Abstract
The insulin-degrading enzyme (IDE) cleaves numerous small peptides, including biologically active hormones and disease-related peptides. The propensity of IDE to degrade neurotoxic Aβ peptides marks IDE as a potential therapeutic target for Alzheimer disease. Using a synthetic reporter based on the yeast a-factor mating pheromone precursor, which is cleaved by multiple IDE orthologs, we identified seven small molecules that stimulate rat IDE activity in vitro. Half-maximal activation of IDE by the compounds is observed in vitro in the range of 43 to 198 µM. All compounds decrease the Km of IDE. Four compounds activate IDE in the presence of the competing substrate insulin, which disproportionately inhibits IDE activity. Two compounds stimulate rat IDE activity in a cell-based assay, indicating that they are cell permeable. The compounds demonstrate specificity for rat IDE since they do not enhance the activities of IDE orthologs, including human IDE, and they appear specific for a-factor–based reporters since they do not enhance rat IDE-mediated cleavage of Aβ-based reporters. Our results suggest that IDE activators function in the context of specific enzyme-substrate pairs, indicating that the choice of substrate must be considered in addition to target validation in IDE activator screens.
Keywords: Aβ, IDE, protease, activator, Alzheimer disease
Introduction
According to the Aβ hypothesis, Aβ peptides contribute to Alzheimer disease (AD) by serving as a source of neurotoxicity. It remains unclear whether neurotoxicity stems from accumulation of the monomeric, oligomeric (ADDLs), and/or fibrillar forms of Aβ.1 Independent of the neurotoxic species, strategies aimed at reducing Aβ levels hold promise for the treatment of AD. Because biosynthetic and clearance activities regulate steady-state Aβ levels, either or both activities could be manipulated to reduce Aβ. Although most research on Aβ has centered on its production (i.e., the α, β, and γ secretases), several mechanisms are known to clear Aβ.2
One prominent Aβ clearance mechanism involves proteolytic cleavage of the Aβ monomer, which eliminates its ability to form higher order structures. The insulin-degrading enzyme (IDE; EC 3.4.24.56) is one of several Aβ-cleaving enzymes; others include neprilysin, plasmin, matrix metalloprotease-9, angiotensin-converting enzyme, and endothelin-converting enzyme.2 In animal models, IDE deficiency correlates with increased Aβ levels and increased risk of AD, whereas IDE overexpression appears to protect against AD.3,4 Genetic linkage and biochemical analyses also strongly support a connection between IDE and AD.5,6 IDE also cleaves other small molecules (e.g., insulin, glucagon, and amylin) that can adopt β secondary structure and form amyloid fibrils.7
IDE belongs to the M16A family of zinc-dependent metalloproteases, which are evolutionarily widespread and highly conserved in sequence and structure.8,9 An inverted zinc-binding motif is characteristic of this family. The recent elucidation of several M16A enzyme structures, including that of human IDE, pitrilysin (Escherichia coli), and the related M16C enzyme PreP1 (Arabidopsis thaliana), reveals a structure resembling that of a clamshell in which substrates form β-sheet contacts with β-strands of IDE.8,9 Members of the M16A family can recognize each others’ substrates as evident by the ability of mammalian IDE and pitrilysin to substitute for the yeast M16A enzymes Axl1p and Ste23p in production of the yeast a-factor mating pheromone10,11 and the ability of Ste23p and pitrilysin to cleave Aβ.12,13
The proposed physiological role of IDE in Aβ clearance has led to the hypothesis that its hyperactivation could be therapeutically beneficial. Studies of IDE involving under-and overexpression in transgenic mice support this hypothesis.3,4 Small-molecule activators of IDE can thus be considered potential therapeutic agents, and several such molecules have been reported.14,15 Nucleotide triphosphates (i.e., adenosine triphosphate [ATP]) activate IDE to cleave certain small substrates other than Aβ. This effect occurs at ATP concentrations (i.e., 0.1–1 mM) much higher than that thought to exist physiologically in the extracellular environment where Aβ is supposedly proteolyzed by IDE (i.e., 5–50 nM) but within the range expected intracellularly (4–5 mM).16,17 ATP-dependent enhancement of IDE activity does not involve ATP hydrolysis; the effect is also buffer dependent.14 High-throughput screening (HTS) has identified two additional small-molecule activators of IDE.15 Curiously, these molecules enhance IDE-mediated cleavage of an Aβ reporter only when the synthetic non-Aβ HTS substrate is also present. The synthetic compound suramin has also been indicated as an IDE activator.18 To date, data pertaining to its effectiveness as an IDE activator or AD therapeutic have not been released.
In this study, we report seven chemical activators of IDE that were identified by HTS from a pharmacophore-rich small-molecule library provided by the National Institutes of Health (NIH) Developmental Therapeutics Program. We demonstrate the ability of identified compounds to enhance the activity of rat IDE toward both synthetic and natural peptides based on the yeast a-factor mating pheromone. We also describe a novel internally quenched Aβ-based reporter useful for direct measurement of Aβ-cleaving activity. The results we present in this study collectively demonstrate the existence of cell-permeable, substrate- and species-specific activators of IDE.
Materials and Methods
Peptides
Two synthetic substrates were used to monitor the in vitro activities of the IDE orthologs evaluated in this study. One was an internally quenched, fluorogenic peptide Abz-SEKKDNYIIKGV-NitroY-OH (AnaSpec, Inc., San Jose, CA; CHI Scientific, Inc., Maynard, MA), where Abz is aminobenzoic acid and NitroY is 3-nitro-tyrosine. The peptide is based on the sequence flanking the M16A cleavage site found in the yeast a-factor precursor. It was used for the activator screen and for monitoring activities since it is recognized by multiple IDE orthologs, including rat IDE, yeast Ste23p, worm IDE, bovine trypsin, and pronase E; human IDE does not recognize this substrate. The second IDE substrate was the internally quenched, fluorogenic peptide H2N-DAEFRHDSGYEVHHQKDABCYLLVFFAE EDANSDVGSNK-OH (CHI Scientific), where KDABCYL is ε-DABCYL-L-lysine and EEDANS is EDANS-L-glutamate. The peptide, based on the Aβ1–28 sequence, was used for monitoring activities of rat, human, and worm IDE. Powder forms of both peptides were resuspended in DMSO (10 mM) and stored at –80 °C. Diluted 2× working stock solutions (100 µM) were heated to 65 °C for 3 min and then cooled to room temperature prior to use. Product formation was measured using a Bio-Tek Synergy microplate reader (Bio-Tek, Winooski, VT) equipped with 320/420-nm and 320/485-nm excitation/emission filter sets, respectively.
The substrates used to monitor activities of yeast Rce1p and yeast Ste24p were internally quenched, fluorogenic, farnesylated peptide substrates that are based on the K-Ras C-terminus.19,20 Product formation for these substrates was measured using a 320/420-nm excitation/emission filter set.
Recombinant Enzymes and Other Reagents
Plasmids encoding rat and yeast Ste23p have been described.13,21 The plasmid encoding human IDE was constructed by amplifying the human IDE open reading frame from plasmid IDE-pSRα using oligos designed to encode a 6X-His tag at the N-terminus and restriction sites for sub-cloning the PCR product into the XbaI and NotI sites of pET30b(+) (Novagen, Madison, WI).22 The plasmid encoding Caenorhabditis elegans IDE was constructed by amplifying the cDNA sequence of F44E7.4 from the RB1 cDNA library by PCR and subcloning into the XbaI and NotI sites of pET30b(+).23 Recombinant rat, human, C. elegans, and yeast IDE were inducibly expressed in BL21 (DE3) E. coli and recovered by immobilized nickel affinity chromatography essentially as previously described.13,21 Purified IDE was stored at –80 °C as 1 mg/mL aliquots in storage buffer (50 mM HEPES, 140 mM NaCl, 20% glycerol [pH 7.4] or 25 mM KPi, 200 mM NaCl, 20% glycerol [pH 7.6]). Membranes containing yeast Rce1p or yeast Ste24p activity were prepared as previously described.20 Bovine trypsin, pronase E, proteinase K, ATP, bovine serum albumin (BSA), and human recombinant insulin were all from Sigma-Aldrich (St. Louis, MO). Ia1 and Ia2 were from Key Organics (London, UK).
Chemical Library
The Diversity Set compound library was obtained through the NIH Developmental Therapeutics Program (DTP).24 This compound library contains 1981 compounds with unique pharmacophore characteristics that were reduced from a parent set of more than 70 000 compounds with the use of the 3D structure generation program Chem-X (Chemical Design Ltd., Mahwah, NJ).
