Abstract
A β-d-glucan exohydrolase was purified from the cell walls of developing maize (Zea mays L.) shoots. The cell wall enzyme preferentially hydrolyzes the non-reducing terminal glucosyl residue from (1→3)-β-d-glucans, but also hydrolyzes (1→2)-, (1→6)-, and (1→4)-β-d-glucosyl units in decreasing order of activity. Polyclonal antisera raised against the purified exo-β-d-glucanase (ExGase) were used to select partial-length cDNA clones, and the complete sequence of 622 amino acid residues was deduced from the nucleotide sequences of the cDNA and a full-length genomic clone. Northern gel-blot analysis revealed what appeared to be a single transcript, but three distinct polypeptides were detected in immunogel-blot analyses of the ExGases extracted from growing coleoptiles. Two polypeptides appear in the cell wall, where one polypeptide is constitutive, and the second appears at the time of the maximum rate of elongation and reaches peak activity after elongation has ceased. The appearance of the second polypeptide coincides with the disappearance of the mixed-linkage (1→3),(1→4)-β-d-glucan, whose accumulation is associated with cell elongation in grasses. The third polypeptide of the ExGase is an extrinsic protein associated with the exterior surface of the plasma membrane. Although the activity of the membrane-associated ExGase is highest against (1→3)-β-d-glucans, the activity against (1→4)-β-d-glucan linkages is severely attenuated and, therefore, the enzyme is unlikely to be involved with turnover of the (1→3),(1→4)-β-d-glucan. We propose three potential functions for this novel ExGase at the membrane-wall interface.
The cell walls of grasses are compositionally different from those of all other flowering plants, and their growth mechanisms may be different as well (Carpita and Gibeaut, 1993; Carpita, 1996). In the commelinoid monocots (Rudall and Caddick, 1994), the major cross-linking polymers are feruloylated glucuronoarabinoxylans, and this framework is interlaced with much lower amounts of pectic substances than in dicots (Carpita, 1996; Jarvis et al., 1988). The (1→3),(1→4)-β-d-glucans (β-glucans) are found only in the Poales, a single order within the commelinoids that includes cereals and other grass species (Dahlgren et al., 1985; Smith and Harris, 1999). β-Glucan is unbranched, and about 90% of the polymer consists of cellotriosyl and cellotetraosyl units in a ratio of about 2.5:1, connected by single (1→3)-β linkages. The remainder comprises longer runs of cellodextrins interspersed along the polymer and also connected by single (1→3)-β linkages (Wood et al., 1994). β-Glucans are not synthesized in dividing cells, but accumulate specifically during cell enlargement and are then hydrolyzed extensively when growth ceases (Carpita and Gibeaut, 1993). Dissociation of cross-linking glycans from cellulose by expansins (Cosgrove, 1997) or the cleavage and religation of xyloglucans by xyloglucan endotransglycosylases (XETs; Nishitani, 1997) have been considered to be the principal mechanical determinants of cell expansion in dicots. However, the appearance and disappearance of the developmental stage-specific β-glucan during seedling growth in grasses indicate that the growth mechanism might be different in these species.
Over 3 decades ago, Lee et al. (1967) demonstrated that the walls of grasses were capable of autolysis, a term they coined to describe the self-hydrolysis of native walls in vitro and release of Glc into an incubation medium. We report here on the cloning of a gene that encodes the exo-β-d-glucanase (ExGase) responsible for this hydrolysis. Because the β-glucan accumulates during cell elongation and becomes the major cellulose cross-linking glycan, the hydrolysis of the β-glucan by the seedling exo- and endoglucanases has been considered to be necessary for wall expansion in the grasses (Hoson and Nevins, 1989). The ExGase and endo-β-d-glucanase activities are located in the seedling cell walls (Huber and Nevins, 1981). The endoglucanase hydrolyzes the β-glucan only at cellodextrin-rich regions containing four or more contiguous (1→4)-β-d-glucosyl linkages (Hatfield and Nevins, 1987). These cellodextrin sites are spaced roughly 50 glucosyl units apart in the β-glucan. Cleavage of the polymer into smaller fragments facilitates the complete hydrolysis to Glc by the seedling ExGase. The ExGase hydrolyzes both (1→4)-β and (1→3)-β linkages from the non-reducing end of β-d-poly- and oligosaccharides (Huber and Nevins, 1982).
In this paper, we describe the purification of the maize (Zea mays) cell wall ExGase and the characterization of its activity against native and artificial substrates. Although a vast majority of the enzyme is associated with the cell walls of developing maize seedlings where it autolyzes β-glucans, affinity-purified antibodies against the cell wall enzyme cross-react with an ExGase associated with the exterior surface of the plasma membrane. Unlike the wall-associated enzymes, the plasma-membrane ExGase is severely attenuated in its ability to hydrolyze non-reducing terminal (1→4)-β linkages, indicating a polysaccharide other than the (1→4)-linkage-rich β-glucan is the target of this ExGase isoform.
RESULTS
Purification of the ExGase
A summary of the purification steps is shown in Table I. A substantial enrichment of ExGase activity was achieved by purifying cell walls from total cytosolic proteins by filtration of the homogenate and by extensive washing with buffer containing 100 mm NaCl. Most of the ExGase was extracted from the cell walls by 3.5 m LiCl. Polysaccharides were also extracted, and the cationic cell wall proteins were isolated by SP-Sephadex chromatography. More than 63% of the original ExGase activity was recovered from the resin. To separate ExGase from other proteins, HPLC on an S-300 cation-exchange column recovered about 29% of the activity compared with the crude cell wall extract, resulting in a 7-fold increase in specific activity. Concanavalin A (Con A) chromatography to enrich for high-Man-containing glycoproteins increased the specific activity of ExGase 20-fold. After subsequent adsorption chromatography on hydroxyapatite, a single major peak of activity was observed. The peak containing ExGase activity exhibited a single band of about 64 kD in SDS-PAGE (Fig. 1). The single band of native protein, with an apparent pI of 7.2 after isoelectric focusing, was verified to be ExGase by in situ hydrolytic activity with p-nitrophenyl-glucopyranoside (not shown). ExGase increased 38-fold in specific activity with respect to the protein preparation from purified cell walls.
Table I.
Purification of the ExGase from the purified cell walls of developing maize seedlings
| Total Protein | Total Activity | Specific Activity | Purification | |
|---|---|---|---|---|
| mg | unitsa | units mg−1 | -fold | |
| Cell wall extractb | 26.2 | 1,845 | 70 | 1 |
| SP-Sephadex | 11.2 | 1,163 | 104 | 1.5 |
| S-300 HPLC | 1.10 | 538 | 489 | 7 |
| Con-A | 0.21 | 305 | 1452 | 21.7 |
| Hydroxyapatite | 0.11 | 290 | 2636 | 37.6 |
One unit is defined as the capacity to produce 1 μmol Glc mg−1 protein min−1.
Cell walls were purified from 1.5 kg of maize seedlings; the cell wall proteins were extracted with 3.5 m LiCl, the protein extract was concentrated to about 50 mL, and dialyzed against 20 mm sodium acetate, pH 5.0, containing 20 mm NaCl.
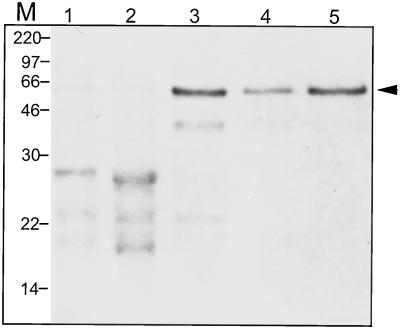
Figure 1.
The enrichment of ExGase by each purification step. Purification after each step as judged by Coomassie Blue-stained polypeptides after SDS-PAGE. M, Marker positions; lane 1, crude cell wall protein; lane 2, SP-Sephadex binding protein; lane 3, ExGase fraction from S-300 HPLC; lane 4, ExGase fraction from Con A column chromatography; lane 5, ExGase fraction from hydroxyapatite column chromatography. The same amount of protein (1 μg) was loaded in each lane.
Total cationic proteins from the cell wall were transferred to nitrocellulose and incubated with polyclonal antisera prepared against the purified ExGase. The polyclonal antisera had no detectable cross-reactivity with other LiCl-soluble cationic cell wall proteins except for the 64-kD polypeptide and a minor band just below it.
Selection of ExGase cDNA Clones
ExGase from maize seedlings was purified sufficiently by S-300 cation-exchange HPLC to be used in N-terminal sequencing after separation from other polypeptides by SDS-PAGE. The enzyme was digested with trypsin, and the resulting peptides were separated by reverse-phase HPLC. Four peptides gave discernible sequence, and the amino acid sequences from the largest of these peptides were used to design oligonucleotide probes to screen cDNA clones selected from an expression library prepared from maize seedling RNA. From a total of six putative cDNA clones selected from the expression library with the polyclonal antisera, two of the cDNAs (1.0 and 1.1 kb) hybridized with a degenerate 17-bp probe constructed to encode a pentapeptide within the largest of the tryptic fragments. These two clones were confirmed, by sequencing of the 3′ region, to be from the same transcript. The larger of the two was completely sequenced and found to contain a peptide sequence of a second tryptic fragment.
