Abstract
Because cell growth and differentiation are regulated by complex interactions among different signaling pathways, a growth defect affects subsequent differentiation. We report on a growth-defective mutant of Arabidopsis, called eld1 (elongation defective 1). Cell elongation was impaired in every organ examined. Later characteristics of the eld1 phenotype include defective vascular tissue differentiation, the inability to grow in soil, ectopic deposition of suberin around twisted vascular bundles, the de-etiolation phenotype, and continuation of shoot development and flowering in the dark. The dwarf phenotype of eld1 could not be rescued by treatment with exogenous growth regulators. Because defective cell elongation is the earliest and most universal feature detected in eld1 mutants, control of or activity in cell elongation may be the primary function of the ELD1 gene. The impaired cell growth results in pleiotropic effects on cell proliferation and differentiation, and the retardation in hypocotyl elongation enables growth and development in darkness.
Higher plants achieve their structural complexity via a precise and orderly control of three cellular processes: division, growth, and differentiation. Changes in cell division and cell growth processes or their control affect the growth and differentiation of the entire plant. Plants with altered cell division and growth often display abnormal morphology and alterations in organogenesis and internal functions.
Plant cell growth and development are controlled by growth regulators and other endogenous factors (Davies, 1995), and by exogenous factors such as light (Chory et al., 1989a; Deng, 1994). Over the last 10 years, many growth-defective mutants of Arabidopsis have been isolated and characterized. Some of these mutants show defective embryonic phenotypes (Meinke, 1985). Others develop normally during embryogenesis but are dwarfed after germination due to failure in cell division at the root tip (root meristemless, rml, Cheng et al., 1995) or inhibition of cell elongation (stp1 [stunted-plant1], Baskin et al., 1995; dim [diminuto] Takahashi et al., 1995). Some growth-defective mutants show additional phenotypes such as photomorphogenesis in darkness. Such photomorphogenic mutants include cop1 (constitutive photomorphogenic; Deng et al., 1991; Deng and Quail, 1992.), det2 (de-etiolated; Chory et al., 1991), and fus6 (fusca; Castle and Meinke, 1994), which also accumulates high levels of anthocyanin in the dark. In some instances, the dwarf phenotype results from defects in the biosynthesis or perception of growth regulators. Examples include bri1 (brassinosteroid-insensitive; Clouse et al., 1996), dwf4 (dwarf4; Azpiroz et al., 1998), dwf1 (dwarf1; Choe et al., 1999a), cpd (constitutive photomorphogenesis and dwarfism; Szekeres et al., 1996), and axr2 (auxin resistant2; Timpte et al., 1992). These diverse mutant phenotypes suggest that different signaling pathways interact in regulating cell division, growth, and differentiation. Many more genes and mutants must be identified and studied to decipher these pathways and their interactions.
We report here a growth-defective mutant of Arabidopsis called eld1 (elongation defective 1). The eld1 mutant exhibits a defect in almost every aspect of cell development but particularly in cell elongation. Defining characteristics of eld1 mutant plants include dwarfism, twisted vascular strands, and, most notably, the abnormal deposition of suberin in vascular cells, a phenotype that has not been reported in any other dwarf mutant. Additionally, the mutant undergoes shoot development and then flowers in complete darkness. Characterization of such a mutant could lead to better understanding of the signal transduction pathways governing the three fundamental cellular processes of plant development and the role of hypocotyl elongation on shoot development in darkness.
RESULTS
Isolation and Genetic Characterization of the eld1 Mutant
The eld1 mutant of the Arabidopsis ecotype Columbia was isolated after gamma ray irradiation as an abnormal seedling with short stature, twisted vascular tissue, and ectopic deposition of suberin around the vascular bundles (Figs. 1B, 3F, and 5E). The mutant phenotype segregated as a single gene recessive trait.
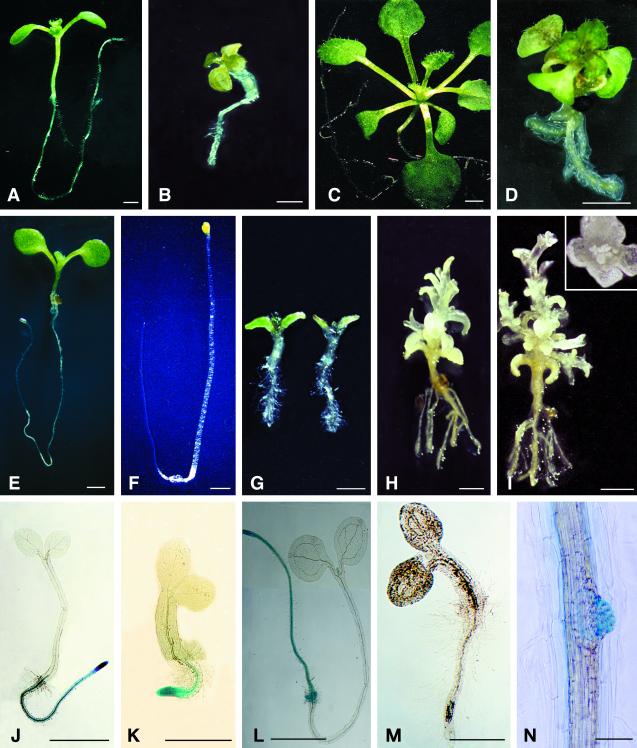
Figure 1.
