Abstract
The myotonic dystrophy-related Cdc42-binding kinases MRCKα and MRCKβ contribute to the regulation of actin-myosin cytoskeleton organization and dynamics, acting in concert with the Rho-associated coiled-coil kinases ROCK1 and ROCK2. The absence of highly potent and selective MRCK inhibitors has resulted in relatively little knowledge of the potential roles of these kinases in cancer. Here we report the discovery of the azaindole compounds BDP8900 and BDP9066 as potent and selective MRCK inhibitors that reduce substrate phosphorylation, leading to morphological changes in cancer cells along with inhibition of their motility and invasive character. In over 750 human cancer cell lines tested, BDP8900 and BDP9066 displayed consistent anti-proliferative effects with greatest activity in hematological cancer cells. Mass spectrometry identified MRCKα S1003 as an autophosphorylation site, enabling development of a phosphorylation-sensitive antibody tool to report on MRCKα status in tumor specimens. In a two-stage chemical carcinogenesis model of murine squamous cell carcinoma, topical treatments reduced MRCKα S1003 autophosphorylation and skin papilloma outgrowth. In parallel work, we validated a phospho-selective antibody with the capability to monitor drug pharmacodynamics. Taken together, our findings establish an important oncogenic role for MRCK in cancer, and they offer an initial preclinical proof of concept for MRCK inhibition as a valid therapeutic strategy.
Introduction
The actin-myosin cytoskeleton provides the structural framework that determines cell shape, and also is the source of physical force which directly powers biological activities including adhesion, migration and cell division. In addition, numerous processes are promoted by the actin-myosin cytoskeleton via less direct routes, such as gene transcription and proliferation, which collectively contribute to cancer (1). Although unlikely to be a primary cancer driver, accumulating evidence indicates that the actin-myosin cytoskeleton provides a critically important ancillary role in tumor growth and spread, which makes actin-myosin cytoskeleton regulators potential targets for cancer chemotherapy (2).
In non-muscle cells, a key event in promoting actin-myosin contractility is the phosphorylation of class 2 regulatory myosin light chains (MLC2) on Thr18 and Ser19 residues, which activates myosin ATP activity to drive the interaction of myosin heavy and light chain complexes with filamentous actin (F-actin) (3). Prominent MLC2 phosphorylating enzymes are the ROCK1 and ROCK2 kinases (4), which act downstream of the RhoA and RhoC small GTPases to regulate cytoskeleton organization and dynamics (5). However, ROCK1 and ROCK2 are not the only kinases regulated by Rho family GTPases; the myotonic dystrophy-related Cdc42-binding kinases (MRCK) interact with Cdc42 and catalyze phosphorylation of a similar set of substrates, including MLC2 (6,7). There are three MRCK kinases; the widely-expressed and closely-related MRCKα and MRCKβ, and the more divergent MRCKγ which is considerably more restricted in its tissue expression.
The roles of MRCK signaling in normal cell function and contributions to cancer are less well characterized than for ROCK, largely due to two historical factors: ROCK kinases were identified before (4) the MRCK kinases (8,9), and because of the discovery in 1997 of the relatively potent and selective small molecule ROCK inhibitor Y27632 (10), which has enabled two decades of research on ROCK biology. The large body of ROCK knowledge also catalyzed small molecule inhibitor discovery efforts, and ROCK inhibitors have been shown to have beneficial therapeutic effects in numerous pre-clinical cancer models (11), which has contributed to their further development for clinical use.
One aspect of cancer with which actin-myosin cytoskeleton regulators, including ROCK and MRCK, are clearly associated is tumor cell invasion and metastasis (12). The metastatic spread of cancer cells is the main cause of cancer mortality, believed to contribute up to 90% of all cancer related deaths (13). It has become increasingly appreciated that the same proteins that enable distant metastasis also contribute to primary tumor growth (14); therefore, drugs that restrict processes which contribute to cancer spread (e.g. motility, local invasion) also have beneficial effects on reducing tumor growth and progression.
It has been demonstrated in several contexts that the concerted inhibition of ROCK and MRCK kinases has greater effects than blocking either ROCK or MRCK alone (15–17). In addition, MRCK knockdown or inhibition alone was sufficient to reduce 3D invasion by squamous cell carcinoma (SCC) cells (18,19). These results suggest that there are likely to be clinical scenarios in which MRCK inhibitors would have therapeutic benefits, either alone or when combined with ROCK inhibition (20). However, the absence of potent and highly selective small molecule inhibitors has restricted research on MRCK relative to the advances made for other kinases for which useful chemical biology tools are readily available.
To determine how MRCK contributes to biological processes, including regulation of cell morphology and motility, and to evaluate MRCK as a cancer drug target, selective and potent MRCK inhibitors were developed, starting from a ligand-efficient fragment that was identified in a focused fragment library screen using an MRCKβ biochemical assay. Structure-guided fragment elaboration led to the novel MRCK inhibitors BDP8900 and BDP9066, which are considerably more potent and selective than the previously described BDP5290 (19), or the mixed ROCK-MRCK inhibitor DJ4 (17) or PKC-MRCK inhibitor chelerythrine (21). Screening of more than 750 human cancer cell lines, from more than 40 different cancer types, for anti-proliferative effects of BDP8900 and BDP9066 identified hematological cancers as the most sensitive malignancy type. Mass spectrometry led to the discovery and validation of MRCKα S1003 autophosphorylation as a pharmacodynamic biomarker that enabled evaluation of on-target drug action in tissues, as well as a biomarker of MRCKα activity in skin tumors. BDP9066 treatment of SCC cells induced changes in cell morphology, motility and invasion, while topical treatment of mice in a two-stage chemical carcinogenesis model of SCC significantly reduced papilloma growth and MRCKα S1003 autophosphorylation. These results demonstrate that the potent and selective inhibitors BDP8900 and BDP9066 are valuable chemical biology tools to identify MRCK biological functions and roles in cancer, and that MRCK inhibition has in vivo therapeutic effects on skin cancer in mice.
Methods
Kinase inhibition assays
MRCKα, MRCKβ, ROCK1 and ROCK2 assays were performed as described (19). Recombinant kinase proteins (Life Technologies) at 8-12 nM were incubated at room temperature for 60 min with 100 nM FAM-S6-ribosomal protein derived peptide (Alta Biosciences) with 1 µM ATP and 0.5 mM MgCl2 in 20 mM Tris buffer (pH 7.4) containing 0.01% (v/v) Tween20, 1 mM DTT for MRCKα and β; or 1 μM ATP, 10 mM MgCl2 in 20 mM Tris buffer (pH 7.5) containing 0.25 mM EGTA, 0.01% (v/v) Triton X-100, 1 mM DTT for ROCK1 and ROCK2. Reactions were stopped by adding 2 volumes 0.25% (v/v) IMAP binding reagent in 1X IMAP binding buffer A (Molecular Devices). After 30 min to allow binding reagent to bind phosphorylated peptide, fluorescence polarization was measured on a Tecan Saphire2 plate reader at excitation (470 nm) and emission (530 nm) wavelengths. Inhibition was calculated using no inhibitor (0%) or no enzyme (100%) controls. MRCKα and MRCKβ kinases assays by out-sourced supplier were carried out in 50 mM HEPES pH 7.5, 0.01% (v/v) BRIJ-35, 10 mM MgCl2, 1 mM EGTA and 2 µM Ser/Thr 13 peptide substrate, with 1 hour incubation before addition of development reagent and quantification by fluorescence resonance energy transfer (FRET). Kinase selectivity profiling was performed by Invitrogen with indicated concentrations of BDP8900 or BDP9066.
Cell culture
MDA MB 231 D3H2LN Luc cells (Caliper LifeScience, obtained 2006), MDA MB 231 D3H2LN Luc TetOn MRCKβ, ROCK1 and ROCK2 inducible cell lines (19), human cancer-associated fibroblasts (obtained 2008) and SCC12 human squamous cell carcinoma cells (18) (gift of Erik Sahai, Crick Institute, London UK; obtained 2006) were cultured as described (19). SCC12 organotypic invasion assays were performed as described (19).
CRISPR/Cas9 mediated MRCKα, MRCKβ, and MRCKαβ knockouts in MDA MB 231 D3H2LN Luc cells were generated using the plasmid lentiCRISPR (gift of Feng Zhang; Addgene plasmid # 52961) (22). Target sequences used were: MRCKα1: GATTGGTCGAGGAGCTTTTG, MRCKα2: TGTCTGGAGAAGTGCGTTTG, MRCKβ1: TGTCGCGGCGCAGGGCCGAG, MRCKβ2: CGTGGCCGAGTTCCTCGAGT. To generate double MRCKαβ knockout MDA MB 231 D3H2LN Luc cells, all 4 targeting sequences were used together.
Western blots
Cell lysates and immunoblot analysis were performed as described in (19). The following antibodies were used: rabbit anti-MRCKα pS1003 antibody raised by Eurogentec using the peptide AcNH-KGCPG-S(PO3H2)-TGFPP-CONH2, rabbit anti-pMLC2 Thr18/Ser19 (#3674; Cell Signaling Technology), mouse anti-MRCL3/MRLC2/MYL9 (sc-28329; Santa Cruz Biotechnology), mouse anti α-tubulin (T9026; Sigma), rabbit anti-ERK2 (Chris Marshall, Institute of Cancer Research, London UK), mouse anti-MRCKα (H00008476-M01; Abnova), mouse anti-MRCKβ (H00009578-M03; Abnova), mouse anti-MRCKαβ (MANDM1 6G8; Glenn Morris, Centre for Inherited Neuromuscular Disease, Oswestry UK) (23), mouse anti-ROCK1 (BD-611136; BD Biosciences), mouse anti-ROCK2 (BD-61062; BD Biosciences), mouse anti-Cas9 (C15200229; Diagenode), mouse anti-FLAG (F4042; Sigma). Secondary antibodies used were: goat anti-mouse IgG Dylight 800 (35521; Thermo Scientific), goat anti-rabbit IgG AlexaFluor 680 (A12076; Invitrogen), goat anti-rabbit IgG IR Dye 800 CW (926-32211; LI-COR Biosciences), goat anti-mouse IgG IR Dye 680 (926-68020; LI-COR Biosciences).
Protein crystallization, data collection and structure determination
Expression, purification and crystallization of the kinase domain of human MRCKβ as well as subsequent ligand soaks were performed as described previously (19). After a brief soak in mother liquor supplemented with 20% ethylene glycol, crystals were flash-frozen to 100K and data were collected at beamlines i24 (BDP8900 complex) and i02 (BDP9066 complex) of Diamond Light Source (Didcot, UK). Data were processed with XDS (24) and scaled using Aimless (25) (via xia2 (26) in the case of the BDP8900 complex) to resolutions of 1.68Å (BDP8900) and 2.00Å (BDP9066), then phased by manual placement of the protein component of the MRCKβ ADP complex structure (PDB ID 4UAK) followed by rigid body refinement with REFMAC (27). Models were improved through cycles of model building with Coot (28) and refinement with REFMAC. Ligand restraints and starting coordinates were generated using PRODRG (29), model validation was carried out using Coot and MolProbity (30). Data collection and final model statistics are presented in Supplemental Table 1. All structural images were rendered using PyMOL (PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
Cell viability
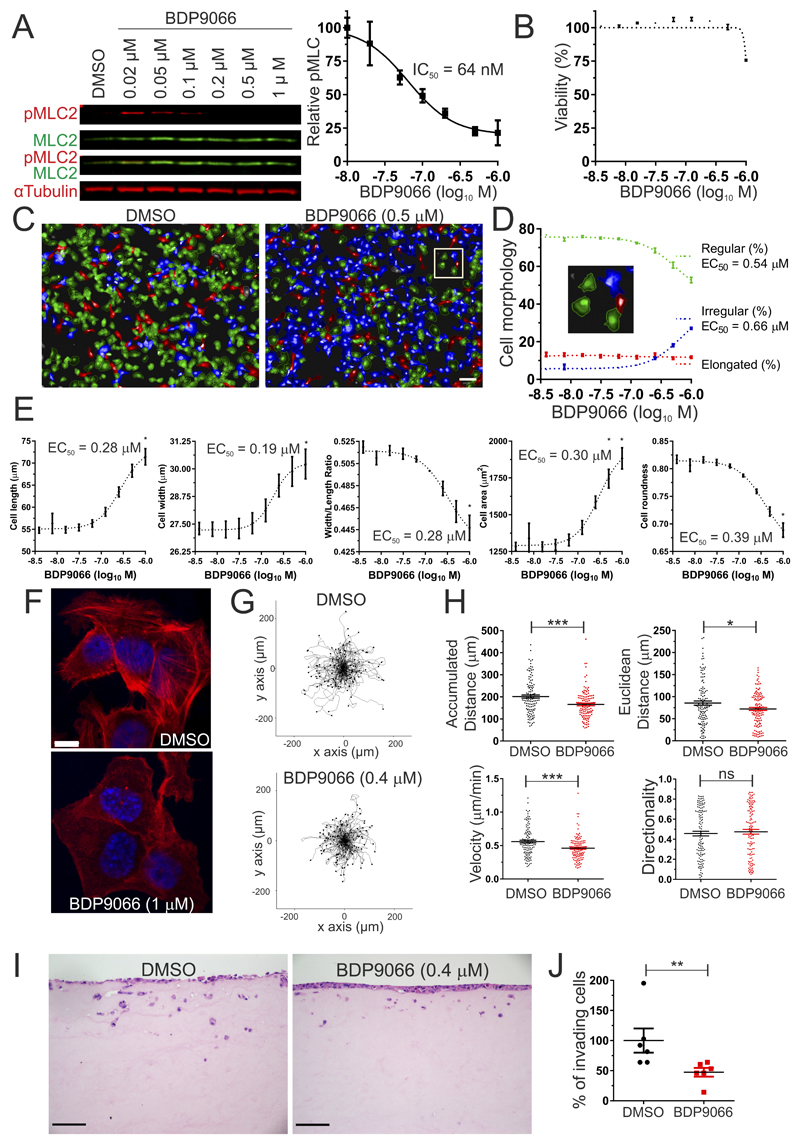
To determine the effect of BDP9066 on SCC12 cell viability, a dose curve was established using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). 1.5 × 104 cells were plated per well of a 96-well plate. After 24 hours, medium was removed and cells cultured for 24 hours with medium supplemented with DMSO vehicle or a dose range of BDP9066. Measurements were taken using a Tecan Safire plate reader.
