Abstract
Assays that can verify full viral eradication are essential in the context of achieving a cure for HIV/AIDS. In vitro quantitative viral out growth assays (qVOA) are currently the gold standard for measuring latent HIV-1 but these assays often fail to detect very low levels of replication-competent virus. Here we investigated an alternative in vivo approach for sensitive viral detection using humanized mice (hmVOA). Peripheral blood CD4+ T cell samples from HIV subjects on stable ART with undetectable viral loads by RT-PCR were first assayed by in vitro qVOA. Corresponding patient samples in which no virus was detected by traditional qVOA were injected into humanized mice to allow viral outgrowth. Of the five in vitro qVOA virus negative samples, four gave positive viral outgrowth in the in vivo hmVOA assay suggesting that it is more sensitive in detecting latent HIV-1.
Keywords: Humanized mouse-based latent HIV-1 viral outgrowth assay, In vivo latent HIV-1 viral outgrowth assay, Comparison of HIV qVOA and in vivo hu-mouse VOA, Ultra-Sensitive latent HIV-1 detection in patient cells, Latent HIV-1 detection by hu-mouse-based VOA
INTRODUCTION
Persistence of latently infected cells even after long-term effective antiretroviral therapy (ART) poses a formidable obstacle for achieving permanent HIV-1 remission and a complete cure (Deeks et al., 2016). Therefore, current strategies are aimed at reactivating the latent virus by various drugs and purging these induced cells by different cytotoxic approaches (Cillo and Mellors, 2016). With regard to latently infected cells, all induced latent virus must arise from integrated HIV-DNA. However, not all integrated HIV-DNA represents inducible replication competent virus making the PCR-based assays undependable to identify cells harboring replication competent virus (RCV) (Rosenbloom et al., 2015; Spina et al., 2013) (Procopio et al., 2015). In this context, other than ATI, quantitative viral out growth assays (qVOA) that screen for latently infected cells by detecting outgrowth of HIV-1 infectious units per million cells (IUPM) have been the gold standard (Spina et al., 2013). To measure the frequency of latently infected resting CD4+ T cells, this assay relies on in vitro reactivation of these cells present in a population with mitogens such as PHA or with antibody co-stimulation of CD3 (T-cell receptor) and CD28 (costimulatory receptor) and expansion of released virus by co-culture with lymphoblasts from HIV negative donors. However, only a few latently infected cells can be measured in one sample due to their rarity (as low as one in a million). Moreover, not all latently infected cells can be successfully stimulated to release virus during the standard qVOA assay’s time frame (Churchill et al., 2016).
Consistent with these observations, the standard qVOA failed to detect the residual virus in several individuals with prolonged undetectable plasma and cell-associated HIV-1, such as the two "Boston patients" who underwent allogeneic HSCT and the "Mississippi child" who initiated ART immediately following birth (Henrich et al., 2014; Luzuriaga et al., 2015; Persaud et al., 2013). Unfortunately, virus eventually rebounded after treatment interruption in these individuals. Thus, it has become important that more sensitive VOAs that employ novel approaches need to be developed and validated. In NHP studies, latent virus was successfully recovered from fully virus suppressed SIV infected macaques (as determined by all standard tests) undergoing intensive ART by adoptive transfer of their resting CD4+ T cells to naive animals (Okoye, 2014). This showed that ultralow levels of otherwise undetectable latently infected cells could be induced and detected using an in vivo system.
At present, humanized mice that harbor a transplanted human immune system with a de novo capacity to continuously generate virus susceptible cells are the only available animal models permissive for HIV-1 infection, viral latency and reactivation (Akkina, 2013). The commonly used new generation hu-mouse models include hu-HSC mice derived by injection of hematopoietic stem cells (HSC) or BLT mice constructed by transplantation of human fetal thymic and liver tissues in addition to HSC. Two recent reports (Metcalf Pate et al., 2015; Yuan Z, 2017) described reconstituting immunodeficient mice with either PBMC or CD4 T cells to detect latently infected cells but the mice used were not previously humanized and thus have some limitations (discussed below). Based on these above observations and considerations, we sought to develop and evaluate an in vivo humanized mouse-based HIV-1 outgrowth assay (hmVOA) to detect latent virus and determine its sensitivity over the traditional in vitro qVOA. Our results from this proof-of-concept study show that the hmVOA is able to detect replication competent HIV-1 in fewer numbers of input cells and in samples with undetectable HIV-1 using the traditional qVOA, and thus is more sensitive.
