Abstract
Tenascin-C is an extracellular matrix molecule that drives progression of many types of human cancer but the basis for its actions remain obscure. In this study, we describe a cell-autonomous signaling mechanism explaining how tenascin-C promotes cancer cell migration in the tumor microenvironment. In a murine xenograft model of advanced human osteosarcoma, tenascin-C and its receptor integrin α9β1 were determined to be essential for lung metastasis of tumor cells. We determined that activation of this pathway also reduced tumor cell-autonomous expression of target genes for the transcription factor YAP. In clinical specimens, a genetic signature comprising four YAP target genes represents prognostic impact. Taken together, our results illuminate how tumor cell deposition of tenascin-C in the tumor microenvironment promotes invasive migration and metastatic progression.
Introduction
The extracellular matrix (ECM) molecule tenascin-C (TNC) that is highly expressed in the tumor microenvironment (TME) represents an active component of cancer tissue. Its high expression correlates with worsened patient survival prognosis in several cancer types (1). TNC promotes multiple events in cancer progression as recently demonstrated in a multi-stage neuroendocrine tumorigenesis model with abundant and no TNC. It was shown that TNC enhances tumor cell survival, proliferation, invasion and lung metastasis. Moreover, TNC increases Notch signaling in breast cancer (2). TNC also promotes stromal events such as the angiogenic switch and the formation of more but leaky blood vessels involving Wnt signaling and inhibition of Dickkopf1 (DKK1) in a neuroendocrine tumor model (3, 4), and Ephrin-B2 signaling in a glioblastoma (GBM) model (5). TNC networks can have similarities with reticular fibers in lymphoid organs (6) and may alter the biomechanical properties of cancer tissue (7), in particular increase tissue stiffening (8). TNC also impairs actin stress fiber formation (9) and regulates gene expression which may impact on cell behaviour and tumor malignancy (10).
The actin polymerization state is interpreted by the cell through two co-transcription factors, megakaryoblastic leukemia 1 (MKL1, myocardin related transcription factor MRTF-A, MAL) (11) and yes activating protein (YAP) (12, 13). Under poorly adhesive conditions, cells fail to polymerize actin and subsequently cannot form actin stress fibers. MKL1 binds to globular G-actin monomers and remains sequestered in the cytoplasm. In consequence MKL1 cannot reach nuclear serum response factor (SRF) or DNA sequences to induce gene transcription (14, 15) and, MKL1-dependent genes remain silent.
YAP and TAZ (transcriptional co-activator with PDZ-binding motif) proteins are integral parts of the Hippo signaling pathway that is important for organ growth control during development and is often found to be deregulated in cancer (16). Recently, YAP and TAZ were demonstrated to transduce mechanical and cytoskeletal cues with actin stress fibers promoting their nuclear translocation (17). Nuclear YAP/TAZ can activate gene expression through binding to the TEAD (TEA domain transcription factors) family of transcription factors (17), thus controlling gene expression upon cell adhesion.
Here, we analyzed the underlying mechanisms and consequences of poor cell adhesion by TNC. We demonstrate that TNC downregulates gene expression through inhibition of actin stress fibers which in turn abolishes MKL1 and YAP activities in tumor cells. TNC itself is downregulated by a negative feedback loop due to inactive MKL1 and YAP. We further show that integrin α9β1 and inactive YAP are instrumental for TNC to promote tumor cell migration in an autocrine and paracrine manner. This has relevance for metastasis as knockdown of TNC or ITGA9 decreases lung metastasis which is associated with increased YAP target gene expression. Finally, poor expression of three YAP target genes (CTGF, CYR61 and CDC42EP3) identifies a group of osteosarcoma and glioblastoma patients with worst prognosis when TNC levels are below the median expression. To our knowledge this is the first report that provides a full view on a signaling pathway initiated by TNC, employing integrin α9β1 subsequently destroying actin stress fibers, inhibiting YAP and abolishing target gene expression thus promoting cell migration and lung metastasis. This information could be of prognostic and therapeutic value.
Material and Methods
More details can be found in the supplemental information section.
Cell culture
Human glioblastoma T98G (ATCC, CRL-169), U87MG (ATCC, HTB-14) and osteosarcoma KRIB (v-Ki-ras transformed human osteosarcoma cells) (18), previously used (9, 19) were cultured up to 10 passages after defrosting in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) 4.5g/l glucose with 10% Fetal Bovine Serum (FBS, Sigma-Aldrich), 100 U/ml penicillin and 100 µg/ml streptomycin and 40u/ml gentamicin at 37°C and 5% CO2. Absence of mycoplasms was regularly checked by quantitative real time polymerase chain reaction (qPCR) according to the manufacturer`s instructions (Venor GeMClassic, Minerva BioLabs). Cells were starved with 1% FBS overnight before drug treatment with 30 μM lysophosphatidic acid (LPA) (H2O, Santa Cruz,), 5 μM Latrunculin B (LB) (DMSO, Calbiochem), 2 μM Jasplakinolide (Jasp) (DMSO, Santa Cruz) and 10 μM Y27632 (DMSO, Selleck Chemicals), respectively or seeding on surfaces coated with purified horse serum-derived fibronectin (FN) or, FN plus purified recombinant human TNC for 24h in DMEM containing 1% FBS.
Animal experiments
KRIB control (shCTRL) and TNC and ITGA9 knockdown cells (shTNC, shα9) (10 x 106), diluted in 100 µl phosphate buffered PBS were subcutaneously injected in the left upper back of nude mice (Charles River) and sacrificed 5 weeks later. The tumor size was measured every 7 days with a digital caliper and was calculated using the formula S = a*b, where b is the longest axis and a is the perpendicular axis to b. Upon extraction the tumor weight was determined with a digital balance. The tumor and the smallest lung lobe of each mouse were directly frozen in liquid nitrogen and further analyzed by qPCR. Experiments with animals were performed according to the guidelines of INSERM and the ethical committee of Alsace, France (CREMEAS) with the reference number of the project AL/73/80/02/13 and the mouse house E67-482-21.
Coating with purified ECM molecules
FN and TNC were coated in 0.01% Tween 20-PBS at 1 μg/cm2 before saturation with 10 mg/ml heat inactivated bovine serum albumen (BSA)/PBS (3, 9).
RNA isolation and qPCR
Total RNA was isolated from cells by using TriReagent (Life Technologies) according to the manufacturer’s instructions, reverse transcribed and used for qPCR with primers listed in Table S1.
Immunoblotting
Cells were lysed in RIPA buffer (150 mM NaCl, 1.0% IGEPAL CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0), separated by polyacrylamide gel electrophoresis, blotted onto nictrocellulose membrane using the Trans-Blot Turbo RTA Mini Nitrocellulose Transfer Kit (BioRad) and incubated with primary and horseradish peroxidase coupled secondary antibodies before signal detection with the Amersham ECL detection reagent.
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde (PFA) for 10 minutes and permeabilized in PBS-Triton 0.1% for 10 minutes, incubated with the anti-YAP antibody and a secondary fluorophore coupled antibody and analyzed with a Zeiss Axio Imager Z2 microscope. At least 150 cells in duplicates per condition were quantified.
Lentiviral transduction of cells
Silencing of MKL1, TNC and ITGA9 was done by short hairpin (sh) mediated gene expression knockdown (see Table S2). MISSION lentiviral transduction particles (Sigma-Aldrich) or MISSION non-target shRNA control transduction particles (SHC002V, Sigma-Aldrich) with a MOI of 1 were used, transduced cells were selected with 2.5, 10 and 1 μg/ml puromycin for MKL1, TNC and ITGA9 knockdown, respectively. Stable knockdown was determined at RNA level by qPCR and protein level by immunoblotting.
Transfection and RNAi
Plasmids encoding YAP (YAP-WT), constitutively active YAP (CA-YAP, S127A mutant (20) and non-TEAD interacting YAP (DN-YAP, S127A-S94A mutant (20)) and, MKL1-WT (pEF full length hemagglutinin-tagged MAL HA) and N-terminal deleted constitutively active CA-MKL1 (pEF HA-ΔN MAL) were provided by Guido Posern (Halle-Wittenberg University, Halle, Germany). Plasmids were transiently transfected (JetPEI, Polyplus) and the siRNA reagent system (sc-45064, Santa Cruz) for reducing expression of YAP, MKL1, ITGA9 and SDC4 were used according to the manufacturer’s instruction. Note, that cells with stable expressing of CA-YAP could not been established.
