Abstract
BACKGROUND
Glyphosate‐resistant goosegrass has recently evolved and is homozygous for the double mutant of EPSPS (T102I, P106S or TIPS). These same mutations combined with EPSPS overexpression, have been used to create transgenic glyphosate‐resistant crops. Arabidopsis thaliana (Wt EPSPS K i ∼ 0.5 μM) was engineered to express a variant AtEPSPS‐T102I, P106A (TIPA K i = 150 μM) to determine the resistance magnitude for a more potent variant EPSPS that might evolve in weeds.
RESULTS
Transgenic A. thaliana plants, homozygous for one, two or four copies of AtEPSPS‐TIPA, had resistance (IC50 values, R/S) as measured by seed production ranging from 4.3‐ to 16‐fold. Plants treated in reproductive stage were male sterile with a range of R/S from 10.1‐ to 40.6‐fold. A significant hormesis (∼ 63% gain in fresh weight) was observed for all genotypes when treated at the initiation of reproductive stage with 0.013 kg ha–1. AtEPSPS‐TIPA enzyme activity was proportional to copy number and correlated with resistance magnitude.
CONCLUSIONS
A. thaliana, as a model weed expressing one copy of AtEPSPS‐TIPA (300‐fold more resistant), had only 4.3‐fold resistance to glyphosate for seed production. Resistance behaved as a single dominant allele. Vegetative tissue resistance was 4.7‐fold greater than reproductive tissue resistance and was linear with gene copy number. © 2017 The Authors. Pest Management Science published by John Wiley & Sons Ltd on behalf of Society of Chemical Industry.
Keywords: mutant EPSPS, glyphosate resistance, Arabidopsis thaliana, gene dosage, hormesis
1. INTRODUCTION
Glyphosate introduced in 1974,1 has become the world's most important herbicide2, 3, 4, 5 since the introduction of glyphosate‐resistant soybeans in 1996.6 As with many herbicides, resistant biotypes of weed species have appeared. Globally, there are now 37 glyphosate‐resistant species, many of which have relatively weak (≤ 2 times) magnitudes of resistance, when fold resistance is compared with the recommended label rate.7 The magnitude of resistance for a weed to a particular herbicide is a key component of the characterization of that resistance because it can impact the subsequent weed control strategy.8 Herbicide resistance can usually be explained by three biochemical mechanisms: target‐site modification, and herbicide metabolism or exclusion. The latter represents active or passive restricted access to the site‐of‐action. Target site resistance has been the most prevalent and biggest concern because, in many cases, it can provide virtual immunity by alteration of the herbicide binding site.
The concept of glyphosate‐resistant crops was made possible with the first reported 5‐enolypyruvylshikimate‐3‐phosphate synthase (EPSPS) mutant resistant to glyphosate9 found in Salmonella typhimurium less than 3 years after the discovery that glyphosate targeted EPSPS.10 The mutation was identified as a target site mutation at Pro106 (using Zea mays mature sequence as a reference). The overexpression of this enzyme in tomato11 resulted in weak glyphosate‐resistant plants.12 However, it was not until a second mutation Thr102Ile (T102I) was added to the Pro106Ser (P106S) that an enzyme significantly more resistant to glyphosate was made.13, 14 This double mutant variant of EPSPS‐TIPS (alias event GA21) with a K i = 50 μM, 100‐fold more resistant than wild‐type,15 was overexpressed constitutively to commercialize glyphosate‐resistant corn in 1998 by Dekalb.16, 17 A distinctly different class of bacterial EPSPSs was discovered and found to be very useful in developing glyphosate‐resistant crops when overexpressed in crop plants18, 19, 20 and the enzyme CP4 is used by the Monsanto Company in all of their glyphosate‐resistant crops.
Evolved EPSPS‐based resistance to glyphosate has now been elaborated to three types: amino acid mutations, gene duplication and gene overexpression.21 Baerson et al.,22 first characterized a P106S in Eleusine indica (goosegrass), and now goosegrass has been first to select the T102I,P106S double mutation.23, 24 Goosegrass has separately been shown to have gene amplification of the native gene.22 Most recently, Mao et al.,25 while studying glyphosate‐resistant lilyturf species (Liriope spicata) identified a unique version of a plant EPSPS that increases the K i for glyphosate, and this EPSPS is overexpressed.25 The resistant lilyturf is both the first case of alternate novel target site mutations contributing to resistance and the first combination with increased expression.
Combinations of point mutation(s) with gene amplification, or overexpression, is the path to full glyphosate resistance. Given the potential importance of double mutants among naturally occurring weed populations our intent was to determine how much glyphosate resistance could be acquired with a double variant EPSPS with native expression of one copy of EPSPS‐T102I, P106A (TIPA) when compared with wild‐type (Wt) Arabidopsis thaliana sp. Columbia. The experiment was complicated because there are two genes for EPSPS in the genome for A. thaliana. Therefore, the goal was to compare the glyphosate susceptibility of Wt At with synthetic glyphosate resistance At lines expressing either At1‐TIPA or At2‐TIPA. Unfortunately, a plant with a single copy of the At1 promoter cassette was not made although a single copy plant of the At2 promoter cassette was found. Therefore, to control for copy number and tissue specific expression, Wt At was compared with three transgenic lines: line 673 with a single copy At2‐TIPA, line 469 with two copies of At1‐TIPA and line 661 with four copies of At2‐TIPA. Glyphosate resistance was measured using the resultant seed production of treated plants.
2. MATERIALS AND METHODS
2.1. Cloning
Arabidopsis thaliana contains two 5‐enolpyruvylshikimate‐3‐phosphate (EPSP) synthases: At1EPSPS (AT1G48860) and At2EPSPS (AT2G45300) located on chromosomes 1 and 2, respectively, both expressing EPSPS with highly similar mature protein primary amino acid sequence. Using the nucleotide sequence information in the NCBI database each of these genes was cloned. The AtEPSPS expression cassettes were defined as the transcribed regions and the adjacent 5′ regulatory regions, and were amplified with primers 1 and 2 (At1EPSPS, 4125 bp) or primers 3 and 4 (At2EPSPS, 3991 bp) and cloned into a pCRTOPO II vector (Invitrogen) (Table 1). Selected clones representing At(1or 2)EPSPS were sequenced to confirm their correctness. Mutations (Thr102Ile and Pro106Ala: TIPA) were introduced to the genes using Quick Change mutagenesis kit (Statagene) using primers 5 and 6 (At1EPSPS) and primers 7 and 8 (At2EPSPS) (Table 1). Selected clones designated At1‐TIPA and At2‐TIPA were digested with NotI and XhoI, and subcloned (for expression in plants) to a TI Agrobacterium tumefaciens transformation binary vector cut with the same restriction enzymes (Fig. 1). The binary vector contained a BAR gene driven by 35S CaMV promoter for the selection of transgenic events.26 The resulting clones At1‐TIPA and At2‐TIPA were transformed to A. thaliana sp. Columbia by the Agrobacterium tumefaciens floral dip method.27
Table 1.
Primers used for cloning and mutagenesis of AtEPSPS expression cassettes (restriction sites are underlined with base changes given in bold)
| Primer | Sequence |
|---|---|
| 1 AtEPSPS1F | 5′‐CAGCGGCCGCTTCAGCTCCATCGAATTTTGGGAGACAA‐3′ |
| 2 AtEPSPS1R | 5′‐AAATCGAGATGATAACTAGAGAGAACCTGAACAAACA‐3′ |
| 3 AtEPSPS2F | 5′‐CAAGCGGCCGCTCCATTTTTGCTTACAAATATGGCACA‐3′ |
| 4 AtEPSPS2R | 5′‐AACTCGAGTCGAACTCAAATGATTGCATCTCAAAACTC‐3′ |
| 5 AtEPSPS1MF | 5′‐TCGGCAATGCAGGAAtcGCAATGCGTgCACTTACCGCCGCAG‐3′ |
| 6 AtEPSPS1MR | 5′‐CTGCGGCGGTAAGTGcACGCATTGCgaTTCCTGCATTGCCGA‐3′ |
| 7 AtEPSPS2MF | 5′‐TCGGTAATGCAGGAAtcGCAATGCGTgCACTTACCGCTGCGG‐3′ |
| 8 AtEPSPS2MR | 5′‐CCGCAGCGGTAAGTGcACGCATTGCgaTTCCTGCATTACCGA‐3′ |
| 9 ZmEPSPSMF | 5'TGGGGAATGCTGGAAtTGCAATGCGGgCATTGACAGCAGCTG 3′ |
| 10 ZmEPSPSMR | 5′CAGCTGCTGTCAATGcCCGCATTGCAaTTCCAGCATTCCCCA 3′ |
Figure 1.
