Abstract
Activation-induced deaminase (AID) introduces nucleotide substitutions within the variable region of immunoglobulin genes to promote antibody diversity. This activity, which is limited to 1.5 kb downstream of the variable gene promoter, mutates both the coding exon and downstream intronic sequences. We recently reported that RNA polymerase II accumulates in these regions during transcription in mice. This build-up directly correlates with the area that is accessible to AID, and manipulation of RNA polymerase II levels alters the mutation frequency. To address whether the intronic DNA sequence by itself can regulate RNA polymerase II accumulation and promote mutagenesis, we deleted 613 bp of DNA downstream of the JH6 intron in the human Ramos B cell line. The loss of this sequence did not alter polymerase abundance or mutagenesis in the variable gene, suggesting that most of the intronic sequence is dispensable for somatic hypermutation.
Keywords: Immunoglobulin, Variable gene, Somatic hypermutation, RNA polymerase II
1. Introduction
During B cell development, variable (V), diversity (D), and joining (J) gene segments recombine to create a functional V(D)J exon. This initial diversity is further expanded after antigen encounter, when B cells mutate their V(D)J genes to increase antibody affinity. The process is initiated by activation-induced deaminase (AID) (Muramatsu et al., 2000; Revy et al., 2000), an enzyme which converts cytosine to uracil in single-strand DNA (Di Noia and Neuberger, 2002; Maul et al., 2011; Petersen-Mahrt et al., 2002). The presence of uracil in DNA initiates a mutagenic cascade, resulting in the introduction of point mutations within the V region during somatic hypermutation (SHM), and DNA double-strand breaks within switch (S) regions preceding constant (C) genes during class switch recombination. The ability of AID to access single-strand DNA is intimately associated with the process of transcription by RNA polymerase II (Pol II) (Bransteitter et al., 2003; Chaudhuri et al., 2003; Dickerson et al., 2003; Pham et al., 2003; Ramiro et al., 2003; Sohail et al., 2003). It has recently been proposed that AID physically interacts with components of the stalled transcription complex to bring AID to the immunoglobulin locus (Pavri et al., 2010; Willmann et al., 2012). In S regions, Pol II stalling is caused by the generation of R-loop structures (Rajagopal et al., 2009; Yu et al., 2003). These structures occur when the newly transcribed RNA remains annealed to the transcribed DNA strand and occludes the non-transcribed strand from reannealing. However, in V regions, there are no R-loop structures that arise during transcription. Nonetheless, we recently described the accumulation of Pol II which correlates with AID activity (Maul et al., 2014), indicating that other DNA sequences may be involved in pausing Pol II.
During SHM, mutagenesis is limited to a 1.5 kb region downstream of the Ig promoter. This mutation window spans both the leader (L) and V(D)J exons and downstream intronic sequences on both heavy and light chain loci. Interestingly, the distance is independent of which V(D)J gene is used, suggesting that any conserved AID targeting element would not be found within the exon itself. Consistent with this hypothesis, replacement of the V(D)J exon with non-immunoglobulin sequences produced normal frequencies of mutagenesis (Yeap et al., 2015; Yelamos et al., 1995). Thus, any cis targeting component might be located within other DNA sequences in the mutation window. The intron sequences downstream of rearranged J gene segments are attractive candidates because they are proximal to V(D)J genes and are shared in every rearrangement. In mice, intronic sequences have frequencies of mutation that are comparable to those in adjacent V(D)J exons (Gearhart and Bogenhagen, 1983; Kim et al., 1981; Lebecque and Gearhart, 1990; Pech et al., 1981). The frequency then diminishes after 500 bp, suggesting that AID activity declines over distance. The universal nature of SHM in these unselected introns is confirmed by noting that the 3′ flanking sequences on three different loci in mice undergo SHM: IgH (Both et al., 1990; Jolly et al., 1997), Igκ (Hackett et al., 1990), and Igλ (Gonzalez-Fernandez et al., 1994). Furthermore, different species such as human (Goossens et al., 1998; Qian et al., 2014) and shark (Zhu and Hsu, 2010) have high frequencies of SHM in introns. Although these disparate loci and animals do not share sequence conservation, their location and/or structure intimates they may play a role in targeting SHM. In fact, the JH intron sequences in germinal center B cells from mice (Maul et al., 2014) and the human Ramos cell line (Wang et al., 2014) retain an abundance of Pol II as detected by ChIP, suggesting that cis DNA sequences in this region might block progression of transcription complexes.
Previous attempts to study the role of introns have produced conflicting results that are complicated by potential alterations in transcription levels. Of interest, Milstein and colleagues generated a transgenic mouse model where a portion of the Jκ intron was removed, and the mice displayed a 60% reduction in mutation frequency (Yelamos et al., 1995). Alternatively, deletion of the entire J-C intron in the IgL locus in the chicken DT40 cell line did not alter the mutation frequency (Kothapalli et al., 2011). In both scenarios, it is unclear whether these deletions affected Pol II accumulation. Thus, to address the long-standing question of a role for the downstream intronic sequence in promoting mutagenesis through transcriptional perturbation, we generated a Ramos cell line which lacked 613 bp in the intron downstream of JH6 (Fig. 1).