Luciferase reporter assay
Cells were transiently transfected (JetPEI, Polyplus, Strasbourg, France) with the pGL3-5 x MCAT(SV)-49 plasmid (provided by I. Farrance, University of Maryland School of Medicine, Baltimore, U.S.A) encoding 5 x MCAT (TEAD binding sites) or 3DA.Luc plasmid (provided by Guido Posern, Halle-Wittenberg University, Halle, Germany) encoding FOS derived SRF binding sites together with the pRL-TK (TK-Renilla) plasmid for normalization. Cells were lysed and analyzed by the Dual-Luciferase reporter assay system (Promega) and a BioTek Luminometer EL800. Firefly luciferase activity was normalized to internal Renilla luciferase control activity.
Migration and invasion assays
For 2D migration, 2 x 105 cells were seeded in a 50 mm lumox dish (SARSTEDT). Real time phase contrast images were taken with a Zeiss microscope (Axiovert observation) every 15 minutes for 24 hours. Migration of individual cells in the first 12 hours (10 cells in each field, 2 fields per condition) was analyzed with the ImageJ software. For Boyden chamber transwell migration or invasion assays, 2 x 104 cells were plated onto the upper chamber of a transwell filter with 8 μm pores (Greiner Bio-one) that had been coated on the upper side with FN and FN/TNC (1 μg/cm2), growth factor reduced Matrigel (0.5 mg/ml, Corning) or rat tail type 1 collagen gel (2.5 mg/ml, BD Biosciences) as described (21, 22). 10% FBS in the lower chamber was used as chemoattractant. Cells at the lower side were fixed with 4% PFA in PBS, stained with DAPI (Sigma D9542), photographed and abundance was quantified using the ZEN Blue software (Zeiss).
Patient survival analysis
The patient dataset GSE21257 (osteosarcoma) and GSE42669 (GBM) available in the Gene Expression Omnibus (GEO) Database (http://www.ncbi.nlm.nih.gov/gds) were used. Microsoft Excel was used to extract the expression values of a small number of genes (probesets) and were compared to the clinical data from GEO. Survival analysis was performed using SPSS23.0 and the Kaplan-Meier survival procedure.
Statistical analysis
All experiments were performed at least three times independently with at least two to three replicates per experiment. For all data, Gaussian distribution was tested by the d’Agostino-Pearson normality test. Statistical differences were analyzed by unpaired t-test (with Welch’s correction in case of unequal variance) or ANOVA one-way with Tukey post-test for Gaussian dataset distribution. Statistical analysis and graphical representation were performed using GraphPad Prism. GSEA (23) was used to analyze enrichment of the YAP/TAZ/TEAD target genes (24) and MKL1 target genes (25) in the TNC specific gene expression signature (10). p-values < 0.05 were considered as statistically significant. Mean ± SEM. p values *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Results
TNC inhibits actin stress fiber formation on a mixed FN/TNC substratum
FN and TNC are often coexpressed and act as accomplices in cell adhesion where TNC counteracts the adhesive properties of FN (9, 26, 27). To set the stage for the subsequent mechanistic analysis, we determined how low cell adhesion to FN implemented by TNC affects actin dynamics and downstream gene expression in two previously used human tumor cell lines derived from GBM (T98G) and osteosarcoma (KRIB) (9, 28). Whereas most experiments were performed with KRIB cells some were reproduced in T98G cells (supplemental figures). We found that both cells were round and adhered less on the FN/TNC substratum (Fig. S1A-C). Western blot upon fractionation into monomeric G-actin and polymerized F-actin revealed less F-actin in both cells grown on FN/TNC compared to FN (Fig. S1D-F). TRITC-phalloidin staining showed no actin stress fibers on FN/TNC (Fig. S1 G, H).
A TNC repression signature negatively correlates with MKL1 and YAP responsive genes
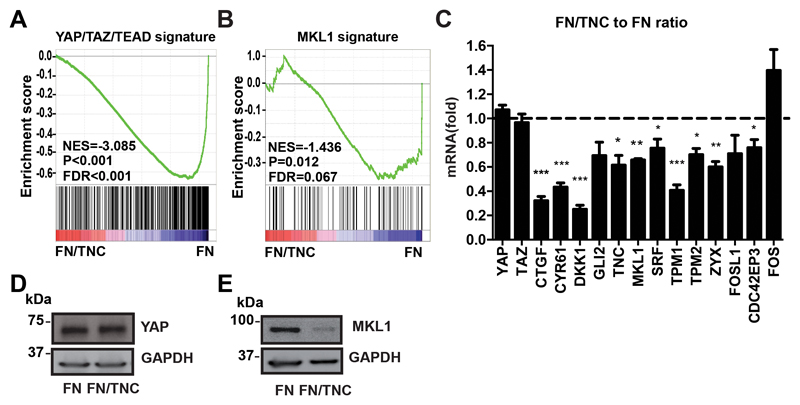
Since TNC inhibits actin stress fibers and, actin stress fibers regulate MKL1 and YAP/TAZ (11 – 13), we asked whether TNC modulates MKL1 and/or YAP activities. Therefore, we searched for a potential correlated expression of genes that are regulated by TNC (10) and genes that are regulated by MKL1/SRF (25) or YAP/TAZ (24), respectively. We used publicly available mRNA expression data and Gene Set Enrichment Analysis (GSEA) and found that both gene sets are significantly negatively correlated with a gene signature that is downregulated by TNC in T98G cells (Fig. 1A, B) (10). By qPCR we evaluated TNC substratum specific gene expression and found that in contrast to FOS, that is increased on FN/TNC, a selection of known MKL1 regulated genes (tropomyosin-1/TPM1, TPM2, ZYX, FOSL1/Fos-related antigen 1, CDC42EP3/CDC42 effector protein-3, TNC) (29 – 31) and YAP regulated genes (CTGF/CCN2, CYR61/CCN1, DKK1/Dickkopf-1, GLI2/GLI family zinc finger 2) (32) was indeed lowered on the FN/TNC substratum in both cells (Fig. 1C, S1I). Whereas TAZ mRNA level was slightly enhanced in T98G (yet not in KRIB), YAP protein levels consistently were not affected by the FN/TNC substratum in either cell (Fig. 1D, S1J). In contrast, MKL1 protein levels were reduced on FN/TNC in both cells suggesting that TNC blocks expression of MKL1 but not of YAP (Fig. 1E, S1K).
Figure 1. Impact of TNC on MKL1 and YAP target gene expression.
Gene Set Enrichment Analysis reveals a significant anti-correlation between TNC and a YAP/TAZ (A) and a MKL1/SRF (B) gene expression signature, respectively. The Normalized Enrichment Score (NES) and the False Discovery Rate (FDR) q-value assessing the significance of enrichment are indicated. (C) Gene expression by qPCR of selected genes in KRIB cells upon growth on FN or FN/TNC (n = 9) is expressed as relative ratio of values on FN/TNC versus FN. (D, E) Immunoblotting for YAP and MKL1 in KRIB cells on FN or FN/TNC. In all figures n = 9 and n = 6 represents 3 independent experiments with 3 replicates and 2 replicates, respectively. Mean ± SEM.
