Abstract
Background
To determine the prognostic value of embryonic origin in patients undergoing resection after chemotherapy for colon cancer liver metastases (CCLM).
Methods
We identified 725 patients with primary colon cancer and known RAS mutation status who underwent hepatic resection after preoperative chemotherapy for CCLM (1990-2015). Survival after resection of CCLM from midgut origin (n=238) and hindgut origin (n=487) was analyzed. Predictors of pathologic response and survival were determined. Prognostic value of embryonic origin was validated with a separate cohort of 252 patients with primary colon cancer who underwent resection of CCLM without preoperative chemotherapy.
Results
Recurrence-free survival (RFS) and overall survival (OS) after hepatic resection were worse in patients with midgut origin tumors (RFS rate at 3 years: 15% vs. 27%, P<0.001; OS rate at 3 years: 46% vs. 68%, P<0.001). Independent factors associated with minor pathologic response were midgut embryonic origin (odds ratio [OR] 1.55, P=0.010), absence of bevacizumab (OR 1.42, P=0.034), and mutant RAS (OR 1.41, P=0.043). Independent factors associated with worse OS were midgut embryonic origin (hazard ratio [HR] 2.04, P<0.001), carcinoembryonic antigen value ≥5 ng/mL at hepatic resection (HR 1.46, P=0.0021), synchronous CCLM (HR 1.45, P=0.012), and mutant RAS (HR 1.43, P=0.0040). In the validation cohort, patients with CCLM of midgut origin had a worse 3-year OS rate (55% vs. 78%, P=0.003).
Conclusion
Compared to CCLM from hindgut origin, CCLM from midgut origin are associated with worse pathologic response to chemotherapy and worse survival after resection. This effect appears to be independent of RAS mutation status.
INTRODUCTION
Colorectal cancer (CRC) is the third most common cancer and the third most common cause of death due to cancer in the United States.1 Colorectal liver metastases occur in 30% of patients with CRC and account for two-thirds of deaths from CRC.2 Hepatectomy offers the best chance for long-term survival in patients with colorectal liver metastases.3
As a result of genetic and molecular evaluation, CRC is no longer regarded as a single entity. Three distinct pathways of CRC development have been described: the chromosomal instability, germline mutation, and serrated/methylator pathways.4 Furthermore, and of particular interest in the current study, midgut origin and hindgut origin CRCs have been demonstrated to follow different pathways of carcinogenesis.5-8 Whereas midgut origin tumors are more likely to be diploid and exhibit mucinous histology, high microsatellite instability, high CpG island methylation, and BRAF mutations,6, 7 hindgut origin tumors are more likely to be aneuploid and exhibit chromosomal instability.6, 8
Previous attempts to evaluate the effect of embryonic origin on survival in patients with CRC have yielded conflicting results because of high heterogeneity between studies with respect to patients’ clinical characteristics (tumor stage, presence/absence and type of chemotherapy, etc) and limited information on molecular features (RAS mutation status, etc).9, 10 Recently, Loupakis et al, in an analysis of three independent studies, demonstrated that midgut origin was independently associated with poor survival after treatment of unresectable metastatic CRC.11 Additionally, a previous German multicenter study including 17,641 CRC patients with Union Internationale Contre le Cancer stage I to III identified midgut origin as an independent predictor of worse survival after resection with curative intent.12 However, the effect of embryonic origin on survival in patients with colorectal liver metastases undergoing preoperative chemotherapy followed by resection of colorectal liver metastases has never been reported. In patients with resectable colorectal liver metastases, pathologic response to preoperative chemotherapy and RAS mutation status are established major predictors of survival.12-14
The aim of the current study was twofold: (1) to determine the prognostic value of embryonic origin in patients undergoing resection after chemotherapy for colon cancer liver metastases (CCLM) and (2) to investigate the correlations among embryonic origin, pathologic response to preoperative chemotherapy, and RAS mutation status in such patients.
