Abstract
Terminal differentiation of many vascular cells involves cell wall changes. Cells first elongate their primary wall, then lay down a lignified secondary wall, which is often followed by digestion of the primary wall. Expansins are wall proteins that regulate wall changes, but little is known about the specific functions of the many individual expansin isoforms. An in vitro cell culture of synchronously differentiating tracheary elements was used to identify three new expansins and to compare their expression kinetics with the timing of wall changes. The genes encoding these expansins from zinnia (Zinnia elegans), designated ZeExp1, ZeExp2, ZeExp3, are expressed during cell elongation. ZeExp1 and ZeExp2 mRNA decrease at the early stage of secondary wall formation, whereas ZeExp3 does not. In planta, all three ZeExp mRNAs are found predominantly in a single flank of cells adjacent to protoxylem and metaxylem vessels and in cells roughly at the radial position of the fasicular and interfasicular cambium. Furthermore, within these cells, Exp mRNA is localized exclusively either to the apical or basipetal end of cells depending on the expansin gene and organ, providing the first evidence for polar localization of mRNA in plant cells. ZeExp1 and ZeExp3 mRNA are localized at the apical tip, whereas ZeExp2 mRNA is found in the basal tip. These observations indicate that these three expansins are xylem cell specific and possibly involved in the intrusive growth of the primary walls of differentiating xylem cells.
Plant cells expand by controlling slippage of the wall network of microfibrils and adhering matrix polysaccharides and proteins (Cosgrove, 1993). A family of cell wall proteins called α-expansins are wall-loosening factors that can promote extension of isolated cell wall, probably by disrupting noncovalent bonds between the cellulose microfibrils and matrix polymers (McQueen-Mason and Cosgrove, 1994; McQueen-Mason, 1995; Cosgrove, 1998). The α-expansins are represented by a large gene family (Cosgrove, 1998, 1999), suggesting that they serve various functions throughout development. A ripening-related expansin in tomato fruit may enhance accessibility of noncovalently bound polymers to endogenous enzyme action during ripening (Rose et al., 1997). It has also been proposed that α-expansins are involved in leaf organogenesis (Fleming et al., 1997; Reinhardt et al., 1998) and vascular cell differentiation (Cho and Kende, 1998), but otherwise little is known about the specific role of the individual expansins.
Tracheary elements (TEs) are functional cell corpses that form the water-conducting vessels in plant xylem. During normal stem development, primary TEs develop from meristematic cells of procambial cells, then later by fascicular and interfascicular cambial cells (Shininger, 1979). Prior to secondary wall formation, procambium cells first expand primarily along the longitudinal axis by intrusive growth (Bannan, 1956; Roberts and Uhnak, 1998). In this type of cell extension found among different xylem cell types, the wall matrix in the apical regions of the cell shears. Since these apical regions do not attenuate during growth, wall precursors and growth-limiting proteins must also be directed to the cell wall-growing zone, probably by longitudinal microtubules.
RESULTS
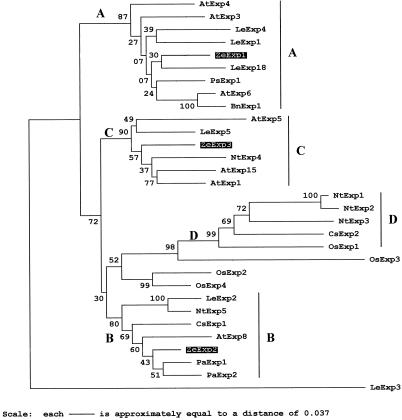
Three α-expansin cDNA clones (ZeExp1, ZeExp2, and ZeExp3) were isolated from a cDNA library constructed from zinnia (Zinnia elegans) cell culture induced for TEs for 48 h (Ye and Varner, 1993) using a mix of Arabidopsis Exp1 and Exp9 probes. The three cDNA sequences were compared with the Arabidopsis Exp1 sequence using the CLUSTAL W algorithm (Thompson et al., 1994). The 949-bp ZeExp1 cDNA represents a partial sequence lacking a signal peptide of presumably 25 to 28 amino-terminal peptides (Fig. 1), but contained a 337-bp 3′-untranslated region (UTR). The 1,088-bp ZeExp2 contained a 5′-UTR of 15 bp, an open reading frame of 738 bp, and a 3′-UTR of 335 bp. The 1,054-bp ZeExp3 contained a 5′-UTR of 23 bp, an open reading frame of 729 bp, and a 3′-UTR of 302 bp. The deduced polypeptides of ZeExp2 and ZeExp3 were 242 and 245 amino acids, respectively, and each polypeptide possessed the conserved Cys and Trp residues that are characteristic of expansins (Shcherban et al., 1995). The deduced mature proteins of zinnia expansins fell into three of the five major phylogenetic branches for α-expansins (Fig. 2). ZeExp1 aligned into a clade with LeExp18, which is expressed in the meristem (Reinhardt et al., 1998), and LeExp1, whose expression is preferentially induced during the ripening process in tomato fruit (Rose et al., 1997). ZeExp2 aligned with PaExp1 cloned from apricot fruit. ZeExp3 fell into a group containing AtExp1, which is expressed in guard cells (D.J. Cosgrove, unpublished data).
