Abstract
The functions of the paralogous transcriptional coactivators Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) in bone are controversial. Each has been observed to promote or inhibit osteogenesis in vitro, with reports of both equivalent and divergent functions. Their combinatorial roles in bone physiology are unknown. We report that combinatorial YAP/TAZ deletion from skeletal lineage cells, using Osterix-Cre, caused an osteogenesis imperfecta-like phenotype with severity dependent on allele dose and greater phenotypic expressivity with homozygous TAZ vs. YAP ablation. YAP/TAZ deletion decreased bone accrual and reduced intrinsic bone material properties through impaired collagen content and organization. These structural and material defects produced spontaneous fractures, particularly in mice with homozygous TAZ deletion and caused neonatal lethality in dual homozygous knockouts. At the cellular level in vivo, YAP/TAZ ablation reduced osteoblast activity and increased osteoclast activity, in an allele dose-dependent manner, impairing bone accrual and remodeling. Transcriptionally, YAP/TAZ deletion and small-molecule inhibition of YAP/TAZ interaction with the transcriptional coeffector TEAD reduced osteogenic and collagen-related gene expression, both in vivo and in vitro. These data demonstrate that YAP and TAZ combinatorially promote bone development through regulation of osteoblast activity, matrix quality, and osteoclastic remodeling.—Kegelman, C. D., Mason, D. E., Dawahare, J. H., Horan, D. J., Vigil, G. D., Howard, S. S., Robling, A. G., Bellido, T. M., Boerckel, J. D. Skeletal cell YAP and TAZ combinatorially promote bone development.
Keywords: osteogenesis, transcriptional regulation, osteoprogenitor cells, osteoblasts
Bone is a living hierarchical composite, with form and function dependent not only on tissue structure but also on matrix composition and organization. Each of these components is controlled during development by skeletal cell lineage progression and by dynamic regulation of bone deposition and remodeling. Various genetic, hormonal, or environmental abnormalities can impair these processes, leading to debilitating diseases including osteoporosis and osteogenesis imperfecta (OI). However, the molecular mechanisms that govern cell fate and matrix production in bone remain poorly understood, limiting therapeutic intervention. Several transcriptional programs have been described as essential regulators of bone development, but current understanding is insufficient to fully explain the heterogeneity found in congenital and acquired bone diseases (1–3). In this study, we sought to define the functions of the paralogous transcriptional coactivators yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) in bone development.
YAP/TAZ functional diversity
YAP and TAZ (also known as WWTR1) display either equivalent or divergent functions, depending on cell type and context (4). YAP and TAZ possess transcription activation domains, but they lack DNA-binding domains and require interaction with cofactors for transcriptional activity (5). Their most potent and well-studied interactions are with the transcriptional enhancer activator domain (TEAD) family proteins, which themselves lack activation domains, providing specificity for YAP/TAZ–TEAD signaling (6). However, other coeffectors are also known, including runt-related transcription factor (Runx)-2 (7), β-catenin (8–10), and Smad2/3 (11, 12), each of which contributes to bone development and osteoprogenitor lineage progression (13–17). Thus, independent pathways that regulate coincident activation of these various binding partners could provide additional layers of contextual specificity in bone. Further, as paralogues, the YAP and TAZ proteins also possess structural differences (reviewed in Ref. 18) that enable distinct protein interactions to confer unique physiologic functions of YAP vs. TAZ. Notably, global YAP deletion in mice is embryonic lethal [embryonic day (E)8.5] because of impaired–yolk sac vasculogenesis (19), whereas the global TAZ knockout lives to maturity with modest skeletal defects and polycystic kidney disease (20), demonstrating conclusive gene-specific functions. However, in other contexts, they exhibit clear functional homology, with either protein capable of compensation for the other (21, 22).
YAP and TAZ function in bone: conflicting evidence
Roles for YAP and TAZ in osteogenesis were first described in 2004 and 2005, respectively (23, 24). YAP was reported to suppress osteoblastic differentiation through sequestration and transcriptional repression of Runx2 (23), whereas TAZ was identified as a Runx2 coactivator and an inhibitor of the adipogenic nuclear receptor, peroxisome proliferator-activated receptor-γ (24, 25). A subsequent study found that overexpression of a constitutively active YAP mutant in marrow stromal cells (MSCs) promoted osteogenic differentiation, even under conditions more favorable for adipogenesis (26). In contrast, another report found that YAP overexpression inhibits osteogenesis in MSCs by suppressing activation of wingless-type (WNT) target genes (27). The role of TAZ in osteogenic differentiation in vitro is similarly complicated, with reports demonstrating both inhibition (28) and induction (29) of osteogenic differentiation by modulating the canonical WNT pathway. In vivo, osteoblast-specific overexpression of TAZ promotes bone formation with higher expression levels of Runx2 expression (30), whereas YAP overexpression in chondrocytes impairs cartilage template formation during endochondral bone development (31). Together, these observations suggest the importance of YAP and TAZ in bone, but the conflicting evidence remains unresolved and their combinatorial roles in bone physiology remain unknown. To address these questions, we implemented a combinatorial deletion approach in vivo to evaluate the influence of allele dose-dependent YAP/TAZ deletion on bone development.
MATERIALS AND METHODS
Animals
All protocols were approved by the Institutional Animal Care and Use Committees at the University of Notre Dame and the University of Pennsylvania and in compliance with the National Research Council’s Guide for the Care and Use of Laboratory Animals. Mice harboring loxP-flanked exon 3 alleles in both YAP and TAZ were kindly provided by Eric Olson (University of Texas Southwestern Medical Center, Dallas, TX, USA). Tetracycline-responsive B6.Cg-Tg(Sp/7-tTA,tetO-EGFP/Cre)1AMc/J (Osterix-Cre) mice from The Jackson Laboratory (Bar Harbor, MA, USA) were raised, bred, and evaluated without tetracycline administration, to induce constitutive gene recombination in osteoprogenitor cells and their progeny (32). Mice with homozygous floxed alleles for both YAP and TAZ (YAPfl/fl;TAZfl/fl) were mated with double heterozygous conditional-knockout (cKO) mice (YAPfl/+;TAZfl/+;Osx-Cre) to produce 8 possible genotypes in each litter, but only Cre+and YAPfl/fl;TAZfl/fl animals were compared (Table 1). Both male and female mice were evaluated, with YAPfl/fl;TAZfl/fl mice serving as littermate wild type (WT) controls. All mice were fed regular chow ad libitum and housed in cages containing 2–5 animals each. Mice were maintained at constant 25°C on a 12 h light–dark cycle. Mice were tail or ear clipped after weaning or before euthanasia and genotyped by an external service (Transnetyx, Cordova, TN, USA).
TABLE 1.
Experimental genotypes and abbreviations
| Genotype | Abbreviation |
|---|---|
| Yapfl/fl;Tazfl/fl | YAPWT;TAZWT |
| Yapfl/+;Tazfl/+;Osx1cre/+ | YAPcHET;TAZcHET |
| Yapfl/fl;Tazfl/+;Osx1cre/+ | YAPcKO;TAZcHET |
| Yapfl/+;Tazfl/fl;Osx1cre/+ | YAPcHET;TAZcKO |
| Yapfl/fl;Tazfl/fl;Osx1cre/+ | YAPcKO;TAZcKO |
Skeletal preparations
Skeletal preparations were stained with Alcian blue (A3157; Millipore-Sigma) and Alizarin red (A5533; both from Millipore-Sigma, Billerica, MA, USA) (33).
Histology and histomorphometric analysis
Bone samples were fixed and decalcified according to standard procedures. Paraffin-embedded sections (5 μm thickness) were processed for either immunohistochemistry or histology. Primary antibodies were compared to negative control sections. Anti-Osterix (ab22552, 1:250; Abcam, Cambridge, United Kingdom), anti-YAP (14074, 1:400; Cell Signaling Technology, Danvers, MA, USA), and anti-TAZ (4883, 1:400; Cell Signaling Technology) were applied overnight. Colorimetric detection with the 3,3'-diaminobenzidine–horseradish peroxidase–linked DAB-HRP Substrate Kit (Vector Laboratories, Burlingame, CA, USA) allowed for immunohistochemical detection of YAP, TAZ, or Osterix+ cells. Hematoxylin and eosin (H&E), safranin-O/fast green (Saf-O), tartrate-resistant acid phosphatase (TRAP), and picrosirius red stains were used. The number of osteoblasts per bone surface, osteoclast surface vs. bone surface, and number of osteocytes per bone area were quantified with Osteomeasure (OsteoMetrics, Decatur, GA, USA) on H&E- and TRAP-stained sections. Hypertrophic chondrocyte zone percentage thickness (percentage HZ thickness was calculated with ImageJ (U.S. National Institutes of Health) by measuring 3 separate lines across the area of positive Saf-O staining, normalized to the respective length of the total growth plate within each line and averaged for each image. Methylmethacrylate–embedded bones from mice injected with Calcein (C0875-25G) and Alizarin Complexone (A3882-25G; both from Millipore-Sigma) were processed for dynamic bone histomorphometry. With a diamond-embedded wire saw (Histo-saw; Delaware Diamond Knives, Wilmington, DE, USA), transverse sections (40 µm) were cut from the midshaft and ground to a final thickness of 20 µm. The sections were mounted on slides, and 3 sections per limb were analyzed with Osteomeasure. The following primary data were collected: total bone surface length (BS); single label perimeter (sL.Pm); double-label perimeter (dL.Pm); and double label width (dL.Ith). From primary data, we derived the mineralizing surface: MS/BS = (1/2sL.Pm + dL.Pm)/B.Pm × 100%; mineral apposition rate: MAR = dL.Ith/5 d (µm/d); and bone formation rate: BFR/BS = MAR × MS/BS (µm3/µm2 per day).
Micro–computed tomography
Harvested femora from 8-wk-old mice were stored at −20°C until evaluation. Frozen specimens were thawed and imaged with a vivaCT 80 scanner (Scanco Medical, Zurich, Switzerland) to determine trabecular and cortical femoral bone architecture before mechanical testing to failure in 3-point bending. The mid diaphysis and distal femur were imaged with an X-ray intensity of 114 μA, energy of 70 kVp, integration time of 300 ms, and resolution of 10 μm. Mid-diaphyseal and distal femoral 2-dimensional tomograms were manually contoured, stacked, and binarized by applying a Gaussian filter (σ = 1, support = 1) at a threshold of 250 mg HA/cm3.