MATERIAL AND METHODS
Study Population
The Institutional Review Board of The University of Texas MD Anderson Cancer Center approved this study protocol (PA16-0162). A prospectively maintained hepatobiliary database of the Department of Surgical Oncology was reviewed to identify patients who underwent curative resection for colorectal liver metastases between November 1990 and February 2015. A total of 2195 patients were identified (Figure 1). Of those patients, we identified patients with known RAS mutation status who underwent preoperative chemotherapy and curative resection. The following exclusion criteria were applied: (1) concomitant radiofrequency ablation, (2) primary rectal cancer, (3) primary transverse colon cancer, (4) absence of preoperative chemotherapy, (5) absence of pathologic description of response to preoperative chemotherapy, (6) use of anti-epidermal growth factor receptor (EGFR) agents before or after surgery, and (7) undetermined RAS mutation status. We excluded patients who had primary rectal cancer because rectal cancer is clinically and molecularly distinct from colon cancer and because rectal cancer treatment (such as preoperative chemoradiation) is different from colon cancer treatment.15 We excluded patients who had primary transverse colon cancer because we could not affirm retrospectively whether such patients had midgut origin or hindgut origin tumors given that the division point between midgut and hindgut is the point separating the first two-thirds from the final third of the transverse colon. The final study set included 725 patients (Figure 1). For our validation set, from the original 2195 patients who underwent curative resection for colorectal liver metastases, we selected the 252 patients who did not receive concomitant radiofrequency ablation, did not have primary rectal or primary transverse colon cancer, and did not receive preoperative chemotherapy (Figure 1).
Fig. 1.
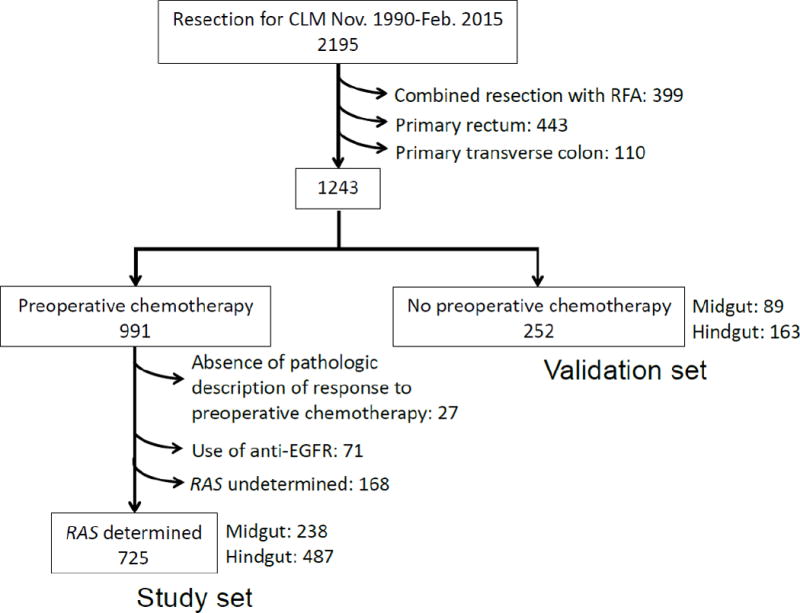
Patient selection.
The following data were extracted from electronic patient medical records: sex, age, body mass index, primary tumor characteristics (embryonic origin, depth of invasion, and lymph node metastases), preoperative chemotherapy characteristics (number of cycles and regimen), preoperative carcinoembryonic antigen (CEA) level, perioperative outcomes (blood loss, red blood cell transfusion, operative time, and surgical procedure [major resection was defined as liver resection including three or more liver segments]), and CCLM characteristics (timing of diagnosis, tumor size, tumor number, differentiation, margin status [R0, defined as no tumor cells at the margin, or R1, defined as tumor cells <1 mm from the margin], pathologic response to preoperative chemotherapy [major response, defined as cancer cells accounting for 0% to 49% of residual cells, or minor response, defined as cancer cells accounting for ≥ 50% of residual cells], and RAS mutation status).16
Perioperative Management
During preoperative chemotherapy, restaging was performed, and CCLM were deemed resectable when a hepatectomy could achieve a negative margin while preserving more than 20-30% of the total estimated liver volume, sparing two continuous hepatic segments, and maintaining vascular inflow and outflow and biliary drainage.17 Second-line chemotherapy was considered for patients with progression of disease or suboptimal tumor response after first-line chemotherapy.18 Decisions regarding the treatment sequence (classic, combined, or reverse) for patients having synchronous CCLM with an intact primary were made at multidisciplinary consultation including hepatobiliary surgeons, colorectal surgeons and medical oncologists. It was primarily based on the extent of the primary tumor and CCLM.20 In patients with an anticipated insufficient future liver remnant, preoperative portal vein embolization and staged hepatectomy were proposed. Postoperative chemotherapy was administered to complete a total of 12 cycles, including those given preoperatively.19 Patients were followed after resection with history, physical examination, laboratory evaluation, and axial imaging every 3-4 months for the first 2 years, and every 4-6 months for the subsequent 3 years.