METHODS
Patient samples and resting CD4+ T cell isolation
To develop and evaluate the hmVOA, we obtained CD4+ T cells from two groups of patients from the Zuckerberg San Francisco General Hospital and the Brigham and Women's Hospital. The ART regimens, CD4+ T cell counts and HIV-1 viral loads and clinical status of donors are summarized in Table 1. One group included HIV-1-infected individuals on ART but with intermittent or persistent low-level viremia (<1000 HIV-1 RNA copies/ml of plasma) and the other included individuals on fully suppressive ART with no detectable VL >50 copies/ml at the time of sampling.
Table 1.
ART regimens, CD4+ T cell and plasma viral load status of HIV-1 + donors
| Donor | Sex | Recent Low Level Detectable Viral Loada |
ART Regimen | CD4+ T Cell Count (cells/µL) |
Plasma HIV-1 RNA (copies/mL) |
|---|---|---|---|---|---|
| 2 | M | Y | DTG/TDF/FTC | 633 | ND |
| 5 | M | Y | ATV/ABC/3TC | 872 | Detected, <20 |
| 8 | M | Y | DRV/ABC/3TC | 744 | Detected, <20 |
| 9 | M | Y | TDF/FTC/RAL | 2020 | ND |
| 13 | M | Y | TDF/FTC/EGV/COB | 1077 | ND |
| 15 | M | Y | ANC/3TC/ATV/R | 702 | Detected, <20 |
| 30 | M | N | ABC/3TC/RAL | 842 | ND |
| 32 | M | N | DTG/RPV | 692 | ND |
| 34 | M | N | EGV/TDF/FTC/COB | 1044 | ND |
| 41 | M | N | DRV/R/ABC/DTG/3TC | 741 | ND |
| 42 | M | N | ABC/DTG/3TC | 716 | ND |
DTG = dolutegravir; TDF = tenofovir; FTC = emtricitabine; ATV = atazanavir; ABC = abacavir; 3TC = lamivudine; RAL = raltegravir; EGV = elvitegravir; COB = cobicistat (boost); R = ritonovir (boost); DTG = dolutegravir; ABC = abacavir; RPV = rilpivirine; ND = not detectable
one or more VL <500 copies/mL within 12 months of sample draw
Institutional approval was obtained from both UCSF and The Brigham and Women's Hospital for collecting samples and conducting this study. Written informed consent was obtained from all study participants. PBMCs were collected from peripheral blood and purified by Ficoll-Hypaque (Sigma-Aldrich) density gradient centrifugation followed by cryopreservation for later testing. Untouched, total CD4+ T cells were purified by negative selection as described previously (95% pure) using antibody coated magnetic beads (Stem Cell Technologies, Vancouver, Canada). A modified version of an in vitro qVOA was performed based on a previously described method using duplicate wells for serial dilutions of 5 million to 0.1 million for each patient sample (Chun et al., 2003; Siliciano and Siliciano, 2005). Exact dilutions used were based on the number of viable CD4+ T cells that were able to be obtained. Cells were stimulated using anti-CD3/anti-CD28 antibodies for up to 21 days in the presence of CD8+ Tcell-depleted lymphoblasts obtained from HIV-uninfected blood donors. Positive wells were determined by increasing HIV-1 unspliced (us) RNA detected in assay supernatants over time. HIV-1 RNA was quantified using a Taqman real-time polymerase chain reaction (PCR) method based on previously described primer and probe sequences that amplify of a conserved region in the LTR/gag that is specific to nearly all group M HIV-1 sequences (Malnati et al., 2008). Briefly, 10 µL of RNA was added to 10 µL of TaqMan® Fast Virus 1-Step Master Mix (ThermoFisher) incorporating forward primer, 5'-TACTGACGCTCTCGCACC, reverse primer, 5'-TCTCGACGCAGGACTCG, and probe, 5'-FAM-CTCTCTCCTTCTAGCCTC-MGB, as per manufacturers protocol followed by thermocycling with an annealing temperature of 60°C using the LightCycler 96 system (Roche). The IUPM were calculated for each of the samples positive for viral outgrowth using a maximum likelihood method and online calculator as described (Rosenbloom et al., 2015). Positive controls in qVOA incorporated serial dilutions of ACH2 cells harboring an integrated HIV-1 subtype B infectious clone and subjected to the same conditions as experiments involving participant samples. Cells from the same collection time points were later subjected to hmVOA as described below.