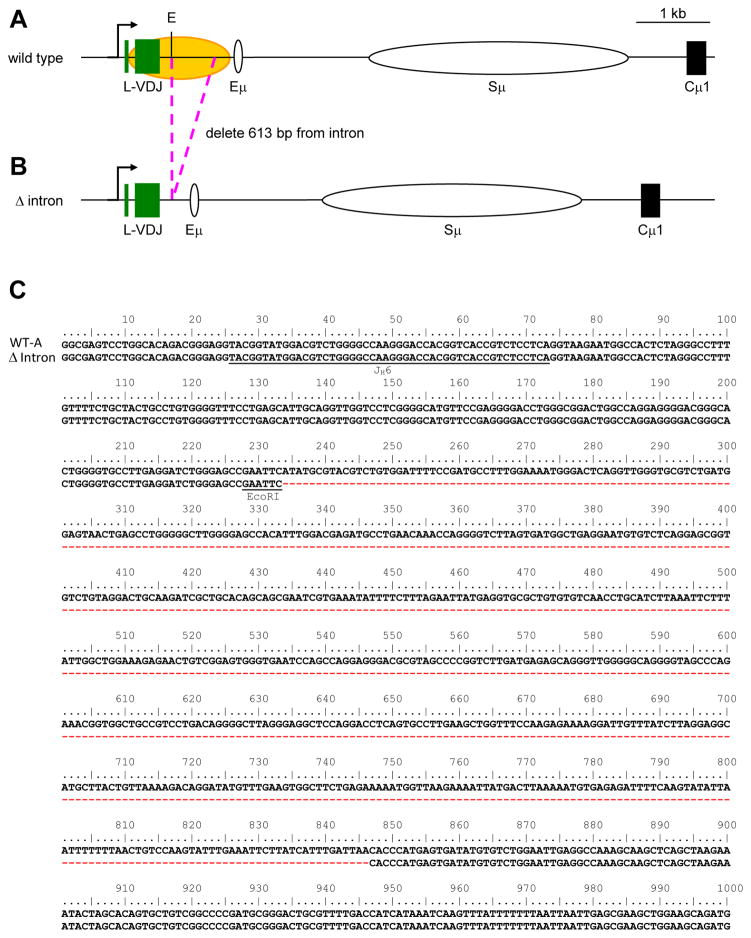
Fig. 1.
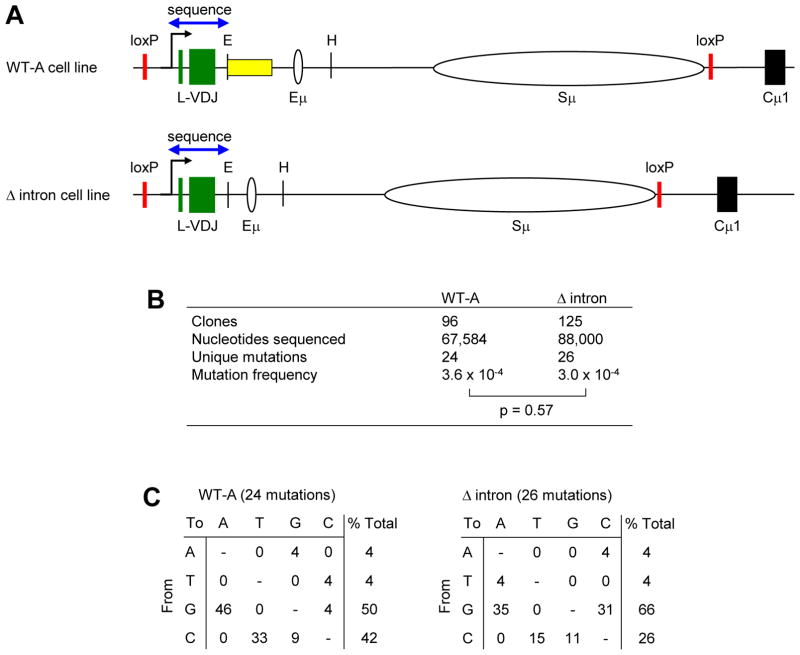
Somatic hypermutation in the IgH locus of Ramos cells. A) Wild type, and B) Δ intron. Arrow marks the transcription start site. Green boxes represent L and VDJ exons; yellow oval depicts the area targeted for SHM (Qian et al., 2014); white ovals signify the Eμ enhancer and Sμ region; and black box represents the Cμ1 exon. Pink dotted lines show the deletion. E, EcoRI. C) Nucleotide sequence (613 bp) that was deleted downstream of the EcoR1 site (underlined) is marked by red dashes. The rearranged JH6 gene segment is underlined for reference.