Mechanical testing
Mechanical analysis of the femurs was carried out by 3-point bend testing. The femurs were loaded with the condyles facing down onto the bending fixtures, with a lower span length of 4.4 mm. The upper fixture was aligned with the mid diaphysis. The femora were loaded to failure at a rate of 0.5 mm/s by the ElectroForce 3220 Series testing system (TA Instruments, New Castle, DE, USA).
Imaging
Histologic and immunohistochemical sections were imaged on a 90i Upright/Widefield Research Microscope (Nikon Instruments, Melville, NY, USA) at the ×4, 10, 20, and 40 objectives. Three-point bend femur sections, stained with Picrosirius red, were imaged under polarized light with an Eclipse ME600 Microscope (Nikon Instruments) at the ×20 objective while second-harmonic image microscopy (SHIM) images were taken on a multiphoton-enabled Fluoview Research Microscope (Olympus, Center Valley, PA, USA) at a fundamental wavelength of 875 nm with the ×25 objective on sections oriented in the same direction for all groups. All SHIM images were quantified by ImageJ and reported as mean pixel intensity within the cortical region relative to WT bone. Mean pixel intensities across 4 separate regions of interest within each image of the cortex were averaged as technical replicates for a given histologic section.
MSC isolation and culture
Mouse MSCs were isolated from either WT or Osterix-conditioned YAP/TAZ-deficient mice and cultured at 37°C and 5% O2 in medium supplemented with fibroblast growth factor (FGF)-2 (34). In brief, mice were anesthetized by isoflurane inhalation (2%) and euthanized via cervical dislocation. Long-bone samples were dissected, and marrow cavities were flushed out into a tissue culture plastic flask for 3–5 d. MSCs were cultured at 5% O2 in DMEM with 10% fetal bovine serum, 10 ng/ml basic FGF (GF-030-5; Austral Biologicals, San Ramon, CA, USA), 1% penicillin-streptomycin, and 1 μg/ml doxycycline. During passaging, culture medium was removed, and the cells were quickly rinsed once with 4 ml TrypLE Express Enzyme (12605036; Thermo Fisher Scientific, Waltham, MA, USA) by rolling the trypsin over the plate to allow the senescent cells from the cultures to initially detach. These senescent cells were then discarded before standard passaging of the MSCs. MSCs were then seeded at 21% O2 into 6-well plates (9 × 103 cells/cm2) containing 30 ml osteogenic induction medium, which included 2 mg/ml β-glycerophosphate, 50 μM dexamethasone, and 3.75 mg/ml ascorbic acid to the previously described medium without doxycycline or FGF-2. The osteogenic medium was changed every other day before RNA isolation.
UMR-106 cell culture
Osteoblast-like UMR-106 cells (UMRs) were cultured in DMEM containing 4 mM l-glutamine, 4500 mg/L glucose, 1 mM sodium pyruvate, 1500 mg/L sodium bicarbonate, and 10% fetal bovine serum according to American Type Culture Collection (30-2002; ATCC, Manassas, VA, USA) recommendations. UMRs at 50 % confluence in 96-well plates were transfected in antibiotic-free medium for 4 h with 4 previously described luciferase reporter constructs: 1) Runx2-responsive 6xOSE2, 2) 657 bp osteocalcin (OCN) promoter (35), 3) TEAD-responsive 8XGTIIC (Addgene, Cambridge, MA, USA), and a control Renilla plasmid, kindly provided by Munir Tanas (University of Iowa, Iowa City, IA). Forty-eight hours after transfection, UMRs were treated with either DMSO or 0.5 or 1 μM verteporfin (VP) in serum-free conditions for 1 h. All VP experiments were performed in the dark to prevent photoactivation. Cells were then lysed immediately using the Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions (Promega, Madison, WI, USA). Luciferase activity was measured on a Victor 3 (PerkinElmer, Waltham, MA, USA) plate reader and normalized to baseline Renilla activity (36). Separately cultured UMR-106 cells were seeded (4 × 103 cells/cm2) onto 6-well plates and simultaneously treated with DMSO or 0.5 or 1 μM VP and cultured under serum-free conditions for 1 h before RNA isolation.
VP delivery in vivo
Six littermate control (4 male and 2 female) mice (YAPWT;TAZWT) were aged 16 wk. Three mice each (2 males and 1 female each) were assigned to VP or vehicle control (DMSO) groups. In brief, DMSO-solubilized VP was diluted in 0.9% saline and injected intraperitoneally every other for day for 2 wk. Control animals received corresponding injections of DMSO in 0.9% saline. Livers and femurs from both VP- and DMSO-treated mice were harvested on the day of the last injection for RNA isolation.
RNA isolation and quantitative PCR
Total bone and liver samples were snap frozen in liquid nitrogen–cooled isopentane for 1 min before storage at −80°C until processing. Tissue was then homogenized in a mortar and pestle, and RNA from the sample was collected with TRIzol Reagent (Thermo Fisher Scientific) followed by centrifugation in chloroform. RNA from both bone tissue samples and in vitro experiments were purified with the RNA Easy Kit (Qiagen, Germantown, MD, USA) and quantified by spectrophotometry with a NanoDrop 2000 (Thermo Fisher Scientific). RT-PCR was performed on 0.5 μg/μl concentration of RNA with the TaqMan Reverse Transcription Kit (Thermo Fisher Scientific). Quantitative PCR (qPCR) assessed RNA amount using a CFX Connect (Bio-Rad, Hercules, CA, USA) relative to the internal control of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Data are presented using the 2−ΔΔCt method. Specific mouse and rat primer sequences are listed (Table 2).
TABLE 2.
Mouse and rat qPCR primers
| Gene | Primer sequence, 5′–3′ | |
|---|---|---|
| Forward | Reverse | |
| Mouse gene | ||
| Gapdh | TCACTGCCACCCAGAAGAC | TGTAGGCCATGAGGTCCAC |
| Yap | TGGACGTGGAGTCTGTGTT | AAGCGGAACAACGATGGACA |
| Taz | GTCCATCACTTCCACCTC | TTGACGCATCCTAATCCT |
| Col1a1 | GCTCCTCTTAGGGGCCACT | CCACGTCTCACCATTGGGG |
| Col1a2 | GTAACTTCGTGCCTAGCAACA | CCTTTGTCAGAATACTGAGCAGC |
| Col2a1 | GACTGAAGGGACACCGAG | CCAGGGATTCCATTAGAG |
| Col10 | ATGCTGCCTCAAATACCCT | TGCCTTGTTCTCCTCTTACT |
| Serpinh1 | AGCCGAGGTGAAGAAACCC | CATCGCCTGATATAGGCTGAAG |
| Runx2 | AGCCTCTTCAGCGCAGTGAC | CTGGTGCTCGGATCCCAA |
| Osx | CTGGGGAAAGGAGGCACAAAGAAG | GGGTTAAGGGAGCAAAGTCAGAT |
| Ocn | TGAGCTTAACCCTGCTTGTG | TAGGGCAGCACAGGTCCTA |
| Alp | GGACAGGACACACACACACA | CAAACAGGAGAGCCACTTCA |
| Bsp | ACAATCCGTGCCACTCACT | TTTCATCGAGAAAGCACAGG |
| Cyr61 | CTGCGCTAAACAACTCAACGA | GCAGATCCCTTTCAGAGCGG |
| Ctgf | GGGCCTCTTCTGCGATTTC | ATCCAGGCAAGTGCATTGGTA |
| Rat gene | ||
| Gapdh | CATGGCCTTCCGTGTTCCTA | GCGGCACGTCAGATCCA |
| Col1a1 | ACAGCGTAGCCTACATGG | AAGTTCCGGTGTGACTCG |
| Col1a2 | ATGGTGGCAGCCAGTTTG | GCTGTTCTTGCAGTGGTAGG |
| Serpinh1 | TCATGGTGACCCGCTCCTAC | GCTTATGGGCCAAGGGCATC |
| Runx2 | CAGGTTCAACGATCTGAGATTTGT | TGAAGACCGTTATGGTCAAAGTGA |
| Osx | CAGCCTGCAGCAAGTTTGG | TTTTCCCAGGGCTGTTGAGT |
| Alp | GAGCAGGAACAGAAGTTTGC | GTTGCAGGGTCTGGAGAGTA |
| Bsp | TCCTCCTCTGAAACGGTTTCC | CGAACTATCGCCATCTCCATT |
| Ctgf | ATCCCTGCGACCCACACAAG | CAACTGCTTTGGAAGGACTCGC |
Statistics and regression
All statistics and regression analyses were performed in Prism (GraphPad. San Diego, CA, USA) or using R (v2.13.1). Comparisons between 2 groups were made using the independent t test, whereas comparisons between 3 or more groups were made with a 1-way ANOVA with post hoc Bonferroni’s multiple comparisons test, if the data were normally distributed according to D’Agostino-Pearson omnibus normality test and homoscedastic according to Bartlett’s test. When parametric test assumptions were not met, data were log transformed, and residuals were evaluated. If necessary, the nonparametric Kruskal-Wallis test with post hoc Dunn’s multiple comparisons was used. Significance was set at P < 0.05 (adjusted for multiple comparisons). Data are represented as individual samples with means ± sem. Multivariate analysis was performed according to a previously described procedure, with some modifications (37). In brief, we used an exhaustive best-subsets algorithm to determine the best predictors of maximum load and stiffness from a subset of morphologic parameters measured, which included a moment of inertia (I) or section modulus (I/c), tissue mineral density (TMD), and second harmonic generated (SHG) intensity based on Akaike’s information criterion (AIC) (38). The lowest AIC selects the best model while giving preference to less complex models (those with fewer explanatory parameters). Finally, the overall best model for each predicted mechanical property was compared to the prediction from only the moment of inertia (I/c or I for maximum load and stiffness, respectively) using type II general linear regression. Sample sizes were selected a priori by power analysis based on effect sizes and population sd taken from published data on YAPfl/fl; TAZfl/fl mice in other tissues (22), assuming a power of 80% and α = 0.05.