Generation of humanized mice
Immunodeficient BALB/c/RAG1 or RAG2−/−γc−/− mice were used to generate humanized mice (hu-mice). Hu-HSC mice were generated by injecting human fetal liver-derived CD34+ hematopoietic stem cells (HSC) intra-hepatically into newborn mice as we described previously (Berges et al., 2010). Mice were maintained at the CSU Painter Animal Center. These studies have been reviewed and approved by the Institutional Animal Care and Use Committee. Human fetal liver-derived CD34+ cells were purified and cultured for 24 hours in cytokine media (Akkina et al., 1994; Hu et al., 2016). Neonatal mice were irradiated with 350 rads and injected intra-hepatically with 0.5–1×106 human CD34 cells. BLT hu-mice were prepared by transplantation of fragments of human fetal liver and thymic tissues under the mouse kidney capsule as described previously followed by tail vein injection of autologous fetal CD34+ HSC (Akkina, 2013; Hu et al., 2017). Transplanted mice were screened for human cell engraftment at 10–12 week post-reconstitution. Peripheral blood was collected and the red blood cells were lysed using the Whole Blood Erythrocyte Lysing Kit (R&D Systems, Minneapolis, MN). Fractioned white blood cells were stained with human CD4+5 FITC marker and FACS analyzed to confirm human cell engraftment (Berges et al., 2006)
Humanized mouse viral outgrowth assay (hmVOA)
Purified and previously frozen CD4+ T cells were used for determining viral outgrowth in hu-mice. Cell batches were thawed, washed twice in RPMI media and were allowed to recover in complete media containing 5ng/ml IL-2 for 4–6 hours at 2×10^6 cells/ml. After recovery, cells were either non-stimulated or stimulated with PHA (2µg/mL) or anti-CD3 and anti-CD28 soluble antibody (100ng/mL) overnight. Later, the cells were aliquoted at desired cell number for each hu-mouse injection. Cell count and viability were determined using an AOPI live/dead stain on the Nexcelom Cellometer Auto 2000. Mice were injected by intraperitoneal route with cell numbers ranging from 0.1 to 20×106 live cells and were followed for 8 weeks. Viral RNA extracted from weekly plasma samples from each of the mice was tested by RT-PCR to detect viremia as described previously (Rouet et al., 2005).
RESULTS
Humanized mice are permissive for patient derived latent HIV-1 viral outgrowth
Our first set of experiments involved standardizing basic parameters for developing the hmVOA assay. We started with injecting and testing samples from participants on ART with low but detectable plasma HIV RNA levels. Replication-competent virus from total CD4+ T cells were detectable in all samples using the traditional qVOA. The available frozen CD4+ T cell numbers as well as viable cell yield after thawing varied for each subject, and the numbers of cells and mice injected were optimized accordingly (Table 2). Viral outgrowth as defined by detectable plasma HIV RNA was measured weekly for 8 weeks. All the above samples gave positive viral outgrowth at some level in hu-mice (irrespective of whether they were hu-HSC or BLT) although the results were dependent on the number of cells injected and method of cell stimulation as described below.
Table 2.
Standardization of hmVOA with qVOA positive samples
| Donor | Total Cells (Millions) |
Range of Cell Number Injected per Mouse (Millions) |
Number of Mice Used |
Number of Mice with Viral Outgrowth |
Cell Number Resulting in hmVOA Viral Outgrowth (Millions) |
|---|---|---|---|---|---|
| 2 | 26 | 0.5–10 | 9 | 2 | 1.25, 5 |
| 5 | 34 | 0.5–20 | 8 | 1 | 1 |
| 8 | 24 | 0.1–10 | 11 | 5 | 0.5, 1, 5, 10 |
| 9 | 54 | 0.1–20 | 13 | 6 | 0.5, 1, 5, 10, 20 |
| 13 | 24 | 0.1–10 | 11 | 3 | 5, 10 |
| 15 | 24 | 0.1–10 | 11 | 4 | 0.1, 0.5, 5, 10 |
With the Donor 2 (D2) sample, viral outgrowth was seen in one of two mice injected with 5 million unstimulated cells and in one of two mice injected with 1.25 million PHA-stimulated cells. Data from the in vitro qVOA showed one of two wells was positive at 5 million cells and none at both wells of 1 million cells. With sample D5, one of 2 mice injected with 1 million unstimulated cells was positive; this was similar to in vitro VOA wherein one of 2 wells was positive at 1 million cells. These initial results suggested that prior stimulation of cells led to detectable virus in fewer number of input cells in the hmVOA viral outgrowth over in vitro VOA.