2. Materials and methods
2.1. Generation of Δ intron Ramos cell line
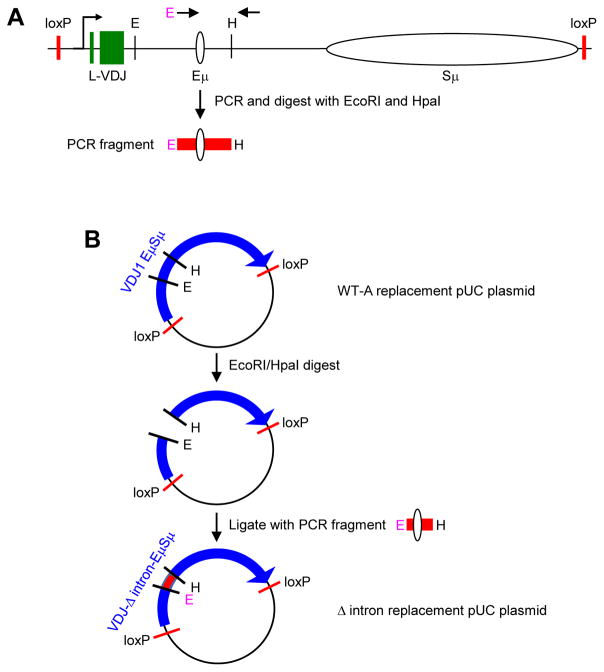
WT-A and Hyg-TK Ramos cell lines were obtained from Matthew Scharff (Albert Einstein College of Medicine, Bronx, NY). Cells were grown in Iscove’s modified Dulbecco’s medium(Gibco) supplemented with 10% FBS (Sigma), 1% glutamax (Gibco), and 100 U/mL penicillin-streptomycin (Gibco) at 37°C with 5% CO2. Δ intron cells were generated using recombinase-mediated cassette exchange (Baughn et al., 2011; Han et al., 2011), beginning with the Hyg-TK Ramos cell line and using a Δ intron replacement construct. To generate the construct, a 1,037 bp fragment was PCR amplified from the plasmid pUC-VDJ1 EμSμ (WT-A) (Baughn et al., 2011) using the primers Intron deletion fwd with an engineered EcoRI site in italics (5′CGCGAATTCCACCCATGAGTGATATGTGTCTGG 3′) and Intron deletion rev (5′GTCCATGGAAGCCACGCATCCCAGCTCTGG 3′) (Fig. 2A). The PCR fragment and the pUC-VDJ1 EμSμ plasmid were then cut with EcoRI and HpaI, followed by ligation of the fragment into the cut plasmid backbone to generate pUC-VDJ-Δ intron-EμSμ (Fig. 2B). Two million Hyg-TK cells were electroporated in 100 μl nucleofector (Lonza) containing 12.5 μg pUC-VDJ-Δ intron-EμSμ and 3.2 μg pCMV-Cre vector obtained from Dr. Scharff. Immediately after electroporation, 500 μl culture media was added, and the cells were plated in 12-well culture plates containing an additional 1 ml of media. Cells were expanded for 1 month, stained with mouse anti-human-IgM antibody (Biolegend, clone MHM-88), and single-cell sorted for surface IgM. Positive clones were expanded for two weeks for genotyping analysis to confirm a knockin of the replacement construct, and further expanded for 12 weeks to analyze SHM events.
Fig. 2.
Schematic of the Δ intron replacement strategy. A) Map of WT-A replacement construct with loxP recombination sites (Baughn et al., 2011). Arrows represent the area amplified by PCR to generate the Δ intron fragment used for subcloning. The 5′ primer had a novel EcoRI (E) site (pink), and the 3′ primer was located near a HpaI (H) site. The PCR fragment containing the Eμ enhancer was then digested with E and H enzymes. B) Stepwise generation of Δ intron replacement plasmid. The WT-A replacement plasmid was cut with E and H, and ligated to the PCR fragment from A) to generate the Δ intron replacement plasmid.
2.2. Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed as previously described (Maul et al., 2014) on cells after two weeks in culture. Briefly, 0.5 million cells were crosslinked with 1% formaldehyde for 10 min at room temperature. Cells were washed in PBS and resuspended in 120 μl lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.0, and protease inhibitor (Roche)) for 10 min on ice. Lysates were sonicated until the genomic DNA was sheared to an average size of 500 bp, and the chromatin was diluted to 1 ml with buffer (10 mM Tris pH 7.4, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, and 0.1% SDS). 100 μl was incubated with protein-G Dyna beads (Invitrogen), that were pre-bound with 2.4 μg mouse-anti-RNA Polymerase II (Millipore, clone CTD4H8) or total mouse IgG control (Sigma) for 2 hr at 4°C. Beads were washed extensively, and the bound fraction was eluted in 200 μl of elution buffer (20 mM Tris pH 7.4, 5 mM EDTA, 50 mM NaCl, and 15 μg proteinase K) at 68°C for 2 hr. After digestion, supernatants were collected, and DNA was isolated. Pulldown levels were analyzed using primer sets located in the JH intron adjacent to the VDJ exon (V) and the Cμ1 exon (C), corresponding respectively to primers F and K in Scharff and colleagues (Wang et al., 2014), and power SYBR green reagent (Invitrogen). Percent input was calculated as previously described (Maul et al., 2014).
2.3. Hypermutation analysis
Independent WT-A and Δ intron clones were plated as single cells in 96-well culture plates and maintained in growth media for 2 weeks. Clones which displayed similar growth rates were further expanded for 10 additional weeks, and genomic DNA was isolated for sequencing analysis. Two WT-A and 4 Δ intron clones were chosen and sequenced using primers SEQ fwd (5′ ACACATTTCCTTAAATTCAGGGTC 3′) and SEQ rev (5′ GGCTCCCAGATCCTCAAGGCAC 3′). Due to clonal expansion, shared mutations were counted only once, and mutation frequencies were calculated as mutations per total nucleotides sequenced.