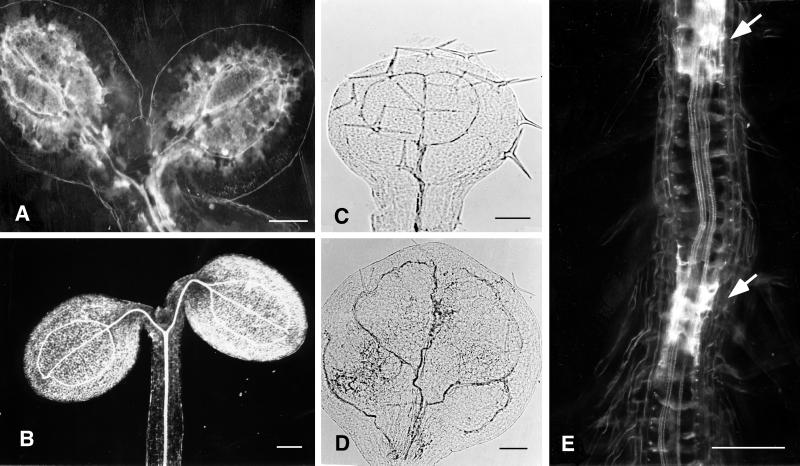
Morphology of eld1 mutant and wild-type plants. A, 10-d-old wild type; B, 10-d-old eld1; C, 30-d-old wild-type shoot grown under short-day conditions; D, 30-d-old eld1 grown under short-day conditions; E, 7-d-old wild type grown in light under short-day conditions; F, 7-d-old wild type grown in darkness; G, 7-d-old eld1 grown in light (left) and in darkness (right); H, 15-d-old eld1 grown in darkness, showing the elongated shoot; I, 30-d-old eld1 grown in darkness, showing inflorescenses and terminal flower. The flower is enlarged and shown in the box. J, 2-d-old wild type showing GUS activity in entire root; K, 2-d-old eld1, showing GUS activity in root meristematic region; L, 5-d-old wild type, showing GUS activity in the primary root, lateral root primodia, and shoot apical; M, 5-d-old eld1, showing no GUS activity in the plant; N, a portion of 10-d-old eld1 root, showing weak GUS activity in lateral root primordium. Bars = 1 mm (A–J, and M), 0.5 mm (K), 3 mm (L), and 150 μm (N).
The ELD1 gene was mapped to 17.2 cM from the top of chromosome 3 via a PCR-based mapping method using primers designed against single sequence length polymorphism microsatellites (Bell and Ecker, 1994). Heterozygous eld1 mutants were crossed with wild-type plants of the Landsberg ecotype. The F2 homozygous eld1 mutants were analyzed. In a sample of 42 mutant plants, all 42 were Columbia at the marker nga 162, showing the close linkage between the ELD1 gene and nga 162.
Because the eld1 mutant mapped near fus9/cop10/emb144 and other growth-defective mutants, e.g. dwf1-1 and dwf7-1 on chromosome 3, complementation tests were carried out between eld1 and these mutants. eld1 was not allelic to any of them, indicating that ELD1 is a new gene.
eld1 Mutant Growth Is Stunted
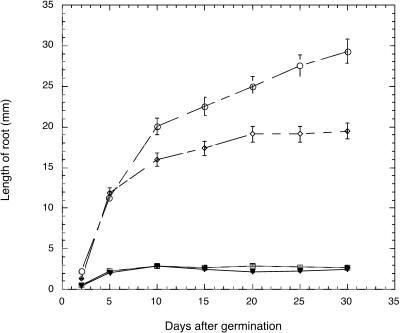
Seed development and embryogenesis in the eld1 mutant were normal. However, shortly after germination, seedling growth was severely inhibited. The first expression of this stunted growth was significant inhibition of root elongation (Fig. 1, A and B). The root of a 5-d-old eld1 root was 2.3 ± 0.5 mm in length and showed limited growth by 30 d (2.7 ± 0.4 mm), whereas a wild-type root grew from 11.4 ± 1.5 mm at 5 d to 29.3 ± 2.2 mm by 30 d (Fig. 2; Table I). Growth inhibition was also observed in the hypocotyl. A 5-d-old eld1 mutant attained a hypocotyl length of 1.2 ± 0.2 mm then ceased to grow, whereas wild-type hypocotyls were 2.9 ± 0.1 mm long on 5 d and grew to 5.0 ± 0.3 mm by 30 d (Fig. 3A; Table I).
Figure 2.
The growth rate of eld1 and wild-type roots in dark- and light-grown conditions. ○, Wild type, light-grown; ⋄, wild type, dark-grown; □, eld1, light-grown; ▾, eld1, dark-grown.
Table I.
Comparison of the organ and cell sizes of wild type and eld1 mutanta
| Organ or Cell Type | Wild Type | eld1 |
|---|---|---|
| mm | ||
| 5 d | ||
| Hypocotyl lengthb | 2.9 ± 0.1 | 1.2 ± 0.1 |
| Primary root length | 11.4 ± 1.5 | 2.3 ± 0.5 |
| Cotyledon blade length | 1.8 ± 0.1 | 0.8 ± 0.1 |
| Cotyledon blade width | 1.5 ± 0.2 | 0.6 ± 0.1 |
| μm | ||
| 7 d | ||
| Hypocotyl cortical cell lengthc | 96 ± 10.0 | 36 ± 10.0 |
| Hypocotyl cortical cell width | 26 ± 3.0 | 56 ± 5.0 |
| Root cortical cell length | 70 ± 10.0 | 26 ± 9.0 |
| Root cortical cell width | 22 ± 2.0 | 35 ± 5.0 |
| mm | ||
| 30 d | ||
| Petiole length of rosette leafd | 4.2 ± 0.5 | 0.4 ± 0.2 |
| Blade length of rosette leaf | 3.6 ± 0.2 | 1.3 ± 0.1 |
| Blade width of rosette leaf | 2.8 ± 0.1 | 1.1 ± 0.1 |
Plants were grown on two-fifths MS medium under 8 h of light at 21°C.
Data represent mean (±se) of 10 plants.
Data represent mean (±se) of 10 cortical cells, randomly taken from longitudinal sections of hypocotyl and root tip.
Measurements taken from the first pair of rosette leaves. Blade length is measured from the leaf tip to leaf base.
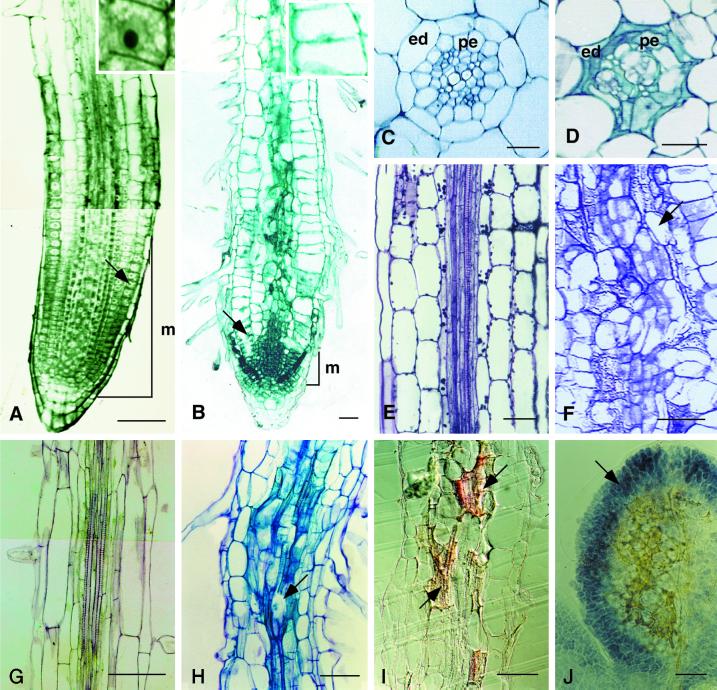
Figure 3.