Operetta high content morphology analysis
SCC12 cells were plated at 1.5 × 104 per well of a 96-well plate. After 24 hours, medium was removed and cells were cultured for 2 hours with medium supplemented with DMSO vehicle or a dose range of BDP9066. Cells were fixed with 4% paraformaldehyde in PBS for 15 minutes, permeabilized with 0.5% (v/v) Triton-X100 in PBS for 15 minutes and blocked in 1% (w/v) BSA in PBS for 30 minutes. Cells were stained with Alexa 568 Phalloidin (A12380; ThermoFisher Scientific), DAPI (D9564; Sigma) and Deep Red Whole Cell Stain (H32721; ThermoFisher Scientific). The cells were imaged and analyzed using the Operetta high content imaging system.
Immunofluorescence
SCC12 cells were seeded onto glass coverslips and allowed to settle and grow overnight. Next day, medium was replaced with medium containing 1 µM BDP9066 or DMSO vehicle. After 1 h of drug treatment, cells were fixed in 4% paraformaldehyde/PBS for 15 min and permeabilized with 0.5% Triton X-100/PBS for 5 min. F-actin was visualized by incubation with Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific). Cells were mounted with ProLong Diamond including DAPI (Molecular Probes). Images were taken on a Zeiss 710 confocal microscope.
Random migration
SCC12 cells were plated at 2× 104 per well of an Essen Bioscience ImageLock 96 well plate. After 24 hours of culture, medium was replaced with medium supplemented with either DMSO vehicle or 0.4 μM BDP9066. After 1 hour, plates were placed in an Incucyte Zoom apparatus to image cells every 15 minutes for 6 hours. Image stacks were analyzed using ImageJ with the Manual Tracking plugin, and 120 cells in 6 different wells per condition were tracked. Data was plotted using ImageJ with the Chemotaxis and Migration tool.
Plasmids and protein production
The pEF-FLAG-MRCKα plasmid containing N-terminal FLAG tagged full length MRCKα isoform b and the pEF-FLAG-MRCKα-K106M plasmid with a mutation of Lysine 106 to Methionine (K106M) were gifts from Simon Wilkinson (University of Edinburgh, Edinburgh UK). The pEF-FLAG-MRCKα-S1003A was generated by site directed mutagenesis of pEF-FLAG-MRCKα using Quickchange II XL (Agilent Technologies) following manufacturer recommendations. Primers used were: S1003A forward: aaaaaggatgtcctggtgcaactggctttccacct, S1003A reverse: aggtggaaagccagttgcaccaggacatccttttt. The pEF-FLAG-MRCKαΔC plasmid was created using sited directed mutagenesis of pEF-FLAG-MRCKα by inserting a stop codon after nucleotide 4860 corresponding to amino acid 1620. The resulting protein is 100 amino acids shorter than full length MRCKα isoform b. The primers used were: MRCKαΔC forward: gagtgctagcagtggctgattgtcagcaaggtcat, MRCKαΔC reverse: atgaccttgctgacaatcagccactgctagcactc.
A pGEX2T-MLC2 plasmid was used for bacterial production of recombinant MLC2. BL21 bacteria were transformed with pGEX2T-MLC2 and grown on ampicillin plates, 1 colony was picked and cultured overnight in 100 mL of LB medium supplemented with ampicillin. The culture was diluted 1 in 10 (30 mL in 300 mL of LB + ampicillin) and cultured for 1 hour, then 50 µM IPTG was added and incubated for 3 hours. Cultures were centrifuged at 4000 rpm for 15 minutes at 4°C and the pellet lysed in Tris buffered saline (TBS) with 5 mM MgCl2, 1 mM DTT, 1 mM PMSF. Lysates were sonicated 3 times for 1 minute on ice and then centrifuged at 10,000 rpm for 10 minutes at 4°C. The supernatant was collected and used for in vitro MLC2 phosphorylation assays.
MRCKα transfection and immunoprecipitation
HEK293 human embryonic kidney cells were plated at 1× 106per 10 cm culture dish. The next day, medium was replaced with OPTIMEM, and after 1 hour cells were transfected in OPTIMEM with 10 μg of pEF-FLAG-MRCKα, pEF-FLAG-MRCKαΔC, pEF-FLAG-MRCKα-K106M or pEF-FLAG-MRCKα-S1003A using Fugene HD (Promega). After 6 hours, medium was replaced with DMEM + 10% fetal calf serum (FCS) + L-glutamine. 2 days after the transfection, cells were placed on ice, washed in ice cold PBS and lysed in 1 mL of ice cold lysis buffer (TBS with 1 mM EDTA, 1% (v/v) Triton-X100, 1mM PMSF, 1X Complete protease inhibitor (Roche), 20 mM NaF, 20 mM β-glycophosphate, 0.2 μM Na3VO4, 20μg/mL Aprotinin). Lysates were then incubated on a rotating wheel for 30 minutes at 4°C, then centrifuged at 13,200 rpm for 10 minutes at 4°C and supernatants collected. The lysates were then either used directly for western blots or MRCKα was immuno-precipitated.
For immunoprecipitation, lysates were incubated with anti-Flag agarose beads (A2220; Sigma) on a rotating wheel for 2 hours at 4°C. Beads were washed 3 times in lysis buffer by successive centrifugations at 3,000 rpm for 1 minute at 4°C. For in vitro MRCKα auto-phosphorylation, phosphatase assays and MLC2 phosphorylation assays, beads were used as described in their respective sections. For mass spectrometry and western blots, beads were boiled at 95°C in 1% (w/v) SDS for 5 minutes, centrifuged at 3,000 rpm for 2 minutes and supernatants collected.
Cell treatment with MRCK inhibitors
SCC12 cells were plated at 5 × 105per well of a 6 well plate. After 24 hours, medium was replaced with medium supplemented with DMSO or a dose range of BDP9066. After 2 hours, cells were washed in PBS and lysed in 1% (w/v) SDS, 50 mM Tris-HCl, pH 7.4. Lysates were passed through QIAshredder spin columns (79654; Qiagen) and analyzed by SDS-PAGE.
HEK293 cells were transfected with pEF-FLAG-MRCKα. After 48 hours, medium was replaced with medium supplemented with either DMSO, 3 μM BDP5290 or 1 μM BDP9066. After 2 hours, cells were washed in PBS and lysed in 1% (w/v) SDS, 50 mM Tris-HCL pH 7.4. Lysates were passed through QIAshredder spin columns and analyzed by SDS-PAGE.
MDA MB 231 cells expressing doxycycline inducible ROCK1, ROCK2 or MRCKβ kinase domains were plated at 1.1 x 105 per well of a 12 well plate. After 24 hours, cells were treated with 1 µg/ml doxycycline for 18 hours to induce kinase domain expression and then tested with BDP8900 or BDP9066 at the concentrations indicated for 60 minutes. Cells were then washed with PBS and lysed with Tris-SDS lysis Buffer (50 mM Tris-HCl Ph 7.4, 0.5% (v/v) SDS, 1x PhosStop Inhibitors (04 906 837 001; Roche) and 1x Complete Protease Inhibitors (04 693 124 001; Roche). Whole cell lysates were clarified by passing through QIAshredder spin columns and analyzed by SDS-PAGE.
Mass spectrometry
Full length MRCKα isoform b and full length MRCKα isoform b K106M were expressed in HEK293 cells and immunoprecipitated with FLAG antibody in triplicate experiments. Samples were run on SDS-PAGE and gels stained using Instant Blue (Expedeon). MRCK bands were excised and digested with trypsin (Promega) as previously described (31)
Protein tryptic digests were separated by nanoscale C18 reverse-phase liquid chromatography using an EASY-nLC II (Thermo Scientific) coupled on-line to a Linear Trap Quadrupole (LTQ) Orbitrap Velos mass spectrometer (Thermo Scientific) via a nanoelectrospray ion source (Sonation). The mass spectrometer was operated in data-dependent acquisition. Fragmentation was performed on the ten most intense ions using collision-induced dissociation (CID) with multistage activation enabled and higher energy collision dissociation (HCD) in two separated acquisitions.
Raw data were processed with MaxQuant version 1.5.5.1 (32). Andromeda (33) peak list files (.apl) were converted to Mascot generic files (.mgf) using APL to MGF Converter (www.wehi.edu.au/people/andrew-webb/1298/apl-mgf-converter). MGF files were searched using Mascot (Matrix Science, version 2.4.1), querying the UniProt Homo sapiens database (09/07/2016; 92,939 entries) plus an in-house database containing common proteomic contaminants and the MRCKα K106M sequence. Mascot was searched assuming trypsin digestion, allowing two maximum missed cleavages, and with fragment ion and parent ion mass tolerances of 0.1 Da and 15 ppm, respectively. The iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. Methionine oxidation, and serine, threonine or tyrosine phosphorylation were specified in Mascot as variable modifications. MS/MS phosphopeptide data were manually curated with Xcalibur Qual Browser version 2.2 (Thermo Scientific), and the MS-Product utility of Protein Prospector v 5.12.4 (prospector.ucsf.edu/) was used to generate theoretical product ions fragmentation series.
Raw msms.txt data files from MaxQuant and Mascot DAT files were imported into Skyline to build a library of MRCKα peptides. Extracted ions chromatograms (XICs) of the 3 main isotopic peaks (30 K resolution at 400 m/z) of precursor ions from unmodified and phosphorylated MRCKα peptides that carried 2+ and 3+ charges were used for quantification of autophosphorylation sites. The peak areas of phosphopeptide 999-1009 that were measured in the 3 replicate experiments were normalized to “Global Standard” areas derived from the unmodified MRCKα tryptic peptides 310-315, 1113-1122 and 1268-1286.
In vitro MRCKα assays
For treatments with phosphatase and in vitro autophosphorylation, full length MRCKα or mutants bound to anti-FLAG beads were resuspended in 100 μL of phosphatase buffer with lambda protein phosphatase (P0753; New England Biolabs) following manufacturer’s instructions and incubated with constant agitation at 30°C for 1 hour. For samples treated for in vitro autophosphorylation and activity assays, beads were washed 3 times in PBS and 1 time in kinase buffer (20 mM Tris-HCl pH 7.4, 0.5 mM MgCl2, 0.01% (v/v) Tween20, 1 mM DTT). For autophosphorylation, the beads were re-suspended in 98 μL of kinase buffer supplemented with 2 μL of 5 mM ATP. For MLC2 phosphorylation assays, the beads were resuspended in 94 μL of kinase buffer supplemented with 2 μL of 5 mM ATP and 4 μL of MLC2. For CDC42 assays, the beads were resuspended in 97 μL of kinase buffer supplemented with 2 μL of 5 mM ATP and 1 μg of constitutively active CDC42 Q61L (Cytoskeleton, Inc. Cat. # C6101). The beads were then incubated with constant agitation at 30°C for 1 hour. To stop the reactions, 100 μL of boiling 2% (w/v) SDS was added to samples, and reactions were incubated for 5 minutes at 95°C. Samples were centrifuged at 3,000 rpm for 2 minutes and supernatants were collected and analyzed by western blot.
In vitro 32P incorporation
Immunoprecipitated full length MRCKα and MRCKα K106M bound to anti-FLAG beads were resuspended in kinase buffer supplemented with γ[32P]-labelled ATP with constant agitation at 30°C for 1 hour. Samples were washed 3 times in kinase buffer. Beads were resuspended in 50 μL of boiling 1% (w/v) SDS and incubated for 5 minutes at 95°C. Samples were then run on SDS-PAGE, gel was stained with Instant Blue (Expedeon) and an autoradiograph collected.
