T4 fibritin (55–58), wherein the major amount of stabilization energy is provided by the external trimerization domain. In the present approach, using a chaperna, the RID remains an independent monomer, and thus, it is not expected to contribute to intermolecular stabilization among HAgDs. Rather, trimer formation is primarily driven by the intrinsic stabilization energy of mutual interactions among HAgDs. Here, an RID, as a small, structurally independent domain, is not expected to interfere with the interactions within HAgD trimers in space, as governed by the thermodynamic stabilization energy in the assembly of trimers.
The RID that was tested in the present study is derived from human LysRS, which can switch spontaneously from an unfolded structure into an α-helical structure after tRNA binding (16). It is noteworthy that the unique ability of intrinsically unfolded domains to increase the solubility of target proteins has been reported, although its effect on biologic activities remains unknown (59). The RID that was used in this study is relatively unfolded, and therefore, the linking of the docking protein with the RID may increase its solubility, in agreement with other reports (14, 59). Therefore, it is intriguing that the hRIDmu2, hRIDmu6, and hRIDmu9 proteins maintained high solubility (even higher than that of the WT hRID; Fig. 4C), and yet, the extent of the trimeric assembly of HAgD was greatly lowered (Fig. 4E). These results confirm that the RNA interaction is indeed crucial for the trimeric assembly of HA. The results also offer a discretionary note on the quality evaluation of recombinant antigens: solubility is necessary but not sufficient to represent a biologically or immunologically relevant conformation.
The mechanism of the chaperna-mediated assembly of monomers should be explored further, although it is functionally similar to that of protein-based molecular chaperones. Molecular chaperones are proven to be crucial for not only the de novo folding but also, the assembly of monomers into multimeric complexes (44, 45). Moreover, in contrast to the foldase type of molecular chaperones, which consume ATP for recycling chaperones from docking proteins, a chaperna is expected to function as a chaperone without the ATP requirement, in a way similar to that of the holdase type of chaperones (60–62). Here, the affinity of RNA may be a determining factor for the capture and folding of the guest proteins. In addition, the potential effect on the tRNA-induced α-helical structural transition of an RID to folding of the downstream docking protein should be investigated further. Overall, our results show that RNA can affect the kinetic landscape of the folding pathway in favor of the productive folding and assembly of vaccine antigens (37, 63).
The potential immunogenicity of the RID docking tag should be taken into account, especially in designing the folding vehicle for vaccine antigens. To ensure that specific antibody responses are directed against the HAgD after immunization, the RID should remain nonimmunogenic. Accordingly, an RID should originate, preferably from the host being immunized, and was of murine origin in this study, wherein immunization of mice and a mouse model of a lethal challenge were used. For human clinical trials, the RID can be of human origin to direct the elicited antibody response predominantly against HA.
The chaperna function may be extended to a supramolecular assembly of viral antigens, which is required for augmenting immunogenicity. We recently confirmed that the present strategy can be successfully applied to the assembly of bacterially synthesized monomers of a norovirus particle, which comprises 180 monomers, into virus-like particles (VLPs; unpublished results). However, in VLP assemblies, prior cleavage of the RID tag was required to enable the compact packing into VLPs. This system may also be applied to preparation of mAb for diagnostic and therapeutic interventions. Basically, the process involves isolation of spleen cells from an immunized mouse, followed by their fusion with immortalized cells and screening of reactive clones. In this case, an RID, originating from the host being immunized (murine or rabbit origin in this study), was used to ensure that most of the positive antibody clones were directed to the target antigens, for instance, the spike protein of the Middle East respiratory syndrome coronavirus (unpublished results).
In summary, the chaperone function of RNA, which is intrinsic to some naturally occurring ribozymes, could be exploited for the folding and assembly of viral antigens in immunologically relevant conformations. The process is expected to facilitate the timely supply of pandemic vaccines with shorter lead time, as well as the design of an immunologically tailored folding vehicle to a repertoire of antigens for prophylactic and diagnostic purposes.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
This work was supported by grants from the Korean Ministry of Health and Welfare (HI13C0826) and the Ministry of Agriculture, Food and Rural Affairs (MAFRA; 716002-7). The authors declare no conflicts of interest.