Somatic Gene Mutation Profiling
The mutational status was assessed using DNA from biopsy or resected specimens of primary tumors or metastases. RAS mutation status was determined as previously described: routine polymerase chain reaction-based primer extension assay was performed to screen for mutations in KRAS codons 12 and 13 in all patients and for mutations in KRAS codons 61 and 146 and NRAS codons 12, 13, and 61 in the majority of patients.20 Single mutations in the various codons of KRAS and NRAS were analyzed together and reported as RAS mutations.
Statistical Analyses
Continuous variables were compared using the Wilcoxon rank-sum test, and categorical variables were compared using the χ2 test. For the evaluation of predictors of pathologic response, univariable and multivariable analyses were performed by logistic regression analysis. Recurrence-free survival (RFS) was measured from the date of hepatic resection until the date of radiographic detection of recurrence or last follow-up. Recurrence was defined as reappearance of a lesion with typical findings on standard imaging modalities (enhanced computed tomography, magnetic resonance imaging, and positron emission tomography, or a combination thereof). Overall survival (OS) was measured from the date of hepatic resection until the date of death or last follow-up. Survival curves were generated using the Kaplan-Meier method, and differences between curves were evaluated with the log-rank test. Univariable and multivariable analyses to identify predictors of survival were performed by using Cox proportional hazards regression models. Variables with P<0.1 in univariable analysis were entered into each multivariable analysis. P<0.05 was considered statistically significant in all analyses. Statistical analyses were performed with IBM SPSS software (version 23.0; SPSS Inc., Chicago, IL, USA).
RESULTS
Patient Characteristics and Survival According to Embryonic Origin in Study Set
Of the 725 patients in the study set, 238 (33%) had primary tumors derived from midgut, and 487 (67%) had primary tumors derived from hindgut (Figure 1). Table 1 lists clinicopathologic and operative data for the midgut and hindgut groups. The rate of major pathologic response after preoperative chemotherapy in CCLM was higher for the hindgut group (71% vs. 62%, P=0.012). There were no significant differences between the two groups with respect to extent of primary tumor, type of preoperative chemotherapy or number of cycles, or proportion with RAS mutation (midgut 39%, hindgut 35%, P=0.324).
Table 1.
Clinicopathologic characteristics by embryonic origin of primary colon cancer*
| Characteristic | Total | Midgut | Hindgut | p value (midgut vs hindgut)* |
|---|---|---|---|---|
| n | 725 | 238 | 487 | |
| Sex, M: F | 422: 303 | 132: 106 | 290: 197 | 0.295 |
| Age, y, median (interquartile range) | 58 (50-65) | 56 (49-64) | 58 (50-66) | 0.137‡ |
| Body mass index ≥25 kg/m2 | 524 (72) | 168 (71) | 356 (73) | 0.478 |
| Primary tumor | ||||
| T1/T2: T3/T4 | 157: 568 | 54: 184 | 103: 384 | 0.637 |
| Lymph node metastases present | 490 (68) | 155 (65) | 335 (69) | 0.323 |
| Pre-hepatic resection chemotherapy | ||||
| ≤6 cycles | 426 (59) | 147 (62) | 279 (57) | 0.250 |
| Fluorouracil-based chemotherapy regimen | ||||