Anatomy of the eld1 mutant and wild-type hypocotyls and roots. A, Longitudinal section of a 7-d-old wild-type root tip stained with TBO. The cell marked by an arrow is enlarged in the adjacent box. B, Longitudinal section of a 7-d-old eld1 mutant root tip was stained with TBO. The cell marked by an arrow is enlarged in the adjacent box. C, Cross-section of wild-type root showing diarch protoxylem in the stele surrounded by normal endodermis (ed) and pericycle (pe). D, Cross section of the eld1 root, showing disordered vessels, aberrant endodermis (ed) and pericycle (pe). E, Longitudinal section of wild-type hypocotyl showing parallel vascular cell files arranged tightly in the stele. F, Longitudinal section of the eld1 mutant hypocotyl showing a distorted vascular bundle and irregularly shaped vascular cells (marked by arrow). G, Longitudinal section of the upper region of the wild-type root, showing the stele. H, Longitudinal section of the upper region of the eld1 mutant root, showing abnormal vascular differentiation (marked by arrow). I, Longitudinal section of 7-d-old eld1 root stained with Sudan red 7B, suberin was stained red, and can be seen scattered randomly in the vascular bundle (marked by arrows). J, Cleared eld1 cotyledon stained with I-IK showing starch accumulated in the leaf periphery (marked by arrow) and suberized vein meshes in the middle. Bars = 100 μm (A and G–J), 50 μm (B, E, and F), and 20 μm (C and D).
In addition, the mature organs of eld1, cotyledons, stems, and leaves were all shorter than those of the wild type (Table I; Fig. 1, C and D). For example, the rosette leaf petiole of a 30-d-old eld1 was 0.4 ± 0.2 mm long and the wild type 4.2 ± 0.5 mm (Table I). eld1 rosette leaf blades were also shorter than wild type. All of these features argue that a general inability to grow results in the short stature of the eld1 mutant.
eld1 Is Impaired in Cell Elongation
Growth inhibition in eld1 results primarily from reduced cell elongation. The eld1 root cortical cells were significantly shorter than wild type (Fig. 3, A and B). Furthermore, eld1 root cortical cells expanded radially in the root tip. The root tip cortical cell of 7-d-old eld1 plant was 26 ± 9.0 μm long and 35 ± 5.0 μm wide, while the corresponding wild-type root tip cortical cell was 70 ± 10.0 μm long and 22 ± 2.0 μm wide (Fig. 3, A and B; Table I).
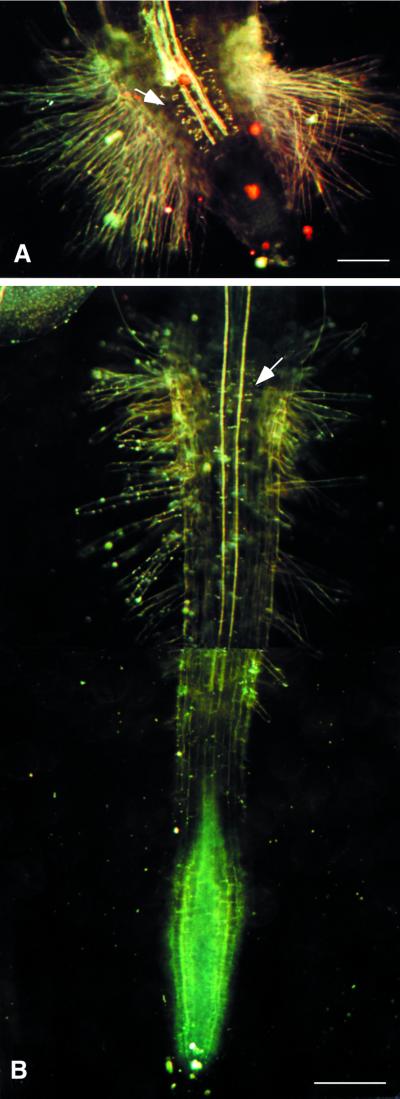
Interestingly, root hair cell elongation does occur in the eld1 mutant. The epidermal cells of the eld1 root do not expand radially, but produce longer root hairs than the wild type (Fig. 4, A and B). Moreover, eld1 root hairs emerged from nearly every epidermal cell, unlike the wild type, which tends to produce root hairs only from epidermal cells that overlie the anticlinal walls between adjacent cortical cells (Schiefelbein et al., 1997). This, together with the short epidermal cells in the mutant, resulted in a hairy root appearance from root hairs growing in close proximity to each other (Fig. 4A).
Figure 4.
eld1 and wild-type roots. A, 2-d-old cleared eld1 root showing the thick Casparian strips (marked by arrow) and dense root hairs. B, 2-d-old cleared wild-type root showing normal Casparian strips (marked by arrow) and root hairs. Bars = 100 μm (A) and 250 μm (B).
The short hypocotyl in eld1 mutants is also characterized by impaired cell elongation. The cortical cell length of a 7-d-old eld1 hypocotyl was only one-third of the wild-type length (36 ± 10.0 μm in eld1 and 96 ± 10.0 μm in wild type), while cell width was twice that of the wild type (56 ± 5.0 μm in eld1 and 26 ± 3.0 μm in wild type; Table I). At 20 d, the number of eld1 hypocotyl epidermal cells/cell files was the same as wild type, around 23 to 24 cells (Cheng et al., 1995). As no obvious change in cell number/cell file is observed, the short eld1 hypocotyl results primarily from reduced cell elongation, not reduced cell proliferation. Inhibition of cell elongation as a cause of dwarfism in the eld1 mutant is also consistent with the finding that organs that grow by cell elongation are the most severely affected.
eld1 Root Meristem Is Impaired in Cell Division
Cell division is conspicuously impaired in the root meristem region of the eld1 mutant, as shown in root longitudinal sections of the wild type and eld1 (Fig. 3, A and B). As early as 2 d after germination, the eld1 root meristem and elongation regions were shorter than the wild type. The eld1 root meristem region was 0.2 ± 0.1 mm in length at 2 d and was reduced to 0.05 ± 0.01 mm by 10 d, while the wild-type root meristem remained at 0.4 ± 0.01 to 0.5 ± 0.1 mm during this same period. The cells of the newly germinated eld1 root tip were isodiametric in shape, rich in cytoplasm, and contained large nuclei, as is characteristic of wild-type meristematic cells (Fig. 3A). However, by 7 to 10 d after germination, the apical cells in the eld1 root tip were flat, vacuolated, and had small nuclei (Fig. 3B). Thus, the eld1 root apical cells may have lost their meristematic activity upon germination.