High-Throughput Screen
Assays were conducted in the wells of a black, clear-bottom, 96-well microplate (Costar, Corning, Inc., New York, NY) in 0.1 M potassium phosphate buffer (KPi), pH 7.6. The fully assembled reactions contained rat IDE (10 µg/mL), compound (100 µM), and a-factor–based substrate (50 µM). The assay protocol involved mixing the enzyme and substrate in a 1:1 ratio after the enzyme was preincubated for 10 min at 37 °C with a unique compound from the DTP library. Product fluorescence was monitored for each well over a 60-min time course. The 15- to 30-min window of the assay was typically used to determine rate of reaction. Compounds yielding IDE activity ≥150% relative to the DMSO-treated control were preliminarily assigned as activators (n = 33; 1.67% hit rate). Detailed examination of hits identified compounds that altered the baseline fluorescence and/or enhanced the fluorescence intensity of the fluorophore, generating false positives. Thus, observed fluorescence values were normalized by first subtracting the baseline fluorescence of the substrate in the presence of compounds and then dividing observed rates of reaction by the maximum fluorescence of the sample observed after complete digestion of the substrate by a mixture of trypsin (5 µg/mL) and pronase E (10 µg/mL). Compounds yielding ≥150% activity relative to a DMSO-treated control after normalization were designated as activators for follow-up studies (n = 8; 0.4% hit rate). One of these compounds (4) was subsequently dropped from analysis due to no observed impact on the kinetic parameters of rat IDE and inconsistency between experiments.
Chemical Analyses
The structures of the identified IDE activators were rendered in ChemBioDraw Ultra (v. 11.0; CambridgeSoft, Cambridge, MA). Molecular weight, Log P, and other quantitative structure-activity relationship (QSAR) values were calculated using the Analysis and Chemical Properties tools of ChemBioDraw. In addition, molecular weights of the compounds were confirmed by electrospray ionization (ESI) analysis (Proteomic and Mass Spectrometry Core Facility, University of Georgia) using an API I Plus, PE Sciex mass spectrometer (Perkin Elmer, Waltham, MA) in positive or negative ion mode depending on the biophysical properties of the compounds. All samples were dissolved in methanol for ESI analysis.
Target Specificity Analyses
The effects of compounds on the activities of other enzymes were assessed using methods similar to that outlined above for HTS. Reaction rates were determined in triplicate after a 10-min preincubation with 100 µM compound. Single-dose effects on rat IDE (10 µg/mL), Ste23p (10 µg/mL), trypsin (0.5 µg/mL), and pronase E (0.5 µg/mL) were determined using the a-factor–based peptide using conditions outlined above, with the exception that Ste23p activity was assayed at 30 °C. Effects on Rce1p and Ste24p activities were determined using yeast-derived microsomes (250 µg/ mL) enriched for these enzymes and a K-Ras–based peptide.20 In all cases, activity values were normalized using the trypsin/pronase approach described above and are reported relative to a DMSO-treated control that was set at 100% within each enzyme set.
Dose-Response, Kinetic, and Other Assays
Values for dose-response curves were determined according to the conditions outlined above (i.e., KPi buffer) using a range of activator concentrations (up to 1 mM) and trypsin/pronase normalization. Half-maximal activating concentration values (AC50) were derived from best-fit curves obtained by plotting the data points in Prism 4.0 (GraphPad Software, Inc., La Jolla, CA). For compounds lacking sigmoidal dose response-curves, the concentration resulting in half-maximal activation ([Max]50) was used.
Data for kinetic analyses were determined over a range of substrate concentrations (0–750 µM) in the absence and presence of activator (100 µM). Data collected were subjected to nonlinear regression analysis in Prism 4.0 to extract kinetic parameters. Curves were fitted using the equation Y = Vmax * X/[Km + X]. Because internal quenching effects were observed with the a-factor–based substrate at concentrations ≥30 µM, activities observed at these concentrations were multiplied by a correction factor for the purposes of kinetic analysis (Suppl. Fig. S1).
The effects of BSA, ATP, insulin, and buffer composition were determined in the context of 50 µM a-factor–based substrate. BSA assays were performed in 0.1 M KPi (pH 7.6) containing 0.01% BSA; dose-response curves contained 0% to 0.5% BSA.25 ATP assays were performed in 50 mM Tris (pH 7.5) containing 1 mM ATP and 0.1% BSA; dose-response curves contained 0 to 10 mM ATP in either 0.1 M KPi (pH 7.6) or 50 mM Tris (pH 7.5) containing 0.1% BSA.15 Insulin competition experiments were performed in 0.1 M KPi (pH 7.6) containing 0.92 µM human recombinant insulin; dose-response curves contained 0 to 17.4 µM insulin. The effect of activators on Aβ1–28 cleavage was determined using 50 µM substrate in the presence of 100 µM activator. In this case, proteinase K was used to obtain complete cleavage of the Aβ1–28 to obtain the maximum fluorescence amplitude. Human and C. elegans IDE were used at a concentration of 100 µg/mL and 10 µg/mL, respectively.
The activating properties of two previously reported IDE activators (Ia1 and Ia2) were also evaluated for comparison to the hits obtained in this study.15 They were evaluated as for the single-dose target specificity studies described above using previously reported concentrations of Ia1 (50 µM) and Ia2 (10 µM).
Enzyme-Chemical Interaction Studies
For determining thermal melt profiles, a solution of recombinant rat IDE (0.5 µM) and SYPRO Orange (5× working solution diluted from a 5000× stock solution; Sigma-Aldrich) was prepared in 0.1 M KPi (pH 7.6) and dispensed as 100-µL aliquots into a 96-well plate (HSP-9601; MJ Research, Waltham, MA) containing DMSO or a candidate activator (100 µM). The wells were overlaid with 10 µL mineral oil and incubated in the dark at room temperature for 60 min. Samples were heated from 28 to 70 °C (Δt increment = 1–3 °C) and fluorescence measured (465 nm excitation, 590 nm emission) at appropriate intervals using a FluoDia T70 high-temperature fluorescence plate reader (Photon Technology International, Birmingham, NJ). Thermal melt mid-points (Tm) were determined from a fluorescence versus temperature plot that was fit to a four-parameter sigmoidal dose-response (variable slope) equation using Prism software. Similar results were obtained when IDE was evaluated in 0.1 M HEPES buffer (pH 7.5) containing 150 mM NaCl.
For native polyacrylamide gel electrophoresis (PAGE) profiles, rat IDE (0.5 µg) was preincubated with compound (100 µM) for 60 min at 37 °C, mixed with equal volume of 2× native gel sample buffer (0.1% bromophenol blue, 20% glycerol, 0.6 M Tris-HCl, pH 6.8), and analyzed by 10% native PAGE at 4 °C. Gels were stained with silver and scanned using a flatbed scanner.
Yeast Strains and Plasmids
The yeast strains used in this study were IH1783 (MATa trp1 leu2 ura3 his4 can1), y272 (MATa trp1 leu2 ura3 his4 can1 axl1::LEU2 ste23::LEU2), and RC757 (MATα sst2-1).26–28 Plasmid-bearing versions of these strains were generated according to published methods.29 Strains were grown at 30 °C using YEPD or synthetic complete dropout (SC−) medium.27 Plasmids pRS316 (CEN URA3), pWS192 (2µ TRP1 MFA1), pWS491 (2µ URA3 PPGK-RnIDE), and pWS496 (2µ URA3 PPGK-RnIDE::2HA) have been reported.10,30,31
Yeast Toxicity Assay
To determine whether compounds could exert their activating effects in vivo, it was first critical to determine whether any were cytotoxic, which would complicate in vivo analyses. The effect of compounds on yeast growth was assessed by determining the density of compound-treated cultures after incubation for 44 h at 30 °C. Briefly, a saturated starter culture of yeast y272 cotransformed with pWS192 and pWS496 was diluted (1:2000) into SC-UW and split into equal-volume aliquots (1 mL), and aliquots were treated with DMSO or compound (100 µM). Each condition was evaluated in triplicate, and the average absorbance observed for each condition after 44 h was calculated as a percentage of that observed for the DMSO-treated condition, which was set at 100%. To obtain accurate measurements of culture densities, saturated cultures were typically diluted 10-fold, and A600 values obtained were expressed as percent of untreated. Where toxicity was observed, a lower dose allowing for saturated growth after 44 h of incubation at 30 °C was empirically determined. For compounds that retained toxicity at these lower concentrations, the yeast cultures were grown for 72 h to allow saturated growth.
a-Factor Production Assay
A bioassay was used to assess the impact of compounds on the ability of IDE to promote a-factor production.10 The assay entails the recovery and enrichment of secreted a-factor from cultures of MATa yeast (y272 cotransformed with pWS192 and pWS496) followed by an assessment of bioactivity to judge the potency of the recovered pheromone. Cultures were grown to saturation (72 h at 30 °C) as described above in the presence of DMSO or compound (100 µM or highest nontoxic dose), except that 5 mL volumes were used. The recovered pheromone was spotted as a twofold serial dilution on a thin lawn of RC757 (MATα sst2-1) yeast. The amount of a-factor activity observed for each sample was normalized to the cell density of the original culture as determined from A600 absorbance measurements of appropriately diluted cultures. a-Factor production for the DMSO-treated control was set at 100%. Culture densities did not vary significantly between compound and DMSO-treated cultures (S. S. Kukday and W. K. Schmidt, unpublished observation).