Dot blots and peptide competition
Two peptides including MRCKα isoform b S1003 were synthetized by Eurogentec. One peptide contained phosphorylated serine (P) = AcNH-KGCPG-S(PO3H2)-TGFPP-CONH2, the other contained non-phosphorylated serine (U) = AcNH-KGCPG-S-TGFPP-CONH2. A dose range of peptides P and U were spotted on Protran nitrocellulose membrane (Sigma) pre-soaked in TBS + 0.01% (v/v) Tween20 (TBST), and the membrane was dried. When dry, the membrane was blocked in TBST + 0.5% (w/v) BSA for 30 minutes and incubated overnight at 4°C with MRCKα-pS1003 antibody at 1:500 in TBST + 0.5% (w/v) BSA. Blots were imaged using SuperSignal™ West Femto Maximum Sensitivity Substrate (34095; Thermo Scientific).
For peptide competition, 2 μL of MRCKα-pS1003 antibody in 1 mL of TBST + 0.5% (w/v) BSA was incubated with 4 ng peptide P or 4 ng peptide U, and incubated for 2 hours. Solutions were centrifuged at 13,200 rpm for 10 minutes, supernatants collected and re-incubated with 4 ng peptide P or 4 ng peptide U for 2 hours. The solutions were centrifuged at 13,200 rpm for 10 minutes. Antibodies were then applied to western blots of immunoprecipitated MRCKα.
Cancer cell line screen
The Translational Cancer Genomics team at the Wellcome Trust Sanger Institute screened 757 cancer cell lines with BDP8900 and BDP9066. Cells were seeded in 1536 well plates and compounds were screened using a 7-pt dose response curve and half-log compound dilution series. Cell viability was measured using CellTitre Glo (Promega). Detailed information about the screening process can be obtained at www.cancerrxgene.org
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were dewaxed and rehydrated. Antigen retrieval was performed in a boiling water bath with 10 mM citric acid buffer at pH 6 for 20 minutes. After cooling, endogenous peroxidase was blocked with 3% (v/v) H2O2 for 15 minutes. Sections were subsequently blocked in PBS + 0.5% (v/v) goat serum for 30 minutes. Primary antibodies were incubated overnight at 4°C in PBS + 0.5% (v/v) goat serum: rabbit anti-MRCKα (ab96659; Abcam) was used at 1:100, rabbit anti-MRCKβ (HPA022821; Sigma) was used at 1:1000, rabbit anti-MRCKα pS1003 was used at 1:50. Envision + System-HRP labeled Polymer (Dako) and Liquid DAB + Substrate Chromogen (Dako) were ]used for visualization. For staining quantifications, sections were imaged using a Leica SCN 400f scanner. Intensity thresholds were defined for absence of staining, low intensity staining, medium intensity staining and high intensity staining of MRCKα pS1003 IHC pictures using Leica Slidepath image analysis software. Areas of normal skin and of papilloma were selected for each sample and analyzed. Automatic cell detection was performed and the cytoplasmic intensities of the MRCKα pS1003 staining were measured. Histoscores were measured by multiplying the percentage of low intensity staining in the area by 100 and adding it to the percentage of medium intensity staining in the area multiplied by 200 and adding it to the percentage of high intensity staining in the area multiplied by 300.
Mouse models
Parental MDA MB 231 D3H2LN Luc cells or MRCKαβ KO cells (3.5 x106) were subcutaneously injected in the right flank of CD-1 immunocompromised nude mice. Mice were culled and sampled 2 weeks post-injection before tumors reached 12 mm diameters.
Mouse genetic skin cancer models expressing activated Hras alone, or with c-Fos plus conditional deletion of Pten, in epidermal keratinocytes form papillomas that convert to carcinoma as described (34–36).
To study the effect of BDP9066 on MRCKα pS1003 staining in mouse skin, 5 FVB mice were treated topically with 50 μL of 80% (v/v) DMSO (vehicle) and 5 FVB mice were treated topically with of 25 μg of BDP9066 in 50 μL 80% (v/v) DMSO. Treatments were applied 4 times per mouse over 2 days. Mice were culled 2 hours after final treatment. Skin and blood BDP9066 concentrations were measured by Pharmidex. To study BDP9066 pharmacokinetics on mouse skin and blood, FVB mice were treated topically once with 10 μg or 25 μg BDP9066 in 80% (v/v) DMSO, 4 times daily with 25 μg BDP9066 in 80% (v/v) DMSO, or 8 times daily (4 successive days on, 2 days off, then 4 days on) with 80% (v/v) DMSO or 25 μg BDP9066 in 80% (v/v) DMSO. Mice were culled 2 hours, 4 hours, 8 hours or 24 hours after final treatment. Skin and blood BDP9066 concentrations were measured by Pharmidex.
For DMBA/TPA experiments, 40 FVB mice were treated topically with 25 μg DMBA in acetone on day 1. From day 5, 4.7 μg TPA in acetone was applied 3 times weekly. From day 5, 20 mice were treated with 25 μg BDP9066 in 50 μL 80% (v/v) DMSO and 20 mice were treated with 50 μL 80% (v/v) DMSO 5 times per week. Experimenters were fully blinded to the identity of treatment groups. Mice weights and general conditions were monitored at least 2 times per week, tumor sizes and numbers were recorded weekly. Mice were culled when papillomas reached 12 mm in diameter. Analysis of tumor volume and papilloma numbers were performed blinded to the identity of treatment groups. Skin and blood BDP9066 concentrations were measured by Pharmidex.
All mouse experiments have been conducted in accordance with UK Home Office regulations, and with the approvals of the Cancer Research UK Beatson Institute Animal Care and Use Committee (IACUC) and University of Glasgow Ethical Review.
Statistics
Data analysis was performed using GraphPad Prism. Statistical tests used for each data set are indicated in respective figure legends.
Results
Discovery and characterization of MRCK inhibitors BDP8900 and BDP9066
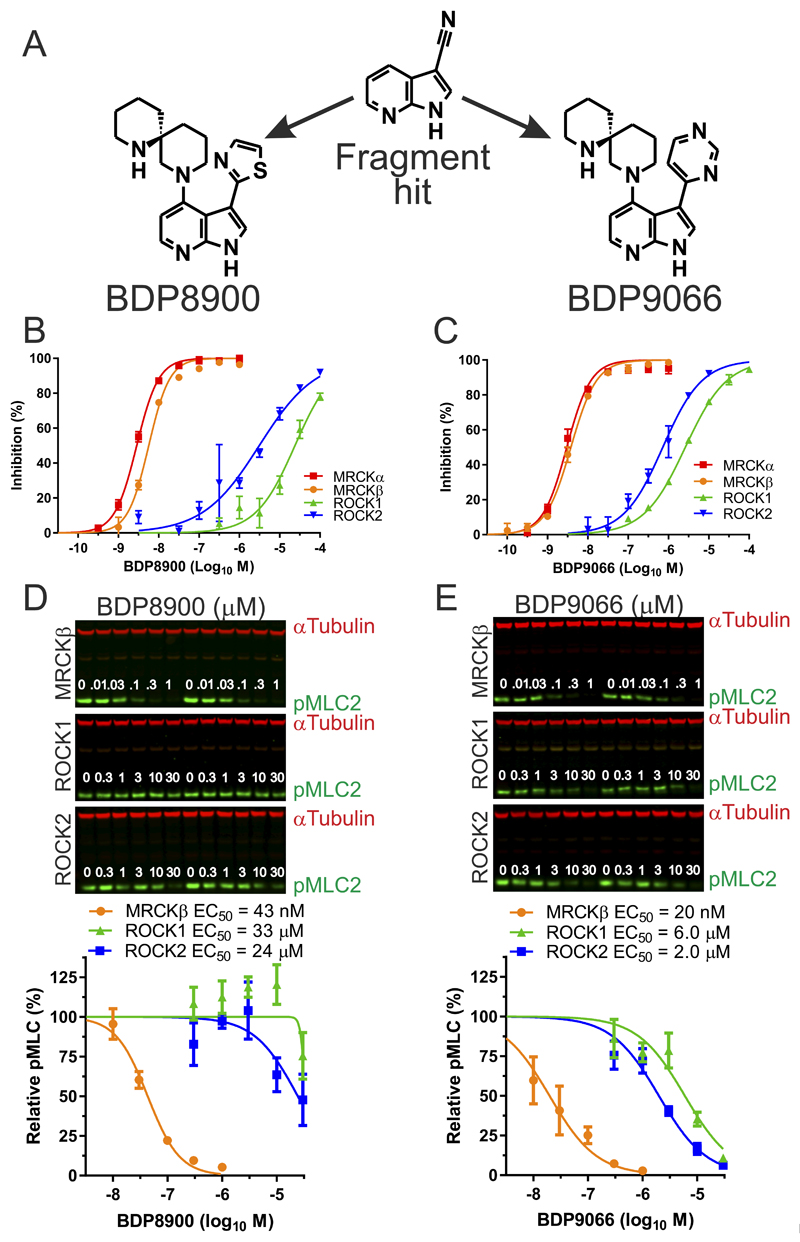
A focused fragment library was screened using an MRCKβ biochemical assay (16,19), and structure-guided elaboration of a ligand-efficient 7-azaindole-3-carbonitrile fragment hit (Figure 1A), prioritizing the identification of MRCK-selective inhibitors, led to the discovery of BDP8900 and BDP9066 (Figure 1A). Synthesis routes and methods for BDP8900 and BDP9066 will be described in a subsequent manuscript (in preparation). When the enzyme activities of MRCKα, MRCKβ, and the closely related AGC family kinases ROCK1 and ROCK2, were assayed in vitro at 1 µM ATP, which approached ATP Km values for these kinases under identical assay conditions (Supplemental Table 2), both compounds induced dose-dependent inhibition with greater selectivity for the MRCK kinases relative to ROCK kinases (Figure 1B and 1C). Using experimentally determined ATP Km values and the Cheng-Prusoff equation (37), calculated Ki values revealed affinities for MRCKα and MRCKβ that were 318 to 2371 fold greater for both compounds than for ROCK1 or ROCK2 (Supplemental Table 2). Inhibition of myosin light chain phosphorylation (pMLC2) in MDA MB 231 human breast cancer cells, engineered to express tetracycline-inducible MRCKβ, ROCK1, or ROCK2 kinase domains (19), by BDP8900 was >562 times more selective for MRCKβ relative to ROCK1 or ROCK2 (Figure 1D), and by BDP9066 was >100 times more selective for MRCKβ relative to ROCK1 or ROCK2 (Figure 1E).
Figure 1. Inhibition of MRCK activity in vitro and in cells by BDP8900 and BDP9066.
(A) Structures of 7-azaindole-3-carbonitrile hit fragment, BD8900 and BDP9066. (B) BDP8900 dose-response curves for inhibition of MRCKα, MRCKβ, ROCK1 and ROCK2 kinase activity in vitro at 1 µM ATP. (C) BDP9066 dose-response curves for inhibition of MRCKα, MRCKβ, ROCK1 and ROCK2 kinase activity in vitro at 1 µM ATP. Results shown are mean ± SD of duplicate independent replicates. (D) Cells expressing doxycycline-induced MRCKβ, ROCK1 or ROCK2 kinase domains were treated with BDP8900 at indicated concentrations for 60 minutes prior to lysis and quantitative western blotting. Inhibition of MLC2 phosphorylation by BDP8900 for each induced kinase domain. Results shown are mean ± SEM of 3-4 independent replicates. (E) Cells expressing doxycycline-induced MRCKβ, ROCK1 or ROCK2 kinase domains were treated with BDP9066 at indicated concentrations for 60 minutes prior to lysis and quantitative western blotting. Inhibition of MLC2 phosphorylation by BDP8900 for each induced kinase domain. Results shown are mean ± SEM of 3-4 independent replicates.
