Abstract
The basic helix-loop-helix (bHLH) transcription factor family is a superfamily found in all eukaryotes that plays important roles in regulating growth and development. Over the past several decades, many bHLH superfamily genes have been identified and characterized in herbaceous and woody plants. However, the genes belonging to the bHLH superfamily in peach (Prunus persica) have not yet been comprehensively identified and characterized. Here, we identified 95 members of the bHLH superfamily in the peach genome, and these genes were classified into 19 subfamilies based on a phylogenetic comparison with bHLH proteins from Arabidopsis. The members within each subfamily were highly conserved according to the analysis of motif compositions and exon/intron organizations. The 95 bHLH genes were unevenly distributed on chromosomes 1 to 8 of the peach genome. We identified 57 pairs of bHLH members that were orthologous between peach and Arabidopsis. Additionally, 48 pairs of paralogous bHLH genes were identified on the eight chromosomes of the peach genome. Coupled with relative expression analysis of bHLH genes in red-fleshed peach fruit at five developmental stages, we identified several bHLH genes that might be involved in fruit development and anthocyanin biosynthesis. This study provides insight into the molecular mechanisms through which these genes are involved in the regulation of biological and biochemical processes in peach and lays the foundation for further studies on these genes.
Introduction
Transcription factors (TFs) play key roles in various physiological and biochemical processes in different tissues at different developmental stages in plants, mainly by repressing or activating related downstream genes to regulate gene expression, thereby controlling the growth, development, and stress response of plants [1,2]. The basic helix-loop-helix (bHLH) superfamily is considered the second largest TF family and is found across eukaryotic kingdoms. Proteins of the bHLH superfamily in all eukaryotic organisms are characterized by a highly conserved bHLH domain, which is approximately 50–60 amino acids in length and is divided into two distinctive regions: the basic region and the HLH region [3,4]. The basic region, which is approximately 10–15 predominant amino acids in length and is located at the N-terminus of the domain, is the DNA-binding interface and is required for DNA binding. The HLH region, which contains two amphipathic α-helices connected by a loop of variable length, acts as dimerization domain [1]. In addition to this highly conserved bHLH domain, other conserved motifs also exist within some bHLH subfamilies of the bHLH superfamily [5,6]. Based on conserved domains and phylogenetic relationships, the bHLH superfamily is usually classified into 15–25 subfamilies in plants [6,7]. As an increasing number of genome sequences are being released, a variety of bHLH superfamily genes have been identified and analyzed in a wide range of plant species, such as peanut [8], tomato [9], apple [10], blueberry [11], Arabidopsis [12], and Chinese cabbage [13]. Furthermore, the functions of many bHLH proteins in plants have been studied in detail. The results from these studies have shown that these proteins are involved in the regulation of various networks or biological and biochemical processes throughout the plant life cycle, including Fe uptake [14], tanshinone biosynthesis [15], petal growth [16], and the response to drought and salt stress [10,17]. Most importantly, many studies have suggested that bHLH proteins are involved in the regulation of fruit development and in anthocyanin biosynthesis in both flowers and fruits [18–21].
Peach (Prunus persica) is a diploid model plant (2n = 2x = 16) of the Rosaceae family and has a relatively small genome size of 265 Mb [22]. Flesh color is an attractive and important trait for the commercial value of peaches. In general, peaches can exhibit four different flesh colors: white, green, yellow and red. Anthocyanin has been reported to contribute to red flesh color, and bHLH genes have been suggested to be involved in the biosynthesis of anthocyanin in some fruit species. Recently, whole-genome sequences of peach were added to the Genome Database for Rosaceae (GDR), providing an important foundation for the genome-wide identification of genes within a given peach family. To the best of our knowledge, genes of the peach bHLH superfamily have not been comprehensively identified and characterized to date. The aims of this study were as follows: (1) identification and characterization of peach bHLH superfamily genes; (2) subfamily classification of peach bHLH superfamily genes; (3) determination of the chromosomal distribution of peach bHLH genes and their syntenic relationships with Arabidopsis; and (4) expression profiling of bHLH genes via qRT-PCR analyses. The findings of this study will be helpful for future functional identification of specific bHLH superfamily genes involved in anthocyanin biosynthesis and fruit development in peach.