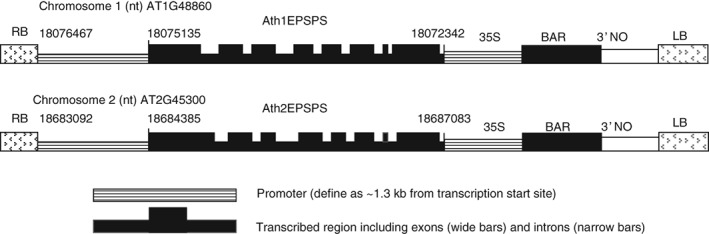
Cartoon of the two cloned AtEPSPSs as a binary vector in the TI plasmid for plant transformation. Nucleotide numbers are found at TAIR or http://www.ncbi.nlm.nih.gov/ .
Mutations (Thr102Ile and Pro106Ala) were introduced to the wild‐type Zea mays EPSPS cDNA (gb|AY106729.1|) using Quick Change mutagenesis kit (Statagene) with primer set (ZmTIPA_F and ZmTIPA_R in Table 1), including a 10× His tag on the N terminus. The wild‐type and Zm‐TIPA EPSPS cDNAs were cloned into pET19b (EMP Millipore) and transformed into Escherichia coli BL21(DE3) for overexpression. The transformants were grown by shaking at 200 rpm at 30 °C in Luria–Bertani medium containing 50 mg ml–1 ampicillin. Reagents were acquired from Sigma‐Aldrich unless specified. When optical density at 600 nm reached 0.6–0.8, isopropyl β‐d‐thiogalactoside (IPTG) was added to a final concentration of 0.75 mM. Cells were harvested by centrifugation at 6000 g for 20 min after a 2 h induction by IPTG. The cells were lysed by sonication and centrifuged at 15 000 g for 20 min. The supernatants containing soluble proteins were purified on a nickel–nitrilotriacetic acid (Ni‐NTA; Qiagen) affinity column and eluted with 250 mM imidazole in 25 mM Tris–HCl pH 8, 0.2 M NaCl, 10% glycerol. The concentration of the protein was analyzed by Bradford method (Bio‐Rad), and then stored at –80 °C.
2.2. Transformation
Arabidopsis thaliana seed embryos were transformed by an Agrobacterium tumefaciens mediated method as described previously.28 Agrobacterium tumefaciens strain ABI containing the DNA construct was prepared as inoculum by growing a culture tube containing 10 ml Luria broth with 1 ml L–1 each of spectinomycin (100 mg ml–1), chloramphenicol (25 mg ml–1), kanamycin (50 mg ml–1), with shaking at 28 °C for 16–20 h. The Agrobacterium tumefaciens inoculum was pelleted at 2700 g and resuspended in 25 ml infiltration medium (MS Basal Salts 0.5%, Gamborg's B‐5 vitamins 1%, sucrose 5%, MES 0.5 g L–1, pH 5.7) 0.05% Silwet L‐77 to an OD600 of 0.8. Mature flowering A. thaliana plants (T0) with open flower buds removed, were inoculated by inverting the pots containing the plants into the inoculum. The vegetation was swirled for 10–20 s and then the pots were placed horizontally to drain, then covered with plastic domes and placed in a growth chamber set to 21 °C, 16 h light and 70% RH. About 2 weeks after inoculation, each plant was covered with a Lawson 511 pollination bag. Approximately 4 weeks post infiltration, watering was stopped to permit dry down and seed harvest. The transgenic A. thaliana plants (T1) produced from the infiltrated seed embryos were selected from the non‐transgenic plants by germination on bialaphos agar. First the seed was surface sterilized using chlorine gas by placing ∼0.4 ml volume of seed in a 15 ml conical tube in a vacuum desiccator containing a beaker of Clorox Ultra (6.15% sodium hypochlorite) with 4 ml of concentrated HCl, a slight vacuum is pulled and incubated at room temperature 16 h. The seeds were then spread onto the surface of selection media plates containing MS Basal Salts 4.3 g L–1, Gamborg's B‐5 (500×) 2.0 g L–1, sucrose 10 g L–1, MES 0.5 g L–1, and 8 g L–1 Phytagar with carbenicillin 250 mg L–1, cefotaxime 100 mg L–1, and bialaphos 10 mg L–1. The plates were vernalized by incubating in the dark at 4 °C for 2–4 days before plates were transferred to a growth chamber with cool white light bulbs with ∼150 μE at a 16: 8 h light/dark cycle and 23 °C. Green plants were visible after 5–10 days at 23 °C. When the bialaphos‐resistant T1 plants had at least one set of true leaves, seedlings were transferred individually as a unique transgenic event to soil, covered with a germination dome, and moved to a growth chamber set for 16 h day length at 130–180 μE at 23 °C light and 21 °C dark with 70% RH and kept covered until new growth was apparent. Seed (T2) was harvested by hand threshing the dry inflorescence and passing the seed through a sieve twice to remove any extra debris or petals. The T1 plant leaf tissue was assayed by Southern blot using a 5′‐end and 3′ untranslated region probes for the AtEPSPS coding region. These T1 plants were also sprayed with a titration of glyphosate (Roundup UltraMAX®, from Monsanto) to insure they were glyphosate resistant. The T2 seed from minimum copy plants was selected for further analysis. The T2 seed were planted in 2.5‐inch square Kord pots in Metro Mix 200 soil‐less media with sub‐irrigation twice a week; once with plain water and once with Peter's 20–20–20 fertilizer at a rate of 800–1000 μS cm–1 in the chamber described above. These T2 plants, were assayed by Taqman29 procedure for BAR copy number and using the respective segregation ratios single locus minimum copy plants were selected for seed production as described above (T3 seed). The T3 generation seeds were grown out again as before and a second Southern analysis was undertaken to confirm that each event had a defined copy number and was single locus and these plants were used to produce the T4 seed that was then used in this study and grown as described.
2.3. EPSPS extraction and assay
The assay used in these studies was adapted from the work of Webb30 and is detailed in a compilation of herbicide target site assays by Dayan et al.31 where the standard extraction procedure is also described. The shikimate 3‐phosphate (S3P) was produced at Monsanto by the procedure described by Castellino et al.,32 which briefly involves using cloned E. coli shikimate kinase to phosphorylate shikimate with ATP [regenerated from ADP with phosphoenolpyruvate (PEP) using pyruvate kinase] to make S3P. The S3P was then purified by anion‐exchange chromatography using a gradient of the volatile buffer triethylammonium bicarbonate. An improved EPSPS assay that has cheaper reagents, is more durable and more sensitive, is now available by adaption of the phosphate assay described by Vazquez et al.33
2.4. Glyphosate resistance
A track‐sprayer fitted with a 9501 Evenflow nozzle was set to a height of 45 cm above the plants and air pressure and track speed adjusted to apply 187 L ha–1. Glyphosate concentrations were varied while maintaining the amount of surfactant constant at the level found in 0.840 kg ha–1 formulation of Roundup UltraMAX®. Fresh weight data were taken at 16 DAT (days after treatment). When determining growth inhibition in the reproductive stage treatment, inflorescences were cut at their base, weighed for fresh weight and dried separately in paper bags. Leaf rosettes were cut from their roots immediately below the oldest leaf and fresh weights collected before drying. These plants were primarily sterile (incomplete silique development) and so no seed weights were obtained. Tissues were dried at 50 °C for at least 2 days and weighed on an analytical balance. Three accessions of each transgenic line were chosen for the resistance determination at each treatment. Line 673 comprised: At_S53673‐37, At_S53673‐38, and At_S53673‐39, all confirmed to be homozygous with one copy of At2‐TIPA and BAR at one locus. Line 469 comprised: At_S53469‐02, At_S53469‐16, and At_S53469‐26, determined to contain two intact copies of At1‐TIPA and BAR at one locus with no fragments. Line 661 comprised: At_S53661‐07, At_S53661‐16, and At_S53661‐30, all determined to have four intact copies of At2‐TIPA with BAR and appeared homozygous with no fragments. A homogeneous sized set of plants was selected to provide three replicates per accession so that nine plants total were used in each treatment. Roundup UltraMAX® was applied at 0.013, 0.026, 0.053, 0.105, 0.210, 0.420, 0.840, 1.260, 1.680 and 3.360 kg ha–1 to plants that had bolts 6–10 cm with no open flowers (reproductive stage treatment). The resistance test used to determine survivor seed production was set up identically except glyphosate treatments were 0.0, 0.013, 0.026, 0.053, 0.105, and 0.210 kg ha–1 and were applied just before bolting (∼ 4–5 days earlier). Surviving plants grew until plants senesced naturally. Seed retainer cups (with bases to catch seed) were attached to each plant and fitted with Aracon tubes (Lehle Seeds). The individual IC50 values in both experiments for these three accessions were not statistically different for each of their three lines and so the results for the nine plants were pooled for each treatment. The 50% effect dose was calculated using the four parameter robust Log‐logistic fit34 in Grafit 4.021 from Erithacus Software where y = a/(1 + exp(s * ln(x/I))) + C. The four parameters are a = (1.05*y maximum – 0.95*y minimum), s = slope, I = IC50 and C = (0.95*y minimum). Standard errors of the mean (SE) are used throughout and are not pooled across treatments.