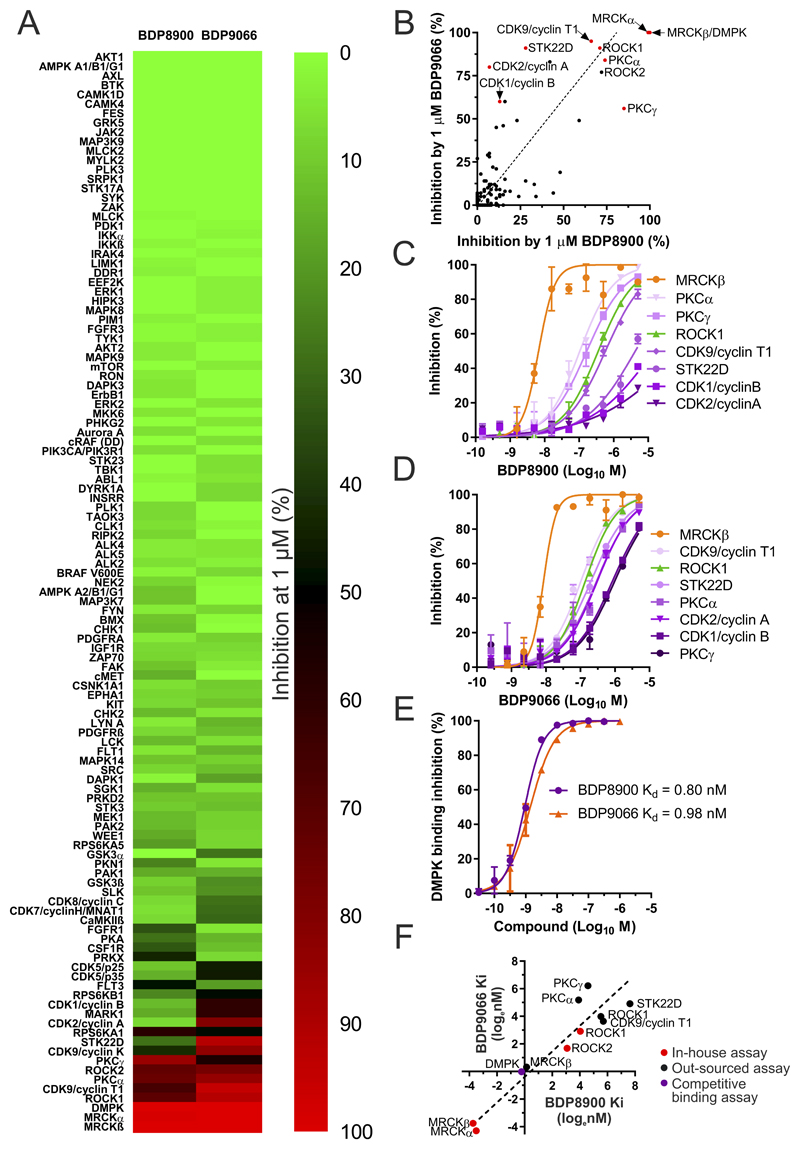
To extend selectivity profiling, a larger panel of 115 kinases was screened with 1 µM BDP8900 or BDP9066 either for inhibition of kinase activity at the apparent Km for ATP of each kinase, or for competitive inhibition of binding to the ATP site (Supplemental Table 3). Kinase inhibition results were mapped onto an annotated human kinase phylogenetic tree (38) using KinMap (39) (Supplemental Figure 1), and displayed as a heat map (Figure 2A). The majority of kinases were unaffected by BDP8900 or BDP9066 (indicated by green circles with labels, with >75% inhibition (red circles) of the AGC kinases PKCγ and DMPK by BDP8900 (Supplemental Figure 1A, Figures 2A and 2B), and ROCK1, ROCK2, CDK2/Cyclin A, CDK9/(Cyclin K or Cyclin T1), PKCα, TSSK1 (STK22D) and DMPK by BDP9066 (Supplemental Figure 1B, Figures 2A and 2B). To more precisely characterize the selectivity of BDP8900 and BDP9066, dose-response assays were performed against kinases identified by the screen as comparatively sensitive (Figure 2B, red dots). Under the assay conditions, BDP8900 and BDP9066 were markedly selective against all kinases tested (Figures 2C and 2D). Using ATP Km values and the Cheng-Prusoff equation (37), calculated Ki values from this screen revealed minimum fold-selectivity for MRCKβ that was 43 times greater for BDP8900 (Supplemental Table 4) and 27 times greater for BDP9066 against all kinases tested (Supplemental Table 5).
Figure 2. Detailed selectivity profiles for BDP8900 and BDP9066.
(A) Percentage kinase inhibition by 1 µM BDP8900 and BDP9066 were ranked and displayed by heat map. (B) Plot of inhibition by BDP8900 and BDP9066, kinases selected for detailed dose-response analysis indicated with red dot. ROCK1 was selected to additionally represent ROCK2 (black dot). Dotted line represents Deming regression (slope deviation from zero, p<0.0001). (C) BDP8900 dose-response curves for inhibition of indicated kinases in vitro. Results shown are mean ± SD of duplicate independent replicates. (D) BDP9066 dose-response curves for inhibition of indicated kinases in vitro. Results shown are mean ± SD of duplicate independent replicates. (E) BDP8900 and BDP9066 dose-response curves for competitive inhibition of tracer binding to DMPK in vitro, with calculated Kd values. Results shown are mean ± SD of duplicate independent replicates. (F) Natural log Ki or Kd (nM) values of BDP8099 and BDP9066 determined using in-house optimized assay conditions (red dots), by outsourced assays (black dots) or competition binding assay (purple dot). Dashed line represents Deming regression (slope deviation from zero, p<0.0001).
The myotonin-protein kinase, also known as DMPK, has high homology to MRCKα and MRCKβ with over 61% amino acid identity in its kinase domain compared to both their kinase domains (80.8% identity between MRCKα and MRCKβ). A competitive LanthaScreen Eu time-resolved fluorescence resonance energy transfer (TR-FRET) kinase binding assay in which BDP8900 and BDP9066 were assayed in vitro for displacement of a fluorescent tracer that binds to the DMPK ATP-binding site with 43 nM Kd (Figure 2E) yielded 0.80 nM (BDP8900) and 0.98 nM (BDP9066) Kd values using the Cheng-Prusoff equation (37).
Given the >1.0 Hill slopes (~1.9) for the MRCKβ inhibition curves for BDP8900 (Figure 2C) and for BDP9066 (Figure 2D), and the lower apparent potencies of both MRCK inhibitors in this assay format relative to the potencies observed assay using optimized conditions established in-house (Figures 1B and 1C), the selectivities of BDP8900 and BDP9066 were likely underestimated. Plotting the natural log Ki values determined in-house (Figure 2F, red dots) against out-sourced values (Figure 2F, black dots) indicated that the differences between in-house and outsourced Ki values for ROCK1 inhibition were only 4.4-fold for BDP8900 and 2.9-fold for BDP9066, but for MRCKβ differed by 41-fold for BDP8900 and 59-fold for BDP9066, consistent with on-target compound potencies being undervalued due to insufficient sensitivity of the out-sourced assays. While numerous parameters were rigorously optimized to maximize the sensitivity of the in-house kinase assays, including detection method, pH, MgCl2 and substrate concentrations, the out-sourced kinase assay conditions were not necessarily optimized for each kinase, but were generic for wide use against many different kinases. When compared against MRCKβ Ki values determined in-house (Figure 2F, red dots), the affinity of BDP8900 was more than 2083 times greater, and of BDP9066 was more than 1635 times greater, relative to all other kinases tested for inhibition of activity (Figure 2F, black dots), and 33-fold higher than the DMPK Kd determined for BDP8900 and 42-fold higher than for BDP9066 (Figure 2F, purple dot). Taken together, these results reveal that BDP8900 and BDP9066 are potent and highly selective inhibitors of MRCK with robust on-target actions in vitro and in whole cells.
Structures of BDP8900 and BDP9066 associated with MRCKβ kinase domain
To investigate the binding mode of these azaindole compounds, crystal structures of MRCKβ in complex with BDP8900 and BDP9066 were determined to 1.68Å and 2.00Å resolutions, respectively (Supplemental Table 1). The overall protein conformations in these complexes were essentially identical to those seen previously, with RMSD values after superposition onto all PDB-deposited MRCKβ structures (16,19) ranging from 0.5Å to 1.5Å for ≈395 Cα atoms. As noted previously (19), the glycine-rich loop (specifically residues 84–88) showed most conformational diversity. While in the MRCKβ·BDP9066 complex structure it was in the closed state observed for other inhibitor complexes (16,19), the MRCKβ·BDP8900 complex showed an open conformation similar to that described for the ADP complex structure (19). This was surprising given the similarity between the two compounds in structure (Figure 1A) and binding mode (see below). It should be noted that in both complexes there were hints of electron density supporting the ‘other’ loop conformation, suggesting that the glycine-rich loop is somewhat flexible and able to switch between closed and open states in the presence of azaindole inhibitors. It is possible that crystal-to-crystal variation, rather than ligand features, determine the apparently preferred state.
A second region of conformational divergence was formed by the C-terminal end of the activation loop (around Val235) and the αEF/αF loop (around Met255), though again both these loops were relatively flexible in all MRCKβ ligand complexes and there is no clear connection between the contents of the ATP-binding site and the major loop conformations.
BDP8900 and BDP9066 adopt highly similar poses in the ATP-binding site (Figures 3A and 3B): the all-atom coordinate RMSD for the two ligands after superposition of the two complex structures on the protein residues highlighted in Figure 3A was 0.105Å for 21 comparable atoms (i.e. excluding the thiazole/pyrimidine). The azaindole acts as the hinge binder, forming hydrogen bonds with the backbone carbonyl of Asp154 and the backbone amine of Tyr156. Both ligands preserved the two ‘pocket waters’ seen in the BDP5290 complex (19) and satisfy the hydrogen bonding potential of the outer (left in Figures 3A and 3B) water by accepting an H-bond to the thiazole (BDP8900) or pyrimidine (BDP9066) nitrogen. In addition, the second pyrimidine nitrogen of BDP9066 accepts a hydrogen bond of relatively poor geometry from Lys105.
Figure 3. Structure of MRCKβ in complex with BDP8900 and BDP9066.
(A) BDP8900 or (B) BDP9066 bound to the MRCKβ ATP-binding site in stereo views. Protein residues (grey) are labelled with the single-letter amino acid code and residue number, selected water molecules are indicated by red spheres. Ligands are shown in purple with σA-weighted |Fo|-|Fc|,ϕc electron density maps calculated prior to the initial inclusion of the ligand in refinement contoured at 3.0σ (dark blue). Potential hydrogen bonds are highlighted by dotted black lines. (C) Detailed view of the BDP9066 spiro moiety. The ligand is shown in cyan with a semitransparent surface. (D) Comparison of the binding modes of BDP9066 (cyan) and BDP5290 (blue, PDB ID 4UAL). Protein residues, pocket waters and potential hydrogen bonds are shown as before and colored to match the ligand.
The spiro moiety of the azaindole ligands provides a ‘plug’ for the ATP-binding site opening, interacting with residues around the glycine-rich loop (Ile82, Gly83, Arg84, Val90), the catalytic loop (Asp204, Asn205, Leu207) as well as Phe370 (Figure 3C). In the MRCKβ·BDP8900 complex, the open conformation of the glycine-rich loop results in a loss of the interaction with Arg84. In addition to the mostly hydrophobic direct protein-ligand contacts, the ring nitrogen of the distal piperidine formed hydrogen bonds with (and orders) two water molecules, one of which (on the right in Figure 3C) provided indirect interactions with the backbone of Asp204 and the side chain of Asp160. The other water was more exposed to bulk solvent and accordingly appears less well bound and less consistent in its interactions, which can involve Asp204 and (via additional ordered waters) Arg84, Asn205 and Asp218.
Comparing the binding modes of the azaindole ligands and the previously described pyrazole amide BDP5290 (19) reveals that most binding interactions are maintained across compound series despite their unrelated chemical structures and binding modes (Figure 3D). The azaindole and pyrazole cores form equivalent interactions with the hinge. Pocket water hydrogen bonding duties fulfilled by the amide linker in BDP5290 are taken over by the heterocyclic 3-substituent (thiazole or pyrimidine), which also partly fills the space occupied by the chloropyrazole of BDP5290. The diazaspiro[5.5]undecane substituent of the azaindole compounds provided an approximate replacement for the piperidine of BDP5290, though it appears that the shift in location and nitrogen position of the terminal piperidine moiety allowed BDP8900/9066 to form more extensive interactions with the protein, which may contribute to the higher affinity of these compounds for MRCKβ. Another notable difference between the two series is that, unlike the 2-pyridylpyrazole of BDP5290, the smaller azaindole of BDP8900/9066 did not displace the Phe370 side chain on binding (Figure 3D), avoiding energetic penalties associated with this conformational change and preserving a more compact ATP-binding site conformation for the spiro moiety to interact with, which could contribute to the improved affinity of the azaindoles compared to BDP5290.
We previously proposed the ligand interaction with the pocket waters as a determinant of selectivity for MRCK over ROCK, and this appears to hold true when comparing BDP8900 and BDP9066. The hydrogen bonding geometry between the thiazole of BDP8900 and the outer pocket water is closer to ideal than that seen for the pyrimidine of BDP9066, and correspondingly the former compound exhibit greater selectivity.