RESULTS
YAP/TAZ expression and deletion in bone
To determine YAP/TAZ expression profiles in bone, we immunostained YAP and TAZ in the growth plate and cancellous and cortical bone of 8-wk-old C57Bl6/J mouse femora. YAP and TAZ immunolocalized in hypertrophic chondrocytes, osteoblasts, and osteocytes with minimal detectable expression in quiescent or proliferating chondrocytes (Supplemental Fig. S1A). Based on these expression patterns, we chose to evaluate the physiologic roles of YAP and TAZ by combinatorial conditional ablation (22) in cells of the skeletal lineage using Osterix-Cre (32). We selected a breeding strategy that yielded littermates with variable YAP/TAZ allele dose. To assess Cre-mediated recombination and deletion of YAP and TAZ, we measured mRNA expression in femoral bone preparations by qPCR (Supplemental Fig. S1B) and verified the absence of protein expression by bone cells in conditional knockout (cKO) mice by immunohistochemistry (Supplemental Fig. S1C). YAP/TAZ expression in skeletal cells was reduced by 50–80% by Osterix-Cre–mediated excision (Supplemental Fig. S1B, C).
Neonatal lethality and hypermineralization
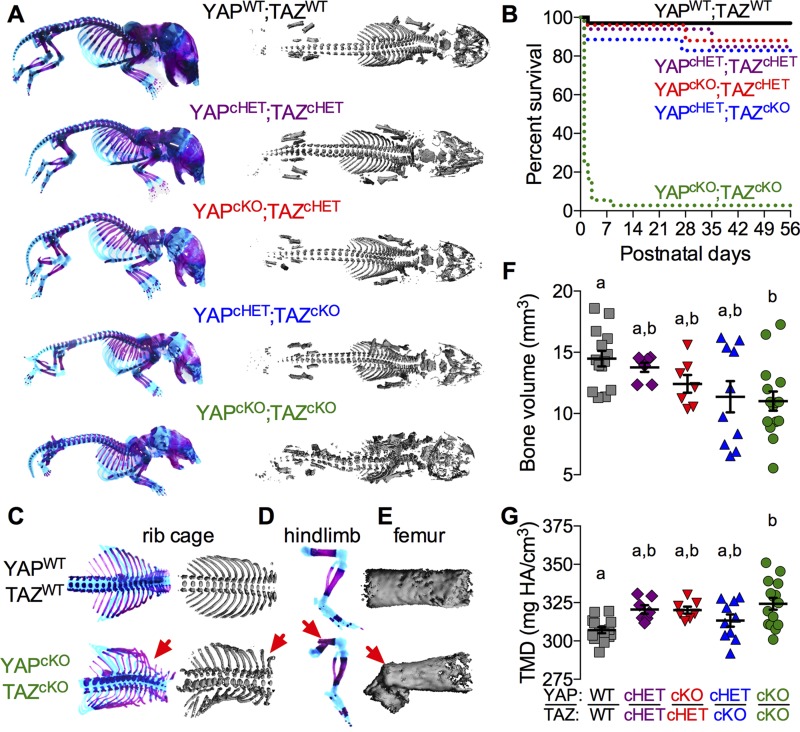
All Osterix-cKOs and littermate controls were born at expected Mendelian ratios, but dual homozygous conditional deletion (YAPcKO;TAZcKO) caused neonatal asphyxiation secondary to ribcage malformation and fracture (Fig. 1A–C), resulting in 75% mortality at postnatal d (P)0 and 99% by P7 (Fig. 1B). Only 1 female YAPcKO;TAZcKO mouse lived to P56 for each endpoint analysis. YAPcKO;TAZcKO neonates exhibited spinal scoliosis, cranial vault deformity, and spontaneous fractures of the ribs, tibia, femur, radius, and ulna (Fig. 1A, C–E). Spontaneous extremity fractures were not present in other genotypes at P0 (Fig. 1A). Littermate neonates displayed reduced whole-skeleton bone volume (Fig. 1F; P < 0.05, ANOVA) and significantly elevated bone TMD (Fig. 1G; P < 0.01, ANOVA) with dual homozygous conditional YAP/TAZ deletion. Osterix-conditional YAP/TAZ deletion also significantly reduced birth weight and intact femoral length in an allele dose-dependent manner (Supplemental Fig. S2). Males and females exhibited similar phenotypes in both growth deficits and P0 skeletal morphology.
Figure 1.
Combinatorial YAP/TAZ ablation from Osterix-expressing cells caused allele dose-dependent perinatal skeletal deformity and lethality. Skeletal structures of littermate mice were evaluated at P0. A) Whole-body skeletal preparations of Osterix-conditional YAP/TAZ knockouts and controls stained with Alcian blue/Alizarin red and micro-CT reconstructions revealed progressive skeletal malformation with decreasing allele dose. B) Survival curves for each genotype show 99% lethality of YAPcKO;TAZcKO mice by P56. C–E) Skeletal preparations and micro-CT reconstructions of rib cages, hindlimbs, and femora, respectively, illustrate spontaneous perinatal fractures in YAPcKO;TAZcKO mice. F) P0 whole-skeleton bone volume was significantly altered by dual homozygous YAP/TAZ deletion. G) P0 whole skeleton TMD increased with YAP/TAZ allele deletion. Data are presented as individual samples with lines corresponding to the mean and sem. Sample sizes, n = 7–15. Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
Spontaneous neonatal long bone fractures and defective endochondral bone formation
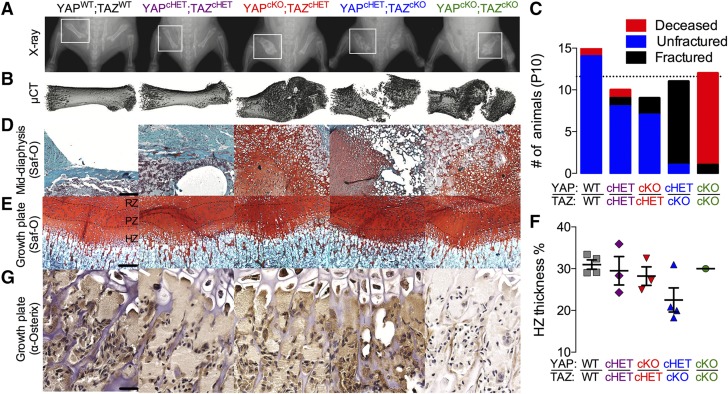
A single copy of either gene rescued neonatal lethality, with 83 and 85% of YAPcHET;TAZcKO and YAPcKO;TAZcHET mice surviving to terminal analysis at P56, respectively. However, between P1 and P10, both YAPcHET;TAZcKO and YAPcKO;TAZcHET mice sustained spontaneous femoral and other bone fractures (Fig. 2A, B), with significantly increased femoral fracture incidence in the YAPcHET;TAZcKO mice (Fig. 2C). Fractures healed by endochondral repair in all groups, though YAPcHET;TAZcKO and YAPcKO;TAZcKO calluses exhibited empty lacunae in the hypertrophic transition zone, suggesting increased hypertrophic chondrocyte death or insufficient progenitor cell recruitment (Fig. 2C c.f. Fig. 2D). Consistently, staining of Osterix+ cells was qualitatively reduced in the transition zone of the YAPcKO;TAZcKO growth plate, but differences in the thickness of resting zone (RZ), proliferating zone (PZ), and hypertrophic zone (HZ) of the growth plate did not reach significance at either P10 (Fig. 2E–G) or P56 (Supplemental Fig. S3A, B).
Figure 2.
YAP/TAZ ablation from Osterix-expressing cells induced spontaneous neonatal femoral fractures and impaired endochondral bone formation. A, B) Representative radiographs (A) with matched micro-CT reconstructions (B) of femoral fracture calluses at P10. C) Quantification of the number of femoral fractures demonstrated significantly increased fracture incidence in YAPcKO;TAZcHET mice. P < 0.01, by χ2 test. D, E) Saf-O/fast green staining of mid diaphysis bone collar (D) and growth plates (E) of matched P10 femora split into RZ, PZ, and HZ. Scale bar, 50 μm. F) Histomorphometric quantification of P10 hypertrophic zone thickness as a percentage of total growth plate thickness (percentage of HZ thickness ). G) Representative micrographs of P10 distal femur growth plates immunostained for Osterix+ cells (brown). Scale bar, 25 μm. Data presented as individual samples with lines corresponding to the mean and sem (sample sizes: n = 3–4, except YAPcKO;TAZcKO, n = 1). Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
Reduced cortical and cancellous microarchitectural properties
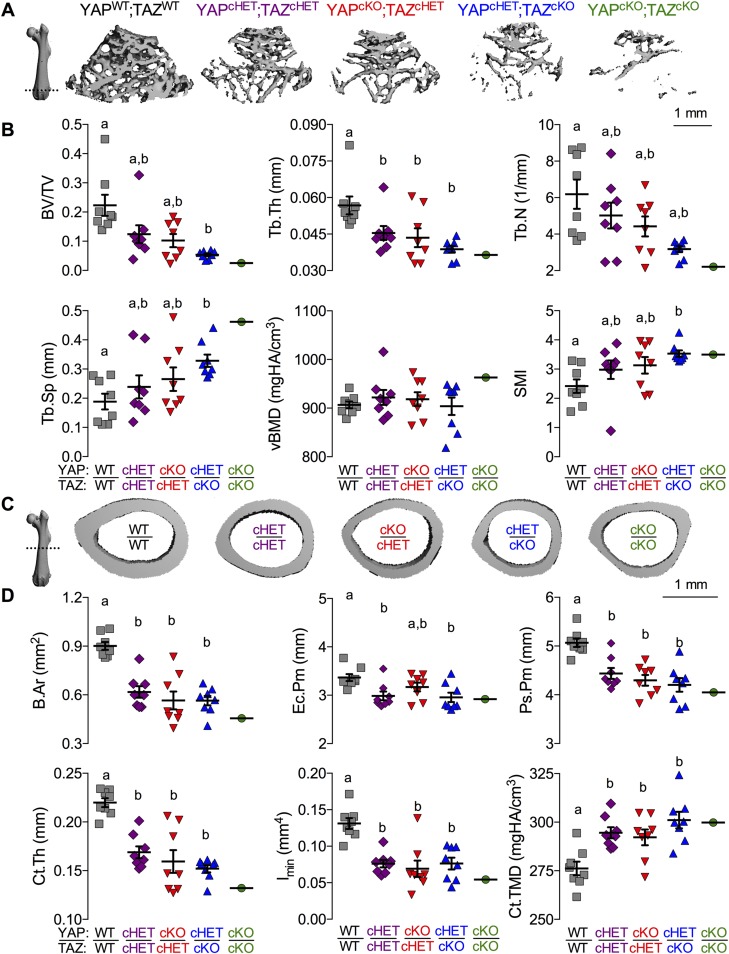
YAP/TAZ deletion from osteoblast precursor cells and their progeny altered cancellous (Fig. 3A, B) and cortical bone (Fig. 3C, D) in adolescent mice (P56) according to allele dose. Distal femur metaphyseal cancellous bone exhibited reduced trabecular bone volume fraction (bone volume/total volume), thickness, and number, and increased spacing and structural model index (indicative of more rod-like trabeculae) (Fig. 3B and Supplemental Fig. S4A–C). The cumulative distribution of trabecular thicknesses shifted in an allele dose-dependent manner, toward reduced numbers of both small and large trabeculae (Supplemental Fig. S4B). Volumetric bone mineral density (vBMD) was not altered, suggesting an increase in local TMD proportional to the decrease in trabecular bond volume (Fig. 3B). The mid diaphyseal femoral cortical bone (Fig. 3C, D and Supplemental Fig. S4D, E) similarly exhibited reduced thickness, area (B.Ar), and moment of inertia (I) in cKO mice, attributable to reduced periosteal and endocortical bone accumulation, as indicated by significant reductions in endocortical perimeter, periosteal perimeter, and B.Ar. Consistent with the observations of vBMD in the cancellous compartment, cortical TMD was significantly increased in an allele dose-dependent manner; however, unlike the cancellous bone, the increase in TMD of the cortical bone was insufficient to normalize bone mass lost by reduced bone volume.
