Abstract
Background
Plasma renin is an important regulator of blood pressure (BP). Plasma renin activity (PRA) has been shown to correlate with variability in BP response to antihypertensive agents. We conducted a genome-wide association study to identify SNPs associated with baseline PRA using data from the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study.
Methods and Results
Multiple linear regression analysis was performed in 461 whites and 297 African Americans using an additive model, adjusting for age, sex, and ancestry-specific principal components. Top SNPs were prioritized by testing the expected direction of association for BP response to atenolol and hydrochlorothiazide. Top regions from the BP response prioritization were tested for functional evidence through differences in gene expression by genotype using RNA-Seq data. Regions with functional evidence were assessed for replication with baseline PRA in an independent study (PEAR-2). Our top SNP rs3784921 was in the SNN-TXNDC11 gene region. The G allele of rs3784921 was associated with higher baseline PRA (β=0.47, p=2.09 x 10−6) and smaller systolic BP reduction in response to hydrochlorothiazide (β=2.97, one-sided p=0.006). Additionally, TXNDC11 expression differed by rs3784921 genotype (p=0.007), and rs1802409, a proxy SNP for rs3784921 (r2=0.98–1.00), replicated in PEAR-2 (β=0.15, one-sided p=0.038). Additional SNPs associated with baseline PRA that passed BP response prioritization were in/near the genes CHD9, XIRP2, and GHR.
Conclusions
We identified multiple regions associated with baseline PRA that were prioritized through BP response signals to two mechanistically different antihypertensive drugs.
Clinical Trial Registration
http://clinicaltrials.gov. Unique identifier: NCT00246519.
Keywords: renin, hypertension, high blood pressure, blood pressure, pharmacogenetics, blood pressure response
Journal Subject Terms: Genetic, Association Studies, Hypertension, High Blood Pressure, Pharmacology
INTRODUCTION
Hypertension is the most prevalent chronic cardiovascular disease, affecting approximately one billion individuals worldwide, and ~80 million adults in the United States.1, 2 Additionally, hypertension contributes to 50% of heart related diseases and stroke, and accounts for ~18% of deaths worldwide.3 These cardiovascular-related complications are largely avoidable with effective blood pressure (BP) management with antihypertensive medications, potentially resulting in a 35–40% reduction in stroke, a 15–25% reduction in myocardial infarction, and a 50% reduction in congestive heart failure.4–6 Approximately, 54% of adults with hypertension have controlled BP (<140/90 mmHg).2 The inter-individual variability in BP response to antihypertensive agents partially accounts for the suboptimal BP control rates.
In addition to known clinical factors associated with BP response, including age, gender, and baseline BP, 7, 8 there is well-documented evidence that variability in components of the renin-angiotensin-aldosterone system (RAAS), including renin, is linked to differences in BP response to antihypertensive agents.9–12 Lower plasma renin activity (PRA) has been linked to better response to thiazide diuretics (hydrochlorothiazide, HCTZ)9, 11 while higher PRA has been associated with better BP response to RAAS-acting agents such as β-blockers (atenolol).11 Collectively, these studies suggest that baseline PRA has reciprocal effects on BP response to antihypertensive drugs that inhibit versus stimulate activity of the RAAS (e.g. diuretics vs. β-blockers).12 Accordingly, because β-blockers inhibit renin release, individuals with higher baseline PRA have better BP response to β-blockers; conversely, because diuretics stimulate renin release, individuals with higher baseline PRA do not respond as well to diuretics.9, 11
While several demographic and clinical factors, including age, race, and PRA are predictive of antihypertensive drug response,11 much of the remaining inter-individual variability in BP response remains unexplained. There have been prior studies on the genetic influence of BP response;13–15 however, the genetic variability of PRA, an important clinical predictor of BP response, is understudied. A recently published genome-wide association study (GWAS) in normotensive individuals documented two independent loci in kininogen 1 (KNG1) and kallikrein B (KLKB1) showing genome-wide significant associations with components of RAAS including PRA, plasma renin concentrations, and aldosterone levels.16 Yet, this phenotype has not been examined in hypertensive individuals, where the results could be used, along with other demographic, clinical, and genetic factors, to predict antihypertensive medication response.
Using a genome-wide association approach, we aimed to identify genetic variants associated with pre-treatment or baseline PRA in hypertensive individuals. For those signals that met the genome-wide or suggestive level of significance, we tested the direction of association with BP response to either the β-blocker atenolol or the thiazide diuretic HCTZ. We also assessed for potential function through gene expression differences by genotype, and tested for replication of the top PRA signal in an independent sample.
METHODS
The genome-wide association data and phenotypes for The Pharmacogenomic Evaluation of Antihypertensive Response (PEAR) study are available on dbGaP (phs000649.v1.p1).
Study Design and Participants
PEAR
The Pharmacogenomic Evaluation of Antihypertensive Response (PEAR) study (Clinical Trial ID NCT00246519, www.clinicaltrials.gov), was a prospective, multicenter, randomized, open-label clinical trial in mild-to-moderate hypertensive participants. Additional details on the PEAR trial have been reported previously.17 Briefly, PEAR enrolled 768 men and women, ages 17–65 years with uncomplicated primary hypertension. After an average washout period of 4 weeks, participants were randomized to either atenolol 50 mg or HCTZ 12.5 mg once daily monotherapy for 3 weeks, followed by dose titration to atenolol 100 mg or HCTZ 25 mg once daily if BP remained above >120/70 mmHg for an additional 6 weeks. After the monotherapy period, drug from the other treatment arm was added if BP remained above goal, followed by a similar dose titration protocol, for another 6–9 weeks of treatment.
