Abstract
Cells have developed lineage-specific mechanisms to control proliferation and drive morphologic changes upon differentiation. A hallmark of differentiation is the assembly of signaling molecules that transduce extracellular signals, such as the production of the G protein–regulated enzyme phospholipase Cβ (PLCβ), which generates calcium signals from sensory stimuli. We found that in most cancerous cell lines there is positive correlation between PLCβ1 levels and cell proliferation. In cells of neuronal lineage, however, reducing PLCβ1 levels increases the rate of proliferation. Using a combination of biochemical and biophysical methods, we find that, in the G1 phase, a cytosolic population of PLCβ1 associates with cyclin-dependent kinase 16 (CDK16), a neuron-specific enzyme that is activated by cyclin Y to inactivate the antioncogenic protein p27Kip1. Binding of PLCβ1 directly inhibits CDK16 activity and in turn reduces the ability of cells to enter the S phase. Activation of Gαq by carbachol causes movement of PLCβ from the cytosol to the plasma membrane, reducing its association with CDK16. Similarly, the overexpression of activated Gαq moves PLCβ1 to the membrane, reverses G1 arrest, and promotes proliferation, thereby connecting external stimuli with cell proliferation. Our results present a model in which the transient high expression of PLCβ1 that occurs at the onset of differentiation arrests cells in the G1 phase through its association with CDK16 and allows CDK16 to transition to its postmitotic function of neurite outgrowth and trafficking of synaptic vesicles. The novel role of PLCβ1 in neuronal cell proliferation offers a unique interaction that can be manipulated to guide cells into a neuronal phenotype or to develop therapies for neuroblastomas.—Garwain, O., Valla, K., Scarlata, S. Phospholipase Cβ1 regulates proliferation of neuronal cells.
Keywords: G protein signaling, cell proliferation, cyclin-dependent kinases
The transition of cells from a rapidly dividing state to a differentiated state requires the arrest of proliferation and the initiation of mechanisms that will define the cell’s specific morphology and function. In the neuronal cell line PC12, differentiation is accompanied by an increased amount of the phospholipase Cβ1 (PLCβ1) (1). PLCβ1 is highly expressed in neuronal cells and is associated with neuronal development (2–6). In differentiated cells, a large population of PLCβ1 localizes on the plasma membrane (7) where it is regulated by the Gαq family of G proteins to transduce calcium signals initiated by extracellular ligands such as serotonin, acetylcholine, histamine, and dopamine (8, 9). In addition, a significant population of PLCβ1 localizes in the cytosol, where it inhibits the nuclease component 3 of RNA-induced silencing complex activity (C3PO) (10, 11), which is composed of translin/translin-associated factor X (TRAX). C3PO plays a role in several neuronal cell functions and promotes RNA-induced silencing (12–14).
In previous work, we found that increased PLCβ1 levels as well a transient interaction with C3PO were necessary for cell differentiation (1). In carrying out these studies, however, we noted that PLCβ1 levels had a direct effect on cell proliferation. Here, we show that PLCβ1 affects proliferation by binding and inhibiting cyclin Y–dependent kinase (CDK16), cyclin Y being a cyclin E homolog. CDK16 is expressed in testis, neuronal, and some cancerous cells (15). In undifferentiated cells, CDK16 promotes transition from the G1 phase to the S phase by phosphorylating the cell-cycle kinase inhibitor p27Kip1 (16). In differentiated cells, CDK16 is required for neurite outgrowth (17), secretary cargo transport (18), and the trafficking of presynaptic components (19). In testis cells, CDK16 is required for spermatogenesis (20). Our studies show that in inhibiting CDK16, PLCβ1 prevents unchecked proliferation by keeping cells in the G1 phase. These studies offer a novel target to control the growth of neuronal cells.
MATERIALS AND METHODS
Cell studies
PC12 cells were cultured in high-glucose DMEM (Thermo Fisher Scientific, Waltham, MA, USA) with 10% heat-inactivated horse serum (Gibco) and 5% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA, USA). SK-N-SH and HeLa cell lines were cultured in high glucose DMEM with 10% fetal bovine serum. All cells were incubated at 37°C in 5% CO2. GαqRC (21) was gifted by Dr. Catherine Berlot (formally of Geisinger Heath Center, Danvllie, PA, USA).
Transfections
Transfection of plasmids and small interfering RNA (siRNA) was performed using Lipofectamine 3000 (Thermo Fisher Scientific) in antibiotic-free medium. Cells were introduced to a new medium containing antibiotics (1% penicillin/streptomycin) 6 h posttransfection. On-TargetPlus SmartPool PLCβ1-siRNA (GE Dharmacon, Lafayette, CO, USA) was used to target 4 sequences (CAACAGAAAUCGUUCGUGA, CAGUUGAAUUUGUCGAAUA, AGAAGAAAUGGUCGAAUA, GAAGGAAAUCUGCGAGAAA). TRAX-siRNA was purchased from Thermo Fisher Scientific.
Carbachol stimulation
Gαq was activated by the addition of 1 µM carbachol directly to the medium.
Measuring protein half-lives
Protein half-lives were determined by adding a final concentration of 5 µg/ml cycloheximide to complete cell culture medium (DMEM) to halt protein synthesis, lysing the cells, and assessing the number of proteins via Western blot.