| Oxaliplatin | 496 (68) | 168 (71) | 328 (67) | 0.379 |
| Irinotecan | 225 (31) | 66 (28) | 159 (33) | 0.179 |
| Use of bevacizumab | 398 (55) | 132 (55) | 266 (55) | 0.831 |
| Pre-hepatic resection CEA value, ng/mL, median (interquartile range) | 2.5 (1.0-7.4) | 2.7 (1.0-6.7) | 2.5 (1.0-7.7) | 0.999‡ |
| Hepatic resection | ||||
| Estimated blood loss, g, median (interquartile range) | 200 (100-400) | 200 (100-390) | 200 (100-400) | 0.460‡ |
| Red blood cell transfusion | 50 (6·9) | 10 (8·0) | 31 (6·4) | 0.420 |
| Operative time, min, median (interquartile range) | 210 (130-251) | 210 (130-240) | 210 (130-259) | 0.331‡ |
| Surgical procedure, major: minor | 401: 324 | 132: 106 | 269: 218 | 0.954 |
| Liver metastases | ||||
| Synchronous | 537 (74) | 181 (76) | 356 (73) | 0.395 |
| Classic/Combined/Reverse | 414/86/37 | 140/31/10 | 274/55/27 | 0.621 |
| Maximum tumor size, mm, median (interquartile range) | 22 (14-35) | 21 (13-33) | 22 (15-35) | 0.737‡ |
| Tumor number, solitary: multiple | 278: 447 | 90: 148 | 188: 299 | 0.838 |
| Residual cancer, R0: R1 | 673: 52 | 218: 20 | 455: 32 | 0.369 |
| Well/moderately/poorly differentiated | 71/545/109 | 25/174/39 | 46/371/70 | 0.666 |
| Pathologic response | ||||
| Major (viable tumor 0-49%) | 496 (68) | 148 (62) | 348 (71) | 0.012 |
| Minor (viable tumor 50-100%) | 229 (32) | 90 (38) | 139 (29) | |
| RAS mutation status | ||||
| Wild-type | 463 (64) | 146 (61) | 317 (65) | 0.324 |
| Mutant | 262 (36) | 92 (39) | 170 (35) | |
| Post-hepatic resection chemotherapy | 480 (66) | 157 (66) | 323 (66) | 0.924 |
Values in table are number of patients (percentage) unless indicated otherwise.
χ2 test unless indicated otherwise.
Wilcoxon rank-sum test.
CEA, carcinoembryonic antigen.
Predictors of Pathologic Response in Study Set
Minor pathologic response was observed in 229 patients (32%). Table 2 lists the results of univariable and multivariable analysis of the predictors of minor pathologic response. Independent predictors of minor pathologic response were midgut origin, absence of bevacizumab, and mutant RAS. The minor pathologic response rate in the patients with all three independent predictors was 53% (25/47), while the minor pathologic response rate in the patients with none of these three independent predictors was 23% (43/185) (P=0.0001).
Table 2.
Univariable and multivariable analyses of minor pathologic response
| Minor pathologic response | ||||||
|---|---|---|---|---|---|---|
| n | n (%) | Univariable p value | Odds ratio | 95% confidence interval | Multivariable p value | |
| All patients | 725 | 229 (32) | ||||
| Primary tumor embryonic origin | ||||||
| Midgut | 238 | 90 (38) | 0.012 | 1.55 | 1.10-2.16 | 0.010 |
| Hindgut | 487 | 139 (29) | ||||
| Use of bevacizumab | ||||||
| Absence | 327 | 119 (36) | 0.012 | 1.42 | 1.03-1.96 | 0.034 |
| Presence | 398 | 110 (28) | ||||
| RAS mutation status | ||||||
| Mutant | 262 | 98 (37) | 0.011 | 1.41 | 1.01-1.95 | 0.043 |
| Wild-type | 463 | 131 (28) | ||||
Predictors of Survival in Study Set
After a median follow-up time of 27 months (range, 1-143), RFS and OS after hepatic resection were significantly worse in patients with midgut origin primary tumors (RFS rate at 3 years: 15% vs. 27%, P<0.0001; OS rate at 3 years: 46% vs. 68%, P<0.0001) (Figure 2A and 2B).
Fig. 2.
