Abstract
Transient receptor potential mucolipin 1 (TRPML1) is an endo/lysosomal cation channel ubiquitously expressed in mammalian cells1,2 and its loss-of-function mutations are the direct cause of Type IV mucolipidosis (MLIV), an autosomal recessive lysosomal storage disease3-6. Here we present the single particle cryo-electron microscopy (cryo-EM) structure of the mouse TRPML1 channel embedded in nanodiscs. Combined with mutagenesis, the TRPML1 structure reveals that phosphatidylinositol bisphosphate (PIP2) binds to the N-terminus of the channel – distal from the pore – and the helix-turn-helix extension between S2 and S3 likely couples ligand binding to pore opening. The tightly packed selectivity filter contains multiple ion binding sites and the conserved acidic residues form the luminal Ca2+ blocking site that confers luminal pH and Ca2+ modulation on channel conductance. A luminal linker domain forms a fenestrated canopy atop the channel, providing multiple luminal ion passages to the pore and also creating a negative electrostatic trap – preferably for divalent cations at the luminal entrance. The structure also reveals two equally distributed S4-S5 linker conformations in the closed channel, providing structural implication for the S4-S5 linker-mediated PIP2 gating mechanism among TRPML channels7,8.
Main Text
The organellar channels of the mucolipin subfamily of transient receptor potential (TRPML) are predominantly localized to the membranes of the late endosomes and lysosomes1,2. Among the three isoforms of TRPML channels (TRPML1-3), TRPML1 is ubiquitously expressed in mammalian cells and has been subjected to extensive studies2,9-14. TRPML1 is a Ca2+-permeable, non-selective, six-transmembrane (6-TM) tetrameric cation channel, and is believed to be the main lysosomal Ca2+ release channel important for lysosomal trafficking and signal transduction15-19. Like most TRP channels, TRPML1 is ligand-gated and can be activated by the endolysosomal specific lipid, phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2)7,8, but is inhibited by the plasma membrane-localized phosphoinositide isoform PI(4,5)P214 (Extended Data Fig. 1). This lipid isoform-dependent gating property defines the compartment specific channel activity of TRPML1. In human, loss-of-function mutations in TRPML1 are the direct cause of Mucolipidosis type IV (MLIV), an autosomal recessive lysosomal storage disease characterized by abnormal neurodevelopment, retinal degeneration and iron-deficiency anemia3-6. Due to its direct linkage to MLIV disease, TRPML1 has been a potential target for small molecule therapeutics and several synthetic small molecule agonists have been developed12,20.
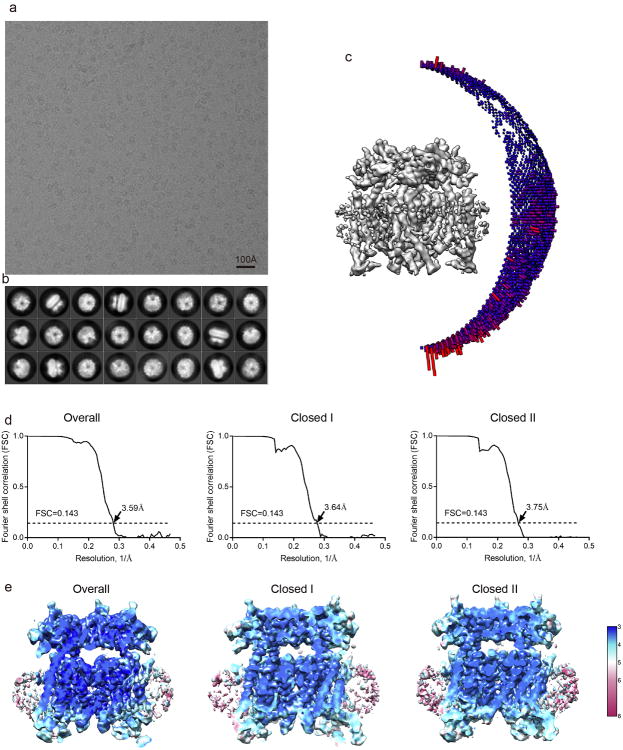
We purified and reconstituted mouse TRPML1 into nanodiscs and determined its structure by single particle cryo-EM to a resolution of 3.59 Å using the gold-standard Fourier shell correlation (FSC) = 0.143 criteria21 (Extended Data Fig. 2-4 and Methods). The cryo-EM density map is of sufficient quality for accurate model building of major parts of the protein (Extended Data Fig. 5 & 6). TRPML1 consists of two structural components: the membrane spanning S1-S6 region embedded in a nanodisc and the luminal S1-S2 linker forming a fenestrated canopy atop the channel domain (Fig. 1). Like other classical 6-TM tetrameric cation channels, the S1-S6 of each subunit is separated into an S1-S4 voltage-sensing domain (VSD) and an S5-S6 pore domain linked by the S4-S5 linker helix. The tetramer assembly is domain swapped, with the S1-S4 domain interacting with the pore-forming S5-S6 of a neighboring subunit. As the structure was determined in a lipid environment, density of poly-acyl chains from closely attached lipid molecules are clearly visible within the grooves between two neighboring S1-S4 domains (Fig. 1a). The 200-residue, luminal-facing linker domain between S1 and S2 can self-assemble into a tetramer and its crystal structure22 fits readily into our EM-density map, facilitating our model building. Asn230 of the luminal linker domain is glycosylated with visible density for the covalently linked N-acetylglucosamine moiety of the sugar (Fig. 1b&c).
Figure 1.
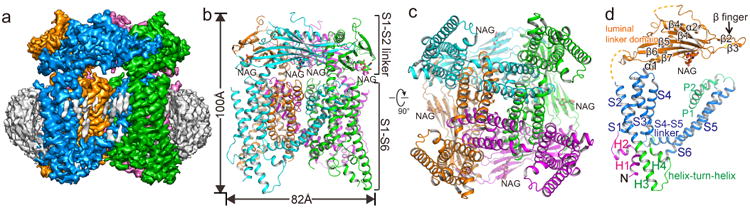
Overall structure of TRPML1. a, Side view of 3-D reconstruction of TRPML1 in nanodisc (grey belt). Channel subunits are colored individually with lipid density in grey. b, Side view of cartoon representation of TRPML1 structure. N-acetylglucosamine (NAG) molecules are rendered as sticks. c, Cytosolic view of the channel. d, Structure of a single subunit in the same orientation as the cyan-colored subunit in b.