Western blot
Western blot was carried out as previously described (1) using the following primary antibodies: PLCβ1 (D-8; Santa Cruz Biotechnology, Dallas, TX, USA), Gαq (E-17; Santa Cruz Biotechnology), β-actin (Abcam, Cambridge, United Kingdom), PO4-Ser10-p27Kip1 (Abcam), and glyceraldehyde 3-phosphate dehydrogenase (Abcam).
Cell synchronization
Cells were divided into 2 groups and left to recover for 24 h. Afterwards, 2 mM thymidine was added to cells and left for another 24 h. The medium was then removed and replaced with fresh complete culture medium for 8 h. Cells were exposed to thymidine (MilliporeSigma, St. Louis, MO, USA) for another 24 h (double thymidine block), after which medium was removed to obtain cells synchronized in the G1 phase. For G2/M synchronization, 40 ng/ml nocodazole (MilliporeSigma) was added after the thymidine-containing medium was removed.
Coimmunoprecipitation
Proteins in synchronized PC12 cells were coimmunoprecipitated using the procedure described in (7).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Thermo Fisher Scientific) was added to cells at a final concentration of 2 mM and incubated at 37°C in 5% CO2 for 3 h. Medium was carefully aspirated from each well and replaced with 200 μl DMSO. The contents of each well were mixed by stirring with a pipette and horizontally shaking the plates until the color was homogeneous. Absorbance data were obtained from a PerkinElmer Victor 3 Plate Reader (Waltham, MA, USA) at 570 nm.
Protein studies
PLCβ1 was purified as previously described (11). CDK16 tagged with human influenza hemagglutinin (HA) was prepared by Sander van den Heuvel (Massachusetts General Hospital Cancer Center, Boston, MA, USA) and purchased from Addgene (Cambridge, MA, USA). CDK16 was purified from PC12 cells synchronized in G1 phase. Cells were lysed in 200 mM 4-(2-hydroxyethyl)-piperazine-1-ethanesulfonic acid, 10 mM MgCl2, 10 mM EDTA, 5 M DTT, 0.1% Triton X-100, 1 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml aprotinin; mechanically homogenized; and centrifuged at 13,000 g for 10 min. The supernatant was separated and incubated with prewashed HA-tagged magnetic beads (Thermo Fisher Scientific) for 4 h. Following incubation, cells were washed twice using lysis buffer, added to an affinity chromatography column, and incubated for 30 min, then washed using an elution buffer containing HA peptide (MilliporeSigma).
Kinase assay
Purified CDK16 activity was measured using a Z’-Lyte Kinase Assay Kit (Thermo Fisher Scientific). The assay uses a synthetic peptide substrate labeled with a Förster resonance energy transfer (FRET) donor (coumarin) and a FRET acceptor (fluorescein). A kinase can phosphorylate the Tyr, Ser or Thr on the peptide. The kit contains a protease that recognizes and cleaves nonphosphorylated peptide substrate at a substantially higher rate than phosphorylated substrate. Cleavage disrupts FRET between the donor and acceptor fluorophores on the nonphosphorylated substrate, while uncleaved, phosphorylated substrate maintains FRET.
We carried out the reactions in 384 well plates and imaged fluorescence on a PerkinElmer Victor 3 Plate Reader using 445 and 520 nm filters (for coumarin and fluorescein, respectively). The progress of the reaction was quantitated by calculating the emission ratio (i.e., amount of FRET) and relating this to phosphorylation percentage.
Plasmid cloning
CDK16 was prepared by Nicola Brugess-Brown (University of Oxford, Oxford, United Kingdom), and obtained from Addgene. The gene was cloned into pmCherry-C1 vector (Addgene) and designing new cloning sites at HINDIII and XbaI and inserting it into the vector using 5′-TTGATCGAATTCTATGCAGTCCGAGATCGCCATGG-3′ (forward) and 5′-AGTTGATCTAGATTAGAACTCGGTGTCCACCAC-3′ (reverse).
Fluorescence imaging
Images of cells in MatTek chambers (MatTek, Ashland, MA, USA) were acquired using a 2-photon Mai Tai laser (Spectra-Physics, Santa Clara, CA, USA) and a Nikon inverted confocal microscope in an ISS Alba System (Champaign, IL, USA). Data were analyzed using ISS VistaVision and ImageJ (National Institutes of Health, Bethesda, MD, USA) software packages. Fluorescence lifetime imaging microscopy (FLIM) was carried out on an ISS instrument. Atto 425 fluorescent dye (τ = 3.61 ns) was used to calibrate the sample lifetimes.
Statistical analysis
Data were analyzed with Student’s t test and 1-way ANOVA using the SigmaPlot v.13.0 statistical software package (Systat Software, San Jose, CA, USA).