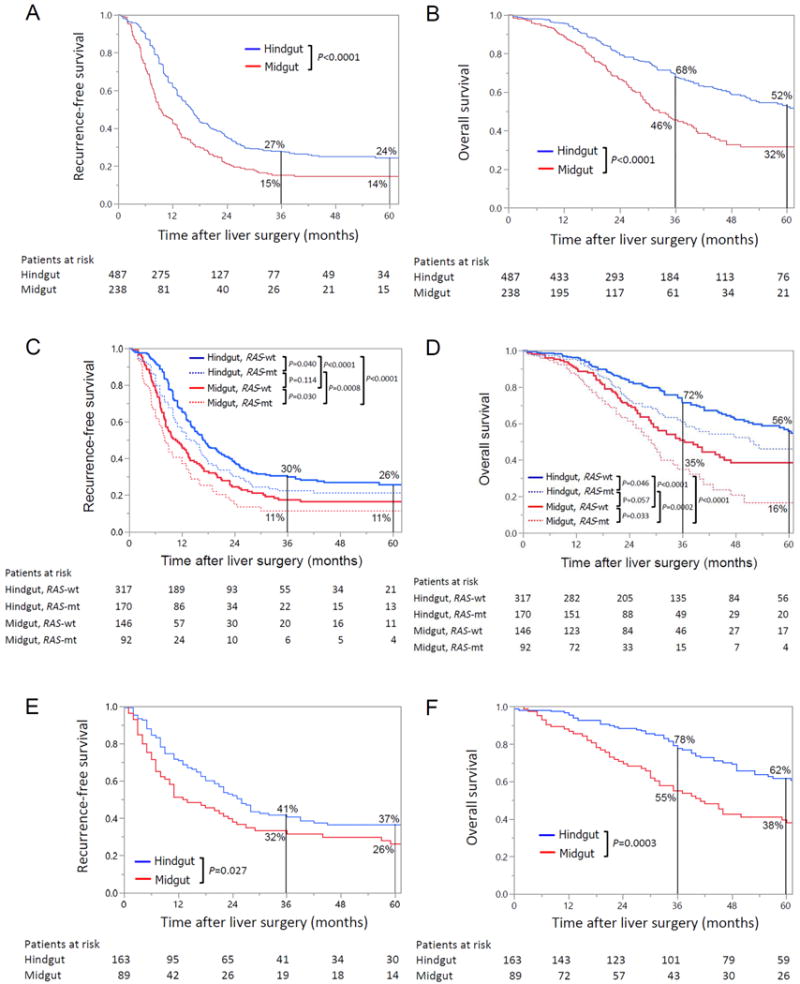
Recurrence-free survival (A) and overall survival (B) by embryonic origin of primary colon cancer in patients with colorectal liver metastases who underwent preoperative chemotherapy followed by curative resection. Recurrence-free survival (C) and overall survival (D) by RAS mutation status and embryonic origin of primary colon cancer in patients with colorectal liver metastases who underwent preoperative chemotherapy followed by curative resection. Recurrence-free survival (E) and overall survival (F) by embryonic origin of primary colon cancer in patients with colorectal liver metastases who underwent curative resection without preoperative chemotherapy.
On multivariable analysis of factors associated with RFS after hepatic resection, independent predictors of worse RFS were midgut origin, CEA level ≥5 ng/mL at hepatic resection, multiple liver tumors, and RAS mutation (Table 3). On multivariable analysis of factors associated with OS after hepatic resection, independent predictors of worse OS were midgut origin, CEA level ≥5 ng/mL at hepatic resection, synchronous CCLM, and RAS mutation (Table 3).
Table 3.
Multivariable Cox regression models for RFS and OS in study set and validation set
| Study set (n=725) | Validation set (n=252) | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| RFS | OS | RFS | OS | |||||
|
| ||||||||
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| Midgut origin | 1.71 (1.41-2.07) | <0.0001 | 2.04 (1.60-2.59) | <0.0001 | 1.48 (1.05-2.08) | 0.026 | 1.90 (1.29-2.77) | 0.0009 |
| CEA value at hepatic resection ≥5 ng/mL | 1.40 (1.15-1.69) | 0.0006 | 1.46 (1.15-1.86) | 0.0021 | NS | NS | ||
| Multiple tumors | 1.33 (1.10-1.61) | 0.0030 | 1.26 (0.99-1.63) | 0.066 | NS | NS | ||
| Synchronous | 1.18 (0.95-1.48) | 0.127 | 1.45 (1.08-1.96) | 0.012 | NS | 1.19 (0.81-1.76) | 0.373 | |
| Maximum size of tumor ≥3 cm | 1.16 (0.96-1.40) | 0.122 | 1.26 (0.99-1.60) | 0.057 | 1.45 (1.04-2.03) | 0.030 | 1.37 (0.94-2.01) | 0.106 |
| Mutant RAS | 1.30 (1.08-1.57) | 0.0063 | 1.43 (1.12-1.82) | 0.0040 | NA | NA | ||
RFS, recurrence-free survival; OS, overall survival; HR, hazard ratio; CI, confidence interval; NA, not applicable; CEA, carcinoembryonic antigen; NS, not significant on univariable analysis.