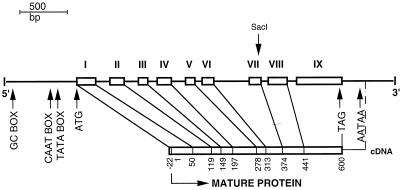
The nucleotide sequence from the cDNA clone represented roughly one-half of an expected full-length cDNA required to encode a 64-kD polypeptide. Reprobing the original cDNA library, as well as a second library obtained from another laboratory, revealed several additional clones, all smaller than or equal in size to the original. Furthermore, no products related to the clone were obtained by reverse transcription-PCR of freshly isolated poly(A+) RNA from maize seedlings using several universal primers in tandem with primers based on the known sequence. We concluded that the secondary structure of the transcript prevented the synthesis of cDNA from the N-terminal portion of the transcript. However, we were able to select genomic clones that provided the full-length sequence, the 5′-untranslated region, and about 1,500 bp of the promoter region (Fig. 2). The genomic sequence contains nine exons, with the eight introns varying between 85 and 304 bp in length. All of the exon/intron junctions follow the GT/AG rule (Goodall and Filipowicz, 1989). The polyadenylation signal AATAA is situated 220 bp from the TAG stop codon. An untranslated region of 682 bp upstream from the 5′ end of the open reading frame contains a putative TATA box situated 126 bp upstream from the ATG start codon. A CAAT box and a GC box, GGGCGG, are located 175 and 471 bp, respectively, upstream from the ATG start signal. A 22-amino acid putative signal peptide of the primary translation product has a predicted cleavage site preceding the first Glu residue to yield a mature protein of 600 amino acids (Fig. 3A). The four tryptic peptides that were microsequenced are represented in the genomic clone, including the peptide EYLK, which represents the N terminus of the mature protein. The mature protein has a monoisotopic molecular mass of 64,613 D.
Figure 2.
Gene organization of ZmEXG1. The boxes represent the approximate size of the nine exons (numbered I–IX), and their end points are indicated in relation to the amino acid of the mature protein. Arrows indicate the positions of GC box (GGGCGG), CAAT box, TATA box, the translational start and end codons, and the potential polyadenylation signal. The position of the SacI restriction site that was exploited to obtain the complete genomic sequence is also indicated. The genomic sequence of the maize ExGase is accession number AF225411.
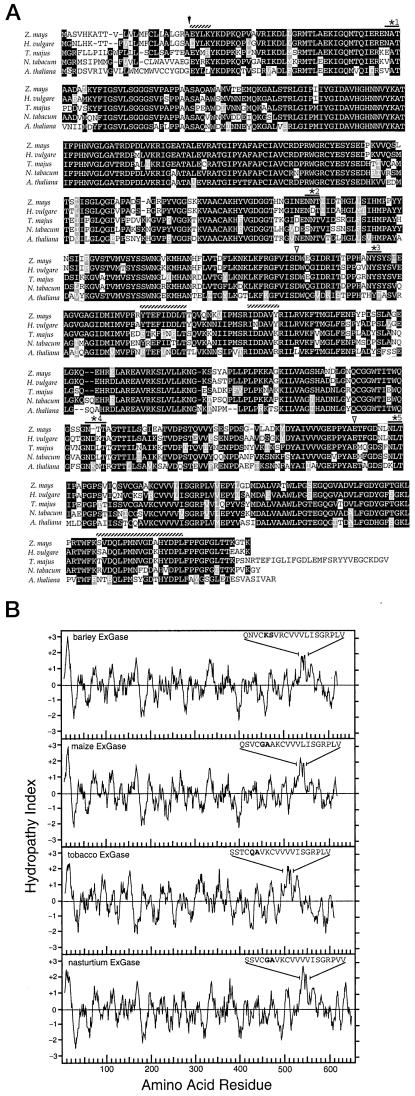
Figure 3.
Comparison of the amino acid sequence and hydropathy indices of the maize, barley, nasturtium, tobacco, and Arabidopsis ExGases. A, The predicted cleavage site of the signal peptide is marked by the black arrow, the sequences of the four tryptic peptides are marked with hatched lines, and the five potential glycosylation sites are starred. The predicted nucleophiles Asp-285 and Glu-492 are marked with white arrows. Whereas the maize ExGase shares potential glycosylation sites 1, 3, and 5 with the barley ExGase HVU46003, it shares only sites 2 and 5 with a barley isoform with the glycosylation sites confirmed by three-dimensional structure (Varghese et al., 1999). B, The hydropathy plots were calculated according to the Kyte-Doolittle parameters with an amino acid range of 11. The sequences of amino acids 531 through 550 for barley ExGase (24; HVU46003) and 529 through 548 for the maize ExGase show the substitution of Gly-533 and Ala-534 in the maize enzyme for Lys-535 and Ser-536 for the barley enzyme in bold. Sequences 496 through 515 encoded by the Arabidopsis homolog (AB008271, clone MUK11) contain Gln-500 and Ala-501, and amino acids 534 through 553 for nasturtium ExGase (AJ006501) contain Gly-538 and Ala-539, in those positions and are also in bold.
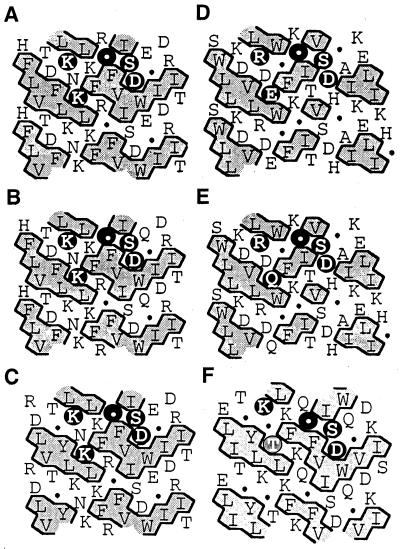
The predicted pI of the mature protein is 6.9. The hydropathy plot of the deduced amino acid sequence of the primary translation product shows, in addition to the strong signal peptide, a 20-amino acid hydrophobic motif between amino acids 528 and 549, 80 amino acids upstream from the C terminus (Fig. 3B). Hydrophobic cluster analysis (HCA) reveals a [KR]-x-[EQK]-xxxx-G-xxx-[ST]-D motif characteristic of family 3 glucosidases (Fig. 4).
Figure 4.
Comparison of HCA of the potential domain containing the aspartyl nucleophile of maize and barley ExGases with those of four family 3 glycosidases with homology to the maize ExGase open reading frame. In the HCA plots, the sequences of proteins are arranged in a duplicated α-helical net, and clusters of contiguous hydrophobic amino acids are shaded. The amino acids conforming to the [KR]-x-[EQK]-xxxx-G-xxx-[ST]-D motif are highlighted with black circles in the upper α-helical net. The Thr residue in the Arabidopsis analysis that does not conform to the family 3 motif is shaded. Gly is represented by a black circle in these representations. A, Maize ExGase; B, barley ExGase (Hrmova et al., 1996); C, nasturtium ExGase (Crombie et al., 1998); D, Salmonella typhimurium β-glucosidase (T-cell inhibitor; D86507); E, Escherichia coli periplasmic β-glucosidase (U15049); and F, Arabidopsis ExGase (AB008271, clone MUK11).
Enzyme Properties of the Cell Wall ExGase
The cell wall ExGase specifically hydrolyzed terminal β-d-glucopyranoside, with little activity toward terminal α-d-glucopyranoside, and no activity against eight other β-glycosides (Table II). The enzyme was active over a broad temperature range, with greater than one-half maximal activity between 25°C and 50°C. The pH dependence of the enzyme activity was relatively broad. Greater than one-half maximal activity of the ExGase was observed in the range pH 3.5 and 6.5, with a pH optimum of 4.8. The Km and Vmax for the hydrolysis of p-nitrophenyl β-d-glucopyranoside by the purified ExGase were derived from the measurement of reaction velocity at different concentrations of substrate. The Km was 0.89 mm, and Vmax was 13.4 μmol mg−1 min−1 (Table II). At substrate concentrations below 1 mm, the ExGase hydrolyzed all four β-d-glucosyl disaccharides but at different rates. The Kms of the 2-, 4-, and 6-linked β-d-glucosyl disaccharides were 66, 53, and 68 μm, respectively, whereas that for laminaribiose was 128 μm. In contrast, the Vmax was highest for laminaribiose at 9.1 μmol mg−1 min−1, with relative Vmax for cellobiose about one-half that of laminaribiose; sophorose and gentiobiose had intermediate rates of 7.8 and 6.3 μmol mg−1 min−1, respectively. Laminaridextrins of tri- to pentasaccharide were hydrolyzed at rates lower than laminaribiose, whereas the cellodextrins were hydrolyzed at rates slightly higher than cellobiose.
Table II.
Substrate specificity and kinetics of the ExGase from developing maize seedlings
| Substrate | Km | Vmax |
|---|---|---|
| μm | μmol mg−1 min−1 | |
| p-Nitrophenyl-β-d-glucoside | 886 | 13.40 |
| p-Nitrophenyl-α-d-glucosidea | n.d.b | 0.04 |
| Sophorose | 66 | 7.75 |
| Laminaribiose | 128 | 9.09 |
| Cellobiose | 53 | 3.96 |
| Gentiobiose | 68 | 6.30 |
| Laminaritriose | n.d. | 6.82 |
| Laminaritetraose | n.d. | 6.25 |
| Laminaripentaose | n.d. | 5.43 |
| Cellotriose | n.d. | 4.49 |
| Cellotetraose | n.d. | 4.33 |
| Cellopentaose | n.d. | 4.41 |
All substrates were tested at initial amounts of saturating concentrations of reducing ends (2 mm). The hydrolyzed Glc was determined by enzymatic assay, whereas the color produced by p-nitrophenyl-β-d-glucoside was corrected for Glc equivalents. Values for Glc in disaccharides was halved, as one hydrolysis event gave rise to two molecules of d-Glc, correction for cleavage of disaccharide products from higher order laminari- and cellodextrin oligosaccharides was based on HPLC of the digestion products as described in Figure 5.
No activity was obtained with p-nitrophenyl-β-d-galactopyranoside, -α-d-galactopyranoside, -β-d-mannopyranoside, -β-d-xylopyranoside, -α-l-arabinopyranoside, -α-l-arabinofuranoside, -β-l-fucopyranoside, or -α-l-rhamnopyranoside.
Not determined.
