Summary
Detection of cytosolic DNA constitutes a central event in the context of numerous infectious and sterile inflammatory conditions. Recent studies have uncovered a bipartite mode of cytosolic DNA recognition, in which the cGAS-STING axis triggers antiviral immunity, whereas AIM2 triggers inflammasome activation. Here, we show that AIM2 is dispensable for DNA-mediated inflammasome activation in human myeloid cells. Instead, detection of cytosolic DNA by the cGAS-STING axis induces a cell death program initiating potassium efflux upstream of NLRP3. Forward genetics identified regulators of lysosomal trafficking to modulate this cell death program, and subsequent studies revealed that activated STING traffics to the lysosome, where it triggers membrane permeabilization and thus lysosomal cell death (LCD). Importantly, the cGAS-STING-NLRP3 pathway constitutes the default inflammasome response during viral and bacterial infections in human myeloid cells. We conclude that targeting the cGAS-STING-LCD-NLRP3 pathway will ameliorate pathology in inflammatory conditions that are associated with cytosolic DNA sensing.
Introduction
The detection of non-self-nucleic acids constitutes an integral sensory function of the innate immune system. Because a variety of pathogens use the cytosol for replication, it is under heavy surveillance by nucleic acid sensing pattern recognition receptors (PRRs). Cytosolic DNA is recognized by both the AIM2 inflammasome (Fernandes-Alnemri et al., 2009, Hornung et al., 2009), inducing a broad pro-inflammatory response and the cGAS-STING signaling axis that drives antiviral immunity by inducing type I interferons (Ishikawa and Barber, 2008, Sun et al., 2013). The inflammasome controls the bioactivity of pro-inflammatory cytokines of the interleukin (IL)-1 family and also a highly pro-inflammatory cell death known as pyroptosis (Broz and Dixit, 2016). Activation of the respective inflammasome sensor triggers the prion-like assembly of the inflammasome adaptor ASC (PYCARD) into a filamentous helical structure called the pyroptosome that recruits the inflammasome effector caspase-1. Consequently, caspase-1 is activated to mature pro-IL-1β, pro-IL-18, and GSDMD, the effector molecule of pyroptosis. Besides cytosolic DNA-mediated AIM2-inflammasome activation, the NOD-like receptor NLRP3 induces inflammasome formation upon various infectious and sterile triggers. Unlike AIM2, NLRP3 may not directly detect a ligand, yet senses perturbations of cellular integrity by a large array of different signals. With a few exceptions, K+ efflux has been identified as the common denominator of NLRP3 activation (Muñoz-Planillo et al., 2013).
Besides inflammasome activation, cytosolic DNA sensing leads to type I interferon production by the cGAS-STING pathway. Here, the cytoplasmic nucleotidyltransferase cGAS (MB21D1) binds dsDNA, which initiates the synthesis of the cyclic dinucleotide 2nd messenger molecule cGAMP (Chen et al., 2016). cGAMP binds to and activates the endoplasmic reticulum (ER)-resident transmembrane receptor STING (TMEM173), which subsequently exits the ER toward the Golgi to initiate signaling. Then, STING recruits and activates TBK1 via its C-terminal tail (CTT). TBK1 in turn phosphorylates a conserved platform within the CTT that then functions as a scaffold to recruit IRF3, which is subsequently phosphorylated in a TBK1-dependent manner (Liu et al., 2015). In this cascade, TBK1 has been reported to also function upstream of nuclear factor κB (NF-kB) activation (Abe and Barber, 2014). In concert, these transcription factors induce antiviral and pro-inflammatory gene expression. Downstream of STING, TBK1 also triggers autophagy, independently of its function to induce gene expression (Watson et al., 2012). In this context, STING has also been shown to co-localize with markers of autophagosomes at late stages following activation (Saitoh et al., 2009).
A series of studies in mice have attributed a critical role for both the cGAS-STING axis and the AIM2 inflammasome in microbial infection, as well as in the context of sterile inflammation. For example, Aim2-deficient mice are highly susceptible toward infection with the intracellular pathogen Francisella tularensis (Fernandes-Alnemri et al., 2010, Jones et al., 2010) or Mycobacterium tuberculosis (Saiga et al., 2012), and display an impaired response toward MCMV (Rathinam et al., 2010). Moreover, in concert with other inflammasome sensors, AIM2 also critically contributes to the host response toward several other microbial pathogens in murine infection models (Man et al., 2016). In addition, a series of studies have demonstrated that AIM2 plays a central role in various sterile self-DNA triggered inflammatory conditions, such as irradiation-induced hematopoietic failure and irradiation-associated gastrointestinal wasting (Hu et al., 2016), arthritis (Baum et al., 2015, Jakobs et al., 2015), and cerebral ischemia (Denes et al., 2015). Altogether, these studies have established AIM2 as an interesting therapeutic target to treat human disease.
Results
DNA-Triggered Inflammasome Activation Depends on NLRP3 in Human Monocytes
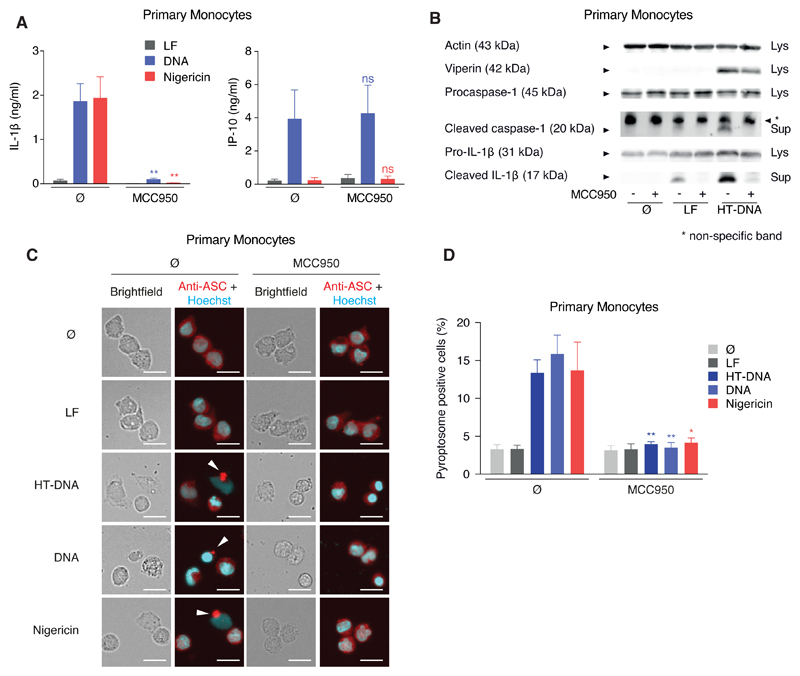
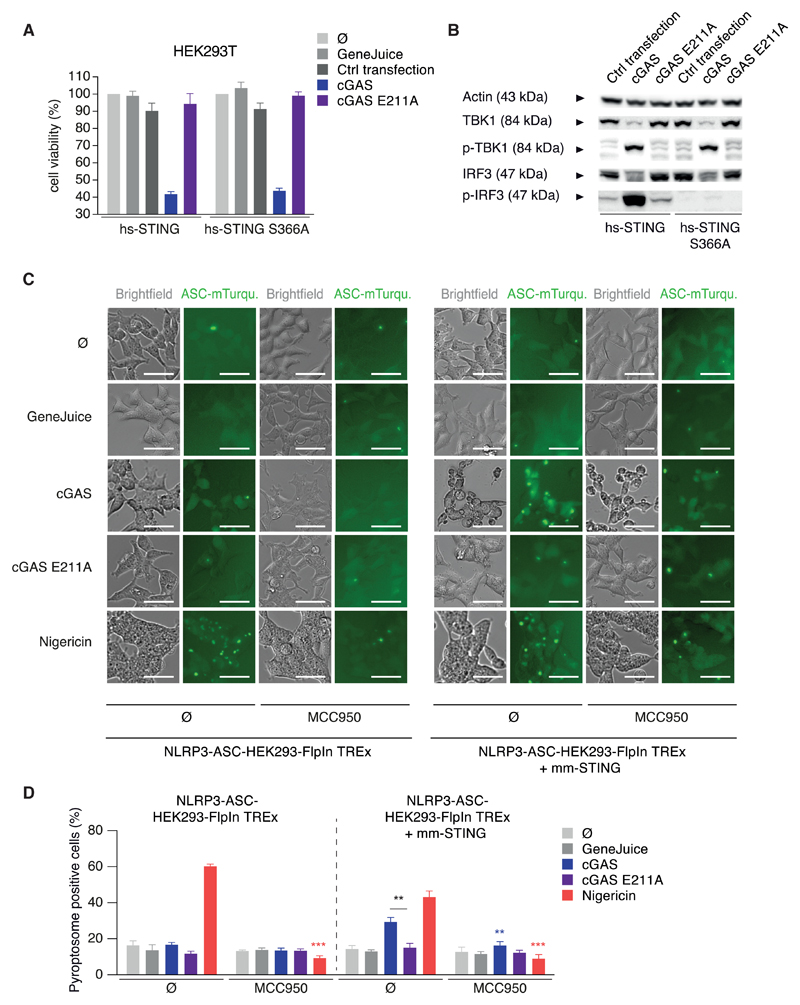
In light of critical importance of the AIM2-inflammasome pathway in the murine system, we turned our attention to inflammasome-dependent DNA recognition in human cells. Studying primary human monocytes from peripheral blood, we observed that pharmacological inhibition of NLRP3 by MCC950 (Coll et al., 2015) not only inhibited IL-1β secretion in response to the classical NLRP3 agonist nigericin, but also in response to transfected dsDNA (Figure 1A). Subsequent experiments revealed that cytosolic DNA led to classical NLRP3 inflammasome activation, as characterized by the secretion of mature caspase-1 and IL-1β and also by pyroptosome formation (Figures 1B and 1C). Moreover, the inflammasome-controlled secretion of IL-18 was also completely inhibited by MCC950 after stimulation with either DNA or nigericin, respectively (Figure S1A). Of note, inhibition of NLRP3 had no impact on DNA-dependent induction of antiviral immunity, as both IP-10 secretion (Figure 1A) and the induction of the interferon-stimulated gene viperin (Figure 1B) were not affected by MCC950 treatment.
Figure 1. Cytosolic DNA Recognition Leads to NLRP3-Dependent Classical Inflammasome Activation in Human Monocytes.
(A and B) Pam3CSK4-primed primary human monocytes were treated with or without MCC950 and stimulated with lipofected DNA, HT-DNA, or lipofection reagent alone (LF) for 8 hr. If indicated, nigericin was added for the last 2 hr of the experiment. Cytokines were quantified by ELISA or samples were analyzed by immunoblotting. Data are depicted as mean + SEM of seven donors (A) or one representative immunoblot of three (B). **p < 0.01; ns, not significant.
(C and D) Primary human monocytes were stimulated as in (A) and pyroptosome formation was analyzed. Representative images (C) and quantifications from four donors are shown as mean + SEM (D). Scale bars denote 15 µm. **p < 0.01, *p < 0.05.
See also Figure S1.
Because the importance of AIM2 in DNA-mediated inflammasome signaling is well established, we set out to confirm the specificities of both our stimuli as well as the NLRP3 inhibitor MCC950 in murine bone marrow-derived macrophages (BMDMs). As expected, DNA stimulation resulted in strong inflammasome activation leading to the secretion of matured IL-1β and caspase-1 (Figures S1B and S1C), which were completely dependent on AIM2, ASC, and caspase-1. Neither genetic deletion of NLRP3 nor its pharmacological inhibition perturbed this AIM2-dependent response.
In order to study the underlying signaling cascade in a model system amenable to genetic manipulation, we turned to the human monocyte-like cell line THP-1, commonly used to model human inflammasome activation. However, IL-1β secretion upon cytosolic DNA sensing was dependent on AIM2, but not NLRP3 in THP-1 cells. At the same time, pharmacologic inhibition of NLRP3 had no impact on DNA-mediated inflammasome activation in these cells (Figure S1D). The fact that the signaling circuits in human THP-1 cells mirrored those observed in murine BMDMs argues against a species-specific difference in inflammasome sensor usage upon DNA-mediated activation. Instead, it appears that cell type-specificities account for the observed differences between primary monocytes and THP-1 cells.
Genetic Dissection of DNA-Mediated Inflammasome Activation in BLaER1 Monocytes
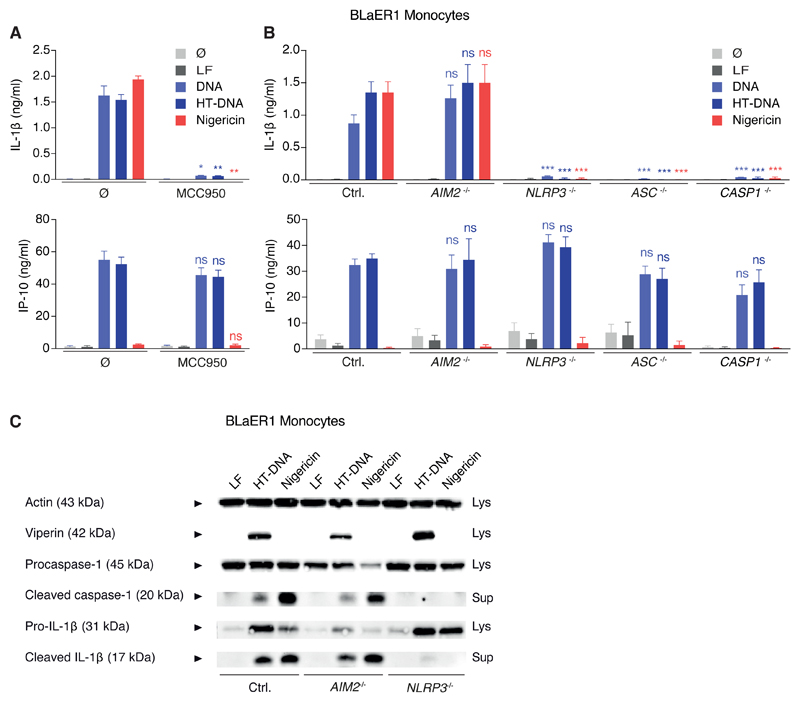
We have recently established a novel cell culture model for inflammasome studies (BLaER1), which relies on the trans-differentiation of proliferative, PAMP-insensitive B cells to postmitotic, PRR-competent monocytes (Gaidt et al., 2016). For technical reasons, all experiments were conducted in CASP4−/− x TRIF−/− (TICAM1) BLaER1 monocytes that will be referred to as Ctrl. (see STAR Methods). In line with the results obtained from primary monocytes, both DNA- and nigericin-mediated inflammasome activation were sensitive to pharmacological inhibition of NLRP3 in BLaER1 monocytes (Figure 2A). Genetic dissection using CRISPR/Cas9 showed that IL-1β secretion required NLRP3, ASC, and CASP1 but was independent of AIM2 (Figure 2B). Immunoblotting for cleaved caspase-1 and IL-1β confirmed these results (Figure 2C). Again, parallel to the situation in primary monocytes, both genetic and pharmacological inhibition of NLRP3 only impaired the inflammasome response to cytosolic DNA, yet left the type I interferon (IFN) response intact (Figures 2A–2C).