RESULTS
PLCβ1 affects cell proliferation in different cell lines
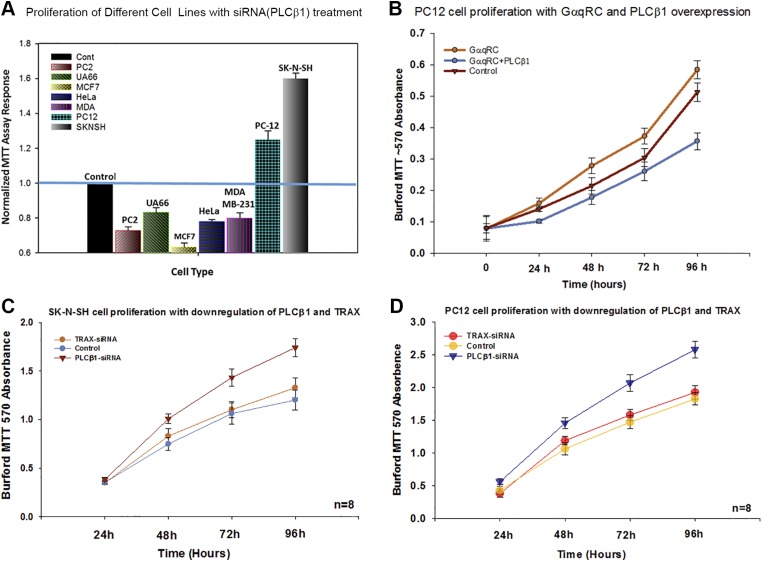
We wanted to determine whether a correlation exists between the amount of PLCβ1 in a cell and proliferation, so we tested this idea by transfecting different cell lines with siRNA (PLCβ1) to obtain a 60–80% reduction of protein level. We found that proliferation decreased in most cell types (Fig. 1A) such as prostate cancer (PC3), melanoma (UACC-62), cervical cancer (HeLa), and 2 breast cancer lines (MCF-7 and MDA-MB-231). We hypothesize that this reduction in proliferation correlates with the diminished ability of cells to respond to extracellular signals linked to Gαq-coupled receptors. Surprisingly, in the 2 neuronal lines tested, PC12 and SK-N-SH, down-regulation of PLCβ1 increases the rate of cell proliferation (Fig. 1A).
Figure 1.
PLCβ1 knockdown increases proliferation in neuronal cell lines. A) Cell proliferation at 72 h for several cell lines, where each value is normalized by setting the control to 1.0 for each cell type. A blue bar is drawn through the plot for easy comparison with controls. In all samples, P < 0.001 compared with control; n = 6–8. B) 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay showing the increase in proliferation of PC12 cells overexpressing constitutively active GαqRC, where the activated G protein recruits cytosolic PLCβ1 to the membrane. This effect is reversed when PLCβ is overexpressed; n = 6, P ≤ 0.001, f = 85.58. C, D) The relationship between PC12 cell proliferation and levels of PLCβ1 and TRAX, where t = 0.254 and P = 0.250 (TRAX-siRNA vs. control) in panel C and t = 2.18 and P = 0.117 (TRAX-siRNA vs. control) (D). As shown, TRAX has no significant effect on PC12 proliferation as PLCβ1.
PLCβ1 localizes on the plasma member with Gαq. To determine whether this cytosolic population of PLCβ1 could be involved in regulating proliferation of the 2 neuronal cell lines, we overexpressed GαqRC in PC12 cells to promote localization of PLCβ1 to the plasma membrane and observed an increase in proliferation compared with the wild type (Fig. 1B). PC12 cell differentiation is associated with an increase in PLCβ1-TRAX complexes and we find that down-regulating TRAX does not affect proliferation. Thus, these studies suggest that PLCβ1 is interacting with a novel neuron-specific protein to reduce proliferation.
PLCβ1 binds to CDK16 in the G1 phase
To determine the specific role that PLCβ1 may play in neuronal cell proliferation, we searched for novel cell cycle–dependent binding partners in PC12 cells. In these experiments, cells were either unsynchronized and thus mostly in the G1 phase or cells were arrested in the G2/M phase by treatment with nocodazole. Following cell disruption, PLCβ1 and its associated proteins were pulled down using a mAb, and the proteins were identified by mass spectrometry.
In unsynchronized cells that are primarily in the G1 phase, we found that one of the most prevalent binding partners associated with PLCβ1 (over 90% certainty) is CDK16. As mentioned previously, CDK16 is a neuron-specific protein that is activated by cyclin Y to allow cells to transition from the G1 phase to the S phase (15). When we repeated the pull-down and mass spectrometry analysis in the G2/M phase, we found that PLCβ1 no longer associated with CDK16.
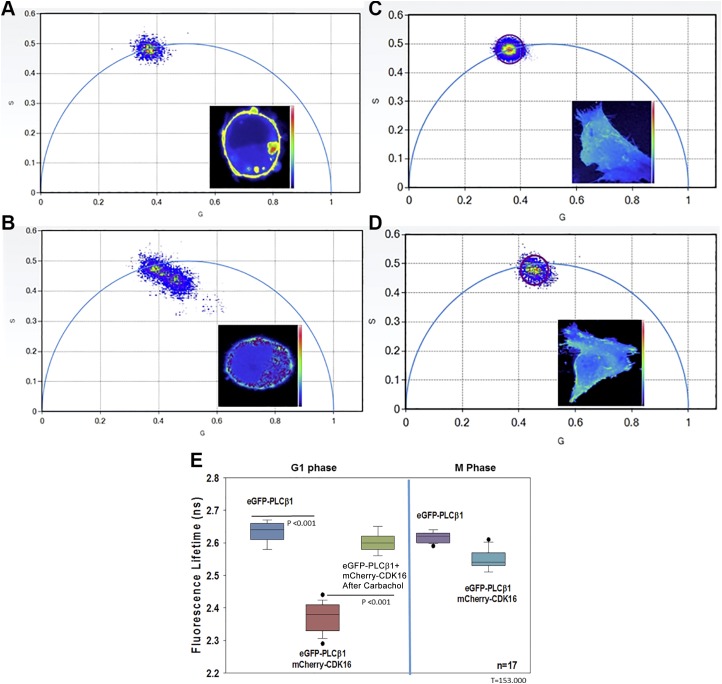
We then determined whether PLCβ1 directly binds to CDK16 in the G1 phase in living cells using FLIM. FLIM measures the amount of FRET by assessing the decrease in donor lifetime in the presence of a FRET acceptor. By exciting the samples with modulated-intensity light, it is possible to determine the lifetimes directly from the phase shift of the emitted light as compared to the excitation, and by the decrease in modulation that occurs as the energy is transferred from the donor to acceptor. By detecting the phase and modulations lifetimes of each pixel in the cell image and viewing the raw data on phasor plots, we can readily see whether the FRET donor population lies on the phasor arc in a single population (i.e., non-FRET), or whether smaller populations are seen inside the phasor arc, which indicates FRET. In this way, FRET is easily seen by viewing raw data without the need to fit each pixel to a model-dependent lifetime value as in pulsed excitation, or to correct for changes in intensity not due to FRET as in intensity measurements (22).