The eld1 root meristem gained few or no cells from new cell divisions, while losing cells to the elongation region. The elongation region was not maintained either. Two-day-old eld1 root had one to two recognizable cells per cell file in the elongation region, while wild type contained five to six cells per cell file. By 5 d, most of the eld1 root cells had matured, as evidenced by the loss of cytoplasm, the presence of root hair immediately above the root cap, and the differentiation of vascular cells close to the columella (Fig. 3B). No undifferentiated cells remained in the elongation region. These phenomena are consistent with the differentiation of root apical cells that lose the ability to divide, which are observed in plants whose root growth is inhibited (Baskin et al., 1995; Cheng et al., 1995).
Differential cdc2a expression between the eld1 mutant and wild-type root cells is further evidence of the lack of cell division in the eld1 root. cdc2a is expressed in cells in the cell division cycle and cells that are competent to divide (Hemerly et al., 1993). The temporal and spatial cdc2a expression in wild-type and eld1 plants was studied by assaying for GUS activity in transgenic plants containing the cdc2a promotor::GUS chimeric constructs (cdc2a::GUS). Two days after germination, the entire wild-type root, but only the meristematic region of the eld1 root, was GUS positive (Fig. 1, J and K). At 5 d, wild-type roots were still GUS positive, but no GUS activity was found in eld1 root cells (Fig. 1, L and M). These results indicate that most eld1 cells in the primary root had lost their competence to divide by 5 d after germination. However, eld1 was able to initiate lateral roots, and the lateral root primordium often exhibited weak GUS activity (Fig. 1N). Lateral root growth soon stopped and GUS activity was lost.
eld1 Cells Are Impaired in Vascular Tissue Differentiation
eld1 mutant cells are also impaired in cell differentiation, most notably in the vascular tissue. The wild-type vascular bundle has a symmetrical diarch in cross-section and is surrounded by a layer of pericycle and endodermal cells (Fig. 3C). The vascular cells of the eld1 mutant root were disorganized; the vessels were twisted and the pericycle and endodermal cells were oddly shaped (Fig. 3D).
Longitudinal sections of eld1 hypocotyl and root showed that vascular bundles were not clearly delineated; xylem and phloem were not readily distinguishable (Fig. 3, F and H). Some of the vessel members displayed incomplete differentiation (Fig. 3H), while wild-type vascular cells in the hypocotyl and root were arranged in a tight, straight fashion (Fig. 3, E and G). Abnormal vascular bundles in the form of twisted veins and veinlets were found in eld1 cotyledons and leaves (Fig. 5D).
Figure 5.
Cleared and whole-mounted organs of eld1 and wild type. A, 5-d-old eld1 cotyledons showing suberin in the vascular bundles. B, 5-d-old wild-type cotyledons, showing no suberin deposited around vein meshes. C, eld1 young cauline leaf grown in the dark, showing no suberin deposition. D, eld1 rosette leaf grown in the light, showing twisted vein. E, A portion of 7-d-old eld1 root, showing suberin deposition in the stele (marked by arrows). Bars = 0.2 mm (A), 0.4 mm (B), and 0.3 mm (C–E).
As procambial cell elongation precedes vascular differentiation, these vascular defects could be the result of poor cell elongation. These defective vascular cells would impair vascular function and the ability to transport nutrients, hormones, and water, contributing to the inhibition of plant growth and the inability of the mutant to grow in soil.
Suberin Is Deposited Ectopically in eld1 Cells
One unique feature of the eld1 mutant is the white and translucent patches in seedling organs when viewed under a dissecting microscope or white patches under dark-field microscopy (Fig. 5E). As early as 2 d after germination, these white translucent patches appeared along the vascular tissue of eld1 roots, hypocotyls, and cotyledons. As the plant aged, the number of these patches increased, and they were eventually found all over the vascular bundles of roots, hypocotyls, cotyledons, and leaves (Fig. 5, A and E). To determine the chemical nature of the white patches, eld1 and wild type were stained with Sudan red 7B, a suberin-specific dye that stains suberin lamellae but not Casparian strips in endodermal cells of roots (Brundrett et al., 1991). Figure 3I shows that the irregularly shaped Sudan red 7B-positive cells were scattered randomly in the eld1 root vascular bundles, indicating that the white patches contained suberin lamellae. Casparian strips on radial and transverse walls of endodermal cells were readily detectable under dark-field microscopy in 2-d-old eld1 and wild-type roots, but were thicker in eld1 than in the wild type (Fig. 4, A and B). Vascular differentiation is impaired in the eld1 mutant, and the endodermal cells and secondary thickening also appeared abnormal (Fig. 3D). This phenomenon of precocious and ectopic suberin deposition was not observed in wild-type organs (Figs. 4B and 5B).
The cotyledons also accumulated high level of starch around the margin and suberin in the middle (Fig. 3J). The cotyledons ceased to grow 5 to 6 d after germination, and had reduced cdc2a::GUS activity. In contrast, wild-type cotyledons continued to grow, and accumulated no starch or suberin by 5 to 6 d after germination (see figure 6A in Kurata and Yamamoto [1998]).
The cause of ectopic suberization is not yet understood. The lack of root meristematic activity could lead to a shortage of auxin or cause a hormonal imbalance, stimulating cell maturation and secondary differentiation. These results suggest that these organs lost cell division competence and switched to cell differentiation by 5 d after germination. Actively growing organs of eld1, such as young green rosette leaves grown in the light or developing etiolated cauline leaves formed in the dark, did not have suberin deposition in their cells (Fig. 5, C and D).