PEAR-2
The Pharmacogenomic Evaluation of Antihypertensive Response-2 (PEAR-2) study (Clinical Trial ID NCT01203852, www.clinicaltrials.gov) was a prospective, randomized, multicenter, sequential, open-label clinical trial in mild-to-moderate hypertensive participants. During PEAR-2, participants were initiated on either the β-blocker metoprolol 50 mg or the thiazide-like diuretic chlorthalidone 15 mg twice daily for 2 weeks, followed by dose titration to metoprolol 100 mg or chlorthalidone 25 mg twice daily for an additional 6 weeks if BP remained >120/70 mmHg. After a minimum 4-week washout period, monotherapy with the other drug followed by a similar dose titration protocol was conducted for another 6–8 weeks of treatment. Inclusion criteria and additional details on the PEAR-2 trial have been previously published.18
All PEAR and PEAR-2 study participants provided written informed consent. Both studies were approved by the Institutional Review Boards at the participating clinical trial sites (University of Florida in Gainesville, FL; Mayo Clinic in Rochester, MN; and Emory University in Atlanta, GA) and were conducted in accordance with the principles of the Declaration of Helsinki and the US Code of the Federal Regulations for Protection of Human Subjects.
Determination of Plasma Renin Activity
PEAR
Laboratory values including PRA were acquired at baseline, after monotherapy, and after combination therapy.17 PRA was measured by radioimmunoassay using reagents purchased from DiaSorin (Stillwater, MN), a 3 hour incubation period for angiotensin I generation, and the method published by Sealey.19 All measurements were conducted at a central laboratory at the Mayo Clinic, Rochester, MN. Samples were tested in duplicate or triplicate, and participants’ mean values were used for analysis.
PEAR-2
Laboratory values, including PRA were acquired at baseline before initiation of each drug treatment, as well as after each monotherapy period. PRA during PEAR-2 was determined at the Mayo Clinic using the same methods and reagents as during PEAR.
Blood Pressure Phenotype
During PEAR, BP measurements were collected at baseline, after monotherapy, and after combination therapy through three different methods: office, home, and ambulatory, as previously described.17 A composite-weighted average of BP response through these different methods (office, home, ambulatory daytime, and ambulatory nighttime) was previously calculated based on row sums of the inverse of the inter-method covariance matrices.20 The composite-weighted average BP response had a higher signal-to-noise ratio compared with any BP measurement method individually.20 The composite BP response measurement is considered the most precise BP response phenotype available in PEAR and was selected for analysis. In its calculation, the composite BP response measurement is already adjusted for age, sex, and baseline BP. The BP phenotype from PEAR-2 was not used for this study.
SNP Genotyping and Quality Control
PEAR
Participants’ genomic DNA was genotyped for >1 million SNPs using the Illumina Human Omni1-Quad BeadChip (Illumina, San Diego, CA). The quality control steps employed have been previously published.13 Principal components for ancestry were identified using the EIGENSTRAT method.21 Genotype imputation was performed using MaCH (version 1.0.16) and HapMap III reference panels.22 SNPs with minor allele frequency (MAF) <3% and/or imputation quality r2 <0.3 were excluded. After imputation and quality control, the final genetic dataset consisted of 758 samples (n=461 white, n=297 African American).
PEAR-2
Participants’ genomic DNA was genotyped for >2.5 million SNPs using the Illumina Human Omni2.5S BeadChip (Illumina, San Diego, CA). Quality control and principal component analysis during PEAR-2 used the same methods as PEAR. For the replication study, only directly typed SNP data were used. Genotype data were available on 303 samples (n=150 white, n=153 African American).
Genome-wide Association Study and SNP Prioritization
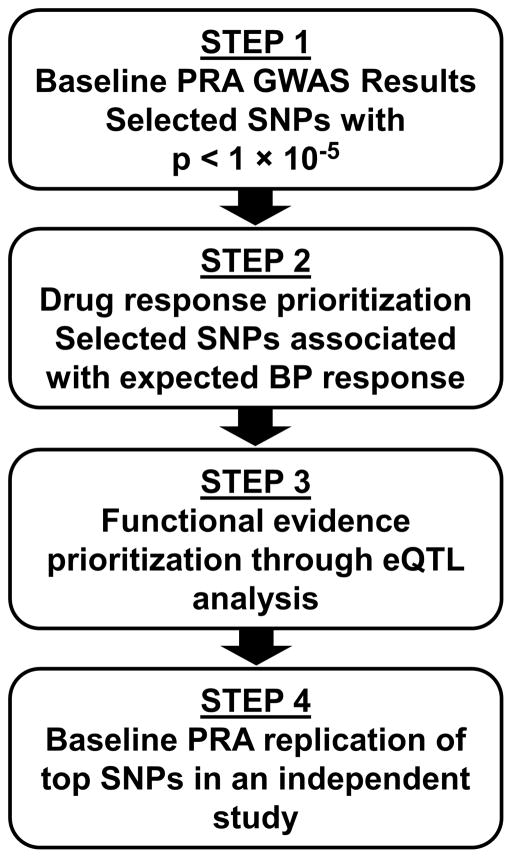
Multiple linear regression was performed using PLINK23 to test the association between genotype dosage and baseline log-transformed PRA in PEAR whites and African Americans separately, adjusting for age, gender and ancestry-specific principal components 1 and 2, under an additive genetic model. Of the top GWAS SNPs associated with baseline PRA (p < 1x10−5), we selected one SNP per gene region to move forward for drug response prioritization (Figure 1, Step 1). During the drug response prioritization step, we assessed whether any of the top SNPs were associated with BP response in the expected direction (Figure 1, Step 2). In this step, SNPs were tested for association with the composite BP response [systolic BP (SBP) and diastolic BP (DBP)] to atenolol or HCTZ monotherapy in PEAR, additionally adjusting for principal components 1 and 2 for ancestry. We expected that SNPs associated with higher baseline PRA would be associated with better BP response to atenolol or reduced response to HCTZ. Conversely, we expected that SNPs associated with lower baseline PRA would be associated with better BP response to HCTZ or reduced response to atenolol.11 The significance threshold for the drug response prioritization step was a one-sided p < 0.05, as we had a one-sided hypothesis for each SNP. All analyses were done separately by ancestry due to race-based differences in allele frequencies, linkage disequilibrium (LD),24 and antihypertensive drug response.9, 10, 25 Haploreg and GTEx were used to assess the potential functional consequences of the SNPs and gene regions that passed the drug response prioritization step.26, 27 Stepwise linear regression was conducted in SAS v. 9.4, by forcing all covariates into the model, in order to determine the partial R2 or variability of the traits that was explained by the top SNP (rs3784921).