3. RESULTS
3.1. Cloning and molecular analysis
A combination of Southern and Taqman analysis selected three events as low copy number single locus insertions. The glyphosate titrations revealed that one accession in line 469 (two copies) and all three accessions in line 661 (four copies) were still segregating, indicating that they were not yet homozygous. Figure S1 shows the single copy null and an apparent heterozygous plant found in accession At_S53469‐16 treated at the 0.42 kg ha–1 rate, similar results were observed for the At_S53661 accessions in the glyphosate rate range of 0.104–0.42 kg ha–1. Nulls were easily identified due to the significantly more sensitive phenotype of Wt plants and could be scored early. The three At_S53661 accessions had a total of two nulls below the 1.64 kg ha–1 treatment (2 of 81 plants). A very few putative heterozygous plants, like the one in Fig. S1 could be excluded as outliers, but otherwise all plants were included. This results in slightly more error at the high rates for the one At_S53469‐16 accession and all three At_S53661 accessions because not all the treated plants were pure two copy or four copy plants respectively and consequently reducing the magnitude of the measured resistance slightly due to the few contaminating plants.
3.2. EPSPS activity
Variants of an enzyme with K i values >100‐fold different are easily distinguished when admixed and challenged with an inhibitor. This is necessary when evaluating extracts of the transgenics for EPSPS activity and to characterize the transgenic levels of TIPA. Figure 2 demonstrates this separation of EPSPSs with the IC50 values measured for Wt and TIPA versions of ZmEPSPS. The Wt EPSPS is fully inhibited by ∼1 mM glyphosate, whereas the TIPA enzyme is still unaffected. Three admixtures based on catalytic activity (not specific activity or molar ratio) of 1: 1 (Wt: TIPA), 2: 1 and 9: 1 are shown in Fig. 2 and illustrate that these mixtures of the two enzymes can be discerned across the full range of glyphosate concentrations from sub‐μM to 30 mM. Figure 3 demonstrates that Wt and TIPA are discernable in extracts of the transgenic At. The 673 one‐copy line however, does not reflect the expected 2: 1 ratio for two homozygous Wt EPSPS native genes to one homozygous transgenic TIPA. The reason for this is the TIPA enzyme like the TIPS, recently kinetically characterized in goosegrass,23 has a fitness penalty for V max which decreases the unit activity per mole of enzyme. This unfit enzyme serves as a bottleneck in the shikimate pathway which results in the goosegrass homozygous TIPS plants being vegetatively smaller. The glyphosate titration in Fig. 3 shows ∼10% TIPA activity at 5 mM glyphosate, which would suggest an ∼80% V max penalty. This is confirmed in the extracts of the 469 two‐copy line and the 661 four‐copy line. The geometric increase in EPSPS activity levels at 5 mM glyphosate correlate with copy number and also confirm the relative expression ratios expected based on the native level of expression of the cloned At (1 and 2) promoters, with no obvious transgene insertion effects. Finally, this also makes it clear that target site mutations that generate glyphosate resistance for this series of double mutants (TIPX) would have a decreased V max.
Figure 2.
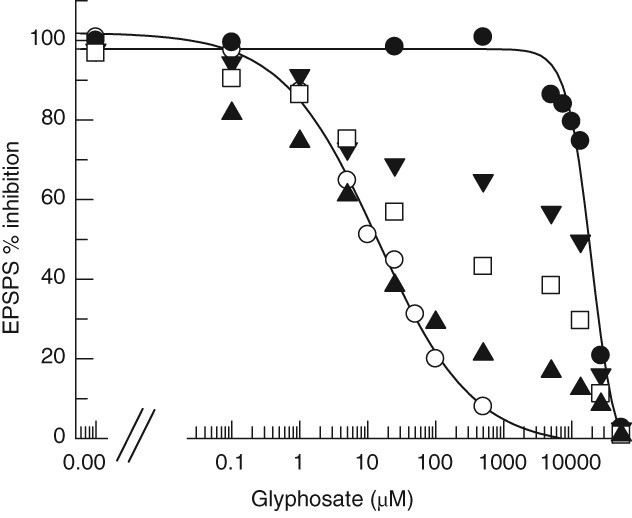
EPSPS enzyme activity for ZmEPSPS and ZmTIPA enzymes either alone or in admixtures of 1: 1, 2: 1 or 9: 1 (Wt: TIPA) where the mixtures were based only on enzyme activity. The wild‐type ZmEPSPS ( ) in this assay has an IC50 (50% inhibitory concentration) of 13.9 ± 3.5 μM. The TIPA ZmEPSPS (
) in this assay has an IC50 (50% inhibitory concentration) of 13.9 ± 3.5 μM. The TIPA ZmEPSPS ( ) has an IC50 of 19.5 ± 2.8 mM. Three mixtures of Wt: TIPA were composed in ratios of 1:1 (
) has an IC50 of 19.5 ± 2.8 mM. Three mixtures of Wt: TIPA were composed in ratios of 1:1 ( ), 2:1(
), 2:1( ) and 9:1 (
) and 9:1 ( ). All rates are averages of two kinetic runs.
). All rates are averages of two kinetic runs.
Figure 3.
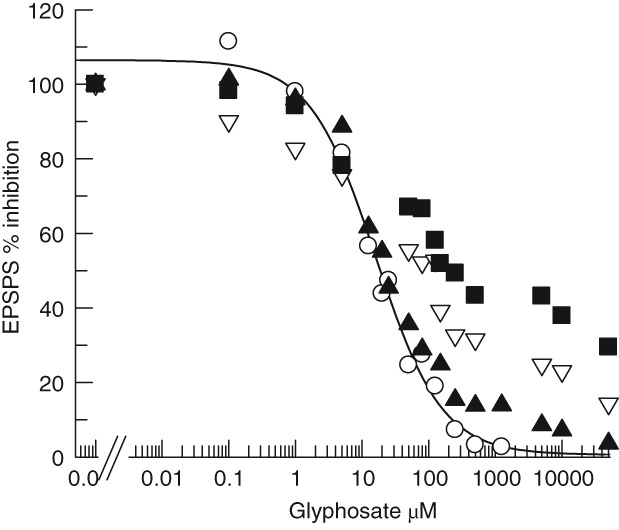
The wild‐type At EPSPS ( ) in this assay has an IC50 of 16.1 ± 2.6 μM. The enzyme activity for EPSPS in the crude extracts of the three TIPA expressing plants for one, two and four copies were At 673 (
) in this assay has an IC50 of 16.1 ± 2.6 μM. The enzyme activity for EPSPS in the crude extracts of the three TIPA expressing plants for one, two and four copies were At 673 ( ), At 469(
), At 469( ), and At 661 (
), and At 661 ( ) respectively.
) respectively.
3.3. Glyphosate resistance
The magnitudes of glyphosate resistance for the three transgenics compared with each other and Wt can only be compared within each experiment. The vegetative stage treatment (Fig. 4) was carried out to seed and is summarized in Table 2 with comparisons of IC50 values. The resistance factors for seed production, R/S, are 4.3 for the one‐copy 673 line, 11.1 for the two‐copy line and 16 for the four‐copy line.
Figure 4.
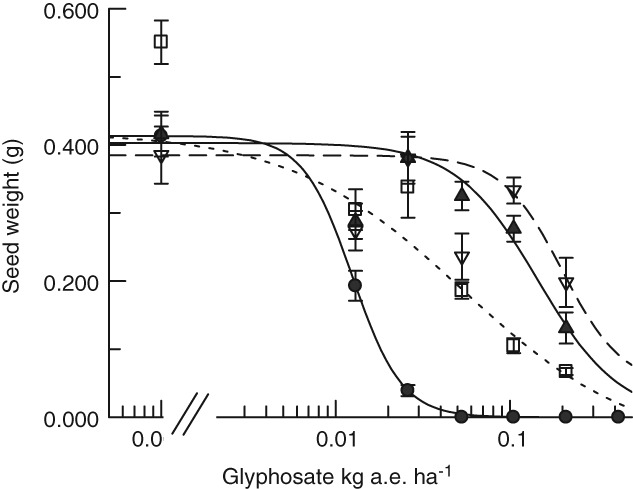
A comparison of seed weight produced in A. thaliana transformed with TIPA after surviving glyphosate treatment applied during the vegetative stage. ( ) Wild‐type At; (
) Wild‐type At; ( ) Dotted line, 673 with one copy of At2TIPA; (
) Dotted line, 673 with one copy of At2TIPA; ( ) Solid line, 469 with two copies of At1TIPA; (
) Solid line, 469 with two copies of At1TIPA; ( ) Dashed line, 661 with four copies of At2TIPA. Error bars are ± standard error (SE) of the mean.
