Summary
To understand the consequences of the complete elimination of E2F regulation we profiled the proteome of Drosophila dDP mutants that lack functional E2F/DP complexes. The results uncovered changes in the larval fatbody, a differentiated tissue that grows via endocycles. We report an unexpected mechanism of E2F/DP action that promotes quiescence in this tissue. In the fatbody, dE2F/dDP limits cell cycle progression by suppressing DNA damage responses. Loss of dDP upregulates dATM, allowing cells to sense and repair DNA damage, and increasing replication of loci that are normally under-replicated in wild-type tissues. Genetic experiments show that ectopic dATM is sufficient to promote DNA synthesis in wild-type fatbody cells. Strikingly, reducing dATM levels in dDP-deficient fatbodies restores cell cycle control, improves tissue morphology and extends animal development. These results show that, in some cellular contexts, dE2F/dDP-dependent suppression of DNA damage signaling is key to cell cycle control and needed for normal development.
Keywords: Endocycle, DNA damage, E2F, DP, quiescence, ATM
Introduction
Correct regulation of E2F-dependent transcription is important for the control of cell cycle progression, cell differentiation and cell survival. E2F is a heterogeneous factor. The term “E2F” refers to the composite activity of multiple transcriptional activator and repressor complexes with similar sequence-specific DNA-binding activities. The canonical form of E2F is a heterodimer, containing proteins encoded by the E2F- and DP-gene families, with both subunits required for high affinity DNA-binding. The human genome contains eight E2F genes, two DP genes, and the diversity of E2F complexes is compounded by alternative transcription and post-translational modifications. Chromatin immunoprecipitation (ChIP) experiments have identified thousands of E2F-binding sites in the human, mouse and Drosophila genomes (Ren et al. 2002; Chicas et al. 2010; Korenjak et al. 2012), and transcription studies show that the activation, or inactivation, of E2F-dependent transcription alters the expression of hundreds of genes in each species.
While studies of mouse mutant alleles have identified unique roles for individual E2F/DP family members, the sheer size of the mammalian E2F family has precluded the genetic elimination of E2F regulation. Although it has not yet been possible to generate mammalian cells that completely lack E2F or DP proteins, understanding the consequences of E2F elimination is an increasingly important topic. Evidence that the deregulation of E2F drives cancer cell proliferation has spurred interest in the potential therapeutic value of E2F inhibitors. Such compounds have given promising results in melanoma cell lines and mouse tumor models (Sangwan et al. 2012; Ma et al. 2008) but further development of E2F inhibitors requires a better understanding of the consequences of the complete elimination of E2F activity in vivo.
Drosophila E2F/DP/RBF proteins provide a streamlined version of the mammalian network (van den Heuvel and Dyson 2008). In flies, as in mammalian cells, E2F-regulation is generated by the interplay between activator and repressor complexes with E2F activation driving cell cycle progression, and E2F inhibition promoting cell cycle arrest. Drosophila encodes two types of dE2F factors: dE2F1 is a potent activator, while dE2F2 acts in repressor complexes (Ohtani and Nevins 1994; Dynlacht et al. 1994; Duronio et al. 1995; Royzman et al. 1997; Frolov et al. 2001; Cayirlioglu et al. 2001; Stevaux et al. 2002; Dimova 2003; Korenjak et al. 2004; Lewis et al. 2004). The products of both dE2F genes function in heterodimeric complexes with dDP (Dynlacht et al. 1994). As dDP is encoded by a single gene, the entire program of dE2F/dDP regulation can be readily eliminated by mutation of dDP (Frolov et al. 2001).
Although E2F is widely viewed as a critical regulator of cell proliferation, studies of dDP and dE2F1;dE2F2 mutant animals show that E2F activity is not essential for mitotic division (Royzman et al. 1997; Frolov et al. 2005; 2001). While maternally supplied dDP may help to support cell cycles in embryos, the extensive larval growth and cell division seen in dDP null larvae appears to occur in the absence of dE2F/dDP regulation.
dDP and dE2F1;dE2F2 mutant animals die during the late stages of fly development. What, then, are the functions of dE2F/dDP that are essential for viability? For many years the answer to this question was unclear. A recent study revealed that dE2F/dDP affects muscle growth and differentiation in pupae and that the absence of this function contributes to the lethality of dDP mutants (Zappia and Frolov 2016). However, given the large numbers of dE2F/dDP target genes, it seems unlikely that muscle defects are the sole phenotype of dDP mutant animals.
Previous studies of E2F function have relied on transcription profiling for molecular insights. Here, to obtain a more complete picture of the overall consequences of eliminating dE2F/dDP regulation in vivo, we integrated proteome and transcriptome profiles of dDP mutant larvae. This approach revealed a novel role for dE2F/dDP in the Drosophila fatbody, a highly differentiated tissue with prominent metabolic functions during larval and pupal development. We report that dDP is important to keep fatbody cells in a quiescent state. From a genetic screen of direct dE2F/dDP targets, we report the unexpected observation that dATM, a central component of the DNA damage response, plays a key role in this phenotype. dE2F/dDP complexes suppress dATM expression in the fatbody. Absence of dDP elevates dATM expression allowing cells to continue DNA replication. Remarkably, reducing the levels of dATM not only suppresses fatbody defects but ameliorates the lethality associated with dDP mutation, resulting in the extension of the development of animals that completely lack E2F/DP complexes. These results highlight the importance of DNA-damage genes as E2F-targets. Although dATM is just one of many hundreds of genes that are directly regulated by E2F/DP complexes, dATM is a crucial factor in the consequences of eliminating dE2F/dDP-regulation in vivo.
Results
Proteome and transcriptome profiling of dDP mutant larvae
To understand the changes that occur when E2F regulation is eliminated we generated proteome profiles of WT and dDP mutant larvae. We used a trans-allelic combination of dDP point mutations (dDPa4/dDPa3) that causes pupal lethality and was previously shown to be defective for the transcriptional regulation of selected E2F target genes (Frolov et al. 2001; 2005). Western blot analysis confirmed that extracts of dDPa3/a4 third instar larvae lack any detectable wild-type dDP protein (Fig 1A). For simplicity, we will refer to these larvae as dDP−/−.
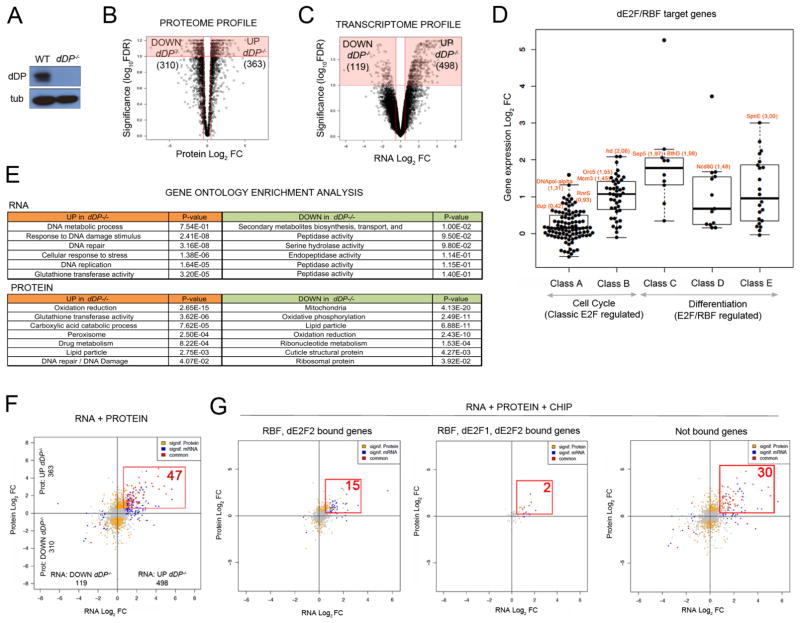
Figure 1. Loss of E2F activity drives proteome and transcriptome changes in vivo.
A: dDP blot of WT and dDP−/− larvae; tubulin, loading control. Volcano plot of Log2 FC between WT and dDP−/− proteomes (B) and transcriptomes (C) (significant changes in red). D: Increased expression of dE2F/RBF targets in dDP−/− larvae. E: GO analysis of RNA and protein datasets. Upregulated (orange) and downregulated (green) representative terms, sorted by descending significance. F: Integrated RNA/Protein data: 47 hits (red) significantly increase in RNA and protein level (compared to 6 significantly decreasing targets). Significant changes in RNA or protein are color coded in blue and yellow, respectively. Not significant changes are plotted in grey. G: ChIP/RNA/Protein plots. Subsets of changing hits in RNA/protein levels (red): RBF/dE2F2-bound (15), RBF/dE2F1/dE2F2-bound (2) and indirect target genes (30).
Multiplexed quantitative mass spectrometry-based proteomics was carried out using tandem mass tag (TMT) technology (McAlister et al. 2014; Edwards and Haas 2016). The levels of 5478 proteins (approximately half of the Drosophila proteome) were quantified in whole larval extracts. This data displayed a correlation of r>0.9 between biological replicates (Fig S1A). Proteins with significantly different levels in WT and dDP−/− larvae were identified by Student’s t tests. Using FDR<10% and Log2FC>0.5 as cut-off values, 363 proteins increased in dDP−/− larvae, and 310 proteins decreased (Fig 1B; Table S1).
Gene expression arrays were used to compare levels of transcripts in WT and dDP−/− larvae. This data showed high correlation values between biological replicates (r>0.95, Fig S1B). Unlike the proteome profiles that had similar numbers of increased and decreased proteins, the transcription profiles displayed a clear bias towards upregulated transcripts in dDP mutants: 498 transcripts showed a statistically significant increase in dDP mutants (log10FDR<1; Log2FC>0.5), while only 119 transcripts were reduced (Fig 1C; Table S2).
In previous work using cultured S2 cells, we subdivided the large program of E2F-regulated genes into 5 classes of targets (A–E) that differ in their dependence on dE2F1 and dE2F2 (with Class A most strongly dependent on dE2F1/dDP-mediated activation, and Class E most dependent on dE2F2/dDP-mediated repression (Dimova 2003)). Classes A and B contain dE2F1/dDP-regulated genes with roles in cell proliferation, and transient expression of these genes is coupled with cell cycle progression. In contrast, Classes C-E contain developmentally regulated genes that are repressed by dE2F2- and dDP-containing complexes in cycling cells. Interestingly, the mRNA profiles of dDP larvae revealed an overall increase in transcription at all five classes of dE2F/dDP targets (Fig 1D). While this does not exclude the idea that E2F-activation is important in specific cellular contexts, it shows that the net effect of E2F regulation in whole larvae is that all classes of E2F target genes are repressed by dE2F/dDP complexes; including sets of target genes that can be activated by dE2F1/dDP.
RNA and proteome data give different perspectives on the dDP mutant phenotype. Gene Ontology (GO) analysis (Fig 1E; FDR<25%, Benjamini p-val<0.05) revealed that the upregulated transcripts in dDP mutants were most strongly enriched for DNA metabolism, response to DNA damage and cellular stress responses. In contrast, the significantly increased proteins in dDP mutants were enriched for other metabolic functions, most notably in oxidation/reduction, glutathione transferase activity, carboxylic acid catabolic process and peroxisome function. While the down-regulated transcripts showed a moderate enrichment in secondary metabolic and peptidase categories, the proteins that significantly decreased in dDP−/− larvae were strongly enriched for roles in energy metabolism and ATP production, including changes in mitochondria, oxidative phosphorylation and lipid particles. Collectively, the changes in the proteome strongly suggest that dDP−/− larvae experience a major change in energy metabolism. These observations extend, and are consistent with, previous studies showing that dE2F/dDP proteins are important for mitochondrial function in the Drosophila eye and during muscle development (Ambrus et al. 2013; Zappia and Frolov 2016). Some deregulated biological processes in dDP−/− larvae, such as DNA repair and DNA Damage Responses (DDR), are evident in both the RNA and protein datasets, but the metabolic changes that showed the strongest enrichment in the proteome data were not enriched in the transcript data.