Results
An a-Factor–Based Internally Quenched Fluorogenic Peptide Is Cleaved by IDE
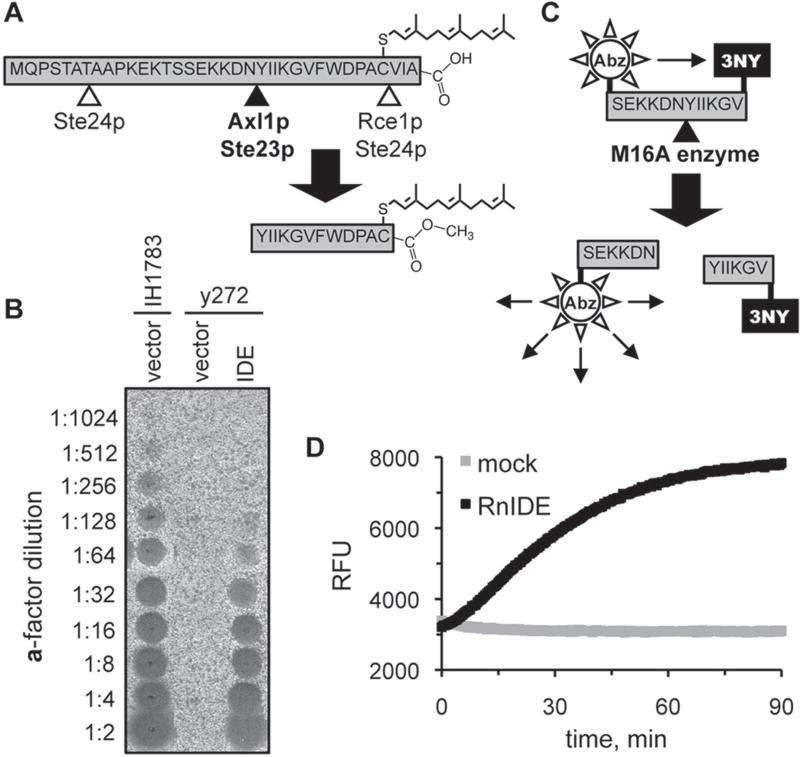
The yeast mating pheromone a-factor is synthesized as a precursor that undergoes extensive posttranslational processing, including isoprenylation, multiple proteolytic cleavages, and carboxylmethylation (Fig. 1A). One of the cleavage events is mediated by the yeast M16A proteases, with Axl1p having a predominant role in this process relative to Ste23p.13,28 We have previously documented that yeast is a convenient system for functional studies of heterologously expressed M16A enzymes.10,11 In this system, the in vivo activity of an M16A enzyme is monitored by the production of the bioactive a-factor mating pheromone, which is measured using straightforward biological assays (Fig. 1B). Given our observations, we hypothesized that an internally quenched fluorogenic peptide centered on the M16A cleavage site found within the a-factor precursor could serve as a suitable substrate for monitoring the in vitro activity of IDE and other M16A enzymes (Fig. 1C). Indeed, recombinant rat IDE cleaves such a peptide with a specific activity of 1.33 nmol/min/mg under our standard assay conditions (Fig. 1D).
Figure 1.
Reporters of M16A enzyme activity. (A) Production of the yeast a-factor mating pheromone is dependent on the action of several proteases, including the M16A enzymes Axl1p and Ste23p. (B) Rat insulin-degrading enzyme (IDE) can substitute for the yeast M16A enzymes in a-factor production in vivo. Yeast strain y272 (MATa axl1 Δ ste23Δ) was transformed with an IDE-encoding plasmid (pWS491) or an empty vector (pRS316), and resultant strains were evaluated for their ability to produce a-factor. A wild-type MATa strain (IH1783) transformed with an empty vector (pRS316) was evaluated in parallel. The appearance of a clear spot (i.e., zone of reduced growth) within the MATα lawn indicates the presence of a-factor. (C) An internally quenched fluorogenic dodecapeptide modeled on the M16A cleavage site in the yeast a-factor precursor. The NH2-terminal fluorophore is aminobenzoic acid (Abz), and the COOH-terminal quenching group is 3-nitro-tyrosine (3NY). (D) Progress curves demonstrating time-dependent fluorescent output in the presence or absence of recombinant rat IDE. The reactions contained rat IDE (10 µg/ml; 87.7 nM) or enzyme storage buffer (mock). RFU, relative fluorescence units.
Identification of IDE Activators by HTS
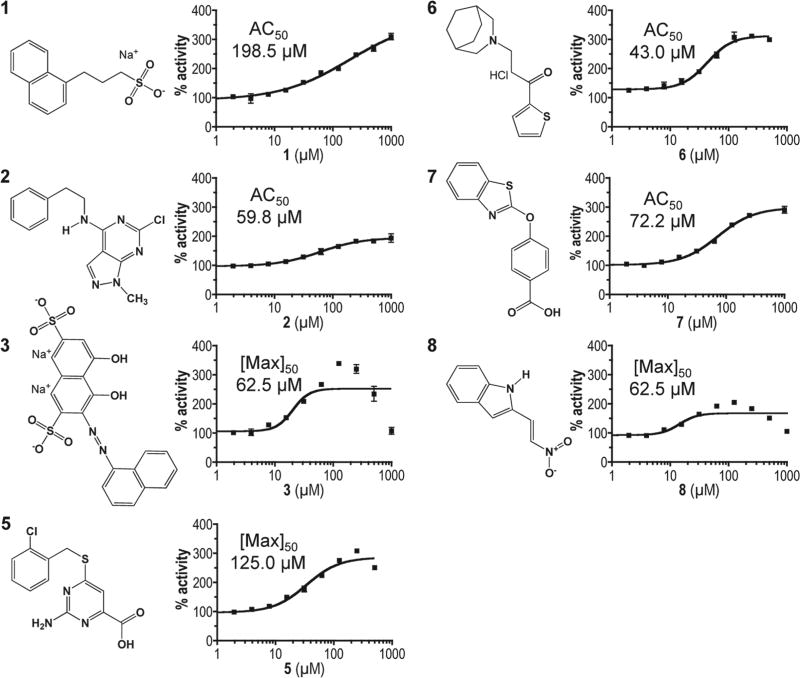
The assay described above was optimized for a 96-well format and used to screen the NIH DTP Diversity Set library for modulators of IDE activity.24 Under the conditions of the screen, the assay had a calculated Z′ factor of 0.86 and a signal-to-noise ratio of 7.0. The compounds of the library had a variety of effects on IDE activity. Of the 1981 compounds within the DTP library, 451 partly inhibited IDE (<50% activity relative to untreated), and 1495 had a minor impact on activity (50%–150% activity observed). Two compounds could not be assessed due to strong quenching effects exerted on the substrate. Of specific interest were 33 compounds (1.7% hit rate) that enhanced the rate of fluorescence output ≥150% relative to that observed for DMSO-treated IDE; DMSO represents the solvent for the library collection and was used as a control. The set of hits was further reduced to eight compounds after elimination of autofluorescing and fluorescence-enhancing compounds. An additional compound (4) was later removed for lack of an effect on the kinetic parameters of rat IDE and inconsistent behavior between experiments (S. P. Manandhar, S. S. Kukday, and W. K. Schmidt, unpublished observation). The structures of the final seven hits (Fig. 2) are represented by a variety of chemical scaffolds incorporating aromatics, heterocycles, charged groups, and aromatic systems.
Figure 2.
Chemical structures and dose-response profiles of insulin-degrading enzyme (IDE) activators. Compounds were identified by their ability to enhance rat IDE-mediated in vitro cleavage of the peptide reporter depicted in Figure 1C. Structures were downloaded from the Developmental Therapeutics Program (DTP) structure database (http://dtp.nci.nih.gov/branches/dscb/diversity_explanation.html). Compound 4 was eliminated due to a lack of measurable effect on IDE kinetic parameters and inconsistent behavior across experiments. Compounds were evaluated for their effectiveness at stimulating rat IDE activity over the indicated dose range using the fluorescence-based IDE activity assay described in Figure 1. A best-fit nonlinear dose-response curve was determined for data points using GraphPad Prism 4.0 and a four-parameter logistic equation (solid line). Where sigmoidal dose-response curves were observed, AC50 values were determined. Where hormetic response curves were observed (i.e., 3, 5, 8), the fitted curves could not be used to determine accurate AC50 values, so the lowest half-maximal activating concentration is reported ([Max]50).