Materials and methods
Plant materials
The fruits from one-year-old fruiting shoots were selected as the experimental materials from the outer southern canopies of six-year-old ‘Zhaoxia’ (ZX, white flesh) and ‘Yejihong’ (YJH, red flesh) peach trees grown under standard field conditions at the National Peach Germplasm Repository in Nanjing, China. The fruits from the two above-mentioned cultivars were sampled 51, 64, 75, 84, and 93 days after full bloom (DAFB), immediately frozen in liquid nitrogen and stored at -80°C.
Identification of peach bHLH superfamily genes
The hidden markov model (HMM) file of the HLH domain (PF00010) was downloaded from the Pfam database (version 30.0; http://pfam.xfam.org/) [23] and was used as a query to scan the peach genome using HMMER software (version 3.1b2; http://hmmer.org/) with a default E-value. The protein sequences for the candidate genes with ID numbers identified from the HMMER results were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov). These protein sequences were further analyzed with the online CD-search tool (https://www.ncbi.nlm.nih.gov/) and InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) to verify the existence of the conserved bHLH domain. Redundant sequences were removed manually.
Motif identification and phylogenetic analysis of bHLH proteins
The online tool Multiple EM for Motif Elicitation (MEME, version 4.8.1) was used to search for conserved motifs among bHLH proteins (http://meme.nbcr.net/meme/cgi-bin/meme.cgi) by uploading the amino acid sequences of the peach bHLH superfamily proteins. The parameter settings were as follows: 0 or 1, occurrence of a single motif per sequence; 2 to 50 amino acids, motif width range; and 3, maximum number of motifs identified. All other parameters were set at the default values. Multiple sequence alignment of peach bHLH proteins (amino acid sequences) was conducted using the ClustalW program [24]. The amino acid sequences and chromosomal location of the 161 bHLH superfamily members of Arabidopsis were downloaded from The Arabidopsis Information Resource (TAIR) database (http://www.arabidopsis.org/). Phylogenetic trees of the bHLH superfamily proteins from Arabidopsis and peach were constructed with the ClustalW tool in conjunction with MEGA 4.1 software [24] using the neighbor-joining method and 1000 bootstrap replicates. Based on the subfamily classification of the bHLH proteins from Arabidopsis, the phylogenetic tree of peach and Arabidopsis constructed in this study, and the characteristics and structures of the genes and proteins, the peach bHLH superfamily was classified into subfamilies.
Analysis of the characteristics and structures of bHLH genes
The genomic sequences, ID numbers (peach v1.0 and v2.0), coding sequences (CDSs), transcript sequences, and genomic locations of peach bHLH genes were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html#). The distribution of each gene on the eight chromosomes of peach was visualized using the circlize software package in R. The start and end positions of each bHLH member on each chromosome in Arabidopsis were obtained from the TAIR database. The orthologous bHLH genes between peach and Arabidopsis as well as the paralogous genes in peach or Arabidopsis were predicted using OrthoMCL software (http://orthomcl.org/orthomcl/), and synteny figures were then drawn using the Circos tool (http://mkweb.bcgsc.ca/circos). The structural features of the peach bHLH genes, including exon and intron numbers as well as locations, were analyzed using the Gene Structure Display Server (GSDS) online tool (http://gsds.cbi.pku.edu.cn/).