Generating a list of candidate drivers of dDP−/− phenotype: integration of gene expression and proteome profiles with E2F and RBF ChIP data
E2F proteins bind to thousands of sites in the Drosophila genome, and transcripts from hundreds of genes are altered in dDP−/− larvae. However, it is unknown which changes functionally contribute to dDP mutant phenotypes. We combined the transcriptomic and proteomic data and identified 53 genes with statistically significantly changes in both RNA and protein levels in dDP−/− larvae (FDR<10%; Log2FC>0.5). The majority of these (47) increased in dDP−/− (Fig 1F; Table S3). These direct RBF/E2F targets were widely distributed in active and repressed chromatin, with a modest enrichment of loci in repressed chromatin domains (Fig S1C).
To identify the subset encoded by direct E2F/DP targets, we integrated this expression data with previously-published genome-wide ChIP data for RBF and dE2F proteins generated from third instar larvae (Korenjak et al. 2012). It is inevitable that any such list will be incomplete; nevertheless, this analysis highlighted 17 genes whose promoters are directly bound by dE2F2 and RBF proteins in wild-type animals, and that displayed robust and statistically significant changes in overall RNA and protein levels in dDP−/− animals (Fig 1G).
dDP is essential in the fatbody for normal control of the endocycle
The major proteomic changes in dDP mutants affect metabolic processes such as mitochondrial function and lipid homeostasis (Fig 1E). The fatbody is the major metabolic tissue and one of the largest tissues of third instar larvae; we therefore examined dDP−/− larvae for fatbody defects.
Oil red staining showed that lipid droplets (organelles where lipids are stored) were reduced in size and irregular in dDP−/− fatbodies compared to WT (Fig 2A). Concordantly, total levels of triacylglycerides (TAG, the major form of stored lipids) were significantly reduced in dDP−/− larvae, relative to total protein levels (Fig 2B). When starved, larvae activate autophagy in the fatbody to mobilize energy stores. To determine how dDP loss influences this process, we starved wild-type and dDP−/− larvae and used Lysotracker to monitor autophagic vesicles (Scott et al. 2004). Interestingly, dDP−/− larvae were unable to activate this program (Fig S2A).
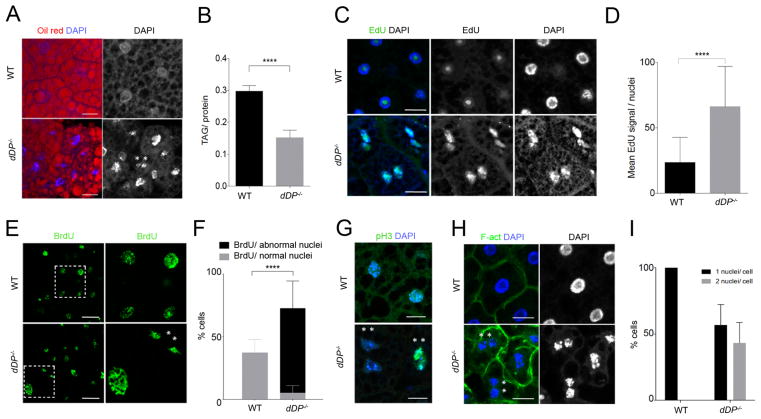
Figure 2. Loss of E2F activity affects fatbody function and endocycle progression.
Oil Red (lipid droplets) and DNA (DAPI) (A) and mean ± SD of total tryacylglicerades (TAG)/ protein ratio (B) in WT and dDP mutant larvae (Unpaired T-test, p<0.0001); “**”: binucleate cells. C: EdU labeled replicating regions in dDP−/− fatbody nuclei (WT cells show a non-specific EdU stain of the nucleolus). D: Quantification of mean ± SD of EdU signal/nuclei increase in FB cells (WT: n=53; dDP−/−: n=30; Mann-Whitney test, p<0.0001). dDP−/− nuclei with normal and abnormal shape incorporate BrdU (E, F; mean ± SD of % BrdU positive cells per category; Two-way ANOVA test, p<0.0001), display abnormal shape (E) and mitotic p-H3 (G); compare to WT nuclei. H: Binucleate cells (membrane labeled with F-actin). I: an average of 50% of dDP−/− FB cells are binucleated (plot of mean ± SD of % cells; WT: n=60; dDP−/−: n=151 nuclei).
The fatbody of wild-type third instar larva contains cells that are highly differentiated and quiescent. During normal development fatbody cells grow via endocycles, a variation of the cell cycle in which cells undergo repeated rounds of DNA replication without an intervening mitosis. At the end of embryogenesis, cells forming the fatbody primordium start endocycling between 14 and 20 hours after egg deposition (Smith and Orr-Weaver 1991). Later in larval stages, fatbody cells stop endocycling and remain quiescent during pupal metamorphosis, when the tissue undergoes extensive remodeling (Nelliot et al. 2006).
In addition to the changes in lipid levels in dDP−/− fatbodies, cell nuclei displayed striking morphological abnormalities (Fig 2A, C, E, H). The nuclei of dDP−/− fatbodies showed irregular shape and size, often contained less condensed DNA, and, unlike the wild-type fatbody, DAPI labeled chromatin was not homogeneously distributed in the nuclei (Fig S2B). EdU and BrdU incorporation assays revealed that most of the nuclei in the dDP−/− fatbody were not quiescent but re-entered S-phase (Fig 2C–F). Most WT endocycling fatbody cells do not strongly incorporate EdU (Fig 2C) and only 37% cells incorporated BrdU during a 24hr pulse (Fig 2E, F). In contrast, in dDP−/− tissue many nuclei stained strongly with EdU (Fig 2C, D) and 67% cells incorporated BrdU (Fig 2E, F), displaying elevated DNA synthesis throughout abnormally-shaped nuclei.
Unexpectedly, dDP−/− fatbodies contain large numbers of binucleate cells (Fig 2), suggesting that many cells not only entered S-phase but also progressed onto mitosis and failed at cytokinesis. Consistent with the notion that fatbody cells have abandoned the endocycle, approximately 5% of cells in dDP−/− fatbodies stained positive for the mitotic mark pH3 (Fig 2G). Remarkably, an average of 50% of fatbody cells form binucleates in dDP−/− larvae (Fig 2H,I) with a distinguishable nuclear membrane (see Lamin A/C in Fig S2C). This defect, however, did not trigger cell death (Fig S2D). Binucleates were observed using different combinations of mutant dDP alleles (Fig S2E). Control experiments confirmed that dE2F1 and dE2F2 proteins were absent from dDP−/− fatbodies (Fig S2F,G), and immunostaining confirmed that SpnE (a previously-described dE2F/dDP target) was upregulated in these dDP mutant cells (Fig S2H, I). We conclude that dDP is needed for fatbody cells to maintain quiescence. Unexpectedly, many dDP−/− fatbody cells not only progress inappropriately through the cell cycle but also attempt to switch to a mitotic cycle.
Depletion of dDP in the fatbody activates DNA Damage Responses and impairs pupal development
In cells that undergo mitotic division, protecting genome integrity from DNA damage is crucial for normal development and cell viability. DNA Damage Responses sense DNA lesions and transmit this information to activate signaling cascades for DNA repair. Drosophila endocycles, however, are associated with elevated DNA damage, underreplication of genomic regions and compromised genomic stability (Edgar and Orr-Weaver 2001; Edgar et al. 2014). Endocycling tissues, such as midgut, salivary glands and fatbody, have characteristic patterns of incomplete replication in which regions that are distant from origins of replication, are rich in heterochromatic marks and have DNA breaks, become under-replicated (Lilly and Duronio 2005; Nordman et al. 2011; Yarosh and Spradling 2014). To permit this, endocycling cells appear to suppress the DNA damage checkpoint pathways that, in diploid cells, would trigger cell cycle arrest in response to DNA breaks (Mehrotra et al. 2008).
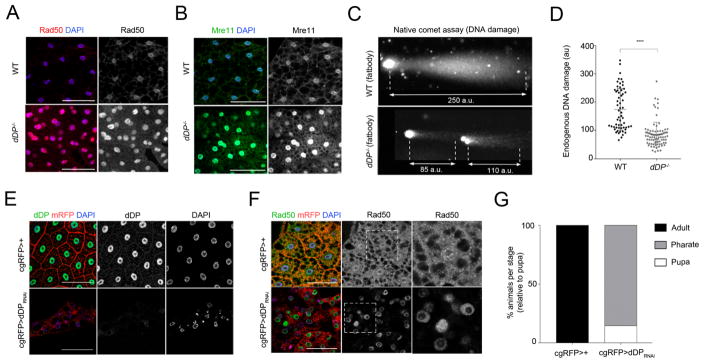
dDP mutant fatbody cells show an abnormal activation of DNA damage responses: DNA repair was one of the significantly enriched categories in GO analysis of the altered RNA and protein profiles of dDP−/− larvae and the list of genes directly regulated by dE2F/dDP/RBF proteins that were significantly upregulated in dDP−/− larvae included both Rad50 and dATM (Fig 1F; Table S3). Rad50 is one of three components of the MRN DNA Damage complex (Rad50-Mre11-Nbs), that senses damaging events such DNA breaks and replication stress and is needed for optimal activation of ATM (Burgess and Misteli 2015). Immunostaining confirmed that Rad50 protein is strongly upregulated in dDP−/− fatbodies (Fig 3A; quantified in Fig S3A). Similar results were seen with antibodies specific for the Rad50-interacting proteins Mre11 (Fig 3B; quantified in Fig S3B) and Nbs (Fig S3C). Native comet assays showed that this upregulation of DDR proteins was accompanied by a reduction in DNA double-strand breaks (Fig 3C, comet lengths from dDP−/− fatbody cells are quantified in Fig 3D; irradiated (IR) and non-IR diploid cells served as controls for induced DNA damage, in Fig S3D).
Figure 3. dDP loss in the fatbody results in high DNA damage activity and pupal lethality.
Increased Rad50 (A) and Mre11 (B) proteins in dDP mutant fatbodies. C: WT fatbody cells (n=59) have a significantly increased chromosome fragmentation (longer DNA comet; au, airy units) compared to dDP−/− FB cells (n=86); (plot showing the mean ± SD comet length in D; Kruskal-Wallis test, p-val<0.001). E: dDP (green) and Rad50 proteins (F, green) in cgRFP>dDPRNAi fatbodies (compare to cgRFP>+). Nuclei are labeled with DAPI and RFP protein (red) is tagged to the membrane (mRFP) under the control of cg-Gal4 transgene (fatbody specific). G: 85.28% of cgRFP>dDPRNAi animals die during pharate stage (n=54; compare to control cgRFP>+).
To ensure that these changes reflect a cell-autonomous function of dDP we used a cg-Gal4 UAS-mRFP transgene (cgRFP>+) to drive expression of UAS-dDP RNAi transgenes specifically in the fatbody (Fig 3E). Similar to dDP mutants, elimination of dDP in the fatbody (cgRFP>dDPRNAi) caused nuclear defects (Fig 3E; Fig S3E), the recruitment of Rad50 protein (Fig 3F), elevated Spn-E protein levels (a known dE2F/dDP target; Fig S3F) and appearance of binucleate cells (Fig S3G). This targeted depletion of dDP in the fatbody severely reduced fly viability. Compared to cgRFP>+ flies, >80% of cgRFP>dDPRNAi flies died as pharates, during late pupal stages (Fig 3G).