Figure 3.
YAP/TAZ ablation altered bone microarchitectural properties in a manner dependent on allele dose. Femora from 8-wk-old Osterix-conditional YAP/TAZ littermates were evaluated by micro-CT analysis. A) Representative micro-CT reconstructions of distal metaphyseal cancellous bone, arranged in decreasing allele dose. B) Cancellous bone microarchitectural parameters were impaired according to YAP/TAZ allele dose: bone volume fraction (BV/TV); trabecular thickness (Tb.Th), number (Tb.N), and spacing (Tb.Sp); and vBMD. C) Representative micro-CT reconstructions of the mid diaphyseal cortex, arranged in decreasing allele dose. D) Cortical cross-sectional properties were reduced in cKO mice: bone area (B.Ar), endocortical perimeter (Ec.Pm), periosteal perimeter (Ps.Pm), cortical thickness (Ct.Th), moment of inertia in the direction of bending (I), and cortical tissue mineral density (Ct.TMD). Data are presented as individual samples with lines corresponding to the mean and sem (sample sizes: n = 8, except YAPcKO;TAZcKO, n = 1). Scale bar, 1 mm for 2D micro-CT slice reconstructions. Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
Reduced intrinsic bone mechanical properties and matrix collagen content and microstructure
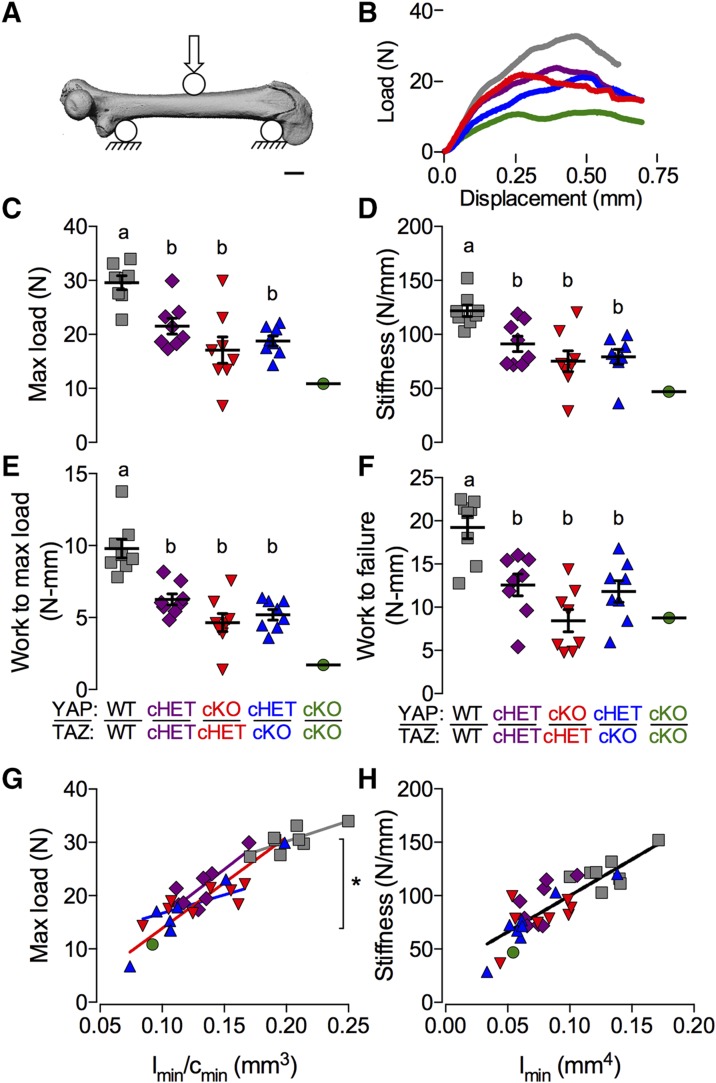
In general, extrinsic bone properties (e.g., failure load, bending stiffness) depend on both the intrinsic mechanical properties of the bone matrix and the bone amount and cross-sectional distribution. To determine whether Osterix-conditional YAP/TAZ deletion impaired bone matrix quality, we performed a 3-point bending test to failure on each femur previously analyzed by micro-CT (Fig. 4A, B). YAP/TAZ deletion reduced stiffness, maximum force at failure, work to maximum load, and work to failure (Fig. 4C–F). Because the assumptions of Euler-Bernoulli beam theory were decidedly not met in the 3-point bending test of mouse long bones (39, 40), we performed an ANCOVA with linear regression (Fig. 4G, H), to decouple the contributions of bone quantity and distribution from the mechanical behavior (41). If the variability in extrinsic mechanical properties is best predicted by individual regression lines for each genotype, this would indicate differences in intrinsic matrix mechanical properties between genotypes; however, a best fit by a single regression line for all groups would indicate that differences in extrinsic behavior are sufficiently described merely by changes in bone geometry. We found that individual regression lines for each genotype best predicted maximum load at failure, indicating significant differences in intrinsic failure properties (Fig. 4G). In contrast, a single regression line best fit the stiffness data (Fig. 4H), indicating that the differences in stiffness can be attributed to changes in moment of inertia rather than intrinsic matrix elastic properties.
Figure 4.
YAP/TAZ ablation reduced intrinsic bone failure properties. A) Femora from 8-wk-old Osterix-conditional YAP/TAZ littermates were tested for 3-point bend to failure. B) Representative load-displacement curves collected during testing. C–F) YAP/TAZ deletion reduced extrinsic mechanical properties measured from the load-displacement curves including maximum load (C), stiffness (D), work to maximum load (E), and work to failure (F). G, H) ANCOVA analysis accounting for bone geometry revealed significant differences in intrinsic failure properties (G), but not intrinsic elastic properties (H). Data are presented as individual samples with lines corresponding to the mean and sem (sample sizes: n = 8, except YAPcKO;TAZcKO, n = 1). Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
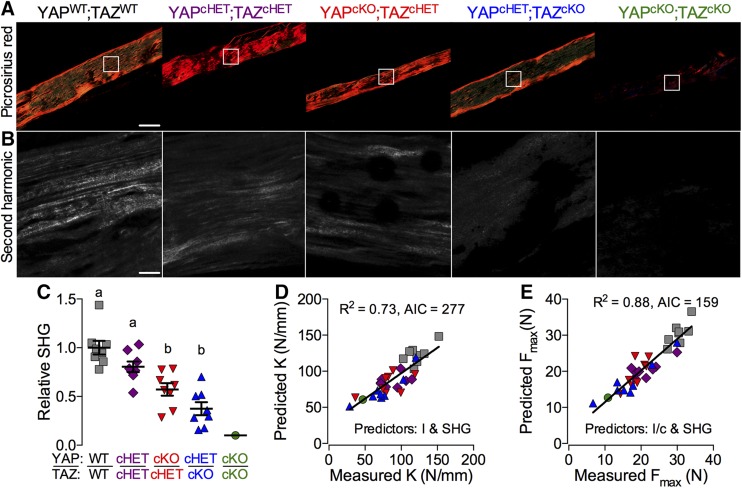
As a composite material, quasi-static bone mechanical behavior is determined predominantly by its 2 primary matrix components: mineral and collagen. We noted above that femora from mice with Osterix-conditional YAP/TAZ deletion exhibited moderate hypermineralization (Fig. 3D). Next, to characterize the bone matrix collagen in these same samples, we performed polarized light microscopy of Picrosirius red–stained sections (Fig. 5A) and SHIM (Fig. 5B) (42). Both approaches revealed that YAP/TAZ deletion significantly reduced local collagen content and organization (Fig. 5C). Therefore, to determine the contributions of geometry, mineralization, and collagen content and microstructure to bone mechanical behavior, we performed a best-subsets correlation analysis to identify significant predictors based on AIC (37, 38). For both elastic (Fig. 5D) and failure (Fig. 5E) properties, bone TMD was not a significant predictor; however, moment of inertia and SHG intensity significantly improved the model’s capability to explain variation (Radj2 = 73 and 88% for stiffness and maximum load, respectively) and reduced AIC (Supplemental Fig. S5). Addition of TMD to the models did not improve predictive power or AIC.
Figure 5.