Figure 1.
Amino acid sequence alignment of three new zinnia expansins. The deduced amino acid sequence alignments for zinnia expansins are compared with the Arabidopsis expansin 1 (AtExp1) using the CLUSTAL W algorithm (Thompson et al., 1994). Identical amino acids are marked with asterisks. Dashes represent gaps that were used to facilitate alignment. Eight Cys residues (+) and four Trp residues (♦) conserved among expansins are indicated. Putative signal peptides are shown in lowercase. Accession numbers for ZeExp1, ZeExp2, and ZeExp3 are AF230331, AF230332, and AF230333, respectively.
Figure 2.
Phylogenetic tree of 32 α-expansin protein sequences. Numbers on the tree indicate bootstrap P values. Large letters denote the subfamily groupings previously identified (Link and Cosgrove, 1998). This tree was rooted using LeExp3 (Lycopersicon esculentum). Sequences and GenBank accession numbers: AtExp1 (U30476), AtExp2 (U30481), AtExp3 (AC004684), AtExp4 (AC003674), AtExp5 (U30478), AtExp6 (U30480), AtExp8 (AC002336); LeExp1 (U82123), LeExp2 (AF096776), LeExp3 (AF059487), LeExp4 (AF059488), LeExp5 (AF059489), and LeExp18 (AJ004997); Nt (Nicotiana tabacum) Exp1 (AF049350), NtExp2 (AF049351), NtExp3 (AF049352), NtExp4 (AF049353), and NtExp5 (AF049354); Os (Oryza sativa) Exp1 (Y07782), OsExp2 (U30477), OsExp3 (U30479), and OsExp4 (U85246); Ps (Pisum sativa) Exp1 (X85187); Bn (Brassica napus) Exp1 (AJ000885); Cs (Cucumis sativus) Exp1 (U30382), and CsExp2 (U30460); Pa (Prunus armeniaca) Exp1 (U93167) and PaExp2 (AF038815). The 21- and 19-amino acid signal peptides were predicted for the ZeExp2 and ZeExp3, respectively, by the method of von Heijne (1986). Mature proteins were aligned with the CLUSTAL algorithm, and γ-corrected distances between amino acid sequences were used to construct this tree by nearest-neighboring joining using the MEGA program (Kumar et al., 1993).
Zinnia expansins belong to a multigene family, as is the case for other plant species, suggesting that expansin isoforms have either specialized functions or cell specificity. The complexity of expansin genes in zinnia was estimated using genomic Southern-blot analysis with a mixture of full-length ZeExp2 and ZeExp3 cDNA probes. After high-stringency washes, two or three bands were detected (data not shown). The single major band in each lane represents the ZeExp3 gene, and two minor bands in each lane represent the ZeExp2 gene because of known internal EcoRI and HindIII restriction sites. We conclude that each of these genes is probably present in the genome as a single copy. After low-stringency washes, multiple bands were detected, indicating that, as in all other tested species, expansins form a multigene family in zinnia.