TNC blocks (non-SRF-mediated) MKL1 target gene expression through repression of MKL1
MKL1 can induce SRF dependent and independent gene expression (11, 15). We addressed whether MKL1/SRF dependent transcription is potentially impaired by TNC in T98G (Fig. S2A-G) and KRIB cells (Fig. S2H-M) by measuring SRF-driven luciferase activity in cells grown on FN or FN/TNC and noticed similar activities suggesting that TNC does not inhibit the SRF-dependent function of MKL1 (Fig. S2A, H). Then, we used loss of function (LOF) and gain of function (GOF) approaches employing shRNAs to reduce MKL1 expression and overexpression of a constitutive active CA-MKL1 molecule, respectively (Fig. S2B, C, I, J). Whereas knockdown of MKL1 caused reduced expression of all tested MKL1 target genes (Fig. S2D, K), CA-MKL1 induced SRF-luciferase activity indicating MKL1 responsiveness (Fig. S2E). CA-MKL1 significantly induced CTGF, CDC42EP3, TNC, DKK1 and TPM1, yet not CYR61 in both cells (Fig. S2F, L). Whereas CTGF and CYR61 remained unaffected, transient expression of CA-MKL1 increased gene expression of CDC42EP3, TNC, DKK1 (only T98G) and TPM1 significantly on FN/TNC in both cells (Fig. S2G, M). These results suggest that TNC downregulates some genes such as TPM1, TNC, CDC42EP3 and DKK1 through impairing MKL1 functions. In contrast, other genes such as CTGF and CYR61 are repressed by TNC through another mechanism.
TNC represses genes in tumor cells through abolishing YAP activity by cytoplasmic retention
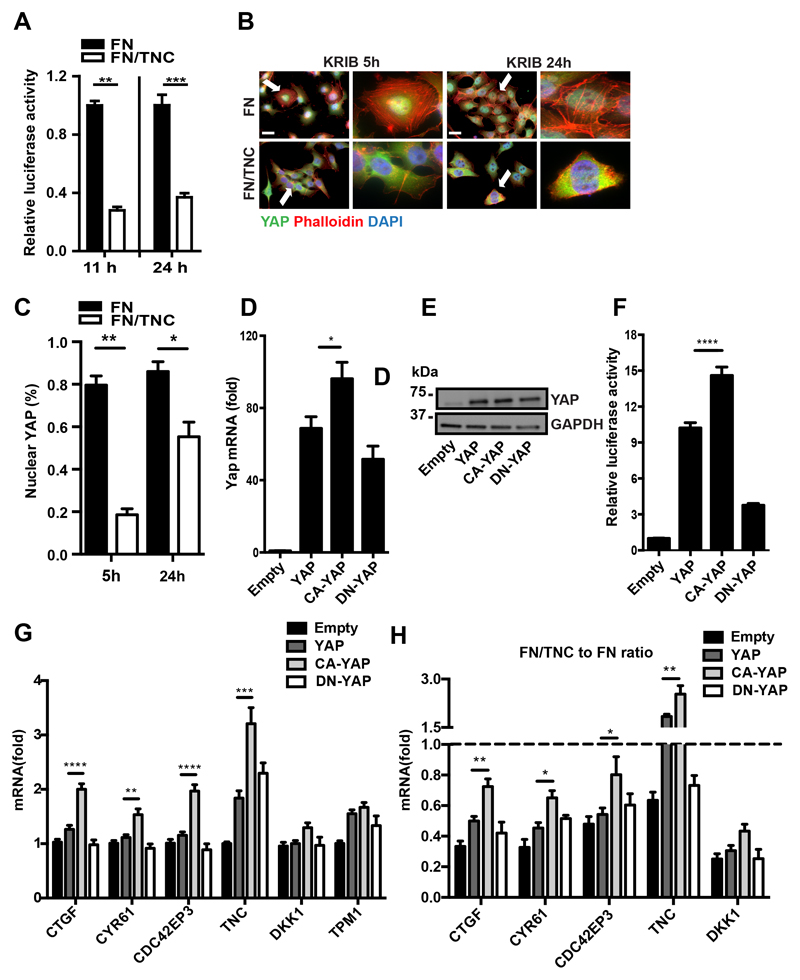
To analyze whether TNC inhibits the YAP co-transcriptional functions, we measured luciferase activity driven by the transcription factor TEAD which requires active YAP (17). Indeed, luciferase activity was reduced in both cells grown on FN/TNC compared to FN (Fig. 2A, S3A). Since YAP protein levels were equal on both substrata (Fig. 1D, S1J), excluding regulation by TNC at expression level, we investigated whether TNC may impair YAP nuclear translocation (17). We assessed YAP subcellular localization by staining cells for YAP. Indeed, whereas YAP was nuclear in the large majority of both cells plated on FN, YAP remained mostly cytoplasmic in cells on FN/TNC even 24 hours after plating which resembles cells in the absence of FBS, a condition that blocks YAP function (Fig. 2B, C, S3B-E) (33). Thus, on FN/TNC, nuclear translocation of YAP is impaired which could explain inactivation of YAP co-transcription function.
Figure 2. A FN/TNC substratum impairs YAP target gene expression by cytoplasmic retention of YAP.
Results for KRIB cells are shown. (A) TEAD luciferase assay of cells grown on FN or FN/TNC. (B) Representative images of YAP (green), polymerized actin (phalloidin, red) and nuclei (DAPI, blue) in cells upon growth on FN or FN/TNC. The arrow points at the cell of higher magnification on the right. Scale bar, 5 µm. (C) Quantification of cells with nuclear YAP on the indicated substrata represented as percentage of all cells.(D) YAP expression in cells by qPCR upon transfection of empty vector (CTRL) or YAP expression constructs. (E) Immunoblotting for YAP and GAPDH upon transient transfection of cells with YAP expression plasmids. (F) TEAD luciferase assay upon transfection of YAP expression plasmids. (G) Gene expression analysis by qPCR upon transient expression of YAP expression plasmids in cells grown on plastic. (H) Ratio of gene expression on FN/TNC versus FN as determined by qPCR upon transient transfection of YAP expression plasmids. n = 9, except for C (n = 6). Mean ± SEM.
To determine regulation of genes by TNC through YAP in more detail we used LOF and GOF approaches by transiently expressing inhibitory (DN-YAP) or activating (CA-YAP) YAP molecules (Fig. 2D, E, S3F, G). We addressed YAP transactivation function with a TEAD-luciferase assay and observed high TEAD-luciferase activity upon transfection of CA-YAP (Fig. 2F, S3H). CA-YAP also significantly increased CTGF, CYR61, CDC42EP3 and, TNC gene expression. In contrast, neither DKK1 nor TPM1 were induced by CA-YAP indicating that these genes are not regulated by YAP (Fig. 2G, S3I). To investigate whether TNC downregulates genes through impairment of YAP we used transient expression of CA-YAP and looked for gene expression on FN/TNC. We noticed in both cells that expression of CTGF, CYR61, CDC42EP3 and TNC was increased. Again, expression of DKK1 was poorly affected in both cells (Fig. 2H, S3J). These results suggest that TNC reduces expression of CTGF, CYR61 and CDC42EP3 by inhibiting YAP. Moreover, we showed for the first time that YAP regulates TNC expression.