Another possibility is that the precocious suberin deposition that usually accompanies secondary wall differentiation might itself cause cell elongation arrest. To investigate the relationship between impaired cell elongation and suberin deposition in the eld1 mutant, we studied the temporal course of suberin deposition in germinating roots. The results indicated that root elongation in eld1 seedlings was inhibited as early as 10 h after germination, while there was no suberin deposition detectable at such time (data not shown). Since root growth was impaired before suberin deposition, the defect in cell elongation was not likely caused by the formation of a rigid secondary wall.
eld1 Undergoes Photomorphogenesis in the Dark
Following several days of growth in darkness, the eld1 mutant seedlings were de-etiolated, with a short hypocotyl, opened and expanded cotyledons, and few leaf primordia (Fig. 1G). In contrast, dark-grown wild-type seedlings were etiolated, with long hypocotyls and cotyledons that were small, closed, and unexpanded (Fig. 1F). Dark-grown eld1 hypocotyls did not elongate more than light-grown eld1: for example, 5-d-old eld1 hypocotyls grown in darkness attained a length of 1.3 ± 0.2 mm, while light-grown hypocotyls were 1.2 ± 0.2 mm. In contrast, wild-type hypocotyls elongated greatly (four times) in darkness compared with growth in light. Five-day-old wild-type hypocotyls grown in darkness attained a length of 11.9 ± 1.1 mm, while light-grown wild-type hypocotyls were 2.9 ± 0.1 mm.
Unlike wild type, eld1 mutant shoots continued to develop in darkness, producing several leaf primordia by 5 to 7 d and displaying internode elongation conspicuously by 10 to 15 d (Fig. 1H). Axillary shoots emerged from nodes, and floral buds were initiated at shoot apical meristems in 30- or 40-d-old plants grown on agar medium (Fig. 1I). The floral organs displayed normal whorl structure. The petals expanded, but the development of anthers and pistils was arrested prematurely (Fig. 1I). eld1 plants grown in the light did not produce flowers at 30 to 40 d, and there was no internodal elongation (Fig. 1D). In contrast, 30- or 40-d-old wild-type plants grown in darkness never bolted under the same growth conditions and displayed only two to three etiolated leaves with long petioles (picture not shown). In light, 30-d-old wild-type plants produced six to eight green rosette leaves (Fig. 1C); many 40-d-old wild-type plants bolted and flowered on agar medium under short-day conditions.
Dark flowering on solid agar medium in eld1 is unusual; it has been reported in cop1-6 (McNellis et al., 1994), but not in many other dwarf mutants. We tested dark flowering of fus9/emb144, det1, det2, and gurke (Torres-Ruiz et al., 1996). These mutants did not flower under the same cultural conditions in the dark. However, further media manipulation may improve dark flowering (see “Discussion”).
eld1 Mutant Phenotypes Cannot Be Rescued by Growth Regulators
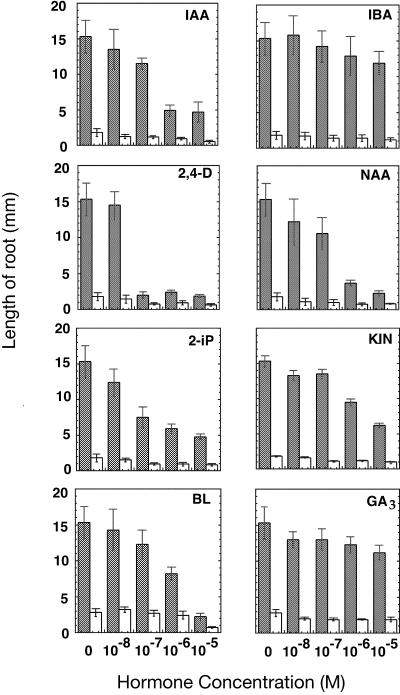
Plant growth and development are partly controlled by growth regulators. Auxin and cytokinin, for instance, stimulate cell division and vascular differentiation (Aloni, 1995). Gibberellic acid (GA3) and brassinosteroids play a critical role in plant cell elongation (Davies, 1995; Clouse et al., 1996). Plants with defects in the synthesis or perception of hormones often exhibit defective seedling development (Reid and Howell, 1995). To investigate the possibility that the dwarf feature of the eld1 mutant is due to hormone impairment, eld1 mutants were grown on media supplemented with various concentrations of the known growth regulators indole-3-acetic acid (IAA), naphthaleneacetic acid (NAA), 2,4-dichlorophenoxyacetic acid (2,4-D), indole-3-butyric acid (IBA), 6(γ,γ-dimethylallylamino)-purine (2iP), kinetin, GA3 and brassinolide (BL). The mutant shoots did not resume normal growth and differentiation patterns when grown in the presence of these growth regulators. None of the hormones could rescue root growth in the mutant (Fig. 6), suggesting that the mutant defect is not in hormone synthesis. Furthermore, the inhibition of mutant root growth observed at a high concentration (10−5 m) of IAA, NAA, IBA, 2,4-D, 2iP, kinetin, GA, and BL indicates that the eld1 mutant is responsive to these hormones.
Figure 6.
Effect of growth regulators on eld1 and wild-type root growth. All measurements were performed as described in “Materials and Methods.” Shaded bars, Wild type; white bars, eld1 mutant. Error bars represent se.
DISCUSSION
Cell Elongation Is Likely the Primary Defect in the eld1 Mutant
eld1 is impaired in cell elongation, cell division, primary (vascular) and secondary (suberization) differentiation, and the inhibition of photomorphogenesis in darkness. Because the earliest and most consistent defect in the mutant was the inhibition of root growth at 10 h after germination, the primary defect is probably the improper regulation of cell growth. The primary cause of reduced eld1 root and hypocotyl length was defective cell elongation. Although the eld1 plant was de-etiolated or constitutively photomorphogenic and displayed ectopic suberization in twisted vasculature, impaired cell elongation preceded these events. Therefore, these phenotypes are likely to be secondary effects of the cell elongation arrest.
Mechanisms Regulating Cell Elongation
Defects in a variety of signaling pathways will affect normal cell elongation. Inability of the growth regulators to rescue the eld1 mutant phenotype indicates that eld1 is not impaired in the synthesis of any of the growth regulators studied. What is the possible role of ELD1 in cell elongation? Proper regulation of cell wall and cytoskeleton organization is important for normal cell elongation. An interesting phenotype of eld1 is that root hair cell elongation is not affected. While most plant cells elongate via diffused growth, root hair cells elongate via tip growth (McClinton and Sung, 1997). The fass mutant is also impaired in cell expansion via diffuse growth, but not tip growth. fass cells are unable to organize the microtubules into the cortical arrays required for proper microfibril organization in the cell wall to allow cell elongation. However, eld1 differs from fass in that fass cells are protoplast like, while eld1 cells are able to maintain the typical rectangular shape of most plant cells. A mutation in the endo-1,4-β-d-glucanase affects the assembly of the cellulose-hemicellulose network in expanding cell walls and causes dwarf seedling phenotype (Nicol et al., 1998). We as yet have no evidence correlating the eld1 phenotype to problems in cell wall metabolism or microtubule organization. Future studies on cell wall and cytoskeleton components may provide information on ELD1's involvement in cell structure.