3. Results
3.1. Deletion of JH6 intron sequence using recombinase-mediated cassette exchange
The Ramos cell line is a human Burkitt B cell lymphoma which undergoes constitutive mutagenesis in the V domain (Sale and Neuberger, 1998; Zhang et al., 2001). This region contains a L exon and VH4-34 gene segment rearranged to the D3-22 and JH6 gene segments, and 1 kb of 3′ intronic DNA prior to an Eμ enhancer site, Sμ switch region, and Cμ constant gene (Fig. 1A). Somatic hypermutation in this cell line occurs for 1.5 kb of DNA downstream of the promoter to include the VDJ exon and intronic DNA (Qian et al., 2014). To test the possibility of cis targeting elements located within the intronic DNA, we created a Δ intron cell line (Fig. 1B) where 613 bp of the intron was deleted at an EcoR1 site located 150 bp downstream of JH6 (Fig. 1C) and ending 200 bp upstream of Eμ, to minimize alterations to enhancer activity.
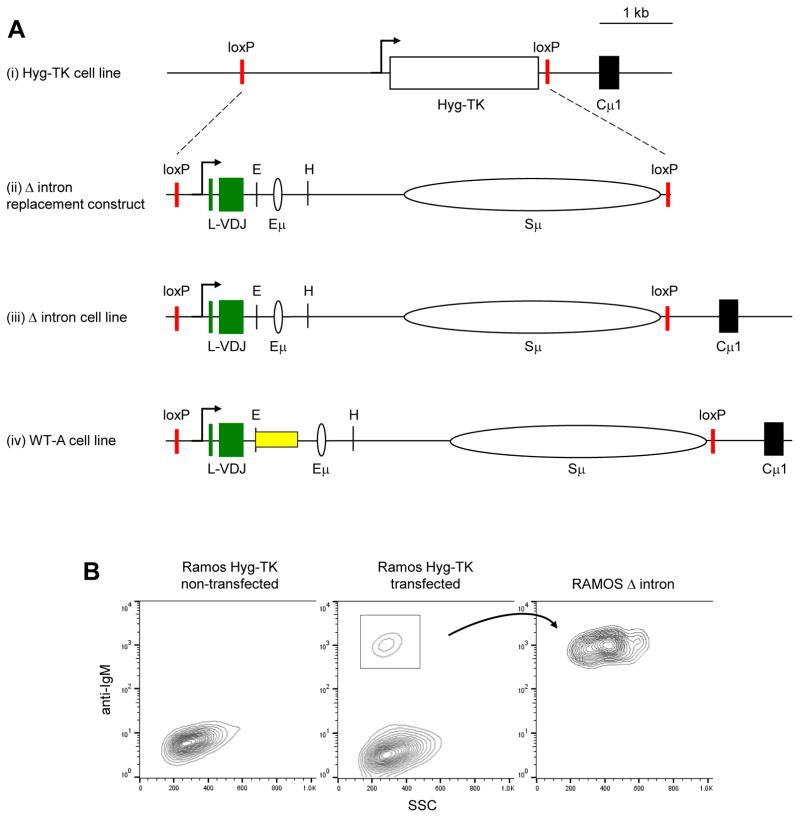
Using recombinase-mediated cassette exchange (Baughn et al., 2011) (Fig. 2), we replaced the Hyg-TK cassette from Hyg-TK Ramos cells (Fig. 3Ai) with a Δ intron replacement cassette (Fig. 3Aii), to create the Δ intron cell line (Fig. 3Aiii). The cells were compared to a wild type cell line, WT-A (Fig. 3Aiv), which contained the full-length intron that was generated via an identical approach by Scharff and colleagues (Baughn et al., 2011). Due to the lack of a VDJ-Cμ sequence in the Hyg-TK line, we screened for recombinants which restored IgM surface expression (Fig. 3B). IgM+ cells were single-cell sorted and sequenced after two weeks to confirm the genotype.
Fig. 3.
Generation of Δ intron cells. A) Scale representation of the genomic rearrangements found in (i) Hyg-TK cell line, (ii) Δ intron replacement construct and (iii) cell line, and (iv) WT-A cell line, with the yellow box showing the location of deleted DNA in the Δ intron line. The Hyg-TK box encodes for hygromycin and thymidine kinase genes. Vertical lines, EcoRI (E) and HpaI (H) sites. B) Representative FACS plot depicting the stages of selection used to generate Δ intron cell lines. The Hyg-TK cell line does not express surface antibody prior to transfection. After introducing the Δ intron cassette, positive recombinants were selected for surface IgM expression, single-cell sorted, and expanded to 100% IgM+ purity.