At concentrations above 2 mm, the cell wall ExGase exhibited glucosyl transferase activity and generated tri- and tetrasaccharides from disaccharides. The primary products of the transglycosylation were (1→6)-linked and (1→4)-linked mixed-linkage oligomers (Fig. 5). Transferase activity with laminaribiose yielded primarily mixed-linkage tri- and tetraglucosides, and after incubations overnight with laminaribiose, gentiobiose and cellobiose were the major disaccharides remaining (Fig. 5B).
Figure 5.
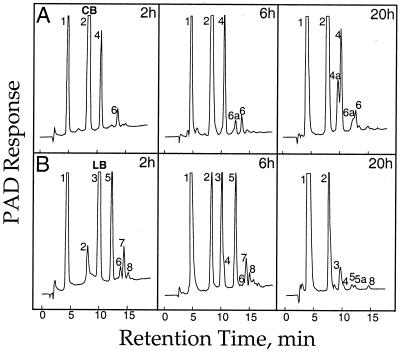
High-pH anion-exchange-HPLC separation of the products of transglucosylase reactions catalyzed by the ExGase. A purified ExGase preparation in 20 mm sodium acetate, pH 5.0, was added to 20 mm laminaribiose or cellobiose, and the reaction products were followed at various intervals. The samples shown here were taken at 2, 6, and 20 h after initiation of the reaction. Relative retention times of the oligomers were based on standards of sophorose, gentiobiose, a cellodextrin series to cellopentaose, a laminaridextrin series to laminaritetraose, and mixed-linkage tri- and tetraglucosides from B. subtilis endoglucanase digests of authentic β-glucan (Sigma). The laminaribiose and cellobiose substrates were >98% pure, with Glc the only contaminant detected by HPLC (not shown). The oligomers were separated by high-pH anion-exchange-HPLC on a 0.4- × 25-cm Dionex PA-1 column in a linear gradient of 0 to 200 mm sodium acetate in 0.5 n NaOH, at 1.0 mL min−1, and detected by pulsed amperometry. Peaks identified by retention time and gas-liquid chromatography-electron impact mass spectrometry of their partly methylated alditol acetates are: 1, Glc; 2, gentiobiose and cellobiose; 3, laminaribiose and sophorose; 4, gentiotriose; 4a, β-d-Glc-(1→6)-β-d-Glc-(1→4)-β-d-Glc; 5, β-d-Glc-(1→6)-β-d-Glc-(1→3)-d-Glc; 5a, not determined; 6, cellotetraose; 6a, β-d-Glc-(1→6)-β-d-Glc-(1→4)-β-d-Glc-(1→4)-d-Glc; 7, β-d-Glc-(1→6)-β-d-Glc-(1→4)-β-d-Glc-(1→3)-d-Glc; 8, laminaritetraose. A, Separation of the reaction products with cellobiose as initial substrate. B, Separation of the reaction products with laminaribiose as initial substrate. Electron-impact mass spectra that verify the linked sugars present in the indicated peaks (as shown in Fig. 11).
Expression of ExGase Genes
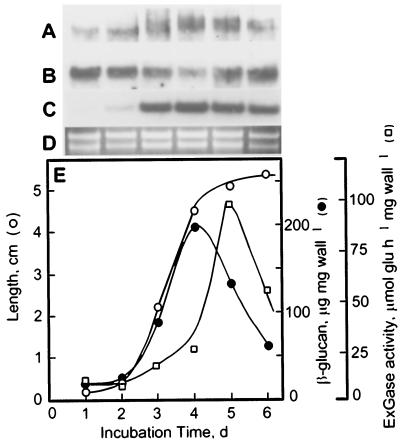
Tissue prints of the coleoptile and mesocotyl immunolabeled with polyclonal antisera against the ExGase showed the enzyme to be concentrated in the basal regions of the coleoptile and in the growing region of the mesocotyl (Fig. 6). The β-glucan content of the coleoptile wall increased to its highest proportion at the maximum rate of elongation and then decreased markedly as growth culminated (Fig. 7E). The loss of β-glucan is correlated with a marked increase in activity of the ExGase extracted from the wall by LiCl. Immunogel-blot analyses of cell wall ExGase indicated that polypeptides of two distinct sizes were present during cell elongation of the coleoptile. The smaller of the two with a molecular mass of 62 kD is constitutive and present immediately after imbibition, whereas a larger polypeptide with a molecular mass of 64 kD appears during later stages of elongation, consistent with the onset of the decrease in β-glucan content (Fig. 7A). Northern gel-blot analyses demonstrated that the ExGase transcripts were abundant soon after imbibition, decreased slightly during the maximum rate of elongation, and increased once again at the late stages of growth (Fig. 7B). In contrast, transcripts of profilin5, an actin-associated protein, displayed an abundance that is expected of a gene whose expression is correlated with growth rate (Fig. 7C).
Figure 6.
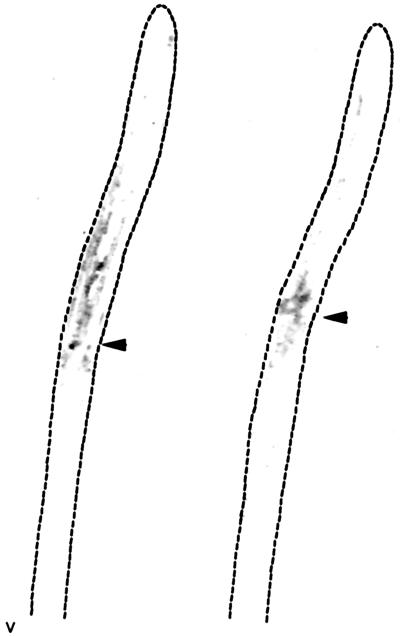
Tissue prints of coleoptile and mesocotyl tissues show presence of ExGase in basal portions of the coleoptile and elongation zone of the mesocotyl. Sections of 2- and 3-d-old etiolated seedlings were pressed against a cellulose nitrate, which had been soaked with 3.5 m LiCl in 20 mm sodium acetate, pH 5.5, and air-dried before blotting. The dotted lines show the outline of the seedling. The ExGase was probed with polyclonal antisera (1:200 dilution) and detected with alkaline phosphatase-conjugated rabbit anti-mouse antisera (1:3,000 dilution). The arrowheads denote the junction of coleoptile and mesocotyl.
Figure 7.
Expression of the ExGase transcripts during the course of coleoptile elongation. The maize seeds were soaked for 24 h in water at 30°C in darkness bubbled with air and then sown in beds of vermiculite for up to 6 d under the same conditions. The unexpanded leaves within the coleoptiles from d 1 and 2 could not be removed. The coleoptiles were frozen in liquid nitrogen and stored at −80°C until RNA, protein, and cell wall material was isolated. A, Immunogel-blot analysis of total ExGase extracted from the cell walls. All lanes contain 1 μg of wall protein and were probed with affinity-purified ExGase antibodies (1:500 dilution) recovered from the smaller molecular mass cell wall polypeptide. B, Northern gel-blot analysis of ExGase. C, Northern gel-blot analysis of profilin5. D, Ethidium bromide-stained blot showing the quality and loading of RNA, based on the intensity of the ribosomal RNAs. All lanes contained 10 μg of total RNA. E, Comparison of the coleoptile length with amount of β-glucan per milligram of cell wall material. The β-glucan was assayed enzymically as described (Gibeaut and Carpita, 1993) and activity of ExGase was determined with laminaribiose substrate from LiCl extracts of the cell wall.
Association of an ExGase with the Plasma Membrane and Cell Wall
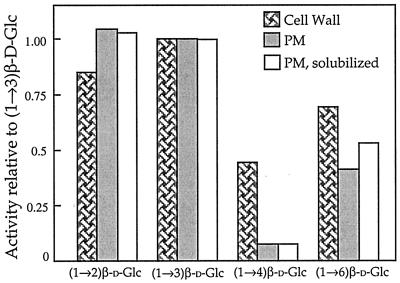
The predicted hydrophobic motifs in the deduced amino acid sequences of ExGases indicated that these enzymes may associate with the plasma membrane as well as the cell wall. ExGase activity was indeed associated with plasma membranes, which were enriched by two-phase aqueous partitioning (Fig. 8), and the enzyme was also detected at the exterior of the plasma membranes of protoplasts with affinity-purified antibodies against the purified cell wall ExGase (Fig. 9, G–L). The membrane-associated ExGase exhibited activity against (1→2)-, (1→3)-, and (1→6)-linked disaccharides in a similar manner as the cell wall ExGase, but activity against (1→4)-linked disaccharides was severely attenuated (Fig. 8). Two-phase detergent partitioning with Triton-X-114 showed that the membrane association was extrinsic (Fig. 10). The molecular mass of the membrane-associated ExGase was similar to the smaller, constitutive polypeptide of ExGase found in the cell wall.
Figure 8.
Disaccharide hydrolysis activities of cell wall and plasma membrane ExGase. Cell wall ExGase activities were measured after extraction from the wall with 3.5 m LiCl and chromatographic purification. The plasma membrane ExGase activities were measured in total membranes from the upper phases collected from the two-phase aqueous system resuspended in sodium acetate, pH 5.5, containing 20 mm NaCl. The plasma membrane soluble ExGase activities were measured in aqueous fractions after two-phase detergent partitioning. Absolute activities were compared relative to β(1→3)-linked (laminaribiose) hydrolase activity, which is the highest activity of cell wall ExGase.
Figure 9.