Figure 2. BLaER1 Monocytes Faithfully Recapitulate DNA-Mediated NLRP3-Inflammasome Activation Independently of AIM2.
(A) LPS-primed BLaER1 monocytes were stimulated as indicated. Data are depicted as mean + SEM of three experiments. **p < 0.01, *p < 0.05, ns, not significant. LF, Lipofectamine.
(B and C) BLaER1 monocytes of indicated genotype were stimulated as in (A). Data are shown as mean + SEM of three experiments (B) or one representative immunoblot from four experiments (C). ***p < 0.001; ns, not significant.
See also Figure S2.
Determining the expression levels of AIM2 in primary monocytes showed that these cells expressed slightly lower protein levels than THP-1 (Figure S2A). Priming with IFNγ, however, restored AIM2 expression in primary monocytes to levels that were comparable to AIM2-competent THP-1 cells (Figure S2A). As the inflammasome response toward transfected DNA still appeared completely sensitive to NLRP3 inhibition after IFNγ priming (Figure S2B), it can be concluded that AIM2 expression is not the factor that limits its activation in primary human monocytes. Consistent with these results, IFNγ also primed AIM2 expression in BLaER1 monocytes (Figure S2C), yet inflammasome activation in response to cytosolic DNA remained completely dependent on NLRP3 in these cells (Figure S2D). From these results, one can conclude that other factors than insufficient expression levels must account for the fact that AIM2 is not functional in human monocytes.
DNA-Mediated NLRP3 Inflammasome Activation Relies on the cGAS-STING Axis
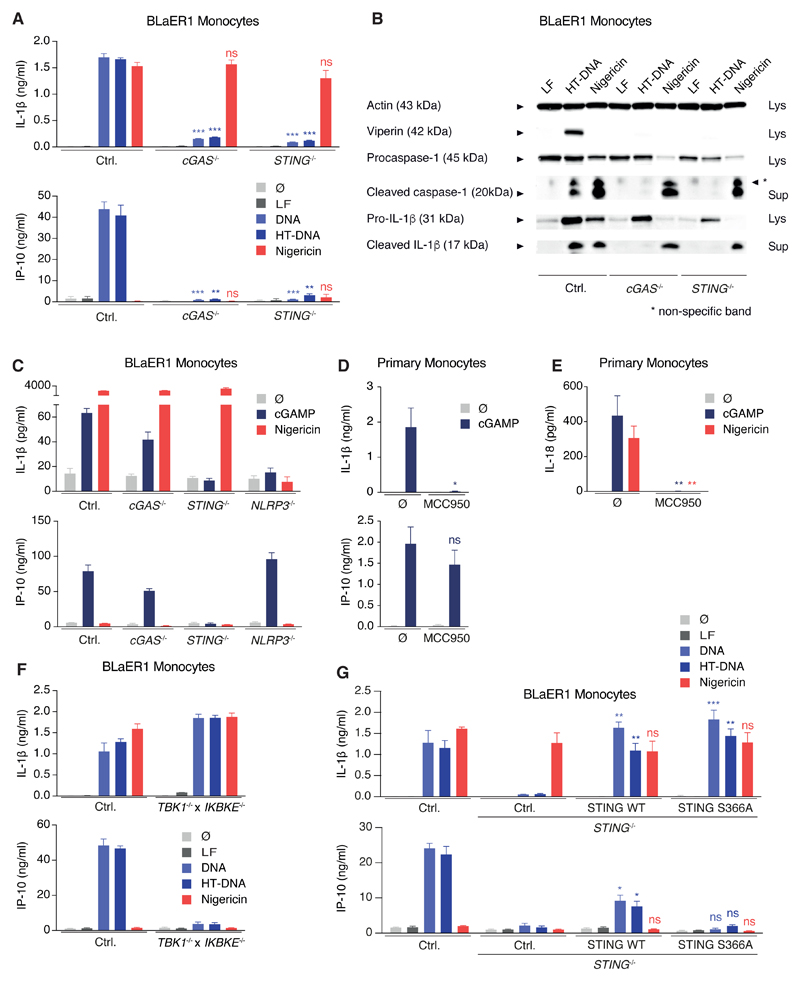
To unravel the activation signal of DNA-mediated inflammasome signaling upstream of NLRP3, we next aimed at identifying the direct DNA-binding sensor. To do so, we first wanted to rule out an involvement of the known DNA-sensor cGAS that drives type I IFN production via STING. As expected, both cGAS and STING were required for IP-10 secretion upon DNA stimulation in BLaER1 monocytes, however, intriguingly IL-1β and caspase-1 activation were also largely abrogated in cGAS and STING-deficient cells (Figures 3A and 3B). On the other hand, cGAS and STING were dispensable for NLRP3 activation by nigericin (Figures 3A and 3B). In concordance with the crucial role for cGAS and STING in NLRP3 activation, direct activation of STING by the endogenous second messenger cGAMP led to STING- and NLRP3-dependent, but cGAS-independent, IL-1β secretion in BLaER1 monocytes (Figure 3C). In keeping with these observations, NLRP3 could be activated by cGAMP in primary monocytes leading to secretion of both IL-1β (Figure 3D) and IL-18 (Figure 3E). To perform further mechanistic studies on this signaling cascade, we made use of the possibility to directly active murine STING (mm-STING) by small molecules such as 10-carboxymethyl-9-acridanone (CMA) (Cavlar et al., 2013). To this effect, BLaER1 monocytes transgenic for mm-STING showed inflammasome activation, as documented by IL-1β secretion and pyroptosome formation (Figures S3A and S3B). This response required both NLRP3 and the mm-STING transgene, whereas the type I IFN response following CMA only relied on mm-STING (Figure S3A). To further dissect type I IFN signaling and inflammasome activation downstream of STING, the role of the adaptor kinases TBK1 and IKKε was investigated. Either one of the kinases was sufficient for type I IFN production (data not shown) and only TBK1−/− x IKBKE−/− BLaER1 monocytes failed to secrete IP-10 upon DNA stimulation. Opposing to this, TBK1 and IKKε were dispensable for STING-mediated inflammasome activation (Figure 3F). Concordantly, the S366A hs-STING mutant that is defective in IRF3 recruitment (Liu et al., 2015) was able to reconstitute inflammasome activation, but not the type I IFN response (Figure 3G). In conclusion, both type I IFN induction and inflammasome activation upon cytosolic DNA sensing in human monocytes share the most upstream signaling components cGAS and STING. Subsequently, the pathways diverge, as STING activates TBK1 and IKKε to drive type I IFN production and NLRP3 to trigger inflammasome activation.
Figure 3. cGAS/STING Signaling Activates NLRP3 Independently of Type I Interferon Induction.
(A and B) LPS-primed BLaER1 monocytes of indicated genotype were stimulated with lipofected DNA for 8 hr. Nigericin was added for the last 2 hr. (A) Cytokine release is depicted as mean + SEM from three experiments. (B) One immunoblot from two experiments is shown. ***p < 0.001, **p < 0.01; ns, not significant.
(C) LPS-primed BLaER1 monocytes were stimulated with 3 µg cGAMP for 16 hr. If indicated, nigericin was added for the last 2 hr of the experiment. Data are depicted as mean + SEM of 3–5 experiments.
(D and E) Pam3CSK4-primed primary human monocytes with or without MCC950 were stimulated with 3 µg cGAMP for 16 hr. Cytokine release from four donors (D) or seven donors (E) is depicted as mean + SEM. **p < 0.01, *p < 0.05; ns, not significant.
(F) LPS-primed BLaER1 monocytes were stimulated as in (A). Cytokine release is depicted as mean + SEM from three experiments.
(G) WT or the S366A mutant of hs-STING were stably expressed in STING−/− BLaER1 monocytes and indicated cells stimulated as in (A). Data are shown as mean + SEM of three independent experiments. **p < 0.01, *p < 0.05; ns, not significant.
See also Figure S3.
STING Activation Triggers a Unique Cell Death Program
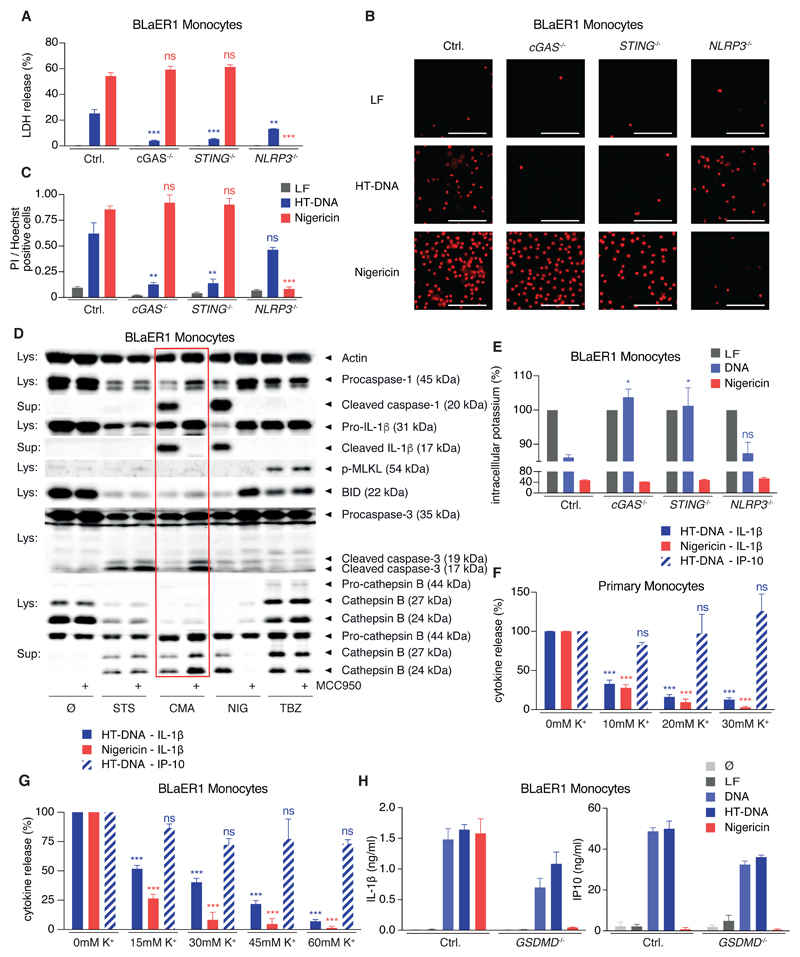
Besides the crucial role in IL-1β maturation, inflammasome activation is closely connected to pyroptotic cell death, which is executed by the caspase-1 substrate GSDMD (Kayagaki et al., 2015, Shi et al., 2015). Surprisingly, cell death after DNA stimulation in BLaER1 monocytes, as quantified by LDH release, was only partially dependent on NLRP3, while both cGAS and STING-deficient cells were largely protected (Figure 4A). Utilizing PI influx as a marker of cell death verified a strong NLRP3-independent but cGAS-/STING-dependent proportion of the cell death (Figures 4B and 4C). With the possibility at hand to directly drive STING-NLRP3 signaling by activating mm-STING with CMA, we set out to further characterize the STING-mediated cell death. The main advantages of this approach are to avoid the side effects of lipofection that is required to deliver DNA into the cytoplasm and the superior STING-agonistic activity of CMA. Under these conditions the NLRP3-independent cell death that was mediated by STING signaling was confirmed by LDH release and PI positivity (Figure S3C). We went on to further characterize CMA-mm-STING-mediated cell death in comparison to apoptosis (induced by staurosporine [STS]), pyroptosis (induced by nigericin [NIG]), and necroptosis (induced by the combination of TNF, birinapant, Z-VAD-FMK [TBZ]). These studies revealed that only CMA/STING- and nigericin-triggered cell death led to NLRP3 inflammasome activation and pyroptosis, as characterized by secretion of mature caspase-1 and IL-1β (Figure 4D). In this setting, pharmacological inhibition confirmed the involvement of NLRP3. Furthermore, the STING-triggered cell death did not bear necroptotic features as the hallmark of necroptosis, phosphorylation of MLKL, could not be detected. However, STING-mediated cell death displayed apoptotic features such as the cleavage of BID or the activation of caspase-3. Importantly, in the context of STING activation, BID cleavage occurred independently of NLRP3 signaling, thereby excluding secondary pyroptosis as the driving force. On the contrary, in the course of nigericin-driven pyroptosis, BID cleavage was sensitive to NLRP3 inhibition and thus an event downstream of NLRP3 activity. Interestingly, the other apoptotic features observed in the context of STING-triggered cell death, such as caspase-3 cleavage and the release of Cathepsin B, could only be observed or were enhanced upon NLRP3 inhibition (Figures 4D and S3D). This suggests that the faster kinetics of secondary pyroptosis prevent this apoptotic-like signaling during STING-cell death. Despite the occurrence of several hallmarks of apoptosis during STING-cell death, two reasons argue against it being primarily executed by the apoptotic machinery. First, only STING-cell death, but not staurosporine-mediated apoptosis, led to NLRP3 activation. And second, pan-caspase inhibition by Z-VAD-FMK could not block the STING-cell death (Figure S3E), while readily inhibiting IL-1β secretion by blocking caspase-1.
Figure 4. STING Activation Leads to a Lytic Cell Death that Drives NLRP3 Activation by Inducing K+ Efflux.
(A–C) LPS-primed BLaER1 monocytes were stimulated with lipofected DNA for 8 hr or with nigericin (last 2 hr). Cell death was quantified by LDH-release or by microscopy for PI-positive cells. Data are depicted as mean + SEM from three experiments (A), representative micrographs from four experiments (B), or quantifications from four experiments as mean + SEM (C). ***p < 0.001, **p < 0.01; ns, not significant. Scale bars denote 100 µm.