Figure 1.
Approach used for selection and prioritization of top SNPs
SNP: single nucleotide polymorphism; PRA: plasma renin activity; GWAS: genome-wide association study; RNA-Seq data was used to conduct association of top SNPs with gene expression (eQTL analysis); PEAR data was used for discovery GWAS analysis; PEAR-2 was used for independent study replication of top SNPs
Gene Expression Analysis for Functional Prioritization
Gene expression analysis by genotype was performed for the top genes and top SNPs associated with PRA that passed the drug response prioritization step, namely, whether the SNP was associated with BP response in the expected direction (Figure 1, Step 3). We used RNA-Seq data from 50 PEAR white participants, who were selected based on the extreme quartiles of BP response after HCTZ treatment: 25 poor BP responders and 25 good BP responders. RNA was extracted from whole blood samples collected at baseline, using the PAXgene Blood RNA kit IVD (Qiagen, Valencia, CA). Libraries were prepared following poly(A) mRNA selection and strand-specific protocol.28 DNA clusters were generated using the Illumina cluster station, followed by 100 cycles of paired-end sequencing on the Illumina HiSeq 2000. Quality per base pair was checked using FastQC. 100 bp paired-end RNA-Seq reads were mapped to the human genome (hg19) using TopHat2,29 and read duplicates were removed using Picard (http://picard.sourceforge.net) MarkDuplicates option. Transcript assembly, and statistical analysis were conducted with Cufflink/Cuffdiff.29 Gene expression levels are reported here in fragments per kilobase per million reads (FPKM). Considering reads mapped to exonic regions of SNN, TXNDC11 and CHD9, eQTL analyses were carried out with genotype calls for rs3784921 (SNN), rs3784921 (TXNDC11), and rs7184292 (CHD9) in R, fitting linear regression models using additive or dominant effects. GHR and XIRP2 were excluded from analysis as they were expressed at extremely low levels in the dataset. The p-value for significance was set at 0.017 (0.05/3 tests) for the eQTL analysis.
Replication of the Top Signal(s)
SNPs from the gene regions that passed the drug response prioritization step (Figure 1, Step 2) and the functional evidence prioritization step (Figure 1, Step 3) were moved forward for replication of the baseline PRA signal in an independent sample of whites from the PEAR-2 study (Figure 1, Step 4). From the SNN-TXNDC11 region, only rs1802409 was available in the replication dataset; the other SNPs were not included on the Illumina Human Omni2.5S BeadChip. As the two SNPs in the SNN-TXNDC11 region are in high LD (r2=0.98–1.00, D′=1), rs1802409 served as a proxy for the region. Multiple linear regression was performed using PLINK23 to test the association between genotype and baseline log-transformed PRA in PEAR-2 whites, adjusting for age, gender and principal component 1 for ancestry, under an additive genetic model. A one-sided significance threshold of p < 0.05 was selected, as only one SNP was tested, and we had a one-sided hypothesis. The results from the discovery and replication were combined in meta-analysis using METAL under a fixed-effects model, weighting by sample size.30
RESULTS
Baseline demographic and clinical characteristics of PEAR study participants are presented in Table 1. A total of 758 study participants were available for analysis after quality control procedures; 61% were white and 39% were African American. Baseline PRA was lower among African Americans than among whites, with an average baseline PRA of 0.59 ± 0.68 ng/mL/hr in African Americans and 1.24 ± 1.26 ng/mL/hr in whites.
Table 1.
Baseline characteristics of PEAR white and African American participants
| Baseline Characteristics | Whites N=461 |
African Americans N=297 |
|---|---|---|
| Age, years | 49.7 ± 9.5 | 47.3 ± 8.6 |
| Female, N (%) | 200 (43.4) | 199 (66.8) |
| Body Mass Index, kg/m2 | 30.3 ± 5.2 | 31.5 ± 5.9 |
| Duration of hypertension, years | 6.5 ± 7.2 | 6.7 ± 7.2 |
| Current smoker, N (%) | 57 (12.4) | 52 (17.4) |
| Baseline SBP, mmHg | 145.5 ± 9.7 | 146.1 ± 11.1 |
| Baseline DBP, mmHg | 93.2 ± 5.5 | 94.5 ± 6.5 |
| Baseline PRA, ng (angiotensin)/mL per hour | 1.24 ± 1.26 | 0.59 ± 0.68 |
Values are presented as mean ± standard deviation unless otherwise noted. SBP: systolic blood pressure; DBP: diastolic blood pressure; PRA: plasma renin activity; PEAR: Pharmacogenomic Evaluation of Antihypertensive Responses
Manhattan plots and corresponding Q-Q plots for the association of SNP dosage with baseline PRA in whites and African Americans are shown in Supplemental Figures S1, S2. Two SNPs (rs7184292 and rs7201620) near CHD9 on chromosome 16 achieved genome-wide significance (p < 5x10−8) in whites. Additionally, 34 SNPs in eight regions among white participants and eight SNPs in seven regions among African American participants met the suggestive level of association with baseline PRA (p < 1x10−5). In total, eight SNPs associated with baseline PRA at the suggestive level of association (p < 1x10−5) also showed association with BP response to atenolol or HCTZ in the expected direction (p < 0.05) during the prioritization stage. All eight SNPs were in Hardy-Weinberg Equilibrium (p > 0.01). Table 2 and Supplemental Table S1 show the top SNPs from each gene region in white and African American participants, respectively; Figure 2 shows the flow of the results.
Table 2.