TNC downregulates YAP target gene expression through blocking actin stress fibers
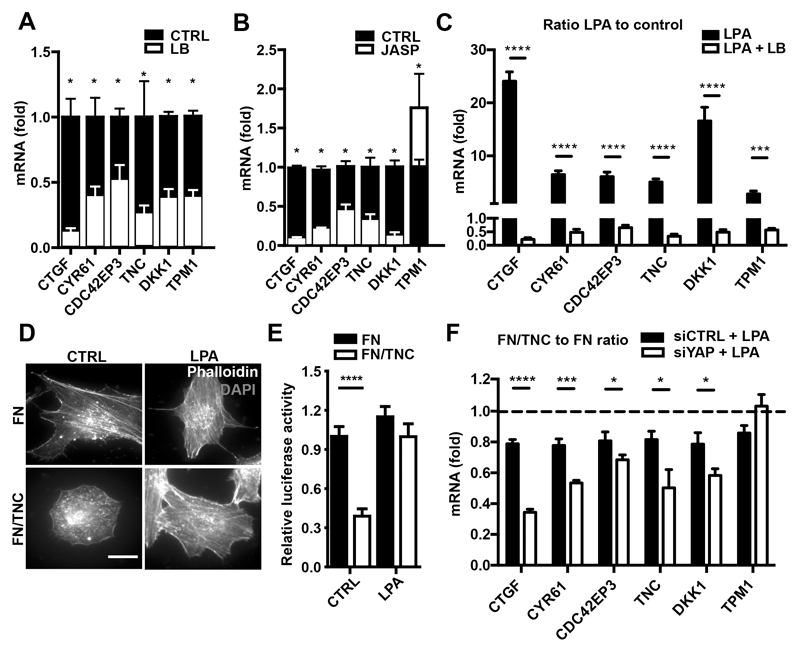
As TNC impacts on the actin cytoskeleton and abolishes MKL1 and YAP target gene expression, we asked whether and how TNC-regulated genes respond to actin dynamics. We treated both cells with Latrunculin B (LB) causing disassembly of actin filaments into monomeric G-actin (34), Jasplakinolide (Jasp) to stabilize F-actin and inhibit stress fibers (35) and, lysophosphatidic acid (LPA) to induce actin stress fibers (36) (Fig. S4A, B) before measuring gene expression. LB blocked expression of all tested genes in both cells (Fig. 3A, S4C). Whereas Jasp blocked CTGF, CYR61, CDC42EP3, TNC and DKK1 expression, TPM1 was even increased over control conditions by Jasp (Fig. 3B, S4D), suggesting that F-actin is sufficient to drive TPM1 expression but not expression of the other five TNC target genes which may require actin stress fibers. Indeed, the actin stress fiber inducer LPA triggered actin stress fiber formation in KRIB cells plated on FN/TNC, as well as TEAD-driven luciferase and expression of all tested genes in both cells and, on a FN/TNC substratum (Fig. 3C - F, S4E - G). These results suggest that actin stress fibers are important regulators of the TNC-repressed genes. To prove that the LPA effect is due to its role in actin stress fiber formation (as LPA can also have other downstream effectors (37)), we treated cells with LPA together with LB, generating G-actin, and measured gene expression (Fig. S4A). We observed that LB abolished LPA-induced expression of all tested genes which indicates that LPA bypasses TNC gene repression through its impact on actin stress fiber formation (Fig. 3C, S4E). Importantly, TNC expression itself is regulated by actin stress fibers as LPA induces and Jasp blocks TNC expression, respectively (Fig. 3A-C, S4C-E).
Figure 3. Actin polymerization dependent expression of TNC downregulated genes.
Results for KRIB cells are shown. (A - C) Gene expression analysis by qPCR of TNC target genes upon treatment with LB (A), Jasp (B) or LPA plus LB (C) after 5 hours (n = 6, 3 experiments in duplicates). (D) Representative images of polymerized actin (phalloidin, white) and nuclei (DAPI, blue) of cells on FN or FN/TNC with or without LPA treatment after 5h. Scale bar, 5 µm. (E) TEAD luciferase assay upon growth on FN or FN/TNC with or without LPA for 24 hours (n = 12, 4 experiments in triplicates). (F) Gene expression analysis by qPCR upon treatment with LPA and siYAP and growth on FN or FN/TNC. Relative expression is depicted as ratio of values on FN/TNC versus FN, n = 9. Mean ± SEM.
We used LPA to induce target gene expression on FN/TNC (Fig. 3D) and then investigated whether inhibition of YAP (Fig. S4H) could revert the LPA effect. Indeed, siYAP abolished expression of all LPA-restored genes on FN/TNC except TPM1 (not a YAP target gene) in both cells (Fig. 3F, S4G, S5A-L). This result suggests that TNC represses YAP target genes through inhibition of actin stress fibers.
TNC promotes 3D migration through integrin α9β1 by blocking actin stress fibers and inactivating YAP
As TNC impairs actin stress fiber formation and YAP-dependent gene expression, we wanted to know whether this has an effect on cell migration. We monitored mobility by time lapse microscopy in KRIB cells and observed that the total migration distance was lower on FN/TNC than on FN (Fig. 4A, B, videos 1 and 2). By using a 3D Boyden chamber migration assay we observed that more KRIB cells moved to the other side of the filter when cells were placed on the FN/TNC substratum in comparison to FN at the 6 and 24 hours time points (Fig. 4C). A similar observation was made for T98G cells (Fig. S6A, B). We demonstrated that the TNC-containing substratum did not affect T98G and KRIB cell proliferation even not upon treatment with LPA (Fig. S6C). Moreover, TNC-induced migration was not affected by proliferation as migration was similar upon treatment with proliferation inhibitory Mitomycin-C (Fig. S6D). Altogether, these observations suggest that TNC promotes transwell migration of KRIB and T98G cells.
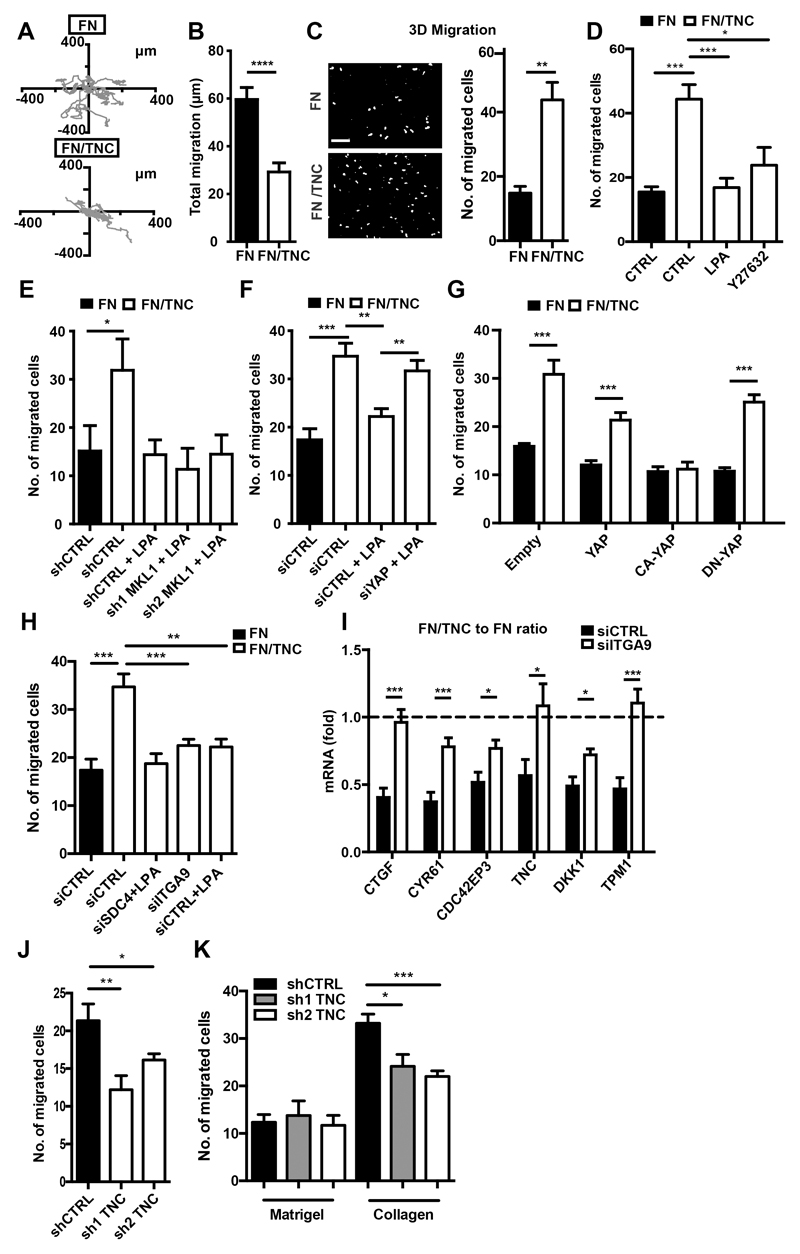
Figure 4. TNC promotes transwell migration through integrin α9β1 and requires inactive YAP.
Results for KRIB cells are shown. Assessment of 2D migration (A, B) showing the movement of individual cells during 12 hours live imaging (A) and transwell migration 24 hours after seeding on FN or FN/TNC (C), and upon treatment with LPA or Y27632 (D), or knockdown of the following genes, MKL1 (E), YAP (F), ITGA9 (H), SDC4 (H) and TNC (J), respectively and, upon overexpression of YAP molecules (G). Scale bar, 20 µm. (I) mRNA levels of the indicated genes upon knockdown of ITGA9 expressed as ratio of values for FN/TNC versus FN. (K) Quantification of invasion of shCTRL and shTNC cells through Matrigel- and collagen gel - coated transwells after 24 hours. n = 6, except for G (n = 7, 3 experiments with at least duplicates) and F, H, I, K (n = 9). Mean ± SEM.