In the subgroup of patients with wild-type RAS (n=463), RFS and OS were significantly worse in patients with midgut origin primary tumors. In the subgroup of patients with RAS mutation (n=262), RFS and OS were also significantly worse in patients with midgut origin primary tumors (Figure 2C and 2D).
Compared to the patients with RAS mutation and midgut origin primary tumors (n=92) the patients with wild-type RAS and hindgut origin primary tumors (n=317) had better RFS rates at 3 years (30% vs. 11%, P<0.0001) and 5 years (26% vs. 11%, P<0.0001) and better OS rates at 3 years (72% vs. 35%, P<0.0001) and 5 years (56% vs. 16%, P<0.0001) (Figure 2C and 2D).
Patient Characteristics and Survival in Validation Set
Among the 252 patients in the validation set, 89 (35%) had midgut origin and 163 (65%) had hindgut origin primary tumors (Figure 1). Supplementary table 1 lists clinicopathologic and operative data for the midgut and hindgut groups. There were no significant differences between the two groups in terms of basic demographics, perioperative outcomes, extent of primary tumor and CCLM, and presence of post-hepatic resection chemotherapy.
After a median follow-up time of 41 months (range, 1-206), RFS and OS after hepatic resection were significantly worse in patients with midgut origin primary tumors (RFS rate at 3 years: 32% vs. 41%, P=0.027; OS rate at 3 years: 55% vs. 78%, P=0.0003) (Figure 2E and 2F).
On multivariable analysis of factors associated with RFS after hepatic resection, independent predictors of worse RFS were midgut origin and largest CCLM lesion at least 3 cm (Table 3). On multivariable analysis of factors associated with OS after hepatic resection, the only independent predictor of worse outcome was midgut origin (Table 3). Subgroup analysis of survival stratified by RAS mutation status and embryonic origin in the validation cohort was not performed since the number of patients with available RAS mutation status was small (hindgut origin and wild-type RAS, n=16; hindgut origin and mutant RAS, n=26; midgut origin and wild-type RAS, n=6; midgut origin and mutant RAS, n=11).
DISCUSSION
In this study, we found that pathologic response to preoperative chemotherapy and survival after resection of CCLM were predominantly predicted by two biologic features: primary tumor embryonic origin and RAS mutation status.
Compared with patients with primary tumors of midgut origin, patients with primary tumors of hindgut origin had a better OS rate at 5 years (52% vs. 32%, P<0.0001), a better RFS rate at 5 years (24% vs. 14%, P<0.0001), and a better rate of major pathologic response (71% vs. 62%, P=0.012). In a separate validation cohort of patients who did not receive preoperative chemotherapy before CCLM resection, hindgut origin was also associated with a better OS rate at 5 years (62% vs. 38%, P=0.0003) and RFS rate at 5 years (37% vs. 26%, P=0.027). Previous reports have shown the prognostic effects of embryonic origin in patients with both primary CRC and unresectable metastatic CRC, but to the best of our knowledge, this is the first report to show the impact of embryonic origin on survival after resection of CCLM.9, 11
We also analyzed the impact of the interaction between embryonic origin of primary colon cancer and RAS mutation status on pathologic response to preoperative chemotherapy and survival after resection of CCLM. We previously reported that RAS-mutant colorectal liver metastases were associated with worse survival and inferior pathologic response to chemotherapy in patients with resectable colorectal liver metastases.13, 14 In the current study, we found that the impact of RAS mutation status on response to chemotherapy and survival after resection of CCLM was independent of embryonic origin. Patients with the combination of wild-type RAS and hindgut primary tumor origin had markedly better survival than patients with the combination of RAS mutation and midgut primary tumor origin (RFS at 5 years: 26% vs. 11%, P<0.0001; OS at 5 years: 56% vs. 16%, P<0.0001).
Our findings suggest that in the era of effective chemotherapy, RAS mutation status and primary tumor embryonic origin have greater prognostic importance than the clinicopathologic and surgery-related factors that have traditionally been reported to be associated with outcome (lymph node metastases of primary tumor, size of liver tumors, R0/R1 resection, etc).19, 21
Previous studies of unresectable colorectal liver metastases suggested that the radiological response to chemotherapy differed according to embryonic origin.22 The current study, which was based on resected CCLM, confirmed that the pathologic response to preoperative chemotherapy, which had previously been demonstrated to be associated with survival,12 also differed according to embryonic origin.