The S1-S4 domain of TRPML1 is similar to that of other TRP channels but differs from the canonical voltage-sensing domain of voltage-gated channel (Fig. 2a). It consists of four straight helices tightly packed against each other. Lacking the canonical voltage-sensing arginines, the S4 helix runs antiparallel to the S5 helix of a neighboring subunit and exhibits extensive hydrophobic interactions (Fig. 2a). The S1-S4 of TRPML likely remains static, similar to TRPV123,24, and does not undergo any conformational change during channel gating. There are two cytosolic extensions to the S1-S4 domain that are unique to TRPML channels (Fig. 2a&b). First, an Arg/Lys-rich (poly-basic) domain from residues 40-62 forms two short clampshaped helices, labeled as H1 and H2, right before S1. Second, the linker between S2 and S3 forms a helix-turn-helix (H3-turn-H4) that extends into the cytosol and also makes direct contact with the N-terminal H1 helix (Fig. 2b). The N-terminal poly-basic domain is important for PIP2 binding7,14 and its deletion abolishes PI(3,5)P2 activation of TRPML1 (Extended Data Fig. 1e). The H1 and H2 helices of the poly-basic domain, along with the N-terminus of S1, create a positively charged pocket directly beneath the cytosolic membrane surface (Fig. 2b), likely to accommodate the inositol trisphosphate (IP3) head group of membrane-anchored PIP2. As the PIP2 site is distant from the inner helix-formed channel gate, we suspect that PIP2 activation is mediated by the H3-turn-H4 extension whose H3 helix interacts with the H1 and H4 helix interacts with the S4-S5 linker in a specific conformation, as will be discussed later.
Figure 2.
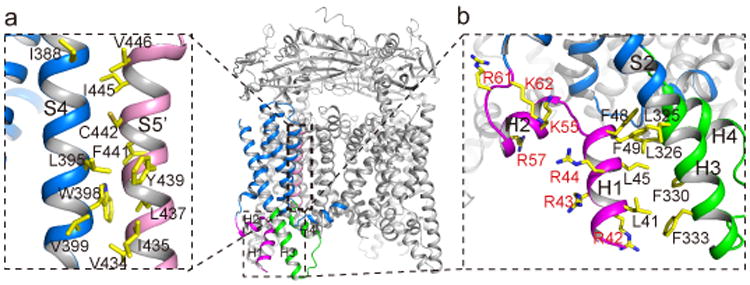
Structure of S1-S4 region and the PIP2 binding site. a, S1-S4 domain (blue) of the front subunit. The two cytosolic extensions are colored differently - magenta for H1 & H2 and green for the H3-turn-H4 between S2 and S3. Inset shows inter-subunit contact between S4 helix (blue) and the neighboring S5 (pink). b, Zoomed-in view of the poly-basic domain and the interactions between H1 and H3 helix.
Since the structure was determined in the absence of ligands, the TRPML1 ion conduction pore, consisting of the S5, S6 and two pore helices (P1 and P2), adopts the closed conformation (Fig. 3a &b). The four pore-lining S6 inner helices form a bundle crossing at the cytosolic side with multiple narrow constriction points that prevent the passage of hydrated cations. Surrounding the cytosolic gate, S6 makes tight contact with the N-terminal part of S5 (Fig. 3a, inset), suggesting a coupled movement between S5 and S6 upon channel gating. The residues of FIYMV on S6 just below the selectivity filter region form a π-helix (Fig. 3a), a structural feature that was also observed in other TRP channel structures23,25 and may facilitate the bending of S6 inner helix for channel gating.
Figure 3.
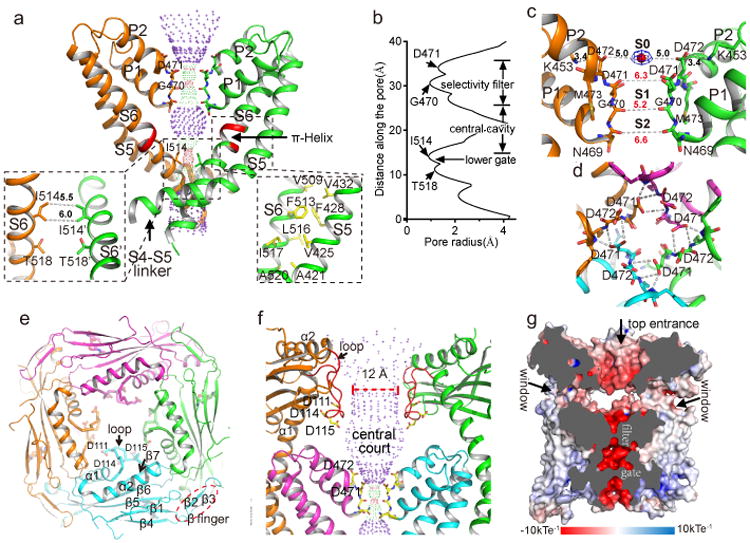
Ion conduction pathway of TRPML1. a, Ion conduction pore of TRPML1 with front and rear subunits removed for clarity. Central pathway is marked with dotted mesh. Insets are zoomed-in views of the bundle crossing with atom-to-atom distances in Å and the packing between S5 and S6. π-helix on S6 is colored in red. b, Pore radius along the central axis. c, Zoomed-in view of the selectivity filter with atom-to-atom distances in Å. S0 density is modeled as Na+ (red sphere). d. Top view of the selectivity filter showing H-bonding networks between Asp471 side chain and backbone nitrogen atoms of neighboring filter residues. e, Top view of the luminal linker domains in tetramer. Three acidic residues on the luminal pore loop (labeled as loop) form the narrowest part of the central hole. f, Side view of the funnel shaped central hole and the open central court just above the filter. Front and rear subunits are removed for clarity. The luminal pore loop is colored red. g, Cross section through the channel showing the surface electrostatic potential along the ion conduction pathway. The open court at the luminal entrance is accessible by multiple luminal ion passages indicated by arrows.
The TRPML1 selectivity filter has the sequence 469NGDDM and adopts a multi-ion configuration (Fig. 3c). The backbone carbonyls of the NGD residues all point towards the central pore axis, creating two contiguous ion binding cages (labeled as S1 and S2) – reminiscent of the two K+ channel sites but with wider dimensions – and likely accommodating partially dehydrated cations. The second Asp (Asp472) has its carboxylate side chain pointed upright and is stabilized laterally by a salt bridge with a highly conserved Lys453 between the S5 and P1 helix. Four Asp472s in a channel tetramer encircle an ion binding site (labeled as S0) at the luminal entrance of the channel where an electron density peak is clearly visible and was modeled as Na+, the only cation in the sample (Fig. 3c). The distance between the S0 ion and the carboxylate oxygen of Asp472 is about 5 Å, and the ion chelation is likely mediated by water. TRPML1 is known to be Ca2+ permeable and Ca2+ permeation lowers channel conductance by blocking monovalent cation currents in a luminal pH-dependent manner (Extended Data Fig. 7a). Surrounded by acidic residues, we suspect that S0 is the Ca2+ blockage site. Indeed, neutralizing Asp472 with Asn results in a significantly diminished luminal Ca2+ block (Extended Data Fig. 7b & c).