Expression of Expansin Genes during TE Development
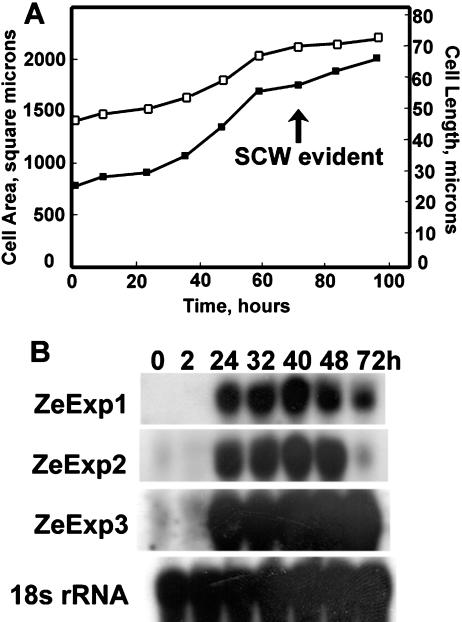
The in vitro system we utilize here to address the role of expansin recapitulates the cellular events of TE differentiation (Seagull and Falconer, 1991). Mesophyll cells were mechanically isolated and induced with auxin and cytokinin to differentiate synchronously into TEs in vitro. After a latent period of approximately 24 h, cells elongate and expand for approximately 24 to 48 h, and then begin to synthesize the secondary cell wall beginning at 72 h, as evident by wall thickening (Fig. 3A). After wall synthesis is complete, developing TEs undergo a programmed cell death (Groover and Jones, 1999) that involves further wall changes such as formation of the perforation plate.
Figure 3.
Expression of three expansin genes during TE induction in zinnia cell culture compared with cell growth. A, Zinnia cells induced to differentiate into TEs were photographed at the indicated times and average lengths (□) and areas (▪) determined. The average se of the mean for cell length and area were 1.8 μm and 57 μm2, respectively. SCW, Secondary cell wall. B, Total RNAs were extracted from cells cultured in TE inductive medium at the indicated times. Total RNA (10 μg) was loaded in each lane and hybridized with gene-specific probes. Numbers above blots indicate the duration of mesophyll cells in inductive medium. The same blot was hybridized with Arabidopsis 18S rRNA probe to check for loading variability.
Expression of the expansin genes was correlated with primary wall expansion and elongation during TE development in vitro. The expression level of all three expansin genes increased after 24 h of inductive levels of hormones; however, the decay patterns were different among the three genes (Fig. 3B). The ZeExp1 expression level did not change from 24 h until 48 h, and decreased moderately at 72 h, a time when secondary cell wall thickenings are detectable by microscopy (Groover et al., 1997; Fig. 3A). ZeExp2 showed a similar expression pattern to that of ZeExp1 until 48 h, however, the mRNA level decreased to nearly the basal level after 72 h of induction. ZeExp3 mRNA remained constant.
Expression of Expansin Genes in Planta
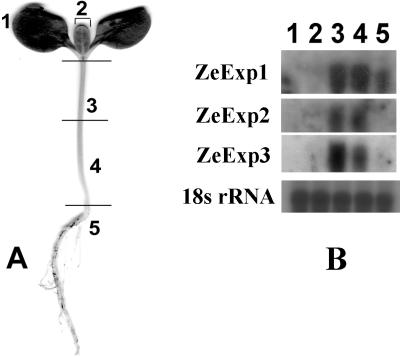
Northern analyses revealed that three expansin mRNAs approximately 1,200 nt in length were expressed in stem tissue and barely detectable in cotyledon and leaf tissues (Fig. 4B). A significant level of ZeExp1 but not ZeExp2 or ZeExp3 mRNA was also detected in roots (Fig. 4B).
Figure 4.
Expression of expansin genes in various organs in zinnia seedlings. A, Ten-day-old zinnia seedling showing the regions used for northern analysis shown in B. 1, Cotyledons without petioles; 2, leaves; 3, upper part of stems; 4, lower part of stems; 5, roots. B, ZeExp1, ZeExp2, and ZeExp3 northern blots showing tissue-specific expression.