(D) LPS-primed BLaER1 mm-STING monocytes were stimulated with staurosporine (STS), CMA, nigericin (NIG) or hsTNF, birinapant, Z-VAD-FMK (TBZ) with or without MCC950. One immunoblot of three experiments is shown.
(E) LPS-primed BLaER1 monocytes were stimulated as in (A) and intracellular K+ content quantified by ICP-MS. Data are shown as mean + SEM from three experiments. *p < 0.05.
(F) Pam3CSK4-primed primary monocytes were stimulated as indicated in presence of increasing concentrations of extracellular K+. Data are shown as mean + SEM of four donors.
(G) LPS-primed BLaER1 monocytes were stimulated as in (A) with increasing concentrations of extracellular K+. Cytokine release was quantified from three experiments and is depicted as mean + SEM.
(H) LPS-primed BLaER1 monocytes were stimulated as in (A). Data are depicted as mean + SEM of three experiments.
See also Figure S3.
In light of the fast kinetics of pyroptosis signaling upon STING activation, we hypothesized that an early event in the lytic STING-cell death drives NLRP3 activation and turned to the efflux of intracellular K+, the necessary and sufficient upstream signaling event in classical NLRP3 activation. Monitoring intracellular levels of K+ during DNA stimulation in BLaER1 monocytes showed a significant drop in intracellular K+ levels that was dependent on cGAS and STING (Figure 4E). Importantly, this K+ efflux was not dependent on NLRP3, suggesting that it occurs upstream of inflammasome activation. Furthermore, blocking K+ efflux prevented inflammasome activation upon DNA stimulation in primary monocytes (Figure 4F) and BLaER1 monocytes (Figure 4G) while leaving the STING-mediated type I IFN response intact, firmly establishing a causative connection between STING-cell death, K+ efflux, and NLRP3 activation. The upstream involvement of a lytic cell death might also explain why the IL-1β secretion upon DNA stimulation appeared independently of GSDMD (Figure 4H) in BLaER1 monocytes, although these cells failed to secrete IL-1β in response to the classical NLRP3 activator nigericin. In summary, STING signaling in human monocytes leads to a lytic cell death with apoptotic-like features, that leads to K+ efflux upstream of NLRP3 activation.
Reconstitution of STING-Dependent NLRP3 Activation in HEK293 Cells
To further dissect the mechanism of the STING-dependent NLRP3 response, we utilized HEK293(T) cells in which cGAS-STING signaling and the NLRP3 inflammasome can be reconstituted. Analogous to BLaER1 monocytes, cGAS-STING signaling led to a profound cell death upon reconstitution in HEK293T cells (Figure 5A), that was dependent on both cGAS activity (Figure 5A) and the STING transgene (data not shown). As in the monocyte system, the STING-mediated cell death was uncoupled from the type I IFN response, because the S366A variant of STING, incapable of IRF3 recruitment, was sufficient in cell death induction, but not IRF3 phosphorylation (Figure 5B). Importantly, also direct activation of mm-STING by CMA led to a dose-dependent cell death in HEK293T cells (Figure S4A) while inducing a type I IFN response (Figure S4B). NLRP3 inflammasome reconstitution assays utilizing NLRP3-ASC-HEK293-Flp-In T-REx cells (“NLRP3-ASC-HEK293”) revealed that both cGAS activation (Figures 5C and 5D) and CMA stimulation (Figures S4C and S4D) led to NLRP3-dependent pyroptosome formation as quantified by light microscopy. In this regard, pyroptosome formation appeared only upon expression of wild-type (WT) mm-cGAS but not the catalytically dead variant mm-cGAS E211A in NLRP3-ASC-HEK293 cells. Importantly, this response required presence of the STING transgene, proving specificity of the cGAS-STING-NLRP3 pathway, and was sensitive to pharmacological inhibition by MCC950, confirming physiological activation of NLRP3 (Figures 5C and 5D).
Figure 5. The cGAS-STING-NLRP3 Inflammasome Can Be Reconstituted in HEK293 Cells.
(A and B) Indicated HEK293T cells were transfected with mm-cGAS or the inactive E211A mutant. Cell viability was quantified by CTB assay and is depicted as mean + SEM of three experiments (A). One immunoblot of two is shown (B).
(C and D) Indicated HEK293 NLRP3 ASC cells were transfected with cGAS or the E211A mutant or stimulated as indicated with or without MCC950. Representative micrographs are shown (C) and were quantified from three experiments (D) and data are depicted as mean + SEM. **p < 0.001, **p < 0.01. Scale bars indicate 50 µm.
See also Figure S4.
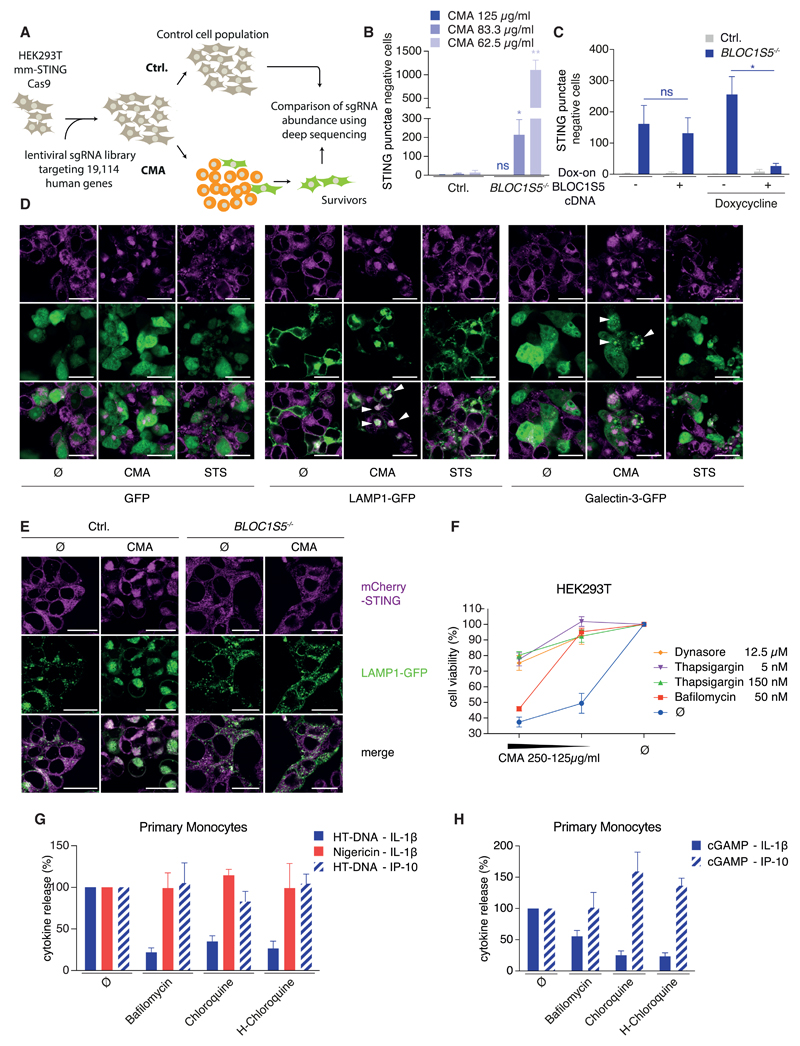
A Genetic Screen Identifies BLOC1 as a Positive Regulator of STING-Induced Cell Death
Taking advantage of the robust and quantitative STING-cell death in HEK293T cells (Figure S4E), a CRISPR/Cas9-based genetic perturbation screen was conducted to further characterize the cell death program (Figure 6A). One of the single guide RNAs (sgRNAs) intended to disrupt hs-STING also targeted the heterologously expressed mm-STING construct, which could be attributed to an off-target effect (Figure S5A). Underscoring the validity of our screening approach, this sgRNA showed the best enrichment over multiple screening replicates. Subsequently, a gene list of potential screening hits displaying 3–4 of 4 sgRNAs in the enriched fraction, were chosen for further validation. After two rounds of validation, several sgRNAs provided reproducible protection upon STING-cell death, targeting the genes CHIT1, LENEP, SMARCA4, BLOC1S5, SLC39A2, and KRTAP2-4. Of note, at high CMA concentrations, the protective effect of these gene disruptions was not visible (see STAR Methods). Of these, we chose BLOC1S5 for further analysis, because cells transduced with its sgRNA were strongly protected (Figure S5B). BLOC1S5 (biogenesis of lysosomal organelles complex 1 subunit 5) encodes a subunit of the BLOC1, a complex that is involved in transport of some but not all cargo to the lysosome and lysosome-related organelles (Setty et al., 2007), a role that might be explained by it being part of an endosomal Rab-GAP (GTPase-activating protein) adaptor complex in yeast (John Peter et al., 2013). Further analysis revealed that two other sgRNAs targeting BLOC1S1 and BLOC1S4, two other subunits of BLOC1, also provided protection against STING-cell death (Figure S5C). Furthermore, thorough analysis of single-cell-derived and sequenced BLOC1S5 knockout clones confirmed a crucial role for BLOC1S5 in STING-mediated cell death upon stimulation with low concentrations of CMA (Figure 6B). Reconstituting respective knockout cells with doxycycline-inducible BLOC1S5 cDNA validated the role of BLOC1S5 in STING-mediated cell death induction (Figure 6C). Of note, despite considerable leakiness of the dox-on system (Figure S5D), only the dox-mediated induction of BLOC1S5 cDNA abolished the survival phenotype of the respective knockout cells (Figure 6C). Collectively, our genetic screen identified BLOC1, a regulator of lysosomal trafficking, as a modulator of STING-cell death, whose deficiency can protect HEK293T cells from STING-cell death upon low concentrations of CMA.
Figure 6. A Genome-wide sgRNA-Screen Identifies BLOC1 as a Modulator of STING-Mediated Cell Death Guiding Its Characterization as Lysosomal Cell Death.
(A) A schematic overview of the screening procedure is shown.
(B) HEK293T mm-STING cells of indicated genotype were stimulated with CMA for 16 hr. STING punctae negative cells per visual field from four experiments are depicted as mean + SEM showing one representative clone of four.
(C) HEK293T mm-STING cells of indicated genotype that were transduced with a dox-on BLOC1S5 cDNA or an empty vector were stimulated with doxycycline for 24 hr and subsequently with 83.3 µg/mL CMA for 16 hr. STING punctae negative cells from three experiment are depicted as mean + SEM. **p < 0.01, *p < 0.05; ns, not significant.
(D) HEK293T mm-STING cells were transfected with GFP, LAMP1-GFP, or Galectin-3-GFP expression constructs. 24 hr later, cells were stimulated with CMA or staurosporine (STS) for 8 hr. Confocal micrographs from one experiment out of three are depicted. Scale bars indicate 25 µm.
(E) HEK293T mm-STING cells of indicated genotype that were transduced with a LAMP1-GFP reporter were stimulated with 62.5 µg/mL CMA for 16 hr. Representative confocal micrographs from one experiment of two are depicted. Scale bars indicate 25 µm.
(F) HEK293T mm-STING cells were stimulated with indicated inhibitors and CMA for 16 hr. CTB assay and is depicted as mean + SEM of three experiments.
(G and H) Pam3CSK4-primed primary monocytes with or without indicated inhibitors were stimulated with HT-DNA for 8 hr (G) or with 3 µg cGAMP (H) for 16 hr. Nigericin was added for the last 2 hr. Normalized cytokine release from 4–7 donors is depicted as mean + SEM.
See also Figures S5 and S6.
STING Induces Lysosomal Cell Death
Given the well-documented role for BLOC1 in the transport of selected cargo to lysosomes and lysosome-related organelles and its crucial role in maintaining endo-lysosomal flux in yeast, the relationship between STING-signaling and lysosomes was investigated. To this end, confocal microscopy revealed that STING trafficked to LAMP1 positive structures following activation, presumably lysosomes (Figure 6D). Lysosomes have been associated with a programed form of cell death known as “lysosomal cell death” (LCD), in which rupture of lysosomes or leakage of lysosomal content into the cytosol leads to cell death. To explore the involvement of LCD downstream of STING, we monitored the rupture of lysosomes following STING activation making use of a galectin-3-GFP reporter. While evenly distributed within the cytosol under steady-state conditions, galectin-3 is recruited to sites of membrane damage (e.g., lysosomal membranes) by binding to polysaccharides of the outer leaflet of the membranes that are not accessible from the cytosol under normal conditions. Indeed, several hours after STING activation, galectin-3 was recruited to punctate structures within the cytosol (Figure 6D). Notably, this was specific for STING-triggered cell death and did not occur in the course of staurosporine-mediated apoptosis. Consistently with the function of BLOC1S5 in transport of selected cargo along the endo-lysosomal route, BLOC1S5-deficient HEK293T cells failed to transport STING to LAMP1-positive structures (Figure 6E). It is important to stress, however, that the operative window for BLOC1S5-dependent STING trafficking is narrow, being limited by signaling strength. As such, stronger activation of STING by higher concentrations of CMA led to its transport to the lysosome in BLOC1S5−/− HEK293T cells (data not shown) and consequently to cell death (Figure 6B). Apart from that BLOC1S5-dependent STING trafficking also appears to be restricted by cell-type specificities, because BLOC1S5−/− BLaER1 monocytes did not display perturbed STING-mediated inflammasome activation (Figure S5E).
In line with the notion of STING-mediated lysosomal damage, pharmacological inhibition of lysosome function by bafilomycin or endo-lysosomal flux by thapsigargin or dynasore (see STAR Methods) potently protected HEK293T cells from STING-cell death (Figure 6F). Additionally, we observed that all three inhibitors prevented STING trafficking into one large complex (Figures S5F and S5G) that had also stained for LAMP1 in previous experiments (Figures 6D and 6E), inferring that the inhibitors prevent cell death by disrupting the lysosomal localization of STING. Importantly, all three inhibitors did not block STING signaling per se, as phosphorylation of TBK1, the immediate downstream event in type I IFN signaling, was largely intact under these conditions (Figure S5H). Translating these findings into STING-mediated inflammasome activation, pharmacological inhibition of LCD by bafilomycin, chloroquine, and hydroxy-chloroquine potently and specifically prevented DNA- and cGAMP-mediated IL-1β secretion from human monocytes (Figures 6G and 6H) or BLaER1 monocytes (Figure S5I), while both IP-10 production as well as direct activation of NLRP3 by nigericin remained intact. Moreover, titration of cGAMP or a stabilized version thereof (thio-cGAMP) on human monocytes could uncouple the signaling strength of the antiviral response (IP-10) and inflammasome activation (IL-1β), as low concentrations of cGAMP induced IP-10 in the absence or with lower amounts of IL-1β (Figure S6A). This additionally confirmed that STING can serve two different signaling purposes in human monocytes and furthermore suggested the existence of a signaling threshold, which STING activation has to pass to trigger LCD-NLRP3 activation.