The compounds all adhere to Lipinski’s rule of 5 and have other drug-like characteristics.32 The compounds range in mass from 188 to 472 Da (associated counterions excluded) (Suppl. Table S1). The most potent in vitro acting compound 3 had the lowest CLogP value (–6.04) and highest tPSA value (179.58). Although the hits were not resynthesized or purchased commercially through other sources, largely due to their unavailability, the activating potential of each hit was further confirmed in samples obtained through an independent request from the DTP. In all cases, high-performance liquid chromatography (HPLC) revealed a single peak fraction, and subsequent liquid chromatography/mass spectrometry (LC/ MS) analyses revealed the major species to be of expected mass, with some minor species detected for compounds 1 and 8 (Suppl. Figs. S2 and S3).
Activators Are IDE Specific
The impact of compounds (100 µM) on the activities of IDE and a select group of other proteases was examined (Table 1). The maximum activation of IDE observed was 356% (compound 3), and several other compounds activated >200% (compounds 5, 6, and 8). By comparison, two previously reported IDE activators (Ia1 and Ia2) enhanced IDE activity to a lesser extent. The specificity of compounds was investigated by examining their ability to stimulate the activities of other proteases. Our analysis revealed no compound-induced activation of the yeast M16A enzyme Ste23p, bovine trypsin, or pronase E. Similarly, no compound-induced activation was observed for the yeast CaaX proteases Rce1p and Ste24p, which catalyze a distinct proteolytic cleavage on the a-factor precursor.33 Although our enzyme panel was clearly not exhaustive, the inability of compounds to activate five other proteases, including a closely related ortholog, suggests that the compounds do not promiscuously activate enzymes. By contrast, partial inhibition (≤75% residual activity) was observed in a limited number of instances for pronase (compound 5) and the CaaX proteases (compounds 3 and 8).
Table 1.
Activities of Various Proteases in the Presence of Rat Insulin-Degrading Enzyme (IDE) Activators
| % Activity
|
|||||||
|---|---|---|---|---|---|---|---|
| Compound | NSC # | IDE | Ste23p | Trypsin | Pronase | Rce1p | Ste24p |
| DMSO | — | 100.0 ± 6.0 | 100.0 ± 2.8 | 100.0 ± 10.6 | 100.0 ± 0.8 | 100.8 ± 0.8 | 100.0 ± 16.0 |
| 1 | 2737 | 195.4 ± 12.2 | 112.0 ± 5.4 | 94.2 ± 0.3 | 100.9 ± 0.1 | 89.7 ± 3.8 | 97.2 ± 12.7 |
| 2 | 19139 | 170.3 ± 16.0 | 117.2 ± 2.8 | 115.1 ± 3.9 | 89.2 ± 0.6 | 83.2 ± 3.4 | 93.0 ± 6.8 |
| 3 | 45208 | 356.0 ± 16.0 | 90.4 ± 0.6 | 99.3 ± 2.2 | 103.8 ± 0.1 | 55.3 ± 1.0 | 74.7 ± 8.6 |
| 5 | 49713 | 263.5 ± 1.0 | 103.4 ± 1.6 | 94.3 ± 5.4 | 65.3 ± 3.6 | 101.2 ± 11.0 | 131.4 ± 18.8 |
| 6 | 95570 | 230.7 ± 13.5 | 82 ± 0.3 | 111.5 ± 2.8 | 104.4 ± 0.0 | 90.1 ± 1.0 | 98.6 ± 9.9 |
| 7 | 122335 | 186.6 ± 7.4 | 102.1 ± 1.7 | 111.0 ± 1.2 | 104.3 ± 0.9 | 89.6 ± 2.4 | 92.8 ± 13.1 |
| 8 | 150982 | 195.0 ± 14.0 | 80.1 ± 6.0 | 101.8 ± 1.8 | 93.3 ± 1.8 | 60.6 ± 2.6 | 89.2 ± 10.7 |
| Ia1 (50 µM) | — | 144.0 ± 5.0 | ND | ND | ND | ND | ND |
| Ia2 (10 µM) | — | 157.0 ± 8.0 | ND | ND | ND | ND | ND |
Compounds (100 µM final, unless otherwise indicated) were evaluated using fluorescence-based in vitro assays. Activities were normalized to that of the appropriate DMSO-treated control, which was set at 100%. Specific reaction conditions are described in the Materials and Methods section. NSC, Cancer Chemotherapy National Service Center; ND, not determined. The dashes indicate that no NSC # is available.
Dose-Response and Kinetic Studies
The HTS screen for IDE activators used a fixed concentration of compound. To determine the optimal activating concentration of compounds, dose-response assays were performed. Compounds 1, 2, 6, and 7 yielded classic sigmoidal dose-responses from which AC50 values were calculated and found to be in the range 43 to 198 µM (Fig. 2). Saturation with compound 1 was not achieved at the highest concentration tested (1 mM), but the data were sufficient for curve fitting. Compounds 3 and 8 and possibly 5 exhibited an inverted bell-shaped activation curve with an observable peak concentration and less activation at both lower and higher doses (i.e., a hormetic dose response). For these compounds, the lowest half-maximal effective dose (i.e., [Max]50) is reported. These values were in the range of 62.5 to 125 µM.
Enzyme kinetic studies were performed in the presence of 100 µM compound. All compounds reduced the Km of IDE, and all but compound 5 increased the Vmax of IDE compared with that observed for a DMSO-treated control (Table 2). Compound 6 had the largest impact on Km, whereas compound 3 had the largest impact on Vmax.
Table 2.
Kinetic Assessment of Activators
| Compound | Km, µM |
Vmax, nmol/min/mg |
|---|---|---|
| DMSO | 183.7 ± 25.94 | 154.8 ± 12.26 |
| 1 | 107.7 ± 6.42 | 194.1 ± 5.85 |
| 2 | 117.9 ± 9.04 | 164.2 ± 6.4 |
| 3 | 122.1 ± 6.08 | 267.9 ± 6.7 |
| 5 | 148.8 ± 23.67 | 127.8 ± 10.41 |
| 6 | 92.2 ± 7.7 | 176.3 ± 7.35 |
| 7 | 148.3 ± 20.07 | 194.1 ± 13.81 |
| 8 | 123.8 ± 13.14 | 185.0 ± 9.73 |
Compounds (100 M final) were evaluated using the fluorescence-based in vitro assay in 0.1 M KPi buffer, pH 7.6. Kinetic parameters were derived using nonlinear regression analysis in GraphPad Prism 4.0.
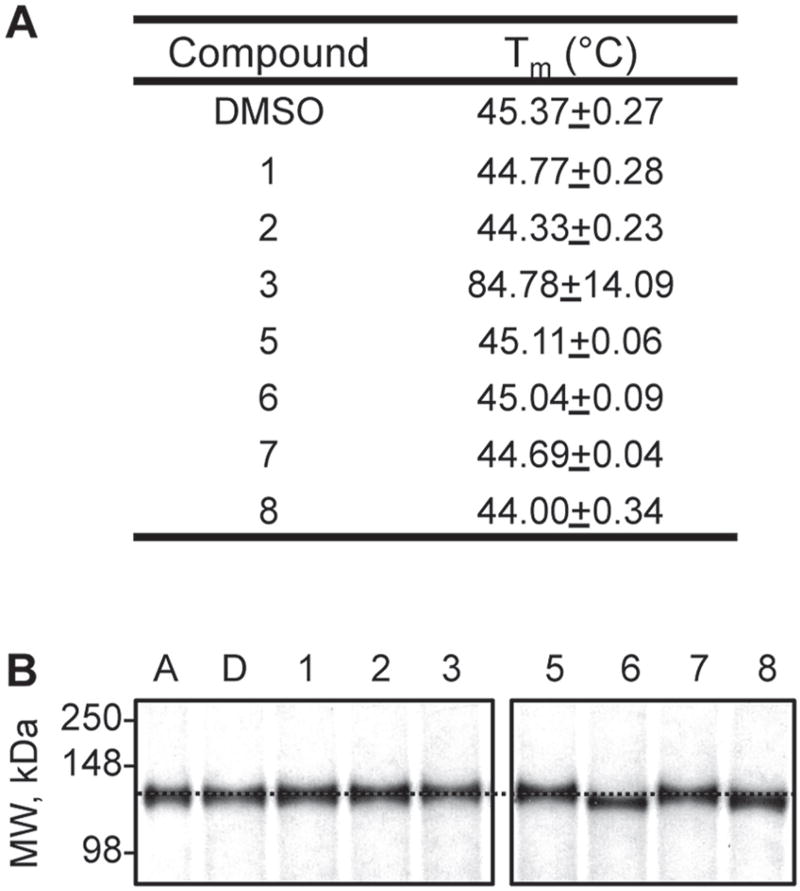
Compounds 3, 6, and 8 Physically Interact with IDE
To determine whether the activators interact with IDE to exert their effects, we performed two types of studies. First, we hypothesized that ligand binding would alter the biophysical properties of IDE in some manner. Thermal melt analyses revealed a substantial shift in the melting profile of IDE in the context of compound 3 only (Fig. 3A). Second, we hypothesized that binding of activators might induce structural changes in the enzyme. Native PAGE analyses did not reveal a mobility shift in the context of compound 3 but did reveal a noticeable shift with compounds 6 and 8, suggestive of a physical interaction (Fig. 3B). The nature and strength of this interaction remain unknown and open for future investigation. We did not observe a mobility shift with ATP, which induces a measurable conformational change in IDE.34 The differing results may be due to differences in the native gel protocols.