Expression analysis of peach bHLH superfamily genes
The total RNA from fruits sampled from the two cultivars was extracted using the Plant RNA Kit (TaKaRa Biotechnology Co. Ltd., Dalian, China). The extracted RNA was then reverse transcribed to cDNA using the PrimeScript™ RT Reagent Kit (TaKaRa Biotechnology Co. Ltd., Dalian, China). All cDNA samples were diluted to 100 ng μl−1 with RNase-free water and stored at -20 °C before being used as templates for quantitative real-time PCR (qRT-PCR).
Gene-specific primers for qRT-PCR of the 22 bHLH genes were designed based on the CDSs of the bHLH genes using Primer Premier 5.0 software (Premier Biosoft) (S1 Table). To guarantee primer specificity, one of each pair of primers was designed to not be located within the conserved domain. RNA polymerase II (RP II, accession number TC1717) and translation elongation factor 2 (TEF2, accession number TC3544) of peach were used as the reference genes for qRT-PCR. qRT-PCR was conducted on an Applied Biosystems 7500 real-time PCR system using SYBR® Premix Ex Taq™ reagent (Tli RNaseH Plus) (TaKaRa Biotechnology Co. Ltd., Dalian, China) according to the manufacturer’s instructions. Each 20-μl reaction contained 1.0 μl of diluted cDNA, 0.4 μl of each primer, 0.4 μl of ROX, 10.0 μl of master mix, and 7.8 μl of RNase-free water. Thermal cycling conditions were set as per the manufacturer’s instructions for SYBR® Premix Ex Taq™. Each assay was repeated three times using replicate fruit samples. The relative expression levels of each gene were calculated using the 2-ΔΔCT method [25].
Results and discussion
Identification of peach bHLH superfamily genes
Based on the HMM results, a total of 95 genes belonging to the peach bHLH superfamily were identified in this study (S2 Table). To verify the reliability of each member, the 95 protein sequences were analyzed with the online CD-search tool and InterProScan, and all 95 bHLH proteins were found to exhibit a typical bHLH domain. To a certain degree, this finding is consistent with the fact that the bHLH superfamily is the second largest gene family in plants. Compared with the number of bHLH genes identified in other plant species in previous studies, fewer bHLH superfamily genes were identified in peach in this study. In previous studies, 132, 124, 188, 225, 155, 167, 146 and 159 bHLH genes were identified in peanut [8], potato [26], apple [10], wheat [27], bean [17], rice [4], carrot [28], and tomato [9], respectively. These values are probably associated with differences in evolution and genome duplication or genome sizes in these plants. The density of bHLH superfamily genes in the peach genome was approximately 0.36, which is lower than those in the genomes of Arabidopsis thaliana (1.11), Chinese cabbage (0.81), and papaya (0.65) but higher than those in the genomes of lower plants, including Volvox carteri (0.024) and Chlorella vulgaris (0.081) [13].
The ID numbers of the 95 peach bHLH genes from both peach genomes v1.0 and v2.0 are listed in S2 Table. The corresponding ID numbers and sequences for peach genome v1.0 were not found for the Prupe.6G159200.1, Prupe.7G040100.1, and Prupe.1G540400.1 genes (ID numbers of peach genome v2.0) in the Phytozome and GDR databases (S2 Table). This disparity is a result of the lower quality of the peach v1.0 genome assembly compared with that of the peach v2.0 genome assembly. The peach v1.0 assembly was improved using a large amount of community molecular mapping data, resulting in the peach v2.0 assembly [29]. Previously, unmapped sequences (7.3 Mb) of the peach v1.0 assembly were integrated within the eight peach chromosomes in the peach v2.0 assembly [29]. The lengths of the CDSs, peptide sequences, genomic sequences, and transcript sequences were recorded and are shown in S2 Table. The lengths of the peptide sequences of the 95 peach bHLH genes ranged from 123 to 729 bp, the lengths of the CDSs of the 95 peach bHLH genes ranged from 372 to 2190 bp, and the lengths of the genomic sequences of the 95 peach bHLH genes ranged from 561 to 7530 bp. The predicted functions of these bHLH genes are also listed in S2 Table.