Genome-wide association results for baseline PRA among PEAR white participants and prioritization with respect to atenolol or HCTZ BP response
| SNP | CHR | Location | Nearest gene region | #SNPs in region | A1 | Freq A1 | Baseline log PRA | SBP Response | DBP Response | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P-value | β (SE) | P-value* | β (SE) | P-value* | |||||||
| Validation through Atenolol BP Response | Atenolol BP Response | |||||||||||
| rs7184292 | 16 | 51,477,067 | CHD9 | 2 | A | 0.42 | 0.30 (0.05) | 4.05E-09 | −1.68 (0.78) | 0.015 | −0.61 (0.51) | 0.113 |
| Validation through HCTZ BP Response | HCTZ BP Response | |||||||||||
| rs3784921 | 16 | 11,681,702 | SNN-TXNDC11 | 3 | G | 0.93 | 0.47 (0.10) | 2.09E-06 | 2.97 (1.19) | 0.006 | 1.94 (0.82) | 0.009 |
P-value is one-sided, based on the one-sided prioritization hypothesis
SNP: Single nucleotide polymorphism; CHR: Chromosome; Location based on NCBI build 36; A1: Reference Allele; HCTZ: hydrochlorothiazide; BP: blood pressure; DBP: diastolic blood pressure; SBP: systolic blood pressure; PRA: plasma renin activity; PEAR: Pharmacogenomic Evaluation of Antihypertensive Responses
N=461 (Baseline PRA), N=233 (Atenolol BP response), N=228 (HCTZ BP response)
Figure 2.
Flow of the results in white and African Americans through the selection and prioritization steps
SNP: single nucleotide polymorphism; PRA: plasma renin activity; ATEN: atenolol; HCTZ: hydrochlorothiazide; PEAR data was used for discovery GWAS analysis; PEAR-2 was used for independent study replication of top SNPs
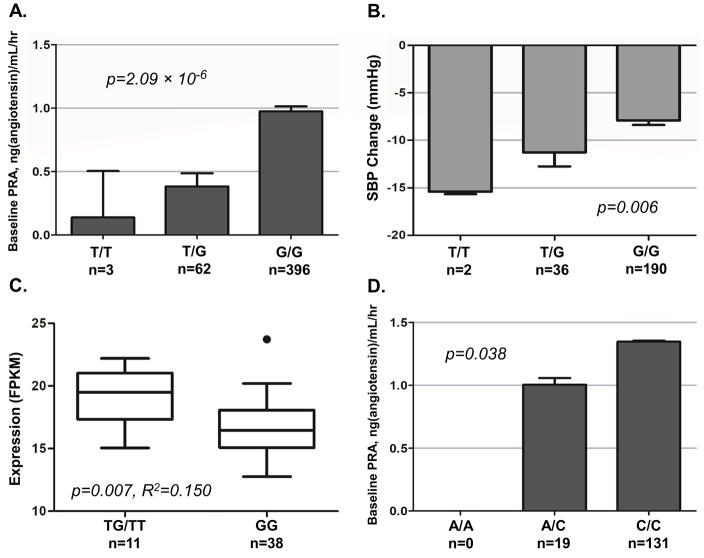
Three SNPs in the SNN-TXNDC11 gene region at 16p13.13 were associated with baseline PRA at a suggestive level (p < 1.0 x 10−5). Of the three SNPs, we selected the top SNP rs3784921 to move forward to prioritization. This SNP passed the drug response prioritization step through association with SBP response to HCTZ in whites, where the G allele of rs3784921 was associated with higher baseline PRA (β= 0.47, p = 2.09 x 10−6; Table 2, Figure 3 panel A), and reduced SBP response to HCTZ (β = 2.97, one-sided p = 0.006; Table 2, Figure 3 panel B), with a mean baseline PRA of 0.14, 0.38, and 0.97 ng/mL/hr and a mean HCTZ-induced SBP change of −15.4, −11.3, and −7.9 mmHg for the TT, TG, and GG genotypes, respectively. Within whites in PEAR1, rs3784921 accounted for 3.8% of the variability in baseline renin. Additionally, rs3784921 accounted for 1.4% and 1.1% of the variability of SBP response to HCTZ and DBP response to HCTZ, respectively.
Figure 3.
Baseline PRA and SBP to HCTZ by TXNDC11 rs3784921 genotype (Panels A, B) and TXNDC11 expression (FPKM) by rs3784921 genotype (Panel C) in PEAR white participants; baseline PRA by SNN-TXNDC11 rs1802409 (proxy SNP) genotype in PEAR-2 white participants (Panel D)
*rs1802409 is a proxy for the top SNP rs3784921 in SNN-TXNDC11 gene region (r2=0.98–1.00); Means and standard error bars are shown (Panels A, B, D); PRA: plasma renin activity; SBP: systolic blood pressure; ATEN: atenolol; HCTZ: hydrochlorothiazide; PEAR: Pharmacogenomic Evaluation of Antihypertensive Responses; FPKM: Fragments per kilobase per million reads RNA-Seq analysis was performed in N=50 PEAR white participants; PEAR white participants: N=461 (Baseline PRA), N=228 (HCTZ response); PEAR-2 white participants: N=150
In the cis-eQTL analysis, rs3784921 in the SNN-TXNDC11 region, which was associated with baseline PRA among PEAR whites, was significantly associated with baseline expression of TXNDC11. T allele carriers showed significantly higher FPKM values for TXNDC11 and SNN compared to GG homozygotes (r2 = 0.150, p = 0.007; Figure 3 panel C). No difference in expression by genotype was observed for the other genes.
Furthermore, a proxy SNP, rs1802409, in high LD with the top SNP rs3784921 in TXNDC11, replicated in PEAR-2 whites for association with baseline PRA. The C allele of rs1802409 was associated with higher baseline PRA (β = 0.15, one-sided p = 0.038), and the mean baseline PRA was 1.01 and 1.35 ng/mL/hr for the AC and CC genotypes, respectively (Figure 3 panel D). There were no individuals with the AA genotype in PEAR-2. Meta-analysis between rs3784921 from PEAR and rs1802409 from PEAR-2, showed a stronger association (meta p = 2.63 x 10−7). Demographic characteristics of PEAR-2 whites are broadly similar to those of PEAR whites. The demographics of the 150 PEAR-2 whites included in the replication study are provided in Supplemental Table S2.