) Dashed line, 661 with four copies of At2TIPA. Error bars are ± standard error (SE) of the mean.
Table 2.
Comparing IC50 values for growth inhibition during reproductive stage treatment (inflorescence and leaf) and vegetative stage treatment (seed) with glyphosate kg ha–1 with the respective R/S resistance magnitudes
| At Line, copy no. | Reproductive stage | Vegetative stage | |||||
|---|---|---|---|---|---|---|---|
| IC50 inflorescence | R/S | IC50 leaf | R/S |
|
IC50 seed | R/S | |
| Wt | 0.014 ± 0.002 | 0.08 ± 0.03 | 5.7 | 0.0125 ± 0.00001 | |||
| 673, 1 | 0.117 ± 0.017 | 8.4 | 0.61 ± 0.12 | 7.6 | 5.2 | 0.054 ± 0.0003 | 4.3 |
| 469, 2 | 0.279 ± 0.036 | 20 | 1.18 ± 0.22 | 14.8 | 4.2 | 0.144 ± 0.013 | 11.1 |
| 661, 4 | 0.524 ± 0.186 | 37 | 1.91 ± 0.35 | 23.9 | 3.6 | 0.200 ± 0.028 | 16 |
The reproductive stage treatment was evaluated as fresh weight for foliar plant growth reduction at 16 DAT and failed to produce seed upon threshing having sterile siliques shown in Fig. S2. The whole plants are depicted in Fig. 5 where sterility is labeled with the individual accession and slight seed production noted for Fig. 5(C,D). The data for foliage are broken down to leaves and inflorescence in Table 2 and the total foliar fresh weight is plotted in Fig. 6. The IC50 values from Table 2 are used in Fig. 7 to depict the linearity of the response proportional to the copy number of the three variant At2‐TIPA lines. The whole‐plant weights showed resistance levels for line 673 with one copy of At2EPSPS‐TIPA R/S = 10.1; for line 469 with two copies of At1EPSPS‐TIPA R/S = 24.6 and for line 661 expressing four copies of At2EPSPS‐TIPA R/S = 40.6. Glyphosate resistance in these sterile plants was on average 4.7‐fold weaker in the inflorescence than the corresponding leaf tissue (Table 2).
Figure 5.
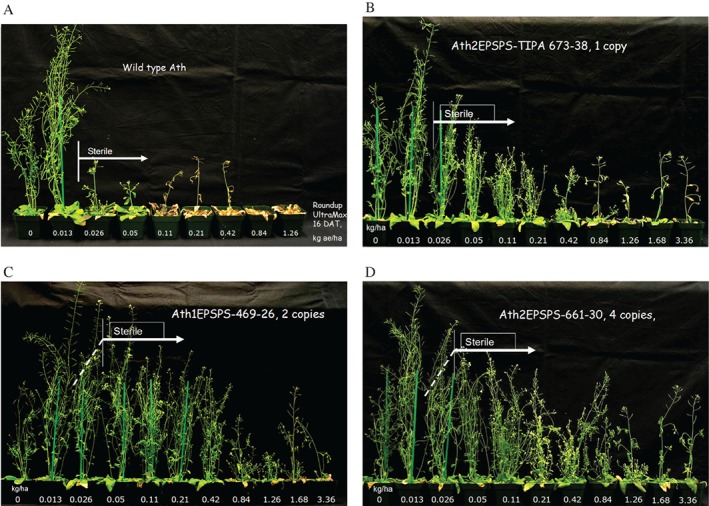
Growth reduction at 16 DAT of Arabidopsis. (A) Wild‐type At; (B) line 673, with one copy of At2TIPA; (C) line 469 with two copies of At1TIPA; (D) line 661 with four copies of At2‐TIPA. Sterile seed production is labeled and dashed line indicates some few seeds were observed.
Figure 6.
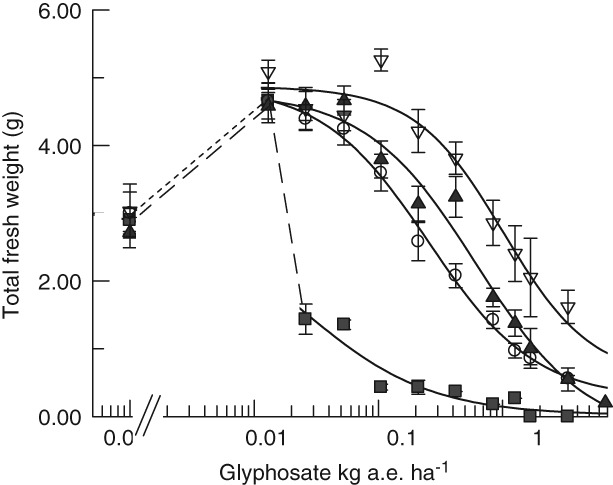
Comparison of growth inhibition by fresh weight in Arabidopsis transformed with TIPA by glyphosate applied in the reproductive stage. Weights include the vegetative portion of the plant for; ( ) wild‐type At; (
) wild‐type At; ( ) 673 with one copy of At2‐TIPA; (
) 673 with one copy of At2‐TIPA; ( ) 469 with two copies of At1‐TIPA; (
) 469 with two copies of At1‐TIPA; ( ) 661 with four copies of At2‐TIPA. Due to hormesis the TIPA plants were fit without the zero‐glyphosate rate and are connected instead with a dotted line. The Wt was fit without the 0.013 kg ha–1 rate and this point is connected by a dashed line. Error bars are ± standard error (SE) of the mean.
) 661 with four copies of At2‐TIPA. Due to hormesis the TIPA plants were fit without the zero‐glyphosate rate and are connected instead with a dotted line. The Wt was fit without the 0.013 kg ha–1 rate and this point is connected by a dashed line. Error bars are ± standard error (SE) of the mean.
Figure 7.
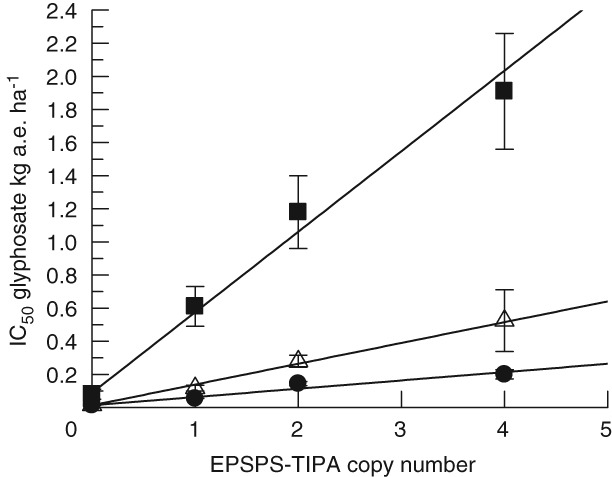
IC50 values (Table 2) for glyphosate vs. TIPA gene copy number are plotted for correlation. Reproductive stage treatment with fresh weights for the foliar portion of the plant broken out into leaves ( ), with linear fit, m = 0.487, r
2 = 0.996, chi2 = 0.023 and inflorescence (
), with linear fit, m = 0.487, r
2 = 0.996, chi2 = 0.023 and inflorescence ( ), with linear fit, m = 0.257, r
2 = 0.995, chi2 = 0.014. The vegetative stage treatment for seed production (
), with linear fit, m = 0.257, r
2 = 0.995, chi2 = 0.014. The vegetative stage treatment for seed production ( ), with linear fit, m = 0.050, r
2 = 0.983, chi2 = 0.009. Error bars are ± standard error (SE) of the mean.
), with linear fit, m = 0.050, r
2 = 0.983, chi2 = 0.009. Error bars are ± standard error (SE) of the mean.