Plant cells attain shape by directional expansion. The shape of the expanding cell often correlates with that of the organ. Therefore elongated organs usually are made of elongated cells; and in normal plants the extent of cell elongation correlates with the state of differentiation. Little is known about the morphogenetic mechanism regulating organ shape; ELD1 could be involved in such mechanisms. eld1 displays additional defects such as twisted vasculature, ectopic suberization, and an inability to grow in soil. These features suggest ELD1 involvement in a new signaling pathway of cell growth that interacts with subsequent differentiation processes.
Suberization Occurs Earlier or at a Faster Pace as eld1 Cells Embark on Differentiation
Suberization is a common feature in roots, as evidenced by Casparian strips on radial and transverse walls of endodermal cells (Fahn, 1990). In the course of cell maturation, the suberin lamellae are deposited on the inner side of walls, rendering the vascular bundle impermeable to water and gases (Fahn, 1990). In woody or herbaceous plants, suberization occurs in exodermal cells tissues and cork cells (Esau, 1977) or in wounded tissues (Soliday et al., 1978). However, in eld1 mutants, suberin deposition occurs precociously and randomly, scattered in the general vicinity of the vascular tissue residing in organs that have ceased to grow (Fig. 5, A and E).
Suberization may be a kind of physiological senescence phenomenon in the eld1 mutant. eld1 mutant cells depart from the cell division cycle at an earlier stage, as evidenced by low cell division competence or reduced cdc2a::GUS activity in roots and cotyledon, perhaps entering the cell differentiation pathway earlier than wild-type cells. Furthermore, as eld1 cells advance from the primary to the secondary differentiation pathway, they begin to accumulate secondary cell wall materials at a faster pace. However, due to aberrant vascular cell differentiation, the site of suberin deposition may be affected, resulting in ectopic suberin accumulation.
Photomorphogenesis in Darkness May Result from Impaired Cell Elongation
Germinating in darkness, wild-type seedlings do not undergo photomorphogenesis; rather, they display etiolated phenotypes, which include elongated hypocotyl, inability to straighten the apical hook and to expand the cotyledons, and lack of chloroplast development and shoot meristematic activity. Mutants impaired in a variety of genes display de-etiolated phenotypes; these mutants include the cop/det/fus (Wei et al., 1994; Chory et al., 1989a, 1991; Miséra et al., 1994), the dwarf (Azpiroz et al., 1998; Choe et al., 1999a, 1999b), the shy2-1D (Kim et al., 1998), and the seedling lethal mutants, such as gurke (J.-C. Cheng and Z.R. Sung, unpublished results). Many of these genes are believed to be involved in regulating cell elongation, for example, DWARF4 is a brassinosteroid-dependent mutant blocked in cell elongation (Azpiroz et al., 1998). Excessive cell elongation in the hypocotyl may deplete the energy needed for cotyledon expansion and shoot meristematic activity. Without photosynthesis, no new energy can be generated to sustain plant growth and development in darkness. Under this scenario, the de-etiolated phenotypes would be the result of a lack of hypocotyl elongation in these mutants, because energy can be directed to support shoot development.
Allelism tests with dwf1, dwf7, fus9/emb144 showed that ELD1 is a new gene. The eld1 mutant is similar to cop/det/fus mutants in its sterility, dwarfism, and photomorphogenesis in the dark. It does not accumulate anthocyanin, but it displays vascular defects and deposits suberin precociously. Both anthocyanin and suberin accumulations are secondary differentiation processes. eld1 and cop/det/fus may be involved in separate signaling pathways of cell elongation that are linked to different secondary differentiation pathways.
Dark Flowering May Be a Consequence of Shoot Development in the Dark
Wild-type plants of Arabidopsis grown in darkness can flower in specific growth conditions, such as in liquid-shaken cultures, a cultural condition that shortens the flowering time in late-flowering mutants grown in darkness (Araki and Komeda, 1993). Some dwarf mutants, such as eld1 and cop1-6, circumvent the need for liquid-shaken culture and display dark flowering on solid agar medium (McNellis et al., 1994). However, Azpiroz et al. (1998) showed that wild-type plants grown on solid medium will also flower if the shoots are grown in proximity to the agar medium in vertically oriented dishes. These cases show that the short hypocotyls caused by mutation or cultural conditions enable photomorphogenesis and flowering in darkness, rather than dwarfism being part of a defect in the control of light-regulated processes.
Wei et al. (1994) proposed that photomorphogenesis is a default pathway of the plants, as gymnosperms and some algae form chloroplasts in the dark. During evolution, mechanisms are evolved to repress photomorphogenesis in the dark. Such mechanisms appear to be channeled through cell elongation. To undergo photomorphogenesis, the repression mechanism must be overcome through mutation or the cultural conditions that inhibit cell elongation. Since dark-grown plants first produce leaves then flowers, it follows that dark flowering is a normal progression of plant shoot development. Thus dark flowering is a consequence of morphogenesis in the dark. On the other hand, some other dark-grown mutants such as emb144/fus9, det2 also can produce leaves in darkness, but never bolt and flower under the same growth conditions as eld1 (data not shown). It is possible that dark flowering in these dwarf mutants occurs under different cultural conditions. Alternatively, the dark-flowering phenomenon seen in the eld1 mutant may be due to its multitude of pleiotropic effects. Molecular characterization of the ELD1 gene and further investigation of its genetic interaction with other dwarf mutants will contribute to the understanding of dwarfism relative to flowering signaling pathways.
MATERIALS AND METHODS
Mutant Isolation and Plant Growth Conditions
Seeds of Arabidopsis ecotype Columbia were mutagenized by γ-irradiation as described previously (Cheng et al., 1995). Selfed progeny from mutagenized seeds were harvested from individual M1 plants. M2 seeds from 3,000 M1 plants were sterilized, kept at 4°C for 2 d, and germinated on two-fifths Murashige and Skoog (MS) agar medium (Murashige and Skoog, 1962) at 21°C under 8 h of light. Progeny of M1 plants were screened under a dissecting microscope for segregating abnormal M2 seedlings. Since homozygous mutants were sterile, the heterozygous seedlings were transplanted into the soil to propagate the mutation. Plants were grown under 16 h of light at 21°C in the greenhouse of the University of California, Berkeley. The mutagenized lines yielded two families with similar phenotypes. We performed allelism tests and confirmed that they were allelic to each other; therefore, we called them eld1-1 and eld1-2. All experiments described in the text were performed on eld1-1. All mutants have been backcrossed for six generations to ensure stable segregation of a single-gene recessive trait.