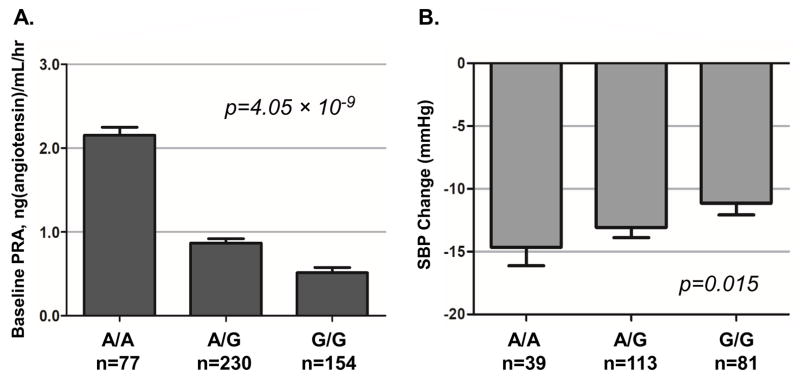
In whites, the SNP rs7184292 near CHD9 at 16q12.2 was associated with baseline PRA at a genome-wide significant level. The A allele of rs7184292 was associated with higher baseline PRA (β = 0.30, p = 4.05 x 10−9; Table 2, Figure 4 panel A) and better SBP response to atenolol (β = −1.68, one-sided p = 0.015; Table 2, Figure 4 panel B), with a mean baseline PRA of 2.15, 0.87, and 0.51 ng/mL/hr and a mean atenolol-induced SBP change of −14.7, −13.1, and −11.1 mmHg for the AA, AG, and GG genotypes, respectively. We did not find additional functional evidence for this SNP.
Figure 4.
Baseline PRA and SBP response to atenolol by CHD9 rs7184292 genotype (Panels A, B) Means and standard error bars are shown; PRA: plasma renin activity; SBP: systolic blood pressure; ATEN: atenolol; HCTZ: hydrochlorothiazide; PEAR: Pharmacogenomic Evaluation of Antihypertensive Responses; N=233 (Atenolol response)
In African Americans, the C allele of rs7606603 in XIRP2, at 2q24.3, was associated with lower baseline PRA (β = −0.42, p = 1.71 x 10−7; Supplemental Table S1 and Figure S3 panel A) and reduced SBP response to atenolol (β = 2.60, one-sided p = 0.022; Supplemental Table S1 and Figure S3 panel B), with a mean baseline PRA of 0.49, 0.33, and 0.13 ng/mL/hr and a mean atenolol-induced SBP change of −12.8, −4.4, and −4.7 mmHg for the TT, TC, and CC genotypes, respectively. Additionally, the C allele of rs16872401 near GHR, at 5p13.1, was associated with lower baseline PRA (β = −0.31, p = 5.51 x 10−6; Supplemental Table S1 and Figure S3 panel C) and better SBP response to HCTZ (β = −1.99, one-sided p = 0.025; Supplemental Table S1 and Figure S3 panel D) among African Americans, with a mean baseline PRA of 0.05, 0.14 and 0.23 ng/mL/hr and a mean HCTZ-induced SBP change of −16.3, −13.1 and −11.9 mmHg for the CC, CT, and TT genotypes, respectively. We did not find additional functional evidence for the top SNPs in African Americans.
Supplemental Table S3 shows the allele frequencies and association results with baseline PRA in the other race group for the top SNPs. There were differences in allele frequencies observed for the top SNPs between whites and African Americans (differences range between 16–24%, Supplemental Table S3). Of the four top SNPs, two were directionally consistent across race groups (rs7606603 and rs3784921; Supplemental Table S3), and two were directionally inconsistent across race groups (rs16872401 and rs7184292; Supplemental Table S3).
DISCUSSION
In this study, among a cohort of uncomplicated hypertensives, we identified SNPs associated with baseline PRA through a GWAS and prioritized these signals through association with BP response to two mechanistically different antihypertensive agents in whites and African Americans. We then prioritized the top SNPs based on functional evidence, followed by replication in an independent study. To our knowledge, this is the first study to identify PRA-associated SNPs with a BP response prioritization method, with additional functional evidence prioritization and replication. Using this approach, we found one region in whites, SNN-TXNDC11, that was associated with baseline PRA in PEAR at the suggestive level (p = 2.09 x 10−6), showed association with reduced BP response to HCTZ, showed association with expression of TXNDC11 as an eQTL, and replicated in an independent study with baseline PRA. Additionally, we found another region in whites that passed SNP prioritization, 16q12.2-CHD9, which was associated with baseline PRA at a genome-wide significant level and also showed association with BP response to atenolol in the expected direction. In African Americans, we found two regions, XIRP2 and GHR, with suggestive evidence of association with baseline PRA and associations with BP response.
The top region from our study spans two genes, SNN and TXNDC11, and is located at chromosome 16p13.13. SNN encodes stannin, a highly conserved protein localized to the mitochondria and endoplasmic reticulum membranes.31 The role of SNN in renin levels and BP regulation through the RAAS is unclear. SNN is upregulated in endothelial cells in response to the inflammatory cytokine tumor necrosis factor (TNF)-alpha, suggesting that SNN may be involved in the inflammatory process leading to atherosclerosis.32 TXNDC11 (also known as EFP1) encodes thioredoxin domain-containing protein 11. The protein primarily exists in the endoplasmic reticulum and was recently identified as being important for proper endoplasmic reticulum-associated degradation (ERAD) of MHC class I molecules.33 Using our baseline RNA-Seq data among PEAR whites, we identified rs3784921 as an eQTL for TXNDC11. Data from Haploreg show that rs3784921 is also an eQTL for SNN in whole blood.27 Furthermore, we were able to replicate the association with baseline PRA and the proxy SNP, rs1802409, in an independent study, PEAR-2. Together, these data suggest that there may be a functional connection between SNPs in this region, SNN and TXNDC11, and renin, the RAAS, and BP regulation, though the basis for this remains unclear. As the results for the PEAR-PEAR-2 meta-analysis were below the genome-wide significance threshold (meta p = 2.63 x 10−7), further validation of this region is needed.