To further test the importance of dDP expression in the fatbody, we built a recombinant stock carrying cg-Gal4 UAS mRFP and a dDP mutant allele (cgRFP-dDPa3). This allowed us to generate dDP mutant larvae (dDPa3/a4; cgRFP/+) that could express UAS transgenes. When combined with a UAS-dDP transgene to re-express dDP specifically in the fatbody of dDP mutant animals, we observed that this restored a normal nuclear morphology and suppressed the appearance of binucleate cells (Fig 4A, 4C). Immunostaining (Fig 4A) and western blot analyses (Fig 4B) confirmed the elimination of dDP protein in dDPa3/a4; cgRFP/+ larvae and the tissue-specific re-expression of dDP protein in dDPa3/a4; cgRFP/UAS-dDP animals. dDP protein was not detectable in salivary glands (Fig S4A) or imaginal discs (Fig S4B), but was detected in the fatbody (Fig S4A, C).
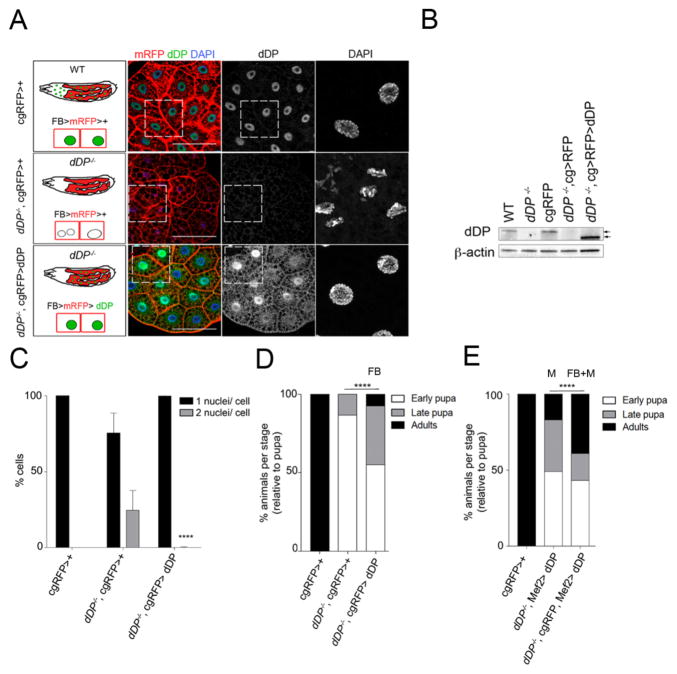
Figure 4. Reintroducing dDP in the fatbody is sufficient to rescue dDP nuclear defects and pupal lethality.
A: Larval genotypes and corresponding fatbodies: cg-Gal4 drives expression of mRFP protein (fatbody) in all genetic combinations. dDP and DNA (DAPI) are shown in control (up), mutant (middle) and rescued fatbodies (low). B: WB detection of dDP in WT, cgRFP>+ and rescued dDP−/−;cgRFP>dDP larvae (absent in dDP−/− and dDP−/−; cgRFP>+). β-actin, loading control. Overexpressed dDP is functional but arrows point its 4 KDa smaller size (Star Methods). C: mean ± SD of % mono- and binucleate cells in all conditions; binucleates are mostly absent in rescued dDP−/−;cgRFP>dDP fatbodies (mean ± SD of 0.02%; p-value<0.0001, ANOVA; n=350). D: % of animals arrested at each developmental stage (relative to total number of pupa). 100% of cgRFP>+ are viable, and dDP−/−;cgRFP >+ animals develop 13% pharates (n=53). Reintroducing dDP in the fatbody (FB) of dDP−/−;cgRFP>dDP larvae significantly rescues pupal lethality: 55.22% pupae, 37.31% pharates, 7.46% adults (n=67, Chi-square test, p-val<0.0001). E: dDP muscle (M) expression in dDP−/−;Mef2>dDP gave 34% of pharates and 17% adults (n=100). Combined re-expression of dDP in fatbody and muscle rescues more efficiently (n=51; 43.14% pupae, 17.65% pharates, 39.22% adults, Chi-square test, p-val<0.0001).
Tissue-specific expression of dDP protein in the fatbody of dDP−/− larvae significantly extended animal development. While dDP−/− animals die as pupa, 37.3% of dDPa3/a4; cgRFP UAS-dDP animals progressed through pupal stages and 7.4% of animals were able to eclose from the puparium (Fig 4D). In most cases, progression through the pupal stages produced a viable animal that was unable to eclose. E2F/DP function has been shown to be important for pupal muscle growth and differentiation, and the muscle-specific re-expression of dDP rescued 29% of animals to adults (Zappia and Frolov 2016). We found that the combined re-expression of dDP in both fatbody and muscle improved the efficiency of rescue to adults to 39.2% (Fig 4E; tissue-specific expression of Gal4 lines and dDP re-expression is shown in Fig S4D, E; the genotype of rescued animals was confirmed by PCR, shown in Fig S4G).
A similar rescue efficiency of 40% was seen with combined fatbody and muscle expression in a different genetic background (dDPa3/Df; cgRFP, Mef2-Gal4, UAS-dDP; Fig S4F). Since tissue-specific depletion of dDP in the fatbody reduces viability, and the tissue-specific restoration of dDP in the fatbody extends the development of dDP−/− animals, we conclude that the role of dDP in the fatbody is important. These results suggest that the functions of dDP in the fatbody and muscle co-operatively promote development to viable adults.
Time controlled depletion of dDP in the fatbody
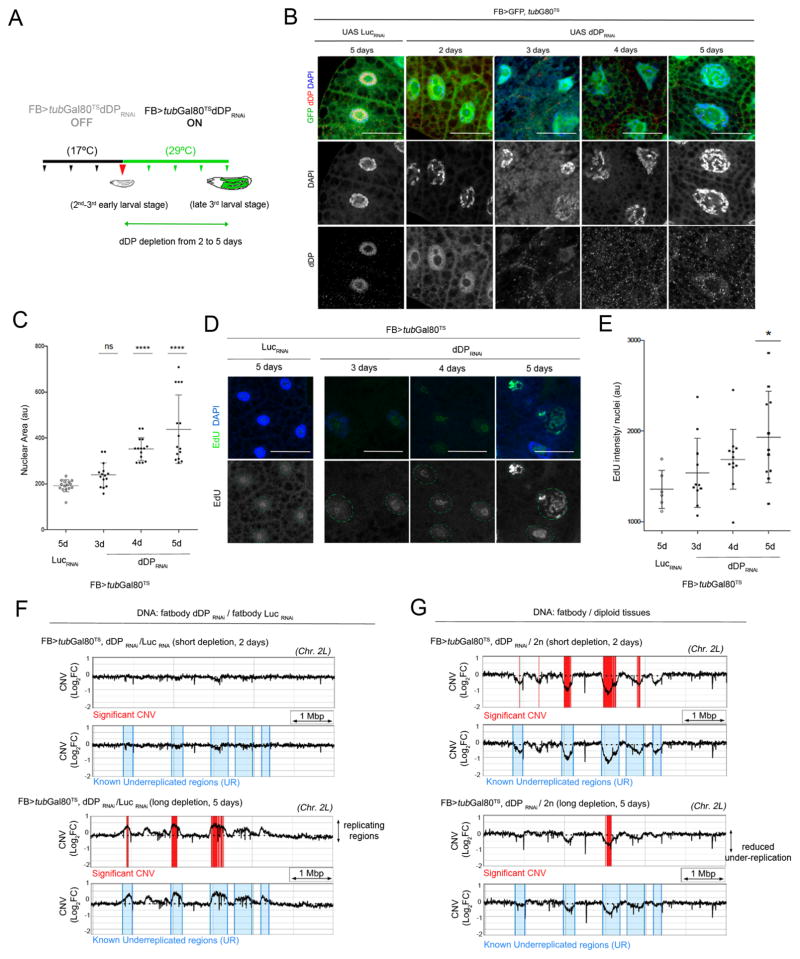
To study the events triggered by depletion of dDP from fatbody cells we generated fatbody-Gal4 tubGal80TS>UAS GFP, UAS dDP RNAi larvae that could be used for tissue-specific and temperature-regulated induction of dDP RNAi. Larvae were transferred from 17°C to 29°C for 2–5 days of larval development to inactivate the Gal80TS protein and to deplete dDP by RNAi in the fatbody before the onset of pupation (Fig 5A). Induction of dDP RNAi eliminated the endogenous dDP protein from fatbody cells, increased nuclear size, activated DNA damage responses and triggered ectopic EdU incorporation. The time course revealed that dDP levels drop quickly (Fig 5B) leading to the transcriptional upregulation of known E2F targets (Fig S5A–C). Increased nuclear size (Fig 5C) and spreading of EdU incorporation (Fig 5D, E) were slow and gradual consequences that started to appear after 3 days of dDP depletion. Cells with widespread and elevated EdU incorporation were only evident when dDP was depleted for 5 days (Fig 5D, E). When EdU intensity and p-H2Av marks of individual cells were compared, we noted that the EdU/p-H2Av ratio switched from low to high after 4 days of dDP depletion (Fig S5D), suggesting that replication occurs when DNA damage has been repaired.
Figure 5. DNA replication is a consequence of dDP depletion in fatbody cells.
A: time controlled depletion of dDP in the fatbody. In all panels, larvae are transferred (during 2nd and 3rd larval stages) from 18°C to 29°C to inactivate Gal80TS protein during 2–5 days and activate FB>GFP, UAS dDPRNAi in the fatbody. FB>tubG80TS>GFP,UAS LucRNAi larvae are used as control (after 5 days at 29°C). B: dDP is eliminated in FB>tubG80TS>GFP,UAS>dDPRNAi fatbodies after 2–5 days at 29°C (compare to LucRNAi control). C: mean ± SD of nuclear area increases upon dDP depletion (2d–5d, days at 29°C): n=15 nuclei/ time-point; one-way ANOVA, p<0.0001. D: EdU-labeled replicating regions are visible after 5 days of dDPRNAi expression (mean ± SD of EdU intensity/nuclear area are shown in E; n=12 nuclei/ time-point; Kruskal-Wallis test, p<0.05). F: Detail of chromosome 2L: significant CNV changes (in red, Log2FC>0.6; FDR<0.001) of DNA of FB>tubG80TS>GFP, UAS dDPRNAi relative to UAS LucRNAi show replicating regions in known under-replicated regions (UR, blue) are visible upon long dDP depletion (during 5 days; compare to short depletion). G: Gradual loss of underreplication (significant CNV, in red) in FB>tubG80TS>GFP, UAS dDPRNAi (relative to diploid WT DNA); known UR domains, in blue. Genomic scale: 1 Million bp (Mbp).