In light of the fact that several cell types other than human monocytes do not succumb to STING-cell death, we wondered at which step they escape the STING-LCD pathway. To this end, we confirmed that e.g., murine macrophages and mouse embryonic fibroblasts (MEFs) did not die upon engagement of the cGAS-STING pathway or by direct STING stimulation (Figure S6B), although STING was still recruited to the lysosome in MEFs (Figure S6C). Because STING has been shown to be degraded upon its activation (Konno et al., 2013) we turned our attention to STING degradation. Interestingly, only cells that did not succumb upon STING activation (MEFs and murine macrophages) were able to degrade STING (Figures S6D–S6F), whereas HEK293T or BLaER1 monocytes cells were unable to do so (Figures S6G and S6H) and consequently died following STING activation. Because surviving cell types degraded STING in a bafilomycin-sensitive manner (Figures S6D–S6F), this likely represents lysosomal degradation. Altogether, these observations support a model in which both signaling strength (Figure S6A) and the cell-type-specific differences in lysosomal constitution dictate if STING can be degraded or whether it induces LCD and subsequent inflammasome activation.
The Role of the STING-NLRP3 Inflammasome Axis in Infection and Sterile Inflammation
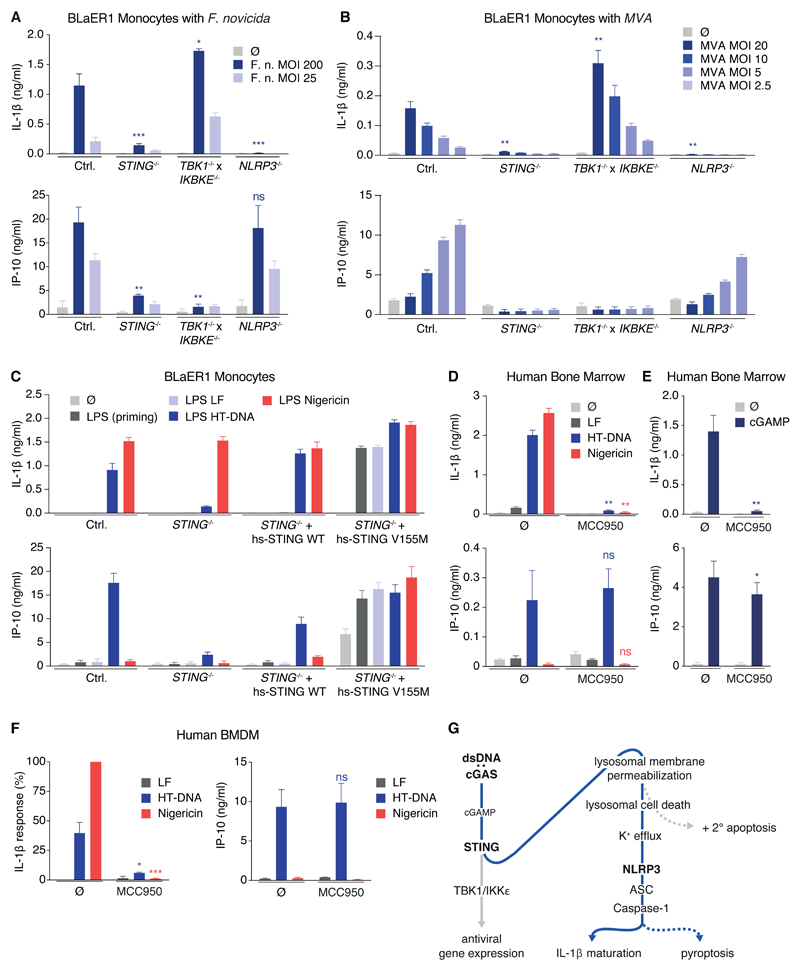
Given the well-documented role of cytosolic DNA sensing in various infection models and sterile inflammatory conditions, we hypothesized that this newly characterized signaling route of DNA-cGAS-STING-LCD-K+ efflux-NLRP3 activation should be responsible for the inflammasome activation in human monocytes for conditions that had previously been associated with AIM2 activation in murine macrophages. To this end, we studied Francisella tularensis subsp. novicida strain U112, a bacterial pathogen that is well known for AIM2-dependent inflammasome activation in mice in vitro and in vivo (Fernandes-Alnemri et al., 2010, Jones et al., 2010). In BLaER1 monocytes, the IL-1β secretion required cytosolic access of bacteria (Figure S7A) and was completely dependent on STING and NLRP3 (Figure 7A). Importantly, the intact IL-1β secretion from TBK1 x IKBKE-deficient BLaER1 monocytes indicated that STING was required to directly activate NLRP3 instead of providing cytosolic access of bacteria and bacterial DNA via induction of GBPs (Man et al., 2015a, Meunier et al., 2015). Importantly, the CASP4 x TRIF deficient genetic background does not exclude other signaling pathways, such as the non-canonical or alternative pathway, to be important for inflammasome activation in response to F. novicida. In analogy to the F. novicida infection experiments, Vaccinia Virus strain Modified Vaccinia Ankara (MVA), a pathogen well known to trigger AIM2-inflammasome activation in mice (Rathinam et al., 2010), triggered IL-1β secretion in a STING-NLRP3-dependent fashion in human monocytes (Figures 7B and S7B). Last, the SAVI (STING-associated vasculopathy with onset in infancy)-associated V155M gain of function variant of STING) (Liu et al., 2014) only required signal 1 inflammasome priming for secretion of IL-1β (Figure 7C) suggesting an involvement of the STING-NLRP3 pathway in sterile inflammatory conditions.
Figure 7. The cGAS-STING-NLRP3 Inflammasome Is Involved in Bacterial and Viral Sensing and Constitutes the Main DNA-Sensing Inflammasome in Human Myeloid Cells.
(A and B) LPS-primed BLaER1 monocytes were stimulated with different MOIs of the respective strain of F. novicida (A) or with different MOIs of MVA (B) for 8 hr. Data are depicted as mean + SEM of three experiments.
(C) Indicated LPS-primed BLaER1 monocytes were stimulated for 8 hr. Data are depicted as mean + SEM of three independent experiments.
(D and E) Mononuclear cells from human bone marrow were primed with Pam3CSK4 for 2 hr and stimulated with lipofected DNA or nigericin for 8 hr or with 3 µg cGAMP for 16 hr in absence or presence of MCC950. Cytokine secretion from three (D) or six (E) donors is depicted as mean + SEM. **p < 0.01, *p < 0.05.
(F) Human BMDM were primed with Pam3CSK4 for 2 hr and stimulated with lipofected DNA for 8 hr in absence or presence of MCC950. Nigericin was added for the last 2 hr. Cytokine secretion from 4 donors is depicted as mean + SEM. IL-1β responses were normalized to nigericin-mediated IL-1β secretion.
(G) A schematic view of the cGAS-STING-LCD-NLRP3 signaling cascade is shown. See also Figure S7.
In order to extend the relevance of the STING-NLRP3 pathway from human peripheral blood monocytes to other myeloid cells, we studied primary human bone marrow obtained from biopsies or surgery (Figure S7C). Containing myeloid cells at various stages of differentiation and representing the precursor cell pool of the adult myeloid lineage, we consider this cell population the most informative ex vivo system to study human myeloid cells. We observed that mononuclear cells from human bone marrow responded to lipofected DNA and cGAMP with NLRP3-dependent IL-1β secretion (Figures 7D and 7E). To further extend the relevance of the STING-NLRP3 pathway as the default DNA-inflammasome in human macrophages, we sorted human macrophages (CD11b+, CD14+, C163+) from in vitro differentiated hematopoietic stem cells. Studying these cells confirmed that the STING-NLRP3 pathway, instead of the AIM2 inflammasome, was triggered upon cytosolic DNA sensing in human macrophages (Figures 7F and S7E). Collectively, these data suggest that in human myeloid cells, STING-mediated inflammasome activation is at play in the course of viral and bacterial infections that activate the AIM2 inflammasome in murine macrophages. Furthermore, the STING-NLRP3 pathway is also active in sterile inflammatory conditions that are driven by STING gain of function variants.
Discussion
Here, we show NLRP3 functionally replaces AIM2 as the bona-fide DNA inflammasome sensor in human myeloid cells. As opposed to AIM2, which directly binds to DNA, NLRP3 is hardwired to the upstream DNA-sensing cGAS-STING pathway to trigger inflammasome activation. Interestingly, the molecular mechanism of STING-mediated NLRP3 activation turned out to be distinct from its canonical signaling cascade that engages TBK1/IKKε to induce antiviral gene expression. In fact, STING translocated to lysosomes upon activation, which resulted in their disruption and thereby induced a lytic form of cell death, also known as lysosomal cell death (Aits and Jäättelä, 2013). The subsequent drop in cytoplasmic K+ then triggered the classical mode of NLRP3 activation (Figure 7G). Importantly, in human myeloid cells, the cGAS-STING-NLRP3 axis turned out to be the default mode of inflammasome activation in response to prokaryotic and viral pathogens that are known to exclusively engage the AIM2 inflammasome activation in the murine system (Man et al., 2016). At the same time, a gain of function mutant within STING that is associated with a severe sterile auto-inflammatory condition resulted in STING-dependent, lysosomal cell death with subsequent NLRP3 activation.
The Myeloid Human DNA-Inflammasome Does Not Depend on AIM2
AIM2 is the only member of the PYHIN gene family that is truly orthologous between the murine and human species (Cridland et al., 2012), and gain- and loss-of-function studies in cell lines have implied that human AIM2 operates as its murine counterpart (Man et al., 2016). As such, it has been generally accepted that studies on AIM2 in the murine system can be extrapolated to the human situation. Our data, however, show that AIM2 is functionally replaced by cGAS-STING-NLRP3 during DNA inflammasome activation in primary human myeloid cells. Although cGAS-STING-NLRP3 constitutes the default inflammasome signaling circuit in human myeloid cells, it is not the only human DNA-inflammasome in sensu stricto, as human myeloid-like THP-1 cells displayed an AIM2-dependent inflammasome response. As this point, it cannot be excluded that certain differentiation- or tissue-dependent cues that are not present in peripheral blood, bone marrow cells, or bone marrow-derived macrophages enable AIM2 responsiveness in the human system in vivo. However, with regards to the fact that the cGAS-STING-NLRP3 axis constitutes the default DNA-inflammasome signaling circuit in inflammasome-competent human bone marrow cells, we consider it unlikely that these cells lose cGAS-STING-NLRP3 signaling competence, while gaining AIM2 signaling competence upon migration into the periphery and differentiation along myeloid lineages. Of note, unlike the cGAS-STING axis, which evolved early in the metazoan lineage (Margolis et al., 2017), AIM2 is a much more recent development, in that it is only present in the mammalian class (Cridland et al., 2012). In addition, various mammalian lineages have independently lost AIM2 or encode for pseudogenes (e.g., Cetartiodactyla), despite the presence of core components of the inflammasome system. As such, it appears likely that the cGAS-STING-NLRP3 axis is the default DNA sensing inflammasome in other mammalian species as well. Given the pivotal relevance of human myeloid cells, especially macrophages, for infectious and sterile inflammatory diseases, we conclude that AIM2 plays a subordinate role in the DNA-inflammasome response of the human system. Indeed, it is possible that the main role of AIM2 in the human system is ascribable to its previously reported inflammasome-independent function in tumorigenesis (Man et al., 2015b, Wilson et al., 2015).
The cGAS-STING-LCD Axis Functions as the Extended Arm of the DNA-NLRP3 Inflammasome
Interrogating the modalities of the cGAS-STING-NLRP3 axis, we observed that STING-triggered LCD operates independently, and thus upstream, of NLRP3-dependent inflammasome activation. This type of cell death has previously been acknowledged as a unique signaling entity, in which leakage of lysosomal content functions as the cell death initiation module. It is well-documented that leakage of lysosomally sequestered hydrolases into the cytoplasm (e.g., cathepsins) results in the degradation of cytoplasmic proteins and consequently cell death (Aits and Jäättelä, 2013). While LCD has been shown to display apoptosis-like features, there seem to be additional cell death executioner modules that operate in a redundant manner to apoptosis. To this end, it also remains unresolved how LCD results in plasma membrane permeabilization. However, it is conceivable that specific substrates exist, whose cleavage perturbs the plasma membrane (e.g., membrane scaffolding proteins) (Ivanova et al., 2011). Consequently, plasma membrane permeability results in K+ efflux and thereby engages the NLRP3 inflammasome. In keeping with the faster kinetics of inflammasome signaling (Shi et al., 2015), NLRP3-dependent caspase-1 activation supersedes the initiation of apoptosis, rendering LCD an inflammatory cell death in myeloid cells. Indeed, the hallmarks of apoptosis only became apparent in STING-activated cells when NLRP3 activation was blocked. In line with the fact that LCD triggered a lytic form of cell death already upstream of inflammasome activation, GSDMD turned out to be dispensable for DNA-inflammasome-dependent IL-1β release, while it was fully required for nigericin-mediated IL-1β secretion (Kayagaki et al., 2015, Shi et al., 2015). In this regard, the mechanism of STING-LCD-dependent NLRP3 activation is reminiscent of non-canonical inflammasome activation, in which a lytic cell death (GSDMD-dependent pore formation) functions upstream of K+ efflux-dependent, cell-autonomous NLRP3 inflammasome activation (Broz and Dixit, 2016). At the same time, STING-mediated NLRP3 activation also bears similarities to crystal- and particulate material-mediated NLRP3 activation, in which lysosomal rupture in the context of phagocytosis induces K+ efflux driving NLRP3 activation (Hornung et al., 2008). However, as LCD downstream of STING constitutes a programmed form of cell death, to which the cell “voluntarily” commits, we consider the comparison to non-canonical inflammasome activation more suitable and conclude that in both circumstances recognition by an unrelated sensing module (cGAS-STING or caspase-4/-11) induces lytic cell death upstream of NLRP3. Altogether, the convergence of two innate sensing modules cGAS-STING axis and NLRP3 inflammasome in primary human myeloid cells dictates the inflammasome response to DNA in these cells.