Figure 3.
Effect of compounds on the biophysical properties of rat insulin-degrading enzyme (IDE). (A) Thermal melt midpoints (Tm) observed in the presence of IDE activators (100 µM) were determined using a thermal shift assay. Compound-treated rat IDE (0.5 µM) was evaluated across the temperature range 28 to 70 °C. (B) Mobility shifts observed in the presence of IDE activators. The indicated compounds were incubated with 1 mg/mL IDE for 60 min at 37 °C and then analyzed by native polyacrylamide gel electrophoresis (10%). Adenosine triphosphate (A) was used at 3 mM; compounds 1 to 8 were used at 100 µM. D, DMSO. A dashed horizontal line has been drawn across the image at the expected mobility of IDE.
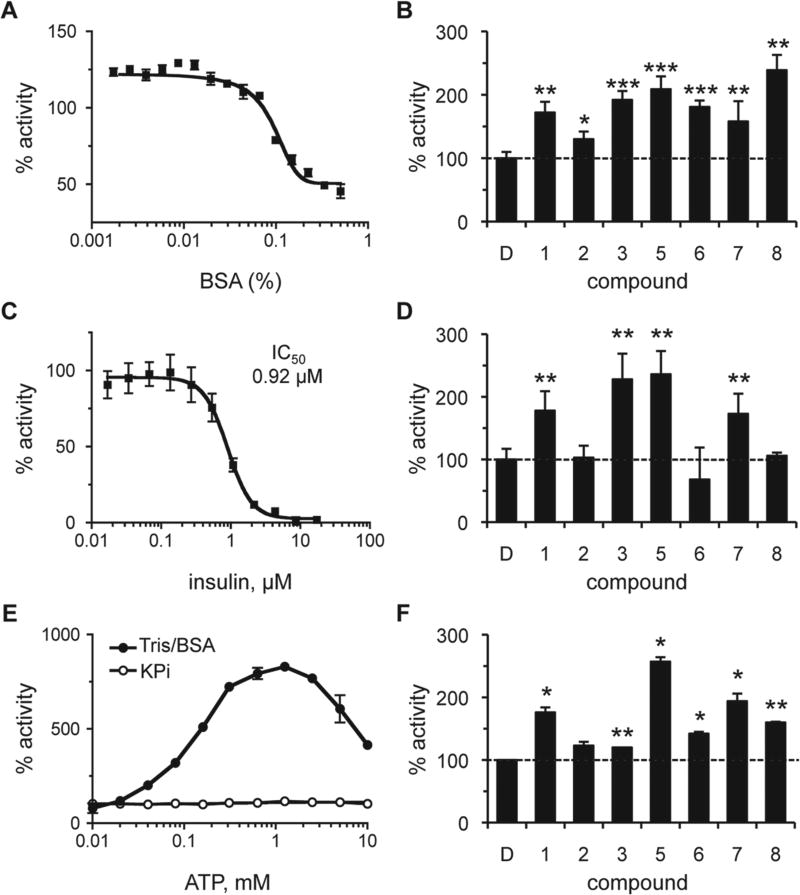
Effect of Assay Conditions on Activator Potency
Promiscuous activation has been reported for certain enzyme/ activator combinations.25 Often, the activating effects of such compounds are mitigated in the presence of BSA or low concentrations of detergent. In the presence of low amounts of BSA (≤0.02%), a modest increase in IDE activity was observed (Fig. 4A). The reason for this effect is unknown and could reflect stabilization of a more active form of IDE or better substrate/enzyme availability (i.e., less nonspecific adsorption by plasticware). Importantly, all of the compounds retained their ability to activate IDE in the presence of an optimal dose of BSA (Fig. 4B). The relative amount of activation observed was less than that observed in the absence of BSA (Table 1), which is due to the higher baseline activity of the BSA-treated control. Similarly, detergent treatment (0.1% CHAPS) also reduced but did not completely neutralize compound-induced activation of IDE (S. P. Manandhar and W. K. Schmidt, unpublished observation).
Figure 4.
Effect of assay conditions on the properties of rat insulin-degrading enzyme (IDE) activators. The effect of bovine serum albumin (BSA), insulin, and adenosine triphosphate (ATP) on the activity of IDE was determined both in the absence and presence of compounds. Rat IDE was used (87.7 nM) in the assay described in Figure 1. Values are reported as percentages relative to a water or DMSO-treated control as appropriate (n = 4); *p < 0.05, **p < 0.01, and ***p < 0.001 relative to the mock-treated control. For dose-response curves, data points were plotted, and a best-fit nonlinear dose-response curve was determined using GraphPad Prism 4.0 as described in Figure 2. (A) Observed activity of IDE in 0.1 M KPi over a range of BSA (0%–0.5% final). (B) Observed activity in the presence of compounds (100 µM) in 0.1 M KPi/0.01% BSA. The dashed line is a visual reference for 100% activity (also present in panels D and F). (C) Observed activity of IDE in 0.1 M KPi over a range of human insulin (0–17.2 µM). (D) Observed activity in the presence of compounds (100 µM) in 0.1 M KPi containing 0.92 µM (IC50) insulin. (E) The effect of ATP (0–10 mM) on IDE activity was evaluated in 0.1 M KPi or 50 mM Tris (pH 7.5) containing 0.01% BSA (Tris/BSA). (F) Observed activity of IDE in the presence of compounds in Tris/BSA containing 1 mM ATP. Compounds were used at optimal concentrations as derived from dose-response curves in Tris/BSA buffer (S. P. Manandhar and W. K. Schmidt, unpublished observations). Compound 1 was used at 1000 µM; compounds 5, 6, and 7 at 500 µM; compound 2 at 250 µM; and compounds 3 and 8 at 125 µM.
On the basis of the fact that IDE can cleave multiple substrates, we investigated the ability of our compounds to hyper-activate IDE under mixed substrate conditions. The compounds were evaluated in the presence of insulin, which disproportionately inhibits IDE-mediated Aβ degradation.21 The effect of insulin on degradation of the a-factor–based substrate was also disproportionate. Dose-response studies revealed that 10 µM insulin was sufficient to fully inhibit IDE activity in the presence of 50 µM a-factor–based substrate (Fig. 4C). The observed IC50 for insulin was approximately 0.92 µM. We evaluated our compounds at the IC50 concentration of insulin and observed that only four compounds (1, 3, 5, 7) retained their ability to stimulate IDE activity (Fig. 4D).
Last, we examined whether our compounds were activating IDE in an ATP-like manner.14 We expected this to be unlikely since ATP-dependent activation was not observed under the assay conditions used for HTS. To resolve whether the lack of ATP-dependent activation was due to our substrate or assay conditions, we evaluated the effect of ATP under assay conditions where effects had been observed.14,15 The switch from a KPi buffering system to one containing Tris/BSA resulted in ATP-dependent activation of IDE, indicating that using KPi buffer indeed neutralizes ATP-dependent activation of IDE (Fig. 4E). The maximum activating concentration in the Tris/BSA buffering system was approximately 1 mM. Evaluation of the activators under these conditions revealed that all the compounds, with the exception of compound 2, displayed a significant additive effect in the presence of ATP (Fig. 4F). Compound 5 was associated with the highest additive effect (>250% activity relative to ATP-only condition). Despite having the highest activating effect under KPi-buffered conditions, compound 3 marginally activated IDE in the presence of ATP (approximately 25% over the ATP-only condition). Since a majority of the compounds displayed additive effects in the presence of ATP, we deduce that these compounds likely activate IDE by a mechanism distinct from that used by ATP. Our results also indicate that our compounds function in a buffer-independent manner, unlike ATP, whose use is restricted to certain buffering systems.
Effect of Activators on Yeast Growth and IDE-Mediated Production of a-Factor
The ability of compounds to enhance IDE-mediated cleavage of an a-factor–based substrate in vitro led to the hypothesis that these compounds would enhance a-factor production in vivo when IDE is expressed heterologously as the only M16A enzyme in yeast. Prior to testing this hypothesis, we first evaluated whether compounds negatively affected the viability of yeast. At a dose of 100 µM, compounds 1 and 3 did not substantially affect yeast saturation density (>90% relative to DMSO-treated control). Compounds 2, 5, and 7 modestly decreased density (61%– 87% relative to DMSO-treated control). Compound 6 and 8 completely inhibited growth. Dose-response studies revealed that treating cultures with lower concentrations of compounds (i.e., 5, 6, and 8) resulted in saturation after 44 h (>90% relative to DMSO-treated culture). Compound 7 did not exhibit a dose-dependent response, and treated cultures did not achieve saturated growth at the lowest concentration evaluated (12.5 µM). In this case, extending the incubation period to 72 h led to near saturation of the treated culture (>85% relative to DMSO-treated culture) (S. S. Kukday, S. P. Manandhar, and W. K. Schmidt, unpublished observation).