Glossary
- 6xHis
hexahistidine tag
- BMP2
bone morphogenetic protein 2
- BSA
bovine serum albumin
- chaperna
RNA as a molecular chaperone
- dpc
days postchallenge
- eGFP
enhanced green fluorescent protein
- GFP
green fluorescent protein
- HA
hemagglutinin
- HAgD
hemagglutinin globular domain
- HI
hemagglutination inhibition
- His tag
histidine tag
- hRID
RNA-interacting domain of lysyl-tRNA synthetase of human origin
- HRP
horseradish peroxidase
- IACUC
Institutional Animal Care and Use Committee
- Kav
partition coefficient
- LysN
N-terminal domain of lysyl-tRNA synthetase
- LysRS
lysyl-tRNA synthetase
- MCS
multiple cloning site
- Ni
nickel
- NT
neutralizing
- OD
optical density
- PBST
PBS/Tween 20
- PFU
plaque-forming unit
- pGE
promiscuous gene expression
- PR8 (H1N1)
A/Puerto Rico/8/34 (H1N1) virus
- RID
RNA-interacting domain of lysyl-tRNA synthetase
- RIDmu
RID mutant form
- SEC
size-exclusion chromatography
- TBST
Tris-buffered saline/Tween 20
- TEV
tobacco etch virus
- VLP
virus-like particle
- WB
Western blot
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
S. W. Yang, S. I. Choi, and B. L. Seong designed the research; S. W. Yang, Y. H. Jang, and B. L. Seong wrote the manuscript; S. W. Yang, S. B. Kwon, Y. J. Lee, W. Chae, Y. H. Byun, and C. Park conducted the research; Y. H. Jang, S. B. Kwon, and Y. J. Lee analyzed the data; S. B. Kwon, C. K. Kim, and Y. S. Kim provided technical assistance; P. Kim developed the necessary software for conducting the experiments and recording the data; and B. L. Seong supervised all of the experiments, as well as preparation of the manuscript.
REFERENCES
- 1.Hendrick J. P., Hartl F. U. (1995) The role of molecular chaperones in protein folding. FASEB J. 9, 1559–1569 [DOI] [PubMed] [Google Scholar]
- 2.Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 10.1126/science.1068408 [DOI] [PubMed] [Google Scholar]
- 3.Bukau B., Horwich A. L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366 10.1016/S0092-8674(00)80928-9 [DOI] [PubMed] [Google Scholar]
- 4.Docter B. E., Horowitz S., Gray M. J., Jakob U., Bardwell J. C. (2016) Do nucleic acids moonlight as molecular chaperones? Nucleic Acids Res. 44, 4835–4845 10.1093/nar/gkw291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horowitz S., Bardwell J. C. (2016) RNAs as chaperones. RNA Biol. 13, 1228–1231 10.1080/15476286.2016.1247147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovachev P. S., Banerjee D., Rangel L. P., Eriksson J., Pedrote M. M., Martins-Dinis M. M. D. C., Edwards K., Cordeiro Y., Silva J. L., Sanyal S. (2017) Distinct modulatory role of RNA in the aggregation of the tumor suppressor protein p53 core domain. J. Biol. Chem. 292, 9345–9357 10.1074/jbc.M116.762096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Son A., Choi S. I., Han G., Seong B. L. (2015) M1 RNA is important for the in-cell solubility of its cognate C5 protein: implications for RNA-mediated protein folding. RNA Biol. 12, 1198–1208 10.1080/15476286.2015.1096487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattopadhyay S., Das B., Dasgupta C. (1996) Reactivation of denatured proteins by 23S ribosomal RNA: role of domain V. Proc. Natl. Acad. Sci. USA 93, 8284–8287 10.1073/pnas.93.16.8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudlicki W., Coffman A., Kramer G., Hardesty B. (1997) Ribosomes and ribosomal RNA as chaperones for folding of proteins. Fold. Des. 2, 101–108 10.1016/S1359-0278(97)00014-X [DOI] [PubMed] [Google Scholar]