Our study also identified a region at chromosome 16q12.2 that included two genome-wide significant SNPs (rs7184292 and rs7201620, located 169kb 3′ of the gene CHD9, which encodes chromodomain helicase DNA binding protein 9). These SNPs have expected regulatory function, show evidence of regulatory chromatin states in DNAse and enhancer histone marks in the brain, and are predicted to alter regulatory motifs.27 Additionally, there have been prior studies on CHD9, the closest gene to the associated region. A previous GWAS showed that a variant in CHD9 is associated with plasma fibrinogen concentration.34 While the role of CHD9 in the cardiovascular system, particularly the RAAS and BP regulation, is unclear, CHD9 is a transcriptional activator of peroxisome proliferator-activated receptor (PPAR)-α, a nuclear receptor that is widely expressed in the vasculature among other tissues and may be involved in BP regulation.35–39
In African Americans, we identified two regions with suggestive evidence of association with baseline PRA and signals with BP response. rs7606603 in XIRP2, also known as CMYA3, was associated with baseline PRA and BP response to atenolol. XIRP2 is a member of the cardiac and skeletal muscle-specific actin-binding Xin gene family and encodes cardiomyopathy-associated protein 3, which is involved in protecting actin filaments from depolymerization. XIRP2 interacts with the RAAS and kallikrein-kininogen system but possibly more so as a mediator of angiotensin II-induced myocardial damage.40, 41 Other studies have found angiotensin II increases XIRP2 expression by inducing transcription factor myocyte enhancer factor 2A or MEF2A.40, 42 The connections between XIRP2, the RAAS, and the kallikrein-kininogen system are interesting given that a recent GWAS among normotensive individuals identified associations between renin activity and SNPs in the kininogen 1 and kallikrein B genes, which are both involved in the production of bradykinin.16 Further, the second region identified among African Americans is on chromosome 5, with the nearest coding gene, GHR, located ~230kb 3′ of rs16872401. GHR is a member of the type I cytokine receptor family and encodes a transmembrane receptor that binds growth hormone. Studies in mice overexpressing growth hormone and individuals with acromegaly have suggested that growth hormone, as well as growth hormone receptor activation, may contribute to hypertension and cardiovascular diseases through interaction with RAAS.43–46
When examining the top SNPs in the other race group, we were unable to replicate our findings. However, this may be driven by the differences in allele frequencies between the two race groups for the top SNPs (differences range between 16–24%). We did still observe a consistent direction of effect for rs3784921 in the SNN-TXNDC11 region in African Americans.
Our study has a few limitations. We have a relatively small sample size. However, through careful measurements of PRA, prioritization based on BP response to two mechanistically different antihypertensive drugs, assessment of functional evidence, and replication, we were able to identify one region: SNN-TXNDC11. Additionally, through BP response prioritization, we found a genome-wide significant locus (CHD9), and two other suggestive loci (XIRP2 and GHR). All of these loci were prioritized through directionally specific associations with BP response to a β-blocker (atenolol) or thiazide diuretic (HCTZ). This method relies on the premise that genetic variants with a positive correlation with baseline PRA will have increased response to atenolol and decreased response to HCTZ. Conversely, we hypothesized that variants associated with lower baseline PRA will be associated with greater BP response to HCTZ and a lower response to atenolol. While there is literature evidence to support our hypothesis, including prior studies in the PEAR population,9, 11 this direct method has never been tested before. As we only focused on SNPs that met the requirements of our prioritization scheme, we could have missed other signals that impact PRA. Additionally, none of variants were associated with BP response to both a β-blocker (atenolol) and thiazide diuretic (HCTZ), in the expected directions. This may be due to our small sample size, or the fact that BP response can be influence by several factors.7–12
There are also other factors, such as urine sodium excretion that could impact PRA. However, when we added baseline urine sodium as a covariate to our linear regression model, the magnitudes of association for our top signals were broadly unchanged (data not shown). Additionally, differences in the mean doses of each antihypertensive agent by genotype could confound the results from our BP prioritization step. Yet, when we examined mean dose by genotype, there were no significant differences seen for three out of four of our top SNPs, with only rs7184292 showing a significantly lower dose in GG homozygotes (Supplemental Table S4). We did not observe signals in the regions previously associated with PRA in normotensive individuals.16 It is possible that differences in the underlying genetic architecture between normotensive and hypertensive individuals, one of which may manifest as altered RAAS signalling for hypertensive individuals, may account for the dissimilar PRA signals across the two studies. Finally, while we have adjusted for multiple comparisons in the current analyses, we have previously published other studies in these cohorts with other phenotypes including BP response.13–15 As we only utilized BP response to prioritize the PRA signals, we observed no overlap between the genes identified in this study, and those identified previously.13–15
In conclusion, we used a genome-wide association approach to identify SNPs associated with baseline PRA, prioritized these signals by testing for their expected direction of association with BP response to two mechanistically different antihypertensive drug classes, a β-blocker and a thiazide diuretic, and then further assessed these regions for functional evidence with an eQTL analysis, followed by replication. Using this method, we identified one region that showed functional evidence and replicated in an independent study (SNN-TXNDC11), one genome-wide significant region that passed the prioritization stage (CHD9), and two other suggestive loci in African Americans that passed the prioritization stage (XIRP2, GHR). Further validation and investigation into the functional roles of the top loci are needed to confirm these findings.
Supplementary Material
CLINICAL PERSPECTIVE.
Only about half of adults with hypertension have controlled blood pressure (BP), which is due in part to inter-individual variability in BP response to antihypertensive agents. In addition to known clinical factors, components of the renin-angiotensin-aldosterone system, including renin, have been associated with these differences in BP response. In order to identify biomarkers that could be used to predict antihypertensive response, we conducted a genome-wide association study with plasma renin activity (PRA) in hypertensive individuals. The top signals were prioritized and validated based on BP response to a β-blocker and thiazide diuretic, gene expression differences by genotype, and replication in an independent sample. Our prioritization and validation strategy identified the SNN-TXNDC11 gene region. The G allele of rs3784921 was associated with higher baseline PRA and reduced systolic BP response to hydrochlorothiazide. In cis-expression quantitative trait locus analysis, T allele carriers at rs3784921 showed significantly higher expression for TXNDC11 and SNN compared to GG homozygotes. A proxy single nucleotide polymorphism (SNP), rs1802409, replicated the PRA signal in an independent sample, and on meta-analysis the region showed a stronger association. Other signals associated with PRA and BP response to a β-blocker or thiazide diuretic were identified in CHD9, XIRP2, and GHR. The regions identified in this study may help personalize selection of antihypertensive agents. These SNPs and regions should be tested in other independent cohorts to validate their role in PRA and BP response. Additionally, investigation into the functional role of these genes may provide further insight into the BP response mechanisms associated with β-blockers and thiazide diuretics.