3.2. Pol II accumulation was similar in V regions of WT-A and Δ intron cells
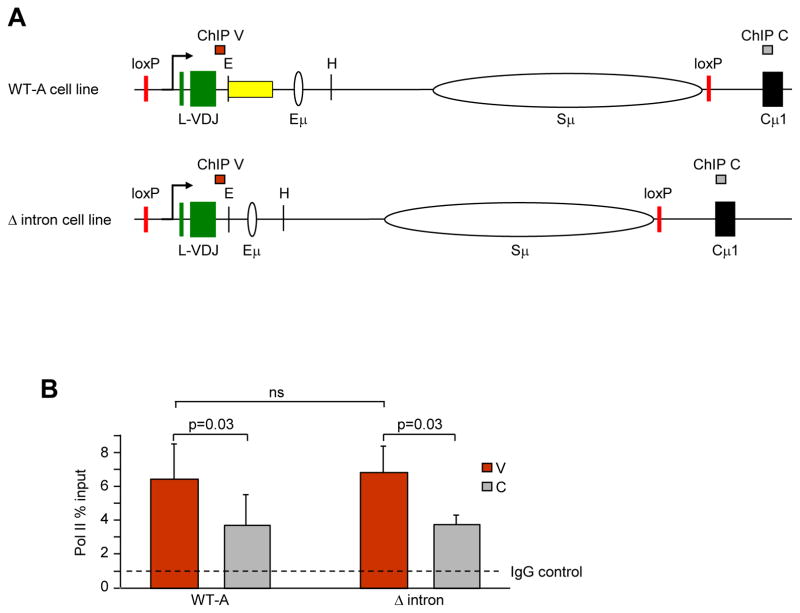
To examine a potential role for JH6 intronic sequences in causing Pol II abundance, the levels were assayed by chromatin immunoprecipitation (ChIP) with anti-Pol II antibody in WT-A and Δ intron cells, in both the V and C regions (Fig. 4A). Consistent with accumulation in mice (Maul et al., 2014), the V region had significantly higher levels of Pol II compared to the C region in WT-A Ramos cells (Fig. 4B, (Wang et al., 2014)). However, this accumulation was independent of the downstream JH6 intronic sequence, as Δ intron cells displayed similar Pol II levels. Control ChIPs with non-specific IgG showed negligible precipitation in both cell lines. Thus, the 613-bp deletion in the JH6 intronic sequence was dispensable for Pol II accumulation.
Fig. 4.
Pol II abundance in V and C regions as detected by ChIP. A) Position of V (red bar) and C (gray bar) primer sets within the WT-A and Δ intron cell lines. B) ChIP assays for Pol II and IgG from 4 independent experiments with 1–2 clones per strain per experiment. The dotted line shows the level of Pol II using nonspecific IgG. Error bars represent the standard error of the mean. Significance was measured using one-tailed paired student t-test; ns, not significant.
3.3. SHM was comparable in V regions from WT-A and Δ intron cells
To address the role of JH6 intronic sequences in promoting mutagenesis, mutation frequencies were determined in WT-A and Δ intron cells after 12 weeks in culture. Frequencies were calculated for all unique mutations that occurred in the 700 bp region between the V region TATA box and the EcoRI site common between both cell lines (Fig. 5A). DNA sequencing revealed that there was no significant change in mutation frequency between WT-A and Δ intron cell lines (Fig. 5B). The spectra of changes for the single nucleotide substitutions are listed in Fig. 5C, and show that over 90% of the mutations were at G and C nucleotides, indicating AID deamination of C on both DNA strands. This is in accord with previous reports indicating a strong G:C bias in Ramos mutations (Sale and Neuberger, 1998), despite the presence of proteins that are associated with A:T mutations, such as functional MSH2/MSH6 repair proteins and DNA polymerase eta in the cells (Xiao et al., 2007). Ramos and another Burkitt lymphoma line, BL2 (Denepoux et al., 1997), may lack the A:T spectra if uracils are repaired earlier in the cell cycle than they are in vivo, leading to more faithful base excision repair and less mutagenic mismatch repair (Kano et al., 2011; Saribasak and Gearhart, 2012).
Fig. 5.
Mutation analysis in WT-A and Δ intron cells. A) Area sequenced for mutations, including exons and flanking DNA. Blue arrow represents 700 bp. B) Average mutation frequency from 2 and 4 independent clones for WT-A and Δ intron cell lines, respectively. P value by Fisher’s exact test. C) Types of substitutions. Data are expressed as % of the number of mutations.
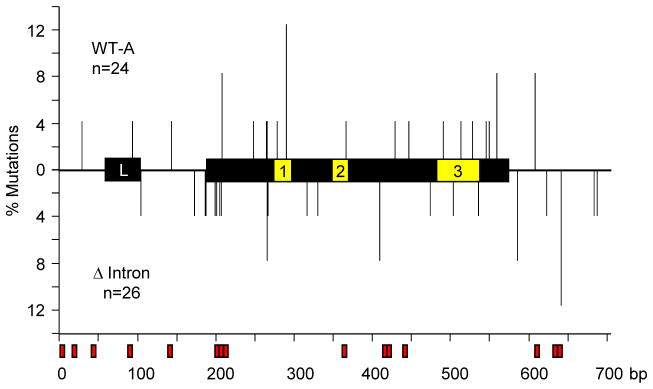
To evaluate the possibility that deletion of a large region of the JH intron might shift the mutations towards the 5′ end of the VDJ exon, we mapped the location of mutations. As shown in Fig. 6, the distribution of mutations was not significantly different between the two cell lines (p = 0.8, Mann-Whitney test). As expected, mutations were not clustered in the complementarity-determining regions in Ramos cells, which mutate constitutively in the absence of antigen selection. There was also no discernible accumulation in the 15 WGCW (W = A/T) hotspots (Martomo et al., 2004; Zarrin et al., 2004) in this limited sample. Although some V genes contain overlapping hotspots in complementarity-determining regions 1 and 2 that generate a high frequency of mutations (Wei et al., 2015), the VH4-34 gene does not have shared motifs in these regions. In summary, like Pol II accumulation, deletion of most of the JH6 intronic sequence had no distinguishable effect on mutagenesis.
Fig. 6.