For general observations and growth measurements, plants were grown on two-fifths MS medium under 8 h of light. The same strength of MS medium was also consistently used in other experiments mentioned in this report. For light-grown tests, 10 MS medium plates containing seeds from heterozygous parents were incubated in the growth chamber with 8 h of light at 21°C. For dark-grown treatment, another 10 plates were placed in a dark box, covered with a black cloth, and kept in the same growth chamber. All plates (light- and dark-grown) were vertically oriented at 45° to allow straight root growth along the agar surface. For comparison of the organ length of the wild type and the eld1 mutant, samples were measured under dissecting microscope. The lengths of cotyledons and rosette leaf blades were measured from leaf tip to the junction of leaf base and petiole.
Genetic Mapping and Complementation Test
For ELD1 mapping, heterozygous eld1 plants of the Columbia ecotype were crossed with the wild-type Landsberg ecotype. F1 plants that segregated eld1 F2 progeny were used for analysis. Seven-day-old homozygous seedlings were transferred into MS liquid medium individually and incubated at 24°C on a shaker at 100 rpm to maximize tissue generation. After 2 to 3 weeks, they had generated about 0.5 to 1 g fresh weight from which to extract DNA. DNA was isolated from individual mutant plants in microcentrifuge tubes. The tubes were frozen briefly in liquid nitrogen. Then, 400 μL of DNA extraction buffer (100 mm Tris-HCl, pH 8.0, 50 mm EDTA, 500 mm NaCl, 1.4% [w/v] SDS, and 10 mm βM-ETOH) was added and the plants were ground with a plastic pestle. The DNA was extracted according to the method of Edwards et al. (1991). The DNA samples extracted from F2 mutant plants were used in single sequence length polymorphism mapping (Bell and Ecker, 1994).
For complementation tests, heterozygous eld1 plants were crossed with heterozygous fus9 and emb144, homozygous dwf1-1 and dwf7-1 plants. Phenotypes of F1 plants were examined to determine allelism. Seeds of fus9 were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus); emb144 was kindly provided by Dr. D.W. Meinke (Oklahoma State University, Stillwater); and dwf1-1 and dwf7-1 by Dr. K.A. Feldmann (University of Arizona, Tucson).
Light Microscopy and Whole Mount Techniques
For histological examination, samples of eld1 and wild type were fixed in 1% (v/v) glutaraldehyde and 4% (v/v) formaldehyde dissolved in 50 mm sodium phosphate buffer, pH 7.2, for 4 to 12 h at 4°C. After fixation, samples were treated with a series of ethanol and monomer solution A (JB-4 Plus Embedding Kit, Polysciences, Warrington, PA) treatments for dehydration and infiltration, and then embedded in solution A and B, and sectioned as described by Cheng et al. (1995). For whole mount preparations, seedlings were fixed in 9 parts of ethanol and 1 part of acetic acid (v/v) for 2 to 4 h at room temperature. After rinsing with 95% (v/v) ethanol, the samples were transferred to 70% (v/v) ethanol, then moved to a glass slide and mounted in a clearing solution of 8 parts of chloral hydrate, 1 part of glycerol, and 2 parts of water (v/v), as described in Berleth and Jürgens (1993) with some modifications. After 1 h of treatment at room temperature, the samples were observed and photographed with a microscope (Axiophot, Zeiss, Jena, Germany). Sections or samples were stained with 0.05% (v/v) toluidine blue O (TBO), Sudan red 7B, or iodine-potassium iodide (I-KI). Sudan red 7B is a nonfluorescent dye that stains lipids and suberin lamellae (Brundrett et al., 1991). 0.1% (w/v) Sudan red 7B (F 1000, Sigma-Aldrich, St. Louis) was dissolved in polyethylene glycol (400 D, Sigma-Aldrich) by heating at 90°C for 1 h, and an equal volume of 90% (v/v) glycerol was added. Sections were stained with Sudan red 7B for 2 to 24 h at room temperature, then rinsed briefly with deionized water, air dried, and mounted with Permount oil.
Histochemical GUS Assays
cdc2a::GUS transgenic Arabidopsis, homozygous for the single-copy pVPC2AGUS construct (Hemerly et al., 1993), was kindly provided by Dr. Dirk Inzé (Universiteit Gent, Gent, Belgium). This transgenic plant was crossed with heterozygous eld1 plants. The F1 plants were self-pollinated and F2 seedlings were used for GUS assays; 2-, 5-, 7-, 10-, 15-, and 20-d-old eld1 transgenic plants were prepared by treating them with X-glucuronide (X-gluc) as described by Jefferson et al. (1987) and Hemerly et al. (1993) with some modifications. The stock X-gluc solution consists of 0.005 g of 5-bromo-4-chloro-3-indolyl β-d-glucuronide (CLONTECH Laboratories, Palo Alto, CA), which was first dissolved in 0.05 mL of N,N-dimethyl formamide (DMF), and then mixed with 0.05 mL of 5 mm potassium ferrocyanide, 0.05 mL of 5 mm potassium ferricyanide, 0.01 mL of Triton X-100, and 4.84 mL of 50 mm sodium phosphate, pH 7.0. Stock X-gluc of 0.33 mL was diluted with 0.47 mL of 50 mm sodium phosphate buffer, pH 7.0, and mixed with 0.2 mL of 100% (v/v) methanol to reduce the background caused by endogenous GUS activity (Kosugi et al., 1990) and then used to incubate with seedlings. After vacuum infiltration in X-gluc solution for 10 min, the seedlings were incubated at 37°C in the dark for a period from 2 to 16 h. Samples were rinsed with 50 mm sodium phosphate buffer, pH 7.0, then fixed in 9 parts of ethanol (v/v) and 1 part of acetic acid (v/v) for 2 to 4 h at room temperature. Samples were whole-mounted on the glass slide, and then observed and photographed with a microscope.