With regard to the method of stimulation overnight, we found that co-stimulation with anti-CD3/anti-CD28 antibodies yielded more viable and healthier cells compared with those treated with PHA. Accordingly, the subsequent experiments employed anti-CD3/anti-CD28 antibody stimulation of cells prior to injection, as was performed in the traditional in vitro qVOA. Based on the numbers of cells available, and for a direct comparison with in vitro qVOA, mice were injected with 20, 10, 5, 1, 0.5 and 0.1 million cells to determine the viral outgrowth end point. Donor samples D8, D9, D13, D15 (4 donors total) followed this scheme. Of the mice injected with different cell numbers, positive viral outgrowth was obtained in those with high numbers of cells, as expected. Parallel samples from D8, D9, and D15 showed positive viral outgrowth from 0.5, 0.5, and 0.1 million cells respectively in contrast to in vitro VOA wherein D8 and D9 were negative suggesting that hmMOA is potentially more sensitive. Successful detection of latent HIV-1 as seen here from all ART patient samples tested demonstrated that the hmVOA model is amenable for viral outgrowth and set the stage for the experiments below.
hmVOA was able detect latent viral outgrowth from ART-suppressed participant samples that were negative by qVOA
Based on the above promising data, we proceeded to ask and confirm whether hmVOA is more sensitive in detecting latent virus from samples wherein no viral outgrowth could be detected by qVOA (subjects D30, D32, D34, D41 and D42, Table 3). Since the test samples were already negative by qVOA, our approach here was to inject largest number/aliquots of cells in replicates (based on the number of available cells) to multiple mice mimicking the wells receiving the largest number of cells in a typical qVOA (5 million cells per well). Of the 5 samples evaluated, 4 yielded positive viral outgrowth in the hmVOA (Table 3). The earliest time point of viral detection was 2 weeks with two different samples followed by weeks 4 and 6 in two other samples. The remaining sample was negative through week 8. Of mice injected with different numbers of cells, those injected with higher numbers of cells (for example 7 million versus 5 million) became virus positive sooner, as expected. These above data suggest that hmVOA can detect replication competent HIV-1 in similar or greater total input cells when the standard qVOA is unable to do so.
Table 3.
hmVOA has increased sensitivity compared to qVOA in detecting latent HIV-1
| Donor | Total Cells (Millions) |
Range of Cell Number Injected per Mouse (Millions) |
Number of Mice Used |
Number of Mice with Viral Outgrowth |
Cell Number Resulting in hmVOA Viral Outgrowth (Millions) |
|---|---|---|---|---|---|
| 30 | 32 | 2–10 | 6 | 1 | 10 |
| 32 | 16 | 4 | 4 | 1 | 4 |
| 34 | 17 | 4.25 | 4 | 1 | 4.25 |
| 41 | 15 | 5 | 3 | 0 | 5 |
| 42 | 27 | 5-7 | 5 | 3 | 5, 7 |
DISCUSSION
Establishment of HIV-1 latency and viral induction from the latent state are shown to involve varied and complex mechanisms (Shan and Siliciano, 2013; Spina et al., 2013). While the in vitro qVOA is currently the gold standard for measuring latent viral burden, a number of factors can play a role in its failure to detect ultralow virus levels (Hill et al., 2016). Here we developed and compared an in vivo humanized mouse based assay (hmVOA) with that of the in vitro qVOA using samples from participants on ART who were either fully suppressed or experienced low-level viremic events during the time of sampling.