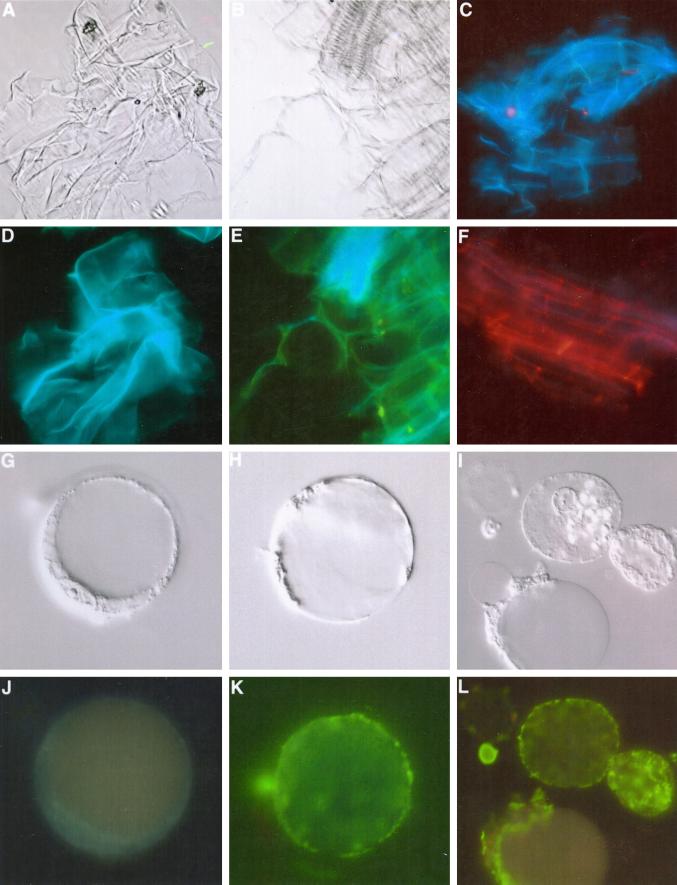
Immunolocalization of the ExGase in cell walls and at the exterior surface of the plasma membrane. Cell walls and protoplasts were probed with affinity-purified ExGase antibodies recovered from the smaller molecular mass cell wall polypeptide (Fig. 7). Isolated cell walls of maize exhibit a bluish-green autofluorescence when irradiated with UV light, whereas the FITC-conjugated secondary antibodies possess a distinctive yellow-green fluorescence when irradiated with blue light. Alternatively, TRITC-conjugated antibodies have a red fluorescence when irradiated with blue-green light. A and B, Isolated walls viewed in differential interference contrast microscopy. C, Walls incubated with preimmune sera or FITC-conjugated antisera alone (shown here) are autofluorescent but unlabeled. D, Walls in A when irradiated with UV light are autofluorescent due to the enrichment of hydroxycinnamic acid esters and ethers in Type II walls (Carpita and Gibeaut, 1993). E, Walls in B when irradiated with UV and blue light after immunolabeling with the ExGase antibodies and FITC-conjugated secondary antibodies exhibit wall autofluorescence and FITC fluorescence. F, Walls immunolabeled with ExGase antibodies and detected by TRITC-conjugated secondary antibodies after irradiation with UV and blue-green light. (Legend continues on facing page.)G through I, Protoplasts isolated from coleoptiles were separated from intact cells and broken protoplasts by flotation centrifugation in Ficoll gradients and resuspended in protoplast medium. Each is viewed by differential interference contrast microscopy. J, Protoplast shown in G probed with FITC-conjugated antisera alone exhibit no fluorescence when irradiated with UV and blue light. K, Protoplast shown in H probed with ExGase antibodies exhibit strong surface labeling with FITC-conjugated secondary antisera when viewed with UV and blue irradiation. L, Three protoplasts of I probed with ExGase antibodies exhibit strong surface labeling with FITC-conjugated secondary antisera. The upper protoplast remains intact, whereas a second is just beginning to crenate but still exhibits only surface labeling, and the third protoplast has just burst to release the vacuoles but the labeling remains with the plasma membrane as the cytoplasm coagulates to one side.
Figure 10.
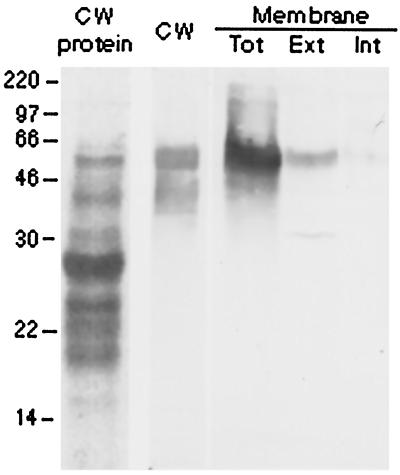
An immunogel-blot analysis of total cell wall proteins from 4-d-old coleoptiles, and plasma membranes from 3-d-old coleoptiles, with affinity-purified ExGase antibodies. Lane 1 (CW protein) shows Coomassie Blue-stained cell wall polypeptides, and lanes 2 through 5 show immunogel blots with anti-ExGase antisera. Lane 2, Cell wall polypeptides (CW); lane 3, total plasma membrane (Tot); lane 4, aqueous-soluble extrinsic polypeptides (Ext); lane 5, detergent-soluble intrinsic polypeptides (Int).
The membrane-associated ExGase retained activity after recovery from the aqueous phase, but showed no increase in relative activity against cellobiose (Fig. 8). Whereas the cell wall enzyme demonstrated substantial transglycosylase activity at substrate concentrations above 2 mm, the membrane-associated activity did not (Fig. 11). The affinity-purified antibodies against the wall ExGase labeled cross walls and longitudinal walls of maize coleoptiles equally (Fig. 9F).
Figure 11.
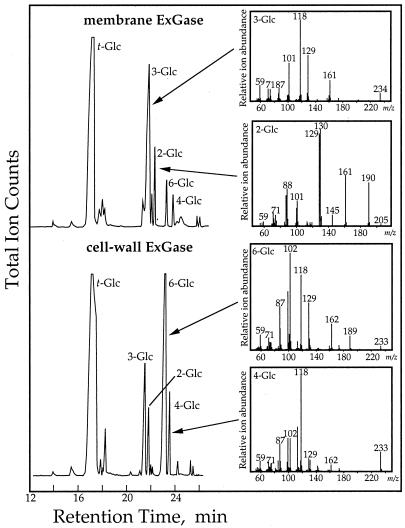
Products of transglycosylase reactions with cell wall- and plasma membrane-associated ExGase. Cell wall and plasma membrane ExGases were incubated in 20 mm laminaribiose at 37°C for 20 h. Portions of the incubations were separated by high-pH anion-exchange-HPLC as illustrated in Figure 5, and the remainders were freeze-dried and subjected to methylation analysis with n-butyllithium and methyl iodide (Gibeaut and Carpita, 1993). Linkage analysis was based on electron-impact mass spectra as described by Carpita and Shea (1989). Key masses that identify the linked Glc units are m/z 190 for 2-linked Glc, m/z 234 for 3-linked Glc, m/z 233 for 4-linked Glc, and m/z 189 and 233 for 6-Glc.
DISCUSSION
The Cell Wall ExGase Has Both Hydrolase and Transferase Activities
The purification of the seedling cell wall ExGase was simplified greatly by virtue of its tenacious binding to the wall. By isolating cell walls, the enrichment of ExGase was at least 20-fold at the onset of the column purifications. High-performance cation-exchange chromatography and Con A affinity chromatography enriched the enzyme to near homogeneity. Although a single major activity associated with hydrolysis of non-reducing terminal β-d-linked glucosyl units was observed in our extractions of the total cationic proteins from the walls of developing shoots, there appeared to be a shoulder of ExGase activity tailing the major peak. The purified enzyme gave a single band by SDS-PAGE after hydroxyapatite chromatography (Fig. 1). Polyclonal antisera recognized primarily a single polypeptide in total protein extracts from the wall but faintly detected a second polypeptide of slightly smaller molecular mass (not shown). In contrast, extraction of freshly isolated walls directly with SDS-PAGE sample buffer revealed two distinct polypeptides of the ExGase (Figs. 7 and 10). Isoelectric focusing revealed a single broad band with an apparent pI of 7.2. These results are consistent with reports of ExGase activity in maize coleoptiles associated with a single polypeptide of about 70 kD in SDS-PAGE and western gel blots (Hoson and Nevins, 1989). Northern gel-blot analyses also show a single broad band that may include several independent and unresolved transcripts, but at the present time we cannot determine if the two polypeptides of the cell wall are from different genes or represent post-translational processing of the same polypeptide. In contrast, Hrmova et al. (1996) reported that at least four barley β-d-glucosidase activities were well resolved by cation-exchange chromatography; they purified two isoforms of ExGase similar to the maize enzyme reported here and two isoforms of β-d-glucosidase. Whereas we extracted cell walls only from developing shoots, Hrmova et al. (1996) purified glucosidases from the entire germinating seedling, including the endosperm, which was undergoing complete digestion of its cell walls. Further, the β-d-glucosidases were similar in activity to those of the germinating seeds of barley as reported by Leah et al. (1995). The maize ExGase reported here is strictly associated with growing cells of the developing seedling and not the endosperm.
The differences in activity between ExGases and the β-d-glucosidases are substantial. The ExGase has a strict requirement for non-reducing terminal β-d-glucosyl units but can cleave the terminal sugar from the four possible β-linkage positions. The enzyme can preferentially hydrolyze 3-linked units over 4-linked units, but the enzyme will completely digest polymeric fragments of β-glucan to Glc once the longer runs of cellodextrins within the macromolecule have been cleaved by a seedling-specific endo-β-d-glucanase. In contrast, the β-d-glucosidase I hydrolyzes both β-d-glucosyl and mannosyl p-nitrophenyl glycosides, and preferentially hydrolyzes the terminal unit of 3- and 4-linked oligomers, but is inactive against 6-linked oligomers (Leah et al., 1995). The second isoform of the β-d-glucosidase is essentially inactive against 3-linked oligomers and preferentially digests the cellodextrins (Hrmova et al., 1996).
Transglycosylase activities at high substrate are well documented for numerous exo- and endohydrolases (Henrissat, 1991), and, consistent with the observations of Hrmova and Fincher (1998), the ExGase can convert oligomers of one linkage type to another, i.e. convert laminaridextrins and sophorose to gentiobiose and cellodextrins, but not vice versa (Fig. 5, A and B). The propensity to accumulate gentio- and cellodextrins may simply arise from the fact that they are the poorest substrates; the transglycosylase activity may be random, but the cellodextrins may accumulate because the enzyme exhibits the lowest turnover rates with these substrates.