Implications for Disease and Therapy
From a therapeutic perspective, these findings put the cGAS-STING axis even more in the focus of attention. Sterile inflammatory conditions, in which cytosolic DNA sensing is at play would benefit from STING inhibition not only to block antiviral gene expression, but also NLRP3-dependent inflammasome activation or cell death-associated inflammation. Conversely, agonists aimed at activating STING also trigger cell death and associated inflammasome activation within the human myeloid compartment, which should be taken into account when developing these compounds for immunotherapeutic applications.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by Veit Hornung.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AIM2 (D5X7K) Rabbit mAb | Cell Signaling Technology | Cat#12948 |
| anti-Caspase-1 (p20) (human), mAb (Bally-1) | Adipogen International | Cat#AG-20B-0048-C100 |
| anti-Caspase-1 (p20) (mouse), mAb (Casper-1) | Adipogen International | Cat#AG-20B-0042-C100 |
| Anti-CD10 (ALB1) | Beckman Coulter | Cat#IM3633 |
| Anti-CD14 (RMO52) | Beckman Coulter | Cat#PNIM2707U |
| Anti-CD19 (J3-119) | Beckman Coulter | Cat#A94681 |
| Anti-CD2 (39C1.5) | Beckman Coulter | Cat#A60794 |
| Anti-CD3 (UCHT1) | Beckman Coulter | Cat#A07726 |
| Anti-CD33 (D3HL60.251) | Beckman Coulter | Cat#A54824 |
| Anti-CD34 (581) | Beckman Coulter | Cat#IMI870 |
| Anti-CD4 (RPA-T4) | Beckman Coulter | Cat#A07726 |
| Anti-CD45 (J33) | Beckman Coulter | Cat#B36294 |
| Anti-CD64 (22) | Beckman Coulter | Cat#IM3601U |
| Anti-CD65 (88H7) | Beckman Coulter | Cat#PNB08164 |
| Anti-CD8 (B9.11) | Beckman Coulter | Cat#A07726 |
| Anti-MLKL (phospho S358) antibody | Abcam | Cat#ab187091 |
| Anti-Viperin Antibody, clone MaP.VIP | Merck Chemicals GmbH | Cat#MABF106 |
| Brilliant Violet 421 anti-human CD163 Antibody | BioLegend | Cat#333611 |
| Caspase-3 Antibody | Cell Signaling Technology | Cat#9662 |
| Caspase-8 (human) monoclonal antibody (12F5) | Enzo Life Sciences | Cat#ALX-804-242-C100 |
| Cathepsin B (D1C7Y) XP® Rabbit mAb | Cell Signaling Technology | Cat#31718 |
| CD11b Monoclonal Antibody (ICRF44), FITC, eBioscience | Thermo Fisher Scientific | Cat#11-0118-42 |
| donkey anti-goat IgG-HRP | Santa Cruz Biotechnology | Cat#sc-2020 |
| F(ab’)2-Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fisher Scientific | Cat#A-11019 |
| FITC anti-human CD68 Antibody | BioLegend | Cat#333806 |
| goat anti-mouse IgG-HRP | Santa Cruz Biotechnology | Cat#sc-2005 |
| goat anti-rabbit IgG-HRP | Santa Cruz Biotechnology | Cat#sc-2004 |
| Human IL-1 beta /IL-1F2 Antibody | R&D Systems | Cat#AF-201-NA |
| Human/Mouse BID Antibody | R&D Systems | Cat#AF860 |
| IRF-3 (D83B9) Rabbit mAb | Cell Signaling Technology | Cat#4302 |
| Mouse IL-1 beta /IL-1F2 Antibody | R&D Systems | Cat#AF-401-NA |
| MUTED Antibody | Proteintech Group | Cat#24015-1-AP |
| PE/Cy7 anti-human CD14 Antibody | BioLegend | Cat#301813 |
| Phospho-IRF-3 (Ser396) (4D4G) Rabbit mAb | Cell Signaling Technology | Cat#4947 |
| Phospho-TBK1/NAK (Ser172) (D52C2) XP® Rabbit mAb | Cell Signaling Technology | Cat#5483 |
| Purified anti-ASC (TMS-1) Antibody | BioLegend | Cat#653902 |
| STING (D2P2F) Rabbit mAb | Cell Signaling Technology | Cat#13647 |
| TBK1/NAK Antibody | Cell Signaling Technology | Cat#3013 |
| β-Actin Antikörper (C4) | Santa Cruz Biotechnology | Cat#sc-47778 |
| Bacterial and Virus Strains | ||
| Francisella tularensis subsp. novicida strain U112 | (Meunier et al., 2015) | N/A |
| Francisella tularensis subsp. novicida strain U112 deltaFPI | (Meunier et al., 2015) | N/A |
| MVA | (Kastenmüller et al., 2012) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 2′3′-cGAM(PS)2 (Rp/Sp) | InvivoGen | Cat#tlrl-nacga2srs |
| 2′3′-cGAMP | InvivoGen | Cat#tlrl-nacga23-1 |
| 9-Oxo-10(9H)-acridineacetic acid (CMA) | Sigma-Aldrich | Cat#17927-250MG |
| Bafilomycin A1 from Streptomyces griseus | Sigma-Aldrich | Cat#B1793-10UG |
| Birinapant | BioVision | Cat#2597-1 |
| Chloroquine diphosphate salt | Sigma-Aldrich | Cat#C6628-25G |
| CP-456773 sodium salt (MCC950) | Sigma-Aldrich | Cat#PZ0280-25MG |
| Deoxyribonucleic acid sodium salt from herring testes | Sigma-Aldrich | Cat#D6898-250MG |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D9891-1G |
| Dynasore hydrate | Sigma-Aldrich | Cat#D7693-5MG |
| GeneJuice® Transfection Reagent | Merck Chemicals GmbH | Cat#70967 |
| Hoechst 34580 | Thermo Fisher Scientific | Cat#H21486 |
| Hydroxychloroquine sulfate | Sigma-Aldrich | Cat#H0915-5MG |
| Lipofectamine 2000 Transfection Reagent | Thermo Fisher Scientific | Cat#11668019 |
| LPS-EB Ultrapure | InvivoGen | Cat#tlrl-3pelps |
| Nigericin sodium salt | Sigma-Aldrich | Cat#N7143-10MG |
| Pam3CSK4 | InvivoGen | Cat#tlrl-pms |
| Propidium Iodide | Thermo Fisher Scientific | Cat#P1304MP |
| Recombinant Human IFN-γ | PeproTech | Cat#300-02 |
| Recombinant Human IL-3 | PeproTech | Cat#200-03 |
| Recombinant Human M-CSF | PeproTech | Cat#300-25 |
| Recombinant Human TNF-α | PeproTech | Cat#300-01A |
| Resazurin sodium salt | Sigma-Aldrich | Cat#R7017-5G |
| Staurosporine (CAS 62996-74-1) | Santa Cruz Biotechnology | Cat#sc-3510A |
| Thapsigargin | Sigma-Aldrich | Cat#T9033-.5MG |
| Z-Val-Ala-Asp(OMe)-CH2F [Z-VAD-FMK] | PEPTIDE INSTITUTE | Cat#3188-v |
| β-Estradiol | Sigma-Aldrich | Cat#E8875-250MG |
| Critical Commercial Assays | ||
| CD14 MicroBeads, human | Miltenyi Biotec | Cat#130-050-201 |
| Human IL-1β ELISA Set II | BD Biosciences | Cat#557953 |
| Human Total IL-18 DuoSet ELISA | R&D Systems | Cat#DY318-05 |
| Mouse CXCL10/IP-10/CRG-2 DuoSet ELISA | R&D Systems | Cat#DY466 |
| CD34 MicroBead Kit UltraPure, human | Miltenyi Biotec | Cat#130-100-453 |
| Human IL1 beta Kit (HTRF) | Cisbio | Cat#62IL1PEC |
| Human IP-10 ELISA Set | BD Biosciences | Cat#550926 |
| Pierce LDH Cytotoxicity Assay Kit | Thermo Fisher Scientific | Cat#88954 |
| Mouse IL-1 β ELISA Set | BD Biosciences | Cat#559603 |
| MiSeq Reagent Kit v2, 300 Cycles | Illumina | Cat#MS-102-2002 |
| Experimental Models: Cell Lines | ||
| BLaER1 human b-cell to monocyte trans-differentiation cell line | (Rapino et al., 2013) | N/A |
| THP-1 | (Schmid-Burgk et al., 2015) | N/A |
| HEK293T-mmSTING | (Ablasser et al., 2013) | N/A |
| Immortalized murine macrophages | (Hornung et al., 2008) | N/A |
| MEF | (Grimm et al., 2014) | N/A |
| Flp-In T-REx 293 Cell Line | Thermo Fisher Scientific | Cat#R78007 |
| Experimental Models: Organisms/Strains | ||
| Nlrp3−/− mice | (Mariathasan et al., 2006) | N/A |
| Asc−/− mice | (Mariathasan et al., 2004) | N/A |
| Casp1−/− x Casp11−/− mice | (Kuida et al., 1995) | N/A |
| Aim2−/− mice | (Jakobs et al., 2015) | N/A |
| Recombinant DNA | ||
| pLenti(p)-EF1-Flag-mm-cGAS | (Gao et al., 2013) | N/A |
| pLenti(p)-EF1-Flag-mm-cGAS E211A | (Gao et al., 2013) | N/A |
| pEGFP-hGal3 | (Maejima et al., 2013) | Addgene plasmid # 73080 |
| LAMP1-mGFP | (Falcón-Pérez et al., 2005) | Addgene plasmid # 34831 |
| pRP mmSTING mCitrine | This study | N/A |
| pRP mmSTING | This study | N/A |
| pLIX mmSTING mCherry | This study | N/A |
| pRP LAMP1 GFP | This study | N/A |
| pcDNA5/FRT CMV-tetO2-NLRP3-tagRFP PGK-ASC-mTurq-Flag | This study | N/A |
| pRP mmSTING mCitrine S366A | This study | N/A |
| pRP mmSTING mCitrine V155M | This study | N/A |
| CMV-mCherry-Cas9 | (Schmid-Burgk et al., 2014) | N/A |
| pLK0.1-gRNA-CMV-GFP | (Schmid-Burgk et al., 2014) | N/A |
| pLIX BLOC1S5 | This study | |
| Software and Algorithms | ||
| Outknocker | (Schmid-Burgk et al., 2014) | N/A |
| GraphPad Prism 6 | GraphPad | N/A |
| Cell Profiler | (Carpenter et al., 2006) | N/A |
| Other | ||
| human Brunello CRISPR knockout pooled library | (Doench et al., 2016) | Addgene #73178 |
Experimental Model and Subject Details
Cell culture
Primary monocytes, human bone marrow cells, BLaER1 and THP-1 cells were cultivated in RPMI Medium 1640 supplemented with L-glutamine, sodium pyruvate, 10% (v/v) FCS and 100 U/mL Penicillin-Streptomycin (all Life Technologies). HEK293(T) cells, murine BMDMs and immortalized murine macrophages were cultivated in DMEM Medium (Life Technologies) containing the same supplements, MEFs were cultivated in DMEM Medium with the same supplements and 0.1 mM 2-mercaptoethanol (Sigma-Aldrich). Primary cells were allowed to recover for several hours from the isolation process before stimulation. BLaER1 cells were trans-differentiated into monocytes for 6-7 days in medium containing 10 ng/mL of hrIL-3, 10 ng/mL hr-CSF-1 (M-CSF) (both PeproTech) and 100 nM β-Estradiol (Sigma-Aldrich). To study the mechanisms of DNA-dependent inflammasome signaling in this model system, we made use of BLaER1 monocytes in which TRIF and CASP4 had been disrupted. TRIF was deleted to avoid LPS-dependent alternative inflammasome activation (use of LPS to prime pro-IL-1β expression), whereas caspase-4 was disrupted to avoid non-canonical inflammasome activation (use of transfection reagents) (Gaidt et al., 2016). For reasons of simplicity the respective CASP4−/− x TRIF−/−genetic background will be referred to as ‘Ctrl.’ throughout this manuscript. THP-1 cells were differentiated over night with 100 ng/mL PMA (Sigma-Aldrich), washed three times with ice-cold PBS and replated.
Isolation of human primary cells
PBMCs were obtained from healthy, informed, and consenting volunteers and monocytes were isolated using CD14 MACS microbeads (Miltenyi Biotec) according to the provider’s protocol. Bone marrow samples were obtained from patients during hip surgery (Technical University Munich) or from lymphoma patients without infiltration of malignant cells into the bone marrow as confirmed by the Laboratory for Leukemia Diagnostics, Department of Internal Medicine III, Klinikum of the LMU Munich. Informed consent was obtained from all subjects according to the Declaration of Helsinki and approval by the Institutional Review Board of the Ludwig-Maximilians-University (Munich, Germany) and the Technical University Munich (Munich, Germany). Information about sex, gender, and age of respective donors were blinded.
Derivation of human bone marrow macrophages (BMDMs)
Hematopoietic stem cells were isolated by CD34+ MACS isolation (Miltenyi Biotec) from human bone marrow and differentiated into macrophages in IMDM Medium supplemented with L-glutamine, sodium pyruvate, 10% (v/v) FCS, 100 U/mL Penicillin-Streptomycin (all Life Technologies) and 50 ng/mL of hrIL-3, 50 ng/mL hr-CSF-1 (M-CSF) (both PeproTech). The medium was replaced every third day for 12 days, after which macrophages were sorted as CD11b+, CD14+, C163+ and 7 × 104 cells per 96-well were re-plated in RPMI Medium 1640 containing the same supplements without IL-3 and M-CSF. After recovery over night the medium was replaced and macrophages stimulated as indicated.