Our initial experiments focused on determining whether hu-mice are permissive for viral outgrowth. Cells from HIV-infected adults with low but detectable plasma HIV RNA levels were first tested. CD4+ T cells from these subjects were also subjected to in vitro qVOA to measure the extent of the replication competent reservoir. Results from the qVOA showed the presence of measurable levels of infected cells from these samples and provided a broad range of IUPM levels (Table 4). When injected into hu-mice, positive viral outgrowth was observed with all these samples, indicating that hu-mice are permissible for detecting latently infected cells. In addition, viral outgrowth could be seen with fewer numbers of input patient-derived cells than in a qVOA (Table 4). We found that mitogenic stimulation of cells prior to injecting mice gave better outgrowth and that anti-CD3 and anti-CD28 antibody stimulation of thawed cells yielded higher numbers of viable cells.
Table 4.
Comparison of hmVOA with qVOA for viral outgrowth
| Donor | qVOA Outgrowth | Lowest # Cells Detected qVOA vs. hmVOA (Millions) |
qVOA IUPM |
Week of hmVOA Viral Outgrowth |
|---|---|---|---|---|
| 2 | + | 5 / 5 unstimulated, 1.25 stimulated PHA | 0.102 | 2 |
| 5 | + | 1 / 1 unstimulated | 1.083 | 3 |
| 8 | + | 8×10−3/ 0.5 | 4.468 | 2 |
| 9 | + | 8×10−3/ 0.5 | 1.171 | 1 |
| 13 | + | 1 / 5 | 3.74 | 2 |
| 15 | No data | N/A / 0.1 | No data | 1 |
| 30 | - | 0/10 | 0 | 4 |
| 32 | - | 0/4 | 0 | 3 |
| 34 | - | 0/4.25 | 0 | 2 |
| 41 | - | N/A / N/A | 0 | N/A |
| 42 | - | 0/5 | 0 | 2 |
This encouraging data with hu-mice for in vivo viral outgrowth formed the basis for evaluating the question if the hmVOA can detect low levels of latent virus undetectable by qVOA. Indeed, of the five qVOA negative samples tested, four gave viral outgrowth in hmVOA with the exception of one sample (D41) using similar input cell numbers (Table 3). We attribute the failure of detection with this particular sample to poor initial quality of cells noticed at the time of injection. In general, mice injected with higher numbers of cells showed viral outgrowth sooner. These above data suggest that hmVOA is more capable of detecting latent inducible HIV-1 from CD4+ T cells than the standard qVOA.
A number of factors might be contributing to the increased sensitivity of hmVOA. First, it is an in vivo assay in which the cells are introduced into a physiological environment wherein conditions for cell survival and maintenance are likely to be more permissive than in vitro. Second, hmVOA permits longer time period for viral detection, i.e., 8 weeks or longer compared to 3 weeks for in vitro qVOA. Third, since the hu-mice continuously generate human hematopoietic HIV-1 targets cells (CD4+ T cells, monocytes/macrophages and dendritic cells) including highly viral susceptible immature thymocytes, this may provide a much broader spectrum of cells for facilitating virus outgrowth versus in vitro VOA. Finally, the xenograft environment in hu-mice may be another contributing factor in the expansion of the low initial number of input cells.
There are two reports of HIV-1 viral outgrowth using murine models (mVOA) which are not humanized (Metcalf Pate et al., 2015; Yuan Z, 2017). Metcalf Pate et al described using immunodeficient NSG mice. Either PBMCs or CD4+ T cells in large numbers (66 million PBMC or 10–26 million CD4+ T cells) from 11 HIV subjects (including 6 elite controllers) with undetectable viral loads (by RT-PCR) were injected into mice (Metcalf Pate et al., 2015). Of these, all were qVOA positive for viral outgrowth with the exception of one sample. Viral outgrowth was noted in mice with all the samples including the one which was qVOA negative. A drawback with this model is eventual GvH, the need to inject anti-CD8 antibodies for CD8 cell depletion from PBMC injected mice, and the required injection of anti-CD3 antibodies for activation of cells in vivo. This study included only a single qVOA negative sample (as compared to the five in our current study). A more recent report by Yuan et al also used NSG mice to detect latent virus but again this study only included samples that had detectable virus by the qVOA (Yuan Z, 2017).
While the non-humanized mVOA and our present hmVOA for viral outgrowth are different and can be put use for latent viral induction and detection based on the research question on hand, the latter is endowed with higher sensitivity as shown here due to the de novo generation of HIV susceptible hematopoietic cells at all stages of differentiation thus providing a more relevant physiological in vivo setting. These promising results form a sound basis for further refinement of the hmVOA, which can be eventually deployed to preclude treatment interruption as a means to confirm full remission.