- 10.Kim J. M., Choi H. S., Seong B. L. (2017) The folding competence of HIV-1 Tat mediated by interaction with TAR RNA. RNA Biol. 14, 926–937 10.1080/15476286.2017.1311455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gräslund S., Nordlund P., Weigelt J., Hallberg B. M., Bray J., Gileadi O., Knapp S., Oppermann U., Arrowsmith C., Hui R., Ming J., dhe-Paganon S., Park H. W., Savchenko A., Yee A., Edwards A., Vincentelli R., Cambillau C., Kim R., Kim S. H., Rao Z., Shi Y., Terwilliger T. C., Kim C. Y., Hung L. W., Waldo G. S., Peleg Y., Albeck S., Unger T., Dym O., Prilusky J., Sussman J. L., Stevens R. C., Lesley S. A., Wilson I. A., Joachimiak A., Collart F., Dementieva I., Donnelly M. I., Eschenfeldt W. H., Kim Y., Stols L., Wu R., Zhou M., Burley S. K., Emtage J. S., Sauder J. M., Thompson D., Bain K., Luz J., Gheyi T., Zhang F., Atwell S., Almo S. C., Bonanno J. B., Fiser A., Swaminathan S., Studier F. W., Chance M. R., Sali A., Acton T. B., Xiao R., Zhao L., Ma L. C., Hunt J. F., Tong L., Cunningham K., Inouye M., Anderson S., Janjua H., Shastry R., Ho C. K., Wang D., Wang H., Jiang M., Montelione G. T., Stuart D. I., Owens R. J., Daenke S., Schütz A., Heinemann U., Yokoyama S., Büssow K., Gunsalus K. C.; Structural Genomics Consortium , China Structural Genomics Consortium , Northeast Structural Genomics Consortium . (2008) Protein production and purification [erratum in Nat. Methods (2008) 5, 369]. Nat. Methods 5, 135–146 10.1038/nmeth.f.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachmann M. F., Rohrer U. H., Kündig T. M., Bürki K., Hengartner H., Zinkernagel R. M. (1993) The influence of antigen organization on B cell responsiveness. Science 262, 1448–1451 10.1126/science.8248784 [DOI] [PubMed] [Google Scholar]
- 13.Bachmann M. F., Jennings G. T. (2010) Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10, 787–796 10.1038/nri2868 [DOI] [PubMed] [Google Scholar]
- 14.Choi S. I., Han K. S., Kim C. W., Ryu K. S., Kim B. H., Kim K. H., Kim S. I., Kang T. H., Shin H. C., Lim K. H., Kim H. K., Hyun J. M., Seong B. L. (2008) Protein solubility and folding enhancement by interaction with RNA. PLoS One 3, e2677 10.1371/journal.pone.0002677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tompa P., Csermely P. (2004) The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 18, 1169–1175 10.1096/fj.04-1584rev [DOI] [PubMed] [Google Scholar]
- 16.Yiadom K. P., Hammamieh R., Ukpabi N., Tsang P., Yang D. C. (2003) A peptide from the extension of Lys-tRNA synthetase binds to transfer RNA and DNA. Peptides 24, 987–998 10.1016/S0196-9781(03)00188-8 [DOI] [PubMed] [Google Scholar]
- 17.Agou F., Yang Y., Gesquière J. C., Waller J. P., Guittet E. (1995) Polyanion-induced alpha-helical structure of a synthetic 23-residue peptide representing the lysine-rich segment of the N-terminal extension of yeast cytoplasmic aspartyl-tRNA synthetase. Biochemistry 34, 569–576 10.1021/bi00002a023 [DOI] [PubMed] [Google Scholar]
- 18.Francin M., Kaminska M., Kerjan P., Mirande M. (2002) The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain. J. Biol. Chem. 277, 1762–1769 10.1074/jbc.M109759200 [DOI] [PubMed] [Google Scholar]
- 19.Nicholson K. G., Wood J. M., Zambon M. (2003) Influenza. Lancet 362, 1733–1745 10.1016/S0140-6736(03)14854-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonsen L., Clarke M. J., Williamson G. D., Stroup D. F., Arden N. H., Schonberger L. B. (1997) The impact of influenza epidemics on mortality: introducing a severity index. Am. J. Public Health 87, 1944–1950 10.2105/AJPH.87.12.1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giersing B. K., Modjarrad K., Kaslow D. C., Okwo-Bele J. M., Moorthy V. S. (2016) The 2016 Vaccine Development Pipeline: a special issue from the World Health Organization Product Development for Vaccine Advisory Committee (PDVAC). Vaccine 34, 2863–2864 10.1016/j.vaccine.2016.04.041 [DOI] [PubMed] [Google Scholar]