The TRPML1 filter is rigidly held in place by two pore helices with tightly packed surroundings. The side chain carboxylate of Asp471 plays an essential role in stabilizing the filter conformation by generating a hydrogen-bonding network with the backbone amide nitrogen atoms of the neighboring filter residues (Fig. 3d). The Met473 sidechain is extended parallel to and fills the void surrounding the filter (Fig. 3c). The tight packing of the TRPML1 filter is quite different from that of the TRPV1 filter, where the equivalent P2 helix is replaced by an extended loop, allowing the filter to undergo significant conformational change upon the spider vanillotoxin binding and gating24. A similar filter conformational change is unlikely to occur in TRPML1.
The four luminal linker domains, each consisting of two long helices and seven strands, form a square shaped canopy with a central opening above the channel pore (Fig. 1b and Fig. 3e), a characteristic feature of group 2 TRP channels including TRPML and TRPP25,26 (Extended Data Fig. 8). The α1 and α2 helices, along with the long downward-facing loop (named luminal pore loop) between them, generate a funnel shaped hole at the center of the canopy. The tip of the luminal pore loop is clustered with acidic residues and forms the narrowest opening at the bottom of the funnel with a diameter of about 12 Å, wide enough for free ion passage (Fig. 3e & f and Extended data Fig. 8). The α1 helix comes from the C-terminal part of an exceptionally long S1 helix, and, consequently, the luminal canopy is firmly supported by four pillar-like S1 helices over the transmembrane channel with a wide gap, creating an open central court just above the filter with four side windows (Fig. 1b and Fig. 3f & g) that provide additional luminal ion passages to the ion conduction pore. The open central court is surrounded by an exceptionally high number of acidic residues from the luminal pore loop and the luminal entrance to the filter, creating an extremely negative environment (Fig. 3f & g). This feature, along with multiple converging ion passages, renders the central court an electrostatic sink that can favorably attract divalent Ca2+ ions. The enrichment of Ca2+ ions can effectively limit the access of monovalent cations to the filter and, thereby, reduce the permeation of monovalent ions. Indeed, it has been shown that neutralizing the acidic residues on the luminal pore loop weakens the Ca2+ blockage22.
The density of the S4-S5 linker in the overall TRPML1 structure is poorly resolved, indicating dynamic motion of the linker. A focused classification at the transmembrane region yielded two sub-sets of particles that were reconstructed individually into two structures at 3.64 and 3.75Å resolution, respectively, each adopting a distinct conformation at the S4-S5 linker (Extended Data Fig. 3). The nearly equal distribution of particles for the two structures (9,000 and 11,000 particles, respectively) suggests that the unliganded TRPML1 channel is stable in two closed states (closed I and II) with the S4-S5 linker undergoing conformational equilibrium.
The S4-S5 linker in the closed I state adopts a straight helix with its N-terminus interacting with the S5 helix from the neighboring subunit (Fig. 4a-c). Four linker helices surround the S6 bundle crossing and stabilize the closed channel pore. It is interesting to note that several gain-of-function mutations similar to the V432P varitint-waddler phenotype15,27-29 have been identified on S5 in a proline-scanning mutagenesis study30, including R427P, C430P and C431P, which are all clustered at the interaction interface between the S5 and S4-S5 linker (Fig. 4c). These mutations likely cause a kink in the S5 helix, disrupt the inter-subunit interactions and destabilize the closed channel pore. In the closed II state, the S4-S5 linker undergoes a significant conformational change as compared to the closed I state (Fig. 4a & b). Pivoting at its C-terminus, the linker swings away from the central axis and unlocks the cuff around the bundle crossing; the N-terminal half of the linker helix becomes partially unfolded and bulges outward; the H3-turn-H4 extension of S2/S3 moves slightly towards the center and engages in direct contact with the bulged linker that is not observed in the closed I state (Fig. 4d). While the S6 inner helices remain in a closed state, the outward movement of the S4-S5 linker starts to pull S5 away from the central axis and any further movement of S5 would result in a concurrent movement of S6 and pore opening. Therefore, the closed II structure likely represents a transition state in which the channel is primed for opening.
Figure 4.
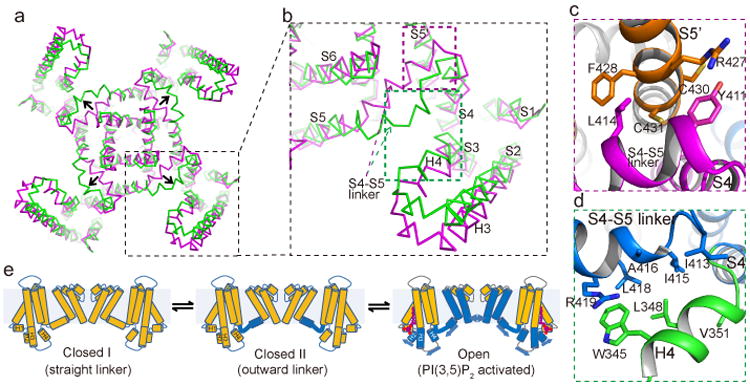
Alternative conformations of TRPML1 in closed state. a, Superposition between -closed I (purple) and closed II (green) states with H1 and H2 helices omitted for clarity. Arrows indicate the movement of S4-S5 linker. b, Zoomed-in view of the superposition for one subunit. c, Inter-subunit interactions between N-terminus of the linker and N-terminus of the neighboring S5 in closed I state at the location marked by the purple box in b. d, Interactions between partially unfolded linker and H4 helix at the location marked by the green box in b. e, Proposed working model for PI(3,5)P2 activation. Arrows indicate proposed movements upon ligand binding.
Since the S4-S5 linker is strategically positioned to couple the voltage- or ligand-induced conformational changes to the opening of the pore in various 6-TM channels, it likely plays a similar role in PIP2 gating of TRPML1. With its binding site at the N-terminus of TRPML1, distal from the S4-S5 linker, the PI(3,5)P2 activation is likely mediated by the H3-turn-H4 of the S2/S3 extension whose H3 helix interacts with the H1 and H4 helix interacts with the bulged S4-S5 linker in the closed II state. We therefore proposed a simple gating mechanism in which the PI(3,5)P2 induced conformational change can be propagated to the outward-bulged S4-S5 linker via the H3-turn-H4 and opens the pore (Fig. 4e). This model implies that PI(3,5)P2 activation can only occur when TRPML1 is in the closed II transition state as the S4-S5 linker helix is decoupled from H3-turn-H4 in the closed I state, and thereby the linker transition between the two closed states is prerequisite for the PIP2 activation of TRPML.