The Cell Wall ExGase Is a Family 3 Glycosidase
The transcript from the maize ZmEXG1 encodes a 622-amino acid polypeptide. The deduced sequence of the maize ExGase shares an 86% amino acid identity with a barley seedling ExGase (Fig. 3A), which is reflected in the similarity of their hydropathy indices (Fig. 3B). Two additional ExGases from Arabidopsis and tobacco with substantial amino acid identity were found in the database, and another from cotyledons of nasturtium seedlings has also been characterized (Crombie et al., 1998). The maize enzyme contains a 22-amino acid signal peptide with a predicted cleavage site between an Ala and Glu residues. This prediction is supported by the presence of a Glu-Tyr-Leu-Lys peptide from N-terminal sequencing of tryptic peptides of the mature protein.
The predicted pIs of these related ExGases vary widely, from greater than 9.5 for the nasturtium enzyme (Crombie et al., 1998) to 7.9 for the barley enzyme to 6.9 for the mature maize enzyme. These predictions are consistent with a pI of 8.0 empirically determined for the barley ExGase by chromatofocusing and a pI of 7.2 for the maize enzyme by isoelectric focusing in polyacrylamide gels (not shown).
Despite the deduced homology between the ZmEXG1 and other plant ExGase cDNAs, the native substrates and relative activities against β-linked substrates are different. The maize and barley ExGase are both able to cleave the non-reducing terminal sugar from the mixed-linkage β-glucan, and can also cleave the terminal β-d-linked glucosyl unit from each of the four possible disaccharides and the cellodextrin and laminaridextrin series. However, cleavage of (1→4)-β-d-glucosyl terminal units is substantially higher in the nasturtium ExGase than in the cereal enzymes. As Hrmova et al. (1996) noted, because the ExGases displayed highest activity toward the (1→3)-β-linkage, they belong to the EC 3.2.1.58 group (Cline and Albersheim, 1981; Labrador and Nevins, 1989).
The maize ExGase shares some sequence similarity with some more primitive extracellular β-d-glucosidases such as an 869-amino acid β-d-glucosidase (EC 3.2.1.74) from Pseudomonas fluorescens (Rixon et al., 1992), an 822-amino acid β-d-glucosidase (EC 3.2.1.21) from Dictyostelium discoideum (Bush et al., 1994), and a β-d-glucosidase (EC 3.2.1.4) from an unidentified bacterium (Healy et al., 1995). Nevertheless, the motif [KR]-x-[EQK]-xxxx-G-xxx-[ST]-D in a hydrophobic cluster places the maize ExGase in the family 3 group of glucosidases (Henrissat, 1991). This conclusion is strengthened by an HCA (Henrissat et al., 1995), which predicts that the Asp-286 of the mature protein is the nucleophile of the active site (Fig. 4). In contrast, the Arabidopsis ExGase contains a similar motif but is missing the consensus [EQK] amino acid in what is otherwise well-conserved sequence among the five ExGases (Figs. 3A and 4). Three-dimensional structural analysis of another barley ExGase isoform shows that the expected nucleophilic Asp-285 and Glu-491 reside in a pocket predicted to be the catalytic domain (Varghese et al., 1999). The three-dimensional structure of a barley ExGase also shows that three of the five potential glycosylation sites bear carbohydrates.
The Membrane-Associated ExGase Is a Distinct Enzyme
The cereal ExGases display hydrophobic runs between amino acids 528 and 549 (Fig. 3), which are predicted to be weak membrane anchors (Horton and Nakai, 1996, 1997). The Arabidopsis and nasturtium ExGases also contain the hydrophobic regions that indicate possible membrane association (Fig. 3). Of the three, nasturtium has the highest hydrophobicity, peaking at over +2.8. Although the prediction of the weak membrane anchor in the deduced amino acid sequence of the ExGase prompted us to look for glucosidase activity in the plasma membrane, the sequence of the gene we cloned encodes the cell wall enzyme. The four peptides sequenced from the cell wall ExGase are each identical to those predicted in the deduced amino acid sequence of the cloned gene (Fig. 3A). Two additional partial-length cDNAs obtained from seedling transcripts were also examined, and neither of them match these four peptide sequences (data not shown). We surmised that the plasma membrane activity could result from transient association of the cell wall ExGase, and that the selective attenuation of the (1→4)-β-d-glucosyl hydrolase activity (Fig. 8) is a result of its membrane-directed conformation. However, the membrane-associated activity is aqueous-soluble after two-phase detergent partitioning (Fig. 10), and the (1→4)-β-d-glucosyl hydrolase activity is not recovered after this extraction (Fig. 8). Whereas the cell wall ExGase exhibits significant transglycosylase activity (Figs. 5 and 11), the membrane-associated ExGase demonstrates negligible activity (Fig. 11). Even though they cross-react with the same affinity-purified antibodies directed against purified cell wall ExGase, we conclude that the membrane-associated enzyme is a different gene product whose gene we have yet to identify.
Wall- and Membrane-Associated ExGases Must Have Different Functions during Cell Growth
Early studies on the enzymatic activities of the primary cell wall associated with elongation in grasses and dicots were made before there was widespread appreciation for the differences in structure of the cross-linking molecules and cell wall architecture (Taiz, 1984). Only species in the Poales, the order containing the grass family, contain the β-glucans (Smith and Harris, 1999). The synthesis of the β-glucans only during cell expansion is a special feature of the grasses, indicating that the polymer is at least associated with the mechanism of cell expansion. Growth in vivo is accompanied by a net accumulation of the β-glucan (Fig. 7) that masks an extremely rapid turnover (Gibeaut and Carpita, 1991). The β-glucan is gradually lost from the wall once growth and synthesis of new β-glucan ceases (Fig. 7). Hence, the synthesis and hydrolysis of the β-glucan are in dynamic equilibrium. The correlation factor for β-glucan and growth is not the relative abundance, but rather the rate of turnover, the balance of synthesis, and degradation (Gibeaut and Carpita, 1991). A long-held idea is that the β-glucans that cross-link the cellulose microfibrils are cleaved by an endo-β-d-glucanase and the turgor pressure of the cell expands the loosened microfibrils (Huber and Nevins, 1981; Taiz, 1984). The appearance of endo-β-d-glucanase and ExGase in the cell walls during cell growth and the ability of antisera directed against these enzymes to inhibit growth strengthen this hypothesis (Hoson and Nevins, 1989).
The discovery of XET (Pritchard et al., 1993; Wu et al., 1994) and expansins (Li et al., 1993) in growing grass tissues has reopened the question of the necessity of glucan hydrolysis during growth and, consequently, in the function of the glucan hydrolases. The activity of neither expansin nor XET involves a net breakage of glycosidic linkages (Cosgrove, 1997; Nishitani, 1997). The endoglucanase- and ExGase-catalyzed turnover of β-glucan may occur subsequent to the physical mechanism of wall loosening required for expansion and may be part of a sugar recycling mechanism that occurs after growth. Our expression and activity studies support this idea. Transcripts are more abundant very early and very late during cell elongation (Fig. 7), but the appearance of the second wall-associated ExGase protein and extractable activity coincide only with the later expression. Hence, the appearance of the enzyme in the wall is not strictly correlated with growth but rather the turnover of the β-glucan that occurs after growth has ceased. In contrast, transcript levels of profilin5, an actin-associated protein, demonstrate the abundance expected of a growth-correlated gene (Fig. 7). The ExGase is also not unique to the grasses. Very similar enzymes appear in species with Type I walls, such as tobacco, Arabidopsis, and nasturtium—species that contain no β-glucan but have xyloglucan as the major cross-linking polysaccharide (Crombie et al., 1998).
The presence of a hydrophobic motif in the ExGase genes is puzzling. The more abundant ExGase is associated with the cell wall and not the plasma membrane. The apparent membrane anchor could partition the ExGase away from its substrate during synthesis of β-glucan in the Golgi apparatus and co-packaging into secretory vesicles. Local pH, ionic environment, or other physiological conditions at the wall-membrane interface may control membrane binding or release to the cell wall. Nevertheless, an ExGase is associated tightly with the plasma membrane (Figs. 9, G–L, and 10) that possesses an alteration that specifically attenuates activity against cellodextrins (Fig. 8) and essentially lacks transglycosylase activity (Fig. 11). These data indicate that the membrane-associated ExGase is a different enzyme with a different function.
We thought of three possible functions of such an enzyme at the cell wall-plasma membrane interface. First, the enzyme may function coordinately with the pathogenesis-related endoglucanase as part of the defense against fungal pathogens (for review, see Boller 1993). Oligomers hydrolyzed from fungal (1→3)-β-d- and (1→6)-β-d-glucans would be converted to Glc, and the lack of (1→4)-β-hydrolase activity would ensure that no plant cell wall glucan is targeted for hydrolysis. As the fungal oligomers have been shown to possess elicitor activity, the ExGase would diminish this activity by removal of the effector. A related phenomenon is the removal of the 3-linked glucan callose that is made transiently during cell plate formation (Samuels et al., 1995) or as callose-plugs that are associated with the blockage of penetration of fungi or in other membrane wounding. The membrane-associated ExGase with high activity toward 3-linked glucans could function in this turnover. Third, the ExGase could be associated with cellulose biosynthesis, which occurs at the wall-membrane interface. Endoglucanases with membrane anchors also have been discovered recently in tomato (Brummell et al., 1997) and Agrobacterium (Matthysse et al., 1995). The bacterial enzyme is required for cellulose synthesis, and a mutation in expression of an Arabidopsis endoglucanase related to the tomato enzyme results in severe dwarfism accompanied by a decrease in the normal cellulose content (Nicol et al., 1998). Brummell et al. (1997) indicated that the membrane-associated endoglucanase functions in cellulose synthesis. Chapple and Carpita (1998) suggested that this endoglucanase may proofread the glucan chains and excise any mistake linkages. The membrane-associated ExGase may assist in this repair by removing any non-reducing terminal Glc linkages other than (1→4)-β. Regardless of function at the membrane, the presence of a unique hydrolase at the cell wall-membrane interface demands an evaluation of the maize and other plant ExGases for functions in addition to depolymerization of growth-related β-glucans and xyloglucans.