Derivation of murine bone marrow macrophages (BMDMs)
Bone marrow was isolated from the femur and tibia of 9 to 10-week-old C57BL/6 mice that were housed under SPF conditions. The gender of the individual mice was not recorded. Nlrp3−/− (Mariathasan et al., 2006) and Asc−/− (Mariathasan et al., 2004) mice were a gift from Vishva Dixit (Genentech), Casp1−/− x Casp11−/− mice (Kuida et al., 1995) were a gift from Richard A. Flavell (Yale University School of Medicine) and Aim2−/− mice (Jakobs et al., 2015) have been previously described. Bone marrow cells were filtered and subjected to erythrocyte lysis (BD Pharm Lyse). For the derivation of macrophages, the bone marrow cells were incubated with 30% L929 supernatant in DMEM for a total of seven days. For stimulation cells were re-plated.
Cell stimulation
If not otherwise indicated, primary monocytes, bone marrow cells and human BMDMs were primed with 2 µg/mL Pam3CSK4 (Invivogen) for 2 hr. 200 ng of pBlueScript Plasmid DNA, isolated from E. coli with a Maxi-prep-kit (Invitrogen) (“DNA”) or DNA from herring testes (“HT-DNA”) (Sigma-Aldrich) were complexed to Lipofectamine 2000 (LF) (Life Technologies) using the supplier’s protocol and cells were stimulated with the mix for 8 hr. 6.5 µM nigericin (Sigma-Aldrich) was added for the last 2 hr of the experiment. BLaER1 monocytes were primed with 200 ng/mL ultrapure E.coli LPS (Invivogen) for 2 hr and stimulated with 400 ng of indicated DNA. THP-1 cells were primed with 2 µg/mL Pam3CSK4 for 30 min and murine BMDM with 200 ng/mL LPS for 2 hr before being stimulated with 200 ng of DNA. If indicated, cells were stimulated after indicated priming steps with 3 µg cGAMP or indicated concentrations of 10-carboxymethyl-9-acridanone (CMA) (Sigma-Aldrich), 0.625 µM Staurosporine (BLaER1 monocytes), 2.5 µM Staurosporine (HEK293T), 100 ng/mL hsTNF (Peprotech), 5 µM Birinapant (BioCAT) and 20 µM) Z-VAD-FMK (Peptide Institute) (TBZ) for indicated period of times. For infections with Francisella tularensis subsp. novicida strain U112 (hereafter F. novicida), and the derivative strains, were grown overnight at 37°C with aeration in brain heart infusion supplemented with 100 µg/mL ampicillin (Sigma-Aldrich). Bacteria were harvested, washed with PBS and LPS-primed BLaER1 monocytes infected with indicated MOIs. The plates were centrifuged for 5 min at 400 g to ensure proper adhesion of the bacteria to the cells and cultivated for 2 hr. Subsequently, the medium was replaced with fresh medium containing 10 µg/mL gentamicin to kill extracellular bacteria and the plates incubated for another 6 hr. Vaccinia Virus strain Modified Vaccinia Ankara (MVA) infections were carried out with primed cells using the indicated MOIs. If not otherwise state, all experiments involving gene deficient monoclonal cell lines are depicted as one representative clone of two.
Inhibitors
If not otherwise indicated, small molecule inhibitors were added to the cells 1 hr before stimulation using the following concentrations: 40 µM MCC950 (HEK293 NLRP3 ASC cells), 5 µM MCC950 (all other cells), 12.5 µM Dynasore, 50nM Bafilomycin A1, 150 nM Thapsigargin, 50 µM Chloroquine, 50 µM Hydroxy-chloroquine (all Sigma-Aldrich), 20 µM Z-VAD-FMK (Peptide Institute). Of note, the inhibition of endo-lysosmal flux is not typically associated with Thapsigargin, commonly referred to as SERCA inhibitor, or Dynasore, commonly referred to as an inhibitor of dynamin-dependent endocytosis. However, it was recently noted, that Thapsigargin efficiently blocks RAB7-dependent trafficking to the lysosome independently of SERCA inhibition (Ganley et al., 2011) and that Dynasore also blocks endo-lysosomal flux (Mesaki et al., 2011). For blocking the efflux of intracellular cells were incubated in medium that was diluted with 135mM KCl (Roth) in sterile water containing 10% FCS to contain indicated concentrations of K+.
CRISPR/Cas9 mediated knockout-cell line generation
Gene deficient BLaER1, THP-1 and HEK293T cells were generated as follows. Briefly, sgRNAs (20 or 18-mer), specific for the indicated genes, were designed to target an early coding exon of the respective gene. BlaER1 or THP-1 cells were electroporated with pLK0.1-gRNA-CMV-GFP and CMV-mCherry-Cas9 expression plasmids using a Biorad GenePulser device and HEK293T cells were transiently transfected with similar plasmids using GeneJuice lipofection. FACSorted mCherry positive cells were cloned by limiting dilution. Monoclonal cell lines were identified, rearranged and duplicated for genotyping using deep sequencing (Illumina’s Miseq-platform). Knockout cell clones contained all-allelic frameshift mutations without any wild-type reads. Two independent knockout single-cell clones were analyzed per genotype, whereas always one representative clone per genotype is shown.
Quantification of pyroptosome formation
Pyroptosome formation in human primary monocytes was analyzed by immunostaining of ASC. To do so, cells were plated in poly-L-ornithin (Sigma-Aldrich) coated Ibidi 8 well µ-Slides (Ibidi) and stimulated as described. Cells were fixed, stained with anti-ASC antibody (10 µg/mL, Biolegend, TMS-1) and appropriate secondary antibody (2.5 µg/mL, Invitrogen, A-11019), stained with Hoechst 34580 (0.2 µg/mL, Invitrogen) and subsequently imaged using a Leica DMi8 inverted microscope. For analysis of pyroptosome formation in BLaER1 cells, a previously generated ASC-TagRFP reporter was used, in which endogenous ASC was tagged by TagRFP (Gaidt et al., 2016). For this, cells were plated per well in poly-L-ornithine coated Ibidi 15 µ-Plates (96 Well). After 4 days, differentiation medium was replaced with fresh medium containing 20 µM Z-YVAD-FMK (Peptide Institute). Indicated cells were primed with 5 µM MCC950 (Sigma-Aldrich) for two hr. After priming, nigericin was added to a final concentration of 6.5 µM and CMA to a final concentration of 0.1 mg/mL. Microscopic images were acquired at two hr and 16 hr after stimulation. For quantification purpose, > 100 cells per condition were manually analyzed to contain punctual or diffuse ASC localization. Ratios between cells and cells containing pyroptosomes were calculated.
Microscopy
If not otherwise indicated, all microscopic images were acquired using a Leica DMi8 inverted microscope equipped with one of the following objectives: H3 PL FLUOTAR 10x/ 0.32; HC PL FLUOTAR L 20x/ 0.40 DRY; HC PL FLUOTAR L 25x/ 0.80 IMM; HC PL APO 63x/ 1.20 W CORR CS2. If not otherwise indicated, micrographs were acquired by live cell imaging. The LAMP1-GFP and Galectin-3-GFP reporter assays were imaged with Zeiss 710 Confocal Microscope equipped with a C-Apochromat 40x/1.2 W Corr M27 objective using PFA-fixed cells.
Immunoblotting
Immunoblotting of precipitated supernatant was performed as follows: Briefly, methanol/chloroform precipitated cell supernatants and whole cell lysates were resuspended in 1x Laemmli buffer, denatured for 10 min at 95°C and separated by tris-glycine denaturing SDS-PAGE. Proteins were blotted onto 0.2 µM nitrocellulose membranes, blocked in 3% milk and incubated with indicated primary and corresponding secondary antibodies (Santa Cruz). Chemiluminescent signals were recorded with a CCD-camera and respective images contrast-enhanced in a linear fashion.
Kits
Pierce LDH Cytotoxicity Assay Kit (Life Technologies) was performed according to the instructions. Relative LDH release was calculated as LDH release [%] = 100 *(measurement – unstimulated control)/ (lysis control – unstimulated control). huIL-1β, huIP-10, mIL-1β ELISA (OptEIA, BD Biosciences) and mIP-10 ELISA (R&D Systems) were performed according to the provider’s protocol. The ELISA for quantification of huIL-1β preferentially recognizes cleaved IL-1β over pro-IL-1β. The IL-1β secretion from human BMDM was quantified using the HTRF assay (Cisbio) due to its superior miniaturization.
Potassium measurement
After stimulation 2 × 106 BLaER1 cells were lysed in water for 1h at room temperature and cells ruptured by freeze/thawing for six rounds. Cell debris was pelleted and K+ measured by ICP-MS (7700 Series, Agilent Technologies) and quantified against an initial calibration verification standard (Agilent Technologies).
Cell Titer Blue resazurin cell viability assay
Cell Titer Blue assay was performed using a home-made resazurin stock solution. For this, resazurin sodium salt (Sigma-Aldrich) was dissolved in sterile PBS to 0.5 mg/mL, filter-sterilized through a 0.2 µm filter and stored protected from light at −20°C. For cell viability assays the resazurin stock was added to the medium to a final concentration of 6.25 µg/mL and incubated for several hours at 37°C. Fluorescence (560Ex/590Em) was recorded using a TECAN Spark 20M microplate reader. Cell viability was calculated by subtraction of the signal recorded from triton-lysed cells and normalized to mock treated control. For inhibitor experiments values were normalized to wells treated with inhibitor without the respective stimulus. Quantification of the STING-cell death in HEK293T cells by measuring reduction of resazurin by viable cells, commonly showed 30%–40% of surviving cells (e.g., Figure 5A). However, this was a gross underestimation of the magnitude of cell death. Quantifying the fraction of cells that could give rise to colonies after CMA stimulation, we observed that the majority (> 97%) of HEK293T cells succumbed upon STING activation (Figure S4E).
Lenti/retroviral expression
Retroviral hs-STING or mm-STING expression construct with or without an N-terminal mCitrine-tag or mCherry-tag were generated by conventional restriction enzyme-based cloning based on previously published constructs (Cavlar et al., 2013). Hs-STING S366A and V155M variants were generated using a loop-out-PCR approach. BLOC1S5 was amplified from cDNA of HEK293T cells and cloned into a dox-on expression construct by conventional restriction enzyme-based cloning. The dox-on expression construct pLIX was a gift from Andreas Pichlmair, MPI of Biochemistry, Munich. LAMP1-GFP was subcloned into a retroviral expression vector by conventional restriction enzyme-based cloning. Respective HEK293(T) or BLaER1 cells of indicated genotype were transduced and selected if possible and sorted if necessary.
Quantification of cell death by PI influx
After BLaER1 monocytes were stimulated as indicated PI (Life Technologies) was added to the cell culture medium at a concentration of 1 µg/mL. Images were acquired by fluorescence microscopy and quantified using Cell Profiler software (Carpenter et al., 2006). Hoechst and PI channels were used to define total and dead cell numbers respectively using a global threshold enhancement, followed by a two-class Otsu algorithm. Data are presented as the ratio of PI positive cells over Hoechst positive cells.
Transient transfection of HEK293T cells
The day before transfection, HEK293T cells were plated at a density of 25 × 103 cells per well of a 96 well plate. The next day, 200ng of indicated plasmid DNA was lipofected using Genejuice (Merck Millipore) as suggested by the provider. Commonly, cells were analyzed 24h later, if not otherwise indicated. Mm-cGAS wt and E211A expression plasmids were a gift from Thomas Zillinger (Gao et al., 2013), pEGFP-hGal3 (Maejima et al., 2013) was a gift from Tamotsu Yoshimori (Addgene plasmid # 73080) and LAMP1-mGFP (Falcón-Pérez et al., 2005) was a gift from Esteban Dell’Angelica (Addgene plasmid # 34831).
Inflammasome reconstitution in HEK293 cells
A plasmid encoding for tetracyclin-inducible NLRP3-tagRFP, ASC-mTurquoise was generated with standard cloning techniques. Using this construct, a HEK293-FlpIn TREx cell line (Invitrogen) with a single insertion of the genes of interest was generated according to manufacturer’s instructions. Subsequently, mm-STING was expressed in these cells by retroviral transduction. Inflammsome activation upon cGAS expression, CMA or Nigericin stimulation was monitored by following formation of the ASC-mTurquoise pyroptosome.
Genetic perturbation screen for STING-mediated cell death
In order to limit the screening approach to STING-dependent cell death that does not require TBK1 signaling (Figure 3F), the genetic perturbation screen was performed in HEK293T mm-STING Cas9 TBK1−/− cells that showed similar cell death sensitivity and kinetics compared to the parental cell line (data not shown). For reasons of simplicity, these cells were called HEK293T mm-STING Cas9 cells throughout this manuscript. The human Brunello CRISPR knockout pooled library, a gift from David Root and John Doench (Addgene #73178) (Doench et al., 2016), was used to perform the genetic perturbation studies. The library targeting 19,114 human genes with 76,441 sgRNAs was amplified and equal distribution of library members verified by deep sequencing. Directly before the screen, HEK293T cells were cultured in medium containing 5 µg/mL puromycin (Sigma-Aldrich) to re-select for strong expression of the mm-STING transgene. For the genetic perturbation screen 10 × 106 HEK293T mm-STING Cas9 cells per technical replicate were infected at an MOI of 1. Since the sgRNA library also contained a puromycin resistance cassette, no selection could be applied for sgRNA positive cells. However, correct MOIs were ensured by careful determination of the viral titer by genotyping single cell HEK293T mm-STING clones for integration of the sgRNA cassette by PCR. After infection and subsequent culture for 5 days allowing genome editing to occur, 10% of cells per replicate were subjected to direct lysis as previous described (Schmid-Burgk et al., 2014), whereas 90% of cells were stimulated with 500 µg/mL CMA. 24 hr later, flasks were carefully washed with warm PBS and fresh medium added to the cells. Survivors were allowed for 10 days to proliferate and then subjected to direct lysis. sgRNA sequences were amplified with Ex Taq Polymerase (Takara) using a nested PCR approach to add Illumina-sequencing adaptor as previously described (Schmid-Burgk et al., 2014). The first PCR utilized a mix of 8 different forward primers to ensure diversity of the deep sequencing library:
lenti guide fwd 0nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTTTGTGGAAAGGACGAAACACCG
lenti guide fwd 1nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTCTTGTGGAAAGGACGAAACACCG
lenti guide fwd 2nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTGCTTGTGGAAAGGACGAAACACCG
lenti guide fwd 3nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTAGCTTGTGGAAAGGACGAAACACCG
lenti guide fwd 4nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTCAACTTGTGGAAAGGACGAAACACCG
lenti guide fwd 6nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTTGCACCTTGTGGAAAGGACGAAACACCG
lenti guide fwd 7nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTACGCAACTTGTGGAAAGGACGAAACACCG
lenti guide fwd 8nt stagger ACACTCTTTCCCTACACGACGCTCTTCCGATCTGAAGACCCTTGTGGAAAGGACGAAACACCG
lentiCRISPRv2 rev TGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCAATTCCCACTCCTTTCAAGACCT
Samples were subjected to deep sequencing using Illumina’s Hi-Seq platform with a 50 base single read. Reads were aligned to the Brunello library and the fold change in read number of every sgRNA in each of the 6 technical replicates was calculated. Given that the sgRNA targeting the mm-STING transgene showed reproducible rather than particularly strong enrichment, sgRNAs were scored first according to the number of replicates showing a fold change > 5 and second to the mean fold change of all replicates. The top 10% of this sgRNA list were used for scoring all targeted genes, first according to the number of sgRNAs in the list and second to the mean rank of the sgRNAs. The arrayed analysis included all genes, of which 3-4 sgRNAs were in the top 10% of the sgRNA list and some of the genes, of which 2 sgRNAs scored with high ranks.