3.4. Hormesis
Arabidopsis thaliana Wt did show a hormesis (growth promotion) at the lowest glyphosate dose, 0.013 kg ha–1. The growth promotion was not durable in the Wt with the foliar plant weights dropping precipitously at the next highest rate 0.026 kg ha–1 (Figs 5A and 6). The TIPA transgenics also displayed the same quantitative hormesis at the lowest rate, but their hormesis persisted relative to increasing glyphosate dose (Figs 5B–D and 6). Transgenics and Wt had very similar total fresh weight for the untreated control 2.98 ± 0.39 g, reflecting no fitness penalty caused by TIPA in normal growth as expected with the underlying native EPSPS. All four At lines increased ∼63 ± 4 % in fresh weight at the 0.013 kg ha–1 treatment (Fig. 6). Hormesis occurred when the glyphosate treatment was timed just after bolting initiated (reproductive stage of development) and was persistent in the EPSPS‐TIPA expressing lines with increasing glyphosate (Figs 5B–D and 6). The hormesis effect was not present in any of the lines when the glyphosate treatment was timed just prior (3–5 days) to bolting (vegetative stage treatment and fertile). The vegetative stage treated plants were more sensitive than the reproductive stage treated plants, and were stunted with increasing dose and produced less seed as a function of increasing dose (Fig. 4).
The fresh weight data for the plants in Fig. 6 were also evaluated for IC50 values using hormesis‐corrected equations,35, 36 which produced statistically similar results to the more common log‐logistic four‐parameter fit presented here and so are not reported. However, the non‐linear curve fits for the transgenic lines exclude the untreated control weights due to the large hormesis (dotted line extension in Fig. 6, represents all three transgenics). The Wt non‐linear curve fit conversely, excludes the plant weights at the lowest rate (0.013 kg ha–1) due to the discontinuity caused by the hormesis (dashed connecting line in Fig. 6).
4. DISCUSSION
4.1. Glyphosate resistance regarding K i
Resistance by target site modification is the most problematic resistance mechanism because it can result in high levels of resistance or even immunity with no antidote. Target site resistance is made much simpler when the herbicide binding site is not substantially overlapping with the enzyme active site37 because the non‐active site amino acids are often less critical to function, and so are more easily substituted and thereby change the inhibitor binding affinity.8 The targeted enzyme must have a modified herbicide binding site without disturbing its functional activity to accomplish resistance. This seems to be easy for several herbicide‐targeted enzymes, e.g., acetolactate synthase38 and acetyl CoA carboxylase,39 although there are reports of Trp574Leu and Ala205Val in acetolactate synthase correlating with a fitness penalty.40, 41 When the herbicide is a transition state inhibitor, a molecule that fits precisely in that fleeting conformation at the point of chemical conversion by the enzyme, modifying amino acids becomes very difficult without ruining the catalytic efficiency of the reaction. Glyphosate is such a transition state inhibitor10, 42 and, as expected, active site mutations are either ineffectual or catalytically ruinous.43 EPSPS has been studied extensively by many researchers and the most prevalent point mutation is the P106 position with substitutions of Ser,22 Thr,44 Ala45 and Leu.46 This is consistent with the published overexpression of the EPSPS‐P106S in tobacco producing plants barely surviving 1.3 kg ha–1.11 It is difficult to assign the glyphosate resistance in goosegrass22, 44 and ryegrass47 entirely to their respective P106 variants, even if they are expressed at slightly higher levels. Nevertheless, the plants do survive low levels of glyphosate and do produce seed suggesting there is more to the explanation of their resistances.
Consider Z. mays EPSPS with a glyphosate K i of 0.5 μM as the starting point for measuring potential resistance levels.15 ' 21 Substitutions at Pro106 have glyphosate K i values <10 μM, and mutations at T102 can provide a significant improvement in glyphosate K i when combined with Pro106.15 The T102I mutation alone is catalytically devastating due to the dramatic decrease in selectivity for PEP (K m = 233 μM), but also induces a larger decrease in glyphosate affinity (K i = 148.6 μM).42 The very poor K m value for PEP may explain why this singular mutation has not been found in nature to date. The two mutations, T102I and P106S, have a combinatorial effect that restores the PEP selectivity (K m = 10.6 μM) while retaining some of the decreased affinity for glyphosate (K i = 58 μM). The double variant TIPS can serve to provide glyphosate resistance in a commercial crop.16, 17 The expression level of TIPS is critical as illustrated in one of the early transgenic tobacco field tests in which a histone promoter construct of TIPS failed to produce substantially glyphosate‐resistant plants because whole‐plant constitutive expression was not achieved.48 CaJacob et al.49 defined two key elements for crop design, a catalytically efficient enzyme that is insensitive to glyphosate and a robust constitutive promoter. Glyphosate‐resistant weeds do not require the resistance level expected in commercial crops to be competitive, but they do need to survive and, more importantly, set seed for selection to occur. The discovery of a TIPS mutation in goosegrass confirms that a catalytically inefficient enzyme can contribute to a high level of glyphosate resistance at native levels of expression.23
The Zm‐TIPA has a glyphosate K i of 150 μM, three times higher than Zm‐TIPS, and seemed a good candidate for evaluating native levels of expression to determine the potential resistant levels when incorporated into a species with a very high degree of sensitivity to glyphosate, i.e., no apparent other resistance mechanisms present. Seed production is the ultimate measure of resistance and in these lines of transgenic A. thaliana expressing one to four copies of TIPA, resistance is relatively weak ranging from 4.3 R/S with one copy of TIPA to 16 R/S with four copies (Table 2). Even though the TIPA variant has a K i 300‐fold more resistant to glyphosate than Wt. These results demonstrate much less glyphosate resistance than expected for such a glyphosate‐resistant enzyme. The idea that glyphosate resistance magnitude would be directly and simply related to K i is not supported in this model system. If this TIPA, with a K i 300‐fold more resistant to glyphosate, provides only approximately fourfold reproductive resistance, it is not clear how the TIPS, only 100 times better than Wt reported in goosegrass, could have such high levels of resistance without an auxiliary mechanism.
Second mechanisms can improve resistance levels substantially. For example, addition of the P106S to vacuole sequestration in ryegrass greatly increases the resistance due to either alone.50, 51 The results here prove that more TIPA is better with the R/S values increasing proportionately to copy number. The linearity of the resistance (IC50 values) correlated to TIPA copy number for seeds, leaves and inflorescence is shown in Fig. 7. The total amount of TIPA is an important factor in the magnitude of glyphosate resistance and conforms to learning in the engineering of glyphosate‐resistant crops.52
4.2. Reproductive penalty
The sensitivity to seed production is dramatically illustrated in the reproductive stage treatment, and this is a hallmark of EPSPS‐only resistance mechanisms. Arabidopsis thaliana is self‐pollinating and if pollen development is blocked, no seed are produced. The application of glyphosate at the beginning of inflorescence development successfully blocked all future pollen development even though the inflorescence itself was only modestly stunted with dose. The vegetative development stage application also impacted seed development, but in proportion to plant stunting and inflorescence stunting as a function of dose. Reproductive resistance has been a major hurdle in the production of glyphosate‐resistant crops.49 Apparently, pollen production has a critical requirement for the shikimate pathway. Several researchers have noticed the effects on pollen and breeding strategies have been implemented relying on the gamete selection possible.53 The idea that different plant tissues could be more or less sensitive to glyphosate (actually well known to those involved in glyphosate drift claims) was first quantified by Feng et al.,54 who showed in velvetleaf that apical meristems were 10× more sensitive than the lower stem or mature source leaves. The concept that pollen mother cells are more sensitive to glyphosate at a particular early stage of development was detailed by Chen et al. for commercial glyphosate‐resistant cotton.55 A study by Ye et al.56 focused on plastid transformation in tobacco and found that tissue sensitivity to glyphosate seemed to correlate with plastid number and function. They reported that reproductive tissues were still more sensitive to glyphosate even with plastid overexpression of the very resistant CP4 EPSPS. This concept of variable tissue sensitivities creates an explanation for the high degree of sterility relative to plant development stage. That is, if mature leaf tissue is less sensitive to glyphosate (being the major entry point), and if phloem function is competent, then that glyphosate can be translocated to and collect in the more sensitive sink tissues as they develop. The IC50 results summarized in Table 2 and Fig. 7 for the reproductive stage treatment, demonstrate that leaves are on average 4.7‐fold more resistant than the inflorescence for the transgenic plants. An average sensitivity difference R/S of 4.3‐fold was still present for the transgenics in the vegetative stage treatment when the IC50 for seed production is compared with its dry inflorescence (not reported). Overall, this differential in resistance between leaves and inflorescence creates the reproductive penalty that comes with insufficient glyphosate resistance. We can make comparisons of the evolved TIPS found in goosegrass to this AtEPSPS‐TIPA system for seed production. The native level of TIPS in goosegrass, as an unfit enzyme (decreased V max), can not maintain the flux in the pathway for normal growth and therefore reveals a vegetative fitness penalty. The resistant goosegrass still makes seed after being treated with glyphosate at field use rates, or they could not have been selected. In this AtEPSPS‐TIPA model system, the V max penalty can be compensated for by Wt EPSPS in the absence of glyphosate. However, in the presence of glyphosate, the Wt enzyme is shut down by 0.11 kg ha–1 (because the Wt plants are dead, Fig. 5A) making the transgenics dependent only on TIPA as the glyphosate increases. The one‐copy TIPA plants (treated on the vegetative schedule) produced no seed >0.2 kg ha–1 (Fig. 4). Therefore, the reproductive penalty from insufficient resistance prevents seed production at field rates when the mechanism is reliant only on mutant EPSPS. Hence, we hypothesize an alternate supporting mechanism to allow seed production in the highly resistant goosegrass because the TIPS plants should otherwise be sterile as a comparison with TIPA in this model system under glyphosate challenge.