The best homolog of each peach bHLH gene in Arabidopsis was extracted from the TAIR database (S2 Table). Some peach bHLH genes shared the highest homology with the corresponding gene in Arabidopsis; for example, both Prupe.5G144400.1 and Prupe.1G217700.1 were homologous to AT5G50915.2, with E-values of 1e-47 and 3e-26, respectively. Other peach genes that shared an Arabidopsis homolog included Prupe.6G022200.1 and Prupe.7G071500.1 as well as Prupe.8G143600.1 and Prupe.1G424100.1. We inferred that a duplication event for peach bHLH genes might have occurred during the evolution of development or resistance to stress.
Phylogenetic analysis, motifs, and multiple sequence alignment of bHLH proteins
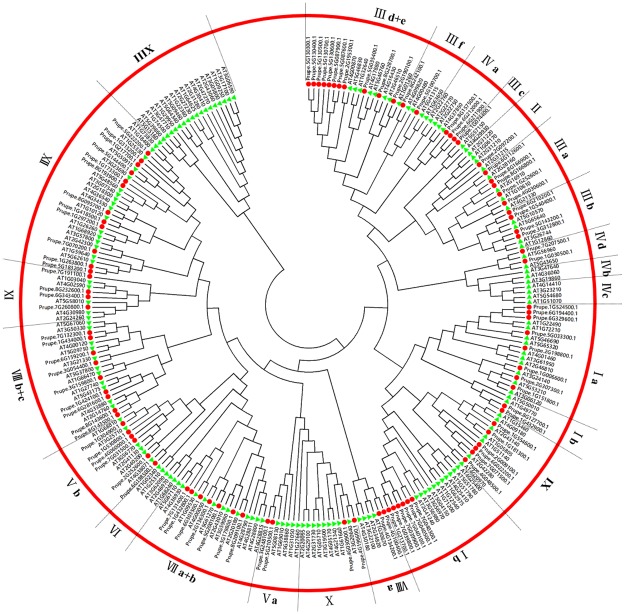
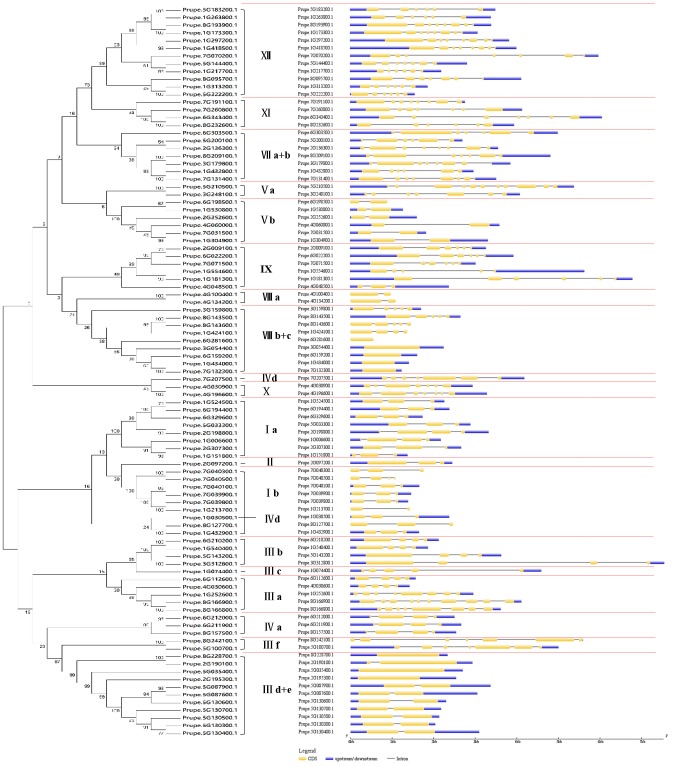
To classify the peach bHLH proteins into subfamilies and identify the evolutionary relationships between the bHLH proteins of peach and Arabidopsis, a phylogenetic tree was constructed using the sequences of the 95 peach bHLH proteins and 161 Arabidopsis bHLH proteins (Fig 1). As a result, the 95 peach bHLH members were clustered into 19 subfamilies based on the topology of the tree and the classification of the bHLH superfamily in Arabidopsis and Chinese cabbage [13]. The 19 subfamilies were designated I a, I b, II, III a, III b, III c, III d+e, III f, IV a, IV d, V a, V b, VII a+b, VIII a, VIII b+c, IX, X, XI, and XII (Fig 1). Most of these subfamilies are common and are consistent with the subfamily classification reported in previous phylogenetic analyses in other plant species, such as tomato [9], peanut [8], Chinese cabbage [13], and wheat [27]. This finding indicated that the bHLH proteins in the conserved subfamilies among plant species may play a fundamental role during plant development and evolution. Additionally, this result is consistent with the earlier finding that the bHLH superfamily in plants can be divided into 15–25 subfamilies [6]. Some previous studies have named the bHLH subfamilies using English letters [4,10], whereas most published reports have named them using Roman numerals [13,9,11,8,27,26,6], and some authors have named each subfamily using Arabic numerals [3,28]. For the peach bHLH subfamilies identified in this study, we adopted the nomenclature using Roman numerals, as employed for Chinese cabbage by Song et al. [13] and for other plants by Pires and Dolan [6]. None of the peach bHLH proteins were grouped into subfamilies IV b, IV c, VI, and XIII, which may be due to the loss of these proteins during the evolution of peach. This finding indicates that some non-conserved bHLH subfamilies among certain plant species may have specifically evolved to meet the developmental needs of the plant or for stress resistance. The number of peach bHLH proteins within each subfamily varied from 1 to 12. Most importantly, we found that some adjacent genes on the same chromosome were clustered together in the phylogenetic tree. For example, Prupe.5G130300.1, Prupe.5G130400.1, Prupe.5G130500.1, Prupe.5G130600.1, and Prupe.5G130700.1 were