We assessed FRET between enhanced green fluorescent protein (eGFP)–PLCβ1 and mCherry-CDK16 by transfecting G1 synchronized PC12 cells and SK-N-SH cells and measuring the reduction of eGFP lifetime in each pixel in the image as energy was transferred to mCherry. We found that in the absence of mCherry-CDK16, or in the presence of free mCherry (data not shown), eGFP-PLCβ1 lifetimes are uniformly distributed on the arc of the phasor plot, indicating a single-lifetime population (e.g., Fig. 2A, C). In the presence of mCherry-CDK16, however, we found a shift toward lower values (e.g., Fig. 2B, D) and movement of the lifetime distribution below the phasor arc. This shift of values inside the phasor corresponds to shorter eGFP-PLCβ1 lifetimes due to transfer of energy to mCherry-CDK16 (i.e., FRET). Overall lifetime values are summarized in Fig. 2E. In the cell images, we can view the shorter lifetime values where the amount of FRET is highest and determine their localization in the cell (see images in the inserts in Fig. 2A–D). We find that PLCβ1-CDK16 complexes are cytosolic, lying close to but not on the plasma membrane. When we repeated this study in PC12 cells arrested in the G2/M phase, we could not detect a reduction in lifetime due to the low production of mCherry-CDK16 in this phase. In addition, differentiating PC12 cells by treating them with nerve growth factor for 24 h resulted in a loss in FRET between eGFP-PLCβ1 and mCherry-CDK16 (data not shown). Together with the mass spectrometry studies, our data show that PLCβ1 binds to CDK16 in the G1 phase but not the G2/M phase.
Figure 2.
PLCβ1 binds to CDK16 assayed by the decrease in eGFP lifetime as it transfers energy to m-Cherry (i.e., FRET). A) eGFP-PLCβ1 alone in PC12 cells shows a uniform species centered on the phasor arc, where each point corresponds to the lifetime of each pixel shown in the inset. B) In the presence of m-Cherry-CDK16, the lifetimes of eGFP-PLCβ1 move inside and to the right of the arc toward lower values. C, D) Corresponding phasors in SK-N-SH. Warmer colors correspond to higher PLCβ1 intensity. E) Compilation of FLIM data for all samples showing a reduction in eGFP-PLCβ1 lifetime in the presence of mCherry-CDK16 (P ≤ 0.001, t = 442.000), and the reversal with carbachol stimulation (P < 0.001, t = 153), where n = 17 over 3 experiments.
PLCβ1 reduces the ability of CDK16 to increase proliferation through expression and activity
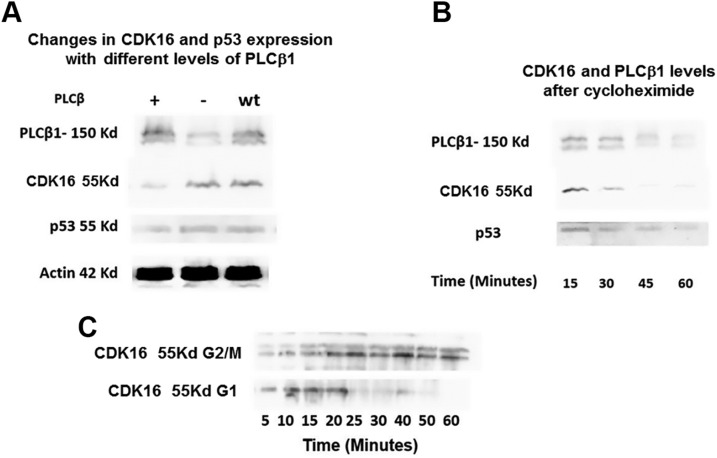
CDK16 has been shown to increase cell proliferation though phosphorylation of p27Kip1, which inhibits cyclin D (CDK2) and cyclin E (CDK4) to hold cells in the G1 phase (23). CDK18 is a close relative to CDK16 and may be correlated to the lethality of tumor protein p53 (24). We tested whether the expression of PLCβ1 altered the levels of CDK16 and p53. In Fig. 3, we show that down-regulating PLCβ1 has little effect on CDK16 and p53, but overexpressing it reduces the levels of both enzymes, which suggests that binding to PLCβ1 increases the degradation of CDK16. To determine whether binding to PLCβ1 changes the longevity of CDK16, we halted protein synthesis with cycloheximide and measured the amount of CDK16 in the G1 phase, where it is bound to PLCβ1, and in the G2/M phase, where it is not. We found that the half-life of CDK16 is much shorter in the G1 phase, but it increases almost 2-fold in the G2/M phase. Thus, an enhanced level of PLCβ1 increases turnover of CDK16.