Methods
Protein expression, purification, and nanodisc reconstitution
Mouse TRPML1 (MmTRPML1, NCBI refseq NM_053177.1) containing a C-terminal thrombin cleavage site followed by a 10× His tag was cloned into a pEZT vector31 and heterologously expressed in HEK293F cells (Life Technologies) using the BacMam system (Thermo Fisher Scientific). The baculovirus was generated in Sf9 cells (Life Technologies) following the standard protocol and used to infect HEK293F cells at a ratio of 1:40 (virus:HEK293F, v:v) and supplemented with 2 mM sodium butyrate to boost protein expression. Cells were cultured in suspension at 37 °C for 48 hours and harvested by centrifugation at 3,000g. All purification procedures were carried out at 4°C unless specified otherwise. The cell pellet was re-suspended in buffer A (50 mM Tris pH 8.0, 200 mM NaCl) supplemented with a protease inhibitor cocktail (containing 1mg/mL each of DNAse, pepstatin, leupeptin, and aprotinin and 1 mM PMSF) and homogenized by sonication on ice. MmTRPML1 was extracted with 1% (w:v) n-Dodecyl-β-D-Maltopyranoside (DDM) (Anatrace) supplemented with 0.02% (w:v) cholesteryl hemisuccinate (CHS; Sigma Aldrich) by gentle agitation for 2 hours. After extraction, the supernatant was collected after a 40-minute centrifugation at 48,000g and incubated with Ni-NTA resin (Qiagen) using gentle agitation. After 2 hours, the resin was collected on a disposable gravity column (Bio-Rad), washed with buffer B (buffer A + 0.1% DDM + 0.02% CHS) with 20 mM imidazole, and then buffer B. The washed resin was left on column in buffer B and digested with thrombin (Roche) overnight. After thrombin digestion, the flow-through containing untagged MmTRPML1 was collected, concentrated, and purified by size exclusion chromatography on a Superose 6 10/300 GL column (GE Heathcare) pre-equilibrated with buffer C (20 mM Tris pH 8.0, 150 mM NaCl 0.1% DDM and 0.02% CHS). The protein peak was collected, concentrated to 40 μM and reconstituted into lipid nanodiscs following the published protocol24. Briefly, MSP1, MmTRPML1, and lipid (POPC:POPG:POPE = 3:1:1) were mixed at a molar ratio of 4:1:10, respectively, and incubated on ice for 30 minutes. Detergents were removed by adding Bio-beads SM2 (Bio-Rad) to a concentration of 100 mg/ml followed by gentle agitation. The used bio-beads were replaced with fresh ones every 4 hours for two times. After detergent removal, the sample was loaded onto a Superose 6 10/300 GL column pre-equilibrated with buffer D (20 mM Tris pH 8.0 and 150 mM NaCl) and the elution peak corresponding to the reconstituted MmTRPML1 was collected for cryo-electron microscopy analysis.
EM data acquisition
The cryo-EM grids were prepared by applying 3 μl of MmTRPML1 in nanodiscs (1.3 mg/ml) to a glow-discharged Quantifoil R1.2/1.3 200-mesh copper holey carbon grid (Quantifoil, Micro Tools GmbH, Germany) and blotted for 4.0 seconds under 100% humidity at 4 °C before being plunged into liquid ethane using a Mark IV Vitrobot (FEI). Micrographs were acquired on a Titan Krios microscope (FEI) operated at 300 kV with a K2 Summit direct electron detector (Gatan), using a slit width of 20 eV on a GIFQuantum energy filter. EPU software (FEI) was used for automated data collection following standard FEI procedure. A calibrated magnification of 46,730X was used for imaging, yielding a pixel size of 1.07 Å on images, with the defocus ranging from −1.2 μm to −3 μm. Each micrograph was dose-fractionated to 30 frames under a dose rate of 4e-/pixel/s, with a total exposure time of 15 s, resulting in a total dose of about 50 e-/Å2.
Image processing
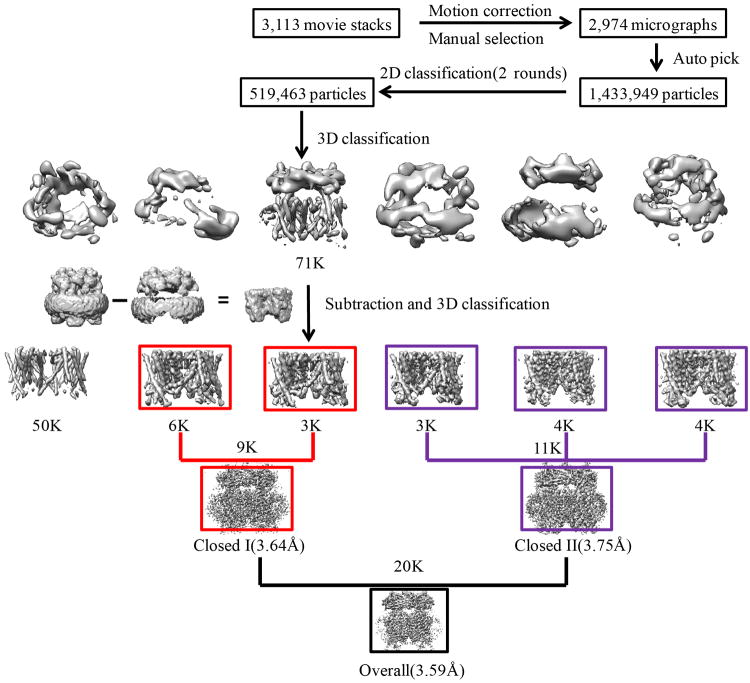
Motion correction was performed using MotionCorr2 program32, and the CTF parameters of the micrographs were estimated using the GCTF program33. All other steps of image processing were performed using RELION34. Initially, ∼ 1,000 particles were manually picked from a few micrographs. Class averages representing projections of MmTRPML1 in different orientations were selected from the 2D classification of the manually picked particles, and used as templates for automatic particle picking from the full data set of 2,974 micrographs. The extracted particles were binned 3 times and subjected to two rounds of 2D classification, and a total of 519,463 particles were finally selected for 3D classification and 3D refinement. For the first round of 3D classification, the TRPV1 structure (EMDB code 5778) was used as the reference. One of the 3D classes showed good secondary structural features and is significantly different from TRPV1. It was then selected as the reference for the second round of 3D classification. One of the resulting 3D reconstructions from about 71,000 particles showed improved cryo-EM density and a clear 4-fold symmetry with a resolution of ∼ 4 Å after 3D refinement using the un-binned particles (C4 symmetry imposed) and particle polishing. However, part of the transmembrane domain showed poor density, indicative of local structural heterogeneity. Therefore, we performed a focused 3D classification with density subtraction35 in order to improve the density of the transmembrane domain. In this approach, the density corresponding to the soluble domain of the channel as well as the belt-like density from the nanodisc was subtracted from the original particles. The subsequent 3D classification on the modified particles was carried out by applying a mask around the transmembrane domain and having all of the orientations fixed at the value determined in the 3D refinement. This round of classification yielded 6 classes of particles, among which one major class (∼50,000 particles) showed poor density in the transmembrane region and was discarded. After a new 3D refinement using the particles from the 5 classes (∼20,000 total particles), the resolution of the map was improved from 4 Å to 3.59 Å. In addition, these five classes of particles can be partitioned into two groups with 9,000 and 11,000 particles, respectively. 3-D refinement of each group yielded structures at 3.64 Å (closed I state) or 3.75 Å (closed II state) resolution, respectively, each showing distinct conformations at the S4-S5 linker. All resolutions were estimated by applying a soft mask around the protein density and the gold-standard Fourier shell correlation (FSC) = 0.143 criterion. ResMap36 was used to calculate the local resolution map.