For arrayed validation, one sgRNA per gene from our arrayed genome-wide library was transduced in an arrayed fashion into HEK293T STING Cas9 cells, generating a polyclonal mix of KO and WT cells. Subsequently, cells were stimulated with indicated concentrations of CMA. However, none of these sgRNAs provided protection from STING-cell death upon CMA stimulation at concentrations used in the initial screen (data not shown). We hypothesized that there might not be a single gene product whose depletion could fully rescue STING-cell death. However, since we consistently observed the enrichment of 3-4 sgRNA for several genes in the survivor population, we considered the screen to be sensitive enough to identify modulators of STING-dependent cell death. Accordingly, to also validate weak modifiers of STING-cell death, we adjusted our validation approach to include lower CMA concentrations and to monitor for long-term survival following STING stimulation, which is comparable to the screening conditions.
Flow Cytometry
Flow cytometric analysis of healthy bone marrow samples were performed in the Laboratory for Leukemia Diagnostics, Department of Internal Medicine III, Klinikum of the LMU Munich (Navios, Beckman Coulter, Krefeld, Germany). The following fluorochrome-labeled antibodies were used for the determination of the immune phenotype: CD45 (J33), CD34 (581), CD33 (D3HL60.251), CD64 (22), CD14 (RMO52), CD2 (39C1.5), CD65 (88H7), CD45 (J33), CD4 (RPA-T4), CD8 (B9.11), CD3 (UCHT1), CD10 (ALB1), CD19 (J3-119) (all antibodies from Beckman Coulter, Krefeld, Germany) and CD68 (Y1/82A) (BioLegend, San Diego, California).
Quantification and Statistical Analysis
If indicated, data were analyzed for statistically significant differences using paired t test if two conditions or groups were to be compared or One-way ANOVA and a post hoc test using Bonferoni’s correction for multiple testing if multiple conditions or experimental groups were to be compared. If normalized data were analyzed a one sample t test was conducted. Gaussian distribution and sphericity of datasets were assumed. Sample sizes were empirically determined and the exact number of repetitions (n) is stated in the respective figure legend. The statistical analysis was performed using GraphPad Prism 6. *** p < 0.001, ** p < 0.01, * p < 0.05, ns = not significant. If not otherwise state, all experiments involving gene deficient monoclonal cell lines are depicted as one representative clone of two independent monoclonal cell lines.
Supplementary Material
(A) Primary human monocytes were primed with Pam3CSK4 for 2 hr in absence or presence of MCC950 and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. IL-18 release from seven donors is depicted as mean + SEM.
(B) Murine BMDMs of indicated genotype were primed with LPS (for IL-1β detection) or left untreated (for IP10 detection) for 2 hr and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. Data are depicted as mean + SEM of three independent experiments.
(C) Murine BMDM were stimulated as in B and analyzed by immunoblot. One representative of three independent experiments is shown.
(D) THP-1 cells of indicated genotype were primed with Pam3CSK4 for 30 min and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. Data from one representative clone of two is shown as mean + SEM of 3-5 independent experiments.
(A) AIM2 expression was quantified by immuno-blot from indicated cells that were left unstimulated or stimulated 100 U/mL IFNγ for 24 hr.
(B) Primary human monocytes were primed as in A and stimulated with Pam3CSK4 in absence of presence of MCC950 for 2 hr and stimulated as indicated for eight hr. Nigericin was added for the last 2 hr of the experiment. Cytokine release is depicted as mean + SD from one representative donor of three.
(C and D) BLaER1 monocytes were primed with 100 U/mL IFNγ for 16 hr or left untreated, primed with LPS for 2 hr and stimulated if indicated for 8 hr. Representative immunoblots showing one clone of two per genotype is depicted (C) and cytokine release is depicted as mean + SD from one representative experiment of two from one representative clone of two (D).
(A) BLaER1 monocytes of indicated genotype that stably expressed mm-STING were primed with LPS for 2 hr and stimulated with CMA for 16 hr. Nigericin was added for the last 2 hr of the experiment. Data are depicted as mean + SEM of three independent experiments from one representative NLRP3−/− clone of two.
(B) ASC-TagRFP BLaER1 monocytes transgenic for mm-STING were stimulated in absence or presence of MCC950 as in A. Representative micrographs and the pyroptosome quantification as mean + SEM of three independent experiments are shown. Scale bars denote 100 µm.
(C) Indicated BLaER1 monocytes were stimulated as in A and cell death was quantified by LDH release and microscopy of PI positive cells. Data are depicted as mean + SEM of three independent experiments from one representative NLRP3−/− clone of two.
(D) LPS-primed BLaER1 mm-STING monocytes were stimulated with 0.625 µM Staurosporine (STS), 100 µg/mL CMA, 6.5 µM Nigericin (NIG) or hsTNF (100 ng/mL), Birinapant (5 µM), Z-VAD-FMK (20 µM) (TBZ) in absence of presence of MCC950 and analyzed by immunoblot. One representative blot of three independent experiments is shown.
(E) LPS-primed BLaER1 mm-STING monocytes were stimulated in absence or presence of 20 µM Z-VAD-FMK as in A. Data are depicted as mean + SEM of three independent experiments.
(A) HEK293T mm-STING cells were stimulated with indicated concentrations of CMA for 16 hr and cell viability was quantified by Cell Titer Blue assay. Data are presented as mean + SEM of three independent experiments.
(B) HEK293T mm-STING cells were stimulated with indicated concentrations of CMA for 4 hr and analyzed by immunoblot. One representative blot of two is depicted.
(C and D) HEK293T cells stably expressing tetracycline inducible NLRP3-tagRFP and ASC-mTurquoise were transduced with an mm-STING construct or left untreated. Subsequently, cells were stimulated with CMA for 16 hr or nigericin for 2 hr in absence or presence of MCC950. Representative micrographs are shown (C) and were quantified from three independent experiments (D). Data are shown as mean ± SEM of these three experiments. Scale bars indicate 50 µm.
(E) HEK293T mm-STING Cas9 cells were stimulated with CMA. After 10 days individual cell colonies were counted and compared to the cell number that was originally plated into the well. 100% recovery indicates that every cell plated into the well gave rise to a colony. Data are shown as mean ± SEM of three experiments
(A) Enrichment of sgRNA abundance in the survivor fraction of HEK293T cells after STING-cell death from the genome-wide sgRNA-screen. Data are pooled from six replicates. Alignment of the best-enriched sgRNA, originally designed to target hs-STING, with its putative off-target binding site in the mm-STING transgene.
(B and C) mm-STING-HEK293T Cas9 cells were transduced with indicated sgRNAs in an arrayed fashion. Several days later, polyclonal cell cultures were stimulated with CMA. Long-term survival was monitored using Cell Titer Blue assay after 10 days. The CTB assay was normalized to wells in which the cells were continuously split over the duration of the assay. Data are shown as mean of three (B) or two (C) independent experiments.
(D) HEK293T mm-STING cells of indicated genotype that were transduced with a dox-on BLOC1S5 cDNA or an empty vector were stimulated with doxycycline for 24h and analyzed by immunoblot. One representative experiment of two is depicted.
(E) LPS-primed BLaER1 monocytes of indicated genotype were primed with LPS for 2 hr and stimulated with lipofected DNA for 8 hr. Nigericin was added for the last 2 hr of the experiment. Cytokine release is depicted as mean + SEM from three independent experiments showing one representative clone of two.
(F and G) HEK293T mm-STING cells were pretreated with indicated inhibitors and stimulated with CMA for 4 hr. Representative images from one of six independent experiments are shown and STING complex formation was quantified from all experiments and is depicted as mean + SEM. Scale bars denote 50 µm.
(H) HEK293T mm-STING cells were pre-treated with indicated inhibitors for 1 hr, stimulated with CMA for 3 hr and analyzed by immunobloting. One representative experiment of two is depicted.
(I) LPS-primed BLaER1 monocytes were pre-treated with indicated concentrations of Bafilomycin and stimulated with lipofected DNA for 8 hr. Nigericin was added for the last 2 hr of the experiment. Cytokine release is depicted as mean + SEM of two independent experiments.
(A) Primary human monocytes were primed with Pam3CSK4 for 2 hr and stimulated with 1 µg, 0.33 or 0.11µg cGAMP or its stabilized version thio-cGAMP for 16 hr. Cytokine secretion was normalized to stimulation with 1µg of respective stimulus and is depicted as mean + SEM of four donors.
(B) Immortalized murine macrophages or immortalized MEFs were stimulated for 24 hr as indicated. Cell viability was quantified by CTB and is depicted as mean + SD of duplicate stimulation from one representative experiment of two.
(C) HEK293T or MEFs that stably expressed dox-on mCherry-mmSTING and LAMP1-GFP reporters were primed for 24 hr with 1 µg/mL doxycycline and stimulated with 125 µg/mL CMA or left untreated for 24 hr. Representative confocal images from one experiment of two are depicted. Scale bars depict 25 µm.
(D–H) Indicated cell lines were stimulated with 125 µg/mL CMA or with lipofected DNA for 8 hr or for 24 hr. If indicated cells were pre-treated with 50 nM or 10 nM Bafilomycin for 1 hr before stimulation. Representative immunoblots from 2-3 independent experiments are depicted.
(A) LPS-primed BLaER1 monocytes of indicated genotype were stimulated with different MOIs of the respective strain of F. novicidafor 8 hr. Data are depicted as mean + SEM of three independent experiments showing one representative clone per genotype. The cytokine secretion upon F. novicida infection is duplicated from Figure 7A.
(B) Primary monocytes were primed with Pam3CSK4 and stimulated in absence or presence of MCC950 with indicated MOIs of MVA for 8 hr. Cytokine secretion is shown as mean + SEM of seven donors.
(C) Human bone marrow was isolated from surgery samples and analyzed by flow cytometry. Data show mean + SEM of three donors.
(D) Human hematopoietic stem cells were differentiated into macrophages for 12 days and analyzed by flow cytometry. Data show mean + SEM of four donors.
(E) Human BMDM were primed with Pam3CSK4 for 2 hr in absence or presence of MCC950 and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. Cytokine secretion from four donors is depicted from a single measurement (Donor 1+2) or mean of duplicate measurements (Donor 3+4). The same data are shown as normalized values in Figure 7F.
Acknowledgments
We kindly thank Andreas Jaensch and Harald Färber for support with K+measurements (Institute for Hygiene and Public Health, University of Bonn); Andreas Wegerer, Kristin Leike, and Larissa Hansbauer for great technical support (Gene Center, LMU); Sara Batelli and Christophe Jung for support with confocal imaging (Gene Center, LMU); the LAFUGA unit and Yiming Cheng for support with deep sequencing (Gene Center, LMU); Thomas Graf for providing BLaER1 cells (CRG, Barcelona); Vishva Dixit for Nlrp3- and Asc-deficient mice (Genentech, USA); Thomas Zillinger for cGAS plasmid constructs (Institute of Clinical Chemistry and Clinical Pharmacology, University of Bonn); Petr Broz for Francisella tularensissubsp. novicida strain U112 (Biozentrum Basel); and Wolfgang Kastenmüller for MVA (Institute of Experimental Immunology, Univ. of Bonn). V.H. serves on the scientific advisory board of Inflazome Ltd. V.H. is a co-founder of RIGONTEC and a member of its scientific advisory board. E.L. is a co-founder of IFM Therapeutics and a member of its scientific advisory board. This work was supported by grants from the ERC (ERC-2013-CoG - 616777 - InflammAct) to E.L., the ERC (ERC-2014-CoG - 647858 - GENESIS) to V.H., and the German Research Foundation (SFB670) to E.L. and V.H.
Footnotes
Author Contributions
M.M.G. and V.H. conceived this study and designed the experiments. M.M.G. performed most of the experiments. T.S.E. and K.R. helped with microscopy. D.C. performed experiments with murine BMDMs and characterization of cell death by immunoblot. F.P. derived human BMDMs. S.Z. performed experiments with THP-1 cells. F.O.D. helped with AIM2 detection. J.L.S.-B. provided mm-STING-HEK293T Cas9 cells. F.H. and E.L. provided NLRP3-ASC-HEK293 cells. R.B. and G.W. provided human PBMCs. M.S. provided human bone marrow and performed flow cytometry. M.M.G. and V.H. wrote the manuscript with the input from all authors. V.H. supervised the study.