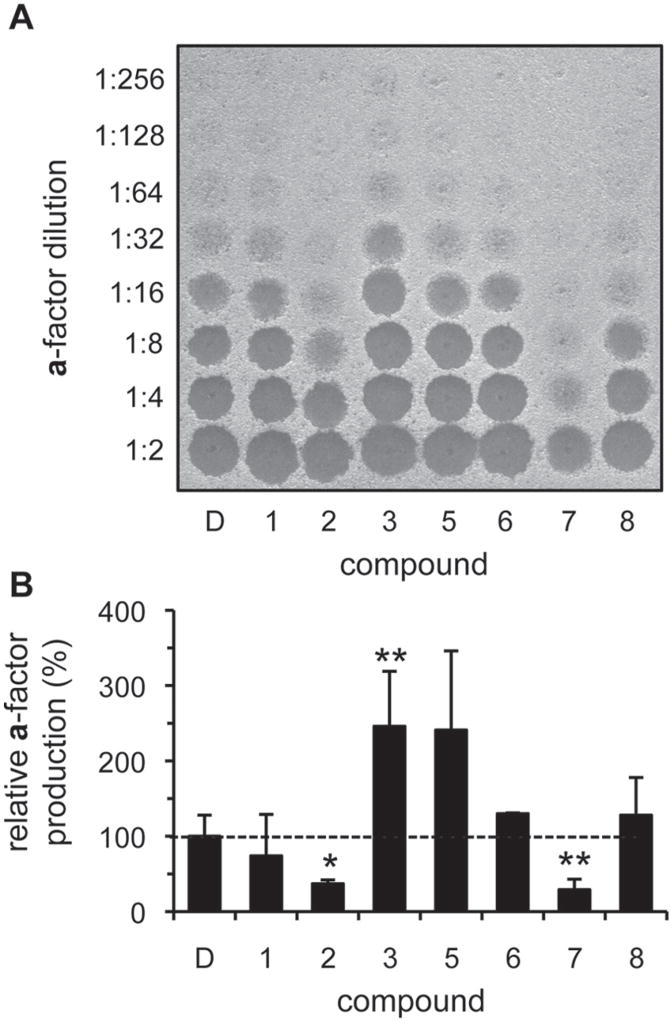
Using conditions guided by the toxicity data, we next addressed whether any of the compounds could stimulate IDE-mediated a-factor production when added to yeast cultures. After 72 h of incubation in the presence of 100 µM compound, most cultures achieved a saturated culture density; compounds 5, 6, and 8 were used at 12.5 µM, 50 µM, and 25 µM, respectively. The a-factor produced was isolated at the end of the treatment period and assayed for biological activity (Fig. 5A). Quantification of amounts of a-factor produced revealed that compounds 3, 5, 6, and 8 enhanced a-factor production in vivo (Fig. 5B), with compound 3 bringing about the most significant change. Unexpectedly, compounds 2 and 7 reduced the yield of a-factor produced.
Figure 5.
Select compounds enhance in vivo insulin-degrading enzyme (IDE)–dependent production of yeast a-factor. Diluted yeast cultures (1:2000; y272 cotransformed with pWS192 and pWS496) were grown to saturation (72 h) in the presence of activators, and the a-factor produced was recovered and analyzed. Compounds were used at 100 µM, with the exception of compounds 5, 6, and 8 (12.5 µM, 50 µM, and 25 µM, respectively). The raw data (A) were quantified and mean values graphed relative to a DMSO-treated control (B). Each value is normalized to the density of the culture at the time a-factor was collected (n = 4 for all compounds, except 6, for which n = 2); *p < 0.05 and **p < 0.01 relative to the DMSO-treated control. A nearly identical graph is observed in the absence of normalization.
Where changes in net a-factor production were observed, there were no significant differences in final culture density, thus ruling out this trivial explanation for the effect of compound treatment (S. S. Kukday and W. K. Schmidt, unpublished observation). To rule out the possibility that the activators were nonspecifically enhancing a-factor recovery or mimicking a-factor biological activity, untreated cultures were grown to saturation and then treated with activators and incubated for an additional 24 h. At the end of this incubation period, the cultures were processed for a-factor recovery. None of the compound-treated cultures showed a net increase in a-factor production by comparison to a DMSO-treated control (S. S. Kukday and W. K. Schmidt, unpublished observation).
Substrate and Species Specificity of IDE Activators
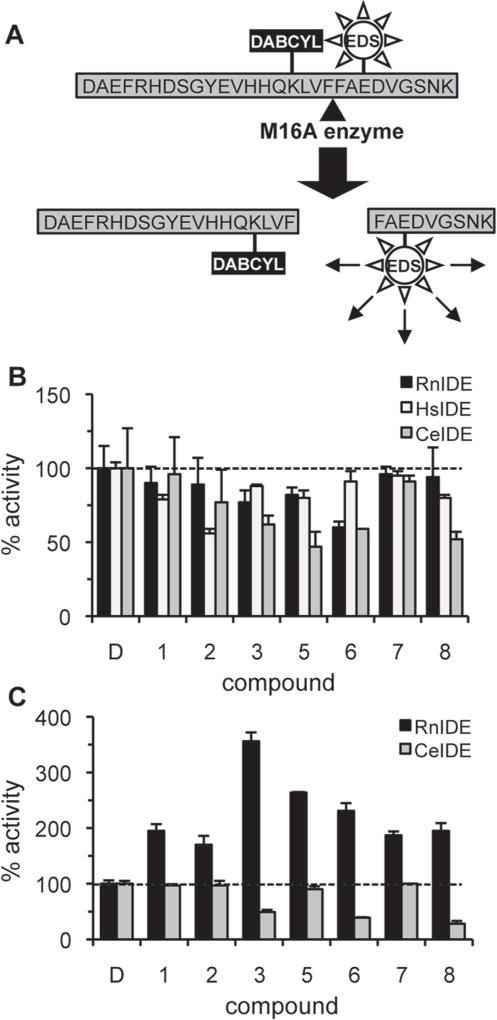
Although the synthetic and in vivo a-factor–based substrates described above are convenient reporters of IDE activity, we sought to examine the ability of compounds to enhance cleavage of a more physiologically relevant substrate. Hence, we designed a novel fluorescence resonance energy transfer (FRET)–based reporter to evaluate in vitro cleavage of Aβ1–28 (Fig. 6A). Under the same reaction conditions used to evaluate our a-factor–based reporter (0.1 M KPi, pH 7.6), the Km observed for rat IDE with the fluorogenic Aβ1–28 substrate was 142.3 µM. All of the compounds identified as rat IDE activators in the context of the a-factor– based reporter, as well as Ia1 and Ia2 identified by Cabrol et al.,15 failed to enhance rat IDE-mediated cleavage of this Aβ reporter (Fig. 6B; S. S. Kukday and W. K. Schmidt, unpublished observation). Our compounds also failed to stimulate human and C. elegans IDE. Moreover, the compounds were unable to enhance rat IDE-mediated degradation of full-length Aβ1–40 (S. S. Kukday and W. K. Schmidt, unpublished observation). The compounds were further unable to enhance CeIDE-mediated cleavage of the a-factor reporter (Fig. 6C). Human IDE did not recognize the a-factor–based substrate and could not be evaluated. Together, these findings indicate that the rat IDE activators reported here display substrate and species specificity.
Figure 6.
Rat insulin-degrading enzyme (IDE) activators are substrate and species specific. (A) An internally quenched fluorogenic peptide was modeled on Aβ1–28. The quencher DABCYL is conjugated to Lys16 and the fluorophore EDANS (EDS) to Glu22. The peptide has unmodified N- and C-termini. (B) Effect of compounds (100 µM) on rat (RnIDE; 10 µg/mL), human (HsIDE; 100 µg/mL), and worm IDE (CeIDE; 10 µg/mL) mediated cleavage of the Aβ1–28 reporter (50 µM) was evaluated in 0.1 M KPi, pH 7.6. Mean activity values are reported as percentages relative to a DMSO-treated control (n = 3). (C) Effect of compounds (100 µM) on the ability of RnIDE and CeIDE (each at 10 µg/mL) to cleave the a-factor reporter was evaluated as in Figure 1 HsIDE does not recognize the a-factor–based reporter and thus was not evaluated. Mean activity values are reported as percentages relative to the DMSO-treated control (n = 3).