Cell-Specific Expression and Intracellular Localization of Expansin Transcripts
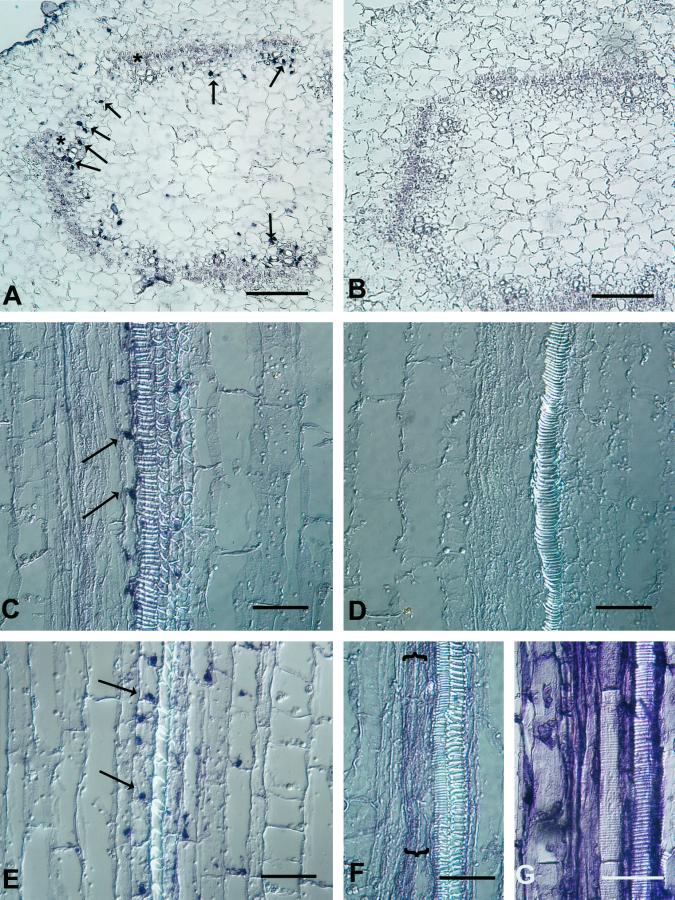
In situ hybridization was used to identify expansin-expressing cells in stem tissues (Figs. 5 and 6). Transverse sections probed with sense control RNAs (Fig. 5B) lacked hybridization signal; however, all three expansin mRNAs were detected at the same subtissue location by antisense probes (Fig. 5A, data for ZeExp2 and ZeExp3 not shown). Expansin mRNA signals were confined to the developing primary xylem cells, predominantly adjacent to vessel members, and in cells at the future position of the interfasicular cambium. Surprisingly, the hybridization signals routinely appeared punctate (arrows in Fig. 5A). Serial transverse sections revealed that these punctate signals were at most 16 μm in depth (data not shown), suggesting that these expansin mRNAs are highly localized at the specific sites. In situ hybridization of longitudinal sections confirmed the size and punctate shape of the hybridization signals. The single flank of elongated cells adjacent to protoxylem or metaxylem vessels showed strong hybridization of the ZeExp riboprobes (Fig. 5, C and E), but occasionally nonadjacent cells located 50 to 100 μm from vessel members also showed clear hybridization.
Figure 5.
In situ localization of ZeExp1 and ZeExp2 mRNA in zinnia stem tissues. A, Cross-section of the upper stem probed with the antisense (A) and sense (B) ZeExp1 3′-UTR RNA. Arrows indicate some of the punctate patterned hybridization signals. Note that the signals are found exclusively in the vascular bundles (two bundles are highlighted by asterisks) and in cells near where the interfascicular cambium forms. Longitudinal sections of the upper stem were probed with the antisense (C) and sense (D) ZeExp1 3′-UTR RNA and the antisense ZeExp2 3′-UTR RNA (E). Arrows indicate the apical (C) and basal (E) location of the punctate hybridization signals. Note that the indicated flank of cells in (C) and (E) appear to begin tip growth from the apical corners. Longitudinal sections probed with the zinnia Ted3 (F) and 18S rRNA (G) antisense probes, respectively, indicate an even hybridization signal in cells associated with the protoxylem (F) or all cells (G). A and B, Bars = 100 μm; C to G, bars = 50 μm.
Figure 6.
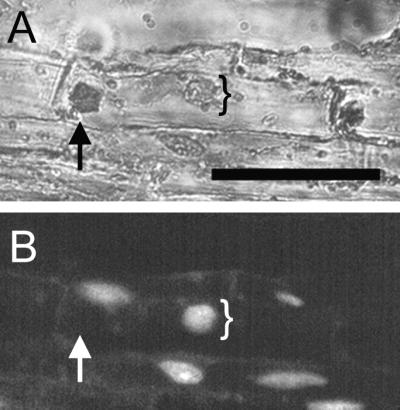
Relative location of expansin mRNA signal and the nucleus in a single cell. Longitudinal section in situ hybridized with the ZeExp3 antisense probe was stained with DAPI to visualize the nucleus under UV light (B). The section was observed under a combination of white and UV light (A) and UV light only (B). Note that the position of the localized expansin transcript indicated by arrows does not colocalize with the position of the nucleus indicated by brackets. Bar = 50 μm.