As LPA restored cell spreading through induction of actin stress fibers we asked whether LPA had an impact on transwell migration. Indeed, LPA reduced migration of KRIB cells on FN/TNC to levels as on FN (Fig. 4D). Thus, actin stress fibers counteract TNC-induced transwell migration suggesting that impairment of stress fibers is important for migration by TNC. Cells with a round cell shape can migrate in an amoeboid manner where active Rho-kinase (ROCK) is crucial (38). We chemically inhibited ROCK and observed that ROCK is required for transwell migration by TNC, as Y27632 blocked migration from FN/TNC in the Boyden chamber experiment (Fig. 4D).
Now, we addressed a potential interdependence with MKL1 and/or YAP. Therefore, we added LPA to KRIB cells with knockdown of MKL1 or YAP and measured Boyden chamber migration. Whereas knockdown of MKL1 did not alter KRIB cell migration on FN/TNC in the presence of LPA, knockdown of YAP restored transwell migration (Fig. 4E, F). To substantiate a link to actin stress fibers we stained KRIB cells with phalloidin upon growth on FN/TNC and addition of LPA and, transfection of siYAP or expression of DN-YAP and CA-YAP, respectively. Whereas LPA induced actin stress fibers on FN/TNC, this did not occur in KRIB cells with siYAP or expressing DN-YAP (Fig. 3D, S6E, F). We conclude that siYAP abolishes the stress fiber inducing effect of LPA. We further noticed that CA-YAP restored actin stress fibers on FN/TNC and, abolished TNC-induced transwell migration. This was not the case with DN-YAP or WT-YAP (Fig. 4G, S6F). We conclude that TNC promotes transwell migration through blocking actin stress fibers and YAP.
Next, we addressed which upstream regulators as e.g. syndecan-4 (9) or integrin α9β1, a receptor for TNC (39, 40) are mediating TNC-induced migration. We lowered gene expression by siRNA and shRNA, respectively and, confirmed reduced expression of SDC4 and the ITGA9 chain (Fig. S6G - J). Reduced levels of SDC4 (mimicking cell rounding by TNC (28)) did not abolish LPA-specific migration on FN/TNC suggesting that inactivation of syndecan-4 by TNC is not relevant for TNC transwell migration (Fig. 4H). In contrast, transient knockdown of ITGA9 induced actin stress fibers on FN/TNC and abolished TNC-specific transwell migration in KRIB cells, pointing at integrin α9β1 as relevant TNC receptor (Fig. 4H, S6E).
As TNC transwell migration occurs in the absence of actin stress fibers and, the knockdown of ITGA9 and of YAP impaired actin stress fibers and TNC-specific migration, we wanted to know whether TNC downregulates YAP target genes through integrin α9β1 By qPCR we indeed observed that the ITGA9 knockdown in KRIB cells increased expression of all tested TNC target genes on FN/TNC, reaching levels close to FN (Fig. 4I).
In addition, we analyzed whether TNC potentially also enhances transwell migration through an autocrine mechanism. Therefore, we measured Boyden chamber migration in control (shCTRL) and TNC knockdown (shTNC) KRIB cells (Fig. S6I) and found less TNC knockdown cells moving through the uncoated filter than shCTRL cells suggesting that endogenously made TNC is important (Fig. 4J). Next, we addressed whether TNC affects invasion through Matrigel and/or a type 1 collagen gel with a pore size that was shown to favor amoeboid migration (21, 22), respectively. We observed that less KRIB cells passed through Matrigel than through the collagen gel - coated substratum, yet Matrigel invasion was independent of TNC. In contrast, 3D migration through the collagen gel was TNC dependent as it was reduced upon TNC knockdown (Fig. 4K). We conclude that endogenously expressed TNC as well as a TNC substratum induces α9β1 signaling and promotes amoeboid-like transwell migration.
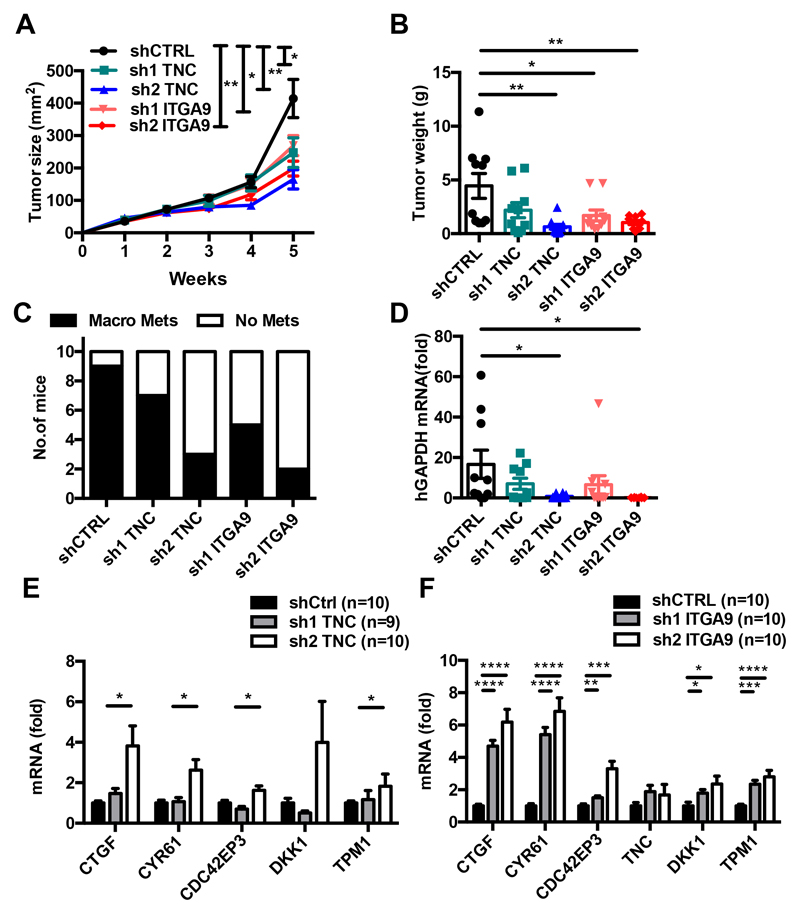
TNC and integrin α9β1 promote lung metastasis of osteosarcoma cells, associated with low levels of YAP target gene expression
We tested whether signaling by TNC and integrin α9β1 influences expression of YAP target genes and migration in vivo by generating KRIB cells with a knockdown of TNC and the ITGA9 chain, respectively and grafted cells subcutaneously into nude mice (Fig. S6I, J). We noticed stable knockdown of both genes in the arising tumors (Fig. S7A, B) and, that knockdown of TNC or ITGA9 reduced tumor growth (Fig. 5A, B). In addition, KRIB cells disseminated and formed lung metastasis, as assessed by the appearance of macrometastasis and expression of human GAPDH by qPCR. We observed that knockdown of either gene, TNC or ITGA9, reduced lung metastasis (Fig. 5C, D, S7C). A potential in vivo effect of TNC and/or integrin α9β1 on YAP target gene expression was addressed by measuring gene expression in KRIB tumors with knockdown of TNC or ITGA9, respectively. We observed that sh2TNC tumors displayed reduced tumor weight and less metastasis and, significantly increased expression of all tested TNC target genes (Fig. 5E). This was not the case for sh1TNC tumors (Fig. 5B, E). Also in human U87MG GBM cell-derived tumors, where TNC promoted tumor growth (4) TNC increased YAP target gene expression (Fig. S7D). Most importantly, in ITGA9 knockdown KRIB tumors gene expression of CTGF, CYR61 and CDC42EP3 was significantly increased (Fig. 5F).
Figure 5. TNC and integrin α9β1 increase subcutaneous tumor growth, enhance lung metastasis, and reduce YAP target gene expression in vivo.