Discussion
IDE cleaves small amyloidogenic peptides.35 Thus, the activation of IDE has been viewed as therapeutic for the clearance of Aβ.36 Indeed, IDE overexpression reduces amyloid deposits in the brain of amyloid precursor protein (APP) transgenic mice.4 IDE can also be rendered hyperactive through mutations that expose the active site for a longer period of time.9 As documented by our findings and related studies, chemical activation of IDE is also feasible.15,18 We have identified seven small-molecule activators of rat IDE that lower the Km of IDE, retain activation in the presence of a competing substrate (insulin), and are effective in an in vivo assay. The activators are structurally distinct from previously reported IDE activators, consistent with the possibility that there may be multiple modes of IDE activation.
An important and critical outcome of our study revolves around the use of nonnative IDE substrates as reporters (i.e., a-factor). The fact that M16A enzymes have reciprocal substrate specificity suggested that any IDE substrate might have utility in identifying IDE activators. This observation drove our initial choice of a-factor–based reporters because of the potential for both in vitro and in vivo assay development. This direction, however, yielded rat IDE activators that did not enhance cleavage of an Aβ1–28 reporter. The finding that Ia1 and Ia2 enhance IDE-mediated cleavage of the a-factor–based reporter but not the Aβ1–28 reporter further lends support to the idea of substrate-specific activation. In the original study, these activators enhanced IDE-mediated Aβ degradation only when the nonnative, high-throughput synthetic substrate was included in the reaction.15 We posit that failure of compounds to enhance Aβ cleavage is an indication that compounds are substrate specific. Should this indeed be the case for IDE activators in general, it may be feasible to identify activators that specifically enhance cleavage of Aβ over that of insulin and other IDE substrates, thereby limiting the potential negative impact on glucose homeostasis and other physiological pathways affected by IDE.
We expect our current findings to provide guidance for future studies aimed at identifying IDE activators. Foremost, the observation that the compounds identified in this study do not generally activate IDE orthologs emphasizes the importance of using the appropriate target enzyme (i.e., human IDE) in future high-throughput screens aimed at generating therapeutic agents. Our study also provides evidence that a hormetic in vitro dose-response profile (e.g., compounds 3 and 5) should not necessarily exclude an agent from being evaluated in cell-based assays. The underlying reason for the observed hormetic effect is unknown but could reflect an issue as straightforward as solubility (i.e., precipitation at high concentrations) or as complicated as multiple binding sites (i.e., a high-affinity activating site and a low-affinity inhibitory site). We have also provided evidence that insulin affects the properties of IDE activators. In our case, several of our hits were adversely affected when insulin was present in the assay mixture. The impact of insulin and possibly other competing substrates on the activating potential of compounds should therefore be considered an important component of future activator screens. We have also provided the first example of potentially nontoxic activators of IDE, albeit toxicity was gauged against yeast cells. Nevertheless, we are encouraged by the observation that relatively nontoxic compounds exert positive effects on IDE activity both in vitro and in a cell-based system (e.g., compound 3). By comparison, suramin is broadly toxic and nonspecific, both activating and inactivating other enzymes.37,38 It also displays a hormetic response under our assay conditions (S. P. Manandhar and W. K. Schmidt, unpublished observation).
Perhaps the most important impact of our study will be on the design of future IDE reporters, which our study suggests should resemble Aβ as closely as possible. A key step toward this goal is our development of a FRET-based Aβ reporter. Our reporter is based on Aβ1–28 with a strategically positioned quencher (DABCYL) and fluorophore (EDANS) on K16 and E22, respectively. Choice of the 1–28 peptide versus a longer Aβ species was driven by the technical issues frequently reported for syntheses and storage of longer forms of Aβ.39 Moreover, residues beyond E23 lack ordered density in the IDE/Aβ1–40 co-crystal structure (PDB 2g47), suggesting that they do not affect substrate binding.9 K16 and E22 were chosen for side-chain modification because our analysis of the IDE/Aβ1–40 co-crystal structure suggested that these residues are in reasonable proximity and could be modified without impairing interactions between Aβ and IDE. Importantly, the Aβ1–28 reporter has a free N-terminus and thus retains an important exosite binding capability that is not found on other IDE HTS-compatible reporters (e.g., FAβB).40 An Aβ1–28 reporter having an N-terminal fluorophore has been reported as a relatively poor IDE substrate, but specific data were not reported, and direct comparison between the reporters is thus not possible.40 We contend that N-terminal modifications may lead to binding interference at the IDE exosite and complicate the determination of kinetic parameters. We also acknowledge that the failure of our activators to enhance cleavage of the FRET-based Aβ1–28 reporter could be indicative of interference by the EDANS and/or DABCYL moieties leading to inappropriate folding of the peptide. It is unknown whether alternative fluorophores and quenchers will yield similar results.
There are many examples of enzymes that are activated by biological small molecules (e.g., PKA by cAMP). There are, however, relatively few examples of enzymes that can be activated through synthetic small molecules. This list of enzymes is growing and includes notable targets such as glucokinase, AMP-activated protein kinase (AMPK), and the sirtuins.41 IDE activators can now be added to the limited set of small-molecule enzyme activators, and it is expected that our development of a FRET-based Aβ1–28 reporter will help drive the identification of species and Aβ-specific activators of IDE.
Supplementary Material
Acknowledgments
We thank the Developmental Therapeutics Program (National Institutes of Health) for providing the Diversity Set compound library and the following (all at the University of Georgia): Dr. Z. A. Wood for help with IDE purification and design of the Aβ1–28 reporter, Dr. J. P. Rose for assistance with thermal shift studies, the Bioexpression and Fermentation Facility for help with HPLC analyses, Dr. T. M. Dore, and members of the Schmidt laboratory for critical discussions and technical assistance.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by grants from the Alzheimer’s Drug Discovery Foundation/Institute for the Study of Aging (grant 271223 to WKS) and the National Institutes of Health (5R01GM067092 to WKS).
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Haass C, Selkoe DJ. Soluble Protein Oligomers in Neurodegeneration: Lessons from the Alzheimer’s Amyloid Beta-Peptide. Nat. Rev. Mol. Cell. Biol. 2007;8(2):101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 2.Nalivaeva NN, Beckett C, Belyaev ND, Turner AJ. Are Amyloid-Degrading Enzymes Viable Therapeutic Targets in Alzheimer’s Disease? J. Neurochem. 2012;120(Suppl. 1):167–185. doi: 10.1111/j.1471-4159.2011.07510.x. [DOI] [PubMed] [Google Scholar]
- 3.Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-Degrading Enzyme Regulates the Levels of Insulin, Amyloid Beta-Protein, and the Beta-Amyloid Precursor Protein Intracellular Domain In Vivo. Proc. Natl. Acad. Sci. U. S. A. 2003;100(7):4162–4167. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leissring MA, Farris W, Chang AY, Walsh DM, Wu X, Sun X, Frosch MP, Selkoe DJ. Enhanced Proteolysis of Beta-Amyloid in APP Transgenic Mice Prevents Plaque Formation, Secondary Pathology, and Premature Death. Neuron. 2003;40(6):1087–1093. doi: 10.1016/s0896-6273(03)00787-6. [DOI] [PubMed] [Google Scholar]
- 5.Kim M, Hersh LB, Leissring MA, Ingelsson M, Matsui T, Farris W, Lu A, Hyman BT, Selkoe DJ, Bertram L, Tanzi RE. Decreased Catalytic Activity of the Insulin Degrading Enzyme in Chromosome 10-Linked Alzheimer’s Disease Families. J. Biol. Chem. 2007;282(11):7825–7832. doi: 10.1074/jbc.M609168200. [DOI] [PubMed] [Google Scholar]