In all cases, hybridization signals were exclusively located at the distal ends of cells (Fig. 5, C and E), but the apical versus basal location was dependent on the riboprobe used and the tissue. Medial sections through the base of the cotyledonary petiole revealed ZeExp1, ZeExp2, and ZeExp3 mRNA at the cell base. In contrast, at a position just below the apical meristem where protoxylem differentiates, ZeExp1 and ZeExp3 mRNAs were found in the apical cytoplasm, while ZeExp2 mRNA was found in the basal cytoplasm (Table I; Fig. 5, C and E). More precisely, hybridization signal was found in a distal corner of the cells flanking the vessels (Fig. 5E), in each case where intrusive growth appeared to occur (note carefully the elongated corners of the cells in the indicated flanks in Fig. 5, C and E).
Table I.
Subcellular location of expansin mRNA in xylem cells
| Expansin | Cotyledon Petiole
|
Hypocotyl
|
||||
|---|---|---|---|---|---|---|
| Apical | Basal | Unclear | Apical | Basal | Unclear | |
| no.a | ||||||
| ZeExp1 | 27 (54%) | 0 | 23 | 42 (84%) | 0 | 8 |
| ZeExp2 | 37 (74%) | 0 | 13 | 0 | 40 (80%) | 10 |
| ZeExp3 | 36 (72%) | 0 | 14 | 41 (82%) | 0 | 9 |
No. of cells scored having the indicated location in this organ. Nos. in parentheses are the percent of the total cells scored. Unclear means it was not possible to locate the end wall.
Several lines of evidence showed that the punctate pattern of ZeExp mRNA hybridization is not an artifact but, rather, is evidence of the cytoplasmic location of the ZeExp message. First, only xylem cells hybridized with ZeExp antisense probes (Fig. 5, A–E), so the pattern was not due to probe aggregation. Second, an 18S rRNA revealed a relatively diffuse cytoplasmic staining even in the xylem cells, where ZeExp probes hybridize in the punctate pattern, indicating that fixation artifacts did not cause cytoplasmic aggregation in these particular cells (Fig. 5G). Third, a TED3 probe, previously shown to hybridize to xylem cells (Demura and Fukuda, 1994), had a hybridization signal that was evenly distributed throughout the cytoplasm (Fig. 5F) in the same cell types in which expansin mRNA hybridization was shown to be punctate. Fourth, the punctate pattern was not due to nuclear sequestration of the ZeExp message (Fig. 6).
The strongest evidence that the punctate hybridization signals are not due to fixation artifacts comes from the observation that the ZeExp1 and ZeExp3 hybridization patterns were apical, whereas for ZeExp2 they were basal in the same cell types from the same fixed tissue (Fig. 5, C versus E). Clearly, fixation or hybridization artifacts cannot explain the sequence specificity of this opposite localization.
DISCUSSION
We introduce three new members of a large gene family of expansins, and indirectly show that they play a specific developmental role in shoot histogenesis. These expansin genes are expressed in developing TEs in vitro, with kinetics and expression patterns linking the function to the regulation of primary wall expansion of xylem-specific cells. In planta, expression of these genes is limited to a specific set of xylem cells.
Several xylem cell types elongate by intrusive growth (Bannan, 1956; Roberts and Uhnak, 1998), whereby wall matrix slips and wall precursors are delivered to these localized regions of the wall by direct secretion through the endomembrane system. However, loosening of the existing wall at the localized site must precede or accompany wall deposition before intrusive growth can ensue. Given what is known about the biochemical properties of expansins, it is reasonable to presume that this loosening is accomplished by the action of expansins. In situ hybridization clearly indicates that expansin mRNA is located exclusively at the end of the cytoplasm in a cell type that appears poised to elongate by intrusive growth. We propose that these expansin mRNAs are subcellularly localized to the site of future growth so as to provide restricted wall loosening by expansins produced there.
The identity and fate of the flanking cells showing localized expansin mRNA is not known with certainty. While their morphology points to their being fully differentiated xylem parenchyma and not precursors to vessel members, their tissue location and signs of intrusive growth (Fig. 5E) suggest they may transdifferentiate into fibers.