- 22.Jang Y. H., Cho S. H., Son A., Lee Y. H., Lee J., Lee K. H., Seong B. L. (2014) High-yield soluble expression of recombinant influenza virus antigens from Escherichia coli and their potential uses in diagnosis. J. Virol. Methods 196, 56–64 10.1016/j.jviromet.2013.10.035 [DOI] [PubMed] [Google Scholar]
- 23.Kim C. W., Han K. S., Ryu K. S., Kim B. H., Kim K. H., Choi S. I., Seong B. L. (2007) N-terminal domains of native multidomain proteins have the potential to assist de novo folding of their downstream domains in vivo by acting as solubility enhancers. Protein Sci. 16, 635–643 10.1110/ps.062330907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Webb B., Sali A. (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 86, 2.9.1–2.9.37 [DOI] [PubMed] [Google Scholar]
- 25.Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 10.1146/annurev.biophys.29.1.291 [DOI] [PubMed] [Google Scholar]
- 26.Kozakov D., Beglov D., Bohnuud T., Mottarella S. E., Xia B., Hall D. R., Vajda S. (2013) How good is automated protein docking? Proteins 81, 2159–2166 10.1002/prot.24403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozakov D., Brenke R., Comeau S. R., Vajda S. (2006) PIPER: an FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 10.1002/prot.21117 [DOI] [PubMed] [Google Scholar]
- 28.Sachdev D., Chirgwin J. M. (1998) Order of fusions between bacterial and mammalian proteins can determine solubility in Escherichia coli. Biochem. Biophys. Res. Commun. 244, 933–937 10.1006/bbrc.1998.8365 [DOI] [PubMed] [Google Scholar]
- 29.Raran-Kurussi S., Keefe K., Waugh D. S. (2015) Positional effects of fusion partners on the yield and solubility of MBP fusion proteins. Protein Expr. Purif. 110, 159–164 10.1016/j.pep.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vera A., González-Montalbán N., Arís A., Villaverde A. (2007) The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol. Bioeng. 96, 1101–1106 10.1002/bit.21218 [DOI] [PubMed] [Google Scholar]
- 31.Francin M., Mirande M. (2003) Functional dissection of the eukaryotic-specific tRNA-interacting factor of lysyl-tRNA synthetase. J. Biol. Chem. 278, 1472–1479 10.1074/jbc.M208802200 [DOI] [PubMed] [Google Scholar]
- 32.Welsh J. P., Lu Y., He X. S., Greenberg H. B., Swartz J. R. (2012) Cell-free production of trimeric influenza hemagglutinin head domain proteins as vaccine antigens. Biotechnol. Bioeng. 109, 2962–2969 10.1002/bit.24581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aguilar-Yáñez J. M., Portillo-Lara R., Mendoza-Ochoa G. I., García-Echauri S. A., López-Pacheco F., Bulnes-Abundis D., Salgado-Gallegos J., Lara-Mayorga I. M., Webb-Vargas Y., León-Angel F. O., Rivero-Aranda R. E., Oropeza-Almazán Y., Ruiz-Palacios G. M., Zertuche-Guerra M. I., DuBois R. M., White S. W., Schultz-Cherry S., Russell C. J., Alvarez M. M. (2010) An influenza A/H1N1/2009 hemagglutinin vaccine produced in Escherichia coli. PLoS One 5, e11694 10.1371/journal.pone.0011694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khurana S., Verma S., Verma N., Crevar C. J., Carter D. M., Manischewitz J., King L. R., Ross T. M., Golding H. (2010) Properly folded bacterially expressed H1N1 hemagglutinin globular head and ectodomain vaccines protect ferrets against H1N1 pandemic influenza virus. PLoS One 5, e11548 10.1371/journal.pone.0011548 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Lee K. H., Seo S. U., Song J. M., Lee C. M., Kim H. A., Seong B. L. (2006) Characterization of live influenza vaccine donor strain derived from cold-adaptation of X-31 virus. Vaccine 24, 1966–1974 10.1016/j.vaccine.2005.10.051 [DOI] [PubMed] [Google Scholar]