- 6.Miller BC, Eckman EA, Sambamurti K, Dobbs N, Chow KM, Eckman CB, Hersh LB, Thiele DL. Amyloid-Beta Peptide Levels in Brain Are Inversely Correlated with Insulysin Activity Levels In Vivo. Proc. Natl. Acad. Sci. U. S. A. 2003;100(10):6221–6226. doi: 10.1073/pnas.1031520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker AB, Roth RA. Insulysin and Pitrilysin: Insulin-Degrading Enzymes of Mammals and Bacteria. Methods Enzymol. 1995;248:693–703. doi: 10.1016/0076-6879(95)48046-3. [DOI] [PubMed] [Google Scholar]
- 8.Johnson KA, Bhushan S, Stahl A, Hallberg BM, Frohn A, Glaser E, Eneqvist T. The Closed Structure of Presequence Protease PreP Forms a Unique 10,000 Angstroms3 Chamber for Proteolysis. Embo J. 2006;25(9):1977–1986. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of Human Insulin-Degrading Enzyme Reveal a New Substrate Recognition Mechanism. Nature. 2006;443(7113):870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Lapham A, Freedman C, Reed T, Schmidt W. Yeast as a Tractable Genetic System for Functional Studies of the Insulin-Degrading Enzyme. J. Biol. Chem. 2005;280(30):27481–27490. doi: 10.1074/jbc.M414192200. [DOI] [PubMed] [Google Scholar]
- 11.Alper BJ, Nienow TE, Schmidt WK. A Common Genetic System for Functional Studies of Pitrilysin and Related M16A Proteases. Biochem. J. 2006;398(1):145–152. doi: 10.1042/BJ20060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornista J, Ikeuchi S, Haruki M, Kohara A, Takano K, Morikawa M, Kanaya S. Cleavage of Various Peptides with Pitrilysin from Escherichia coli: Kinetic Analyses Using Beta-Endorphin and Its Derivatives. Biosci. Biotechnol. Biochem. 2004;68(10):2128–2137. doi: 10.1271/bbb.68.2128. [DOI] [PubMed] [Google Scholar]
- 13.Alper B, Rowse J, Schmidt W. Yeast Ste23p Shares Functional Similarities with Mammalian Insulin-Degrading Enzymes. Yeast. 2009;26(11):595–610. doi: 10.1002/yea.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song ES, Juliano MA, Juliano L, Fried MG, Wagner SL, Hersh LB. ATP Effects on Insulin-Degrading Enzyme Are Mediated Primarily through Its Triphosphate Moiety. J. Biol. Chem. 2004;279(52):54216–54220. doi: 10.1074/jbc.M411177200. [DOI] [PubMed] [Google Scholar]
- 15.Cabrol C, Huzarska MA, Dinolfo C, Rodriguez MC, Reinstatler L, Ni J, Yeh LA, Cuny GD, Stein RL, Selkoe DJ, Leissring MA. Small-Molecule Activators of Insulin-Degrading Enzyme Discovered through High-Throughput Compound Screening. PLoS ONE. 2009;4(4):e5274. doi: 10.1371/journal.pone.0005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farias M, III, Gorman MW, Savage MV, Feigl EO. Plasma ATP during Exercise: Possible Role in Regulation of Coronary Blood Flow. Am. J. Physiol. Heart Circ. Physiol. 2005;288(4):H1586–H1590. doi: 10.1152/ajpheart.00983.2004. [DOI] [PubMed] [Google Scholar]
- 17.Kwak J, Wang MH, Hwang SW, Kim TY, Lee SY, Oh U. Intracellular ATP Increases Capsaicin-Activated Channel Activity by Interacting with Nucleotide-Binding Domains. J. Neurosci. 2000;20(22):8298–8304. doi: 10.1523/JNEUROSCI.20-22-08298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adessi C, Enderle T, Grueninger F, Roth D. Activator for Insulin Degrading Enzyme. WO/2006/066847 U.S. Patent. 2006
- 19.Hollander I, Frommer E, Mallon R. Human Ras-Converting Enzyme (hRCE1) Endoproteolytic Activity on K-Ras-Derived Peptides. Anal. Biochem. 2000;286(1):129–137. doi: 10.1006/abio.2000.4795. [DOI] [PubMed] [Google Scholar]
- 20.Porter SB, Hildebrandt ER, Breevoort SR, Mokry DZ, Dore TM, Schmidt WK. Inhibition of the CaaX Proteases Rce1p and Ste24p by Peptidyl (Acyloxy)methyl Ketones. Biochim. Biophys. Acta. 2007;1773(6):853–862. doi: 10.1016/j.bbamcr.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alper B, Schmidt W. A Capillary Electrophoresis Method for Evaluation of Aβ Proteolysis In Vitro. J. Neurosci. Methods. 2009;178:40–45. doi: 10.1016/j.jneumeth.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Affholter JA, Hsieh CL, Francke U, Roth RA. Insulin-Degrading Enzyme: Stable Expression of the Human Complementary DNA, Characterization of Its Protein Product, and Chromosomal Mapping of the Human and Mouse Genes. Mol. Endocrinol. 1990;4(8):1125–1135. doi: 10.1210/mend-4-8-1125. [DOI] [PubMed] [Google Scholar]
- 23.Barstead RJ, Waterston RH. The Basal Component of the Nematode Dense-Body Is Vinculin. J. Biol. Chem. 1989;264(17):10177–10185. [PubMed] [Google Scholar]
- 24.Holbeck SL. Update on NCI In Vitro Drug Screen Utilities. Eur. J. Cancer. 2004;40(6):785–793. doi: 10.1016/j.ejca.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 25.Goode DR, Totten RK, Heeres JT, Hergenrother PJ. Identification of Promiscuous Small Molecule Activators in High-Throughput Enzyme Activation Screens. J. Med. Chem. 2008;51(8):2346–2349. doi: 10.1021/jm701583b. [DOI] [PubMed] [Google Scholar]
- 26.Chan RK, Otte CA. Isolation and Genetic Analysis of Saccharomyces cerevisiae Mutants Supersensitive to G1 Arrest by a Factor and Alpha Factor Pheromones. Mol. Cell. Biol. 1982;2(1):11–20. doi: 10.1128/mcb.2.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michaelis S, Herskowitz I. The a-Factor Pheromone of Saccharomyces cerevisiae Is Essential for Mating. Mol. Cell. Biol. 1988;8(3):1309–1318. doi: 10.1128/mcb.8.3.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adames N, Blundell K, Ashby MN, Boone C. Role of Yeast Insulin-Degrading Enzyme Homologs in Propheromone Processing and Bud Site Selection. Science. 1995;270:464–467. doi: 10.1126/science.270.5235.464. [DOI] [PubMed] [Google Scholar]
- 29.Elble R. A Simple and Efficient Procedure for Transformation of Yeasts. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 30.Sikorski RS, Hieter P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen P, Sapperstein SK, Choi JD, Michaelis S. Biogenesis of the Saccharomyces cerevisiae Mating Pheromone a-Factor. J. Cell. Biol. 1997;136(2):251–269. doi: 10.1083/jcb.136.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipinski C, Lombardo F, Dominy B, Feeney P. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Del. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 33.Tam A, Schmidt WK, Michaelis S. The Multispanning Membrane Protein Ste24p Catalyzes CAAX Proteolysis and NH2-Terminal Processing of the Yeast a-Factor Precursor. J. Biol. Chem. 2001;276(50):46798–46806. doi: 10.1074/jbc.M106150200. [DOI] [PubMed] [Google Scholar]
- 34.Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, Sun CY, Meredith SC, Sisodia SS, Leissring MA, Tang WJ. Structure of Substrate-Free Human Insulin-Degrading Enzyme (IDE) and Biophysical Analysis of ATP-Induced Conformational Switch of IDE. J. Biol. Chem. 2007;282(35):25453–25463. doi: 10.1074/jbc.M701590200. [DOI] [PubMed] [Google Scholar]
- 35.Kurochkin IV. Amyloidogenic Determinant as a Substrate Recognition Motif of Insulin-Degrading Enzyme. FEBS Lett. 1998;427(2):153–156. doi: 10.1016/s0014-5793(98)00422-0. [DOI] [PubMed] [Google Scholar]
- 36.Selkoe DJ. Clearing the Brain’s Amyloid Cobwebs. Neuron. 2001;32(2):177–180. doi: 10.1016/s0896-6273(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 37.Kaur M, Reed E, Sartor O, Dahut W, Figg WD. Suramin’s Development: What Did We Learn? Invest. New Drugs. 2002;20(2):209–219. doi: 10.1023/a:1015666024386. [DOI] [PubMed] [Google Scholar]
- 38.McCain DF, Wu L, Nickel P, Kassack MU, Kreimeyer A, Gagliardi A, Collins DC, Zhang ZY. Suramin Derivatives as Inhibitors and Activators of Protein-Tyrosine Phosphatases. J. Biol. Chem. 2004;279(15):14713–14725. doi: 10.1074/jbc.M312488200. [DOI] [PubMed] [Google Scholar]
- 39.Zagorski M, Yang J, Shao H, Ma K, Zeng H, Hong A. Methodological and Chemical Factors Affecting Amyloid Beta Peptide Amyloidogenicity. Methods Enzymol. 1999;309:189–204. doi: 10.1016/s0076-6879(99)09015-1. [DOI] [PubMed] [Google Scholar]
- 40.Leissring MA, Lu A, Condron MM, Teplow DB, Stein RL, Farris W, Selkoe DJ. Kinetics of Amyloid Beta-Protein Degradation Determined by Novel Fluorescence- and Fluorescence Polarization-Based Assays. J. Biol. Chem. 2003;278(39):37314–37320. doi: 10.1074/jbc.M305627200. [DOI] [PubMed] [Google Scholar]
- 41.Zorn JA, Wells JA. Turning Enzymes ON with Small Molecules. Nat. Chem. Biol. 2010;6(3):179–187. doi: 10.1038/nchembio.318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.