Figure 3.
Cellular expression of PLCβ1 and CDK16. A) The relation between PLCβ1 expression and the expression of CDK16 and p53. B) Half-lives of the proteins in G1 phase determine whether protein synthesis is halted by the addition of cycloheximide. We note that the actin band disappears 25 min after cycloheximide treatment. C) Comparison between the longevity of CDK16 in the G1 vs. in the G2/M phase.
PLCβ1 inhibits CDK16 kinase activity in vitro and in vivo
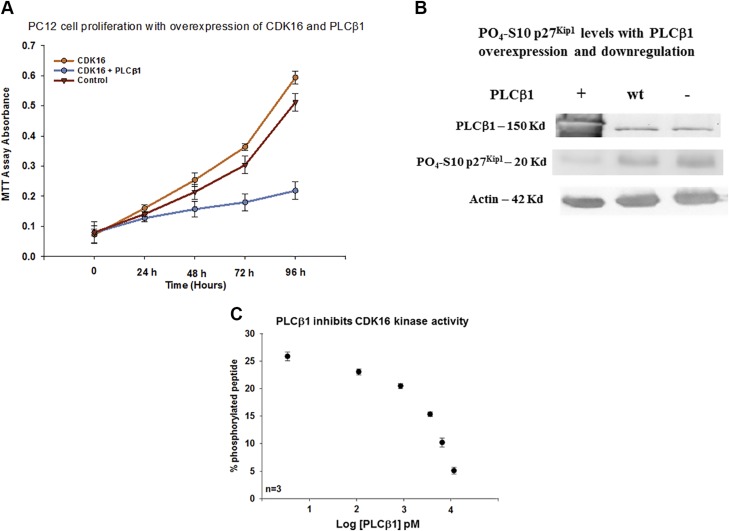
We tested whether PLCβ1 increases cell proliferation by inhibiting the catalytic activity of CDK16. We transfected PC12 cells with CDK16 to observe increased proliferation, and verified that cotransfecting with PLCβ1 reversed proliferation to a point of almost complete arrest (Fig. 4A).
Figure 4.
PLCβ1 inhibits CDK16 activity. A) Increased PC12 cell proliferation with CDK16 overexpression and its reversal with increased PLCβ1 expression; n = 6; control vs. PLCβ1 + CDK16: t = 14.023, P ≤ 0.001; CDK16 + PLCβ1 vs. CDK16: t = 3.887, P = 0.009. B) Western blot of SK-N-SH cell showing the elimination of inactive PO4-Ser10- p27Kip1 when PLCβ1 is overexpressed (n = 6), or slightly down-regulated [i.e., 12-h treatment with siRNA (PLCβ1)] as shown in the third lane. We note that here we overexpressed yellow fluorescent protein–PLCβ (160 kDa) to visually monitor the level of overexpression; this construct has slightly reduced mobility compared with that of the wild-type protein (160 vs. 140 kDa). C) FRET-based commercial assay showing the relative phosphorylation of ∼40 nM CDK16 with increasing PLCβ1 where data is presented. The PLCβ1 concentration is presented in picomolars in logarithmic form so that the full range of PLCβ1 concentrations can be readily viewed (P ≤ 0.001; f = 88.441).
CDK16 moves cells into the S phase by phosphorylating Ser10 on p27Kip1 to inhibit its activity (15). We used an antibody specific to PO4-Ser10 p27Kip1 and probed for the inhibited enzyme in untreated SK-N-SH and PC12. We found that nonphosphorylated p27Kip1 is absent when PLCβ1 is overexpressed, but is readily seen at low PLCβ1 levels (Fig. 4B). This finding suggests that PLCβ1 inhibits the ability of CDK16 to phosphorylate p27, thus halting proliferation.
To verify that PLCβ1 directly inhibits CDK16, we purified the 2 proteins and measured kinase activity of CDK16 using a commercial in vitro kinase assay kit. As described in the Materials and Methods section, this assay uses a peptide with an attached FRET donor and acceptor, and a kinase that primarily cleaves nonphosphorylated peptides. Thus, the kinase activity of CDK16 can be directly assessed by the change in FRET. In Fig. 4C, we plot the amount of phosphorylation as a function of PLCβ1 concentration and find that PLCβ1 significantly inhibits the kinase activity of CDK16.
PLCβ1 dissociates from CDK16 after carbachol stimulation
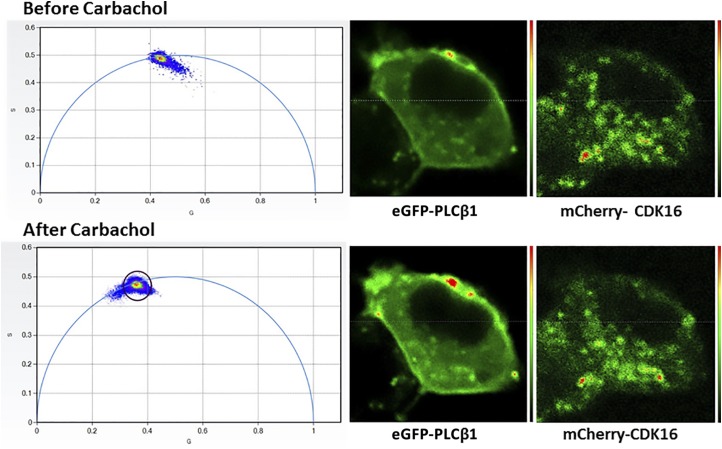
The finding that the cytosolic population of PLCβ1 associates with CDK16 to inhibit proliferation suggests that reducing this population by cell stimulation will promote proliferation, as indicated in Fig. 1B. We verified this hypothesis by stimulating cells transfected with eGFP-PLCβ1 and mCherry-CDK16, and monitored FRET using FLIM (Fig. 5). We found a reduction of FRET after carbachol stimulation, evidenced by the elimination of shorter-lifetime FRET populations (see Fig. 2E). In addition, we found that eGFP-PLCβ1 moved toward the plasma membrane when stimulated (Fig. 5).