Model building, refinement, and validation
For the soluble domain, the structure of HsTRPML1 I-II liner (PDB code: 5TJC) was docked into the cryo-EM map of MmTRPML1 followed by manual adjustment and mutation of residues that differed between MmTRPML1 and HsTRPML1. For the transmembrane domain, de novo atomic model building was conducted in Coot37. Amino acid assignment was achieved based mainly on the clearly defined densities for bulky residues (Phe, Trp, Tyr, and Arg). Models were refined against summed maps using phenix.real_space_refine38 implemented in PHENIX39. The model was validated using previously described methods40 to avoid overfitting. The final structure models include residues 40–198, 215-285, and 296–527. About 40 residues at the amino-terminus and 50 residues at the carboxyl-terminus are disordered and not modeled. The statistic of the models' geometries was generated using MolProbity41. Pore radii were calculated using the HOLE program42. All the figures were prepared in PyMol43 or Chimera44.
Electrophysiology
By replacing the two di-leucine motifs (L15L and L577L) responsible for lysosomal targeting with alanines13,45, mouse TRPML1 (MmTRPML1-4A) can be over-expressed and trafficked to the plasma membrane in HEK293 cells, allowing for direct measurement of channel activity by patching the plasma membrane. In this setting, the extracellular side is equivalent to the luminal side of TRPML1 in endo/lysosomes. The constitutively active V432P mutant, equivalent to the gain-of-function mutation of the mouse TRPML3 in the varitint-waddler (va) phenotype15,27-29, can be trafficked to the plasma membrane without replacing the di-leucine motifs and therefore V432P and V432P/D472N mutants were both constructed on the background of the wild type channel. The N-terminus truncation mutant was constructed on the background of MmTRPML1-4A. DNA fragments encoding MmTRPML1 and its mutants were cloned into a pEGFP-C2 vector using xhoI and sacII restriction sites, as described2. N-terminal eGFP-tagged MmTRPML1 or its mutant were expressed in HEK293 cells grown in a six-well tissue culture dish by transiently transfecting 2 μg of DNA plasmid using Lipofectamine 2000 (Thermo Fisher Scientific). 24-48 hours after transfection, cells were dissociated by trypsin treatment and kept in complete serum-containing medium; the cells were re-plated onto 35 mm tissue culture dishes and kept in a tissue culture incubator until recording. Patch clamp in the whole-cell or inside-out configuration was used to measure TRPML1 activity on HEK plasma membrane. The standard bath solution for whole cell current recording contained (in mM): 145 sodium methanesulfonate (Na-MS), 5 NaCl, 1 MgCl2, 1 CaCl2 or nominal Ca2+ free, 10 HEPES buffered with Tris, pH 7.4 or 10 MES buffered with Tris, pH=4.6; and the pipette solution contained (in mM): 140 Cs-MS, 5 NaCl, 5 MgCl2, 10 EGTA, 10 HEPES buffered with Tris, pH=7.4. For the measurement of luminal [a2+] dependent blockage, the bath solution contained (in mM): 145 sodium methanesulfonate (Na-MS), 5 NaCl, 1 MgCl2, 10 HEPES buffered with Tris, pH 7.4, 0.5 EGTA (for 0 Ca2+) or 0.1, 1, 10 Ca2+ without EGTA. The bath solution for inside-out configuration contained (in mM): 140 K-MS, 5 NaCl, 2 MgCl2, 0.4 CaCl2, 1 EGTA, 10 HEPES buffered with Tris, pH=7.4; and the pipette solution contained (in mM): 145 Na-MS, 5 NaCl, 1 MgCl2, 0.5 EGTA, 10 MES buffered with Tris, pH=4.6. The patch pipettes were pulled from Borosilicate glass (Harvard Apparatus) and heat polished to resistance of 2-5 MΩ (2-3 MΩ for inside out patch, and 3-5 MΩ for whole cell current recoding). Data was acquired using an AxoPatch 200B amplifier (Molecular Devices) and a low-pass analogue filter set to 1 kHz. The current signal was sampled at a rate of 20 kHz using a Digidata 1322A digitizer (Molecular Devices) and further analyzed with pClamp 9 software (Molecular Devices). After the patch pipette attached the cell membrane, the giga seal (5-10 GΩ) was formed by gentle suction. The whole cell configuration was formed by short zap or suction to rupture the patch. The holding potential was set to 0 mV. The whole cell and inside-out macroscopic current recordings were obtained using voltage pulses ramp from −140 to +50 mV over 800 ms duration. The sample traces for single channel recording or the I-V curves of macroscopic currents shown in each figure were obtained from recordings on the same patch. The lipid ligands used in this study are phosphatidylinositol 3,5-bisphosphate diC8 (PI(3,5)P2 diC8, Echelon), phosphatidylinositol 4,5-bisphosphate diC8 (PI(4,5)P2 diC8, Echelon) and mucolipin synthetic agonist 1 (ML-SA1, Sigma). All data points are mean ± SEM (n ≥5).