YAP/TAZ ablation reduced bone matrix collagen content and organization. Imaging of matrix collagen was performed on femora from 8-wk-old Osterix-conditional YAP/TAZ littermates. A, B) Representative polarized light (A) and SHG (B) microscopy images from cortical bone tissue sections of 3-point bend–tested femora. C) SHG intensity, relative to WT, was reduced according to allele dose. D, E) Best subsets regression analyses indicating significant contributions of both bone geometry and collagen content and microstructure, but not TMD, to both elastic (D) and failure (E) mechanical properties (sample sizes: n = 8, except YAPcKO;TAZcKO, n = 1). Scale bars, 100 and 25 μm in Picrosirius red and SHG images, respectively. Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
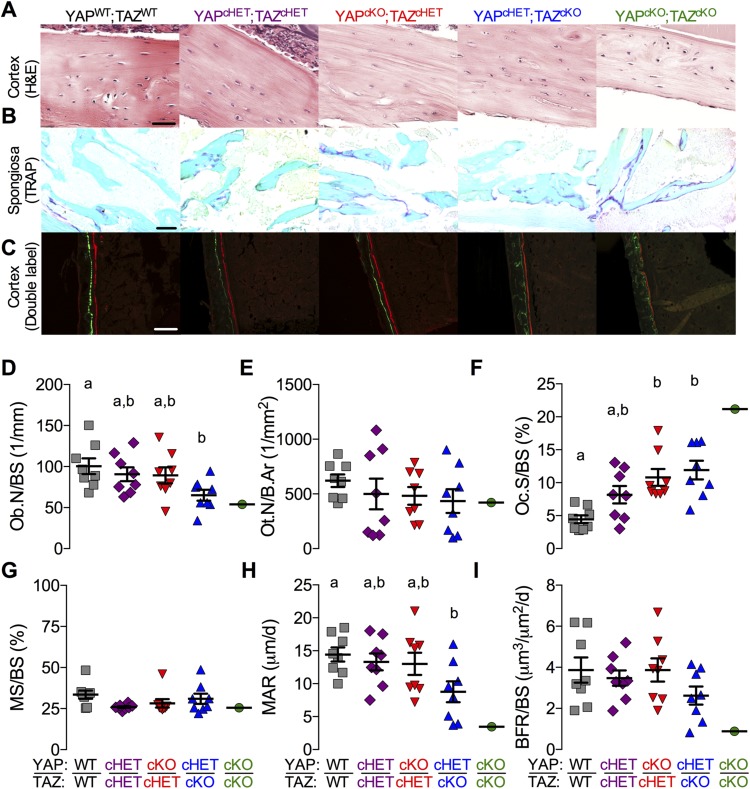
Static histomorphometric analysis of Osterix-conditional YAP/TAZ-deficient P56 femora revealed an allele dose-dependent decrease in the number of periosteal osteoblasts per bone surface (Ob.N/BS), but a dose-dependent increase in osteoclast surface vs. bone surface (Oc.S/BS) in the metaphyseal secondary spongiosa (Fig. 6A, B, D, E). Cortical osteocyte density (Ot.N/B.Ar) was not significantly altered (Fig. 6A, F). Dynamic histomorphometric analysis of Osterix-conditional YAP/TAZ deficient P28 femora revealed no significant differences in mineralizing surface percentage (MS/BS), whereas the mineral apposition rate (MAR) was significantly reduced, according to allele dose (Fig. 6C, G, H). Differences in bone formation rate (BFR/BS) did not reach statistical significance (Fig. 6C, I). YAP/TAZ deletion similarly altered fluorescent labeling of epiphyseal and metaphyseal cancellous bone compartments (Supplemental Fig. S6).
Figure 6.
YAP/TAZ ablation reduced the number of osteoblasts and increased osteoclast activity. Femora were evaluated by static and dynamic histomorphometry from Osterix-conditional YAP/TAZ deletion. A) Representative micrographs of P56 middiaphyseal cortical bone stained by H&E from the Osterix-conditional YAP/TAZ deletion. B) Representative micrographs of P56 metaphyseal cancellous bone stained by TRAP from the Osterix-conditional YAP/TAZ deletion. C) Representative micrographs of double fluorochrome–labeled P28 femoral cortices. C–F) Static histomorphometric quantification of osteoblast per bone surface (Ob.N/BS) (D), osteocyte number per bone area (Ot.N/B.Ar) (E), and osteoclast surface per bone surface (Oc.S/BS) (F). G–I) Dynamic histomorphometric quantification of mineralizing surface percentage (MS/BS) (G), mineral apposition rate (MAR) (H), and bone formation rate (BFR/BS) (I). Data are presented as individual samples with lines corresponding to the mean and sem (sample sizes: n = 8, except YAPcKO;TAZcKO;Osterix-Cre, n = 1). Scale bars, 25, 50, and 100 μm in H&E, TRAP, and double-labeled micrographs, respectively. Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
YAP/TAZ deletion and acute YAP/TAZ–TEAD inhibition reduced osteogenic and collagen-related gene expression
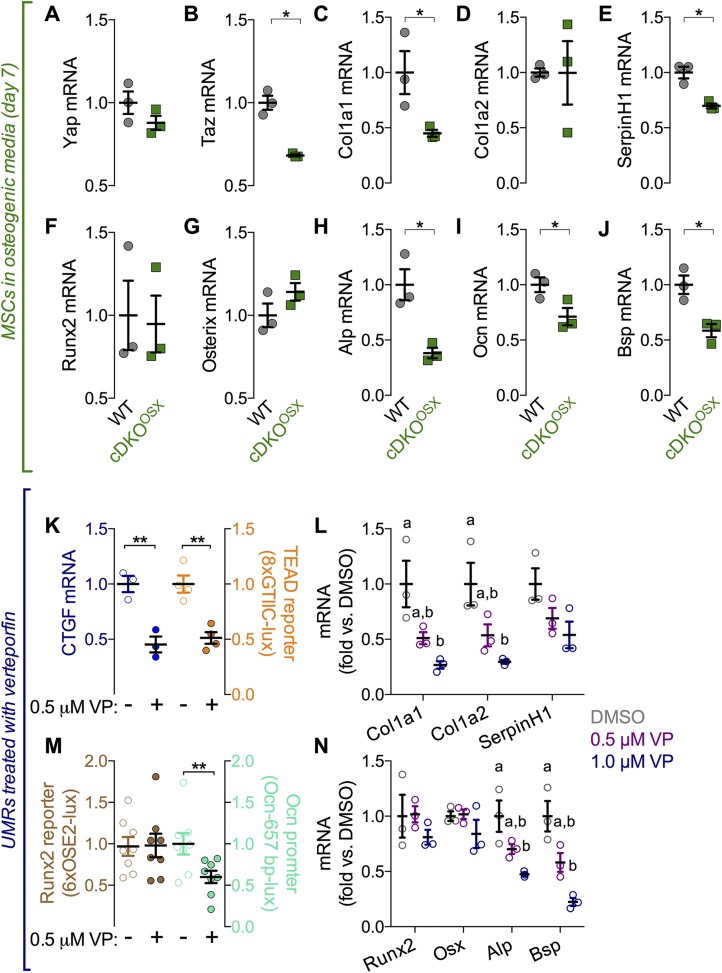
To identify potential YAP/TAZ transcriptional targets, we evaluated expression of candidate genes known to regulate osteogenesis or whose mutations cause OI in MSCs isolated from WT and Osterix-conditional YAPcKO;TAZcKO mice. For all tested genes, mRNA expression levels were equivalent before osteogenic induction, verifying Osterix-dependence of gene recombination (Supplemental Fig. S7). However, after 7 d in osteogenic medium, Osterix-conditional Cre-mediated recombination significantly reduced TAZ mRNA expression, whereas the reduction in YAP expression did not reach statistical significance (Fig. 7A, B). However, mRNA expression of canonical YAP/TAZ target genes, cysteine-rich angiogenic inducer 61 and connective tissue growth factor (Ctgf), was significantly reduced (Supplemental Fig. S8A, B). Of the collagen-related genes, mRNA expression of Col1a1and serine proteinase inhibitor clade H (SerpinH)-1, but not Col1a2, Col2, or Col10 was significantly reduced in YAP/TAZ cKO cells (Fig. 7C–E and Supplemental Fig. S8C, D). Of the osteogenic genes, mRNA expression of Ocn, alkaline phosphatase (Alp), and bone sialoprotein (Bsp) were significantly reduced, but expression of Runx2 and Osterix were not altered (Fig. 7F–J).
Figure 7.
YAP/TAZ-deletion and acute inhibition of YAP/TAZ–TEAD with VP-reduced osteogenic and collagen-related gene expression in vitro. A–J) MSCs were isolated from Osterix-conditional YAPcKO;TAZcKO mice and cultured under osteogenic conditions. Expression levels, normalized to GAPDH, were evaluated for YAP (A) and TAZ (B) along with collagen–related genes, Col1a1(C), Col1a2 (D), and SerpinH1 (E); key upstream osteogenic transcription factors Runx2 (F) and Osterix (G) and the downstream osteogenic genes Ocn (H), Alp (I), and Bsp (J). Osteoblast-like UMR-106 cells were treated with the inhibitor VP, to block interaction of YAP/TAZ with TEAD. K) Effectiveness was assessed by mRNA expression of the canonical YAP/TAZ–TEAD target gene CTGF and synthetic TEAD (8xGTIIC) reporter activity. 8xGTIIC reporter activity was normalized to Renilla luciferase expression and is expressed as fold vs. DMSO. L) VP treatment dose dependently reduced mRNA levels of Col1a1, Col1a2, and SerpinH1 in UMR-106 cells in comparison to DMSO. M) The activity of the Runx2 (6xOSE2) reporter activity was not altered after VP treatment, but the activity of the 657 bp Ocn promoter was reduced after VP treatment. N) VP treatment dose dependently reduced mRNA levels of Bsp and Alp, but not Runx2 and Osterix (Osx) in UMR-106 cells in comparison to DMSO (sample sizes: n = 3–8). Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
We next sought to determine whether this gene regulation was dependent on YAP/TAZ–TEAD in osteoblast-like cells by using a small molecule inhibitor, VP, which blocks YAP/TAZ interaction with TEAD (43). We found that VP treatment of osteoblast-like UMR-106 cells reduced expression of the known YAP/TAZ–TEAD target gene CTGF, concomitant with reduced YAP/TAZ–TEAD–sensitive synthetic promoter activity (8xGTIIC-lux) (Fig. 7K). mRNA expression of OI-related genes Col1a1 and Col1a2 was reduced by YAP/TAZ–TEAD inhibition, whereas differences in SerpinH1 expression did not reach statistical significance (Fig. 7L). VP treatment did not alter Runx2 transcriptional activity (OSE2-lux) or Runx2 and Osterix expression levels, but reduced Ocn promoter activity (Ocn-657 bp-lux) concomitant with reduced expression of Bsp and Alp mRNA (Fig. 7M, N).