In the present study, we used a validation set of patients who had never received preoperative chemotherapy. Even in this validation set, the 5-year OS rate was better in the patients with hindgut origin tumors than in the patients with midgut origin tumors (62% vs. 38%), suggesting that the effect of embryonic origin on prognosis was independent of the effect of preoperative chemotherapy. In previous reports on studies of unresectable colorectal liver metastases and primary CRC, multiple reasons have been proposed for the worse prognosis of patients with midgut origin tumors, including the high rate of BRAF mutations, CpG island methylation, and ERCC1 expression, all of which were regarded as predictors of worse sensitivity to chemotherapy.7, 11, 23-25 However, because colorectal liver metastases with mutant BRAF are rarely resected given their extremely rapid growth and aggressive behavior, BRAF mutations are not likely to be a main reason for prognostic differences between resectable colorectal liver metastases with midgut and hindgut origin.20 Furthermore, CpG island methylation is also not likely to be a main reason for such differences because a previous study demonstrated that the proportion of tumors with CpG island methylation was lower in resected colorectal liver metastases (up to 10%) than in primary CRC (15-35%).26 Further investigations regarding reasons for the differences of oncologic outcomes between midgut and hindgut origin in resectable colorectal liver metastases patients are needed using colorectal liver metastases specimens.
This study has several limitations. It is a retrospective study, but the data were prospectively collected. Additionally, the characteristics of the patients with midgut origin and hindgut origin primary tumors were equivalent in terms of extent of primary tumor and extent of CCLM. Second, the study did not include patients with transverse colon and rectal cancer, because the embryonic origin could not be determined with certainty in the patients with primary transverse colon cancer, and rectal cancers originate from hindgut are evaluated and treated separately in practice from colon cancers.15 Third, RAS mutational status was determined either on resected CCLM or on the primary tumor and the two tumor sites might differ in mutational status. However, a growing body of evidence suggests a high rate of concordance (> 90%) in somatic gene mutational status between primary tumor and related metastases.27 Finally, the current study excluded a small number of patients treated with anti-EGFR agents, as such treatment could have affected the comparison between patients with wild-type RAS and RAS mutation. The present patients with hindgut and midgut origin received similar preoperative chemotherapy regimens.
In conclusion, beyond RAS mutation status, emerging data highlight the potential relevance of embryonic origin in predicting pathological response to preoperative chemotherapy and survival after resection in the era of effective chemotherapy and individualized management of resectable CCLM. In research practice, this knowledge has implications for future study design, interpretation of data, and analysis.
Supplementary Material
Acknowledgments
The authors would like to recognize Ms. Ruth Haynes for the administrative support in the preparation of this manuscript and thank Stephanie Deming, an employee of the Department of Scientific Publications at MD Anderson Cancer Center, for copyediting the manuscript. This research was supported in part by the National Institutes of Health through MD Anderson Cancer Center’s Support Grant, CA016672.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Kopetz S, Chang GJ, Overman MJ, et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. 2009;27:3677–83. doi: 10.1200/JCO.2008.20.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdalla EK, Vauthey JN, Ellis LM, et al. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg. 2004;239:818–25. doi: 10.1097/01.sla.0000128305.90650.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 5.Yamauchi M, Morikawa T, Kuchiba A, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–54. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113:779–88. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 7.Hutchins G, Southward K, Handley K, et al. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–70. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 8.Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403–8. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 9.Price TJ, Beeke C, Ullah S, et al. Does the primary site of colorectal cancer impact outcomes for patients with metastatic disease? Cancer. 2015;121:830–5. doi: 10.1002/cncr.29129. [DOI] [PubMed] [Google Scholar]