Data availability
The cryo-EM density maps of the mouse TRPML1 in nanodisc have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-8883 (overall), EMD-8881 (closed I) and EMD-8882 (closed II). Atomic coordinates have been deposited in the Protein Data Bank under accession numbers 5WPV (overall structure), 5WPQ (closed I state) and 5WPT (closed II state).
Extended Data
Extended Data Figure 1. Ligand activation of TRPML1 over-expressed in HEK cell.
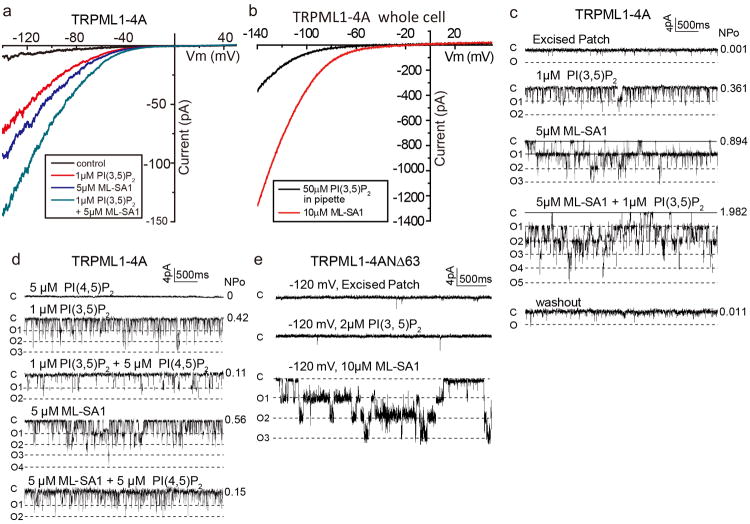
a, Macroscopic currents of plasma membrane-localized TRPML1-4A in an inside-out patch in the presence and absence of ligands in bath (cytosolic). TRPML1 can be activated by PI(3,5)P2 and mucolipin synthetic agonist 1 (ML-SA1), yielding inwardly rectified cation currents. PI(3,5)P2 and ML-SA1 activations are synergistic, indicating non-overlapping activation sites between the two ligands. b, Macroscopic currents of TRPML1-4A in the whole-cell configuration. The pipette (cytosolic) solution contained 50 μM PIP(3,5)P2 (black trace). Addition of 10μM ML-SA1 in the bath (extracellular/luminal side) yielded a much larger current, suggesting that ML-SA1 can also activate the channel from the luminal side. c, Sample traces of single channel currents recorded at −120 mV in an inside-out excised patch showing PIP(3,5)P2 and ML-SA1 activation. The patch contained multiple channels (N ≥ 5). N, total number of channels; Po, single channel open probability. d, Sample traces of single channel currents recorded at −120 mV in an inside-out excised patch showing PI(4,5)P2 (in bath) inhibition of TRPML1. e, Single channel recordings of N-terminal truncation mutation of TRPML1-4A in an inside-out patch. Deletion of the poly-basic domain abolishes PI(3,5)P2 activation confirming its participation in PIP2 binding; the ML-SA1 activation remains intact in this mutant, confirming distinct activation sites between PIP2 and the small molecule agonist.
Extended Data Figure 2. Structure determination of mouse TRPML1 in nanodiscs.
a, Representative micrograph of TRPML1 in nanodisc. b, Two-dimensional class averages. c, Euler angle distribution of particles used in the final three-dimensional reconstruction, with the heights of the cylinders corresponding to the number of particles. d, Gold-standard FSC curves of the final 3-D reconstructions. e, Final density maps colored by local resolution.
Extended Data Figure 3. Flowchart of image processing.
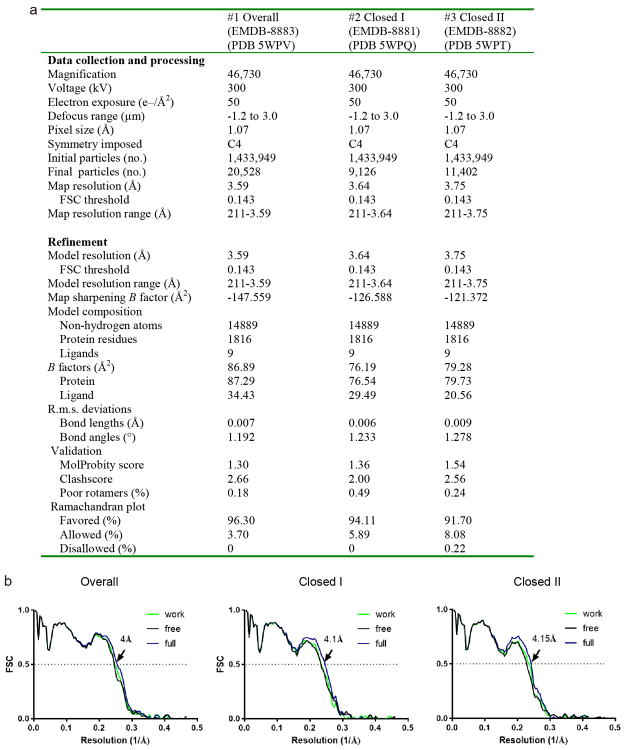
Extended Data Figure 4. Data collection, structure refinement and model validation.
a, Data collection and model refinement statistics. b, FSC curves for cross-validation between the maps and the models. Curves for model vs. summed map in blue (full), for model vs. half map in green (work), and for model vs. half map not used for refinement in black (free).
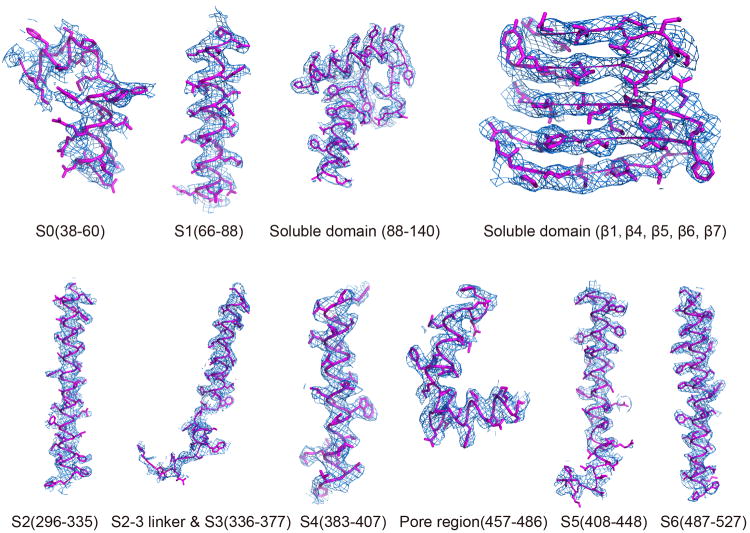
Extended Data Figure 5. Sample EM density maps (blue mesh) for various parts of the channel.