Acknowledgments
We acknowledge and thank the participants of the PEAR study and the PEAR support staff and physicians.
SOURCES OF FUNDING: PEAR was supported by the National Institute of Health (NIH) Pharmacogenetics Research Network grant U01-GM074492 and the National Center for Advancing Translational Sciences under the award number UL1 TR000064 (University of Florida); UL1 TR000454 (Emory University) and UL1 TR000135 (Mayo Clinic). PEAR was also supported by funds from the Mayo Foundation. Additional support for this project comes from NIH grants KL2 TR001429 (CWM), T32 DK104721 (OM), and T32 HL083810 (NEL).
Footnotes
DISCLOSURES: None
References
- 1.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: Analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics-2016 update: A report from the american heart association. Circulation. 2016;133:e38–60. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 3.Campbell NR, Lackland DT, Niebylski ML, Committee WHL Committee ISoHE. High blood pressure: Why prevention and control are urgent and important: A 2014 fact sheet from the world hypertension league and the international society of hypertension. J Clin Hypertens (Greenwich) 2014;16:551–553. doi: 10.1111/jch.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alderman MH. Jnc 7: Brief summary and critique. Clin Exp Hypertens. 2004;26:753–761. doi: 10.1081/ceh-200032158. [DOI] [PubMed] [Google Scholar]
- 5.Neal B, MacMahon S, Chapman N Collaboration BPLTT. Effects of ace inhibitors, calcium antagonists, and other blood-pressure-lowering drugs: Results of prospectively designed overviews of randomised trials. Blood pressure lowering treatment trialists’ collaboration. Lancet. 2000;356:1955–1964. doi: 10.1016/s0140-6736(00)03307-9. [DOI] [PubMed] [Google Scholar]
- 6.Psaty BM, Smith NL, Siscovick DS, Koepsell TD, Weiss NS, Heckbert SR, et al. Health outcomes associated with antihypertensive therapies used as first-line agents. A systematic review and meta-analysis. JAMA. 1997;277:739–745. [PubMed] [Google Scholar]
- 7.Filigheddu F, Argiolas G, Bulla E, Troffa C, Bulla P, Fadda S, et al. Clinical variables, not raas polymorphisms, predict blood pressure response to ace inhibitors in sardinians. Pharmacogenomics. 2008;9:1419–1427. doi: 10.2217/14622416.9.10.1419. [DOI] [PubMed] [Google Scholar]
- 8.Gill JS, Zezulka AV, Beevers DG, Davies P. Relation between initial blood pressure and its fall with treatment. Lancet. 1985;1:567–569. doi: 10.1016/s0140-6736(85)91219-x. [DOI] [PubMed] [Google Scholar]
- 9.Chapman AB, Schwartz GL, Boerwinkle E, Turner ST. Predictors of antihypertensive response to a standard dose of hydrochlorothiazide for essential hypertension. Kidney Int. 2002;61:1047–1055. doi: 10.1046/j.1523-1755.2002.00200.x. [DOI] [PubMed] [Google Scholar]
- 10.Canzanello VJ, Baranco-Pryor E, Rahbari-Oskoui F, Schwartz GL, Boerwinkle E, Turner ST, Chapman AB. Predictors of blood pressure response to the angiotensin receptor blocker candesartan in essential hypertension. Am J Hypertens. 2008;21:61–66. doi: 10.1038/ajh.2007.24. [DOI] [PubMed] [Google Scholar]
- 11.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-DeHoff RM, et al. Plasma renin activity predicts blood pressure responses to beta-blocker and thiazide diuretic as monotherapy and add-on therapy for hypertension. Am J Hypertens. 2010;23:1014–1022. doi: 10.1038/ajh.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laragh JH, Lamport B, Sealey J, Alderman MH. Diagnosis ex juvantibus. Individual response patterns to drugs reveal hypertension mechanisms and simplify treatment. Hypertension. 1988;12:223–226. doi: 10.1161/01.hyp.12.3.223. [DOI] [PubMed] [Google Scholar]
- 13.Turner ST, Boerwinkle E, O’Connell JR, Bailey KR, Gong Y, Chapman AB, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension. 2013;62:391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong Y, McDonough CW, Beitelshees AL, El Rouby N, Hiltunen TP, O’Connell JR, et al. Ptprd gene associated with blood pressure response to atenolol and resistant hypertension. J Hypertens. 2015;33:2278–2285. doi: 10.1097/HJH.0000000000000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong Y, Wang Z, Beitelshees AL, McDonough CW, Langaee TY, Hall K, et al. Pharmacogenomic genome-wide meta-analysis of blood pressure response to β-blockers in hypertensive african americans. Hypertension. 2016;67:556–563. doi: 10.1161/HYPERTENSIONAHA.115.06345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieb W, Chen MH, Teumer A, de Boer RA, Lin H, Fox ER, et al. Genome-wide meta-analyses of plasma renin activity and concentration reveal association with the kininogen 1 and prekallikrein genes. Circ Cardiovasc Genet. 2015;8:131–140. doi: 10.1161/CIRCGENETICS.114.000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, et al. Pharmacogenomics of antihypertensive drugs: Rationale and design of the pharmacogenomic evaluation of antihypertensive responses (pear) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamadeh IS, Langaee TY, Dwivedi R, Garcia S, Burkley BM, Skaar TC, et al. Impact of cyp2d6 polymorphisms on clinical efficacy and tolerability of metoprolol tartrate. Clin Pharmacol Ther. 2014;96:175–181. doi: 10.1038/clpt.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sealey JE. Plasma renin activity and plasma prorenin assays. Clin Chem. 1991;37:1811–1819. [PubMed] [Google Scholar]