To locate the genomic regions that are replicated upon following dDP depletion, we performed DNA sequencing and calculated the Copy Number Variation (CNV). CNV Log2 fold changes were calculated between fatbodies of fatbody-Gal4 tubGal80TS>UAS GFP, UAS dDP RNAi and >UAS Luc RNAi larvae (after 2 and 5 days of depletion; WT diploid cells (CNS, discs) and endocycling tissues (fatbody, midgut) were used for reference; Fig S5E). A set of genomic loci increased significantly in copy number following the depletion of dDP (Fig 5F, compare CNV peaks between dDP RNAi/ Luc RNAi after short and long depletion). Strikingly most of these loci, and all of the loci showing a statistically significant increase, fell within the regions that have previously been reported to be under-replicated in the fatbody (Nordman et al., 2011; Fig 5F). This indicates that the depletion of dDP increases replication within regions of the genome that are normally under-replicated. Confirming this, when the DNA of dDP-depleted fatbodies was compared with diploid tissues, the sustained depletion of dDP greatly reduced the overall level of under-replication evident in the fatbody (Fig 5G; Control UR regions are shown in Fig S5F). Interestingly, dDP depletion reduced under-replication at loci scattered along 2L, 2R and 3L chromosome arms, but not at pericentromeric or heterochromatic regions (Fig S5G).
Taken together, this data suggests a temporal sequence following dDP depletion in the fatbody in which transcriptional changes and the sensing of DNA damage eventually lead to repair of DNA damage and replication. Remarkably, the replication seen after dDP depletion in the fatbody has the greatest impact on genomic regions that are normally under-replicated.
A screen identifies ATM as a key target of dE2F/dDP in the fatbody
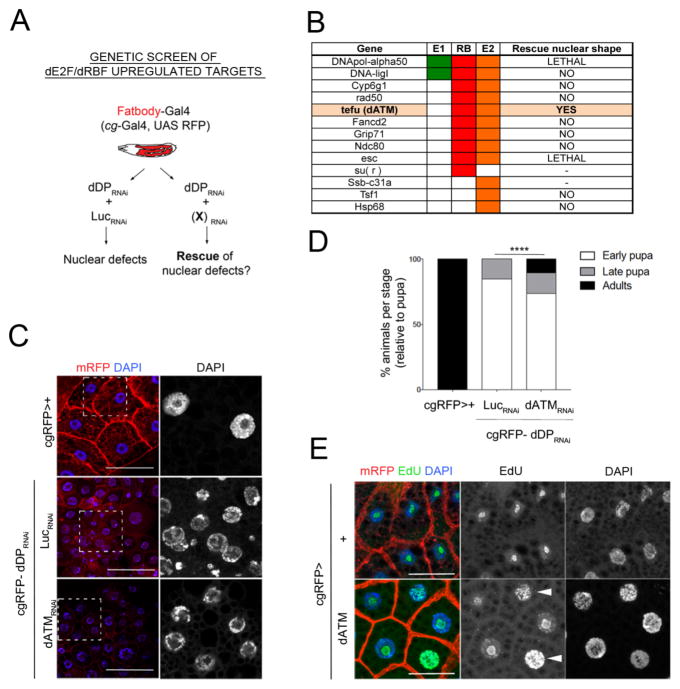
To identify the deregulated E2F/DP targets that are critical for the dDP mutant phenotypes in the fatbody, we obtained RNAi transgenes targeting the set of candidate genes previously identified (Fig 1G) as being directly regulated by dE2F/RBF proteins and significantly upregulated in both RNA and protein in dDP−/− larvae. To ask whether reducing the expression of any of these individual genes was sufficient to suppress the defects generated by removing dDP from the fatbody we constructed a w; cgGal4 UAS mRFP- UAS dDP RNAi/CyO; sb/TM6B-Gal80 flystock (referred as cgRFP-dDPRNAi) and crossed it to RNAi lines that were available for 11 out of 17 candidate genes (Fig 6A, B). As a control, the nuclear defects caused by depletion of dDP were unaffected by co-depletion of Luciferase RNAi (Fig S6A).
Figure 6. dATM plays a key role in the dDP depleted fatbody cells.
A: Genetic screen performed in the fatbody. Parallel crosses of cgRFP-UAS dDPRNAi were set with UAS LucRNAi and UAS (X)RNAi candidate lines. B: List of dE2F regulated candidates (columns indicate direct ChIP binding of E2F1 (E1), E2F2 (E2) and RBF (RB)) and screen result (rescue of nuclear shape defects). C: Representative fatbody nuclei from cgRFP>+ (control), cgRFP-UAS dDPRNAi>UAS LucRNAi (defective), and cgRFP-UAS dDPRNAi>UAS ATMRNAi (TRIP) (rescued). D: % animals arrested at each developmental stage in cgRFP>+, cgRFP-UAS dDPRNAi>UAS LucRNAi (n=190) and UAS ATMRNAi (TRIP) animals (n=100, Chi-square test, p-val<0.0001). E: Induced DNA replication in cgRFP>dATM fatbodies (arrowheads: whole replicating nuclei. EdU, green; control, cgRFP>+).
This mini-screen unexpectedly revealed that the Drosophila orthologue ATM gene, tefu (dATM), a central component of DDR, has a key role in the fatbody. The combined depletion of dATM and dDP strongly suppressed the defects in nuclear morphology generated by dDP depletion alone. Fatbodies from cgRFP-dDPRNAi>dATMRNAi larvae lacked the binucleated cells seen in cgRFP-dDPRNAi>LucRNAi controls (Fig S6B) and showed a notably restored nuclear morphology (Fig 6C). Similar effects in nuclear morphology were observed when dATM was depleted using an alternative dATMRNAi line that targeted different sequences and dDP depletion remained unaffected (Fig S6C,D). None of the RNAi lines targeting other direct E2F/DP target genes gave a similar rescue (Fig 6B). Surprisingly, depleting dATM in cgRFP-dDPRNAi>dATMRNAi larvae resulted in a significant extension of pupal development, as indicated by 26% of animals developing as pharates and 8% of adult flies (Fig 6D; compared to 15.26% of pharates and 0% adult flies obtained from cgRFP-dDPRNAi>LucRNAi animals).
Supporting the conclusion that upregulation of dATM is functionally important in dDP−/− fatbodies, we found that overexpression of dATM (cgRFP>dATM) was sufficient to promote DNA synthesis in the fatbody of larvae with a wild-type background (increased EdU incorporation; Fig 6E). This change was accompanied by a significant increase in nuclear area and DNA content of cgRFP>dATM fatbody cells (Fig S6E).
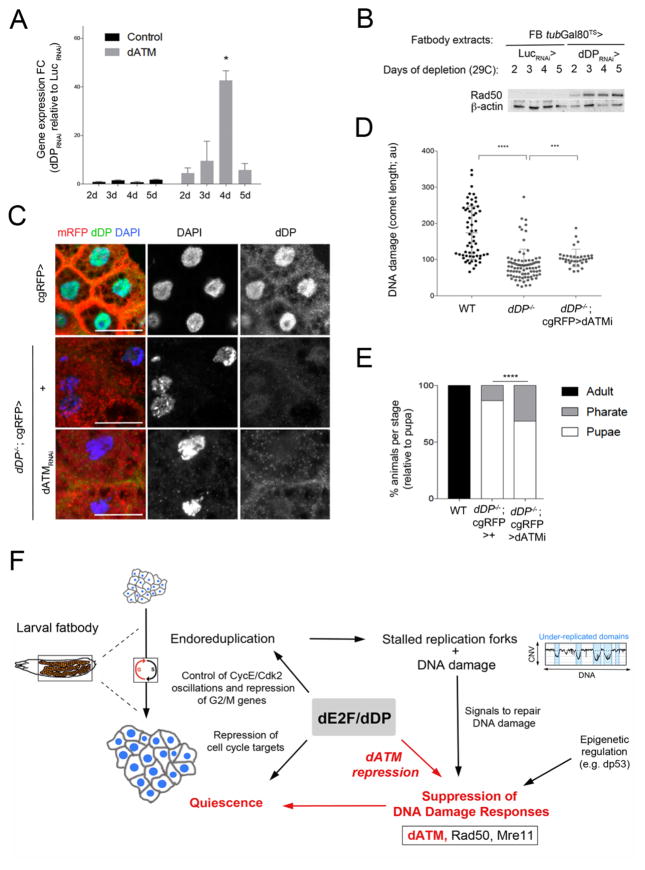
DNA Damage responses involve the coordinated action of multiple protein complexes: it has been suggested that MRN positively regulates dATM in flies (Ciapponi et al. 2006), similar to mammalian cells, where ATM activation requires the MRN complex, and creates a regulatory loop that maintains ATM activity (Shiloh and Ziv 2013). Overexpression of Mre11 and Rad50 (but not Nbs), were also sufficient to allow DNA replication in wild-type fatbody cells, providing additional evidence that elevated DNA damage signals can trigger DNA synthesis in this tissue (Fig S6F,G). Although increased dATM, Mre11 and Rad50 proteins are able to induce DNA replication, we note that only RNAi lines targeting dATM suppressed the phenotypes resulting from dDP depletion in the fatbody. Time-course experiments confirmed that increased transcription of dATM is an early consequence of dDP deleption (Fig 7A), and accompanied by a de-repression of Rad50 and Mre11 (Fig 7B; Fig S7A–C; depletion of dDP had no effect on ATR expression).
Figure 7. dATM depletion is sufficient to rescue dDP mutant phenotypes.
A: Mean ± SD of dATM expression fold changes of in the fatbody (grey) of FB>tubG80TS>UAS GFP, dDPRNAi relative to > UAS LucRNAi (plot of calculated FC during 2–5 days of dDP depletion; ANOVA, p<0.05). The mean of Gapdh and Rpl32 is used as control. B: Increase of Rad50 protein in fatbody extracts upon time-controlled dDP depletion during 2–5 days (FB>tubG80TS>GFP, >dDPRNAi; compare to UAS LucRNAi). C: Rescued nuclear defects in dDP−/−; cgRFP> dATMRNAi fatbody cells (compare to dDP−/−; cgRFP>+; control, cgRFP>+; dDP is only present in control, in green). D: Plot showing the mean ± SD of comet length in dDP−/−;cgRFP>dATMRNAi fatbodies is significantly increased (n=36) than dDP−/− fatbodies (n= 86; WT, n=59; Kruskal-Wallis test, p-val<0.001). E: % of rescued dDP−/−; cgRFP>dATMRNAi(TRIP) animals (n=89; 68.54% pupae, 31.46% pharates, Chi-square test, p-val<0.0001; compare to 13.21% of pharates and 0% adults in dDP−/−; cgRFP>+ (n=53) and control cgRFP>+ animals). F: Model: The ability of dE2F/dDP to repress dATM and limit DNA Damage responses (DDR) in fatbody cells helps to maintain a quiescent state. dE2F/dDP function promotes the oscillation of key cell cycle regulators during the endocycle (Zielke et al., 2011). During endocycles differential replication creates under-replicated regions that contain persistent DNA damage and stalled replication forks (Yarosh and Spradling, 2014, Andreyeveva et al., 2008). Cells tolerate incomplete replication by suppressing apoptotic pathways and downregulating DDR (Mehrotra et al., 2008). Our results (in red) show that dE2F/dDP-mediated suppression of dATM is critical for the downregulation of DDR, for the persistence of under-replicated regions, and for suppression of cell cycle progression in fatbody.
Encouraged by the ability of dATM RNAi to suppress the consequences of dDP RNAi in the fatbody, we examined the effects of dATM RNAi on dDP mutant animals. Remarkably, expression of the dATM RNAi transgene not only suppressed the nuclear defects (Fig 7C) and the endogenous damage (Fig 7D) in the fatbody of cgRFP-dDP−/−; dATM RNAi/+ larvae, but this tissue-specific targeting also gave a mild but significant extension of animal development (Fig 7E). Strikingly, reducing dATM expression in this dDP mutant background resulted in 31.46% dDP mutant pharates (the genotype of rescued pharates was confirmed by PCR, Fig S7F). A similar rescue in nuclear shape and absence of binucleate cells was seen using an independent dATM RNAi construct (Fig S7D, E). We conclude that dATM is upregulated in dDP−/− larvae, it plays a critical role in the fatbody defects of dDP−/− larvae, and its expression contributes to the lethality of dDP mutant animals.