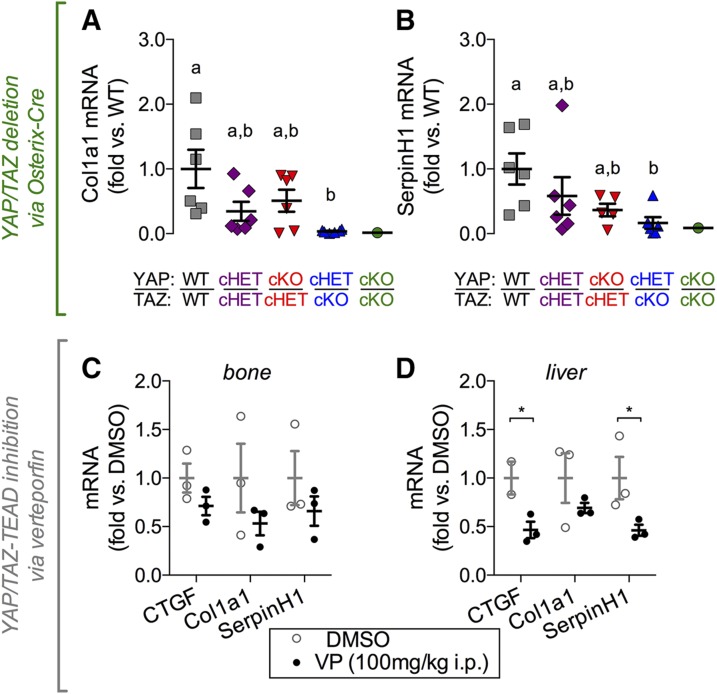
To determine whether YAP and TAZ regulate expression of these genes in vivo, we performed real-time qPCR amplification of mRNA transcripts isolated from femoral cortical bone preparations. Osterix-conditional YAP/TAZ deletion significantly reduced Col1a1 and SerpinH1 expression in a manner dependent on allele dose (Fig. 8A, B). No differences in Col1a2 (Supplemental Fig. S9A), Col2a1, or Col10 (Supplemental Fig. S9B, C) expression were observed. Similarly, gene expression of osteogenic transcripts, Runx2, Osx, Ocn, Alp, and Bsp did not exhibit any significant differences in expression levels in vivo (Supplemental Fig. S9D–G). Next, we evaluated whether YAP/TAZ–TEAD regulate the identified collagen-related candidate genes in vivo by acute YAP/TAZ inhibition in WT mice by VP injection. VP delivery (100 mg/kg i.p. injection every other day for 2 wk) significantly reduced expression of CTGF and SerpinH1 in liver tissue in vivo, but reductions in Col1a1 expression in liver and CTGF, Col1a1, and SerpinH1 in bone did not reach statistical significance (Fig. 8C, D).
Figure 8.
YAP/TAZ-deletion and acute inhibition of YAP/TAZ–TEAD with VP reduced collagen-related gene expression in vivo. Femoral cortical bones from Osterix-conditional YAP/TAZ-deficient mice were harvested to quantify mRNA expression. Expression levels, normalized to GAPDH, were evaluated in Osterix-conditional YAP/TAZ-deficient cortical bone for Col1a1 (A) and SerpinH1 (B). The 16-wk-old WT mice received intraperitoneal injections of VP (100 mg/kg) every 2 d for 2 wk. VP treatment did not significantly reduce mRNA expression levels in bone (C), but differences in CTGF and SerpinH1 were detected in VP-treated livers (D). Repeated significance indicator letters (a, b) signify P > 0.05, while groups with distinct indicators signify P < 0.05 by ANOVA with Bonferroni post hoc test.
DISCUSSION
Reports on the roles of YAP and TAZ in bone are contradictory (23–30, 44–46). To resolve these apparent conflicts in a physiologic context, we performed combinatorial conditional YAP/TAZ deletion in mice to dissect the roles of YAP and TAZ in the cells of the osteoblast lineage, from the precursors to terminal osteocytes, using Osterix-Cre. Our data reveal that YAP and TAZ have combinatorial roles in promoting osteogenesis by regulating bone formation, remodeling, and matrix mechanical properties.
YAP/TAZ deletion from skeletal cells phenocopies OI
Bone cell-conditional YAP/TAZ deletion caused skeletal defects similar to OI, with severity dependent on allele dose. OI is a highly heterogeneous group of inherited genetic diseases characterized by bone fragility and deformity, whose severity varies from mildly increased fracture risk to perinatal lethality (47). YAP/TAZ cKO mice mimicked clinical OI (48) and several established OI mouse models (49–51) with reduced bone volume in both cancellous and cortical compartments. For example, the human Col1a1 minigene mouse (49, 50), which expresses a human transgene containing a clinically observed mutation in proα1(I) collagen, dose-dependently reproduces the phenotypes seen in Osterix-conditional YAP/TAZ knockouts, including neonatal lethality at high transgene dose and spontaneous femoral fractures and reduced failure, but not elastic, bone material properties at moderate dose. Similarly, the naturally occurring oim mouse, caused by a frameshift mutation in proα2(I) collagen, also features reduced bone mechanical properties and increased fracture incidence with elevated mineral density (51, 52), a product of increased mean tissue age. In addition, multivariate regression analyses revealed intrinsic matrix mechanical property deficiencies in YAP/TAZ cKO mice similar to the oim mouse, also attributable to defects in local collagen content and organization (52, 53). Similarly, conditioned medium from osteoprogenitor cells isolated from oim mice increased osteoclast formation in vitro (54), consistent with our observation of increased osteoclast activity in Osterix-conditional YAP/TAZ knockout bone. This finding suggests altered osteoclast recruitment and activation as a result of defective skeletal cell communication. Because global YAP deletion is embryonic lethal in animal models, loss-of-function mutations in YAP/TAZ are unlikely to be a cause of human OI; however, many pathways including TGF-β–Smad2/3 (11, 12) and WNT–β-catenin (9, 10) converge on YAP/TAZ, which could place this signaling axis upstream of the human disease. Further research is needed to evaluate whether YAP/TAZ signaling is causally linked to clinical OI. The elucidation of this pathway in bone may contribute new insights into the heterogeneity and etiology of the disease.
YAP/TAZ compensatory function
Mice possessing a single copy of either gene in Osterix-expressing cells rescued the lethality found in dual homozygous knockouts, indicating mutual compensatory function. However, mice with homozygous deletion of TAZ (i.e., YAPcHET;TAZcKO) exhibited consistently increased phenotypic expressivity compared with YAPcKO;TAZcHET for all outcome measures, including bone formation, osteoclast activity, and bone quality. This result suggests that either TAZ is the more potent of the 2 paralogues in bone or that the 2 floxed loci exhibited differential efficiency of Cre-mediated excision. This latter possibility is supported by the greater reduction in TAZ expression observed in differentiating MSCs isolated from YAPfl/fl;TAZfl/fl;Osx-Cre mice; however, in vivo, mRNA and protein levels of YAP and TAZ were similarly reduced. Thus, further study is necessary to elucidate potentially distinct coeffectors or transcriptional efficiency for YAP vs. TAZ in bone. A recent report demonstrated a unique binding mode of TAZ to TEAD4 based on crystal structure, suggesting a potential difference in regulatory function of TAZ vs. YAP (55). In addition, the Osterix-Cre transgene exhibits some non–skeletal-cell targeting, including potential recombination in muscle (56) and causes defects in craniofacial development (57); however, we did not observe differential YAP/TAZ expression in skeletal muscle, and the allele dose-dependent response establishes YAP/TAZ specificity. These data demonstrate a critical combinatorial role for both YAP and TAZ in bone development and combinatorial function, evidenced by the rescue of neonatal lethality by a single intact allele of either gene.
A recent study found that YAP overexpression in developing chondrocytes, under control of the Col2a1 promoter, impairs bone development (31). This finding appears to contradict our results; however, this study was not designed to isolate the role of YAP in the skeletal lineage and featured YAP overexpression in the cartilaginous anlage, as well as the osteoblast precursors. YAP has elsewhere been reported to negatively regulate chondrogenesis (58), and changes in anlage formation may therefore alter bone development independent of defects in osteogenic cells. Further, the developmental phenotype appeared only in homozygously overexpressed transgenics, which points to the limitations of overexpression approaches for tightly regulated transcriptional regulators that may exhibit nonphysiologic transcriptional activity at high concentrations. Consistent with our observations that YAP and TAZ have compensatory roles, they did not observe statistically significant effects of Col2a1-conditional YAP deletion on skeletal development (31). Similarly, Yang et al. (30) overexpressed TAZ in collagen I-expressing cells and observed increased bone formation, consistent with the present data and the phenotype of the global homozygous TAZ knockout, which also presents bone development defects (20). Synthesis of these studies indicates the importance of dual and combinatorial loss-of-function approaches to interrogate YAP/TAZ compensatory function.
YAP/TAZ-dependent gene expression
Bone cell–conditional YAP/TAZ deletion produced an allele dose-dependent phenotype characterized by defects in both osteogenesis and matrix composition, associated with reduced osteogenic and collagen-related gene expression. These transcriptional patterns were consistent in vitro and in vivo. YAP/TAZ deletion reduced osteogenic gene induction in isolated osteoprogenitors and reduced osteoblast numbers and mineral apposition rates in vivo, indicating that YAP/TAZ deletion impaired osteoblast differentiation and activation. Further, reduced collagen content and organization in vivo and impaired expression of Col1a1 and the endoplasmic reticulum–associated collagen chaperone, SerpinH1, suggest that YAP/TAZ regulate collagen production. These findings support a convergent, pro-osteogenic function for both YAP and TAZ (24–26, 29, 30).