Results for KRIB cells are shown. Growth curves (A) and weight (B) of subcutaneous tumors arising from control, TNC and ITGA9 knockdown cells are shown. (C) Number of mice with and without lung macrometastasis in each group. (D) Metastatic burden is determined by measuring human GAPDH in lung tissue of tumor bearing mice (fold change, qPCR). (E, F) Gene expression levels (qPCR) of the indicated genes in tumors derived from shCTRL, shTNC (E) and shITGA9 (F) cells. 10 tumors per group (A - E), except for (F) sh1TNC (9 tumors). Mean ± SEM.
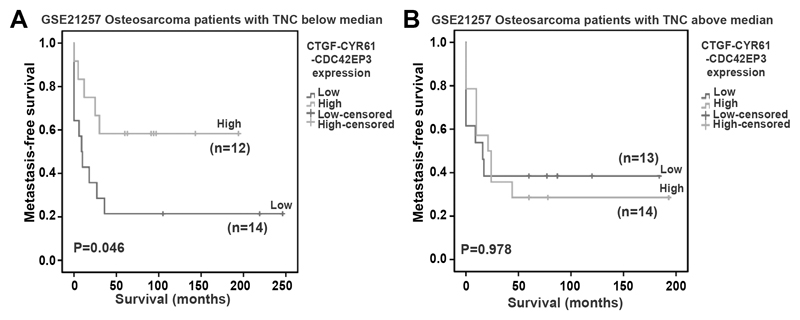
Predictive value of TNC regulated genes CTGF, CYR61 and CDC42EP3 for cancer patient survival
By having established a link of TNC to enhanced migration through abolishing YAP activity and increasing osteosarcoma metastasis we asked now whether this information could be of relevance for cancer patient survival. We analyzed expression of a YAP signature (41) that was downregulated by TNC (10) in a publicly available mRNA expression data set of osteosarcoma patients (n = 53) (GSE 21257) and patient survival (42), but noticed no link (unpublished observation). Yet, when we used the three genes CTGF, CYR61 and CDC42EP3 together, that are strongly repressed by TNC in our cellular and two animal models, we noticed a shorter metastasis-free survival of patients with tumors exhibiting abundant TNC yet below the median tumor level (Fig. 6A) (28). No correlation was seen in tumors with TNC levels above the median (Fig. 6B). Moreover, neither low nor high expression of each gene alone or in different combinations had any predictive value (Fig. S8). We also analyzed expression levels of the three-gene signature in a cohort of 46 GBM patient-derived tumor xenografts (PDX) where the gene signature of the experimental tumors correlated with invasiveness and worsened overall GBM patient survival (43). We observed that PDX tumors that had lower expression of CTGF, CYR61 and CDC42EP3 in a context of abundant but TNC expression below the median, represent a group of GBM patients with worsened progression-free survival (Fig. S9A, B). Low expression of either gene alone or in combinations of three had no relevance for patient prognosis (Fig. S10). Altogether we identified a short list of TNC-downregulated YAP target genes with correlation to worse prognosis in osteosarcoma and GBM patients.
Figure 6. Kaplan Meier survival analysis in osteosarcoma patients.
Kaplan-Meier survival analysis of patients with osteosarcoma upon stratification into tumors with abundant TNC expression below the median (A) and above the median (B) in combination with low (below the median) and high (above the median) expression of CTGF, CYR61 and CDC42EP3. The number of patients in each group is indicated within brackets, and p values indicate the significance of survival differences between the groups of individuals by log-rank test.
Discussion
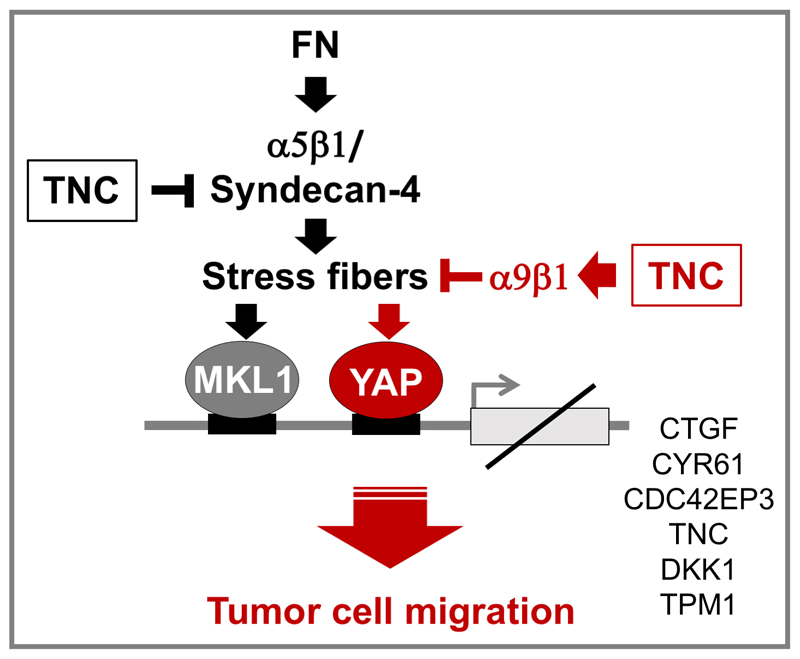
By using LOF and GOF approaches (Table S3, Fig. S9C), here we have shown a novel function of TNC in cancer. Our results suggest that TNC/integrin α9β1 signaling destroys actin stress fibers thus inhibiting YAP which promotes migration with amoeboid-like properties and metastasis. In addition to surface adsorbed TNC, also endogenously expressed TNC promotes transwell migration suggesting an autocrine, in addition to a paracrine, TNC/integrin α9β1 signaling loop. This mechanism may be relevant in tumors as we observed an increased expression of YAP target genes in grafted osteosarcoma cell derived tumors upon knockdown of TNC and ITGA9, respectively. Remarkably, knockdown tumors also caused less lung metastasis suggesting that TNC/integrin α9β1 signaling is enhancing lung metastasis. Our observations suggest that TNC matters in tumors as soon as it is expressed where promotion of tumor cell migration may be an important and early mechanism driving tumor malignancy (Fig. 7).
Figure 7. Summary of TNC effects on actin polymerization, gene expression and tumor cell migration.
Upon cell adhesion to FN through integrin α5β1/syndecan-4 cells establish actin stress fibers. MKL1 and YAP are two sensors of actin dynamics. In the presence of actin stress fibers both molecules are translocated to the nucleus where they act as co-transcription factors. TNC impairs actin polymerization and actin stress fiber formation in cells grown on FN by inhibiting integrin α5β1/syndecan-4 signaling (10). As we showed here, TNC also inhibits actin stress fiber formation through integrin α9β1. By GOF and LOF experiments we discovered that TNC downregulates some genes through impairing MKL1 (TPM1), or YAP (CTGF, CYR61) or MKL1 and YAP (TNC, CDC42EP3 and DKK1). TNC impairs MKL1 expression and nuclear translocation of YAP, respectively. Integrin α9β1 signaling is induced by a TNC substratum as well as by tumor cell-expressed TNC suggesting an autocrine and paracrine mechanism of action. TNC/integrin α9β1 signaling causes YAP impairment and repression of YAP target genes CTGF, CYR61 and CDC42EP3 thus, promoting transwell migration. Our results indicate that inhibition of YAP is a prerequisite for TNC-induced amoeboid-like migration. This mechanism may have clinical relevance as patients with osteosarcoma that have abundant yet TNC levels below the median together with low levels of CTGF, CYR61 and CDC42EP3 have worst prognosis.
As it was incompletely understood how TNC regulates gene expression and migration through cell adhesion, here we have revisited the effect of TNC on cell adhesion in context of FN. TNC competes syndecan-4 binding to FN, thus blocking integrin α5β1-mediated cell adhesion and actin stress fiber formation (9) which results in a pro-tumorigenic gene expression profile and repression of multiple cell adhesion associated genes (10). Here, we have identified the two actin cytoskeleton sensors MKL1 and YAP to be impaired by TNC which leads to repression of target genes. We identified three groups of genes that TNC represses through its impact on MKL1 (TPM1), YAP (CTGF, CYR61) or MKL1 and YAP (CDC42EP3, TNC and DKK1). Most importantly, through inhibition of YAP, TNC promotes transwell migration (Fig. 7, S9C).