MATERIALS AND METHODS
Purification of the Cell Wall ExGase
Maize (Zea mays L.) seeds were soaked overnight in running deionized water, sown in trays of moist vermiculite, and incubated in the dark at 30°C. The shoots of 4-d-old etiolated seedlings, including mesocotyl, coleoptile, and primary leaves, were harvested and used for enzyme isolation. The maize shoots were homogenized in ice-cold extraction buffer (50 mm sodium citrate, 50 mm NaCl, 30 mm ascorbic acid, 1 mm dithiothreitol [DTT], and 0.1 mm phenylmethylsulfonyl fluoride, pH 6.5), filtered through four layers of cheesecloth, and the walls were washed extensively with citrate buffer (50 mm sodium citrate and 50 mm NaCl, pH 5.5), followed by 20 mm NaCl, and then deionized water. Cell walls were purified from cytosolic and membrane contaminants by sequential homogenizations in acetone, 100 mm NaCl, and water, and the walls were collected on nylon cloth after each step. The walls were washed with 20 mm NaCl and deionized H2O until the effluent was completely clear. The cell walls were then suspended, stirred overnight in ice-cold 3.5 m LiCl in 20 mm sodium acetate, pH 5.0, and 20 mm NaCl to release the basic cell wall proteins, and the extract was filtered. The filtrate was concentrated to about 50 mL in dialysis bags, overlaid with polyethylene glycol 8,000, and then dialyzed against 20 mm sodium acetate and 20 mm NaCl, pH 5.0. The filtrate was centrifuged at 10,000g for 10 min to remove precipitated material, and SP-Sephadex cation-exchange resin equilibrated with 20 mm sodium acetate and 20 mm NaCl, pH 5.0, was added to the cleared filtrate. The resin with bound proteins was washed with 20 mm sodium acetate and 20 mm NaCl, pH 5.0, and the cell wall proteins were eluted with 0.7 m NaCl in 20 mm sodium acetate and dialyzed against 20 mm sodium acetate and 20 mm NaCl, pH 5.0.
The cell wall proteins binding to SP-Sephadex (15 mg in 50 mL) were loaded onto an S-300 cation-exchange column (Synchropak, Lafayette, IN); the bound proteins were washed with 50 mL of loading buffer and eluted in a 30-mL linear gradient to 0.6 m NaCl in the 20 mm sodium acetate buffer, pH 5.0. Fractions with ExGase activity were pooled and applied to a Con A-Sepharose column (1 × 4 cm) equilibrated in 50 mm sodium acetate, pH 7.0, 2 mm MnCl2, and 2 mm CaCl2. The column was washed with the same buffer and the bound proteins were eluted in a 100-mL linear gradient of methyl β-d-mannopyranoside to 2 mm. The fractions with ExGase activities were pooled and loaded onto a hydroxyapatite column equilibrated with 10 mm sodium phosphate, pH 7.0, and washed with the same buffer. The absorbed proteins were eluted in a two-step linear gradient of sodium phosphate, pH 7.0, from 10 to 50 mm, and then from 50 to 300 mm. After each chromatographic step, the fractions with ExGase activity were dialyzed against 20 mm sodium acetate, pH 5.0, and 20 mm NaCl, and protein was measured with either bicinchoninic acid (Pierce Chemical, Rockford, IL) or Coomassie protein assay reagents (Pierce), with BSA as the standard.
One-dimensional isoelectric focusing of proteins was performed in polyacrylamide slab gels under non-denaturing conditions (Menteur et al., 1995). The gel was sliced, one-half was stained with Coomassie Blue for detection of protein, and the other half was incubated with 0.1% (w/v) p-nitrophenyl glucoside in 20 mm sodium acetate (pH 4.8) and incubated at 35°C for up to 1 h. The reaction product in the gel was detected by the addition of 0.1 m NaOH.
Coleoptile cell walls from seedlings up to 6-d-old in darkness at 30°C were prepared by homogenization of the frozen tissues in ice-cold 50 mm sodium acetate, pH 5.5, containing 50 mm NaCl and 30 mm sodium ascorbate. The settled cell walls were washed several times with additional homogenization buffer, followed by 100 mm NaCl, and water. After each wash, the walls were allowed to settle at ambient gravity. Samples were taken for extraction of the ExGase with 3.5 m LiCl, dialysis, and concentration by SP-Sephadex as described above. β-Glucan in additional portions of the wall sample were digested by Bacillus subtilis endoglucanase and quantified by high-pH anion-exchange HPLC (Gibeaut and Carpita, 1993). The remainder of the wall preparations was concentrated by centrifugation and suspended in an equal volume of PAGE sample buffer containing 20% (w/v) SDS and 100 mm DDT and heated to 65°C for 10 min followed by boiling for 5 min.
Characterization of the Cell Wall ExGase Activity
ExGase activity was measured in column fractions by the determination of Glc released from a barley β-glucan (Sigma, St. Louis) and p-nitrophenyl β-d-glucoside (Sigma). The reaction mixture contained 100 μL of 0.5% (w/v) β-glucan, 10 μL of each protein fraction, and 1 drop of toluene to prevent microbial contamination. The mixture was incubated at 30°C for 2 h, and Glc from the β-glucan was measured enzymatically with hexokinase and Glc-6-P dehydrogenase (Carpita and Kanabus, 1987). One unit of ExGase was defined as the capacity to produce 1 nmol Glc mg−1 min−1.
Several p-nitrophenyl monosaccharides were used as substrate for the enzyme to identify ExGase specificity (Sun and Henson, 1990). The substrate was prepared to 0.1% (w/v) in 20 mm sodium acetate (pH 4.8) and 20 mm NaCl. The reaction mixtures containing 1 mL of p-nitrophenyl monosaccharide and 10 μL (0.1 μg) of the enzyme were incubated at 37°C for up to 1 h. Reactions were stopped with 100 μL of 1 m NaOH, and the A420 was measured. The β-glucan (0.5% [w/v]) was prepared in a pH series from pH 3 to 10 in 20 mm sodium acetate with 20 mm NaCl or mixtures of 10 mm sodium citrate and 20 mm sodium phosphate (McIlvaine, 1921). The substrate (100 μL) in each different pH and enzyme (0.1 μg) was incubated at 30°C and d-Glc was measured as described above. To study temperature dependence, reactions with β-glucan were made with sodium acetate or sodium citrate-phosphate at the optimal pH 4.8. The mixture of substrate (100 μL) and enzyme (0.1 μg) was incubated at different temperatures and the d-Glc was assayed as described above.
p-Nitrophenyl β-d-glucoside (Sigma), sophorose [β-d-glucosyl-(1→2)-d-Glc] (Sigma), laminaribiose [β-d-glucosyl-(1→3)-d-Glc] (Seigakawa Sugar, Falmouth, MA), cellobiose [β-d-glucosyl-(1→4)-d-Glc] (Seigakawa Sugar), and gentiobiose [β-d-glucosyl-(1→6)-d-Glc] (Sigma) were used as substrate to examine the kinetics of hydrolysis for terminal-linked β-d-Glc from different linkage positions. Longer oligomers of the laminari- and cellodextrin series (Seigakawa Sugar) were also used to determine whether degree of polymerization affected the rate of Glc release. The substrates were prepared to various concentrations in 20 mm sodium acetate, pH 4.8 or 5.0, and 20 mm NaCl. The Km and Vmax were determined from Lineweaver-Burk plots for ExGase measured at the optimum pH and temperature and time from apparent first order reaction kinetics.
To determine degree of transglucosylation, the reaction products were separated by HPLC on a PA-1 anion-exchange column (Dionex, Sunnyvale, CA). The column was equilibrated in 0.5 m NaOH and a non-linear programmed gradient to 0.2 m sodium acetate in 0.5 m NaOH, which we designed to optimize separation of glucosyl oligomers to a degree of polymerization of 6 (Bancal et al., 1993). Sugars were detected by pulsed amperometry (Dionex). Oligosaccharides appearing as a result of transglycosylation were collected, desalted, and neutralized by passing through proton-exchange mini-columns (H-columns, Dionex). Residual acetic acid was evaporated under a stream of nitrogen gas. The residue was methylated with n-butyllithium and methyl iodide as described by Gibeaut and Carpita (1993). Partly methylated alditol acetates were separated by gas-liquid chromatography and identified by electron-impact mass spectrometry as described by Carpita and Shea (1989).
Sequencing of the ExGase Peptides
The ExGase in gel slices after SDS-PAGE was washed twice with 200 mm ammonium carbonate in 50% (v/v) acetonitrile and semi-dried on Parafilm (Neenah, WI) under glass. The dried slices were rehydrated with 5 μL of 200 mm ammonium carbonate, pH 8.9, containing 0.02% (w/v) Tween 20; 2 μL of trypsin (200 μg mL−1) in the same buffer was added, and the protein was digested at 30°C for 4 h (Ferrera et al., 1993). The reaction was stopped by addition of 1.5 μL of trifluoroacetic acid (TFA), and the peptides were extracted with 60% (v/v) acetonitrile in 0.1% (v/v) TFA. The peptides were separated by reverse-phase chromatography on a C18 Vydac 218TP52 column (Separations Group, Hesperia, CA) in a non-linear gradient of 0.1% (v/v) TFA in water to 0.8% (v/v) TFA in 70% (v/v) acetonitrile in water. Four major peptides were collected and subjected to automated Edman sequence analysis.