Figure 5.
PLCβ1 moves to the plasma membrane with carbachol stimulation. PC12 cells were transfected with eGFP-PLCβ1 from mCherry-CDK16 and FLIM images were taken before (upper panels) and after (lower panels) the addition of 1 µM carbachol. The phasor plots (left panels) show a reduction in FRET after carbachol stimulation, indicated by the movement of lower eGFP-PLCβ1 lifetime values within the phasor arc toward points on or close to the arc.
DISCUSSION
Understanding the underlying protein associations that drive cell proliferation and differentiation is needed to develop therapeutic agents for disease treatment as well as stem cell therapies. Here, we have uncovered a novel neuron-specific interaction that may be used to target neuroblastomas and other diseases based on abnormal proliferation. CDK16, a member of the PCTAIRE kinase family, has homology to cell division cycle protein 2 (25), which plays a central role in the eukaryotic cell cycle. CDK16, which is activated by cyclin Y, is implicated in the development of the CNS as well as other developmental functions (26). CDK16 has been shown to move cells from the G1 phase to the S phase by deactivating p27Kip1 via phosphorylation of Ser10, and is considered an important target in neuronal cancers (15, 27).
Both PLCβ1 and CDK16 are highly expressed in neuronal cells. CDK16 is expressed in cancer cells (28), whereas PLCβ1 expression is diminished in some cancers (29) and is very low in cancer cell lines such as HeLa and HEK293 (11); however, overexpressing these proteins promotes their association (unpublished results). The disparity in expression between CDK16 and PLCβ1 and the competition with Gαq in proliferative cells will impact the amount of time cells spend in the G1 phase. It is important to note that CDK16 moves to the plasma membrane after differentiation, resulting in a loss in FRET with PLCβ1.
Our studies show that PLCβ1 is a novel and unexpected player in neuronal cell proliferation. In most cell types, reduction of PLCβ1 slows proliferation most likely by reducing Gαq-mediated calcium signals and impacting RNA-induced gene silencing (30). Our studies reveal that PC12 and SK-N-SH cells expressing PLCβ1 have prolonged G1 phases due to both the reduction of CDK16 levels and the inhibition of CDK16 activity (Figs. 3 and 4). The importance of this finding is that inhibition of CDK16 via PLCβ1 allows cancerous cells to terminally differentiate and therefore potentially reverses the metastatic nature of certain cancers.
When PC12 cells are treated with nerve growth factor, proliferation ceases and neurites form (31, 32). Our previous studies show that the levels of PLCβ1 increase dramatically in the first 24 h, long before the levels of Gαq rise (1). Thus, our studies give rise to a model in which the bolus of PLCβ1 produced at the onset of differentiation binds to CDK16 to prevent entry into the S phase. Arrest of cells in the G1 phase allows transition to the production of proteins involved in differentiation, as well as proteins involved in relaying sensory information such as Gαq. The large increase of Gαq levels at longer times coupled with reduced PLCβ1 production limits the amount of cytosolic PLCβ1, which aids in the reassignment of CDK16 to its postmitotic function of neurite growth. One of the most interesting aspects of this study is that, before differentiation, PLCβ1 may act as a sensor that surveys cells’ external conditions to inhibit or promote proliferation.
PLCβ1 plays an important role in cell signaling through Gαq but we have also identified 2 other binding partners: α-synuclein and C3PO. α-Synuclein, which binds strongly to PLCβ1 in vitro, competes with Gαq, thus reducing extracellular responses (33). The association between the 2 proteins occurs on the plasma membrane and may not have a significant effect on the cytosolic population of PLCβ1 that interacts with CDK16. We note that because neurologic diseases attributed to α-synuclein overproduction occur in differentiated cells, it is unlikely to affect the CDK16-PLCβ1 association observed here.
C3PO is a complex consisting of translin and TRAX that plays many roles in RNA metabolism (34), including RNA-induced gene silencing (14). The previous work that identified PLCβ1 as a cellular inhibitor of C3PO studied the endogenous proteins in nonneuronal cells, in cells that overexpressed these proteins, or in cells that formed active RNA-induced silencing complexes. These previous studies showed a dynamic exchange between PLCβ and Gαq during activation and C3PO during RNA silencing (11, 35). Our studies showing that Gαq overexpression reverses enhanced proliferation due to PLCβ1 down-regulation are in accord with this exchange of PLCβ between the cytoplasm and the membrane. Mass spectrometry studies show a high number of C3PO-PLCβ1 complexes when PC12 cells are arrested in the G2/M phase but these complexes are absent in the G1 phase. Thus, the impact of PLCβ on proliferation only impacts cells in the G1 phase.
It is interesting that neuronal cells have adapted PLCβ1, which has been closely linked to neurologic disorders, to act as a proliferation sensor. Our studies indicate that if levels of activated Gαq are elevated due to increased expression or high levels of extracellular agonists, then cells will remain proliferative as cytosolic PLCβ1 detaches from CDK16 and moves to the membrane. Thus, the relative levels of these proteins and the composition of the extracellular milieu must be considered to control proliferation.