Location of mutations. Graph depicts the position of mutations as vertical lines relative to the L and VDJ exons, containing complementarity-determining regions 1, 2, and 3. The % mutations is plotted against the length of the region (bp). WGCW hotspots are outlined in red boxes below the x-axis. Upper half, mutations in WT-A; bottom half, mutations in Δ intron; n, number.
4. Discussion
During affinity maturation, Pol II dynamics within the immunoglobulin gene are a critically important regulator of AID activity. Within switch regions, the repetitive sequences are sufficient to stall Pol II and give AID abundant access to DNA (Rajagopal et al., 2009; Wang et al., 2009). However, the mechanism that targets AID to the V region remains elusive. Recently, we showed that Pol II accumulates within the V region in a manner that is reminiscent of the necessity for pausing in the switch region (Maul et al., 2014). To identify potential cis DNA sequences that may cause Pol II accumulation, we manipulated DNA within the IgH locus of the Ramos cell line. Ramos cells have been utilized for a plethora of somatic hypermutation studies, beginning with experiments by Sale and Neuberger, who found strand breaks in the V gene that were tagged by terminal deoxynucleotidyl transferase (Sale and Neuberger, 1998). Furthermore, Ramos cells have been utilized to address the role of SHM-associated features including new specificities (Cumbers et al., 2002), V gene alterations (Bemark and Neuberger, 2003), single-stand DNA abundance (Ronai et al., 2007), AID recruitment (Singh et al., 2013), Pol II dynamics (Parsa et al., 2012; Wang et al., 2014), AID hotspot dynamics (MacCarthy et al., 2009; Wei et al., 2015), and DNA repair (Frieder et al., 2009). Finally, the cells have been used to probe genome-wide AID targeting (Meng et al., 2014).
Using this adaptable line, we examined sequences that may slow Pol II progression and attract AID. We removed 613 bp of the JH6 intron downstream of the VDJ gene and measured Pol II and SHM levels. This region was chosen because it sustains high levels of mutation and Pol II accumulation. Analysis of Pol II abundance confirmed the finding that V regions accumulate significantly higher levels of Pol II than C regions (Maul et al., 2014); however, this accumulation was unaltered in the Δ intron cells, and the frequency of mutation was unchanged. It is possible that the remaining 150 bp adjacent to JH6 and 200 bp adjacent to Eμ could modulate SHM activity. Nonetheless, our data show that most of the intron does not contain regulatory information for targeting SHM to the VDJ gene. Taken together with published reports that the V region promoter, VDJ exon, and intronic enhancer sequences are dispensable (Betz et al., 1994; Fukita et al., 1998; Inlay et al., 2006; Li et al., 2010; Perlot et al., 2005; Peters and Storb, 1996; Shen et al., 2001; Tumas-Brundage and Manser, 1997; Yeap et al., 2015; Yelamos et al., 1995), it is reasonable to conclude that most of the V region is superfluous for SHM. The finding that the 3′ regulatory region has a significant role in controlling immunoglobulin transcription, chromatin structure, and subsequent mutagenesis (Rouaud et al., 2013), suggests that V region transcriptional accumulation and mutagenesis is regulated through contact between the promoter and 3′ regulatory region, and not through local DNA sequence.
Highlights.
JH downstream intron sequences may recruit hypermutation to variable genes.
Deletion of DNA in Ramos cells did not affect RNA Pol II or mutation accumulation.
Most of the 3′ intron sequence is dispensable for somatic hypermutation.
Acknowledgments
We gratefully thank Matthew Scharff for the Hyg-TK and WT-A cell lines, the WT-A replacement construct, Cre vector, and advice. Juan Alvarez-Gonzalez and William Yang assisted in experiments.
Funding
This work was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging, and by an NIH Intramural AIDS Research fellowship to D.P.C.