Asymmetric RNA localization is well documented in animal cells and invertebrates (Bashirullah et al., 1998), where this mechanism accounts for asymmetric distribution of proteins in polarized somatic cells and embryos. Fewer cases are found in lower organisms, although there is one example each for localized mRNA in yeast (Takizawa et al., 1997) and a brown algae (Bouget et al., 1996). However, the work here presents the first evidence, to our knowledge, for mRNA localization in land plants (Embryophyta).
RNA localization requires an intact cytoskeleton (Oleynikov and Singer, 1998), and 3′-UTR of localized mRNA plays an important role in RNA localization (Kimberly et al., 1992; Goldspink et al., 1997). It has been reported that almost one-tenth of randomly collected Drosophila ovarian mRNA shows asymmetric localization (Dubowy and Macdonald, 1998). The function of RNA localization may prevent random movement of RNA within the cytoplasm and bring it into contact with regulatory factors (Bassell and Singer, 1997). In the case of mRNA encoding co-translated proteins such as the expansins described here, targeting message to specific microdomains on the endoplasmic reticulum may be involved in directed trafficking of the secreted protein (Weeks and Melton, 1987).
Longitudinally oriented microtubules may direct expansin mRNA to a specific intracellular location, where expansin mRNA is translated and secreted to the growing or neighboring cell wall to cause localized cell wall loosening, although expansin protein was not localized in the present study. A previous notion was that microtubules direct transverse deposition of cellulose microfibrils (Fukuda, 1996), but this is not supported by the recent evidence that cortical microtubules are actually oriented parallel to the long axis of elongating cells during TE differentiation, and that overall cell elongation is caused by localized intrusive growth of cells (Roberts and Uhnak, 1998). Parallel orientation of microtubules in these cells is consistent with a role of apical trafficking of mRNA and wall precursors. In light of the current evidence for tip-localized expansin mRNA and the known role for microtubules in mRNA trafficking (Bassell and Singer, 1997), we propose that the seemingly paradoxical longitudinal distribution of microtubules in elongating TE occurs to direct growth-limiting expansins to the cell tip rather than to direct cellulose deposition.
MATERIALS AND METHODS
Plant Material and Culture Media
Zinnia (Zinnia elegans L. cv Envy) was purchased from Stokes Seed (Buffalo, NY). Plants were grown in a growth chamber at 25°C with 14 h of light (110 μmol photons m−2 s−2) and 60% relative humidity. Mesophyll cells of 8- to 16-d-old seedlings were isolated and cultured as described in Groover et al. (1997). Cells were cultured in inductive medium as described by Fukuda and Komamine (1980) with 0.1 mg/L α-naphthalene-acetic acid and 0.2 mg/L 6-benzylamino-purine.
Isolation and Sequencing of cDNA Clones
The zinnia cDNA library, provided by Dr. Z-H Ye (University of Georgia), was constructed from leaf mesophyll cells cultured for 48 h in TE induction medium (Ye and Varner, 1993). The library was screened with a mixture of 32P-labeled full-length AtExp1 and AtExp9 cDNAs at 60°C, followed by two 60°C 1× SSC washes. Three independent cDNA phage clones were subcloned into pBluescript II KS+ phagemid vectors (Stratagene, La Jolla, CA), and sequenced by the Automated DNA Sequencing Facility at The University of North Carolina at Chapel Hill.
Genomic DNA Extraction and Southern-Blot Analyses
Genomic DNA was isolated from 10-d-old seedlings as described previously (Dellaporta et al., 1983). Ten micrograms of genomic DNA was digested with EcoRI and HindIII and fractionated in a 0.6% (w/v) agarose gel, followed by a Southern transfer to a Nytran membrane (Schleicher & Schuell, Keene, NH). The blots were hybridized with a mixture of full-length ZeExp2 and ZeExp3 cDNA probes in 50 mm PIPES (1,4-piperazinediethanesulfonic acid) (pH 6.5), 100 mm NaCl, 50 mm sodium phosphate, 1 mm EDTA, and 5% (w/v) SDS solution for 16 h at 65°C, and then washed. The first two washes in 5× SSC/5% (w/v) SDS at 65°C were followed by two more washes in 2× SSC/5% (w/v) SDS for low stringency and 0.2× SSC/5% (w/v) SDS for high stringency, respectively.