clustered into the branch of subfamily III d+e, and Prupe.8G166900.1 and Prupe.8G166800.1 were clustered together in subfamily III a. These results could be explained by the observation made in other plant species that members within the same clade might have recent common evolutionary origins and conserved molecular functions. The genes of a given bHLH subfamily of a plant are probably involved in the same pathway or biological process. Frequently, these proteins have overlapping functions, making them partially or totally redundant [6].
Fig 1. Phylogenetic tree constructed using the sequences of bHLH proteins from both peach and Arabidopsis.
The small red circles represent the peach bHLH proteins; the small green triangles represent the bHLH proteins from Arabidopsis. The Roman numeral outside the large red circle indicates the name of each subfamily of the peach bHLH superfamily. The black dotted line represents the initial or final boundary of each subfamily of the peach bHLH superfamily.
Some good examples of functional conservation of members within the same subfamily among different plant species have been provided. For example, AtSPCH (AT5G53210), AtMUTE (AT3G06120), and AtFMA (AT3G24140) and corresponding rice orthologs (Os053SPC1, Os055MUTE, and Os051FMA), which are members of subfamily I a, have been reported to function in controlling stomatal development [30,31]. In this study, AT5G53210, AT3G06120, and AT3G24140 and the corresponding peach orthologs (Prupe.2G307300.1, Prupe.1G151800.1, Prupe.1G006600.1) were clustered within one clade of subfamily I a (Fig 1). Based on the functional conservation of orthologs within a given subfamily, we deduced that Prupe.2G307300.1, Prupe.1G151800.1, and Prupe.1G006600.1 may also be associated with stomatal development. Three peach bHLH orthologs (Prupe.7G132300.1, Prupe.1G434000.1, and Prupe.6G159200.1) were found to cluster into the same clade of subfamily VIII b with four corresponding members from Arabidopsis, At037HEC2 (AT3G50330), At088HEC1 (AT5G67060), At040IND (AT4G00120), and At043HEC3 (AT5G09750), which have been reported to play important roles in flower and fruit development [12]. Based on this observation, we deduced that Prupe.7G132300.1, Prupe.1G434000.1, and Prupe.6G159200.1 likely coordinately regulate the development of peach flowers and fruits. In tomato, five genes (SlbHLH073, SlbHLH078, SlbHLH008, SlbHLH127, and SlbHLH069) belonging to subfamily IX have been inferred to be involved in fruit development and fruit ripening based on the expression profile [9]. Furthermore, SlbHLH09 has been previously demonstrated to play a role in fruit ripening [32]. Thus, peach bHLHs in subfamily IX might also be associated with fruit ripening. Additionally, the AtbHLHs of subfamily VIII a have been found to be functionally replaceable by the bHLHs of subfamily VIII a of Marchantia polymorpha, which have been demonstrated to function as core regulators of reproductive development [33]. Therefore, peach bHLHs in subfamily VIII a may also play key roles in the reproductive process.