In summary, the results described here outline a molecular activity of dDP that is critical for fly development (Fig 7F). The differentiated cells of wild-type larval fatbody are quiescent, have incompletely replicated DNA and marks of DNA damage. The sensing and cellular response to this damage is curtailed, at least in part, by dE2F/dDP-dependent repression of dATM. Upregulation of dATM allows cells to sense DNA damage, ultimately leading to DNA replication.
Discussion
Here we outline a new mechanism of E2F/DP-mediated regulation in which E2F/DP-mediated suppression of DDR blocks cell cycle progression. We discovered that it is possible to suppress the appearance of dDP mutant phenotypes in the larval fatbody by decreasing dATM expression. This manipulation is sufficient to extend the development of animals that lack E2F/DP complexes.
These observations highlight an important unresolved issue in E2F/DP research. E2F/DP proteins alter the transcription of many genes but it is uncertain which mRNA changes are functionally significant. It is unlikely that E2F complexes have a single key target, and it is probable that the rate-limiting targets will differ in different cellular contexts. In almost all in vivo studies of E2F function, precisely which target is most critical for the mutant phenotype is not known and there was no previous indication that the regulation of dATM would play such a major role when E2F/DP-mediated regulation was eliminated.
E2F-binding sites are present in the promoters of many genes with functions in DNA damage and repair pathways. This regulation has previously been proposed to be a mechanism by which elevated E2F activity can activate cell death signals (Berkovich and Ginsberg 2003; Bi et al. 2005). This connection has also been interpreted to mean that, during G1/S cell cycle progression, E2F-activation helps to promote the efficiency of DNA repair and, perhaps by sustaining efficient DNA replication, it enhances cancer cells proliferation (Ren et al. 2002). The results described here show, in essence, the complete reverse of this concept. Since E2F complexes mediate both activation and repression, they have the potential to suppress the sensing and repair of DNA damage in quiescent cells. Our results indicate, unexpectedly, that dDP-mediated suppression of DDR pathways is critical for the Drosophila fatbody to stop cell cycle progression. This role is seen in cells that have entered S-phase and later become quiescent, but are capable of continued DNA replication. This role is important for fly development. Indeed, the fact that the development of dDP−/− animals can be extended by reducing dATM levels illustrates that E2F/DP regulation is not absolutely essential for larval growth or cell proliferation. However, E2F/DP regulation is needed to limit DDR signals in a differentiated quiescent tissue.
The reason for the genetic interaction between dDP and dATM stems, in part, from the features of the fly endocycle. Drosophila larval growth is achieved primarily via endoreplication (Lilly and Duronio 2005; Edgar and Orr-Weaver 2001; Edgar et al. 2014). During endocycles the Drosophila genome undergoes differential replication, creating under-replicated regions that contain stalled replication forks and double strand breaks (Yarosh and Spradling 2014; Andreyeva et al. 2008). As a result, endocycling cells accumulate persistent DNA damage and down-regulate DNA damage responses (DDR) to escape from DNA damage induced cell death (Mehrotra et al. 2008; Fox and Duronio 2013; Zhang et al. 2014).
dE2F/dDP proteins have an important role in this process: dATM and Rad50 promoters are bound by E2F/DP/RBF proteins, their transcripts are upregulated in dDP mutants, and quantitative proteomics shows significantly increased levels of dATM and MRN proteins in dDP−/− larvae (Fig 1) and dissected fatbodies (data not shown). In time-course experiments, the increased transcription of dATM and the activation of DDR proteins are relatively early effects of dDP-depletion, while replication (including replication of normally under-replicated regions) is a later consequence. In diploid cells, ATM activity is expected to limit cell cycle progression and to allow DNA repair activities. However, in the fatbody elevated dATM did not appear to activate cell cycle checkpoints or induce apoptosis; instead, we observed increased DNA synthesis, increased copy number variation in under-replicated regions, increased nuclear size, and reduced chromatin breaks in dDP mutant fatbodies. Collectively, these observations suggest that the predominant effect of elevated dATM in this tissue is improved repair of DSB at stalled replication forks in under-replicated regions, and that this permits DNA replication. We appreciate that there may be several reasons why dATM levels are so significant in dDP mutants. dATM plays a central role in the coordination of cellular responses to DNA damage (Shiloh and Ziv 2013; Paull 2015). ATM has also been shown to be activated by oxidative stress (Guo et al. 2010) and the proteome profiles of dDP mutants include increased expression of oxidation/reduction related proteins and decreased levels of mitochondrial proteins, suggesting that dDP mutant cells may provide a metabolic environment that heightens the activity of dATM.
Changes in DNA repair and replication are only part of the dDP−/− fatbody phenotype: these tissues also have large numbers of binucleate cells. Binucleates were not generated by the overexpression of dATM, suggesting that features of the dDP mutant, beyond DNA repair and DNA replication, cause dDP−/− fatbody cells to try to divide. E2F regulation is known to be important for the switch from mitotic division to endocycle in both mice and flies (Chen et al. 2012; Edgar et al. 2014; Ouseph et al. 2012; Pandit et al. 2012) and to control G1/S oscillations during the endocycle (Zielke et al. 2011). Mutation of Dp1 loss causes endoreplication defects in mouse trophoblast giant cells (Kohn et al. 2003). Also, both canonical and atypical E2F members regulate endocycles in placental and liver cells through the regulation of cell cycle and DNA repair genes (Chen et al. 2012; Pandit et al. 2012). E2F/DP proteins are known to affect the transcription of genes needed for both G1/S and G2/M progression (Neufeld et al. 1998; Ren et al. 2002; Chicas et al. 2010; Korenjak et al. 2012; Chen et al. 2012). The differential regulation of these two suites of genes is thought to explain how E2F controls the switch between mitotic cycles and endocycles. The binucleates in dDP−/− fatbodies are presumably the result of the abnormal transcription of multiple G2/M regulators in cells that have been able to repair DNA damage and to resume S-phase. Curiously, however, computational analysis highlighted the changes in DNA repair and DNA Damage related proteins, rather than mitotic proteins, as significantly altered features of the dDP−/− proteome (Fig 1E).
Intriguingly, dDP loss does not generate binucleates in all endocycling tissues; binucleates were not seen in salivary glands and midgut cells (data not shown). This suggests that fatbody cells have a special property that makes them particularly prone to this change. Previous research has shown that under-replicated genomic regions also undergo transcriptional silencing in salivary gland and midgut cells, but not in the fatbody (Nordman et al. 2011). This suggests that genes that become replicated in dDP−/− larvae in the normally under-replicated regions may be more readily transcribed in the fatbody than in other tissues. Interestingly, overexpression of dATM, Rad50 and Mre11 in the salivary glands increases local EdU incorporation but was not sufficient to induce a broad genome-wide incorporation of EdU, suggesting that only a few specific regions can be replicated when DDR is elevated in this tissue (data not shown). Slightly different regions of the genome are under-replicated in the fatbody, salivary gland and midgut (Nordman et al. 2011); we note that we do not know if the under-replicated regions that are specific for salivary glands or midgut that can be efficiently replicated in dDP−/− animals. To the best of our knowledge, Drosophila papilar rectal cells are the only reported example in which endocycling cells revert to a mitotic division during development (Schoenfelder et al. 2014). Interestingly, papillar cells exploit specific DNA damage proteins to facilitate this change in cell cycle (Fox and Duronio 2013).
dDP mutant fatbodies are defective in the storage of lipids, and in starvation induced autophagy. Profiling of dDP mutant larvae shows that changes in levels of metabolic proteins are the major feature of the dDP mutant proteome. These changes may result from altered transcription of genes directly regulated by E2F/DP proteins, but they may also be indirect consequences of the cell cycle changes and the accumulation of binucleate cells. The fact that dDP depletion from the fatbody caused incomplete pupation, and the finding that re-expression of dDP in the fatbody was sufficient to allow dDP mutants to extend pupal development, is consistent with evidence that the fatbody has a key role of the in endocrine communication between growing tissues that sustains tissue remodeling and differentiation during pupation (Nelliot et al. 2006).
Our observations fit a simple model in which the replication seen in dDP mutants is a consequence of activation of DDR, DNA repair and the restart of stalled replication forks. In a broad sense, these observations add to a literature that has described roles for dDP and the dDP-containing repressor complex DREAM in orchestrating developmental patterns of DNA replication (Royzman et al. 1999; Calvi et al. 1998; Beall et al. 2007; Maqbool et al. 2010). dDP is important for follicle cells to repress broad genome endoreplication and to allow dE2F1 to promote selective DNA replication (Royzman et al. 1999). It is conceivable that E2F/DP complexes may also help to cause incomplete genome replication in the fatbody by acting as a barrier to efficient replication. However, examination of E2F1, E2F2 and RBF-binding sites detected by genome-wide ChIP revealed that these binding sites are almost completely absent from the genomic regions that are under-replicated in wild-type fatbodies (data not shown).
The complete elimination of E2F function has not yet been achieved in mammalian models and the results described here illustrate that E2F/DP proteins have important roles in vivo that are different from the activities traditionally studied in cancer cells. Understanding the functions of E2F proteins in normal tissues is important for multiple reasons, not least because it will likely be necessary for E2F-inhibitors to be able to distinguish between the different activities of E2F before they can be useful in the clinic. Future studies are needed to investigate E2F/DP-mediated suppression of DDR in mammalian cells, and to test whether this is used to control quiescence. We note that E2F1, E2F3, E2F4 and E2F6 all bind to the ATM promoter in mammalian cells (Liu et al. 2015; Fischer et al. 2016), that ATM is one of the genes upregulated when RB is inactivated in the mouse small intestine (Liu et al. 2015), that polyploid mouse trophoblast giant cells are reported to reduce E2F dependent gene expression and acquire resistance to apoptosis (Soloveva and Linzer 2004), and that mammalian hematopoietic stem cells down-regulate DNA damage pathways during quiescence leading to the accumulation of DNA damage during aging that needs to be repaired when cells are driven into the cell cycle (Rossi et al. 2007; Beerman et al. 2014). Thus, there are good reasons to think that the ability of E2F/DP proteins to suppress DDR is not only important in Drosophila, but may also be relevant in mammals. Furthermore, elevated expression of chromatin modifiers in human cancer cells results in site-specific copy number variations, a mechanism that has been recently proposed as a driver of early stages of tumorigenesis (Black et al. 2013).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead contact, Nick Dyson (dyson@helix.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Larval tissues
Fatbodies, midgut and diploid tissues (imaginal discs and CNS) were collected from third instar larvae (all tissues came from a mix of male and female larvae).
Fly stocks
Flies were raised in regular Drosophila medium at 25° C (unless indicated). w1118 flies were used as wild-type (WT) control flies. dDPa3 and dDPa4 null alleles of dDP were used in this work to obtain transheterozygous dDP mutant larvae (Royzman et al. 1997; Frolov et al. 2005). dDPa3/a2 and dDPa3/Df(2R) were used to validate dDPa3/a4 phenotype. UAS dDP transgene is described in Dynlacht et al., 1994 and is a validated tool that was previously shown to overexpress a fully functional dDP protein in spite of its 4 KDa smaller size (Zappia and Frolov, 2016).