Effect of MRCK inhibitors on cancer cell line viability
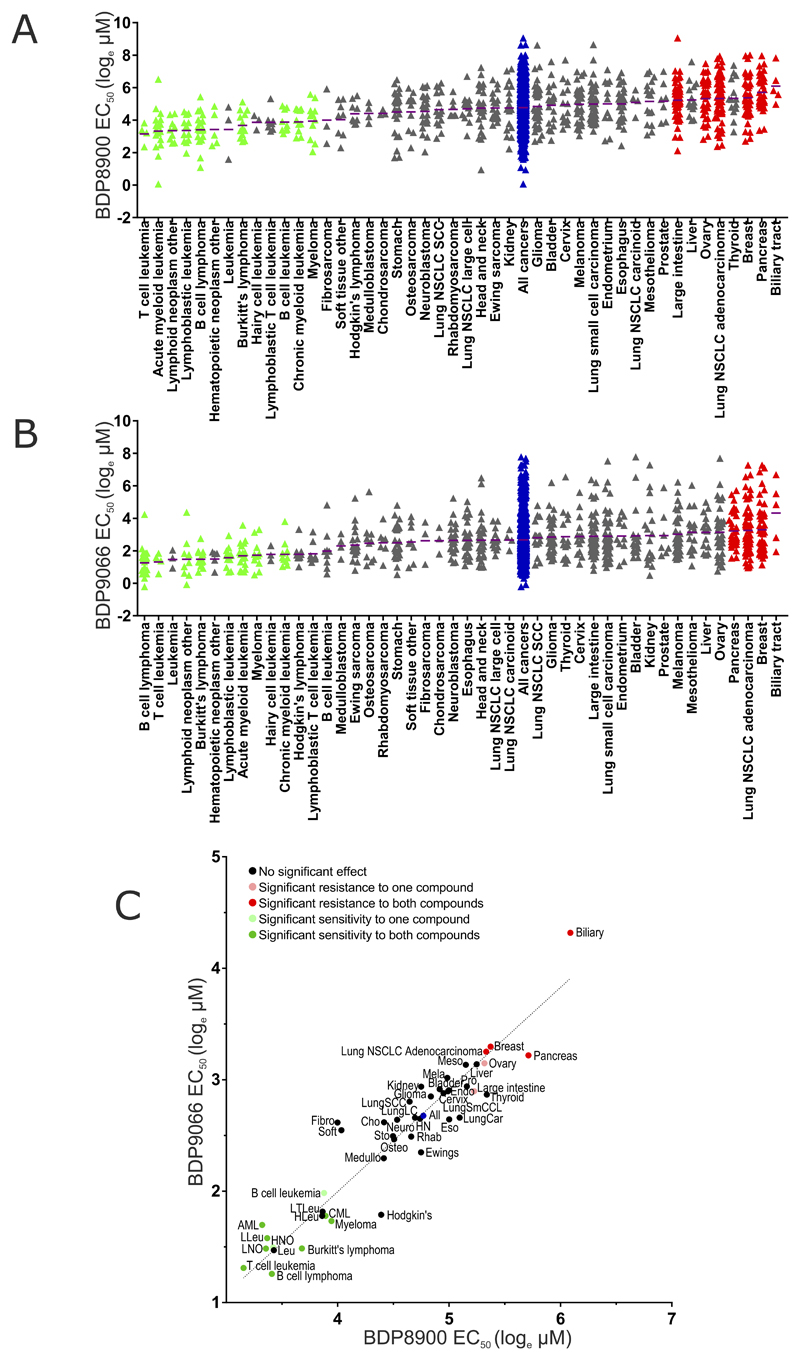
To identify cancer cell lines with significant sensitivity to the anti-proliferative effects of MRCK inhibitors as single agents, 757 cancer cell lines were screened using a 7-point dose range up to 10 µM of either BDP8900 or BDP9066, and viability monitored after 72 hours as described in (40). By comparing cell line EC50 values within a given cancer type against all cancer cell lines (Supplemental Table 6), significantly (p<0.05) sensitive (green) and resistant (red) cancer types were identified. Figure 4A shows natural log EC50 values (µM) of BDP8900 for each cell line within each cancer type, with purple lines indicating mean EC50, arranged from low to high mean EC50. Relative to the effect of BDP8900 on all cancer cell lines together (blue), there were 10 cancer types significantly sensitive and 6 cancer types significantly resistant. Similarly, 8 cancer types were significantly sensitive and 4 were significantly resistant to BDP9066 relative to all cancer cells (Figure 4B). When the mean EC50 values of BDP8900 and BDP9066 for each cancer type were plotted, Deming regression analysis revealed a strong correlation (p<0.001) and a line slope that approached 1.0, with common sensitive cancer types (green dots) and resistant cancer types (red dots) relative to all cancers (blue dots) indicated (Figure 4C). Plotting the EC50 values of BDP8900 and BDP9066 for each cell line also revealed a strong correlation (p<0.0001) by Deming regression and a line slope that approached 1.0 (Supplemental Figure 2). These results strongly support the conclusion that the effect of both inhibitors on cancer cell proliferation is due to a common mechanism of action that is highly likely to be on-target inhibition of MRCK activity. Interestingly, all 8 cancer types sensitive to both BDP8900 and BDP9066 were hematological cancers, with 2 additional blood cancers being significantly sensitive to one compound and others clustering, albeit not reaching statistical significance, to the sensitive end of the rankings.
Figure 4. Evaluation of cancer cell line sensitivities to MRCK inhibitors.
(A) Cancer cell lines (n = 757), divided into 45 cancer types, were treated with 7 concentrations of a half-log dilution series of BDP8900 (top concentration = 10 μM). After 72 hours, cell density was measured, EC50 values calculated and MANOVA performed to identify cancer types significantly (p<0.05) sensitive (green triangles) or resistant (red triangles) to BDP8900 relative to all cancers (blue triangles) considered together. Purple lines indicate mean EC50 for each cancer type. (B) Sensitivity or resistance to BDP9066 was determined as described above. Cancer types significantly (p<0.05) sensitive (red triangles) or resistant (red triangles) from all cancers (blue triangles) are indicated. (Abbreviations: NSCLC, Non-small cell lung cancer; SCC, Squamous cell carcinoma). (C) Mean EC50 values for each cancer type to BDP8900 and BDP9066 were plotted, dark green dots indicate cancer types significantly sensitive to both inhibitors, light green dots indicate significant sensitivity to one inhibitor, dark red dots indicate significant resistance to both inhibitors, pink dots indicate significant resistance to one inhibitor (all p<0.05), relative to all cancers (bleu dot). Deming regression indicates that the there is a significant (p<0.0001) relationship between the responses of cancer types to both drugs. (Abbreviations for sensitive cancer types: AML, Acute myeloid leukemia; CML, Chronic myeloid leukemia; HNO, Hematopoietic neoplasm other; LLeu, Lymphoblastic leukemia; LNO, Lymphoid neoplasm other).
MRCKα S1003 autophosphorylation as a biomarker of activity
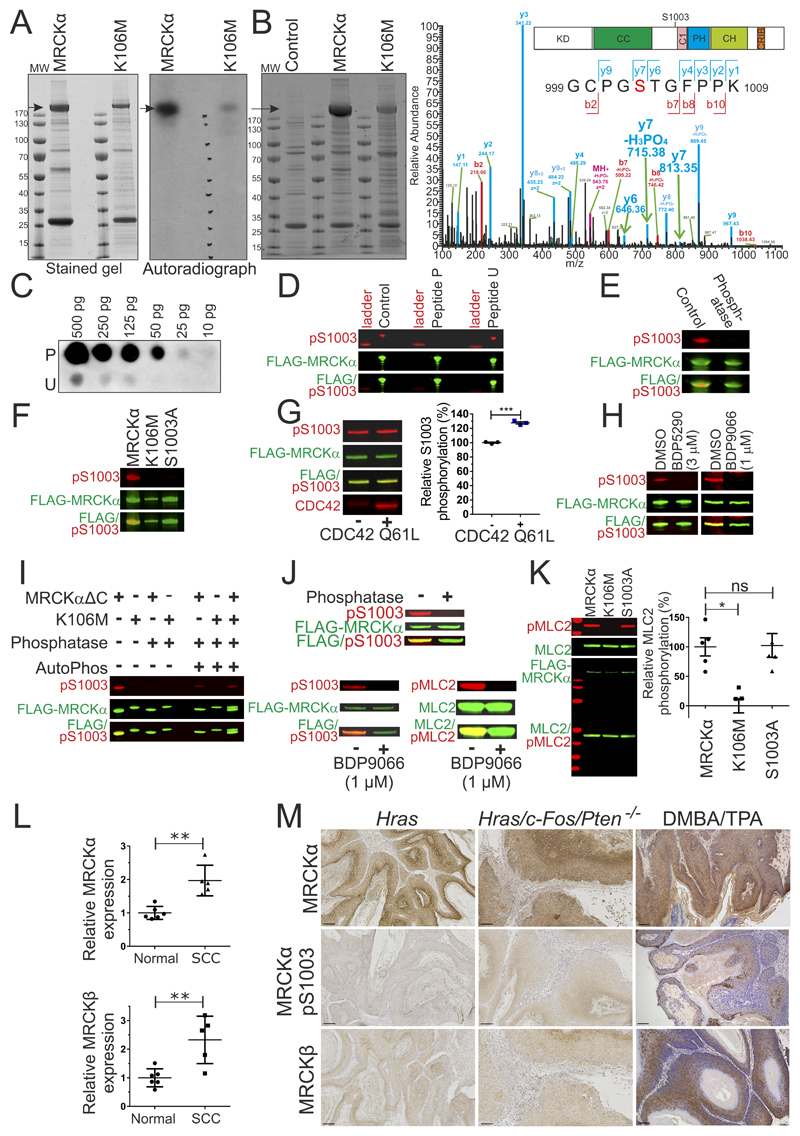
An additional strategy to identify cancer types that potentially would benefit from MRCK inhibitor treatment is to search for indications of increased MRCK signaling. By identifying phosphorylation sites that could be used as surrogate markers of activity, it was previously shown that ROCK1 S1333 phosphorylation (41) and ROCK2 S1366 phosphorylation (42) were associated with kinase activity, and that phosphorylation-state sensitive antibodies could be used to assess ROCK activity in clinical cancer samples (43). When wild-type MRCKα, or a kinase-dead version with a K106M mutation that prevents ATP binding, were expressed and immunoprecipitated from HEK293 cells (Figure 5A, left panel), in vitro kinase assays with [γ-32P] ATP revealed considerable 32P labelling of wild-type MRCKα, but not MRCKα K106M (Figure 5A, right panel). To identify autophosphorylation sites, wild-type MRCKα and MRCKα K106M were expressed and immunoprecipitated from HEK293 cells (Figure 5B, left panel), and phosphorylations measured by mass spectrometry (Figure 5B, right panel). A total of 22 phosphorylations were identified on MRCKα (Supplemental Table 7), of which 8 were not detected in MRCKα K106M, including S1003 (Figure 5B, y7 m/z peaks in right panel; Supplemental Table 8), consistent with these phosphorylations being autocatalyzed. S1003 is located between the coiled-coil (CC) and phorbol ester/diacylglycerol-binding C1 domains (Figure 5B, right panel inset). A comparison with MRCKβ revealed no conservation of MRCKα autophosphorylated residues (Supplemental Figure 3).
Figure 5. MRCKα S1003 autophosphorylation as a biomarker of activity.
(A) Stained SDS-PAGE gel (left) of FLAG immunoprecipitated FLAG-MRCKα and FLAG-MRCKα K106M after in vitro 32P incorporation assay. Arrow indicates MRCKα. Gel autoradiograph (right) shows 32P labelling of MRCKα. MW = molecular weight. (B). Stained SDS-PAGE gel (left) of FLAG-immunoprecipitates from control HEK293 cells, or cells transfected with pEF-FLAG-MRCKα or pEF-FLAG-MRCKα K106M. Arrow indicates MRCKα. MW = molecular weight. Higher energy collision dissociation (HCD) MS/MS fragmentation spectra (right) of MRCKα tryptic peptide 999-1009 containing S1003 (red lettering) in MRCKα or MRCKα K106M (phosphorylation inferred from y7, y7-H3PO4 and y6 signals indicated with green arrows). See Supplemental Table 8 for theoretical y and b fragmentation ion series masses. Diagram of MRCKα protein domains (inset) illustrating location of S1003. (KD = kinase domain, CC = Coiled-coiled domain, C1 = C1 diacylglycerol binding domain, PH = Pleckstrin homology domain, CH = Citron Homology domain, CRIB = Cdc42/Rac interactive binding motif). (C). Dot blot of MRCKα peptides (10 to 500 pg) containing phosphorylated S1003 (peptide P) or unphosphorylated S1003 (peptide U), stained with pS1003 antibody (D). Western blots of FLAG-immunoprecipitated FLAG-MRCKα stained with untreated pS1003 antibody, or pS1003 antibody pre-incubated with phosphorylated S1003 peptide P or unphosphorylated peptide U (E). Western blots of FLAG-immunoprecipitated FLAG-MRCKα, incubated in phosphatase buffer without (control) or with lambda phosphatase (phosphatase) (F). Western blot of FLAG-immunoprecipitated FLAG-MRCKα, FLAG-MRCKα K106M or FLAG-MRCKα S1003A. (G). Western blot (left) of FLAG-immunoprecipitated FLAG-MRCKα incubated in kinase buffer in the absence or presence of recombinant CDC42 Q61L protein. Relative S1003 phosphorylation revealed that CDC42 significantly increased autophosphorylation. Results shown are means ± SEM from 3 experiments. Student’s t-test (*** = p<0.001). (H). Western blots of HEK293 cells expressing FLAG-MRCKα treated with DMSO, 3 µM BDP5290 or 1 µM BDP9066 (I). Western blots of FLAG immunoprecipitated FLAG-MRCKαΔC or FLAG-MRCKα K106M untreated or treated with lambda phosphatase, or treated with lambda phosphatase and then incubated in kinase buffer to allow autophosphorylation (AutoPhos). (J) Western blots of FLAG-immunoprecipitated FLAG-MRCKα, untreated or treated with lambda phosphatase (upper panel). Incubation with kinase buffer resulted in autophosphorylation (pS1003; lower left panel) and recombinant MLC2 substrate phosphorylation (pMLC2; lower right panel) (K). Western blots of FLAG-immunoprecipitated FLAG-MRCKα, FLAG-MRCKα K106M or FLAG-MRCKα S1003A incubated in vitro with recombinant MLC2 in a kinase buffer (left). Relative MLC2 phosphorylation by each kinase revealed that K106M significantly reduced activity, while S1003A was not different from wild-type. Results shown are means ± SEM from 5 independent experiments. One way Kruskal-Wallis ANOVA followed by post-hoc Dunn’s multiple comparison (* = p<0.05). (L) Relative MRCKα (upper) and MRCKβ (lower) expression was significantly higher in cutaneous SCC relative to normal human skin determined by microarray analysis (45). Results shown are means ± SD from 6 normal skin and 5 SCC patients. Two-tailed Mann-Whitney test of significance (** = p<0.01). (M) Immunohistochemistry of MRCKα, MRCKα pS1003 or MRCKβ in benign papillomas from mice expressing oncogenic Hras in epidermal keratinocytes, or in well-differentiated squamous cell carcinomas resulting from Hras/c-Fos/Pten-/- genetic modifications or DMBA/TPA chemical carcinogenesis. Scale bars = 100 µm.