Abbreviations
- AID
activation-induced deaminase
- Pol II
RNA polymerase II
- SHM
somatic hypermutation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baughn LB, Kalis SL, MacCarthy T, Wei L, Fan M, Bergman A, Scharff MD. Recombinase-mediated cassette exchange as a novel method to study somatic hypermutation in Ramos cells. MBio. 2011;2:1–9. doi: 10.1128/mBio.00186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Neuberger MS. By-products of immunoglobulin somatic hypermutation. Genes, chromosomes & cancer. 2003;38:32–39. doi: 10.1002/gcc.10241. [DOI] [PubMed] [Google Scholar]
- Betz AG, Milstein C, Gonzalez-Fernandez A, Pannell R, Larson T, Neuberger MS. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- Both GW, Taylor L, Pollard JW, Steele EJ. Distribution of mutations around rearranged heavy-chain antibody variable-region genes. Mol Cell Biol. 1990;10:5187–5196. doi: 10.1128/mcb.10.10.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci USA. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- Cumbers SJ, Williams GT, Davies SL, Grenfell RL, Takeda S, Batista FD, Sale JE, Neuberger MS. Generation and iterative affinity maturation of antibodies in vitro using hypermutating B-cell lines. Nature biotechnology. 2002;20:1129–1134. doi: 10.1038/nbt752. [DOI] [PubMed] [Google Scholar]
- Denepoux S, Razanajaona D, Blanchard D, Meffre G, Capra JD, Banchereau J, Lebecque S. Induction of somatic mutation in a human B cell line in vitro. Immunity. 1997;6:35–46. doi: 10.1016/s1074-7613(00)80240-x. [DOI] [PubMed] [Google Scholar]
- Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieder D, Larijani M, Collins C, Shulman M, Martin A. The concerted action of Msh2 and UNG stimulates somatic hypermutation at A. T base pairs. Mol Cell Biol. 2009;29:5148–5157. doi: 10.1128/MCB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukita Y, Jacobs H, Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- Gearhart PJ, Bogenhagen DF. Clusters of point mutations are found exclusively around rearranged antibody variable genes. Proc Natl Acad Sci U S A. 1983;80:3439–3443. doi: 10.1073/pnas.80.11.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Fernandez A, Gupta SK, Pannell R, Neuberger MS, Milstein C. Somatic mutation of immunoglobulin lambda chains: a segment of the major intron hypermutates as much as the complementarity-determining regions. Proc Natl Acad Sci USA. 1994;91:12614–12618. doi: 10.1073/pnas.91.26.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens T, Klein U, Kuppers R. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci U S A. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J, Jr, Rogerson BJ, O’Brien RL, Storb U. Analysis of somatic mutations in kappa transgenes. J Exp Med. 1990;172:131–137. doi: 10.1084/jem.172.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Masani S, Yu K. Overlapping activation-induced cytidine deaminase hotspot motifs in Ig class-switch recombination. Proc Natl Acad Sci U S A. 2011;108:11584–11589. doi: 10.1073/pnas.1018726108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlay MA, Gao HH, Odegard VH, Lin T, Schatz DG, Xu Y. Roles of the Ig kappa light chain intronic and 3′ enhancers in Igk somatic hypermutation. J Immunol. 2006;177:1146–1151. doi: 10.4049/jimmunol.177.2.1146. [DOI] [PubMed] [Google Scholar]
- Jolly CJ, Klix N, Neuberger MS. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 1997;25:1913–1919. doi: 10.1093/nar/25.10.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano C, Ouchida R, Kokubo T, Wang JY. Rapid cell division contributes to efficient induction of A/T mutations during Ig gene hypermutation. Mol Immunol. 2011;48:1993–1999. doi: 10.1016/j.molimm.2011.06.218. [DOI] [PubMed] [Google Scholar]
- Kim S, Davis M, Sinn E, Patten P, Hood L. Antibody diversity: somatic hypermutation of rearranged VH genes. Cell. 1981;27:573–581. doi: 10.1016/0092-8674(81)90399-8. [DOI] [PubMed] [Google Scholar]
- Kothapalli NR, Collura KM, Norton DD, Fugmann SD. Separation of mutational and transcriptional enhancers in Ig genes. J Immunol. 2011;187:3247–3255. doi: 10.4049/jimmunol.1101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebecque SG, Gearhart PJ. Boundaries of somatic mutation in rearranged immunoglobulin genes: 5′ boundary is near the promoter, and 3′ boundary is approximately 1 kb from V(D)J gene. J Exp Med. 1990;172:1717–1727. doi: 10.1084/jem.172.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Yan Y, Pieretti J, Feldman DA, Eckhardt LA. Comparison of identical and functional Igh alleles reveals a nonessential role for Emu in somatic hypermutation and class-switch recombination. J Immunol. 2010;185:6049–6057. doi: 10.4049/jimmunol.0902992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCarthy T, Kalis SL, Roa S, Pham P, Goodman MF, Scharff MD, Bergman A. V-region mutation in vitro, in vivo, and in silico reveal the importance of the enzymatic properties of AID and the sequence environment. Proc Natl Acad Sci U S A. 2009;106:8629–8634. doi: 10.1073/pnas.0903803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Cao Z, Venkataraman L, Giorgetti CA, Press JL, Denizot Y, Du H, Sen R, Gearhart PJ. Spt5 accumulation at variable genes distinguishes somatic hypermutation in germinal center B cells from ex vivo-activated cells. J Exp Med. 2014;211:2297–2306. doi: 10.1084/jem.20131512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Saribasak H, Martomo SA, McClure RL, Yang W, Vaisman A, Gramlich HS, Schatz DG, Woodgate R, Wilson DM, 3rd, Gearhart PJ. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat Immunol. 2011;12:70–76. doi: 10.1038/ni.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng FL, Du Z, Federation A, Hu J, Wang Q, Kieffer-Kwon KR, Meyers RM, Amor C, Wasserman CR, Neuberg D, Casellas R, Nussenzweig MC, Bradner JE, Liu XS, Alt FW. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 2014;159:1538–1548. doi: 10.1016/j.cell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Parsa JY, Ramachandran S, Zaheen A, Nepal RM, Kapelnikov A, Belcheva A, Berru M, Ronai D, Martin A. Negative supercoiling creates single-stranded patches of DNA that are substrates for AID-mediated mutagenesis. PLoS Genet. 2012;8:e1002518. doi: 10.1371/journal.pgen.1002518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, Nieland TJ, Root DE, Casellas R, Nussenzweig MC. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M, Hochtl J, Schnell H, Zachau HG. Differences between germ-line and rearranged immunoglobulin V kappa coding sequences suggest a localized mutation mechanism. Nature. 1981;291:668–670. doi: 10.1038/291668a0. [DOI] [PubMed] [Google Scholar]
- Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci U S A. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Harris RS, Neuberger MS. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 2002;418:99–103. doi: 10.1038/nature00862. [DOI] [PubMed] [Google Scholar]
- Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- Qian J, Wang Q, Dose M, Pruett N, Kieffer-Kwon KR, Resch W, Liang G, Tang Z, Mathe E, Benner C, Dubois W, Nelson S, Vian L, Oliveira TY, Jankovic M, Hakim O, Gazumyan A, Pavri R, Awasthi P, Song B, Liu G, Chen L, Zhu S, Feigenbaum L, Staudt L, Murre C, Ruan Y, Robbiani DF, Pan-Hammarstrom Q, Nussenzweig MC, Casellas R. B cell super-enhancers and regulatory clusters recruit AID tumorigenic activity. Cell. 2014;159:1524–1537. doi: 10.1016/j.cell.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal D, Maul RW, Ghosh A, Chakraborty T, Khamlichi AA, Sen R, Gearhart PJ. Immunoglobulin switch mu sequence causes RNA polymerase II accumulation and reduces dA hypermutation. J Exp Med. 2009;206:1237–1244. doi: 10.1084/jem.20082514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat Immunol. 2003;4:452–456. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Ronai D, Iglesias-Ussel MD, Fan M, Li Z, Martin A, Scharff MD. Detection of chromatin-associated single-stranded DNA in regions targeted for somatic hypermutation. J Exp Med. 2007;204:181–190. doi: 10.1084/jem.20062032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouaud P, Vincent-Fabert C, Saintamand A, Fiancette R, Marquet M, Robert I, Reina-San-Martin B, Pinaud E, Cogne M, Denizot Y. The IgH 3′ regulatory region controls somatic hypermutation in germinal center B cells. J Exp Med. 2013;210:1501–1507. doi: 10.1084/jem.20130072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale JE, Neuberger MS. TdT-accessible breaks are scattered over the immunoglobulin V domain in a constitutively hypermutating B cell line. Immunity. 1998;9:859–869. doi: 10.1016/s1074-7613(00)80651-2. [DOI] [PubMed] [Google Scholar]
- Saribasak H, Gearhart PJ. Does DNA repair occur during somatic hypermutation? Semin Immunol. 2012;24:287–292. doi: 10.1016/j.smim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Peters A, Kao D, Storb U. The 3′ Igkappa enhancer contains RNA polymerase II promoters: implications for endogenous and transgenic kappa gene expression. Int Immunol. 2001;13:665–674. doi: 10.1093/intimm/13.5.665. [DOI] [PubMed] [Google Scholar]
- Singh SK, Maeda K, Eid MM, Almofty SA, Ono M, Pham P, Goodman MF, Sakaguchi N. GANP regulates recruitment of AID to immunoglobulin variable regions by modulating transcription and nucleosome occupancy. Nature communications. 2013;4:1830. doi: 10.1038/ncomms2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohail A, Klapacz J, Samaranayake M, Ullah A, Bhagwat AS. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res (Amst) 2003;31:2990–2994. doi: 10.1093/nar/gkg464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumas-Brundage K, Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J Exp Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J Exp Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fan M, Kalis S, Wei L, Scharff MD. A source of the single-stranded DNA substrate for activation-induced deaminase during somatic hypermutation. Nature communications. 2014;5:4137. doi: 10.1038/ncomms5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Chahwan R, Wang S, Wang X, Pham PT, Goodman MF, Bergman A, Scharff MD, MacCarthy T. Overlapping hotspots in CDRs are critical sites for V region diversification. Proc Natl Acad Sci U S A. 2015;112:E728–737. doi: 10.1073/pnas.1500788112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann KL, Milosevic S, Pauklin S, Schmitz KM, Rangam G, Simon MT, Maslen S, Skehel M, Robert I, Heyer V, Schiavo E, Reina-San-Martin B, Petersen-Mahrt SK. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J Exp Med. 2012;209:2099–2111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Ray M, Jiang C, Clark AB, Rogozin IB, Diaz M. Known components of the immunoglobulin A:T mutational machinery are intact in Burkitt lymphoma cell lines with G:C bias. Mol Immunol. 2007;44:2659–2666. doi: 10.1016/j.molimm.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap LS, Hwang JK, Du Z, Meyers RM, Meng FL, Jakubauskaite A, Liu M, Mani V, Neuberg D, Kepler TB, Wang JH, Alt FW. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 2015;163:1124–1137. doi: 10.1016/j.cell.2015.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelamos J, Klix N, Goyenechea B, Lozano F, Chui YL, Gonzalez Fernandez A, Pannell R, Neuberger MS, Milstein C. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- Zarrin AA, Alt FW, Chaudhuri J, Stokes N, Kaushal D, Du Pasquier L, Tian M. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat Immunol. 2004;5:1275–1281. doi: 10.1038/ni1137. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bardwell PD, Woo CJ, Poltoratsky V, Scharff MD, Martin A. Clonal instability of V region hypermutation in the Ramos Burkitt’s lymphoma cell line. Int Immunol. 2001;13:1175–1184. doi: 10.1093/intimm/13.9.1175. [DOI] [PubMed] [Google Scholar]
- Zhu C, Hsu E. Error-prone DNA repair activity during somatic hypermutation in shark B lymphocytes. J Immunol. 2010;185:5336–5347. doi: 10.4049/jimmunol.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]