4.3. Correlating resistance magnitude to gene dosage
The comparison of IC50 values for these transgenics with one, two and four copies of EPSPS‐TIPA reveals a linear correlation of resistance to gene dosage (Fig. 7). It also creates the opportunity to examine the potential for differential expression by the two different promoters and introns, At1EPSPS and At2EPSPS for EPSPS. The design of this experiment used both promoters separately to express the double variant TIPA, primarily because of the concern that there might be differential tissue specific expression. No single‐copy At1EPSPS plants were produced preventing direct comparison with a single copy of the At2EPSPS gene. Hence, a correlation with copy number was incorporated in these experiments using a one‐copy At2TIPA (line 673), a two‐copy At1TIPA (line 469) with a third event with four copies of At2TIPA (line 661) to help resolve the issue of different promoters. The expression level of the At2 promoter seemed to be functioning at the native level overall given the problems with quantifying one equivalent of EPSPS. Enzyme‐linked immunosorbent assay (ELISA) data (not presented) for the one‐copy line support native levels of expression also confirmed in the EPSPS assay. The protein extracts of the At lines evaluated for EPSPS catalytic activity (Fig. 3) demonstrate the proportionality of one‐, two‐ and four‐copy expression. Further, there was no inconsistency in the reproductive penalty or stunting data to suggest that tissue‐specific expression was somehow different for the two promoters. A plot of the IC50 values versus gene copy in Fig. 7 shows a linear gene–dose relationship to glyphosate resistance in both experiments for inflorescence, leaves and seed. The interpolation of the At1 promoter driven cassette to make the linear correlation plotted in Fig. 7 supports the idea the At1 promoter is performing similarly to At2 regarding gene‐dosage and tissue expression. The linear dose relationship for the one to four copies of the At promoter‐driven constructs is consistent with the simplest interpretation of more resistance with more TIPA. Therefore, the linear response of these three model EPSPS‐TIPA plants for glyphosate resistance is consistent with both promoters performing equivalently and unaffected by their genomic insertion sites. The impact of an increase in expression is demonstrated now in several examples. First, in the work for transgenics described by CaJacob et al.,49 and second, in weeds by the discovery of EPSPS gene duplication by Gaines et al.57 Gene duplication is now found in a number of glyphosate‐resistant species21 with more species appearing.58, 59, 60, 61 This creates an overwhelming notion that gene duplication is sufficient for effective glyphosate resistance and is relatively easily evolved.
TIPS in goosegrass with a very high resistance level was selected and does make seed, but demonstrates the fitness penalty of the decreased V max for the enzyme. TIPS probably has another mechanism that multiplies the magnitude of resistance, particularly since there is not a complete reproductive penalty. These supporting mechanisms could be very difficult to single out, but several mechanisms might be possible. For example, investigations by the Duke lab62 where the alternate aromatic pathway proposed by Haslam63, 64 might be functioning to siphon off shikimic acid through quinic acid as the pathway is inhibited by glyphosate. They showed that protocatechuic, gallic and 4‐hydroxybenzoic acids were higher in four of five species when pretreated with glyphosate. This then proposes that species able to divert shikimic acid to other metabolites, possibly directed to other secondary metabolite compounds derived from these hydroxybenzoates, may be able to partially avoid the toxic consequences of EPSPS inhibition occurring with the rapid shikimate build up. The rapid accumulation of shikimic acid was first recognized by Amrhein's lab65 where they demonstrated that S3P actually accumulates first, as required, and then shikimate is formed and stored in the vacuole.66 The ability to export shikimate from the chloroplast before the chloroplast is damaged could be important. Other situations might be envisaged, like limiting glyphosate entry to the chloroplast and thereby reducing the effective glyphosate concentration allowing the variant EPSPSs to function. This could be the most efficient mechanism and would enable overexpression of EPSPS by gene duplication since the native enzyme would be expected to provide very little resistance.
4.4. Hormesis
The unexpected observation of hormesis with glyphosate treatment when the A. thaliana was just bolting (reproductive stage) confirms the observations of Velini et al. and others that hormesis can occur with glyphosate.67, 68 Hormesis has only been reported once previously for A. thaliana where (±)‐catechin (produced from spotted knapweed, Centaurea maculosa) causes a > 100 % growth enhancement for plants grown in agar plates.69 Catechin is a flavin‐3‐ol which derives from dihydroquercetin a well‐known secondary metabolite of the phenylpropanoid pathway.70 It is curious that catechin should be related to the pathway inhibited by glyphosate.
Hormesis was observed in this study when the plants were treated just after bolting (reproductive stage) and not in the just before bolting (vegetative stage) treatment, which suggests that a particular developmental stage is required to see the hormesis, unlike the previous case for catechin.69 The magnitude of the hormesis was a 63 ± 4% (SE) fresh weight gain. The total fresh weight is comprised of 32.6 ± 1.3% leaf and 67.4 ± 1.3% inflorescence in the untreated plants. The total fresh weight gain due to hormesis in the 0.013 kg ha–1 treated plants was 26.3 ± 1.2% leaf and 73.7 ± 1.2% inflorescence. It is not clear whether this slight over representation in weight gain by the inflorescence is related to the differential IC50 values for the leaves and inflorescence where the inflorescence is on average 4.7 times more sensitive to glyphosate (Table 2). Hormesis in the Wt plants is not retained with increasing glyphosate dose. Surprisingly, the hormesis titration is evident in the transgenics even as glyphosate inhibition of growth is observed at higher doses and is persistent throughout the titration. Velini's team did not observe glyphosate‐induced hormesis in glyphosate‐resistant crops and so it is not clear if the non‐toxicity of glyphosate precludes hormesis development or whether the dose and stage of development opportunity was not identified by them. If non‐toxicity precludes hormesis then a decrease in hormetic effect would be observed with increasing glyphosate resistance, but this does not seem to be the case. Further, the fact that both wild‐type and transgenic plants had the same amount of hormesis at 0.013 kg ha–1 suggests that the plants perceive glyphosate similarly at that dose with no toxicity involved in a growth reduction. In any case, these novel observations need more scrutiny.
5. CONCLUSIONS
This is the first report of a defined whole‐plant model system designed to measure the magnitude of resistance possible from a target‐site mutation where a variant enzyme is re‐inserted with the native expression element. TIPA, which is 300 times more resistant then the native EPSPS, was expressed with each of the two promoters known for EPSPS in A. thaliana. This very glyphosate‐insensitive enzyme provides only an R/S of 4.3‐fold as measured by seed production. The three model plants employed each expressed one, two or four copies of TIPA, and a linear correlation with glyphosate resistance was obtained with increasing gene copy number producing a maximum value for R/S of 16 for four copies, but the IC50 value of 0.2 kg ha–1 is still well below the minimum requirement for crop tolerance of at least two times the label rate. Treating plants in the reproductive phase required a higher glyphosate dose resulting in a R/S = 10.1 for one copy and up to R/S = 40.6 for the four‐copy line, but these plants were sterile at all but the lowest doses and represent the vegetative resistance often seen. This level of reproductive penalty places obvious limitations on the selection of resistant weeds.
The observations of hormesis quantified here and discussed by Velini et al.67 are consistent with a host of anecdotal observations of growth enhancement by glyphosate at low doses. How these growth enhancements participate in the selection of glyphosate‐resistant weeds remains to be seen, but minimally, it is a strike against the notion of using reduced rates of glyphosate. Hence, glyphosate‐resistant weeds, using only a target‐site based mechanism of resistance, should be characterized by reduced fertility with increasing glyphosate dose. Target site resistant EPSPSs have a dominant gene effect.
Supporting information
Figure S1. Line 469 accession At_S53469‐16 with two copies of At1EPSPS‐TIPA showing apparent segregation at 0.42 kg ha‐1 of glyphosate, left to right, test response, a putative heterozygous plant and an apparent null.
Figure S2. Comparison of typical siliques on plants in Figure 5 for the reproductive stage treatment. (A.) Surfactant only controls; (B.) Normal siliques at the lowest rate 0.013 kg ha‐1 for all three transgenic lines and wild type; (C.) Aborted siliques starting at 0.026 kg ha‐1 for all four plant lines.