Northern Analyses
Ten-day-old green seedlings were dissected into cotyledon, leaf, upper stem half, lower stem half, and root (Fig. 4A). Total RNA was isolated from each organ and cells in culture as described previously (Wadsworth et al., 1988). RNA (10 μg) was fractionated on 1.2% (w/v) agarose gels in the presence of 3% (w/v) formaldehyde and transferred to Nytran membrane. Hybridization and washing were as described in Southern-blot analyses, with final washes in 0.2× SSC/5% (w/v) SDS. Gene-specific DNA fragments were prepared by PCR amplification from cDNA clones. For the ZeExp1-specific probe, the 340 bp of 3′-UTR was PCR amplified using oligonucleotides 5′CCCGGGAATTCGAGTGTAAAACATGAT3′ and 5′TTTTTTTGGATCCAAAATCAATCCATT3′ as forward and reverse primers, respectively. For the ZeExp2-specific probe, the 293-bp 3′ end was subcloned into EcoRI site of pBS KS phagemid and subsequently PCR amplified using SK and KS primers. For the ZeExp3-specific probe, the 293-bp 3′ region was PCR amplified with forward primer (5′AAGCACATCTATGTTCT3′) and reverse primer (SK). Probes were prepared by 32P labeling using a random primer kit (Boehringer Mannheim/Roche, Basel).
In Situ Hybridization
In situ hybridization was performed as described previously (Cho and Kende, 1998) except higher concentrations of RNaseA (5–10 μg/mL) were used. Hybridization of the riboprobes was detected with anti-digoxigenin antibodies conjugated to alkaline phosphatase, and visualized by color development according to the manufacturer's instructions (Boehringer Mannheim/Roche). Sense and antisense ZeExp1- and ZeExp2-specific riboprobes were generated by in vitro transcription of the subcloned 3′-UTR using a digoxigenin labeling kit (Boehringer Mannheim/Roche). The antisense ZeExp3-specific, 300-bp riboprobe was prepared by transcription of a Nar1 restriction of the full-length ZeExp3 clone. All probes were prepared from 3′-UTR except the ZeExp3 sense probe, which was generated from the full-length cDNA. Probes were hydrolyzed to 100 nt by alkaline hydrolysis prior to hybridization. Each hybridization with a specific sense or antisense probe was performed at least four times, with the same results. To stain nuclei, 4′, 6-diamidino-2-phenyl-indole diacetate (DAPI) was dissolved in water at 1 mg/mL and used at a working concentration of 0.05 mg/mL.
ACKNOWLEDGMENTS
We thank Sabine Hardenack for helpful discussion on in situ hybridization, Jheng-Hua Ye for the cDNA library, Andrew Groover for technical assistance, W. Soh, W.C. Dickison, and J. Romberger for discussions on primary xylem, and Susan Whitfield for help with figures. Daniel M. Durachko assisted with preliminary experiments.
Footnotes
This work was supported by the Department of Energy (grant no. DE–FG02–93ER13179 to D.J.C.), by the National Science Foundation (grant no. IBN–9807801 to A.M.J.), and by the U.S. Department of Agriculture.
This paper is dedicated to and in memory of Prof. William C. Dickison.
LITERATURE CITED
- Bannan MW. Some aspect of the elongation of fusiform cambial cells in Thuza occidentalis. Can J Bot. 1956;34:175–196. [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. RNA localization in development. Annu Rev Biochem. 1998;67:335–394. doi: 10.1146/annurev.biochem.67.1.335. [DOI] [PubMed] [Google Scholar]
- Bassell G, Singer RH. mRNA and cytoskeletal filaments. Curr Opin Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- Bouget F, Gerttula S, Shaw SL, Quatrano RS. Localization of actin mRNA during the establishment of cell polarity and early cell division in Fucus embryo. Plant Cell. 1996;8:189–201. doi: 10.1105/tpc.8.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H. Tissue localization of expansins in deepwater rice. Plant J. 1998;15:805–812. doi: 10.1046/j.1365-313x.1998.00258.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Wall extensibility: its nature, measurement, and relationship to plant cell growth. New Phytol. 1993;124:1–23. doi: 10.1111/j.1469-8137.1993.tb03795.x. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Cell wall loosening by expansins. Plant Physiol. 1998;118:333–339. doi: 10.1104/pp.118.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:391–417. doi: 10.1146/annurev.arplant.50.1.391. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA mini preparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Demura T, Fukuda H. Novel vascular cell-specific genes whose expression is regulated temporally and spatially during vascular system development. Plant Cell. 1994;6:967–981. doi: 10.1105/tpc.6.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy J, Macdonald PM. Localization of mRNAs to the oocyte is common in Drosophila ovaries. Mech Dev. 1998;70:193–195. doi: 10.1016/s0925-4773(97)00185-8. [DOI] [PubMed] [Google Scholar]