- 20.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-Dehoff RM, et al. Power to identify a genetic predictor of antihypertensive drug response using different methods to measure blood pressure response. J Transl Med. 2012;10:47. doi: 10.1186/1479-5876-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 25.Preston RA, Materson BJ, Reda DJ, Williams DW, Hamburger RJ, Cushman WC, Anderson RJ. Age-race subgroup compared with renin profile as predictors of blood pressure response to antihypertensive therapy. Department of veterans affairs cooperative study group on antihypertensive agents. JAMA. 1998;280:1168–1172. doi: 10.1001/jama.280.13.1168. [DOI] [PubMed] [Google Scholar]
- 26.Consortium G. Human genomics. The genotype-tissue expression (gtex) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward LD, Kellis M. Haploreg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, et al. Comprehensive comparative analysis of strand-specific rna sequencing methods. Nat Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with rna-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willer CJ, Li Y, Abecasis GR. Metal: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billingsley ML, Yun J, Reese BE, Davidson CE, Buck-Koehntop BA, Veglia G. Functional and structural properties of stannin: Roles in cellular growth, selective toxicity, and mitochondrial responses to injury. J Cell Biochem. 2006;98:243–250. doi: 10.1002/jcb.20809. [DOI] [PubMed] [Google Scholar]
- 32.Horrevoets AJ, Fontijn RD, van Zonneveld AJ, de Vries CJ, ten Cate JW, Pannekoek H. Vascular endothelial genes that are responsive to tumor necrosis factor-alpha in vitro are expressed in atherosclerotic lesions, including inhibitor of apoptosis protein-1, stannin, and two novel genes. Blood. 1999;93:3418–3431. [PubMed] [Google Scholar]
- 33.Timms RT, Menzies SA, Tchasovnikarova IA, Christensen LC, Williamson JC, Antrobus R, et al. Genetic dissection of mammalian erad through comparative haploid and crispr forward genetic screens. Nat Commun. 2016;7:11786. doi: 10.1038/ncomms11786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabater-Lleal M, Huang J, Chasman D, Naitza S, Dehghan A, Johnson AD, et al. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128:1310–1324. doi: 10.1161/CIRCULATIONAHA.113.002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yagil C, Yagil Y. Peroxisome proliferator-activated receptor alpha: Friend or foe? Hypertension. 2007;50:847–850. doi: 10.1161/HYPERTENSIONAHA.107.100461. [DOI] [PubMed] [Google Scholar]
- 36.Shimamoto Y, Hirota K, Fukamizu A. Effect of peroxisome proliferator-activated receptor alpha on human angiotensinogen promoter. Int J Mol Med. 2004;13:729–733. [PubMed] [Google Scholar]
- 37.Wilson JL, Duan R, El-Marakby A, Alhashim A, Lee DL. Peroxisome proliferator activated receptor-alpha agonist slows the progression of hypertension, attenuates plasma interleukin-6 levels and renal inflammatory markers in angiotensin ii infused mice. PPAR Res. 2012;2012:645969. doi: 10.1155/2012/645969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phelps LE, Peuler JD. Evidence of direct smooth muscle relaxant effects of the fibrate gemfibrozil. J Smooth Muscle Res. 2010;46:125–142. doi: 10.1540/jsmr.46.125. [DOI] [PubMed] [Google Scholar]
- 39.Lee DL, Wilson JL, Duan R, Hudson T, El-Marakby A. Peroxisome proliferator-activated receptor-alpha activation decreases mean arterial pressure, plasma interleukin-6, and cox-2 while increasing renal cyp4a expression in an acute model of doca-salt hypertension. PPAR Res. 2011;2011:502631. doi: 10.1155/2011/502631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duka A, Schwartz F, Duka I, Johns C, Melista E, Gavras I, Gavras H. A novel gene (cmya3) induced in the heart by angiotensin ii-dependent but not salt-dependent hypertension in mice. Am J Hypertens. 2006;19:275–281. doi: 10.1016/j.amjhyper.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 41.Marketou M, Kintsurashvili E, Papanicolaou KN, Lucero HA, Gavras I, Gavras H. Cardioprotective effects of a selective b(2) receptor agonist of bradykinin post-acute myocardial infarct. Am J Hypertens. 2010;23:562–568. doi: 10.1038/ajh.2010.20. [DOI] [PubMed] [Google Scholar]
- 42.McCalmon SA, Desjardins DM, Ahmad S, Davidoff KS, Snyder CM, Sato K, et al. Modulation of angiotensin ii-mediated cardiac remodeling by the mef2a target gene xirp2. Circ Res. 2010;106:952–960. doi: 10.1161/CIRCRESAHA.109.209007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munoz MC, Burghi V, Miquet JG, Giani JF, Banegas RD, Toblli JE, et al. Downregulation of the ace2/ang-(1-7)/mas axis in transgenic mice overexpressing gh. J Endocrinol. 2014;221:215–227. doi: 10.1530/JOE-13-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jara A, Benner CM, Sim D, Liu X, List EO, Householder LA, et al. Elevated systolic blood pressure in male gh transgenic mice is age dependent. Endocrinology. 2014;155:975–986. doi: 10.1210/en.2013-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bielohuby M, Roemmler J, Manolopoulou J, Johnsen I, Sawitzky M, Schopohl J, et al. Chronic growth hormone excess is associated with increased aldosterone: A study in patients with acromegaly and in growth hormone transgenic mice. Exp Biol Med (Maywood) 2009;234:1002–1009. doi: 10.3181/0901-RM-34. [DOI] [PubMed] [Google Scholar]
- 46.Egecioglu E, Andersson IJ, Bollano E, Palsdottir V, Gabrielsson BG, Kopchick JJ, et al. Growth hormone receptor deficiency in mice results in reduced systolic blood pressure and plasma renin, increased aortic enos expression, and altered cardiovascular structure and function. Am J Physiol Endocrinol Metab. 2007;292:E1418–1425. doi: 10.1152/ajpendo.00335.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.