A polyclonal phosphospecific antibody against pS1003 was raised using the phospho-peptide (P) AcNH-KGCPGpSTGFPP-CONH2. Dot blots with varying quantities (10-500 pg) of phosphorylated (P) or unphosphorylated peptides (U) confirmed the phosphorylation-selective binding of this antibody (Figure 5C). Detection of immunoprecipitated FLAG-tagged MRCKα by western blotting was blocked by pre-incubation of the antibody with phosphopeptide P, but not peptide U (Figure 5D). Incubation of immunoprecipitated FLAG-tagged MRCKα with lambda phosphatase also led to loss of pS1003 antibody reactivity (Figure 5E). Immunoreactivity with the pS1003 antibody was detected on immunoprecipitated FLAG-tagged MRCKα, but not kinase-dead K106M or non-phosphorylatable S1003A mutants, consistent with this site being autophosphorylated (Figure 5F).
To establish if MRCKα S1003 autophosphorylation could be used as a MRCKα activity biomarker, recombinant constitutively-active CDC42 Q61L was incubated with immunoprecipitated FLAG-tagged MRCKα, which resulted in a significant ~30% increase in S1003 autophosphorylation (Figure 5G). Treatment of HEK293 cells expressing FLAG-tagged MRCKα with DMSO vehicle, 3 µM BDP5290 (19) or 1 µM BDP9066 MRCK inhibitors blocked pS1003 immunoreactivity (Figure 5H), indicating that the phospho-selective antibody could be used as a surrogate marker of MRCKα activity.
To determine the mechanism of MRCKα S1003 autophosphorylation, a FLAG-tagged MRCKα mutant lacking the last 100 amino acids (MRCKαΔC), or the kinase-dead K106M mutant, were expressed and purified from HEK293 cells. Following FLAG-immunoprecipitation and lambda phosphatase treatment to reverse S1003 phosphorylation, purified proteins were incubated with Mg2+-ATP to allow autophosphorylation. While the faster-migrating MRCKαΔC was able to autophosphorylate S1003 in cis when alone or with K106M, there was no evidence of trans phosphorylation of the larger K106M mutant by active MRCKαΔC (Figure 5I). Therefore, MRCKα utilizes an intra-molecular mechanism for S1003 autophosphorylation. When dephosphorylated MRCKα (Figure 5J, upper panel) was allowed to phosphorylate itself and recombinant MLC2 in the same in vitro reaction, both autophosphorylation (Figure 5J, lower left panel) and MLC2 phosphorylation (Figure 5J, lower right panel) were blocked by BDP9066, supporting the relationship between MRCKα S1003 autophosphorylation and kinase activity.
To characterize whether S1003 autophosphorylation is required for substrate phosphorylation, FLAG-tagged MRCKα, kinase-dead K106M and non-phosphorylatable S1003A mutants were immunoprecipitated from HEK293 cells and assayed for MLC2 phosphorylating activity in vitro. While kinase-dead K106M was significantly less active than wild-type MRCKα or the S1003A mutant, there was no significant difference in activity between wild-type MRCKα or the S1003A mutant (Figure 5K). These results indicate that S1003 autophosphorylation is not required for MRCKα activation and substrate phosphorylation.
MRCK expression and activation in mouse models of skin cancer
Analysis of publicly available human microarray data using Oncomine (44) revealed significantly elevated levels of both MRCKα and MRCKβ in human epidermal squamous cell carcinoma (SCC) relative to normal skin (Figure 5L) (45). Previous studies revealed a role for MRCKα in contributing to SCC in genetically modified (GM) mice expressing an oncogenic Hras transgene combined with Notch1 inhibition (46). To determine whether MRCKα and MRCKβ expression or activation were associated with SCC in additional GM mouse models, antibodies against MRCKα, MRCKα pS1003 and MRCKβ were first validated for immunohistochemistry (IHC) using MDA MB 231 D3H2LN human breast cancer cells (47) knocked out for MRCKα and MRCKβ by CRISPR/Cas9 (Supplemental Figure 4A). Parental and MRCKαβ knockout (KO) cells were grown subcutaneously in immunocompromised nude mice for 2 weeks. Sections of parental cell xenografts stained with the 3 antibodies were positive, but staining was abrogated in sections of KO cell xenografts (Supplemental Figure 4B). Antibody phosphoselectivity was validated when MRCKα pS1003 staining of MDA MB 231 subcutaneous tumors was blocked by antibody preincubation with phosphopeptide P, but not unphosphorylated peptide U (Supplemental Figure 4C).
MRCKα, MRCKα pS1003 and MRCKβ immunoreactivity were examined in GM mouse SCC models expressing epidermis-targeted activated Hras alone (34), or in combination with epidermis-targeted transgenic c-Fos and conditional epidermal deletion of Pten (35), which give rise to stable papillomas that may stochastically convert to malignancy via additional events such as p53 loss (36). MRCKα was expressed at moderately high levels in benign Hras and Hras/c-Fos/Pten-/- tumors, which increased further following malignant conversion to become strong and uniform in invasive SCC layers (Figure 5M). MRCKα pS1003 and MRCKβ were expressed at low levels in benign tumors with slight increases in early stage carcinomas (Figure 5M). MRCKα pS1003 expression was mainly confined to post-mitotic, differentiating SCC layers, while MRCKβ displayed weak expression maintained in proliferative layers and strong expression in occasional invasive cells. The observations were similar in the 7,12-Dimethylbenz[a]anthracene (DMBA)/12-O-tetradecanoylphorbol-13-acetate (TPA) (DMBA/TPA) two-stage chemical carcinogenesis SCC model, with moderate to high levels of MRCKα and MRCKβ, and a restricted pattern of MRCKα pS1003 staining in post-mitotic differentiating SCC cells (Figure 5M). These results indicate that MRCKα and MRCKβ expression, and MRCKα activation are associated with mouse skin tumors induced by genetic modifications and chemical carcinogenesis.
BDP9066 inhibits MLC phosphorylation and blocks SCC12 squamous cell carcinoma motility and invasion
It was previously determined that the phosphorylation of MLC and invasion of human SCC12 cells in an organotypic 3D collagen invasion model was sensitive to combined MRCKα/MRCKβ knockdown (18). Furthermore, the MRCK inhibitor BDP5290 also reduced MLC phosphorylation and inhibited SCC12 organotypic invasion (19). To investigate the effect of BDP9066 on pMLC2 in SCC12 cells, which express MRCK but no detectable DMPK (Supplemental Figure 5), varying BDP9066 concentrations incubated with cells for 2 hours led to dose-dependent inhibition of MLC2 phosphorylation (Figure 6A, left) with an EC50 = 64 nM (Figure 6A, right), corresponding to a 5-fold increase in cellular potency relative to BDP5290 (19). SCC12 cell viability following 24 hour treatment was unaffected by all BDP9066 concentrations up to 0.5 µM (Figure 6B), despite maximal pMLC2 inhibition at this concentration (Figure 6A), with a 25% decrease in viability at 1 μM (Figure 6B). These results indicate that BDP9066 is relatively non-toxic at concentrations that profoundly inhibit substrate phosphorylation.
Figure 6. BDP9066 affects MLC2 phosphorylation, morphology, migration and invasion of human SCC12 squamous cell carcinoma.
(A) Western blots (left) showing dose dependent inhibition (right) of MLC2 phosphorylation by BDP9066 in SCC12 cells. Results shown are means ± SEM from 3 independent replicates. (B). SCC12 viability as a function of BDP9066 concentration after 24 hours treatment. Results shown are means ± SEM from 3 independent replicates. (C). High content analysis of SCC12 morphology following treatment with DMSO vehicle or 0.5 µM BDP9066 for 2 hours. Multiparametric analysis was used to define regular (green), elongated (red) and irregular (blue) SCC12 cell morphologies. Scale bar represents 100 µm. (D) Percentages of each SCC12 cell shape as functions of BDP9066 concentration after 2 hours treatment. Results shown are means ± SEM from 3 independent replicates. (E) Individual SCC12 cell morphology parameters (cell length, cell width, width/length ratio, cell area, cell roundness) as functions of BDP9066 concentration after 2 hours treatment. Results shown are means ± SEM from 3 independent replicates. (F). Representative images of phalloidin-stained filamentous actin structures in SCC12 cells treated with DMSO or 1 μM BDP9066 for ~18 hours. Scale bar = 10 µm. (G). Tracks of SCC12 cell migration (120 cells) treated with DMSO (left) or 0.4 μM BDP9066 (right) over 6 hours. Each line represents the path of a single cell from the origin, with final position indicated by a dot. (H). Accumulated distance, Euclidean distance, velocity and directionality of SCC12 cells treated with DMSO or 0.4 μM BDP9066 for each tracked cell over 6 hours. Results shown are means ± SEM from 120 individual cells. Two tailed unpaired t-tests were used to determine significance (* = p<0.05, *** = p<0.001). (I). Representative images of organotypic invasion by SCC12 cells cultured with medium supplemented with DMSO (left) or 0.4 μM BDP9066 (right). Scale bars represent 100 µm. (J). Percentage of invading SCC12 cells in organotypic invasion assays following treatment with DMSO or 0.4 μM BDP9066. Results shown are means ± SEM from 6 independent replicates. Two-tailed Mann-Whitney test of significance (** = p<0.01).
Treatment of SCC12 cells with non-toxic 0.5 μM BDP9066 led to changes in cell morphology, with reduced proportions of regularly (rounded, few protrusions) shaped cells (Figure 6C, green cells), and increased irregularly (greater spreading, increased protrusions) shaped cells (Figure 6C, blue cells) as determined by high content imaging. By varying BDP9066 concentrations, it was determined that there was a parallel re-distribution from regular to irregular cell morphologies with similar EC50 values (Figure 6D). The proportion of elongated cells (Figure 6D, red cells) did not vary with BDP9066 concentrations (Figure 6D). When individual morphological parameters were examined, there were dose-dependent increases in cell length and width (Figure 6E, left panels), with dose-responsive changes in length exceeding width (Figure 6E, middle panel). The collective effect of these BDP9066 induced morphological alterations resulted in increased 2D cell area and decreased roundness (Figure 6E, right panels).
Consistent with our previous observations using the MRCK inhibitor BDP5290 (19), BDP9066 similarly reduced the bundling of filamentous actin at cortical and cytoplasmic regions (Figure 6F). To determine how MRCK inhibition affected cell motility, SCC12 cells were treated with DMSO vehicle control or non-toxic 0.4 μM BDP9066, and individual cells were tracked over 6 hours to quantify random migration. Scatterplots of cell paths showed marked reduction of cell motility induced by BDP9066 (Figure 6G). Mean cell velocity and accumulated distance travelled were significantly reduced by 21%, while mean Euclidean distance travelled was significantly reduced by 18% (Figure 6H). No differences were observed for directionality of cell motility.