TNC migration has amoeboid-like properties (38) as cells migrate through a collagen gel in a TNC-dependent manner whereas invasion through Matrigel is unaffected by TNC. Moreover, cells display an amoeboid-like phenotype such as a round morphology, lack of actin stress fibers and focal adhesions, inactive FAK and paxillin (9, 10, 28, 44, 19), and ROCK dependence (38), as inhibition of ROCK blocked TNC-mediated transwell migration. We have identified integrin α9β1 as novel upstream regulator of TNC-induced migration. Integrin α9β1 is known as receptor for TNC (39), and the TNC/integrin α9β1 interaction was recently shown to play a role in attraction of prostate cancer cells to bone tissue (45). Yet nothing was known how this interaction affects gene expression, cell migration or metastasis. Here, we have demonstrated for the first time that integrin α9β1 is promoting amoeboid-like migration by TNC. Moreover, we link migration by TNC through integrin α9β1 to destruction of actin stress fibers and inhibition of YAP which may be relevant for metastasis, as knockdown of either molecule reduces lung metastasis of grafted osteosarcoma cells.
In tumor tissue TNC is often co-expressed together with FN and other ECM molecules forming matrix tracks that serve as niches for tumor and stromal cells (6). These matrix dense areas may increase tissue stiffness and cellular tension due to multiple integrin binding opportunities. Indeed, in GBM high TNC levels were correlated with increased tissue stiffness (8). TNC may locally reduce cellular tension by counteracting adhesive signals by inhibiting syndecan-4 or activating integrin α9β1 (Fig. 7). In addition, we have shown that TNC downregulates its own expression. Thus, TNC is an ideal candidate to balancing cellular tension in cancer tissue.
We had investigated whether expression of TNC-downregulated genes correlate with cancer patient survival. Indeed low expression of three YAP target genes, CTGF, CYR61, and CDC42EP3 (that are strongly repressed by TNC in our in vitro and in vivo models) correlates with worst prognosis of patients with osteosarcoma and GBM when TNC is below the median expression. It has to be stressed that these TNC levels are still considerably high in comparison to normal tissue that poorly if at all expresses TNC (28). High TNC levels are correlated with bad patient survival (46) and, lower TNC levels are presumed to indicate a better prognosis (10, 47). Yet, some patients with lower TNC levels are still at high risk to die of their cancer, suggestive of a subgroup of yet unidentified patients with bad prognosis. Our result provides an opportunity to predict prognosis of osteosarcoma and glioma patients with moderate TNC expression, in particular when PDX expression data for GBM are available. Although tumors grown in a patient and in a mouse obviously differ, it is remarkable that the expression data from the PDX tumors have predictive value for GBM patient prognosis. Altogether, GBM patients with moderate TNC expression below the median, that usually are not considered to have a bad prognosis, may be recognized thanks to combined low expression of CTGF, CYR61 and CDC42EP3 in their PDX. Similarly, our predictive gene expression signature may allow identifying osteosarcoma patients with worse prognosis and in need of more forceful treatment.
As TNC expression is regulated by MKL1 and YAP, ablation of these activities may be considered for targeting TNC expression and its tumor promoting effects. Yet, our results suggest that inhibition of YAP may be detrimental, as cells with inactive YAP may be highly motile and metastatic in a TNC context. We believe that integrin α9β1 provides a better targeting opportunity, as inhibiting integrin α9β1 reduces tumor cell migration and metastasis.
Supplementary Material
Statement of Significance.
Results illuminate how the extracellular matrix glycoprotein tenascin-C in the tumor microenvironment promotes invasive migration and metastatic progression, by employing integrin α9β1, abolishing actin stress fiber formation, inhibiting YAP and its target gene expression, with potential implications for cancer prognosis and therapy.
Acknowledgement
We are grateful to G. Posern (Halle-Wittenberg University, Halle, Germany), R. Hynes (MIT, Cambridge, USA) for MKL1 molecules and SRF reporter plasmids, and YAP and TEAD reporter plasmids, respectively and, M. van der Heyden for technical assistance. This work was supported by grants from Worldwide Cancer Research (14-1070), INSERM, University Strasbourg, ANR (AngioMatrix), INCa, and Ligue contre le Cancer to GO and fellowship grants from the Chinese Scholarship Council (ZS), Ligue contre le Cancer (TR) and Fondation ARC, Association pour la recherche sur le cancer (AS, DM).
List of abbreviations
- CDC42EP3
CDC42 effector protein 3
- CTGF
connective tissue growth factor
- CYR61
cysteine rich angiogenic inducer 61
- DKK1
Dickkopf1
- ECM
extracellular matrix
- FN
fibronectin
- FOSL1
FOS like 1
- GBM
glioblastoma
- GLI2
GLI family zinc finger 2
- GOF
gain of function
- GSEA
Gene Set Enrichment Analysis
- ITGA9
integrin subunit alpha 9
- Jasp
Jasplakinolide
- LB
Latrunculin B
- LOF
loss of function
- LPA
Lysophosphatidic acid
- MKL1
megakaryoblastic leukemia 1
- MRTF-A
myocardin related transcription factor
- PDX
patient-derived tumor xenografts
- ROCK
Rho-kinase
- SDC4
syndecan 4
- SRF
serum response factor
- TAZ
transcriptional co-activator with PDZ-binding motif
- TEAD
TEA domain transcription factors
- TME
tumor microenvironment
- TNC
tenascin-C
- TPM1
tropomyosin 1
- TPM2
tropomyosin 1
- YAP
yes-associated protein
- ZYX
Zyxin
Footnotes
The authors declare no potential conflicts of interest
References
- 1.Midwood KS, Chiquet M, Tucker RP, Orend G. Tenascin-C at a glance. J Cell Sci. 2016;129:4321–4327. doi: 10.1242/jcs.190546. [DOI] [PubMed] [Google Scholar]
- 2.Oskarsson T, Acharyya S, Zhang XH-F, Vanharanta S, Tavazoi SF, Morris PG, et al. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saupe F, Schwenzer A, Jia Y, Gasser I, Spenlé C, Langlois B, et al. Tenascin-C Downregulates Wnt Inhibitor Dickkopf-1, Promoting Tumorigenesis in a Neuroendocrine Tumor Model. Cell Rep. 2013;5:482–492. doi: 10.1016/j.celrep.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Langlois B, Saupe F, Rupp T, Arnold C, Van der Heyden M, Orend G, et al. AngioMatrix, a signature of the tumor angiogenic switch-specific matrisome, correlates with poor prognosis for glioma and colorectal cancer patients. Oncotarget. 2014;5:10529–10545. doi: 10.18632/oncotarget.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupp T, Langlois B, Koczorowska MM, Radwanska A, Sun Z, Hussenet T, et al. Tenascin-C Orchestrates Glioblastoma Angiogenesis by Modulation of Pro- and Anti-angiogenic Signaling. Cell Rep. 2016;17:2607–2619. doi: 10.1016/j.celrep.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Spenlé C, Gasser I, Saupe F, Janssen KP, Arnold C, Klein A, et al. Spatial organization of the tenascin-C microenvironment in experimental and human cancer. Cell Adh Migr. 2015;9:4–13. doi: 10.1080/19336918.2015.1005452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imanaka-Yoshida K, Aoki H. Tenascin-C and mechanotransduction in the development and diseases of cardiovascular system. Front Physiol. 2014;5 doi: 10.3389/fphys.2014.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miroshnikova YA, Mouw JK, Barnes JM, Pickup MW, Lakins JN, Kim Y, et al. Tissue mechanics promote IDH1-dependent HIF1α-tenascin C feedback to regulate glioblastoma aggression. Nat Cell Biol. 2016;18:1336–1345. doi: 10.1038/ncb3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang W, Chiquet-Ehrismann R, Moyano JV, Garcia-Pardo A, Orend G. Interference of tenascin-C with syndecan-4 binding to fibronectin blocks cell adhesion and stimulates tumor cell proliferation. Cancer Res. 2001;61:8586–8594. [PubMed] [Google Scholar]