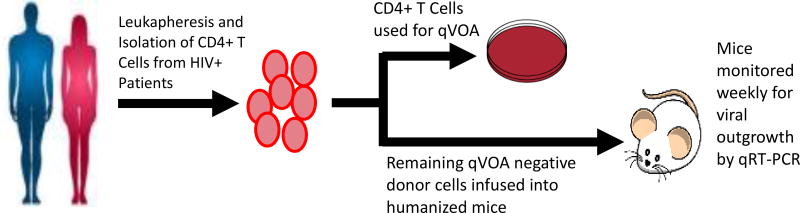
Figure 1.
Schematic of experimental design
Research Highlights.
Development of a humanized mouse-based HIV-1 viral out growth assay (hmVOA)
Comparison of in vitro qVOA with in vivo hmVOA in detecting viral latency
Proof of concept study showing the high sensitivity of in vivo hmVOA over qVOA
Acknowledgments
Work reported here was supported in part by NIH, USA grant RO1 AI100845 and an amfAR grant to RA. The SCOPE cohort was supported the UCSF/Gladstone Institute of Virology & Immunology CFAR (P30 AI027763) and the CFAR Network of Integrated Systems (R24 AI067039). Additional support was provided by the Delaney AIDS Research Enterprise (DARE; AI096109, A127966) and the amfAR Institute for HIV Cure Research (amfAR 109301). We would like to thank Dr. Daniel Kuritzkes of Harvard Medical School for encouragement and Ms. Molly Price for her assistance during this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akkina R. New generation humanized mice for virus research: comparative aspects and future prospects. Virology. 2013;435(1):14–28. doi: 10.1016/j.virol.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkina RK, Rosenblatt JD, Campbell AG, Chen IS, Zack JA. Modeling human lymphoid precursor cell gene therapy in the SCID-hu mouse. Blood. 1994;84(5):1393–8. [PubMed] [Google Scholar]
- Berges BK, Akkina SR, Remling L, Akkina R. Humanized Rag2(−/−)gammac(−/−) (RAG-hu) mice can sustain long-term chronic HIV-1 infection lasting more than a year. Virology. 2010;397(1):100–3. doi: 10.1016/j.virol.2009.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berges BK, Wheat WH, Palmer BE, Connick E, Akkina R. HIV-1 infection and CD4 T cell depletion in the humanized Rag2−/−gamma c−/− (RAG-hu) mouse model. Retrovirology. 2006;3:76. doi: 10.1186/1742-4690-3-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Justement JS, Lempicki RA, Yang J, Dennis G, Jr, Hallahan CW, Sanford C, Pandya P, Liu S, McLaughlin M, Ehler LA, Moir S, Fauci AS. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(4):1908–13. doi: 10.1073/pnas.0437640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: what, where and how to target them. Nature reviews. Microbiology. 2016;14(1):55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- Cillo AR, Mellors JW. Which therapeutic strategy will achieve a cure for HIV-1? Current opinion in virology. 2016;18:14–9. doi: 10.1016/j.coviro.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Deeks SG, Lewin SR, Ross AL, Ananworanich J, Benkirane M, Cannon P, Chomont N, Douek D, Lifson JD, Lo YR, Kuritzkes D, Margolis D, Mellors J, Persaud D, Tucker JD, Barre-Sinoussi F, Alter G, Auerbach J, Autran B, Barouch DH, Behrens G, Cavazzana M, Chen Z, Cohen EA, Corbelli GM, Eholie S, Eyal N, Fidler S, Garcia L, Grossman C, Henderson G, Henrich TJ, Jefferys R, Kiem HP, McCune J, Moodley K, Newman PA, Nijhuis M, Nsubuga MS, Ott M, Palmer S, Richman D, Saez-Cirion A, Sharp M, Siliciano J, Silvestri G, Singh J, Spire B, Taylor J, Tolstrup M, Valente S, van Lunzen J, Walensky R, Wilson I, Zack J. International AIDS Society global scientific strategy: towards an HIV cure 2016. Nature medicine. 2016;22(8):839–50. doi: 10.1038/nm.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, Robles YP, Davis BT, Li JZ, Heisey A, Hill AL, Busch MP, Armand P, Soiffer RJ, Altfeld M, Kuritzkes DR. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161(5):319–27. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AL, Rosenbloom DI, Goldstein E, Hanhauser E, Kuritzkes DR, Siliciano RF, Henrich TJ. Real-Time Predictions of Reservoir Size and Rebound Time during Antiretroviral Therapy Interruption Trials for HIV. PLoS pathogens. 2016;12(4):e1005535. doi: 10.1371/journal.ppat.1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Mohan Kumar D, Sax C, Schuler C, Akkina R. Pseudotyping of lentiviral vector with novel vesiculovirus envelope glycoproteins derived from Chandipura and Piry viruses. Virology. 2016;488:162–8. doi: 10.1016/j.virol.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Neff CP, Kumar DM, Habu Y, Akkina SR, Seki T, Akkina R. A humanized mouse model for HIV-2 infection and efficacy testing of a single-pill triple-drug combination anti-retroviral therapy. Virology. 2017;501:115–118. doi: 10.1016/j.virol.2016.11.013. [DOI] [PubMed] [Google Scholar]