The maps are low-pass filtered to 3.59 Å and sharpened with a temperature factor of -120 Å2.
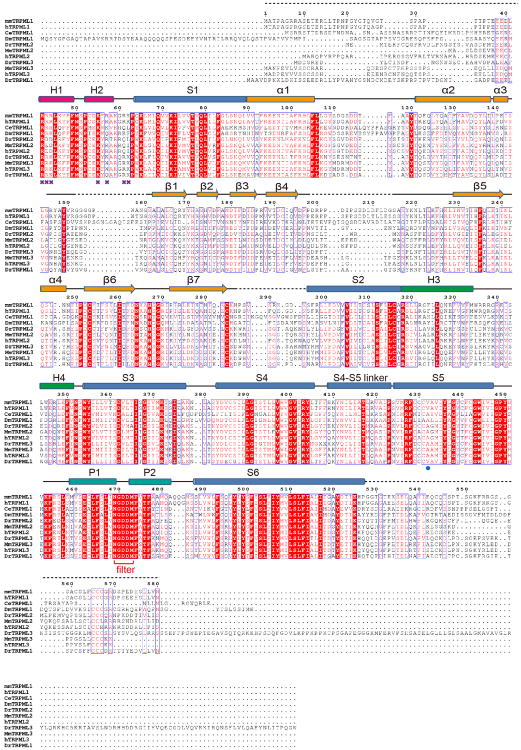
Extended Data Figure 6. Sequence alignment of TRPML channels.
Secondary structure assignments are based on the mouse TRPML1 structure.
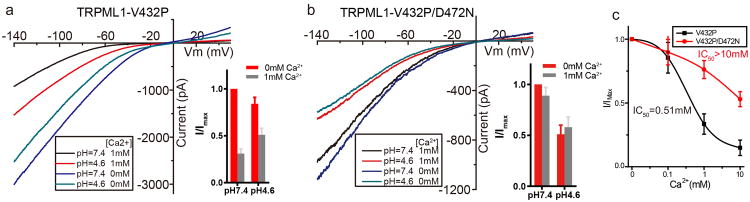
Extended Data Figure 7. Luminal Ca2+ and pH modulation of TRPML1.
a, Sample traces (I-V curves) of whole-cell currents from the constitutively active TRPML1-V432P mutant recorded with various luminal (bath) [a2+] and pH. Inset shows normalized channel currents at −120 mV. Data are mean ± SEM of five measurements. 0 mM [a2+] here refers to nominally Ca2+ free in the recording. b, Sample traces of whole-cell currents of TRPML1-V432P/D472N mutant recorded at various luminal (bath) [a2+] and pH. Inset shows normalized channel currents at −120 mV. Data are mean ± SEM of five measurements. c, Luminal [a2+] dependent blockage of inward currents in TRPML1-V432P and TRPML1-V432P/D472N measured at −120 mV and pH 7.4. In summary, 1 mM Ca2+ – close to the lysosomal Ca2+ concentration – can significantly reduce the channel current; lowering the pH can partially alleviate Ca2+ blockage, likely by protonating the Ca2+ binding acidic residues; in the absence of Ca2+, however, lower pH by itself has an inhibitory effect on channel conductance. Neutralizing Asp472 with Asn diminishes luminal Ca2+ block.
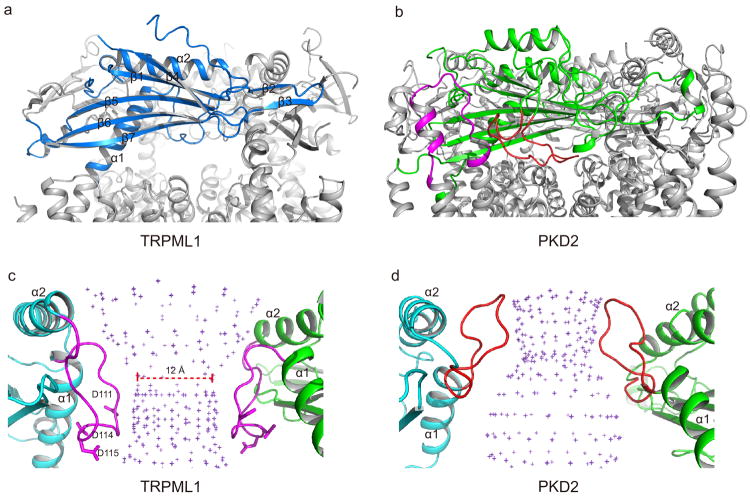
Extended Data Figure 8. Structural comparison between the S1-S2 linker domains of TRPML1 and PKD2, a member of the TRPP family.
a, Side view of TRPML1 luminal linker domain atop the channel with open side windows. The front subunit is highlighted in blue. b, Side view of PKD2 polycystin domain. The polycystin domain has an extra hairpin loop (red) that clogs the side window, making the central hole the only extracellular passage to the filter. There is an extra helix-turn motif (magenta) between S3 and S4 in PKD2 that extend upright and provides extra contact between the polycystin domain and the transmembrane domain. c, The luminal pore loop (magenta) between α1 and 2 points downward in TRPML1, generating a funnel shaped central hole with a constriction of 12 Å. d, The luminal pore loop (red) in PKD2 points upward and generates a central hole with an inverted funnel shape. Front and rear subunits are removed c and d for clarity.
Supplementary Material
Acknowledgments
We thank N. Nguyen for manuscript preparation. Single particle cryo-EM data were collected at the UT Southwestern Medical Center (UTSW) Cryo-Electron Microscopy Facility. We thank D. Nicastro and Z. Chen for support in facility access and data acquisition. Negatively stained sample screening was performed at UTSW Electron Microscopy core. This work was supported in part by the Howard Hughes Medical Institute (Y.J.) and by grants from the National Institute of Health (GM079179 to Y. J; NS062792 and AR060837 to H.X) and the Welch Foundation (Grant I-1578 to Y. J.). X.B. is supported by the Cancer Prevention and Research Initiative of Texas and Virginia Murchison Linthicum Scholar in Medical Research fund. The authors declare no competing financial interests.