- Fleming AJ, McQueen-Mason SJ, Mandel T, Kuhlemeier C. Induction of leaf primordia by the cell wall protein expansin. Science. 1997;276:1415–1418. [Google Scholar]
- Fukuda H. Xylogenesis: initiation, progression, and cell death. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:299–325. doi: 10.1146/annurev.arplant.47.1.299. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Komamine A. Establishment of an experimental system for the tracheary elements differentiation from single cells isolated from the mesophyll of Zinnia elegans. Plant Physiol. 1980;52:57–60. doi: 10.1104/pp.65.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink P, Sharp W, Russell B. Localization of cardiac (alpha)-myosin heavy chain mRNA is regulated by its 3′ untranslated region via mechanical activity and translational block. J Cell Sci. 1997;110:2969–2978. doi: 10.1242/jcs.110.23.2969. [DOI] [PubMed] [Google Scholar]
- Groover A, DeWitt N, Heidel A, Jones AM. Programmed cell death of plant tracheary elements differentiating in vitro. Protoplasma. 1997;196:197–211. [Google Scholar]
- Groover A, Jones AM. Tracheary element differentiation uses a novel mechanism coordinating programmed cell death and secondary cell wall synthesis. Plant Physiol. 1999;119:375–384. doi: 10.1104/pp.119.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberly L, Melton M, Melton D. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Kumar S, Koichiro T, Nei M. MEGA: Molecular Evolutionary Genetics Analysis System Version 1.01. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- Link BM, Cosgrove DJ. Acid-growth response and α-expansins in suspension cultures of Bright Yellow 2 tobacco. Plant Physiol. 1998;118:907–916. doi: 10.1104/pp.118.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason SJ. Expansins and cell wall expansion. J Exp Bot. 1995;46:1639–1650. [Google Scholar]
- McQueen-Mason SJ, Cosgrove DJ. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA. 1994;91:6574–6578. doi: 10.1073/pnas.91.14.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH. RNA localization: different zipcodes, same postman? Trends Cell Biol. 1998;8:381–383. doi: 10.1016/s0962-8924(98)01348-8. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Wittwer F, Mandel T, Kuhlemeier C. Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell. 1998;10:1427–1437. doi: 10.1105/tpc.10.9.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Uhnak KS. Tip growth in xylogenic suspension cultures of Zinnia elegans: implication for the relationship between cell shape and secondary cell wall pattern in tracheary elements. Protoplasma. 1998;204:103–113. [Google Scholar]
- Rose JKC, Lee HH, Bennett AB. Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA. 1997;94:5955–5960. doi: 10.1073/pnas.94.11.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seagull RW, Falconer MM. In vitro xylogenesis. In: Lloyd CW, editor. The Cytoskeletal Basis of Plant Growth and Form. London: Academic Press; 1991. pp. 183–194. [Google Scholar]
- Shcherban TY, Shi J, Durachko DM, Guiltinan MJ, McQueen-Mason SJ, Shieh M, Cosgrove DJ. Molecular cloning and sequence analysis of expansins: a highly conserved multi-gene family of proteins that mediate cell wall expansion in plants. Proc Natl Acad Sci USA. 1995;92:9245–9249. doi: 10.1073/pnas.92.20.9245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shininger TL. The control of vascular development. Annu Rev Plant Physiol. 1979;30:313–337. [Google Scholar]
- Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence waiting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth GJ, Redinbaugh MG, Scandalios JG. A procedure for the small scale isolation of plant RNA suitable for RNA blot analysis. Anal Biochem. 1988;172:279–283. doi: 10.1016/0003-2697(88)90443-5. [DOI] [PubMed] [Google Scholar]
- Weeks DL, Melton DA. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-beta. Cell. 1987;51:861–867. doi: 10.1016/0092-8674(87)90109-7. [DOI] [PubMed] [Google Scholar]
- Ye ZH, Varner JE. Gene expression patterns associated with in vitro tracheary element formation in isolated single mesophyll cell of Zinnia elegans. Plant Physiol. 1993;103:805–814. doi: 10.1104/pp.103.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]