YAP and TAZ control gene expression through formation of transcriptional complexes with other transcription factors. These include TEAD1-4 and Runx2, among others (8–12). Runx2 has been identified as a YAP/TAZ coeffector in osteogenesis in vitro (23–25), but the role of TEAD in bone is unclear. To determine whether TEAD could be involved in YAP/TAZ regulation of osteogenesis- and collagen-related genes, we evaluated the effects of disrupting the YAP/TAZ–TEAD interaction, by using the small-molecule inhibitor VP, in vitro and in vivo. Quantification of YAP/TAZ–TEAD transcriptional activity and canonical downstream gene expression showed that VP treatment significantly inhibited YAP/TAZ–TEAD activity. Analysis of published chromatin immunoprecipitation sequencing data on the UCSC Genome Browser (University of California, Santa Cruz, CA, USA) (59) revealed that TEAD is capable of binding its canonical recognition sequence (3′-ACATTCCA-5′) in the promoter region of both Col1a1 and SerpinH1, suggesting the possibility of direct regulation. However, as YAP/TAZ are known to regulate gene expression through both promoter and enhancer binding, further research using chromatin immunoprecipitation combined with targeted mutagenesis is necessary, to isolate the binding domains and associated coeffectors. In contrast, VP treatment had no effect on Runx2 transcriptional reporter activity or direct Runx2 target genes (i.e., autoregulatory Runx2 or Osterix), either in vitro or in vivo. Despite this finding, expression of mature osteoblast markers was decreased by VP treatment, concomitant with Ocn promoter activity, suggesting that YAP/TAZ–TEAD may be involved in both osteogenic and collagen-related gene regulation. In vivo, VP treatment significantly reduced Col1a1 in the liver, but did not significantly alter gene expression in bone, most likely because of the small sample size and the 4-fold less efficient biodistribution of porphyrins to bone compared to liver (60, 61). VP may also exhibit off-target effects (62), but both VP treatment and YAP/TAZ-conditional deletion produced consistent gene expression profiles.
These data demonstrate that YAP and TAZ have combinatorial roles in promoting skeletal development by regulating osteoblast activity, osteoclast-mediated remodeling, and matrix composition.
Supplementary Material
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
ACKNOWLEDGMENTS
YAPfl/fl;TAZfl/fl mice were provided by Eric Olson (University of Texas Southwestern Medical Center, Dallas, TX, USA); mouse husbandry and maintenance were performed by Theresa Sikorski (University of Notre Dame). 8xGTIIC luciferase and Renilla constructs were provided by Dr. Munir Tanas (University of Iowa, Iowa City, IA, USA); and 6xOSE2 and 657 bp osteocalcin promoter luciferase constructs were provided by Dr. Ling Qin (University of Pennsylvania). This project was supported in part by U.S. National Institutes of Health (NIH) National Center for Advancing Translational Sciences Grant UL1TR001108 (to J.D.B.) and NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant T32-AR007132 (to C.D.K.), by National Science Foundation Grant 1435467 (to J.D.B.), and by American Heart Association Grant 16SDG31230034 (to J.D.B.). The authors declare no conflicts of interest.
Glossary
- AIC
Akaike’s information criterion
- Alp
alkaline phosphatase
- Bsp
bone sialoprotein
- B.Ar
bone area
- BS
bone surface
- Col1a1
collagen type Iα1
- CTGF
connective tissue growth factor
- FGF
fibroblast growth factor
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- H&E
hematoxylin and eosin
- HZ
hypertrophic zone
- I/c
section modulus
- MAR
mineral apposition rate
- micro-CT
micro–computed tomography
- MSC
marrow stromal cell
- OCN
osteocalcin
- OI
osteogenesis imperfecta
- PZ
proliferating zone
- qPCR
quantitative PCR
- Runx2
runt-related transcription factor 2
- RZ
resting zone
- Saf-O
safranin-o/fast green
- SerpinH1
serine proteinase inhibitor, clade H
- SHG
second-harmonic generation
- SHIM
second-harmonic image microscopy
- TAZ
transcriptional coactivator with PDZ-binding motif
- TEAD
transcriptional enhancer activator domain
- TMD
tissue mineral density
- TRAP
tartrate-resistant acid phosphatase
- vBMD
volumetric bone mineral density
- VP
verteporfin
- WNT
wingless-type
- YAP
yes-associated protein
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
C. D. Kegelman, D. E. Mason, and J. D. Boerckel designed the research; C. D. Kegelman, D. E. Mason, A. G. Robling, T. M. Bellido, and J. D. Boerckel analyzed the data; C. D. Kegelman, D. E. Mason, J. H. Dawahare, D. J. Horan, and G. D. Vigil performed the research; D. J. Horan, G. D. Vigil, S. S. Howard, A. G. Robling, and T. M. Bellido contributed new reagents and analytic tools; C. D. Kegelman and J. D. Boerckel wrote the paper; and all authors reviewed the paper.
REFERENCES
- 1.De Crombrugghe B., Lefebvre V., Nakashima K. (2001) Regulatory mechanisms in the pathways of cartilage and bone formation. Curr. Opin. Cell Biol. 13, 721–727 10.1016/S0955-0674(00)00276-3 [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G. (2008) Transcriptional control of skeletogenesis. Annu. Rev. Genomics Hum. Genet. 9, 183–196 10.1146/annurev.genom.9.081307.164437 [DOI] [PubMed] [Google Scholar]
- 3.Javed A., Chen H., Ghori F. Y. (2010) Genetic and transcriptional control of bone formation. Oral Maxillofac. Surg. Clin. North Am. 22, 283–293, v 10.1016/j.coms.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varelas X. (2014) The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development 141, 1614–1626 10.1242/dev.102376 [DOI] [PubMed] [Google Scholar]
- 5.Vassilev A., Kaneko K. J., Shu H., Zhao Y., DePamphilis M. L. (2001) TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229–1241 10.1101/gad.888601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yagi R., Chen L.-F., Shigesada K., Murakami Y., Ito Y. (1999) A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18, 2551–2562 10.1093/emboj/18.9.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbluh J., Nijhawan D., Cox A. G., Li X., Neal J. T., Schafer E. J., Zack T. I., Wang X., Tsherniak A., Schinzel A. C., Shao D. D., Schumacher S. E., Weir B. A., Vazquez F., Cowley G. S., Root D. E., Mesirov J. P., Beroukhim R., Kuo C. J., Goessling W., Hahn W. C. (2012) β-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis [Published correction in Cell (2013) 153, 267–270]. Cell 151, 1457–1473 10.1016/j.cell.2012.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heallen T., Zhang M., Wang J., Bonilla-Claudio M., Klysik E., Johnson R. L., Martin J. F. (2011) Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science 332, 458–461 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azzolin L., Panciera T., Soligo S., Enzo E., Bicciato S., Dupont S., Bresolin S., Frasson C., Basso G., Guzzardo V., Fassina A., Cordenonsi M., Piccolo S. (2014) YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell 158, 157–170 10.1016/j.cell.2014.06.013 [DOI] [PubMed] [Google Scholar]
- 11.Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B. M., Dembowy J., Yaffe M. B., Zandstra P. W., Wrana J. L. (2008) TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 10, 837–848 10.1038/ncb1748 [DOI] [PubMed] [Google Scholar]
- 12.Varelas X., Samavarchi-Tehrani P., Narimatsu M., Weiss A., Cockburn K., Larsen B. G., Rossant J., Wrana J. L. (2010) The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev. Cell 19, 831–844 10.1016/j.devcel.2010.11.012 [DOI] [PubMed] [Google Scholar]
- 13.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 10.1016/S0092-8674(00)80258-5 [DOI] [PubMed] [Google Scholar]
- 14.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., Owen M. J. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 10.1016/S0092-8674(00)80259-7 [DOI] [PubMed] [Google Scholar]
- 15.Mundlos S., Otto F., Mundlos C., Mulliken J. B., Aylsworth A. S., Albright S., Lindhout D., Cole W. G., Henn W., Knoll J. H., Owen M. J., Mertelsmann R., Zabel B. U., Olsen B. R. (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89, 773–779 10.1016/S0092-8674(00)80260-3 [DOI] [PubMed] [Google Scholar]
- 16.Kato M., Patel M. S., Levasseur R., Lobov I., Chang B. H., Glass D. A., II, Hartmann C., Li L., Hwang T. H., Brayton C. F., Lang R. A., Karsenty G., Chan L. (2002) Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J. Cell Biol. 157, 303–314 10.1083/jcb.200201089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afzal F., Pratap J., Ito K., Ito Y., Stein J. L., van Wijnen A. J., Stein G. S., Lian J. B., Javed A. (2005) Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J. Cell. Physiol. 204, 63–72 10.1002/jcp.20258 [DOI] [PubMed] [Google Scholar]
- 18.Hong W., Guan K. L. (2012) The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol. 23, 785–793 10.1016/j.semcdb.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin-Kensicki E. M., Boone B. N., Howell M., Stonebraker J. R., Teed J., Alb J. G., Magnuson T. R., O’Neal W., Milgram S. L. (2006) Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol. Cell. Biol. 26, 77–87 10.1128/MCB.26.1.77-87.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hossain Z., Ali S. M., Ko H. L., Xu J., Ng C. P., Guo K., Qi Z., Ponniah S., Hong W., Hunziker W. (2007) Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc. Natl. Acad. Sci. USA 104, 1631–1636 10.1073/pnas.0605266104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miesfeld J. B., Gestri G., Clark B. S., Flinn M. A., Poole R. J., Bader J. R., Besharse J. C., Wilson S. W., Link B. A. (2015) Yap and Taz regulate retinal pigment epithelial cell fate. Development 142, 3021–3032 10.1242/dev.119008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xin M., Kim Y., Sutherland L. B., Murakami M., Qi X., McAnally J., Porrello E. R., Mahmoud A. I., Tan W., Shelton J. M., Richardson J. A., Sadek H. A., Bassel-Duby R., Olson E. N. (2013) Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. USA 110, 13839–13844 10.1073/pnas.1313192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaidi S. K., Sullivan A. J., Medina R., Ito Y., van Wijnen A. J., Stein J. L., Lian J. B., Stein G. S. (2004) Tyrosine phosphorylation controls Runx2-mediated subnuclear targeting of YAP to repress transcription. EMBO J. 23, 790–799 10.1038/sj.emboj.7600073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong J. H., Hwang E. S., McManus M. T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B. M., Sharp P. A., Hopkins N., Yaffe M. B. (2005) TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science 309, 1074–1078. [DOI] [PubMed] [Google Scholar]