ACKNOWLEDGEMENTS
This research was performed years ago and many small groups in Monsanto participated in one form or another. Their contributions also made this publication possible.
REFERENCES
- 1. Baird DD, Upchurch RP, Homesly WB and Franz JE, Introduction of a new broad spectrum post emergece herbicide of a new broad spectrum post emergence herbicide class with ultility for herbaceous perennial weed control, in Proceedings 26th North Central Weed Control Conference, pp. 64–68 (1971). [Google Scholar]
- 2. James C, Global status of commercialized biotech/GM crops: 2015. ISAAA Brief No. 51. ISAAA, Ithaca, NY: (2015). [Google Scholar]
- 3. Gianessi LP, Economic and herbicide use impacts of glyphosate‐resistant crops. Pest Manag Sci 61:241–245 (2005). [DOI] [PubMed] [Google Scholar]
- 4. USDA , NASS Agricultural Chemical Use Program. https://www.nass.usda.gov/Surveys/Guide_to_NASS_Surveys/Chemical_Use
- 5. Duke SO, Perspectives on transgenic, herbicide‐resistant crops in the United States almost 20 years after introduction. Pest Manag Sci 71:652–657 (2015). [DOI] [PubMed] [Google Scholar]
- 6. Delannay X, Bauman TT, Beighley DH, Buettner MJ, Coble HD, Defelice MS et al, Yield evaluation of a glyphosate‐tolerant soybean line after treatment with glyphosate. Crop Science 35:1461–1467 (1995). [Google Scholar]
- 7. Heap I, International Survey of Herbicide Resistant Weeds http://weedscience.org/ [accessed 1 March 2017].
- 8. Sammons RD, Heering DC, DiNicola N, Glick H and Elmore GA, Sustainability and stewardship of glyphosate and glyphosate‐resistant crops. Weed Technol 21:347–354 (2007). [Google Scholar]
- 9. Comai L, Sen LC and Stalker DM, An altered aroA gene product confers resistance to the herbicide glyphosate Science (Washington DC) 221:370–371 (1983). [DOI] [PubMed] [Google Scholar]
- 10. Steinrucken HC and Amrhein N, The herbicide glyphosate is a potent inhibitor of 5‐enolpyruvylshikimic acid‐3‐phosphate synthase. Biochem Biophys Res Commun 94:1207–1212 (1980). [DOI] [PubMed] [Google Scholar]
- 11. Larson‐Kelly N, Comai L, Kiser J, Mau C, Pokalsky AR, McBride K et al, Chloroplast delivery of a bacterial EPSP synthase in transgenic plants and tolerance to glyphosate. SAAS Bull Biochem Biotechnol 1:37–40 (1988). [Google Scholar]
- 12. Stalker DM, Producing herbicide‐resistant plants by gene transfer technology, in Target Sites of Herbicide Action, ed. by Boger P. and Sandmann G. CRC Press, Boca Raton, FL, pp. 147–163 (1989). [Google Scholar]
- 13. LeBrun M, Leroux B and Sailland A, New double transit peptide coding sequences for use in genes coding for increased herbicide tolerance in plants. European Union Patent 0507698 (1992). [Google Scholar]
- 14. Arnaud L, Sailland A, Lebrun M, Pallett K, Ravanel P, Nurit F et al, Physiological behavior of two tobacco lines expressing EPSP synthase resistant to glyphosate. Pest Biochem Physiol 62:27–39 (1998). [Google Scholar]
- 15. Alibhai MF, Cajacob C, Feng PCC, Heck GR, Qi Y, Flasinski S et al, Glyphosate resistant class I 5‐enolpyruvylshikimate‐3‐phosphate synthase (EPSPS) US2004/004636 (2004).
- 16. Spencer M, Mumm R and Gwyn J, Glyphosate resistant maize lines US Patent US 6040497 (2000).
- 17. Sidhu RS, Hammond BG, Fuchs RL, Mutz JN, Holden LR, George B and Olson T, Glyphosate‐tolerant corn: the composition and feeding value of grain from glyphosate‐tolerant corn is equivalent to that of conventional corn (Zea mays L.). J Agric Food Chem 48:2305–2312 (2000). [DOI] [PubMed] [Google Scholar]
- 18. Heck GR, Armstrong CL, Astwood JD, Behr CF, Bookout JT, Brown SM et al, Development and characterization of a CP4 EPSPS‐based, glyphosate‐tolerant corn event. Crop Sci 45:329–339 (2005). [Google Scholar]
- 19. Kishore GM, Padgette SR and Fraley RT, History of herbicide‐tolerant crops, methods of development and current state‐of‐the‐art – emphasis on glyphosate tolerance. Weed Technol 6:626–634 (1992). [Google Scholar]
- 20. Barry GF, Kishore GM, Padgette SR and Stallings WC, Glyphosate‐tolerant 5‐enolpyruvylshikimate‐3‐phosphate synthases US RE039247 (2006).
- 21. Sammons RD and Gaines TA, Glyphosate resistance: state of knowledge. Pest Manag Sci 70:1367–1377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baerson SR, Rodriguez DJ, Tran M, Feng YM, Biest NA and Dill GM, Glyphosate‐resistant goosegrass. Identification of a mutation in the target enzyme 5‐enolpyruvylshikimate‐3‐phosphate synthase. Plant Physiol 129:1265–1275 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu Q, Jalaludin A, Han HP, Chen M, Sammons RD and Powles SB, Evolution of a double amino acid substitution in the 5‐enolpyruvylshikimate‐3‐phosphate synthase in Eleusine indica conferring high‐level glyphosate resistance. Plant Physiol 167:1440–1447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen JC, Huang HJ, Zhang CX, Wei SH, Huang ZF, Chen JY et al, Mutations and amplification of EPSPS gene confer resistance to glyphosate in goosegrass (Eleusine indica). Planta 242:859–868 (2015). [DOI] [PubMed] [Google Scholar]
- 25. Mao CJ, Xie HJ, Chen SG, Valverde BE and Qiang S, Multiple mechanism confers natural tolerance of three lilyturf species to glyphosate. Planta 243:321–335 (2016). [DOI] [PubMed] [Google Scholar]
- 26. Odell JT, Nagy F and Chua NH, Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature (Lond) 313:810–812 (1985). [DOI] [PubMed] [Google Scholar]
- 27. Clough S and Bent A, Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana . Plant J 16:735–743 (1998). [DOI] [PubMed] [Google Scholar]
- 28. Bechtold N, Ellis J and Pelletier G, In planta Agrobacterium‐mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. CR Acad Sci Paris Sci 316:1194–1199 (1993). [Google Scholar]
- 29. Bubner B and Baldwin IT, Use of real‐time PCR for determining copy number and zygosity in transgenic plants. Plant Cell Report 23:263–271 (2004). [DOI] [PubMed] [Google Scholar]
- 30. Webb MR A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA 89:4884–4887 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dayan FE, Owens DK, Corniani N, Silva FML, Watson SB, Howell J et al, Biochemical markers and enzyme assays for herbicide mode of action and resistance studies. Weed Sci 63:23–63 (2015). [Google Scholar]
- 32. Castellino S, Leo GC, Sammons RD and Sikorski JA, Solution conformations of two shikimate 3‐phosphates determination by NMR and molecular mechanics calculations. J Org Chem 56:5176–5181 (1991). [Google Scholar]
- 33. Vazquez MJ, Rodriguez B, Zapatero C and Tew DG, Determination of phosphate in nanomolar range by an enzyme‐coupling fluorescent method. Anal Biochem 320: 292–298 (2003). [DOI] [PubMed] [Google Scholar]
- 34. Seefeldt SS, Jensen JE and Fuerst PE, Log‐logistic analysis of herbicide dose–response relationships. Weed Technol 9:218–225 (1995). [Google Scholar]
- 35. Brain P and Cousens R, An equation to describe dose responses where there is stimulation of growth at low doses. Weed Res 29:93–96 (1989). [Google Scholar]
- 36. Schabenberger O, Tharp BE, Kells JJ and Penner D, Statistical tests for hormesis and effective dosages in herbicide dose response. Agron J 91:713–721 (1999). [Google Scholar]
- 37. McCourt JA, Pang SS, King‐Scott J, Guddat LW and Duggleby RG, Herbicide‐binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc Natl Acad Sci USA 103:569–573 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tranel PJ and Wright TR, Resistance of weeds to ALS‐inhibiting herbicides: what have we learned? Weed Sci 50:700–712 (2002). [Google Scholar]
- 39. Christoffers MJ, Berg ML and Messersmith CG, An isoleucine to leucine mutation in acetyl‐CoA carboxylase confers herbicide resistance in wild oat. Genome 45:1049–1056 (2002). [DOI] [PubMed] [Google Scholar]