- 10.Ruiz C, Huang W, Hegi ME, Lange K, Hamou MF, Fluri E. Differential gene expression analysis reveals activation of growth promoting signaling pathways by tenascin-C. Cancer Res. 2004;64:7377–7385. doi: 10.1158/0008-5472.CAN-04-1234. [DOI] [PubMed] [Google Scholar]
- 11.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 12.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 13.Wada KI, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 14.Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurbuz I, Ferralli J, Roloff T, Chiquet-Ehrismann R, Asparuhova MB. SAP domain-dependent Mkl1 signaling stimulates proliferation and cell migration by induction of a distinct gene set indicative of poor prognosis in breast cancer patients. Mol Cancer. 2014;13:22. doi: 10.1186/1476-4598-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 18.Berlin Ö, Samid D, Donthineni-Rao R, Akeson W, Amiel D, Woods VL. Development of a novel spontaneous metastasis model of human osteosarcoma transplanted orthotopically into bone of athymic mice. Cancer Res. 1993;53:4890–4895. [PubMed] [Google Scholar]
- 19.Lange K, Kammerer M, Hegi ME, Grotegut S, Dittmann A, Huang W, et al. Endothelin receptor type B counteracts tenascin-C-induced endothelin receptor type A-dependent focal adhesion and actin stress fiber disorganization. Cancer Res. 2007;67:6163–6173. doi: 10.1158/0008-5472.CAN-06-3348. [DOI] [PubMed] [Google Scholar]
- 20.Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, Hynes RO. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci USA. 2012;109:E2441–E2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehmann S, te Boekhorst V, Odenthal J, Bianchi R, van Helvert S, Ikenberg K, et al. Hypoxia Induces a HIF-1-Dependent Transition from Collective-to-Amoeboid Dissemination in Epithelial Cancer Cells. Curr Biol. 2017;27:392–400. doi: 10.1016/j.cub.2016.11.057. [DOI] [PubMed] [Google Scholar]
- 22.Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, et al. Collagen-based cell migration models in vitro and in vivo. Semin Cell Dev Biol. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao B, Ye X, Yu J, Li L, Li W, Li S, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Descot A, Hoffmann R, Shaposhnikov D, Reschke M, Ullrich A, Posern G. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol Cell. 2009;35:291–304. doi: 10.1016/j.molcel.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Chiquet-Ehrismann R, Kalla P, Pearson CA, Beck K, Chiquet M. Tenascin interferes with fibronectin action. Cell. 1988;53:383–390. doi: 10.1016/0092-8674(88)90158-4. [DOI] [PubMed] [Google Scholar]
- 27.Van Obberghen-Schilling E, Tucker RP, Saupe F, Gasser I, Cseh B, Orend G. Fibronectin and tenascin-C: accomplices in vascular morphogenesis during development and tumor growth. Int J Dev Biol. 2011;55:511–525. doi: 10.1387/ijdb.103243eo. [DOI] [PubMed] [Google Scholar]
- 28.Lange K, Kammerer M, Saupe F, Hegi ME, Grotegut S, Fluri E, et al. Combined lysophosphatidic acid/platelet-derived growth factor signaling triggers glioma cell migration in a tenascin-C microenvironment. Cancer Res. 2008;68:6942–6952. doi: 10.1158/0008-5472.CAN-08-0347. [DOI] [PubMed] [Google Scholar]
- 29.Selvaraj A, Prywes R. Expression profiling of serum inducible genes identifies a subset of SRF target genes that are MKL dependent. BMC Mol Biol. 2004;5:13. doi: 10.1186/1471-2199-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SM, Vasishtha M, Prywes R. Activation and Repression of Cellular Immediate Early Genes by Serum Response Factor Cofactors. J Biol Chem. 2010;285:22036–22049. doi: 10.1074/jbc.M110.108878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asparuhova MB, Ferralli J, Chiquet M, Chiquet-Ehrismann R. The transcriptional regulator megakaryoblastic leukemia-1 mediates serum response factor-independent activation of tenascin-C transcription by mechanical stress. FASEB J. 2011;25:3477–3488. doi: 10.1096/fj.11-187310. [DOI] [PubMed] [Google Scholar]
- 32.Seo E, Basu-Roy U, Gunaratne PH, Coarfa C, Lim DS, Basilico C, et al. SOX2 Regulates YAP1 to Maintain Stemness and Determine Cell Fate in the Osteo-Adipo Lineage. Cell Rep. 2013;3:2075–2087. doi: 10.1016/j.celrep.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calvo F, Ege N, Grande-Garcia A, Hooper S, Jenkins RP, Chaudhry SI, et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat Cell Biol. 2013;15:637–646. doi: 10.1038/ncb2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wakatsuki T, Schwab B, Thompson NC, Elson EL. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J Cell Sci. 2001;114:1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 35.Visegrády B, Lőrinczy D, Hild G, Somogyi B, Nyitrai M. The effect of phalloidin and jasplakinolide on the flexibility and thermal stability of actin filaments. FEBS Lett. 2004;565:163–166. doi: 10.1016/j.febslet.2004.03.096. [DOI] [PubMed] [Google Scholar]
- 36.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 37.Willier S, Butt E, Grunewald TGP. Lysophosphatidic acid (LPA) signalling in cell migration and cancer invasion: a focussed review and analysis of LPA receptor gene expression on the basis of more than 1700 cancer microarrays. Biol Cell. 2013;105:317–333. doi: 10.1111/boc.201300011. [DOI] [PubMed] [Google Scholar]
- 38.Paňková K, Rösel D, Novotný M, Brábek J. The molecular mechanisms of transition between mesenchymal and amoeboid invasiveness in tumor cells. Cell Mol Life Sci. 2010;67:63–71. doi: 10.1007/s00018-009-0132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokosaki Y, Matsuura N, Higashiyama S, Murakami I, Obara M, Yamakido M. Identification of the ligand binding site for the integrin alpha9 beta1 in the third fibronectin type III repeat of tenascin-C. J Biol Chem. 1998;273:11423–11428. doi: 10.1074/jbc.273.19.11423. [DOI] [PubMed] [Google Scholar]
- 40.Kon S, Uede T. The role of α9β1 integrin and its ligands in the development of autoimmune diseases. J Cell Commun Signal. 2017 doi: 10.1007/s12079-017-0413-7. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buddingh EP, Kuijjer ML, Duim RAJ, Bürger H, Agelopoulos K, Myklebost O, et al. Tumor-infiltrating macrophages are associated with metastasis suppression in high-grade osteosarcoma: a rationale for treatment with macrophage activating agents. Clin Cancer Res. 2011;17:2110–2119. doi: 10.1158/1078-0432.CCR-10-2047. [DOI] [PubMed] [Google Scholar]
- 43.Joo KM, Kim J, Jin J, Kim M, Seol HJ, Muradov J, et al. Patient-Specific Orthotopic Glioblastoma Xenograft Models Recapitulate the Histopathology and Biology of Human Glioblastomas In Situ. Cell Rep. 2013;3:260–273. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 44.Orend G, Huang W, Olayioye MA, Hynes NE, Chiquet-Ehrismann R. Tenascin-C blocks cell-cycle progression of anchorage-dependent fibroblasts on fibronectin through inhibition of syndecan-4. Oncogene. 2003;22:3917–3926. doi: 10.1038/sj.onc.1206618. [DOI] [PubMed] [Google Scholar]
- 45.San Martin R, Pathak R, Jain A, Jung SY, Hilsenbeck SG, Piňa-Barba MC, et al. Tenascin-C and integrin α9 mediate interactions of prostate cancer with the bone microenvironment. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-0064. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Midwood KS, Hussenet T, Langlois B, Orend G. Advances in tenascin-C biology. Cell Mol Life Sci. 2011;68:3175–3199. doi: 10.1007/s00018-011-0783-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishihara A, Yoshida T, Tamaki H, Sakakura T. Tenascin expression in cancer cells and stroma of human breast cancer and its prognostic significance. Clin Cancer Res. 1995;1:1035–1041. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.