Growth Regulator Treatment
Seeds from heterozygous plants were germinated on plates containing MS medium supplemented with different types of growth regulators at various concentrations. The growth regulators included auxin (IAA, IBA, 2, 4-D, NAA), cytokinin (kinetin, 2iP), and GA3, which were dissolved in DMSO, and BL, which was dissolved in ethanol. BL was kindly provided by Joanne Chory (Salk Institute, La Jolla, CA). The plates were incubated in the same growth conditions described above, and positioned horizontally. Seven days after germination, 10 wild-type and 10 eld1 seedlings were selected from each of the growth regulator treatments for root length measurement and general observation under a dissecting microscope.
ACKNOWLEDGMENTS
We would like to thank David Martin for assisting with the mapping of eld1; Mary Alice Yund, Jack Chang, Edgar Moctezuma, Lingjing Chen, and Dominique Aubert for critically reading the manuscript; and Steve Ruzin and Denise Schichnes at College of Natural Resources Biological Imaging facility, University of California, Berkeley, for computer help.
Footnotes
This work was supported by the National Science Foundation (grant no. IBN–9513522).
LITERATURE CITED
- Aloni R. The induction of vascular tissues by auxin and cytokinin. In: Davies PJ, editor. Plant Hormones Physiology, Biochemistry and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 531–546. [Google Scholar]
- Araki T, Komeda Y. Flowering in darkness in Arabidopsis thaliana. Plant J. 1993;4:801–811. [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell. 1998;10:219–230. doi: 10.1105/tpc.10.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin TI, Cork A, Williamson RE, Gorst JR. STUNTED PLANT1, a gene required for expansion in rapidly elongating but not in dividing cells and mediating root growth responses to applied cytokinin. Plant Physiol. 1995;107:233–243. doi: 10.1104/pp.107.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Berleth T, Jürgens G. The role of MONOPTEROUS gene in organizing the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- Brundrett MC, Kendrick B, Peterson CA. Efficient lipid staining in plant material with Sudan red 7B or Floral yellow 088 in polyethylene glycol-glycerol. Biotech Histochem. 1991;66:111–116. doi: 10.3109/10520299109110562. [DOI] [PubMed] [Google Scholar]
- Castle LA, Meinke DW. A FUSCA gene of Arabidopsis encodes a novel protein essential for plant development. Plant Cell. 1994;6:25–41. doi: 10.1105/tpc.6.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J-C, Seeley KA, Sung ZR. RML1 and RML2, Arabidopsis genes required for cell proliferation at the root tip. Plant Physiol. 1995;107:365–376. doi: 10.1104/pp.107.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, Tax FE, Feldmann KA. The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, Feldmann KA. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell. 1991;3:445–459. doi: 10.1105/tpc.3.5.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto CA, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989a;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. The plant hormones: their nature, occurrence and functions. In: Davies PJ, editor. Plant Hormones Physiology, Biochemistry and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–12. [Google Scholar]
- Deng X-W. Fresh view of light signal transduction in plants. Cell. 1994;76:423–426. doi: 10.1016/0092-8674(94)90107-4. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Caspar T, Quail PH. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- Deng X-W, Quail PH. Genetic and phenotypic characterization of cop1 mutants of Arabidopsis thaliana. Plant J. 1992;2:83–95. [Google Scholar]
- Edwards K, Johnstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. Anatomy of Seed Plants. New York: John Wiley; 1977. [Google Scholar]
- Fahn A. Plant Anatomy. Ed 4. New York: Pergamon Press; 1990. [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell. 1993;5:1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Soh MS, Hong SH, Furuya M, Nam HG. Photomorphogenic development of the Arabidopsis shy2-1D mutation and its interaction with phytochromes in darkness. Plant J. 1998;1:61–68. doi: 10.1046/j.1365-313x.1998.00179.x. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y. An improved assay for β-glucuronidase in transformed cells: methanol almost completely suppresses a putative endogenous β-glucuronidase activity. Plant Sci. 1990;70:133–140. [Google Scholar]
- Kurata T, Yamamoto KT. petit1, a conditional growth mutant of Arabidopsis defective in sucrose-dependent elongation growth. Plant Physiol. 1998;118:793–801. doi: 10.1104/pp.118.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClinton RS, Sung ZR. Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta. 1997;201:252–260. doi: 10.1007/s004250050064. [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng X-W. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6:487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW. Embryo-lethal mutants of Arabidopsis thaliana: analysis of mutants with a wide range of lethal phases. Appl Genet. 1985;69:543–552. doi: 10.1007/BF00251102. [DOI] [PubMed] [Google Scholar]
- Miséra S, Müller AJ, Weiland-Heidecker U, Jürgens G. The FUSCA genes of Arabidopsis: negative regulators of light responses. Mol Gen Genet. 1994;244:242–252. doi: 10.1007/BF00285451. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–479. [Google Scholar]
- Nicol F, His I, Jauneau A, Vernhettes S, Canut H, Höfte H. A plasma membrane-bound putative endo-1,4-β-d glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 1998;17:5563–5576. doi: 10.1093/emboj/17.19.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JR, Howell SH. Hormone mutants and plant development. In: Davies PJ, editor. Plant Hormones Physiology, Biochemistry and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 448–485. [Google Scholar]
- Schiefelbein JW, Masucci JD, Wang H. Building a root: the control of patterning and morphogenesis during root development. Plant Cell. 1997;9:1089–1098. doi: 10.1105/tpc.9.7.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliday CL, Dean BB, Kolattukudy PE. Suberization: inhibition by washing and stimulation by abscisic acid in potato disks and tissue culture. Plant Physiol. 1978;61:170–174. doi: 10.1104/pp.61.2.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua N-H. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 1995;9:97–107. doi: 10.1101/gad.9.1.97. [DOI] [PubMed] [Google Scholar]
- Timpte C, Wilson AK, Estelle M. Effect of the axr2 mutation of Arabidopsis on cell shape in hypocotyl and inflorescence. Planta. 1992;188:271–278. doi: 10.1007/BF00216824. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Lohner A, Jürgens G. The GURKE gene is required for normal organization of the apical region in the Arabidopsis embryo. Plant J. 1996;10:1005–1016. doi: 10.1046/j.1365-313x.1996.10061005.x. [DOI] [PubMed] [Google Scholar]
- Wei N, Kwok SF, von Arnim AG, Lee A, McNellis TW, Piekos B, Deng X-W. Arabidopsis COP8, COP10, and COP11 genes are involved in repression of photomorphogenic development in darkness. Plant Cell. 1994;6:629–643. doi: 10.1105/tpc.6.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]