Fatbody-Gal4 UASGFP UAStub-Gal80TS (kindly provided by Michael Welte, University of Rochester, originally from Ronald Kühnlein lab at Max Plank, Göttingen, Germany; Baumbach et al. 2014) and UAS dDPRNAi line (12722, Vienna Drosophila Resource Center) were used for conditional transgene induction. 0–24h FB-Gal4 UASGFP UAStub-Gal80TS UAS dDPRNAi embryos were grown until 2nd and 3rd instar larvae and transferred from 18° C to 29° C during 2–5 days, before the onset of pupariation. Two different recombinant w; cg-Ga4>myrRFP UAS dDPRNAi VDRC/CyO; sb/ TM6B-Gal80 candidates were generated, tested by PCR, and validated genetically. To perform the fatbody screening, cg-Gal4>myrRFP UAS dDPRNAi VDRC/CyO; sb/ TM6BGal80 recombinant virgins were crossed independently with a set of UAS RNAi lines (indicated below). Control crosses were set with all RNAi lines; control and experimental crosses were raised simultaneously at 25° C. A full description of fly stocks from the Bloomington Drosophila Stock Center (BDSC) and Vienna Drosophila Resource Center (VDRC) is included in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-E2F1 (IF:1/100) | Korenjak et al., 2012 | N/A |
| Rabbit anti-E2F2 (IF:1/100) | Korenjak et al., 2012 | N/A |
| Mouse anti dDP Yun6 (IF:1/15; WB: 1/10) | Stevaux et al., 2002 | N/A |
| Mouse beta-tubulin (WB:1/500) | DSHB | E7 |
| Mouse beta-actin (WB:1/1000) | Abcam | ab8224 |
| Mouse lamin A/C (IF:1/50) | DSHB | 4A7 |
| Rabbit anti-Rad50 (IF:1/100; WB: 1/1000) | Gao et al., 2009 | N/A |
| Guinea pig anti-Rad50 (IF:1/100; WB: 1/1000) | Gao et al., 2009 | N/A |
| Guinea pig anti-Mre11 (IF:1/100; WB: 1/1000) | Gao et al., 2009 | N/A |
| Guinea pig anti-Nbs (IF:1/100; WB: 1/1000) | Gao et al., 2009 | N/A |
| Rabbit anti- p-S137 H2Av (IF:1/100) | Rockland | 600-401-914 |
| Mouse anti-SpnE (IF: 1/50) | Nishida et al., 2009 | N/A |
| Mouse anti-BrdU | BD Biosciences | B44 |
| Chemicals | ||
| Triglyceride Reagent | Sigma | T2449 |
| Free Glycerol Reagent | Sigma | F6428 |
| Oil red O solution | Sigma | O1516 |
| LysoTracker Red DND 99 | Molecular Probes | L7528 |
| BrdU | Sigma | B5002 |
| EdU | Thermo Scientific | A10044 |
| Alexa fluor Azide 488 | Thermo Scientific | A10266 |
| Accumax cell detachment solution | Millipore | SCR0006 |
| Critical Commercial Assays | ||
| TMT-10plex | Thermo Scientific | 90110 |
| Gene expression microarray | Roche Nimbelgen | 12 × 135 K |
| Illumina TruSeq DNA PCR-Free LT | Illumina | FC-121-3002 |
| KAPA Library Quantification Kit | Kappa Biosystems | KK4854 |
| Native Comet Assay | Trevigen | 4250-050-K |
| Trizol | Life Technologies | 15596026 |
| microRNeasy columns | Qiagen | 74004 |
| Taqman reverse transcription kit | Applied Biosystems | 167924 |
| SYBR Green reaction mix | Roche | 04707516001 |
| Deposited Data | ||
| Mass spectrometry proteomics data | MassIVE proteomics | MSV000081383 |
| Gene expression microarray data | NCBI GEO | GSE104544 |
| DNA sequencing data from fatbodies and control tissues | NCBI Sequence Read Archive | PRJNA412980 |
| Experimental Models | ||
| Drosophila melanogaster strains | ||
| dDPa3 | (Royzman et al. 1997; Frolov et al. 2005). | N/A |
| dDPa4 | (Royzman et al. 1997; Frolov et al. 2005). | N/A |
| Df(2R) | BDSC | N/A |
| UAS dDP | Dynlacht et al., 1994 | N/A |
| Fatbody-Gal4 UASGFP UAStub-Gal80TS | Baumbach et al., 2014 | N/A |
| cg-Gal4 UASmyrRFP | BDSC | 44411 |
| cg-Ga4>myrRFP UAS dDPRNAi VDRC/CyO; sb/ TM6B-Gal80 | This paper | N/A |
| UAS dDP RNAi | VDRC | 12722 |
| UAS dATM/tefu RNAi | VDRC | 22502 |
| UAS Mre11 RNAi | VDRC | 30476,30474 |
| UAS Rad50 RNAi | VDRC | 103394 |
| UAS Fancd2 RNAi | VDRC | 106210, 45433, 29518 |
| UAS Luciferase RNAi | BDSC | 31603 |
| UAS dATM/tefu RNAi | BDSC | 44073 |
| UAS DNApol alpha RNAi | BDSC | 44584 |
| UAS DNA ligl RNAi | BDSC | 34564 |
| UAS Cyp6g1 RNAi | BDSC | 42006 |
| UAS Mre11 RNAi | BDSC | 39028 |
| UAS Fancd2 RNAi | BDSC | 53329, 56141 |
| UAS Grip71 RNAi | BDSC | 41978 |
| UAS Ncd80 RNAi | BDSC | 38260 |
| UAS E(z) RNAi | BDSC | 33659 |
| Oligonucleotides | ||
| qPCR oligonucleotides are described in Table S4 | FlyPrimerBank | N/A |
| Genotyping primers are described in Table S4 | Zappia and Frolov, 2016 | N/A |
| Software and Algorithms | ||
| Drosophila melanogaster proteome: Flybase | Attrill et al,, 2016 | N/A |
| Gene expression data processing: Nimblescan data software | Roche Nimbelgen | N/A |
| Gene expression analysis (I): Oligo R package | Carvalho, 2016 | N/A |
| Gene expression analysis (II): Limma R package | Boldstad et al., 2003 | N/A |
| DNA sequencing read quality filter and trimming: Trimmomatic | Bolger et al., 2014 | N/A |
| DNA sequencing alignment: BWA | Li and Durbin, 2010 | N/A |
| Mark duplicates, sort and merge BAMs: Picard tools v2.0.1 | http://broadinstitute.github.io/picard | N/A |
| Samtools | Li et al., 2009 | N/A |
| R: A language and environment for statistical computing | https://www.R-project.org/ | N/A |
| CNV-seq R package | Xie and Tammi, 2009 | N/A |
METHOD DETAILS
Quantitative proteomics
Per sample, 30–40 third instar larvae were snap-frozen. Whole larval protein extracts were prepared in HoB buffer (25 mM HEPES, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], Roche protease inhibitors) and resuspended in 4 M urea/50 mM HEPES (pH 8.5). Multiplexed quantitative mass spectrometry-based proteome mappings on WT and dDP−/− whole larval lysates were done in duplicate using TMT-10 plex reagents and the SPS-MS3 method on an Orbitrap Fusion mass spectrometer (Thermo Scientific) (McAlister et al. 2014; Edwards and Haas 2016). As described previously (Edwards and Haas 2016), disulfide bonds were reduced, free thiols were alkylated with iodoacetamide; proteins were purified by MeOH/CHCl3 precipitation and digested with Lys-C and trypsin, and peptides were labeled with TMT-10plex reagents (Thermo Scientific) (McAlister et al. 2014). Labeled peptide mixtures were pooled and fractionated by basic reversed-phase HPLC as described previously (Edwards and Haas, 2016). Four fractions were analyzed by multiplexed quantitative proteomics performed on an Orbitrap Fusion mass spectrometer (Thermo Scientifc) using a Simultaneous Precursor Selection (SPS) based MS3 method (McAlister et al., 2014). MS2 spectra were assigned using a SEQUEST-based proteomics analysis platform (Huttlin et al. 2010). The protein sequence database for matching the MS2 spectra was based on v5.57 of the D. melanogaster proteome retrieved from Flybase (Attrill et al. 2016). Peptide and protein assignments were filtered to a false discovery rate of < 1 % employing the target-decoy database search strategy (Elias and Gygi, 2007) and using linear discriminant analysis and posterior error histogram sorting (Huttlin et al. 2010). Peptides with sequences contained in more than one protein sequence from the UniProt database were assigned to the protein with most matching peptides (Huttlin et al. 2010). We extracted TMT reporter ion intensities as those of the most intense ions within a 0.03 Th window around the predicted reporter ion intensities in the collected MS3 spectra. Only MS3 with an average signal-to-noise value of larger than 20 per reporter ion as well as with an isolation specificity (Ting et al. 2011) of larger than 0.75 were considered for quantification. A two-step normalization of the protein TMT-intensities was performed by first normalizing the protein intensities over all acquired TMT channels for each protein based on the median average protein intensity calculated for all proteins. To correct for slight mixing errors of the peptide mixture from each sample a median of the normalized intensities was calculated from all protein intensities in each TMT channel and the protein intensities were normalized to the median value of these median intensities.
Proteomics Analysis
A total of 6380 proteins were assayed across all experimental conditions, and proteins with low intensity variance across samples were excluded, leaving 5474 proteins available for downstream analyses. Differential protein expression between WT and dDP−/− proteomes was calculated using a T-test. The Benjamini-Hochberg multiple hypothesis correction was applied to calculate corrected p-values (FDR). Differential expression of proteins between WT and dDP−/− samples was considered significant with an FDR<10% and a minimum absolute log2 FC < −0.5 and FC > 0.5.
Gene expression microarray analysis
Total RNA from Drosophila larvae was extracted with Trizol and quality tested in a Bioanalyzer. cDNA of coding transcripts were Cy3-labeled following manufacturer’s instructions to hybridize 12 × 135K Nimblegen Drosophila Gene expression array. Array analysis was performed with Nimblescan Data Analysis Software. Sample raw data was preprocessed and RMA normalized using the Oligo R package (Carvalho 2016). Among biological triplicates, one WT control sample was excluded from the differential expression analysis because of poor quality resulting in relatively low correlation with the remaining control samples. Differentially expressed genes were identified using the Limma R package (Bolstad et al. 2003). P-values were corrected for multiple hypotheses testing using the Benjamini-Hochberg method. Significant genes were defined as those genes with an FDR<10% and a minimum absolute log2 FC > 0.5.
Gene Ontology Analysis
Significant differentially expressed genes and proteins were assayed for enriched gene ontology terms using DAVID website tool (https://david.ncifcrf.gov/) (Dennis et al. 2003; Huang et al. 2009a; 2009b). DAVID bioinformatics is an integrated resource that analyzes the biological meaning of a genome-scale lists of transcripts/proteins. Gene expression and proteome datasets were uploaded to DAVID and functional terms related to biological process (BP) and molecular function (MF) were identified with a FDR<25%.
DNA Sequencing
Total DNA was isolated from dissected tissues (WT midgut, fatbody, CNS/discs and FB>LucRNAi and FB>dDPRNAi larval fatbodies from 20,40,80 and 40 larvae per replicate, respectively). Genomic DNA was isolated using standard procedures. gDNA was sonicated to an average of 300bp (QSonica Q700). One microgram of sonicated DNA was used for the library preparation (Illumina TruSeq DNA PCR-Free LT kit, FC-121-3002), according to the protocol. Libraries were quantified using the KAPA Library Quantification Kit (Kappa Biosystems KK4854). Single-end sequencing was performed with the Illumina NextSeq 500.