- 10.Pugh SA, Shinkins B, Fuller A, Mellor J, Mant D, Primrose JN. Site and Stage of Colorectal Cancer Influence the Likelihood and Distribution of Disease Recurrence and Postrecurrence Survival: Data From the FACS Randomized Controlled Trial. Ann Surg. 2016;263:1143–7. doi: 10.1097/SLA.0000000000001351. [DOI] [PubMed] [Google Scholar]
- 11.Loupakis F, Yang D, Yau L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst. 2015;107:3. doi: 10.1093/jnci/dju427. pii: dju427. Print 2015 Mar. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blazer DG, 3rd, Kishi Y, Maru DM, et al. Pathologic response to preoperative chemotherapy: a new outcome end point after resection of hepatic colorectal metastases. J Clin Oncol. 2008;26:5344–51. doi: 10.1200/JCO.2008.17.5299. [DOI] [PubMed] [Google Scholar]
- 13.Mise Y, Zimmitti G, Shindoh J, et al. RAS mutations predict radiologic and pathologic response in patients treated with chemotherapy before resection of colorectal liver metastases. Ann Surg Oncol. 2015;22:834–42. doi: 10.1245/s10434-014-4042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brudvik KW, Kopetz SE, Li L, Conrad C, Aloia TA, Vauthey JN. Meta-analysis of KRAS mutations and survival after resection of colorectal liver metastases. Br J Surg. 2015;102:1175–83. doi: 10.1002/bjs.9870. [DOI] [PubMed] [Google Scholar]
- 15.Bosset JF, Collette L, Calais G, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355:1114–23. doi: 10.1056/NEJMoa060829. [DOI] [PubMed] [Google Scholar]
- 16.Pawlik TM, Scoggins CR, Zorzi D, et al. Effect of surgical margin status on survival and site of recurrence after hepatic resection for colorectal metastases. Ann Surg. 2005;241:715–22. doi: 10.1097/01.sla.0000160703.75808.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kishi Y, Abdalla EK, Chun YS, et al. Three hundred and one consecutive extended right hepatectomies: evaluation of outcome based on systematic liver volumetry. Ann Surg. 2009;250:540–8. doi: 10.1097/SLA.0b013e3181b674df. [DOI] [PubMed] [Google Scholar]
- 18.Chun YS, Vauthey JN, Boonsirikamchai P, et al. Association of computed tomography morphologic criteria with pathologic response and survival in patients treated with bevacizumab for colorectal liver metastases. JAMA. 2009;302:2338–44. doi: 10.1001/jama.2009.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brouquet A, Abdalla EK, Kopetz S, et al. High survival rate after two-stage resection of advanced colorectal liver metastases: response-based selection and complete resection define outcome. J Clin Oncol. 2011;29:1083–90. doi: 10.1200/JCO.2010.32.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg. 2013;258:619–26. doi: 10.1097/SLA.0b013e3182a5025a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreou A, Aloia TA, Brouquet A, et al. Margin status remains an important determinant of survival after surgical resection of colorectal liver metastases in the era of modern chemotherapy. Ann Surg. 2013;257:1079–88. doi: 10.1097/SLA.0b013e318283a4d1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boisen MK, Johansen JS, Dehlendorff C, et al. Primary tumor location and bevacizumab effectiveness in patients with metastatic colorectal cancer. Ann Oncol. 2013;24:2554–9. doi: 10.1093/annonc/mdt253. [DOI] [PubMed] [Google Scholar]
- 23.Kopetz S, Desai J, Chan E, et al. Phase II pilot study of vemurafenib in patients with metastatic BRAF-mutated colorectal cancer. J Clin Oncol. 2015;33:4032–8. doi: 10.1200/JCO.2015.63.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiovitz S, Bertagnolli MM, Renfro LA, et al. CpG island methylator phenotype is associated with response to adjuvant irinotecan-based therapy for stage III colon cancer. Gastroenterology. 2014;147:637–45. doi: 10.1053/j.gastro.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viguier J, Boige V, Miquel C, et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res. 2005;11:6212–7. doi: 10.1158/1078-0432.CCR-04-2216. [DOI] [PubMed] [Google Scholar]
- 26.Messick CA, Sanchez J, Dejulius KL, Church JM, Kalady MF. Genetic and molecular diversity of colon cancer hepatic metastases. Surgery. 2009;146:227–31. doi: 10.1016/j.surg.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 27.Knijn N, Mekenkamp LJ, Klomp M, et al. KRAS mutation analysis: a comparison between primary tumours and matched liver metastases in 305 colorectal cancer patients. Br J Cancer. 2011;104:1020–6. doi: 10.1038/bjc.2011.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