References
- Abe T, Barber GN. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-kappaB activation through TBK1. J Virol. 2014;88:5328–5341. doi: 10.1128/JVI.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Schmid-Burgk JL, Hemmerling I, Horvath GL, Schmidt T, Latz E, Hornung V. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature. 2013;503:530–534. doi: 10.1038/nature12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aits S, Jaattela M. Lysosomal cell death at a glance. J Cell Sci. 2013;126:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- Baum R, Sharma S, Carpenter S, Li QZ, Busto P, Fitzgerald KA, Marshak-Rothstein A, Gravallese EM. Cutting edge: AIM2 and endosomal TLRs differentially regulate arthritis and autoantibody production in DNase II-deficient mice. J Immunol. 2015;194:873–877. doi: 10.4049/jimmunol.1402573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol. 2016;17:1142–1149. doi: 10.1038/ni.3558. [DOI] [PubMed] [Google Scholar]
- Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, Vetter I, Dungan LS, Monks BG, Stutz A, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JA, Curley EZ, Wykes MN, Schroder K, Sweet MJ, Roberts TL, Ragan MA, Kassahn KS, Stacey KJ. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol Biol. 2012;12:140. doi: 10.1186/1471-2148-12-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denes A, Coutts G, Lenart N, Cruickshank SM, Pelegrin P, Skinner J, Rothwell N, Allan SM, Brough D. AIM2 and NLRC4 inflammasomes contribute with ASC to acute brain injury independently of NLRP3. Proc Natl Acad Sci U S A. 2015;112:4050–4055. doi: 10.1073/pnas.1419090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon-Perez JM, Nazarian R, Sabatti C, Dell'Angelica EC. Distribution and dynamics of Lamp1-containing endocytic organelles in fibroblasts deficient in BLOC-3. J Cell Sci. 2005;118:5243–5255. doi: 10.1242/jcs.02633. [DOI] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458:509–513. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T, Yu JW, Juliana C, Solorzano L, Kang S, Wu J, Datta P, McCormick M, Huang L, McDermott E, et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol. 2010;11:385–393. doi: 10.1038/ni.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt MM, Ebert TS, Chauhan D, Schmidt T, Schmid-Burgk JL, Rapino F, Robertson AA, Cooper MA, Graf T, Hornung V. Human Monocytes Engage an Alternative Inflammasome Pathway. Immunity. 2016;44:833–846. doi: 10.1016/j.immuni.2016.01.012. [DOI] [PubMed] [Google Scholar]
- Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, et al. Structure-function analysis of STING activation by c[G(2',5')pA(3',5')p] and targeting by antiviral DMXAA. Cell. 2013;154:748–762. doi: 10.1016/j.cell.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Holdt LM, Chen CC, Hassan S, Muller C, Jors S, Cuny H, Kissing S, Schroder B, Butz E, et al. High susceptibility to fatty liver disease in two-pore channel 2-deficient mice. Nat Commun. 2014;5:4699. doi: 10.1038/ncomms5699. [DOI] [PubMed] [Google Scholar]
- Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Jin C, Li HB, Tong J, Ouyang X, Cetinbas NM, Zhu S, Strowig T, Lam FC, Zhao C, et al. The DNA-sensing AIM2 inflammasome controls radiation-induced cell death and tissue injury. Science. 2016;354:765–768. doi: 10.1126/science.aaf7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova S, Gregorc U, Vidergar N, Javier R, Bredt DS, Vandenabeele P, Pardo J, Simon MM, Turk V, Banks L, et al. MAGUKs, scaffolding proteins at cell junctions, are substrates of different proteases during apoptosis. Cell Death Dis. 2011;2:e116. doi: 10.1038/cddis.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs C, Perner S, Hornung V. AIM2 Drives Joint Inflammation in a Self-DNA Triggered Model of Chronic Polyarthritis. PLoS One. 2015;10:e0131702. doi: 10.1371/journal.pone.0131702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John Peter AT, Lachmann J, Rana M, Bunge M, Cabrera M, Ungermann C. The BLOC-1 complex promotes endosomal maturation by recruiting the Rab5 GTPase-activating protein Msb3. J Cell Biol. 2013;201:97–111. doi: 10.1083/jcb.201210038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JW, Kayagaki N, Broz P, Henry T, Newton K, O’Rourke K, Chan S, Dong J, Qu Y, Roose-Girma M, et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc Natl Acad Sci U S A. 2010;107:9771–9776. doi: 10.1073/pnas.1003738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastenmuller W, Torabi-Parizi P, Subramanian N, Lammermann T, Germain RN. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell. 2012;150:1235–1248. doi: 10.1016/j.cell.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S, Cuellar T, Haley B, Roose-Girma M, Phung QT, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K, Lippke JA, Ku G, Harding MW, Livingston DJ, Su MS, Flavell RA. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science. 1995;267:2000–2003. doi: 10.1126/science.7535475. [DOI] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347 doi: 10.1126/science.aaa2630. aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur J Immunol. 2016;46:269–280. doi: 10.1002/eji.201545839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Karki R, Malireddi RK, Neale G, Vogel P, Yamamoto M, Lamkanfi M, Kanneganti TD. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat Immunol. 2015a;16:467–475. doi: 10.1038/ni.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man SM, Zhu Q, Zhu L, Liu Z, Karki R, Malik A, Sharma D, Li L, Malireddi RK, Gurung P, et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell. 2015b;162:45–58. doi: 10.1016/j.cell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis SR, Wilson SC, Vance RE. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017 doi: 10.1016/j.it.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Mesaki K, Tanabe K, Obayashi M, Oe N, Takei K. Fission of tubular endosomes triggers endosomal acidification and movement. PLoS One. 2011;6:e19764. doi: 10.1371/journal.pone.0019764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015a;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E, Wallet P, Dreier RF, Costanzo S, Anton L, Ruhl S, Dussurgey S, Dick MS, Kistner A, Rigard M, et al. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat Immunol. 2015b;16:476–484. doi: 10.1038/ni.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Planillo R, Kuffa P, Martinez-Colon G, Smith BL, Rajendiran TM, Nunez G. K(+) efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapino F, Robles EF, Richter-Larrea JA, Kallin EM, Martinez-Climent JA, Graf T. C/EBPalpha induces highly efficient macrophage transdifferentiation of B lymphoma and leukemia cell lines and impairs their tumorigenicity. Cell Rep. 2013;3:1153–1163. doi: 10.1016/j.celrep.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiga H, Kitada S, Shimada Y, Kamiyama N, Okuyama M, Makino M, Yamamoto M, Takeda K. Critical role of AIM2 in Mycobacterium tuberculosis infection. Int Immunol. 2012;24:637–644. doi: 10.1093/intimm/dxs062. [DOI] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Gaidt MM, Schmidt T, Ebert TS, Bartok E, Hornung V. Caspase-4 mediates non-canonical activation of the NLRP3 inflammasome in human myeloid cells. Eur J Immunol. 2015;45:2911–2917. doi: 10.1002/eji.201545523. [DOI] [PubMed] [Google Scholar]
- Schmid-Burgk JL, Schmidt T, Gaidt MM, Pelka K, Latz E, Ebert TS, Hornung V. OutKnocker: a web tool for rapid and simple genotyping of designer nuclease edited cell lines. Genome Res. 2014;24:1719–1723. doi: 10.1101/gr.176701.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC, et al. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Petrucelli AS, Chen L, Koblansky AA, Truax AD, Oyama Y, Rogers AB, Brickey WJ, Wang Y, Schneider M, et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nat Med. 2015;21:906–913. doi: 10.1038/nm.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Primary human monocytes were primed with Pam3CSK4 for 2 hr in absence or presence of MCC950 and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. IL-18 release from seven donors is depicted as mean + SEM.
(B) Murine BMDMs of indicated genotype were primed with LPS (for IL-1β detection) or left untreated (for IP10 detection) for 2 hr and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. Data are depicted as mean + SEM of three independent experiments.
(C) Murine BMDM were stimulated as in B and analyzed by immunoblot. One representative of three independent experiments is shown.
(D) THP-1 cells of indicated genotype were primed with Pam3CSK4 for 30 min and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. Data from one representative clone of two is shown as mean + SEM of 3-5 independent experiments.
(A) AIM2 expression was quantified by immuno-blot from indicated cells that were left unstimulated or stimulated 100 U/mL IFNγ for 24 hr.
(B) Primary human monocytes were primed as in A and stimulated with Pam3CSK4 in absence of presence of MCC950 for 2 hr and stimulated as indicated for eight hr. Nigericin was added for the last 2 hr of the experiment. Cytokine release is depicted as mean + SD from one representative donor of three.
(C and D) BLaER1 monocytes were primed with 100 U/mL IFNγ for 16 hr or left untreated, primed with LPS for 2 hr and stimulated if indicated for 8 hr. Representative immunoblots showing one clone of two per genotype is depicted (C) and cytokine release is depicted as mean + SD from one representative experiment of two from one representative clone of two (D).
(A) BLaER1 monocytes of indicated genotype that stably expressed mm-STING were primed with LPS for 2 hr and stimulated with CMA for 16 hr. Nigericin was added for the last 2 hr of the experiment. Data are depicted as mean + SEM of three independent experiments from one representative NLRP3−/− clone of two.
(B) ASC-TagRFP BLaER1 monocytes transgenic for mm-STING were stimulated in absence or presence of MCC950 as in A. Representative micrographs and the pyroptosome quantification as mean + SEM of three independent experiments are shown. Scale bars denote 100 µm.
(C) Indicated BLaER1 monocytes were stimulated as in A and cell death was quantified by LDH release and microscopy of PI positive cells. Data are depicted as mean + SEM of three independent experiments from one representative NLRP3−/− clone of two.
(D) LPS-primed BLaER1 mm-STING monocytes were stimulated with 0.625 µM Staurosporine (STS), 100 µg/mL CMA, 6.5 µM Nigericin (NIG) or hsTNF (100 ng/mL), Birinapant (5 µM), Z-VAD-FMK (20 µM) (TBZ) in absence of presence of MCC950 and analyzed by immunoblot. One representative blot of three independent experiments is shown.
(E) LPS-primed BLaER1 mm-STING monocytes were stimulated in absence or presence of 20 µM Z-VAD-FMK as in A. Data are depicted as mean + SEM of three independent experiments.
(A) HEK293T mm-STING cells were stimulated with indicated concentrations of CMA for 16 hr and cell viability was quantified by Cell Titer Blue assay. Data are presented as mean + SEM of three independent experiments.
(B) HEK293T mm-STING cells were stimulated with indicated concentrations of CMA for 4 hr and analyzed by immunoblot. One representative blot of two is depicted.
(C and D) HEK293T cells stably expressing tetracycline inducible NLRP3-tagRFP and ASC-mTurquoise were transduced with an mm-STING construct or left untreated. Subsequently, cells were stimulated with CMA for 16 hr or nigericin for 2 hr in absence or presence of MCC950. Representative micrographs are shown (C) and were quantified from three independent experiments (D). Data are shown as mean ± SEM of these three experiments. Scale bars indicate 50 µm.
(E) HEK293T mm-STING Cas9 cells were stimulated with CMA. After 10 days individual cell colonies were counted and compared to the cell number that was originally plated into the well. 100% recovery indicates that every cell plated into the well gave rise to a colony. Data are shown as mean ± SEM of three experiments
(A) Enrichment of sgRNA abundance in the survivor fraction of HEK293T cells after STING-cell death from the genome-wide sgRNA-screen. Data are pooled from six replicates. Alignment of the best-enriched sgRNA, originally designed to target hs-STING, with its putative off-target binding site in the mm-STING transgene.
(B and C) mm-STING-HEK293T Cas9 cells were transduced with indicated sgRNAs in an arrayed fashion. Several days later, polyclonal cell cultures were stimulated with CMA. Long-term survival was monitored using Cell Titer Blue assay after 10 days. The CTB assay was normalized to wells in which the cells were continuously split over the duration of the assay. Data are shown as mean of three (B) or two (C) independent experiments.
(D) HEK293T mm-STING cells of indicated genotype that were transduced with a dox-on BLOC1S5 cDNA or an empty vector were stimulated with doxycycline for 24h and analyzed by immunoblot. One representative experiment of two is depicted.
(E) LPS-primed BLaER1 monocytes of indicated genotype were primed with LPS for 2 hr and stimulated with lipofected DNA for 8 hr. Nigericin was added for the last 2 hr of the experiment. Cytokine release is depicted as mean + SEM from three independent experiments showing one representative clone of two.
(F and G) HEK293T mm-STING cells were pretreated with indicated inhibitors and stimulated with CMA for 4 hr. Representative images from one of six independent experiments are shown and STING complex formation was quantified from all experiments and is depicted as mean + SEM. Scale bars denote 50 µm.
(H) HEK293T mm-STING cells were pre-treated with indicated inhibitors for 1 hr, stimulated with CMA for 3 hr and analyzed by immunobloting. One representative experiment of two is depicted.
(I) LPS-primed BLaER1 monocytes were pre-treated with indicated concentrations of Bafilomycin and stimulated with lipofected DNA for 8 hr. Nigericin was added for the last 2 hr of the experiment. Cytokine release is depicted as mean + SEM of two independent experiments.
(A) Primary human monocytes were primed with Pam3CSK4 for 2 hr and stimulated with 1 µg, 0.33 or 0.11µg cGAMP or its stabilized version thio-cGAMP for 16 hr. Cytokine secretion was normalized to stimulation with 1µg of respective stimulus and is depicted as mean + SEM of four donors.
(B) Immortalized murine macrophages or immortalized MEFs were stimulated for 24 hr as indicated. Cell viability was quantified by CTB and is depicted as mean + SD of duplicate stimulation from one representative experiment of two.
(C) HEK293T or MEFs that stably expressed dox-on mCherry-mmSTING and LAMP1-GFP reporters were primed for 24 hr with 1 µg/mL doxycycline and stimulated with 125 µg/mL CMA or left untreated for 24 hr. Representative confocal images from one experiment of two are depicted. Scale bars depict 25 µm.
(D–H) Indicated cell lines were stimulated with 125 µg/mL CMA or with lipofected DNA for 8 hr or for 24 hr. If indicated cells were pre-treated with 50 nM or 10 nM Bafilomycin for 1 hr before stimulation. Representative immunoblots from 2-3 independent experiments are depicted.
(A) LPS-primed BLaER1 monocytes of indicated genotype were stimulated with different MOIs of the respective strain of F. novicidafor 8 hr. Data are depicted as mean + SEM of three independent experiments showing one representative clone per genotype. The cytokine secretion upon F. novicida infection is duplicated from Figure 7A.
(B) Primary monocytes were primed with Pam3CSK4 and stimulated in absence or presence of MCC950 with indicated MOIs of MVA for 8 hr. Cytokine secretion is shown as mean + SEM of seven donors.
(C) Human bone marrow was isolated from surgery samples and analyzed by flow cytometry. Data show mean + SEM of three donors.
(D) Human hematopoietic stem cells were differentiated into macrophages for 12 days and analyzed by flow cytometry. Data show mean + SEM of four donors.
(E) Human BMDM were primed with Pam3CSK4 for 2 hr in absence or presence of MCC950 and stimulated as indicated for 8 hr. Nigericin was added for the last 2 hr of the experiment. Cytokine secretion from four donors is depicted from a single measurement (Donor 1+2) or mean of duplicate measurements (Donor 3+4). The same data are shown as normalized values in Figure 7F.