ACKNOWLEDGMENTS
The authors thank Dr. Elizabeth Bavaro (Worcester Polytechnic Institute) for her help in cloning mCherry-CDK16. The authors are grateful for the support of U.S. National Institutes of Health Grant GM116187. O.G. is supported by a Richard Whitcomb Fellowship (to S.S.). The authors declare no conflicts of interest.
Glossary
- CDK
cyclin-dependent kinase
- C3PO
component 3 of RNA-induced silencing complex activity
- eGFP
enhanced green fluorescent protein
- FLIM
fluorescence lifetime imaging microscopy
- FRET
Förster resonance energy transfer
- HA
human influenza hemagglutinin
- PLCβ
phospholipase Cβ
- siRNA
small interfering RNA
- TRAX
translin-associated factor X
AUTHOR CONTRIBUTIONS
O. Garwain designed and carried out the experiments, and analyzed the data; K. Valla assisted with the experiments and helped write the manuscript; and S. Scarlata designed the studies, analyzed the data and wrote the manuscript.
REFERENCES
- 1.Garwain O., Scarlata S. (2016) Phospholipase Cβ-TRAX association is required for PC12 cell differentiation. J. Biol. Chem. 291, 22970–22976 10.1074/jbc.M116.744953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albasanz J. L., Dalfó E., Ferrer I., Martín M. (2005) Impaired metabotropic glutamate receptor/phospholipase C signaling pathway in the cerebral cortex in Alzheimer’s disease and dementia with Lewy bodies correlates with stage of Alzheimer’s-disease-related changes. Neurobiol. Dis. 20, 685–693 10.1016/j.nbd.2005.05.001 [DOI] [PubMed] [Google Scholar]
- 3.Dwivedi Y., Agrawal A. K., Rizavi H. S., Pandey G. N. (2002) Antidepressants reduce phosphoinositide-specific phospholipase C (PI-PLC) activity and the mRNA and protein expression of selective PLC β1 isozyme in rat brain. Neuropharmacology 43, 1269–1279 10.1016/S0028-3908(02)00253-8 [DOI] [PubMed] [Google Scholar]
- 4.Hannan A. J., Kind P. C., Blakemore C. (1998) Phospholipase C-β1 expression correlates with neuronal differentiation and synaptic plasticity in rat somatosensory cortex. Neuropharmacology 37, 593–605 10.1016/S0028-3908(98)00056-2 [DOI] [PubMed] [Google Scholar]
- 5.Manning E. E., Ransome M. I., Burrows E. L., Hannan A. J. (2012) Increased adult hippocampal neurogenesis and abnormal migration of adult-born granule neurons is associated with hippocampal-specific cognitive deficits in phospholipase C-β1 knockout mice. Hippocampus 22, 309–319 10.1002/hipo.20900 [DOI] [PubMed] [Google Scholar]
- 6.Yamada M., Mizuguchi M., Rhee S. G., Kim S. U. (1991) Developmental changes of three phosphoinositide-specific phospholipase C isozymes in the rat nervous system. Brain Res. Dev. Brain Res. 59, 7–16 10.1016/0165-3806(91)90023-C [DOI] [PubMed] [Google Scholar]
- 7.Dowal L., Provitera P., Scarlata S. (2006) Stable association between Gαq and phospholipase Cβ1 in living cells. J. Biol. Chem. 281, 23999–24014 10.1074/jbc.M512330200 [DOI] [PubMed] [Google Scholar]
- 8.Suh P.-G., Park J.-I., Manzoli L., Cocco L., Peak J. C., Katan M., Fukami K., Kataoka T., Yun S.-U., Ryu S.-H. (2008) Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 41, 415–434 10.5483/BMBRep.2008.41.6.415 [DOI] [PubMed] [Google Scholar]
- 9.Hepler J. R., Gilman A. G. (1992) G proteins. Trends Biochem. Sci. 17, 383–387 10.1016/0968-0004(92)90005-T [DOI] [PubMed] [Google Scholar]
- 10.Aisiku O. R., Runnels L. W., Scarlata S. (2010) Identification of a novel binding partner of phospholipase Cβ1: translin-associated factor X. PLoS One 5, e15001 10.1371/journal.pone.0015001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philip F., Guo Y., Aisiku O., Scarlata S. (2012) Phospholipase Cβ1 is linked to RNA interference of specific genes through translin-associated factor X. FASEB J. 26, 4903–4913 10.1096/fj.12-213934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoki K., Suzuki K., Sugano T., Tasaka T., Nakahara K., Kuge O., Omori A., Kasai M. (1995) A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat. Genet. 10, 167–174 10.1038/ng0695-167 [DOI] [PubMed] [Google Scholar]
- 13.Li Z., Wu Y., Baraban J. M. (2008) The Translin/Trax RNA binding complex: clues to function in the nervous system. Biochim. Biophys. Acta 1779, 479–485 10.1016/j.bbagrm.2008.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., Ye X., Jiang F., Liang C., Chen D., Peng J., Kinch L. N., Grishin N. V., Liu Q. (2009) C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325, 750–753 10.1126/science.1176325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yanagi T., Matsuzawa S. (2015) PCTAIRE1/PCTK1/CDK16: a new oncotarget? Cell Cycle 14, 463–464 10.1080/15384101.2015.1006539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han W., Takano T., He J., Ding J., Gao S., Noda C., Yanagi S., Yamamura H. (2004) Role of BLNK in oxidative stress signaling in B cells. Antioxid. Redox Signal. 3, 1065–1073 10.1089/152308601317203576 [DOI] [PubMed] [Google Scholar]