The sequencing data was trimmed for quality and sequencing primer contamination with Trimmomatic (Bolger et al. 2014) using the default settings except for following parameters (SLIDINGWINDOW:4:15 MINLEN:25). Trimmed reads were then aligned to the Drosophila genome (Dm6 Ensembl v78) using BWA v 0.7.5 (default settings; (Li and Durbin 2010)). BWA SAM files were converted to BAM format using Samtools v1.14 (Li et al. 2009). Sample read data was merged across lanes, sorted and removed duplicate mapped reads were removed using Picard tools v2.0.1 (http://broadinstitute.github.io/picard).
CNV identification
Regions of genomic instability were identified using the CNV-seq R package (Xie and Tammi 2009). This tool uses depth of mapped reads with a walking window of 2500bp to identify regions with copy number variation (CNV) between samples. The CNV-seq tool assumes that uneven coverage of sequenced reads can approximated by the Poisson distribution dependent upon the total number of mapped reads in a sample, the size of the genome and the walking window size. The tool calculates the probability that an observed copy number ratio between treatment and control samples for each window is not equal to 1 due to random chance. To improve the power of detecting CNV regions we increased the read depth of the compared conditions by merging duplicate sample mapped reads. We identified the set of regions between samples for each time point by comparing the dDP and Luciferase RNAi samples to the diploid control as well as dDP RNAi vs Luciferase RNAi. Significant region of CNV were defined by a Log2FC> 0.6 and a FDR < 1×10−3. All analytics and plots were generated using R.
Lipid measurements
TAG levels were measured in triplicates as (Hildebrandt et al. 2011) with modifications. Ten larvae were kept on ice and homogeneized in 100 μl PBS Tween 0.1%, and debris were pelleted at 4° C (13K RPM). We added 60 μl of Triglyceride Reagent (Lipase, Sigma T2449) to each 60 μl of homogenate (blanks of each replicate were prepared with 20 μl of homogenate and 20 μl of PBS) and incubated at 37° C. Samples were centrifuged at RT for 2 min. 100 μl of Free Glycerol Reagent (Glycerol Kinase, Glycerol Phosphate Oxidase and Peroxidase, Sigma F6428) was added to 30 μl of each reaction on a flat 96 well plate. Total absorbance at 540 nm was measured and corrected with their correspondent blank. Total larval protein was used to normalize TAG values.
Oil-red lipid detection
Fatbodies were fixed 1h in PFA 4% at RT, washed twice in PBS 1X and Propylen Glycol 100% each and incubated in 0,5% Oil Red in Propylene Glycol for 30 minutes at 37° C. Tissues were washed in 80% Propylene Glycol and PBS, and mounted in Vectashield with DAPI (Vector Laboratories).
Lysotracker labeling
Larvae were dissected in PBS 1X and incubated for 2 min in 100 μM LysoTracker Red DND 99 (Molecular Probes) in DAPI 0,5% Schneider Medium (Gibco). Fatbodies were transferred to glass slides, covered, and immediately photographed.
EdU and BrdU detection
Larvae were fed during 24h in instant food vials (Carolina) with 0.025 mg/ml BrdU (Sigma). BrdU fed larvae were fixed during 15 min in Formaldehyde 2%, washed in PBT. Tissues were treated with 2N HCl during 30 min at 37° C, and processed to detect BrdU by inmunofluorescence (mouse anti-BrdU, BD biosciences). Freshly dissected fatbodies were incubated in a 50 μM EdU in PBS 1X during 30 min at room temperature, fixed during 1h RT in PFA 4 %, permeabilized for 10 min in PBS Triton 0,1% (PBT). Tissues were incubated 1h RT Click-it Buffer and Azide 488 (Alexa), washed 3x in PBS (Triton was not used to avoid lipid leak) and stored in Vectashield with DAPI at 4° C (Vector Laboratories). Note that EdU labeling abolishes GFP fluorescence.
Native comet assay
Larval fatbodies were dissected in PBS, and incubated 10 min in 500 μl of Accumax cell detachment solution at RT (SCR0006, Millipore). Cells were centrifuged 20 min at 2000 rpm to remove Accumax and resuspended in 10 μl PBS. Cells were homogenized in 100 μl of low melting agarose; two replicates of 50 μl gels were prepared on each slide and processed in neutral electrophoresis buffer following manufacturer’s instructions (Trevigen Comet Assay). Comet length quantifications were done in a minimum of n=40 cells/ genotype.
Western blot, immunofluorescence analysis
Western blotting was carried out using standard procedures. Fatbodies were collected in cold PBS 1X and centrifuged at 4° C (13K RPM). PBS was removed prior to snap-freezing of tissues. Fatbodies were thawed on ice, and homogeneized in modified HoB lysis buffer (50 mM HEPES, pH 8, 100 mM NaCl, 2 mM EDTA, 5 mM NaF, 10% Glycerol, 0.1% Triton X-100, 1 mM dithiothreitol [DTT], 1 mM PMSF and Roche protease inhibitors).
Immunofluorescence was performed in fixed fatbodies (1h at RT in 4% PFA in PBS). Only if immunodetection was performed in other tissues than fatbody, tissues were permeabilized after fixation during 15 minutes in PBS Triton 0,1% (fatbodies were washed without detergent in PBS 1X during 15min). Tissues were blocked in 1% BSA PBS and primary antibodies were incubated at 4° C O/N. Larval tissues were washed three times at RT in PBS 1X, incubated 1h at RT with secondary antibodies (Alexa Fluor conjugated secondary antibodies, Life Technologies), washed three times in PBS 1X and mounted in Vectashield with DAPI (Vector Laboratories).
Image acquisition
Images were acquired in a 710 Zeiss Confocal microscope at MGH Cancer Center, taken under the same magnification. Fluorescence measurements were done in raw single-channel micrographs in Fiji (https://fiji.sc/).
RT-qPCR
RNA was extracted from fatbodies of 20–40 larvae in Trizol (Life Technologies). qPCR primers are listed in Table S4 (related to Key Resources Table). Tissues were thawed in ice, homogenized in Trizol following manufacturer’s instructions (Life Technologies) and purified in microRNeasy columns (Qiagen). cDNA was obtained from 1 ug of RNA per RT PCR reaction (Applied Biosystems). Triplicates of each cDNA were diluted 1/5, prepared with SYBR Green reaction mix 2X (Roche). qPCR reactions were run in a Roche 480 LightCycler. qPCR primers were designed with FlyPrimerBank (Hu et al. 2013), and gene expression changes were calculated using the mean of RpL32 and Gapdh as housekeeping gene expression.
Animal genotyping
The genotypes of the rescued animals were confirmed using single fly DNA extraction followed by PCR. The dDPa3 and dDPa4 allele were detected by sanger sequencing. Genotyping primers are described in Table S4 (related to Key Resources Table). Mutant genotype of all dDPa3/Def rescued animals was PCR confirmed using the same procedure (data not shown).
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistics and plots generated for Proteomic, Gene expression array, Gene Ontology analysis and Copy number variation were generated using R. The rest of statistical analysis and corresponding plots were performed using Prism6 software. Data is represented as mean ± SD, and the normality of each sample was assessed to choose the appropriate statistical analysis (ANOVA, t-test, Mann-Whitney or Kruskall-Wallis test; details are described in Figure legends). The calculated statistical significance is indicated as: **** p<0.0001; *** p<0.001; ** p<0.01; * p<0.05. In the survival measurements, data is represented as % of animals (relative to pupa), and Chi-square tests were used to test the significant differences between the observed % of rescued animals and the expected % of mutant animals.
DATA AND SOFTWARE AVAILABILITY
Mass spectrometry proteomics data is deposited in MassIVE proteomics (MSV000081383), gene expression microarray data generated is deposited in NCBI GEO (GSE104544) and DNA sequencing data is deposited in NCBI Sequence Read Archive (PRJNA412980).
Supplementary Material
Table S1, related to Figure 1. Proteome data. Related to the significantly increasing and decreasing proteins in WT and dDP−/− larvae. Calculated Log2FC and FDR with ChIP (E2F1-, E2F2- and RBF- bound genes, “TRUE”; “FALSE”: not bound). Protein IDs are indicated in columns: Uniprot ID/KB/Refseq/Flybase protein for proteins; Flybase transcript, flybase gene and Gene name for transcripts. Significantly increased (Up in DP, Log2FC >0.5) and decreased proteins (Down in DP, Log2FC >0.5) are separated into tabs.
Table S2, related to Figure 1. Gene expression data. Related to the significantly increasing and decreasing transcripts in WT and dDP−/− larvae. Calculated Log2FC and FDR with ChIP binding data (see E2F1, E2F2 and RBF ChIP columns). Synonym IDs are indicated as in Table S1. Significantly increased (Up in DP, Log2FC >0.5) and decreased transcripts (Down in DP, Log2FC >0.5) are separated into tabs.
Table S3, related to Figure 1. Integrated ChIP/RNA/Protein data. Related to the list of candidate drivers of the dDP−/− phenotype. Direct and indirect target genes are separated into subsets (E2F1, E2F2 and RBF ChIP columns). Calculated Log2FC (PROTEIN Log2FC, RNA Log2FC) and statistical FDR values (PROTEIN FDR, RNA FDR) are shown for each candidate. Synonym IDs are indicated as in Table S1.
Acknowledgments
We thank K. White, I. Sanidas and JM. González-Rosa for critical reading of the manuscript; members from White, Zou and Dyson laboratories for helpful discussions; MM. Genois, S. Martin and B. Nicolay for technical help; M. Welte, R. Kühnlein, Rong laboratories for sharing reagents; the Vienna Drosophila Resource Center and the Bloomington Stock Center for providing fly stocks, and the Developmental Studies Hybridoma Bank (DSHB) for providing antibodies. M.K. was supported by a Tosteson Postdoctoral Fellowship Award and a DFG Forschungsstipendium. This study was supported by the following NIH grants: R01G117413 and R01CA163698 to N.J.D., GM093827 and GM110018 to MVF, R01GM097360 to JRW, GM076388 to LZ, NCI R01CA185086 to SR. JRW is the Tepper Family MGH Research Scholar and Leukemia and Lymphoma Society Scholar and N.J.D is the James and Shirley Curvey MGH Research Scholar. Johnathan R. Whetstine is a consultant for QSonica and Celgene.
Footnotes
Supplemental information includes seven figures and four tables and can be found in this article online.
AUTHOR CONTRIBUTIONS
Conceptualization, AGP and NJD; Investigation, AGP, MK, MPZ; Methodology and formal analysis, AGP, RM, MB, WH, CVR, SR, LZ; Funding acquisition, NJD, WH, JW, MVF, SR, LZ; writing – original draft, AGP and NJD; writing- review and editing, all authors; supervision, AGP and NJD.