In subfamily I b, we found that six peach bHLHs (Prupe.7G040300.1, Prupe.7G040500.1, Prupe.7G040100.1, Prupe.7G039900.1, Prupe.7G039800.1, Prupe.1G213700.1) were clustered together with four AtbHLHs (AtbHLH38/AT3G56970, AtbHLH39/AT3G56980, AtbHLH100/AT2G41240, AtbHLH101/AT5G04150), which are involved in iron deficiency responses and homeostasis [34]. Tomato also exhibits four SlbHLHs (SlbHLH025, SlbHLH066, SlbHLH067, SlbHLH068) that cluster correspondingly with five AtbHLHs (AtbHLH47, AtbHLH38, AtbHLH39, AtbHLH100, AtbHLH101) and are predicted to show a similar function to the five AtbHLHs in relation to iron deficiency and uptake based on their expression in roots [9]. These results imply that the six peach bHLHs (Prupe.7G040300.1, Prupe.7G040500.1, Prupe.7G040100.1, Prupe.7G039900.1, Prupe.7G039800.1, and Prupe.1G213700.1) may also function in the response to iron deficiency and uptake.
CmbHLH2, which clusters into subfamily III f, has been demonstrated to be positively correlated with the anthocyanin content of cultivars with red, pink and yellow flowers [19]. In this study on peach, Prupe.8G242100.1, the best ortholog of CmbHLH2, was also found to be located within the subfamily III f (Fig 1). Similarly, AmDEL, one of bHLH superfamily genes of Antirrhinum majus, has been reported to induce anthocyanin biosynthesis in the hairy roots of transformed tobacco and Ipomea tricolor [35]. A Blast search against the GDR database using the CDS sequence of AmDEL revealed that the best ortholog of AmDEL in peach was Prupe.5G100700.1, which is the other member of the two members of subfamily III f (Fig 1). These findings suggested that Prupe.8G242100.1 and Prupe.5G100700.1 of subfamily III f may be involved in the regulation of anthocyanin biosynthesis. Therefore, additional experiments are needed to explore and verify their functions in peach.