The effect of non-toxic 0.4 μM BDP9066 was tested on organotypic invasion, in which the movement of SCC12 cells was measured from the surface downwards into a dense 3D rat tail collagen matrix that had been conditioned by cancer-associated fibroblasts for 7 days prior to their removal (Figure 6I) (18). A significant 50% decrease in the percentage of invading cells was observed for cells treated with 0.4 μM BDP9066 relative to DMSO vehicle (Figure 6J). Taken together, the results indicate that in SCC12 cells BDP9066 potently inhibits MRCK at sub-micromolar levels, leading to inhibition of MLC phosphorylation, changes in cell morphology, and reduced cell motility and 3D collagen invasion, without significantly affecting cell viability. These findings are consistent with the previously reported inhibitory effects of MRCKα + MRCKβ knockdown by siRNA (18), and the MRCK inhibitor BDP5290 (19) on SCC12 organotypic invasion.
In vivo action of BDP9066 on mouse skin cancer
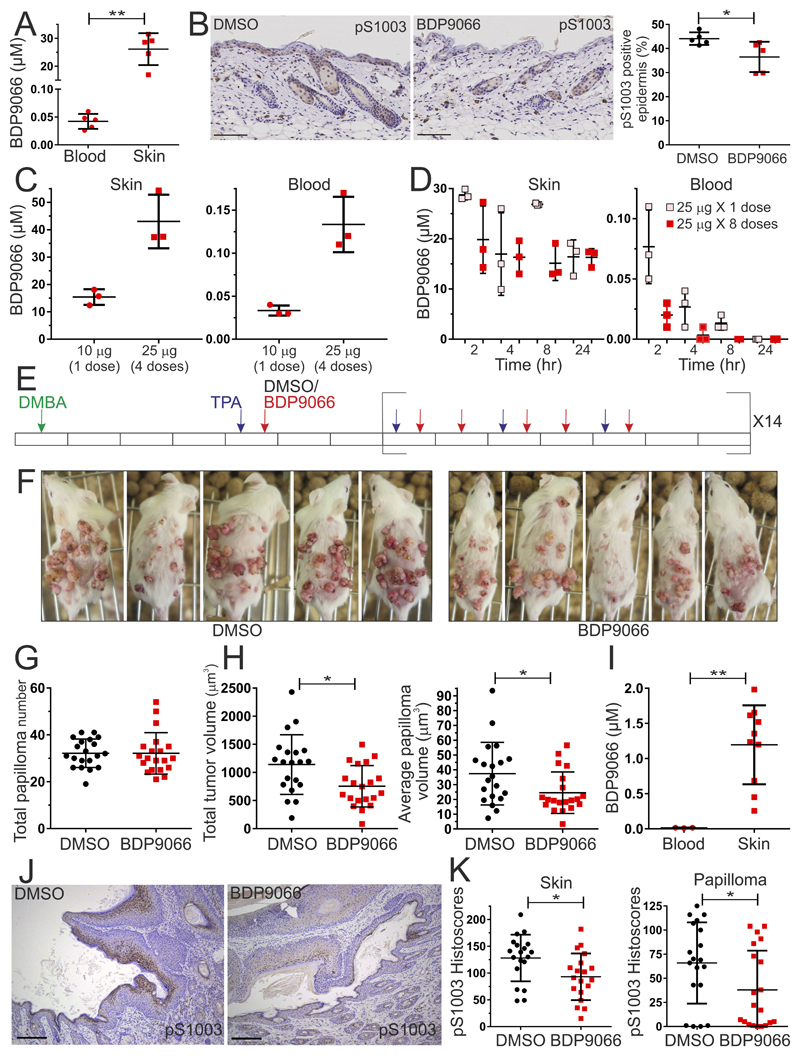
Having established that BDP9066 affects SCC tumor cell motility and invasion at sub-micromolar concentrations and viability at 1 µM in vitro (Figure 6), and that MRCKα, MRCKα pS1003 and MRCKβ levels were elevated in GM and chemically-induced mouse models of skin cancer (Figure 5L), BDP9066 was evaluated for in vivo pharmacological proof-of-concept as an SCC chemotherapeutic agent. Topical application of 25 µg BDP9066 on FVB mouse skin twice per day for 2 days led to 26 µM mean BDP9066 concentration in skin, but only 0.04 µM in blood (Figure 7A). BDP9066 application led to significantly reduced epidermal MRCKα pS1003 positive staining (Figure 7B). To determine how repeated dosing would affect BDP9066 accumulation and distribution, skin and blood concentrations were determined after 10 μg was administered once, or 25 μg was repeated over 4 days (Figure 7C). Although relative to the single 10 μg dose, repeated 25 μg doses did result in 2.8 fold higher concentrations in skin (Figure 7C, left panel), and 4 fold higher concentrations in blood (Figure 7C, right panel), these differences were less than the 10 fold difference in total BDP9066 administered, indicating that compound accumulation was less than additive. To characterize clearance, BDP9066 was either given once at a 25 μg dose, or 8 times on a 4 days on, 2 days off, 4 days on schedule, and then compound concentrations were determined in skin and blood at 2, 4, 8 and 24 hours following the final dose. In either dosing regimen, over 16 μM BDP9066 was detected in skin 24 hours after final dosing (Figure 7D, left panel), while the low levels detected in blood were undetectable after 24 hours (Figure 7D, right panel). These results indicated that it was possible to achieve sustainable BDP9066 levels in mouse skin by repeated topical application, which were sufficient to induce phenotypic responses in squamous cell carcinoma cells in vitro, without significant compound accumulation following sequential administration.
Figure 7. In vivo application of BDP9066 in a DMBA/TPA mouse model of SCC.
(A) Topical application of 4 X 25 µg BDP9066 over 2 days to dorsal skin led to measurable drug levels in the skin that were significantly higher than in blood. Results shown are means ± SD from 5 independent mice, each indicated by a data point. Two-tailed Mann-Whitney test of significance (** = p<0.01). (B) Representative MRCKα pS1003 immunohistochemical staining of mouse skin sections (left) topically treated with DMSO or 4 X 25 µg BDP9066 over 2 days. Scale bars represent 100 µm. Topical administration of BDP9066 (red dots) led to significant reduction in positive epidermal staining for MRCKα pS1003 (right). Results shown are means ± SD from 5 independent mice per condition, each indicated by a data point. Two-tailed Mann-Whitney test of significance (* = p<0.05). (C). Skin (left) or blood (right) BDP9066 concentrations following topical administration of a single 10 µg dose or 4 X 25 µg doses of BDP9066 over 2 days. Results shown are means ± SD from 3 independent mice per condition, each indicated by a data point. (D) Skin (left) or blood (right) BDP9066 concentrations following topical administration of a single 25 µg dose (pink dots) or 8 X 25 µg doses of BDP9066 (red dots; 4 days on, 2 days off, 4 days on) at indicated times after final administration. Results shown are means ± SD from 3 independent mice per condition, each indicated by a data point. (E) Timeline of DMBA (green arrow), TPA (purple arrow), DMSO (black arrow) or BDP9066 (red arrow) administration, with each day represented by a rectangle. (F) Representative images of cagemate DMBA/TPA treated mice with topical application of DMSO or BDP9066 as indicated. (G) Total endpoint papilloma numbers per mouse. Results shown are means ± SD from 20 independent mice per condition, each indicated by a data point. (H) Total tumor volume (left) and average papilloma volume (right) per mouse. Results shown are means ± SD from 20 independent mice per condition, each indicated by a data point. Two tailed unpaired t-tests were used to determine significance (* = p<0.05). (I) Blood and skin BDP9066 concentrations at endpoint. Results shown are means ± SD from 3 independent mice for blood and 10 mice for skin, each indicated by a data point. Two-tailed Mann-Whitney test of significance (** = p<0.001). (J) Representative MRCKα pS1003 staining of DMBA/TPA skin sections treated topically with DMSO or BDP9066. Scale bars represent 100 µm. (K). Cytoplasmic histoscores of MRCKα pS1003 staining of DMBA/TPA treated skin (left) or papilloma (right) sections that had been administered DMSO (black dots) or BDP9066 (red dots). Results shown are means ± SD from 19 independent mice per condition, each indicated by a data point. Two tailed unpaired t-tests were used to determine significance (* = p<0.05).
Given the MRCKα, MRCKα pS1003 and MRCKβ levels observed in skin tumors induced by the DMBA/TPA two stage chemical carcinogenesis protocol (Figure 5L), this model was selected to evaluate the in vivo efficacy of BDP9066. FVB mice were topically treated with 25 μg DMBA (Figure 7E, green arrow), then with 4.7 μg TPA 3 times per week (Figure 7E, purple arrow). In addition, mice were topically treated with 25 μg BDP9066, or an equal volume of DMSO vehicle control (Figure 7E, red arrow), 5 times per week (5 days on, 2 days off) for 14 weeks. At study end, skin papillomas in the BDP9066 treated group appeared visually smaller than in the DMSO treated group (Figure 7F). Although total papilloma numbers per mouse were not different between the DMSO and BDP9066 treatment groups (Figure 7G), both the total tumor volume (Figure 7H, left panel) and average papilloma volume (Figure 7H, right panel) per mouse were significantly reduced in the BDP9066 treatment group. Topical BDP9066 application resulted in undetectable compound in blood, and >1 µM mean BDP9066 concentration in skin at experimental endpoint (Figure 7I), which was associated with significant decreases in MRCKα pS1003 immunohistochemistry staining (Figure 7J) and reduced histoscores both in the treated skin area (Figure 7K, left panel) and in papillomas (Figure 7K, right panel). These results provide pharmacological proof-of-concept evidence indicating in vivo therapeutic actions of BDP9066.
Discussion
The small molecule inhibitors BDP8900 and BDP9066 are the most potent and selective MRCK inhibitors discovered to date. Structure-guided medicinal chemistry facilitated compound optimization for potency and selectivity over closely related kinases including ROCK1 and ROCK2 (Figures 1 and 2, Supplemental Figure 1, and Supplemental Tables 1-4). Consistent with the previously characterized cell biological roles of these proteins, MRCK inhibition led to dose-dependent decreases in MLC2 phosphorylation, morphological changes, and reduced cell motility and invasion into 3D matrices at concentrations below cytotoxic levels (Figure 5). Inhibition of proliferation (e.g. Figures 4 and 6B) was observed at BDP9066 concentrations higher than required for significant substrate phosphorylation, likely indicating a need for near complete inhibition of activity to provoke anti-proliferative responses. Furthermore, topical application of BDP9066 to mice undergoing a DMBA/TPA chemical carcinogenesis protocol resulted in the therapeutic effect of significantly reduced papilloma volumes associated with micromolar compound levels in skin and reduced MRCKα S1003 autophosphorylation (Figure 7). The discovery of these compounds will enable further exploration of MRCK functions and roles in cancer, which will be important for determining how MRCK inhibition would be clinically applicable.
The Rho GTPase family effectors ROCK1, ROCK2, MRCKα and MRKCβ phosphorylate common substrates including MLC2, the myosin-binding subunit (MYPT1) of the MLC2 phosphatase complex, and the LIM kinases 1 and 2 (6,7,48). However, the ROCK and MRCK kinases appear to operate in different subcellular locales, with ROCK actions being more dispersed and MRCK acting primarily close to the plasma membrane. These differing sites of action likely explain why the combined inhibition of ROCK and MRCK together is more effective at blocking tumor cell invasion (15,16). It has been proposed that compounds that inhibit both ROCK and MRCK kinases would be more effective as anti-metastatic therapeutics than agents that were selective for one or the other (20). This hypothesis may be correct, but less selective inhibitors would not be useful as chemical biology tools to explore kinase-specific biological roles. In addition, in situations where only inhibition of MRCK was necessary for therapeutic effect, the additional ROCK inhibitory actions might lead to undesirable responses, such as dose-limiting hypotension. The highly potent and selective inhibitors BDP8900 and BDP9066 will enable studies focused on discovery of MRCK actions and roles in cancer.
Compounds that block tumor cell invasion are likely to be beneficial through a combination of effects, including reduced primary tumor growth and local invasion, as well as decreased dissemination and metastasis. For example, there were multiple beneficial effects on pancreatic tumor growth and spread when mice were treated with ROCK inhibitors, including sensitization of tumors to standard of care therapeutics (49,50). Given that compounds which target processes that contribute to tumor cell invasion do not directly lead to cytotoxicity, they are unlikely to induce easily monitored responses such as tumor shrinkage in patients with advanced cancers, making the clinical development process more challenging than for “classical” cytotoxic type chemotherapeutics. Instead, clinical benefit from anti-invasive drug administration would more likely be observed in alternative scenarios, such as long-term adjuvant treatment as part of post-therapy management, or in situations where local tissue invasion rather than distant metastasis is the cause of morbidity and mortality, as is the case for invasive gliobastoma (51). Additional pre-clinical studies will help to define the contexts in which anti-invasive compounds, such as the MRCK inhibitors BDP8900 and 9066, are most likely to be beneficial for cancer patient therapy.