- Luzuriaga K, Gay H, Ziemniak C, Sanborn KB, Somasundaran M, Rainwater-Lovett K, Mellors JW, Rosenbloom D, Persaud D. Viremic relapse after HIV-1 remission in a perinatally infected child. New England Journal of Medicine. 2015;372(8):786–788. doi: 10.1056/NEJMc1413931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nature protocols. 2008;3(7):1240–8. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- Metcalf Pate KA, Pohlmeyer CW, Walker-Sperling VE, Foote JB, Najarro KM, Cryer CG, Salgado M, Gama L, Engle EL, Shirk EN, Queen SE, Chioma S, Vermillion MS, Bullock B, Li M, Lyons CE, Adams RJ, Zink MC, Clements JE, Mankowski JL, Blankson JN. A Murine Viral Outgrowth Assay to Detect Residual HIV Type 1 in Patients With Undetectable Viral Loads. The Journal of infectious diseases. 2015;212(9):1387–96. doi: 10.1093/infdis/jiv230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye A. NIH Meeting: Strategies for an HIV Cure. Bethesda, Maryland: 2014. Early Antiretroviral Therapy Limits Viral Reservoir in SIV-infected Macaques. [Google Scholar]
- Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M, Jr, Chun TW, Strain M, Richman D, Luzuriaga K. Absence of detectable HIV-1 viremia after treatment cessation in an infant. The New England journal of medicine. 2013;369(19):1828–35. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, Richman DD, O'Doherty U, Palmer S, Hecht FM, Hoh R, Barnard RJ, Miller MD, Hazuda DJ, Deeks SG, Sekaly RP, Chomont N. A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine. 2015;2(8):874–83. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom DI, Elliott O, Hill AL, Henrich TJ, Siliciano JM, Siliciano RF. Designing and Interpreting Limiting Dilution Assays: General Principles and Applications to the Latent Reservoir for Human Immunodeficiency Virus-1. Open Forum Infect Dis. 2015;2(4) doi: 10.1093/ofid/ofv123. ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet F, Ekouevi DK, Chaix ML, Burgard M, Inwoley A, Tony TD, Danel C, Anglaret X, Leroy V, Msellati P, Dabis F, Rouzioux C. Transfer and evaluation of an automated, low-cost real-time reverse transcription-PCR test for diagnosis and monitoring of human immunodeficiency virus type 1 infection in a West African resource-limited setting. Journal of clinical microbiology. 2005;43(6):2709–17. doi: 10.1128/JCM.43.6.2709-2717.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Siliciano RF. From reactivation of latent HIV-1 to elimination of the latent reservoir: the presence of multiple barriers to viral eradication. BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35(6):544–52. doi: 10.1002/bies.201200170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods in molecular biology. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, Mau M, Ruelas D, Saleh S, Shirakawa K, Siliciano RF, Singhania A, Soto PC, Terry VH, Verdin E, Woelk C, Wooden S, Xing S, Planelles V. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS pathogens. 2013;9(12):e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, K G, Lu W, Li Q. Reactivation of HIV-1 provirses in immune-compromised mice engrafted with human VOA-negative CD4+ T cells. Journal of Virus Eradication. 2017;3:61–65. doi: 10.1016/S2055-6640(20)30298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]