- 36.Jang Y. H., Jung E. J., Lee K. H., Byun Y. H., Yang S. W., Seong B. L. (2016) Genetic analysis of attenuation markers of cold-adapted X-31 influenza live vaccine donor strain. Vaccine 34, 1343–1349 10.1016/j.vaccine.2016.01.053 [DOI] [PubMed] [Google Scholar]
- 37.Choi S. I., Ryu K., Seong B. L. (2009) RNA-mediated chaperone type for de novo protein folding. RNA Biol. 6, 21–24 10.4161/rna.6.1.7441 [DOI] [PubMed] [Google Scholar]
- 38.Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem. 270, 20871–20874 10.1074/jbc.270.36.20871 [DOI] [PubMed] [Google Scholar]
- 39.Kim Y. E., Hipp M. S., Bracher A., Hayer-Hartl M., Hartl F. U. (2013) Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82, 323–355 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- 40.Weldon W. C., Wang B. Z., Martin M. P., Koutsonanos D. G., Skountzou I., Compans R. W. (2010) Enhanced immunogenicity of stabilized trimeric soluble influenza hemagglutinin. PLoS One 5, e12466 10.1371/journal.pone.0012466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiley D. C., Wilson I. A., Skehel J. J. (1981) Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289, 373–378 10.1038/289373a0 [DOI] [PubMed] [Google Scholar]
- 42.DuBois R. M., Aguilar-Yañez J. M., Mendoza-Ochoa G. I., Oropeza-Almazán Y., Schultz-Cherry S., Alvarez M. M., White S. W., Russell C. J. (2011) The receptor-binding domain of influenza virus hemagglutinin produced in Escherichia coli folds into its native, immunogenic structure. J. Virol. 85, 865–872 10.1128/JVI.01412-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khurana S., Verma S., Verma N., Crevar C. J., Carter D. M., Manischewitz J., King L. R., Ross T. M., Golding H. (2011) Bacterial HA1 vaccine against pandemic H5N1 influenza virus: evidence of oligomerization, hemagglutination, and cross-protective immunity in ferrets. J. Virol. 85, 1246–1256 10.1128/JVI.02107-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C., Young A. L., Starling-Windhof A., Bracher A., Saschenbrecker S., Rao B. V., Rao K. V., Berninghausen O., Mielke T., Hartl F. U., Beckmann R., Hayer-Hartl M. (2010) Coupled chaperone action in folding and assembly of hexadecameric rubisco. Nature 463, 197–202 10.1038/nature08651 [DOI] [PubMed] [Google Scholar]
- 45.Saschenbrecker S., Bracher A., Rao K. V., Rao B. V., Hartl F. U., Hayer-Hartl M. (2007) Structure and function of RbcX, an assembly chaperone for hexadecameric rubisco. Cell 129, 1189–1200 10.1016/j.cell.2007.04.025 [DOI] [PubMed] [Google Scholar]
- 46.Korshun L. N., Moĭsa L. N., Ganova L. A., Vudmaska M. I., Kovtoniuk G. V., Kiseleva E. K., Spivak NIa. (2011) [Influence of physiological state of Escherichia coli cells on the expression of soluble protein--recombinant analog of glycoprotein G of herpes simplex virus of type 2]. Mikrobiol. Z. 73, 36–46 [PubMed] [Google Scholar]
- 47.Cusack S. (1997) Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 7, 881–889 10.1016/S0959-440X(97)80161-3 [DOI] [PubMed] [Google Scholar]
- 48.Townsend A. R., Bastin J., Gould K., Brownlee G. G. (1986) Cytotoxic T lymphocytes recognize influenza haemagglutinin that lacks a signal sequence. Nature 324, 575–577 10.1038/324575a0 [DOI] [PubMed] [Google Scholar]