Footnotes
Author Contributions: Q.C. and J.S. prepared the samples; Q.C., J.S,.J.G. and X.B. performed data acquisition, image processing and structure determination; W.Z. performed electrophysiology; H.X. provided DNA materials and constructive advice; all authors participated in research design, data analysis, and manuscript preparation.
Author Information: The authors declare no competing financial interests.
References
- 1.Cheng X, Shen D, Samie M, Xu H. Mucolipins: Intracellular TRPML1-3 channels. FEBS Lett. 2010;584:2013–2021. doi: 10.1016/j.febslet.2009.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong XP, et al. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargal R, et al. Identification of the gene causing mucolipidosis type IV. Nat Genet. 2000;26:118–123. doi: 10.1038/79095. [DOI] [PubMed] [Google Scholar]
- 4.Bassi MT, et al. Cloning of the gene encoding a novel integral membrane protein, mucolipidin and identification of the two major founder mutations causing mucolipidosis type IV. Am J Hum Genet. 2000;67:1110–1120. doi: 10.1016/S0002-9297(07)62941-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nilius B, Owsianik G, Voets T, Peters JA. Transient receptor potential cation channels in disease. Physiol Rev. 2007;87:165–217. doi: 10.1152/physrev.00021.2006. [DOI] [PubMed] [Google Scholar]
- 6.Sun M, et al. Mucolipidosis type IV is caused by mutations in a gene encoding a novel transient receptor potential channel. Hum Mol Genet. 2000;9:2471–2478. doi: 10.1093/hmg/9.17.2471. [DOI] [PubMed] [Google Scholar]
- 7.Dong XP, et al. PI(3,5)P(2) controls membrane trafficking by direct activation of mucolipin Ca(2+) release channels in the endolysosome. Nat Commun. 2010;1:38. doi: 10.1038/ncomms1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng X, et al. Drosophila TRPML forms PI(3,5) P2-activated cation channels in both endolysosomes and plasma membrane. J Biol Chem. 2014;289:4262–4272. doi: 10.1074/jbc.M113.506501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra M, et al. A role for the Ca2+ channel TRPML1 in gastric acid secretion, based on analysis of knockout mice. Gastroenterology. 2011;140:857–867. doi: 10.1053/j.gastro.2010.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng X, et al. The intracellular Ca(2)(+) channel MCOLN1 is required for sarcolemma repair to prevent muscular dystrophy. Nat Med. 2014;20:1187–1192. doi: 10.1038/nm.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puertollano R, Kiselyov K. TRPMLs: in sickness and in health. Am J Physiol Renal Physiol. 2009;296:F1245–1254. doi: 10.1152/ajprenal.90522.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen D, et al. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat Commun. 2012;3:731. doi: 10.1038/ncomms1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vergarajauregui S, Puertollano R. Two di-leucine motifs regulate trafficking of mucolipin-1 to lysosomes. Traffic. 2006;7:337–353. doi: 10.1111/j.1600-0854.2006.00387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Li X, Xu H. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc Natl Acad Sci U S A. 2012;109:11384–11389. doi: 10.1073/pnas.1202194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Delling M, Li L, Dong X, Clapham DE. Activating mutation in a mucolipin transient receptor potential channel leads to melanocyte loss in varitint-waddler mice. Proc Natl Acad Sci U S A. 2007;104:18321–18326. doi: 10.1073/pnas.0709096104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatachalam K, Wong CO, Zhu MX. The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium. 2015;58:48–56. doi: 10.1016/j.ceca.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, et al. Presenilin 1 Maintains Lysosomal Ca(2+) Homeostasis via TRPML1 by Regulating vATPase-Mediated Lysosome Acidification. Cell Rep. 2015;12:1430–1444. doi: 10.1016/j.celrep.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilpatrick BS, Yates E, Grimm C, Schapira AH, Patel S. Endo-lysosomal TRP mucolipin-1 channels trigger global ER Ca2+ release and Ca2+ influx. J Cell Sci. 2016;129:3859–3867. doi: 10.1242/jcs.190322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Q, et al. BK Channels Alleviate Lysosomal Storage Diseases by Providing Positive Feedback Regulation of Lysosomal Ca2+ Release. Dev Cell. 2015;33:427–441. doi: 10.1016/j.devcel.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Chen CC, et al. A small molecule restores function to TRPML1 mutant isoforms responsible for mucolipidosis type IV. Nat Commun. 2014;5:4681. doi: 10.1038/ncomms5681. [DOI] [PubMed] [Google Scholar]
- 21.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Li M, et al. Structural basis of dual Ca2+/pH regulation of the endolysosomal TRPML1 channel. Nat Struct Mol Biol. 2017;24:205–213. doi: 10.1038/nsmb.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen PS, et al. The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell. 2016;167:763–773 e711. doi: 10.1016/j.cell.2016.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilius B, Owsianik G. The transient receptor potential family of ion channels. Genome Biol. 2011;12:218. doi: 10.1186/gb-2011-12-3-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm C, et al. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint-waddler mouse. Proc Natl Acad Sci U S A. 2007;104:19583–19588. doi: 10.1073/pnas.0709846104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HJ, et al. Gain-of-function mutation in TRPML3 causes the mouse Varitint-Waddler phenotype. J Biol Chem. 2007;282:36138–36142. doi: 10.1074/jbc.C700190200. [DOI] [PubMed] [Google Scholar]
- 29.Nagata K, et al. The varitint-waddler (Va) deafness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc Natl Acad Sci U S A. 2008;105:353–358. doi: 10.1073/pnas.0707963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong XP, et al. Activating mutations of the TRPML1 channel revealed by proline-scanning mutagenesis. J Biol Chem. 2009;284:32040–32052. doi: 10.1074/jbc.M109.037184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morales-Perez CL, Noviello CM, Hibbs RE. Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure. 2016;24:797–805. doi: 10.1016/j.str.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 32.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH. Sampling the conformational space of the catalytic subunit of human gamma-secretase. Elife. 2015;4 doi: 10.7554/eLife.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonine PV, Headd JJ, Terwilliger TC, A PD. New tool: phenix.real_space_refine. Computational Crystallography Newsletter. 2013;4:43–44. [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amunts A, et al. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE : a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14376:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 43.Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.8. 2015 [Google Scholar]
- 44.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Grimm C, et al. Small molecule activators of TRPML3. Chem Biol. 2010;17:135–148. doi: 10.1016/j.chembiol.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps of the mouse TRPML1 in nanodisc have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-8883 (overall), EMD-8881 (closed I) and EMD-8882 (closed II). Atomic coordinates have been deposited in the Protein Data Bank under accession numbers 5WPV (overall structure), 5WPQ (closed I state) and 5WPT (closed II state).