- 25.Hong J. H., Yaffe M. B. (2006) TAZ: a beta-catenin-like molecule that regulates mesenchymal stem cell differentiation. Cell Cycle 5, 176–179 10.4161/cc.5.2.2362 [DOI] [PubMed] [Google Scholar]
- 26.Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- 27.Seo E., Basu-Roy U., Gunaratne P. H., Coarfa C., Lim D. S., Basilico C., Mansukhani A. (2013) SOX2 regulates YAP1 to maintain stemness and determine cell fate in the osteo-adipo lineage. Cell Reports 3, 2075–2087 10.1016/j.celrep.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park H. W., Kim Y. C., Yu B., Moroishi T., Mo J. S., Plouffe S. W., Meng Z., Lin K. C., Yu F. X., Alexander C. M., Wang C. Y., Guan K. L. (2015) Alternative Wnt signaling activates YAP/TAZ. Cell 162, 780–794 10.1016/j.cell.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byun M. R., Hwang J. H., Kim A. R., Kim K. M., Hwang E. S., Yaffe M. B., Hong J. H. (2014) Canonical Wnt signalling activates TAZ through PP1A during osteogenic differentiation. Cell Death Differ. 21, 854–863 10.1038/cdd.2014.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J. Y., Cho S. W., An J. H., Jung J. Y., Kim S. W., Kim S. Y., Kim J. E., Shin C. S. (2013) Osteoblast-targeted overexpression of TAZ increases bone mass in vivo. PLoS One 8, e56585 10.1371/journal.pone.0056585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng Y., Wu A., Li P., Li G., Qin L., Song H., Mak K. K. (2016) Yap1 regulates multiple steps of chondrocyte differentiation during skeletal development and bone repair. Cell Reports 14, 2224–2237 10.1016/j.celrep.2016.02.021 [DOI] [PubMed] [Google Scholar]
- 32.Rodda S. J., McMahon A. P. (2006) Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244 10.1242/dev.02480 [DOI] [PubMed] [Google Scholar]
- 33.McLeod M. J. (1980) Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology 22, 299–301 10.1002/tera.1420220306 [DOI] [PubMed] [Google Scholar]
- 34.Caroti C. M., Ahn H., Salazar H. F., Joseph G., Sankar S. B., Willett N. J., Wood L. B., Taylor W. R., Lyle A. N. (2017) A novel technique for accelerated culture of murine mesenchymal stem cells that allows for sustained multipotency. Sci. Rep. 7, 13334 10.1038/s41598-017-13477-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ducy P., Karsenty G. (1995) Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell. Biol. 15, 1858–1869 10.1128/MCB.15.4.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanas M. R., Ma S., Jadaan F. O., Ng C. K., Weigelt B., Reis-Filho J. S., Rubin B. P. (2016) Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 35, 929–938 10.1038/onc.2015.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider P., Voide R., Stampanoni M., Donahue L. R., Müller R. (2013) The importance of the intracortical canal network for murine bone mechanics. Bone 53, 120–128 10.1016/j.bone.2012.11.024 [DOI] [PubMed] [Google Scholar]
- 38.Akaike H. (1974) A new look at the statistical model identification. IEEE Trans. Automat. Contr. 19, 716–723 10.1109/TAC.1974.1100705 [DOI] [Google Scholar]
- 39.Jepsen K. J., Silva M. J., Vashishth D., Guo X. E., van der Meulen M. C. H. (2015) Establishing biomechanical mechanisms in mouse models: practical guidelines for systematically evaluating phenotypic changes in the diaphyses of long bones. J. Bone Miner. Res. 30, 951–966 10.1002/jbmr.2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kourtis L. C., Carter D. R., Beaupre G. S. (2014) Improving the estimate of the effective elastic modulus derived from three-point bending tests of long bones. Ann. Biomed. Eng. 42, 1773–1780 10.1007/s10439-014-1027-3 [DOI] [PubMed] [Google Scholar]
- 41.Guss J. D., Horsfield M. W., Fontenele F. F., Sandoval T. N., Luna M., Apoorva F., Lima S. F., Bicalho R. C., Singh A., Ley R. E., van der Meulen M. C. H., Goldring S. R., Hernandez C. J. (2017) Alterations to the gut microbiome impair bone strength and tissue material properties. J. Bone Miner. Res. 32, 1343–1353 10.1002/jbmr.3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen X., Nadiarynkh O., Plotnikov S., Campagnola P. J. (2012) Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 7, 654–669 10.1038/nprot.2012.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu-Chittenden Y., Huang B., Shim J. S., Chen Q., Lee S. J., Anders R. A., Liu J. O., Pan D. (2012) Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26, 1300–1305 10.1101/gad.192856.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang Y., Rowe R. G., Botvinick E. L., Kurup A., Putnam A. J., Seiki M., Weaver V. M., Keller E. T., Goldstein S., Dai J., Begun D., Saunders T., Weiss S. J. (2013) MT1-MMP-dependent control of skeletal stem cell commitment via a β1-integrin/YAP/TAZ signaling axis. Dev. Cell 25, 402–416 10.1016/j.devcel.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim K. M., Choi Y. J., Hwang J. H., Kim A. R., Cho H. J., Hwang E. S., Park J. Y., Lee S. H., Hong J. H. (2014) Shear stress induced by an interstitial level of slow flow increases the osteogenic differentiation of mesenchymal stem cells through TAZ activation. PLoS One 9, e92427 10.1371/journal.pone.0092427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim M., Kim T., Johnson R. L., Lim D. S. (2015) Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Reports 11, 270–282 10.1016/j.celrep.2015.03.015 [DOI] [PubMed] [Google Scholar]
- 47.Forlino A., Marini J. C. (2016) Osteogenesis imperfecta. Lancet 387, 1657–1671 10.1016/S0140-6736(15)00728-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rauch F., Travers R., Parfitt A. M., Glorieux F. H. (2000) Static and dynamic bone histomorphometry in children with osteogenesis imperfecta. Bone 26, 581–589 10.1016/S8756-3282(00)00269-6 [DOI] [PubMed] [Google Scholar]
- 49.Khillan J. S., Olsen A. S., Kontusaari S., Sokolov B., Prockop D. J. (1991) Transgenic mice that express a mini-gene version of the human gene for type I procollagen (COL1A1) develop a phenotype resembling a lethal form of osteogenesis imperfecta. J. Biol. Chem. 266, 23373–23379 [PubMed] [Google Scholar]
- 50.Pereira R., Khillan J. S., Helminen H. J., Hume E. L., Prockop D. J. (1993) Transgenic mice expressing a partially deleted gene for type I procollagen (COL1A1): a breeding line with a phenotype of spontaneous fractures and decreased bone collagen and mineral. J. Clin. Invest. 91, 709–716 10.1172/JCI116252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chipman S. D., Sweet H. O., McBride D. J., Jr., Davisson M. T., Marks S. C., Jr., Shuldiner A. R., Wenstrup R. J., Rowe D. W., Shapiro J. R. (1993) Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta. Proc. Natl. Acad. Sci. USA 90, 1701–1705 10.1073/pnas.90.5.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanleene M., Porter A., Guillot P. V., Boyde A., Oyen M., Shefelbine S. (2012) Ultra-structural defects cause low bone matrix stiffness despite high mineralization in osteogenesis imperfecta mice. Bone 50, 1317–1323 10.1016/j.bone.2012.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nadiarnykh O., Plotnikov S., Mohler W. A., Kalajzic I., Redford-Badwal D., Campagnola P. J. (2007) Second harmonic generation imaging microscopy studies of osteogenesis imperfecta. J. Biomed. Opt. 12, 051805 10.1117/1.2799538 [DOI] [PubMed] [Google Scholar]
- 54.Li H., Jiang X., Delaney J., Franceschetti T., Bilic-Curcic I., Kalinovsky J., Lorenzo J. A., Grcevic D., Rowe D. W., Kalajzic I. (2010) Immature osteoblast lineage cells increase osteoclastogenesis in osteogenesis imperfecta murine. Am. J. Pathol. 176, 2405–2413 10.2353/ajpath.2010.090704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaan H. Y. K., Chan S. W., Tan S. K. J., Guo F., Lim C. J., Hong W., Song H. (2017) Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex. Sci. Rep. 7, 2035 10.1038/s41598-017-02219-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen J., Shi Y., Regan J., Karuppaiah K., Ornitz D. M., Long F. (2014) Osx-Cre targets multiple cell types besides osteoblast lineage in postnatal mice. PLoS One 9, e85161 10.1371/journal.pone.0085161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Mishina Y., Liu F. (2015) Osterix-Cre transgene causes craniofacial bone development defect. Calcif. Tissue Int. 96, 129–137 10.1007/s00223-014-9945-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karystinou A., Roelofs A. J., Neve A., Cantatore F. P., Wackerhage H., De Bari C. (2015) Yes-associated protein (YAP) is a negative regulator of chondrogenesis in mesenchymal stem cells. Arthritis Res. Ther. 17, 147 10.1186/s13075-015-0639-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., Zahler A. M., Haussler D. (2002) The human genome browser at UCSC. Genome Res. 12, 996–1006 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richter A. M., Cerruti-Sola S., Sternberg E. D., Dolphin D., Levy J. G. (1990) Biodistribution of tritiated benzoporphyrin derivative (3H-BPD-MA), a new potent photosensitizer, in normal and tumor-bearing mice. J. Photochem. Photobiol. B 5, 231–244 10.1016/1011-1344(90)80008-L [DOI] [PubMed] [Google Scholar]
- 61.Akens M. K., Yee A. J., Wilson B. C., Burch S., Johnson C. L., Lilge L., Bisland S. K. (2007) Photodynamic therapy of vertebral metastases: evaluating tumor-to-neural tissue uptake of BPD-MA and ALA-PpIX in a murine model of metastatic human breast carcinoma. Photochem. Photobiol. 83, 1034–1039 10.1111/j.1751-1097.2007.00172.x [DOI] [PubMed] [Google Scholar]
- 62.Zhang H., Ramakrishnan S. K., Triner D., Centofanti B., Maitra D., Győrffy B., Sebolt-Leopold J. S., Dame M. K., Varani J., Brenner D. E., Fearon E. R., Omary M. B., Shah Y. M. (2015) Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Sci. Signal. 8, ra98 10.1126/scisignal.aac5418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.