- 49.Gould K. G., Scotney H., Brownlee G. G. (1991) Characterization of two distinct major histocompatibility complex class I Kk-restricted T-cell epitopes within the influenza A/PR/8/34 virus hemagglutinin. J. Virol. 65, 5401–5409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gould K. G., Scotney H., Townsend A. R., Bastin J., Brownlee G. G. (1987) Mouse H-2k-restricted cytotoxic T cells recognize antigenic determinants in both the HA1 and HA2 subunits of the influenza A/PR/8/34 hemagglutinin. J. Exp. Med. 166, 693–701 10.1084/jem.166.3.693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clark E. D. B. (1998) Refolding of recombinant proteins. Curr. Opin. Biotechnol. 9, 157–163 10.1016/S0958-1669(98)80109-2 [DOI] [PubMed] [Google Scholar]
- 52.Dormitzer P. R. (2015) Rapid production of synthetic influenza vaccines. Curr. Top. Microbiol. Immunol. 386, 237–273 [DOI] [PubMed] [Google Scholar]
- 53.Fineberg H. V. (2014) Pandemic preparedness and response--lessons from the H1N1 influenza of 2009. N. Engl. J. Med. 370, 1335–1342 10.1056/NEJMra1208802 [DOI] [PubMed] [Google Scholar]
- 54.D’Aoust M. A., Couture M. M., Charland N., Trépanier S., Landry N., Ors F., Vézina L. P. (2010) The production of hemagglutinin-based virus-like particles in plants: a rapid, efficient and safe response to pandemic influenza. Plant Biotechnol. J. 8, 607–619 10.1111/j.1467-7652.2009.00496.x [DOI] [PubMed] [Google Scholar]
- 55.Kanekiyo M., Wei C. J., Yassine H. M., McTamney P. M., Boyington J. C., Whittle J. R., Rao S. S., Kong W. P., Wang L., Nabel G. J. (2013) Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 10.1038/nature12202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mallajosyula V. V., Citron M., Ferrara F., Lu X., Callahan C., Heidecker G. J., Sarma S. P., Flynn J. A., Temperton N. J., Liang X., Varadarajan R. (2014) Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl. Acad. Sci. USA 111, E2514–E2523 10.1073/pnas.1402766111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki K., Hiroaki H., Kohda D., Tanaka T. (1998) An isoleucine zipper peptide forms a native-like triple stranded coiled coil in solution. Protein Eng. 11, 1051–1055 10.1093/protein/11.11.1051 [DOI] [PubMed] [Google Scholar]
- 58.Güthe S., Kapinos L., Möglich A., Meier S., Grzesiek S., Kiefhaber T. (2004) Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J. Mol. Biol. 337, 905–915 10.1016/j.jmb.2004.02.020 [DOI] [PubMed] [Google Scholar]
- 59.Santner A. A., Croy C. H., Vasanwala F. H., Uversky V. N., Van Y. Y., Dunker A. K. (2012) Sweeping away protein aggregation with entropic bristles: intrinsically disordered protein fusions enhance soluble expression. Biochemistry 51, 7250–7262 10.1021/bi300653m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slepenkov S. V., Witt S. N. (2002) The unfolding story of the Escherichia coli Hsp70 DnaK: is DnaK a holdase or an unfoldase? Mol. Microbiol. 45, 1197–1206 10.1046/j.1365-2958.2002.03093.x [DOI] [PubMed] [Google Scholar]
- 61.Thoma J., Burmann B. M., Hiller S., Müller D. J. (2015) Impact of holdase chaperones Skp and SurA on the folding of β-barrel outer-membrane proteins. Nat. Struct. Mol. Biol. 22, 795–802 10.1038/nsmb.3087 [DOI] [PubMed] [Google Scholar]
- 62.Stull F., Koldewey P., Humes J. R., Radford S. E., Bardwell J. C. A. (2016) Substrate protein folds while it is bound to the ATP-independent chaperone spy. Nat. Struct. Mol. Biol. 23, 53–58 10.1038/nsmb.3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koldewey P., Stull F., Horowitz S., Martin R., Bardwell J. C. A. (2016) Forces driving chaperone action. Cell 166, 369–379 10.1016/j.cell.2016.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.