Preparation of Affinity-Purified Antibodies and Immunogel-Blot Detection of ExGase
The Con A affinity-purified ExGase (100 μg) was isolated as a single band by SDS-PAGE and electroblotted from the gel onto cellulose nitrate. Polyclonal antibodies were raised in Balb-c strain mice after subcutaneous insertion of slices of cellulose nitrate containing the blotted ExGase. The cell wall proteins binding to SP-Sephadex were separated by SDS-PAGE, and the ExGase was detected by western gel blotting about 5 to 6 weeks after inoculation. The blot was routinely incubated with the primary antibody (1:1,000 dilution) overnight at 4°C, and alkaline-phosphatase-conjugated goat anti-mouse antiserum (Bio-Rad, Hercules, CA) was used as secondary antibody (1:3,000 dilution). Affinity-purified antibodies were prepared by incubation of the blotted proteins with the polyclonal ExGase antisera, and elution of the antibodies from slices of the cellulose nitrate containing the ExGase band was as described by Chan et al. (1996). Western gel blotting with the affinity-purified antibodies was with 1:500 dilution of the final preparation.
Isolation of ExGase cDNA Clones
A cDNA expression library was constructed from poly(A+)-enriched RNA from 5-d-old etiolated maize seedlings. The cDNA was ligated into a λUni-ZAP XR vector (Stratagene, La Jolla, CA), and the library was amplified in SURE cells (Stratagene). After induction of cells with isopropyl-1-thio-β-d-galactoside, the library was plated and screened with polyclonal antisera prepared against the purified ExGase. Six independent clones were selected after two rounds of immunoscreening, and the cDNA inserts were rescued into plasmid pBSII SK(−). A low degeneracy oligonucleotide [5′-GA(T/C) GC(T/C) CA(T/C/G/A) TA(T/C) GA(T/C)-3′] based on the amino acid sequence DAHYDP from the 17-mer peptide of ExGase was end-labeled with [γ-32P]ATP and used to probe the cDNA inserts from the putative clones. Two of the six clones were positive, and the larger of the two was sequenced.
Isolation of Genomic Clones Encoding the ExGase
A genomic library of maize W-23 was obtained from Dr. Surinder Chopra (Iowa State University, Ames). The library was constructed in a λFIX II vector using partially filled XhoI/Sau3A digested DNA. About 5 × 105 clones were screened with a randomly primed cDNA clone as probe. Putative clones were purified by secondary and tertiary screening, and DNA was prepared from liquid lysates. The cDNA clone contained a SacI site 940 bp upstream from the polyadenylation site, and this site was exploited to digest the genomic clone into two fragments of 3.5 and 7 kb that both hybridized with the cDNA probe. These fragments were subcloned into pBluescript II KS and sequenced in both directions from the SacI cleavage sites. Protein and nucleotide databank searches were performed with the BIONET National Computer Resource for Molecular Biology with Wisconsin Genetics software.
Northern Gel-Blot Analyses
Maize coleoptiles at various stages of elongation were harvested in liquid nitrogen. Total RNA was prepared with an RNeasy Plant Mini Kit (Qiagen USA, Valencia, CA) and separated in a 1% (w/v) agarose-formaldehyde gel. The RNAs from the gels were transferred onto nylon membranes (Hybond N, Amersham, Buckinghamshire, UK) by capillary blotting and fixed by UV cross-linking. The ExGase cDNA clone and a profilin5 cDNA clone, provided by Dr. Chris Staiger (Purdue University) used as probes were labeled with [32P]dCTP by random priming (Redivue, Amersham). The RNA gel-blot hybridization at high stringency was performed according to Sambrook et al. (1989).
Tissue Printing of Coleoptile and Mesocotyl Seedlings
Cellulose nitrate membranes were soaked in 3.5 m LiCl in 20 mm sodium acetate and 20 mm NaCl, pH 5.5, and air-dried. Two- and 3-d-old etiolated maize seedlings comprising coleoptile and mesocotyl were cut longitudinally, and the cut faces of the tissues were pressed against the membranes for 2 h. The membranes were probed with ExGase antisera (1:200 dilution) as described for western gel-blot analyses.
Identification of ExGase at the Plasma Membrane
Three-day-old etiolated maize coleoptiles were isolated and frozen in liquid nitrogen and gently mashed in a chilled mortar and pestle in an equal volume of ice-cold 50 mm HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid]-Bis Tris Propane [1,3-bis(Tris[hydroxymethyl]methylamino) propane], 10 mm KCl, 5 mm EDTA, 5 mm EGTA, 10 mm DTT, and 0.2 mm phenylmethylsulfonyl fluoride, pH 7.2. The mashings were squeezed through nylon cloth (Nitex, Tetko, Briarcliff Manor, NY), 47-μm2 pores), and centrifuged at 3,000g for 10 min to pellet fine debris. The supernatant liquid was then centrifuged at 140,000g for 20 min in a SW28 rotor (Beckman Instruments, Fullerton, CA). The membrane pellet was gently suspended in 6.5% (w/v) polyethylene glycol 3,350 and 6.5% (w/v) Dextran 500T in 5 mm potassium-phosphate and 3 mm KCl, pH 7.2 (two-phase buffer). The pellet was then frozen, thawed, and gently sheared by six strokes in a smooth glass tube with a Teflon piston (LabGlass, Vineland, NJ). Additional two-phase buffer was added, and plasma membranes were prepared by two rounds of partitioning (100 inversions followed by centrifugation at 400g for 5 min at 4°C). The membranes were washed twice in 20 mm HEPES [KOH], pH 7.2, containing 10 mm KCl, and pelleted by centrifugation at 100,000g. Marker assays were performed after resuspension in the dilution buffer. Membrane proteins were dissolved by water-bath sonication in a PAGE sample buffer containing 10% (w/v) SDS, 100 mm DTT, and 4 m urea, and separated by SDS-PAGE. The ExGase was detected by western gel blotting.
The membrane-associated ExGase was then subjected to two-phase detergent partitioning of extrinsic and intrinsic proteins with Triton-X-114 (Bordier, 1981). Briefly, the plasma membrane preparation was brought up to 10% (v/v) prepartitioned Triton-X-114. The membranes were dissolved by two transitions to 37°C from 4°C, and then returned to 4°C. Undissolved material was removed by centrifugation, and the supernatant was then brought to 37°C for 10 min to achieve phase separation. The supernatant was tested for enzyme activity against β-linked disaccharides as described above. Extrinsic and intrinsic membrane proteins were precipitated in 0.1 m ammonium acetate in methanol at −20°C overnight. The detergent phase pellet was also dissolved in ethanol and the protein therein precipitated overnight.
Preparation of Protoplasts
Two-day-old etiolated maize coleoptiles were isolated, sliced into sections about 1 mm thick, and incubated in Murashige-Skoog salts (Murashige and Skoog, 1962), pH 6.0, containing 0.25 m sorbitol (protoplast medium). The sections were transferred to protoplast medium containing 1% (w/v) cellulase (CELF, Worthington Biochemical, Lakewood, NJ) and 0.2% (w/v) pectinase (PEL, Worthington Biochemical), and incubated with gentle shaking at 30°C. The protoplasts were passed through nylon cloth (Nitex, 47-μm2 pores), washed twice with protoplast isolation medium, and, after gentle pelleting, mixed with 20% (w/v) Ficoll in protoplast medium. The Ficoll-protoplast mixture was overlaid with 10% (w/v) and 0% Ficoll in protoplast medium, and the protoplasts were purified by flotation centrifugation at 100g to the 10% (w/v) Ficoll:0% Ficoll interface. The protoplasts were collected by gentle centrifugation and washed free of Ficoll with additional protoplast medium.
The ExGase antisera and affinity-purified antibodies (1:10 dilution) were added to suspensions of cell walls and protoplasts, incubated at ambient temperature for 30 min, and then the suspensions were diluted several times with protoplast medium, and detected with fluorescein isothiocyanate (FITC)- or tetramethylrhodamine isothiocyanate (TRITC)-conjugated rabbit anti-mouse antiserum secondary antibodies (1:20 dilution, Bio-Rad). Samples were viewed in a research microscope fitted with a digital camera (SPOT, Leica Microsystems, Wetzlar, Germany), and frames were captured in Adobe Photoshop at 300 dpi (Adobe Systems, Mountain View, CA).
ACKNOWLEDGMENTS
We thank John Wilder (Purdue Hybridoma Facility) for the preparation of the polyclonal antisera, Alan Mahrenholz (Purdue Laboratory of Macromolecular Structure) for the peptide sequencing, and undergraduate research assistants Matt McConnell and Joseph Pong for their help with hydrolase and transglycosylase assays. We thank Joe Ogas (Purdue University) for his help with the microscopy, Surinder Chopra (Iowa State University) for the maize genomic library, and Chris Staiger (Purdue University) for the cDNA clone of profilin5. We also thank Larry Dunkle, Mark Hermodsen, Maureen McCann, and Charles Woloshuk for their valuable comments on the manuscript.
Footnotes
This work was supported in part by the Division of Energy Biosciences, U.S. Department of Energy (contract no. DE–FG02–88ER13903). This is journal paper no. 16,003 of the Purdue Agricultural Experiment Station.