References
- Ambrus AM, Islam ABMMK, Holmes KB, Moon N-S, Lopez-Bigas N, Benevolenskaya EV, Frolov MV. Loss of dE2F compromises mitochondrial function. Developmental Cell. 2013;27:438–451. doi: 10.1016/j.devcel.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreyeva EN, Kolesnikova TD, Belyaeva ES, Glaser RL, Zhimulev IF. Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res. 2008;16:851–862. doi: 10.1007/s10577-008-1244-4. [DOI] [PubMed] [Google Scholar]
- Attrill H, Falls K, Goodman JL, Millburn GH, Antonazzo G, Rey AJ, Marygold SJ FlyBase Consortium. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Research. 2016;44:D786–92. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach J, Xu Y, Hehlert P, Kühnlein RP. Gαq, Gγ1 and Plc21C control Drosophila body fat storage. J Genet Genomics. 2014;41:283–292. doi: 10.1016/j.jgg.2014.03.005. [DOI] [PubMed] [Google Scholar]
- Beall EL, Lewis PW, Bell M, Rocha M, Jones DL, Botchan MR. Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes & Development. 2007;21:904–919. doi: 10.1101/gad.1516607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15:37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Ginsberg D. ATM is a target for positive regulation by E2F-1. Oncogene. 2003;22:161–167. doi: 10.1038/sj.onc.1206144. [DOI] [PubMed] [Google Scholar]
- Bi X, Gong M, Srikanta D, Rong YS. Drosophila ATM and Mre11 are essential for the G2/M checkpoint induced by low-dose irradiation. Genetics. 2005;171:845–847. doi: 10.1534/genetics.105.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Manning AL, Van Rechem C, Kim J, Ladd B, Cho J, Pineda CM, Murphy N, Daniels DL, Montagna C, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154:541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Misteli T. Not All DDRs Are Created Equal: Non-Canonical DNA Damage Responses. Cell. 2015;162:944–947. doi: 10.1016/j.cell.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes & Development. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BS. Working with Oligonucleotide Arrays. Methods Mol Biol. 2016;1418:145–159. doi: 10.1007/978-1-4939-3578-9_7. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P, Bonnette PC, Dickson MR, Duronio RJ. Drosophila E2f2 promotes the conversion from genomic DNA replication to gene amplification in ovarian follicle cells. Development. 2001;128:5085–5098. doi: 10.1242/dev.128.24.5085. [DOI] [PubMed] [Google Scholar]
- Chen H-Z, Ouseph MM, Li J, Pécot T, Chokshi V, Kent L, Bae S, Byrne M, Duran C, Comstock G, et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol. 2012a;14:1192–1202. doi: 10.1038/ncb2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas A, Wang X, Zhang C, McCurrach M, Zhao Z, Mert O, Dickins RA, Narita M, Zhang M, Lowe SW. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciapponi L, Cenci G, Gatti M. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics. 2006;173:1447–1454. doi: 10.1534/genetics.106.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dimova DK. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes & Development. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duronio RJ, O’Farrell PH, Xie JE, Brook A, Dyson N. The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes & Development. 1995;9:1445–1455. doi: 10.1101/gad.9.12.1445. [DOI] [PubMed] [Google Scholar]
- Dynlacht BD, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc Natl Acad Sci USA. 1994;91:6359–6363. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- Edgar BA, Zielke N, Gutierrez C. Endocycles: a recurrent evolutionary innovation for post-mitotic cell growth. Nat Rev Mol Cell Biol. 2014;15:197–210. doi: 10.1038/nrm3756. [DOI] [PubMed] [Google Scholar]
- Edwards A, Haas W. Multiplexed Quantitative Proteomics for High-Throughput Comprehensive Proteome Comparisons of Human Cell Lines. Methods Mol Biol. 2016;1394:1–13. doi: 10.1007/978-1-4939-3341-9_1. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic Protein Location Mapping Reveals Five Principal Chromatin Types in Drosophila Cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Grossmann P, Padi M, DeCaprio JA. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Research. 2016;44:6070–6086. doi: 10.1093/nar/gkw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, Dyson NJ. Functional antagonism between E2F family members. Genes & Development. 2001;15:2146–2160. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Moon N-S, Dyson NJ. dDP is needed for normal cell proliferation. Molecular and Cellular Biology. 2005;25:3027–3039. doi: 10.1128/MCB.25.8.3027-3039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Bi X, Chen J, Srikanta D, Rong YS. Mre11-Rad50-Nbs complex is required to cap telomeres during Drosophila embryogenesis. Proc Natl Acad Sci USA. 2009;106:10728–10733. doi: 10.1073/pnas.0902707106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–521. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- Hildebrandt A, Bickmeyer I, Kühnlein RP. Reliable Drosophila body fat quantification by a coupled colorimetric assay. In: Samakovlis C, editor. PLoS ONE. Vol. 6. 2011. p. e23796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Sopko R, Foos M, Kelley C, Flockhart I, Ammeux N, Wang X, Perkins L, Perrimon N, Mohr SE. FlyPrimerBank: an online database for Drosophila melanogaster gene expression analysis and knockdown evaluation of RNAi reagents. G3: Genes|Genomes|Genetics. 2013;3:1607–1616. doi: 10.1534/g3.113.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn MJ, Bronson RT, Harlow E, Dyson NJ, Yamasaki L. Dp1 is required for extra-embryonic development. Development. 2003;130:1295–1305. doi: 10.1242/dev.00355. [DOI] [PubMed] [Google Scholar]
- Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, Dyson NJ. RBF Binding to both Canonical E2F Targets and Noncanonical Targets Depends on Functional dE2F/dDP Complexes. Molecular and Cellular Biology. 2012;32:4375–4387. doi: 10.1128/MCB.00536-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A. Native E2F/RBF Complexes Contain Myb-Interacting Proteins and Repress Transcription of Developmentally Controlled E2F Target Genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes & Development. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly MA, Duronio RJ. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005;24:2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- Liu H, Tang X, Srivastava A, Pécot T, Daniel P, Hemmelgarn B, Reyes S, Fackler N, Bajwa A, Kladney R, et al. Redeployment of Myc and E2f1-3 drives Rb-deficient cell cycles. Nat Cell Biol. 2015;17:1036–1048. doi: 10.1038/ncb3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Kurtyka CA, Boyapalle S, Sung S-S, Lawrence H, Guida W, Cress WD. A small-molecule E2F inhibitor blocks growth in a melanoma culture model. Cancer Research. 2008;68:6292–6299. doi: 10.1158/0008-5472.CAN-08-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maqbool SB, Mehrotra S, Kolpakas A, Durden C, Zhang B, Zhong H, Calvi BR. Dampened activity of E2F1-DP and Myb-MuvB transcription factors in Drosophila endocycling cells. Journal of Cell Science. 2010;123:4095–4106. doi: 10.1242/jcs.064519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra S, Maqbool SB, Kolpakas A, Murnen K, Calvi BR. Endocycling cells do not apoptose in response to DNA rereplication genotoxic stress. Genes & Development. 2008;22:3158–3171. doi: 10.1101/gad.1710208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelliot A, Bond N, Hoshizaki DK. Fat-body remodeling in Drosophila melanogaster. Genesis. 2006;44:396–400. doi: 10.1002/dvg.20229. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, la Cruz de AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. The EMBO Journal. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman J, Li S, Eng T, Macalpine D, Orr-Weaver TL. Developmental control of the DNA replication and transcription programs. Genome Res. 2011;21:175–181. doi: 10.1101/gr.114611.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani K, Nevins JR. Functional properties of a Drosophila homolog of the E2F1 gene. Molecular and Cellular Biology. 1994;14:1603–1612. doi: 10.1128/mcb.14.3.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouseph MM, Li J, Chen H-Z, Pécot T, Wenzel P, Thompson JC, Comstock G, Chokshi V, Byrne M, Forde B, et al. Atypical E2F repressors and activators coordinate placental development. Developmental Cell. 2012;22:849–862. doi: 10.1016/j.devcel.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit SK, Westendorp B, Nantasanti S, van Liere E, Tooten PCJ, Cornelissen PWA, Toussaint MJM, Lamers WH, de Bruin A. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol. 2012;14:1181–1191. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem. 2015;84:711–738. doi: 10.1146/annurev-biochem-060614-034335. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes & Development. 2002a;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Royzman I, Austin RJ, Bosco G, Bell SP, Orr-Weaver TL. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes & Development. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I, Whittaker AJ, Orr-Weaver TL. Mutations in Drosophila DP and E2F distinguish G1–S progression from an associated transcriptional program. Genes & Development. 1997;11:1999–13. doi: 10.1101/gad.11.15.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangwan M, McCurdy SR, Livne-Bar I, Ahmad M, Wrana JL, Chen D, Bremner R. Established and new mouse models reveal E2f1 and Cdk2 dependency of retinoblastoma, and expose effective strategies to block tumor initiation. Oncogene. 2012;31:5019–5028. doi: 10.1038/onc.2011.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Montague RA, Paramore SV, Lennox AL, Mahowald AP, Fox DT. Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development. 2014;141:3551–3560. doi: 10.1242/dev.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Schuldiner O, Neufeld TP. Role and Regulation of Starvation-Induced Autophagy in the Drosophila Fat Body. Developmental Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- Smith AV, Orr-Weaver TL. The regulation of the cell cycle during Drosophila embryogenesis: the transition to polyteny. Development. 1991;112:997–1008. doi: 10.1242/dev.112.4.997. [DOI] [PubMed] [Google Scholar]
- Soloveva V, Linzer DIH. Differentiation of placental trophoblast giant cells requires downregulation of p53 and Rb. Placenta. 2004;25:29–36. doi: 10.1016/S0143-4004(03)00215-7. [DOI] [PubMed] [Google Scholar]
- Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. The EMBO Journal. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ. Conserved functions of the pRB and E2F families. Nature Publishing Group. 2008;9:713–724. doi: 10.1038/nrm2469. [DOI] [PubMed] [Google Scholar]
- Xie C, Tammi MT. CNV-seq, a new method to detect copy number variation using high-throughput sequencing. BMC Bioinformatics. 2009;10:80. doi: 10.1186/1471-2105-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarosh W, Spradling AC. Incomplete replication generates somatic DNA alterations within Drosophila polytene salivary gland cells. Genes & Development. 2014;28:1840–1855. doi: 10.1101/gad.245811.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia MP, Frolov MV. E2F function in muscle growth is necessary and sufficient for viability in Drosophila. Nat Comms. 2016;7:10509. doi: 10.1038/ncomms10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Mehrotra S, Ng WL, Calvi BR. Low Levels of p53 Protein and Chromatin Silencing of p53 Target Genes Repress Apoptosis in Drosophila Endocycling Cells. In: Duronio RJ, editor. PLoS Genet. Vol. 10. 2014. p. e1004581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N, Kim KJ, Tran V, Shibutani ST, Bravo M-J, Nagarajan S, van Straaten M, Woods B, Dassow von G, Rottig C, et al. Control of Drosophila endocycles by E2F and CRL4(CDT2) Nature. 2011;480:123–127. doi: 10.1038/nature10579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, related to Figure 1. Proteome data. Related to the significantly increasing and decreasing proteins in WT and dDP−/− larvae. Calculated Log2FC and FDR with ChIP (E2F1-, E2F2- and RBF- bound genes, “TRUE”; “FALSE”: not bound). Protein IDs are indicated in columns: Uniprot ID/KB/Refseq/Flybase protein for proteins; Flybase transcript, flybase gene and Gene name for transcripts. Significantly increased (Up in DP, Log2FC >0.5) and decreased proteins (Down in DP, Log2FC >0.5) are separated into tabs.
Table S2, related to Figure 1. Gene expression data. Related to the significantly increasing and decreasing transcripts in WT and dDP−/− larvae. Calculated Log2FC and FDR with ChIP binding data (see E2F1, E2F2 and RBF ChIP columns). Synonym IDs are indicated as in Table S1. Significantly increased (Up in DP, Log2FC >0.5) and decreased transcripts (Down in DP, Log2FC >0.5) are separated into tabs.
Table S3, related to Figure 1. Integrated ChIP/RNA/Protein data. Related to the list of candidate drivers of the dDP−/− phenotype. Direct and indirect target genes are separated into subsets (E2F1, E2F2 and RBF ChIP columns). Calculated Log2FC (PROTEIN Log2FC, RNA Log2FC) and statistical FDR values (PROTEIN FDR, RNA FDR) are shown for each candidate. Synonym IDs are indicated as in Table S1.