- 40. Tardif FJ, Rajcan I and Costea M, A mutation in the herbicide target site acetohydroxyacid synthase produces morphological and structural alterations and reduces fitness in Amaranthus powellii . New Phytol 169:251–264 (2006). [DOI] [PubMed] [Google Scholar]
- 41. Ashigh J and Tardif FJ, Water and temperature stress impact fitness of acetohydroxyacid synthase‐inhibiting herbicide‐resistant populations of eastern black nightshade (Solanum ptychanthum). Weed Sci 59:341–348 (2011). [Google Scholar]
- 42. Schonbrunn E, Eschenburg S, Shuttleworth WA, Schloss JV, Amrhein N, Evans JNS et al, Interaction of the herbicide glyphosate with its target enzyme 5‐enolpyvuvylshikimate 3‐phosphate synthase in atomic detail. Proc Natl Acad Sci USA 98:1376–1380 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Eschenburg S, Healy ML, Priestman MA, Lushington GH and Schonbrunn E, How the mutation glycine96 to alanine confers glyphosate insensitivity to 5‐enolpyruvyl shikimate‐3‐phosphate synthase from Escherichia coli . Planta 216:129–135 (2002). [DOI] [PubMed] [Google Scholar]
- 44. Ng CH, Wickneswari R, Salmijah S, Teng YT and Ismail BS, Gene polymorphisms in glyphosate‐resistant and ‐susceptible biotypes of Eleusine indica from Malaysia. Weed Res 43:108–115 (2003). [Google Scholar]
- 45. Yuan CI, Hsieh YC and Chiang MY, Glyphosate‐resistant goosegrass in Taiwan: cloning of target enzyme (EPSPS) and molecular assay of field populations. Plant Protect Bull (Taipei) 47:251–261 (2005). [Google Scholar]
- 46. Ngo TD, Krishnan M, Boutsalis P, Gill G and Preston C, Target site mutations conferring resistance to glyphosate in feathertop Rhodes grass (Chloris virgata) populations in Australia. Pest Manag Sci doi:https://doi.org/10.1002/ps.4512 (2017). [DOI] [PubMed] [Google Scholar]
- 47. Wakelin AM and Preston C, A target‐site mutation is present in a glyphosate‐resistant Lolium rigidum population. Weed Res (Oxford) 46:432–440 (2006). [Google Scholar]
- 48. Arnaud L, Sailland A, Lebrun M, Pallett K, Ravanel P, Nurit F et al, Physiological behavior of two tobacco lines expressing EPSP synthase resistant to glyphosate. Pestic Biochem Physiol 62:27–39 (1998). [Google Scholar]
- 49. CaJacob CA, Feng PCC, Heck GR, Alibhai MF, Sammons RD and Padgette SR, Engineering resistance to herbicides, in Handbook of Plant Biotechnology, ed. by Christou P. and Klee H. Wiley, Chichester, pp. 353–372 (2004). [Google Scholar]
- 50. Ge X, d'Avignon DA, Ackerman JJH, Collavo A, Sattin M, Ostrander EL et al, Vacuolar glyphosate‐sequestration correlates with glyphosate resistance in ryegrass (Lolium spp.) from Australia, South America, and Europe: a 31P NMR investigation. J Agric Food Chem 60:1243–1250 (2012). [DOI] [PubMed] [Google Scholar]
- 51. Collavo A and Sattin M, Resistance to glyphosate in Lolium rigidum selected in Italian perennial crops: bioevaluation, management and molecular bases of target‐site resistance. Weed Res (Oxford) 52:16–24 (2012). [Google Scholar]
- 52. Shah DM, Horsch RB, Klee HJ, Kishore GM, Winter JA, Tumer NE et al, Engineering herbicide tolerance in transgenic plants. Science USA 233:478–481 (1986). [DOI] [PubMed] [Google Scholar]
- 53. Walker DR, Walker AK, Wood ED, Talevera MEB, Fernandez FE, Rowan GB et al, Gametic selection by glyphosate in soybean plants hemizygous for the CP4 EPSPS transgene. Crop Sci 46:30–35 (2006). [Google Scholar]
- 54. Feng PCC, Chiu T and Sammons RD, Glyphosate efficacy is contributed by its tissue concentration and sensitivity in velvetleaf (Abutilon theophrasti). Pestic Biochem Physiol 77:83–91 (2003). [Google Scholar]
- 55. Chen YCS, Hubmeier C, Tran M, Martens A, Cerny RE, Sammons RD et al, Expression of CP4 EPSPS in microspores and tapetum cells of cotton (Gossypium hirsutum) is critical for male reproductive development in response to late‐stage glyphosate applications. Plant Biotechnol J 4:477–487 (2006). [DOI] [PubMed] [Google Scholar]
- 56. Ye GN, Hajdukiewicz PTJ, Broyles D, Rodriguez D, Xu CW, Nehra N et al, Plastid‐expressed 5‐enolpyruvylshikimate‐3‐phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J 25:261–270 (2001). [DOI] [PubMed] [Google Scholar]
- 57. Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL et al, Gene amplification confers glyphosate resistance in Amaranthus palmeri . Proc Natl Acad Sci USA 107:1029–1034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dillon A, Varanasi VK, Danilova TV, Koo D‐H, Nakka S, Peterson DE et al, Physical mapping of amplified copies of the 5‐enolpyruvylshikimate‐3‐phosphate synthase gene in glyphosate‐resistant Amaranthus tuberculatus . Plant Physiol 173:1226–1234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gaines TA, Barker AL, Patterson EL, Westra P, Westra EP, Wilson RG et al, EPSPS gene copy number and whole‐plant glyphosate resistance level in Kochia scoparia . PLoS One 11: e0168295 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Molin WT, Wright AA, Lawton‐Rauh A and Saski CA, The unique genomic landscape surrounding the EPSPS gene in glyphosate resistant Amaranthus palmeri: a repetitive path to resistance. BMC Genom 18:91–91 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chatham LA, Bradley KW, Kruger GR, Martin JR, Owen MDK, Peterson DE et al, A multistate study of the association between glyphosate resistance and EPSPS gene amplification in waterhemp (Amaranthus tuberculatus). Weed Sci 63:569–577 (2015). [Google Scholar]
- 62. Lydon J and Duke SO, Glyphosate induction of elevated levels of hydroxybenzoic acids in higher plants. J Agric Food Chem 36:813–818 (1988). [Google Scholar]
- 63. Haslam E, Hydroxybenzoic acids and the enigma of gallic acid, in Recent Advances in Phytochemistry, ed. by Cohn EE. Plenum Press, New York, pp. 163–200 (1986). [Google Scholar]
- 64. Haslam E, Shikimic Acid Metabolism and Metabolites. Wiley, New York, (1993). [Google Scholar]
- 65. Hollander H and Amrhein N, Inhibition by glyphosate of phenylpropanoid synthesis in buckwheat. Plant Physiol 63(Supplement):41 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hollander‐Czytko H and Amrhein N, Sub cellular compartmentation of shikimic‐acid and phenyl alanine in buckwheat Fagopyrum esculentum cell suspension cultures grown in the presence of shikimate pathway inhibitors. Plant Sci Lett 29:89–96 (1983). [Google Scholar]
- 67. Velini ED, Alves E, Godoy MC, Meschede DK, Souza RT and Duke SO, Glyphosate applied at low doses can stimulate plant growth. Pest Manag Sci 64:489–496 (2008). [DOI] [PubMed] [Google Scholar]
- 68. Velini ED, Alves E, Godoy MC, Meschede DK and Duke SO. Glyphosate and hormesis: Environmental implications, in 233rd ACS National Meeting. AGRO‐056. American Chemical Society, Washington, DC: (2007). [Google Scholar]
- 69. Prithiviraj B, Perry LG, Badri DV and Vivanco JM, Chemical facilitation and induced pathogen resistance mediated by a root‐secreted phytotoxin. New Phytol 173:852–860 (2007). [DOI] [PubMed] [Google Scholar]
- 70. Kristiansen KN, Biosynthesis of proanthocyanidins in barley: genetic control of the conversion of dihydroquercetin to catechin and procyanidins. Carlsberg Res Commun 49:503–524 (1984). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Line 469 accession At_S53469‐16 with two copies of At1EPSPS‐TIPA showing apparent segregation at 0.42 kg ha‐1 of glyphosate, left to right, test response, a putative heterozygous plant and an apparent null.
Figure S2. Comparison of typical siliques on plants in Figure 5 for the reproductive stage treatment. (A.) Surfactant only controls; (B.) Normal siliques at the lowest rate 0.013 kg ha‐1 for all three transgenic lines and wild type; (C.) Aborted siliques starting at 0.026 kg ha‐1 for all four plant lines.