LITERATURE CITED
- Bancal P, Gibeaut DM, Carpita NC. Analytical methods for the determinations of fructan structure and biosynthesis. In: Suzuki M, Chatterton NJ, editors. Science and Technology of Fructans. Boca Raton, FL: CRC Press; 1993. pp. 83–118. [Google Scholar]
- Boller T. Antimicrobial functions of the plant hydrolase, chitinase and β-1,3-glucanase. In: Fritig B, Legrand M, editors. Mechanisms of Plant Defense Responses. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 391–400. [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton-X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Brummell DA, Catala C, Lashbrook CC, Bennett AB. A membrane-anchored E-type endo-1,4-β-glucanase is localized on Golgi and plasma membranes of higher plants. Proc Natl Acad Sci USA. 1997;94:4794–4799. doi: 10.1073/pnas.94.9.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush J, Richardson J, Cardelli J. Molecular cloning and characterization of the full-length cDNA encoding the developmentally regulated lysosomal enzyme β-glucosidase in Dictyostelium discoideum. J Biol Chem. 1994;269:1468–1476. [PubMed] [Google Scholar]
- Carpita NC. Structure and biogenesis of the cell walls of grasses. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Kanabus J. Extraction of starch by dimethyl sulfoxide and quantitation by enzymic assay. Anal Biochem. 1987;161:132–139. doi: 10.1016/0003-2697(87)90662-2. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Shea EM. Linkage structure of carbohydrates by gas chromatography-mass spectrometry (GC-MS) of partially methylated alditol acetates. In: Biermann CJ, McGinnis GD, editors. Analysis of Carbohydrates by GLC and MS. Boca Raton, FL: CRC Press; 1989. pp. 157–216. [Google Scholar]
- Chan J, Rutten T, Lloyd C. Isolation of microtubule-associated proteins from carrot cytoskeletons: a 120-kDa map decorates all four microtubule arrays and the nucleus. Plant J. 1996;10:251–259. doi: 10.1046/j.1365-313x.1996.10020251.x. [DOI] [PubMed] [Google Scholar]
- Chapple C, Carpita N. Plant cell walls as targets for biotechnology. Curr Opin Plant Biol. 1998;1:179–185. doi: 10.1016/s1369-5266(98)80022-8. [DOI] [PubMed] [Google Scholar]
- Cline K, Albersheim P. Host-pathogen interactions: XVI. Purification and characterization of a β-glucosyl hydrolase/transferase present in the walls of soybean cells. Plant Physiol. 1981;68:207–220. doi: 10.1104/pp.68.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. Relaxation in a high-stress environment: the molecular bases of extensible cells and cell enlargement. Plant Cell. 1997;9:1031–1041. doi: 10.1105/tpc.9.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombie HJ, Chengappa S, Hellyer A, Reid JSG. A xyloglucan oligosaccharide-active, transglycosylating β-d-glucosidase from the cotyledons of nasturtium (Tropaeolum majus L.) seedlings: purification, properties and characterization of a cDNA clone. Plant J. 1998;15:27–38. doi: 10.1046/j.1365-313x.1998.00182.x. [DOI] [PubMed] [Google Scholar]
- Dahlgren RMT, Clifford HT, Yeo PF. The Families of the Monocotyledons: Structure, Evolution, and Taxonomy. New York: Springer-Verlag; 1985. [Google Scholar]
- Ferrera P, Rosenfeld J, Guillemet JC, Capdevielle J. Techniques in Protein Chemistry. San Diego: Academic Press; 1993. [Google Scholar]
- Gibeaut DM, Carpita NC. Tracing cell wall biogenesis in intact cells and plants: selective turnover and alteration of soluble and cell wall polysaccharides in grasses. Plant Physiol. 1991;97:551–561. doi: 10.1104/pp.97.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Carpita NC. Synthesis of (1→3), (1→4)-β-d-glucan in the Golgi apparatus of maize coleoptiles. Proc Natl Acad Sci USA. 1993;90:3850–3854. doi: 10.1073/pnas.90.9.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall GJ, Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-messenger RNAs are required for splicing. Cell. 1989;58:473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Hatfield R, Nevins DJ. Hydrolytic activity and substrate specificity of an endoglucanase from Zea mays seedling cell walls. Plant Physiol. 1987;83:203–207. doi: 10.1104/pp.83.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy FG, Ray RM, Aldrich HC, Wilkie AC, Ingram LO, Shanmugam KT. Direct isolation of functional genes encoding cellulases from the microbial consortia in a thermophilic, anaerobic digester maintained on lignocellulose. Appl Microbiol Biotechnol. 1995;43:667–674. doi: 10.1007/BF00164771. [DOI] [PubMed] [Google Scholar]
- Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon J-P, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA. 1995;92:7090–7094. doi: 10.1073/pnas.92.15.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P, Nakai K. A probabilistic classification system for predicting the cellular localization sites of proteins. Intellig Syst Mol Biol. 1996;4:109–115. [PubMed] [Google Scholar]
- Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbor classifier. Intellig Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- Hoson T, Nevins DJ. β-d-Glucan antibodies inhibit auxin-induced cell elongation and changes in cell wall autohydrolysis. Plant Physiol. 1989;90:1353–1358. doi: 10.1104/pp.90.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrmova M, Fincher GB. Barley β-d-glucan exohydrolases substrate specificity and kinetic properties. Carbohydr Res. 1998;305:209–221. [Google Scholar]
- Hrmova M, Harvey AJ, Wang J, Shirley NJ, Jones GP, Stone BA, Høj PB, Fincher GB. Barley β-d-glucan exohydrolases with β-d-glucosidase activity: purification, characterization, and determination of primary structure from a cDNA clone. J Biol Chem. 1996;271:5277–5286. doi: 10.1074/jbc.271.9.5277. [DOI] [PubMed] [Google Scholar]
- Huber DJ, Nevins DJ. Partial purification of endo- and exo-β-d-glucanase enzymes from Zea mays L. seedlings and their involvement in cell wall autohydrolysis. Physiol Plant. 1981;53:533–539. doi: 10.1007/BF00395171. [DOI] [PubMed] [Google Scholar]
- Huber DJ, Nevins DJ. Exoglucanases from Zea mays L. seedlings: their role in β-glucan hydrolysis and their potential role in extension growth. Planta. 1982;155:467–472. doi: 10.1007/BF01607569. [DOI] [PubMed] [Google Scholar]
- Jarvis MC, Forsyth W, Duncan HJ. A survey of the pectic content of non-lignified monocot cell walls. Plant Physiol. 1988;88:309–314. doi: 10.1104/pp.88.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrador E, Nevins DJ. An exo-β-d-glucanase derived from Zea coleoptile walls with a capacity to elicit cell elongation. Physiol Plant. 1989;77:479–486. [Google Scholar]
- Leah R, Kigel J, Svendsen I, Mundy J. Biochemical and molecular characterization of a barley seed β-glucosidase. J Biol Chem. 1995;270:15789–15797. doi: 10.1074/jbc.270.26.15789. [DOI] [PubMed] [Google Scholar]
- Lee S-H, Kivilaan A, Bandurski RS. In vitro autolysis of plant cell walls. Plant Physiol. 1967;42:968–972. doi: 10.1104/pp.42.7.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZC, Durachko DM, Cosgrove DJ. An oat coleoptile wall protein that induces wall extension in vitro and that is antigenically related to a similar protein from cucumber hypocotyls. Planta. 1993;191:349–356. [Google Scholar]
- Matthysse AG, White S, Lightfoot R. Genes required for cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol. 1995;177:1069–1075. doi: 10.1128/jb.177.4.1069-1075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlvaine TC. A buffer solution for colorimetric comparison. J Biol Chem. 1921;43:183–186. [Google Scholar]
- Menteur S, Jestin L, Risacher T, Branlard G. Visualization of barley β-glucan degrading isozymes after gel isoelectric-focusing. Electrophoresis. 1995;16:1019–1023. doi: 10.1002/elps.11501601171. [DOI] [PubMed] [Google Scholar]
- Murashige TR, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhette S, Canut H, Höfte H. A plasma membrane-bound putative endo-1,4-β-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani K. The role of endoxyloglucan transferase in the organization of plant cell walls. Int Rev Cytol. 1997;173:157–206. doi: 10.1016/s0074-7696(08)62477-8. [DOI] [PubMed] [Google Scholar]
- Pritchard J, Hetherington PR, Fry SC, Tomos AD. Xyloglucan endotransglycosylase activity, microfibril orientation and the profiles of cell wall properties along growing regions of maize roots. J Exp Bot. 1993;44:1281–1289. [Google Scholar]
- Rixon JE, Ferreira LM, Durrant AJ, Laurie JI, Hazlewood GP, Gilbert HJ. Characterization of the gene celD and its encoded product 1,4-β-d-glucan glucohydrolase D from Pseudomonas fluorescens subsp. cellulosa. Biochem J. 1992;285:947–955. doi: 10.1042/bj2850947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall PJ, Caddick LR. Investigation of the presence of phenolic compounds in monocotyledonous cell walls, using UV fluorescence microscopy. Ann Bot. 1994;74:483–491. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Samuels AL, Giddings TH, Jr, Staehelin LA. Cytokinesis in tobacco BY-2 and root tip cells: a new model of cell plate formation in higher plants. J Cell Biol. 1995;130:1345–1357. doi: 10.1083/jcb.130.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BG, Harris PJ. The polysaccharide composition of Poales cell walls: Poaceae cell walls are not unique. Biochem Syst Ecol. 1999;27:33–53. [Google Scholar]
- Sun Z, Henson CA. Degradation of native starch granules by barley α-glucosidases. Plant Physiol. 1990;94:320–327. doi: 10.1104/pp.94.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz L. Plant cell expansion: regulation of cell-wall mechanical properties. Annu Rev Plant Physiol. 1984;35:585–657. [Google Scholar]
- Varghese JN, Hrmova M, Fincher GB. Three-dimensional structure of a barley β-d-glucan exohydrolase, a family 3 glycosyl hydrolase. Struct Fold Design. 1999;7:179–190. doi: 10.1016/s0969-2126(99)80024-0. [DOI] [PubMed] [Google Scholar]
- Wood P, Weisz J, Blackwell BA. Structural studies of (1→3),(1→4)-β-d-glucans by 13C-nuclear magnetic resonance spectroscopy and by rapid analysis of cellulose-like regions using high-performance anion-exchange chromatography of oligosaccharides released by lichenase. Cereal Chem. 1994;71:301–307. [Google Scholar]
- Wu Y, Spollen WG, Sharp RE, Hetherington PR, Fry SC. Root growth maintenance at low water potentials: increased activity of xyloglucan endotransglycosylase and its possible regulation by abscisic acid. Plant Physiol. 1994;106:607–615. doi: 10.1104/pp.106.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]