- 17.Graeser R., Gannon J., Poon R. Y. C., Dubois T., Aitken A., Hunt T. (2002) Regulation of the CDK-related protein kinase PCTAIRE-1 and its possible role in neurite outgrowth in Neuro-2A cells. J. Cell Sci. 115, 3479–3490 [DOI] [PubMed] [Google Scholar]
- 18.Palmer K. J., Konkel J. E., Stephens D. J. (2005) PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J. Cell Sci. 118, 3839–3847 10.1242/jcs.02496 [DOI] [PubMed] [Google Scholar]
- 19.Ou C.-Y., Poon V. Y., Maeder C. I., Watanabe S., Lehrman E. K., Fu A. K. Y., Park M., Fu W.-Y., Jorgensen E. M., Ip N. Y., Shen K. (2010) Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell 141, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikolcevic P., Sigl R., Rauch V., Hess M. W., Pfaller K., Barisic M., Pelliniemi L. J., Boesl M., Geley S. (2012) Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Mol. Cell. Biol. 32, 868–879 10.1128/MCB.06261-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes T. E., Zhang H., Logothetis D. E., Berlot C. H. (2001) Visualization of a functional Gαq-green fluorescent protein fusion in living cells: association with the plasma membrane is disrupted by mutational activation and by elimination of palmitoylation sites, but not be activation mediated by receptors or AlF4−. J. Biol. Chem. 276, 4227–4235 10.1074/jbc.M007608200 [DOI] [PubMed] [Google Scholar]
- 22.Digman M. A., Caiolfa V. R., Zamai M., Gratton E. (2008) The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 94, L14–L16 10.1529/biophysj.107.120154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polyak K., Lee M.-H., Erdjument-Bromage H., Koff A., Roberts J. M., Tempst P., Massagué J. (1994) Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78, 59–66 10.1016/0092-8674(94)90572-X [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Simon R. (2013) Identification of potential synthetic lethal genes to p53 using a computational biology approach. BMC Med. Genomics 6, 30 10.1186/1755-8794-6-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okuda T., Cleveland J. L., Downing J. R. (1992) PCTAIRE-1 and PCTAIRE-3, two members of a novel cdc2/CDC28-related protein kinase gene family. Oncogene 7, 2249–2258 [PubMed] [Google Scholar]
- 26.Liu D., Guest S., Finley R. L. (2010) Why cyclin Y? A highly conserved cyclin with essential functions. Fly (Austin) 4, 278–282 10.4161/fly.4.4.12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ćwiek P., Leni Z., Salm F., Dimitrova V., Styp-Rekowska B., Chiriano G., Carroll M., Höland K., Djonov V., Scapozza L., Guiry P., Arcaro A. (2015) RNA interference screening identifies a novel role for PCTK1/CDK16 in medulloblastoma with c-Myc amplification. Oncotarget 6, 116–129 10.18632/oncotarget.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yanagi T., Shi R., Aza-Blanc P., Reed J. C., Matsuzawa S. (2015) PCTAIRE1-knockdown sensitizes cancer cells to TNF family cytokines. PLoS One 10, e0119404 10.1371/journal.pone.0119404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Follo M. Y., Finelli C., Clissa C., Mongiorgi S., Bosi C., Martinelli G., Baccarani M., Manzoli L., Martelli A. M., Cocco L. (2009) Phosphoinositide-phospholipase C β1 mono-allelic deletion is associated with myelodysplastic syndromes evolution into acute myeloid leukemia. J. Clin. Oncol. 27, 782–790 10.1200/JCO.2008.19.3748 [DOI] [PubMed] [Google Scholar]
- 30.Scarlata S., Garwain O., Williams L., Gonzalez Burguera I., Rosati B., Sahu S., Guo Y., Philip F., Golebiewska U. (2016) Phospholipase Cβ connects G protein signaling with RNA interference. Adv. Biol. Regul. 61, 51–57 10.1016/j.jbior.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das K. P., Freudenrich T. M., Mundy W. R. (2004) Assessment of PC12 cell differentiation and neurite growth: a comparison of morphological and neurochemical measures. Neurotoxicol. Teratol. 26, 397–406 10.1016/j.ntt.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 32.Westerink R. H. S., Ewing A. G. (2008) The PC12 cell as model for neurosecretion. Acta Physiol. (Oxf.) 192, 273–285 10.1111/j.1748-1716.2007.01805.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narayanan V., Guo Y., Scarlata S. (2005) Fluorescence studies suggest a role for α-synuclein in the phosphatidylinositol lipid signaling pathway. Biochemistry 44, 462–470 10.1021/bi0487140 [DOI] [PubMed] [Google Scholar]
- 34.Jaendling A., McFarlane R. J. (2010) Biological roles of translin and translin-associated factor-X: RNA metabolism comes to the fore. Biochem. J. 429, 225–234 10.1042/BJ20100273 [DOI] [PubMed] [Google Scholar]
- 35.Philip F., Sahu S., Golebiewska U., Scarlata S. (2016) RNA-induced silencing attenuates G protein–mediated calcium signals. FASEB J. 30, 1958–1967 10.1096/fj.201500140 [DOI] [PMC free article] [PubMed] [Google Scholar]