Indications consistent with a role of MRCKα or MRCKβ in cancer include elevated expression and gene copy number variations. A “poor prognosis gene” expression signature, associated with increased incidence of distant metastasis in fewer than 5 years, included MRCKβ (indicated as PK428) (52), while the MRCKα gene was found to be amplified in 491/2051 (24%) of breast cancer samples (53). However, an additional possibility is that increased MRCK activity is a common mechanism contributing to cancer, which would not necessarily be associated with changes in MRCK expression. Increased MRCK activity might be trigged from upstream signals via CDC42, which could be activated through elevated expression, loss of inhibitory GTPase-activating proteins (GAPs), increased expression or activating mutation of guanine-nucleotide exchange factors (GEFs), or aberrant activity of regulators further upstream, such as receptor tyrosine kinases (54). Furthermore, mechanical stimulation of CDC42 by the extracellular microenvironment (55) may be another pathway leading to MRCK activation. A direct way to assess MRCK activation state is through the identification of autophosphorylation sites that report on activity status. A total of 8 phosphorylation sites were identified in wild-type MRCKα, but not the kinase dead K106M mutant, consistent with their being autophosphorylation events (Figure 5, Supplemental Tables 7 and 8). A phospho-sensitive pS1003 confirmed that phosphorylation of this site was mediated by an intramolecular mechanism, and was dependent on MRCK activity, supporting the conclusion that S1003 was autophosphorylated (Figure 5). Furthermore, the antibody reagent could be used in formalin-fixed paraffin embedded mouse tissue samples to interrogate MRCKα activity in tumors, or following BDP9066 treatment (Figures 5 and 7). Similarly, ROCK1 S1333 phosphorylation (41) and ROCK2 S1366 phosphorylation (42) were shown to be associated with kinase activity, and that phosphorylation-state sensitive antibodies could be used to assess ROCK activity in clinical cancer samples (43). Therefore, MRCKα and MRCKβ phospho-selective antibodies raised against autophosphorylation sites are likely to be very useful tools to identify cancer types that would benefit from MRCK inhibitor treatment. Although MRCKα S1003 autophosphorylation was validated and the phospho-sensitive antibody revealed increased staining in mouse skin tumors (Figure 5), other sites that have been identified in broad-scale phosphoproteomics studies might be better candidates for further evaluation, such as the putative autophosphorylation site S1629.
A role for MRCK in squamous cell carcinoma was previously indicated by microarray expression analysis of normal human skin versus epidermal SCC (Figure 5K) (44) and by a GM mouse model of epidermal SCC induced by oncogenic Hras expression combined with Notch1 inhibition (46). Using validated antibodies for MRCKα, phosphorylated MRCKα S1003 and MRCKβ, there was increased staining of each in two GM skin cancer models, and especially increased staining in the DMBA/TPA chemical carcinogenesis model (Figure 5L). Furthermore, BDP9066 topical application significantly decreased phosphorylated MRCKα S1003 staining and tumor volumes (Figure 7), consistent with antibody staining for MRCK autophosphorylation being both an indicator of an association between MRCK activity with cancer, and a predictor of therapeutic response to inhibitor treatment. The approach described, in which differences between the phosphorylation of wild-type versus kinase-dead versions were used to identify autophosphorylation events that could be used to monitor activation states, has the potential to be applied to additional kinases to evaluate the associations between activity and cancer, and to predict therapeutic responses. Monitoring kinase activity by immunohistochemical approaches would be a complementary or alternative approach that could be applied to signaling pathways in which activating mutations are not major factors.
Supplementary Material
Significance.
The development of selective small molecule inhibitors of the Cdc42-binding MRCK kinases reveal its essential roles in cancer cell viability, migration and invasive character.
Acknowledgements
Funding provided by Cancer Research UK (A18276), Medical Research Council (MR/J005126/1) and Worldwide Cancer Research (14-0223) to MFO. Work in the MJG lab is supported by funding from CRUK (C44943/A22536), SU2C (SU2C-AACR-DT1213) and the Wellcome Trust (102696). The authors would like to thank Diamond Light Source for beamtime (proposal mx8659) and the staff of beamlines i02 and i24 for their assistance. Thanks to Sara Zanivan and Catherine Winchester (Beatson Institute) for critical reading of the manuscript.
Footnotes
Competing interests
No potential conflicts of interest were disclosed by the authors.
References
- 1.Hall A. The cytoskeleton and cancer. Cancer Metastasis Rev. 2009;28:5–14. doi: 10.1007/s10555-008-9166-3. [DOI] [PubMed] [Google Scholar]
- 2.Mardilovich K, Olson MF, Baugh M. Targeting Rho GTPase signaling for cancer therapy. Future Oncol. 2012;8:165–77. doi: 10.2217/fon.11.143. [DOI] [PubMed] [Google Scholar]
- 3.Heissler SM, Sellers JR. Various Themes of Myosin Regulation. J Mol Biol. 2016;428:1927–46. doi: 10.1016/j.jmb.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK) Small GTPases. 2014;5:e29846. doi: 10.4161/sgtp.29846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olson MF. Rho GTPases, their post-translational modifications, disease-associated mutations and pharmacological inhibitors. Small GTPases. 2016 doi: 10.1080/21541248.2016.1218407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unbekandt M, Olson MF. The actin-myosin regulatory MRCK kinases: regulation, biological functions and associations with human cancer. J Mol Med. 2014;92:217–25. doi: 10.1007/s00109-014-1133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Z, Manser E. Myotonic dystrophy kinase-related Cdc42-binding kinases (MRCK), the ROCK-like effectors of Cdc42 and Rac1. Small GTPases. 2015;6:81–8. doi: 10.1080/21541248.2014.1000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung T, Chen XQ, Tan I, Manser E, Lim L. Myotonic dystrophy kinase-related Cdc42-binding kinase acts as a Cdc42 effector in promoting cytoskeletal reorganization. Mol Cell Biol. 1998;18:130–40. doi: 10.1128/mcb.18.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng Y, Tan I, Lim L, Leung T. Expression of the human myotonic dystrophy kinase-related Cdc42-binding kinase gamma is regulated by promoter DNA methylation and Sp1 binding. J Biol Chem. 2004;279:34156–64. doi: 10.1074/jbc.M405252200. [DOI] [PubMed] [Google Scholar]
- 10.Uehata M, Ishizaki T, Satoh H, Ono T, Kawahara T, Morishita T, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–4. doi: 10.1038/40187. [DOI] [PubMed] [Google Scholar]
- 11.Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13:900–8. doi: 10.1038/embor.2012.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–87. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 13.Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–64. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 14.Waclaw B, Bozic I, Pittman ME, Hruban RH, Vogelstein B, Nowak MA. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–4. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilkinson S, Paterson HF, Marshall CJ. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat Cell Biol. 2005;7:255–61. doi: 10.1038/ncb1230. [DOI] [PubMed] [Google Scholar]
- 16.Heikkila T, Wheatley E, Crighton D, Schroder E, Boakes A, Kaye SJ, et al. Co-crystal structures of inhibitors with MRCKbeta, a key regulator of tumor cell invasion. PLoS One. 2011;6:e24825. doi: 10.1371/journal.pone.0024825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kale VP, Hengst JA, Desai DH, Dick TE, Choe KN, Colledge AL, et al. A novel selective multikinase inhibitor of ROCK and MRCK effectively blocks cancer cell migration and invasion. Cancer Letters. 2014;354:299–310. doi: 10.1016/j.canlet.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaggioli C, Hooper S, Hidalgo-Carcedo C, Grosse R, Marshall JF, Harrington K, et al. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat Cell Biol. 2007;9:1392–400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- 19.Unbekandt M, Croft DR, Crighton D, Mezna M, McArthur D, McConnell P, et al. A novel small-molecule MRCK inhibitor blocks cancer cell invasion. Cell Commun Signal. 2014;12:54. doi: 10.1186/s12964-014-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kale VP, Hengst JA, Desai DH, Amin SG, Yun JK. The regulatory roles of ROCK and MRCK kinases in the plasticity of cancer cell migration. Cancer Lett. 2015;361:185–96. doi: 10.1016/j.canlet.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 21.Tan I, Lai J, Yong J, Li SF, Leung T. Chelerythrine perturbs lamellar actomyosin filaments by selective inhibition of myotonic dystrophy kinase-related Cdc42-binding kinase. FEBS Lett. 2011;585:1260–8. doi: 10.1016/j.febslet.2011.03.054. [DOI] [PubMed] [Google Scholar]
- 22.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pham YC, Man N, Lam LT, Morris GE. Localization of myotonic dystrophy protein kinase in human and rabbit tissues using a new panel of monoclonal antibodies. Hum Mol Genet. 1998;7:1957–65. doi: 10.1093/hmg/7.12.1957. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–14. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr. 2010;43:186–90. [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schüttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–63. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 30.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGarry DJ, Shchepinova MM, Lilla S, Hartley RC, Olson MF. A Cell-Permeable Biscyclooctyne As a Novel Probe for the Identification of Protein Sulfenic Acids. ACS Chem Biol. 2016;11:3300–4. doi: 10.1021/acschembio.6b00742. [DOI] [PubMed] [Google Scholar]
- 32.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 33.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 34.Greenhalgh DA, Rothnagel JA, Quintanilla MI, Orengo CC, Gagne TA, Bundman DS, et al. Induction of epidermal hyperplasia, hyperkeratosis, and papillomas in transgenic mice by a targeted v-Ha-ras oncogene. Mol Carcinog. 1993;7:99–110. doi: 10.1002/mc.2940070208. [DOI] [PubMed] [Google Scholar]
- 35.Yao D, Alexander CL, Quinn JA, Chan WC, Wu H, Greenhalgh DA. Fos cooperation with PTEN loss elicits keratoacanthoma not carcinoma, owing to p53/p21 WAF-induced differentiation triggered by GSK3beta inactivation and reduced AKT activity. J Cell Sci. 2008;121:1758–69. doi: 10.1242/jcs.021147. [DOI] [PubMed] [Google Scholar]
- 36.Macdonald FH, Yao D, Quinn JA, Greenhalgh DA. PTEN ablation in Ras(Ha)/Fos skin carcinogenesis invokes p53-dependent p21 to delay conversion while p53-independent p21 limits progression via cyclin D1/E2 inhibition. Oncogene. 2014;33:4132–43. doi: 10.1038/onc.2013.372. [DOI] [PubMed] [Google Scholar]
- 37.Yung-Chi C, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 38.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The Protein Kinase Complement of the Human Genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 39.Eid S, Turk S, Volkamer A, Rippmann F, Fulle S. KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinformatics. 2017;18:16. doi: 10.1186/s12859-016-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–54. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chuang HH, Liang SW, Chang ZF, Lee HH. Ser1333 phosphorylation indicates ROCKI activation. J Biomed Sci. 2013;20:83. doi: 10.1186/1423-0127-20-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chuang HH, Yang CH, Tsay YG, Hsu CY, Tseng LM, Chang ZF, et al. ROCKII Ser1366 phosphorylation reflects the activation status. Biochem J. 2012;443:145–51. doi: 10.1042/BJ20111839. [DOI] [PubMed] [Google Scholar]
- 43.Hsu C-Y, Chang Z-F, Lee H-H. Immunohistochemical evaluation of ROCK activation in invasive breast cancer. BMC Cancer. 2015;15:943. doi: 10.1186/s12885-015-1948-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nindl I, Dang C, Forschner T, Kuban RJ, Meyer T, Sterry W, et al. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol Cancer. 2006;5:30. doi: 10.1186/1476-4598-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes Dev. 2007;21:562–77. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jenkins DE, Hornig YS, Oei Y, Dusich J, Purchio T. Bioluminescent human breast cancer cell lines that permit rapid and sensitive in vivo detection of mammary tumors and multiple metastases in immune deficient mice. Breast Cancer Res. 2005;7:R444–54. doi: 10.1186/bcr1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott RW, Olson MF. LIM kinases: function, regulation and association with human disease. J Mol Med. 2007;85:555–68. doi: 10.1007/s00109-007-0165-6. [DOI] [PubMed] [Google Scholar]
- 49.Rath N, Morton JP, Julian L, Helbig L, Kadir S, McGhee EJ, et al. ROCK signaling promotes collagen remodeling to facilitate invasive pancreatic ductal adenocarcinoma tumor cell growth. EMBO Mol Med. 2017;9:198–218. doi: 10.15252/emmm.201606743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vennin C, Chin VT, Warren SC, Lucas MC, Herrmann D, Magenau A, et al. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai8504. eaai8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paw I, Carpenter RC, Watabe K, Debinski W, Lo HW. Mechanisms regulating glioma invasion. Cancer Lett. 2015;362:1–7. doi: 10.1016/j.canlet.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 53.Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nature Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arias-Romero LE, Chernoff J. Targeting Cdc42 in cancer. Expert Opin Ther Targets. 2013;17:1263–73. doi: 10.1517/14728222.2013.828037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z, Wu H, Jiang K, Wang Y, Zhang W, Chu Q, et al. MAPK-Mediated YAP Activation Controls Mechanical-Tension-Induced Pulmonary Alveolar Regeneration. Cell Reports. 2016;16:1810–9. doi: 10.1016/j.celrep.2016.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.