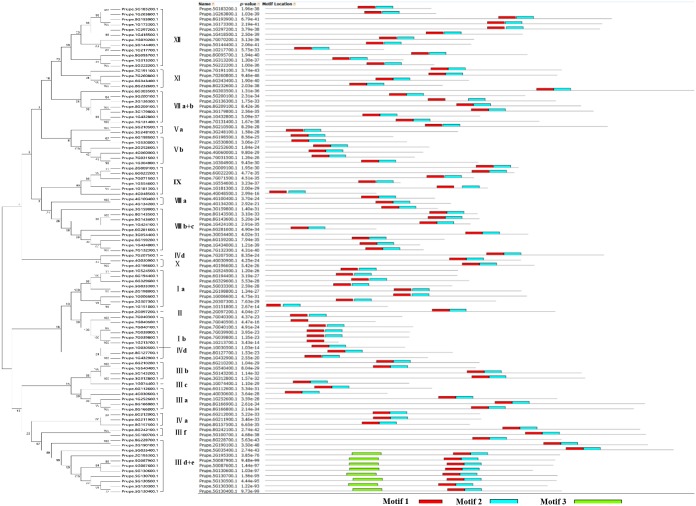
Additionally, to combine the phylogenetic tree and the features of the subfamily members, a phylogenetic tree was constructed using only the 95 sequences of the peach bHLH proteins (Fig 2). Although the ordering of the subfamilies in the phylogenetic tree is slightly different from that in the phylogenetic tree circled above (Fig 1), the member composition of each subfamily is the same as in the phylogenetic tree of the peach bHLH proteins circled above. The online MEME tool (version 4.8.1) was used to search for conserved motifs shared by bHLH proteins by uploading the 95 amino acid sequences of the peach bHLH superfamily [36]. Almost all the sequences exhibited two highly conserved motifs with the same width (29 amino acids), which are shown in the red and blue blocks, respectively (Fig 2 and S3 Table). Most of the motifs are located near the C-terminus of the peach bHLH proteins, such as the peach bHLH proteins in subfamilies III d+e, III a, III f, and IV a. Some motifs are located near the N-terminus, such as in the peach bHLH protein subfamily V a. The two motifs are very close to each other in 63 of the peach bHLH proteins but are slightly farther apart in a few of the peach proteins. Among the 95 peach bHLH proteins, the distance between the two motifs is widest in Prupe.2G236300.1. Based on a review of the literature regarding the bHLH domains of wheat [27] and tomato [9], in the present study, motif 1 (red block) was found to be composed of basic residues and helix 1, and motif 2 (blue block) was found to be composed of a loop and helix 2. In some bHLH proteins, the space between motifs 1 and 2 consists of a loop of variable length. Motif 1 (in red) and motif 2 (in blue) are composed of logos 1 and 2, respectively (Figs 2 and 3, S3 Table). The backbones of motifs 1 and 2 are also conserved at the amino acid level in the majority of plant species reported to date [9,27,37]. It has been deduced that the highly conserved residues in bHLH domains are responsible for dimerization [37].
Fig 2. Motif distribution of peach bHLH superfamily proteins.
The motifs of the bHLH superfamily proteins were analyzed using the MEME web server. The red, blue, and green blocks represent motifs 1, 2 and 3, respectively. The length of the gray line indicates the length of a sequence relative to all the other sequences. The position of each block indicates the location of a motif with a matching sequence. The phylogenetic tree on the right was constructed using only the sequences of bHLH proteins from peach.
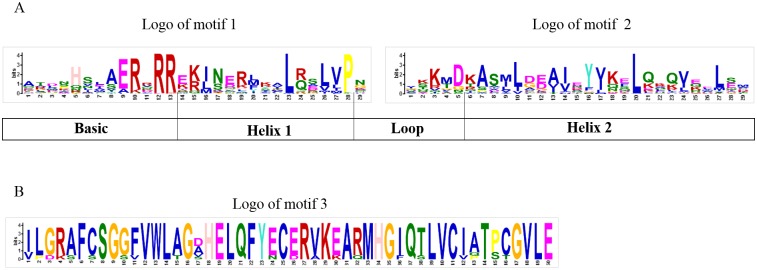
Fig 3. Motif composition and logos of peach bHLH proteins.
A. The logos of motifs 1 and 2, which together constitute the bHLH domain in peach. The overall height of each stack represents the conservation of the sequence at that position. The capital letters indicate over 50% conservation of amino acids among the bHLH domains. The Arabic numerals under the colored capital letters indicate the width of the motif. Each color of the English letters represents a type of amino acid residue. B. The logo of motif 3, which constitutes another conserved motif, is present only in eight members of subfamily III e.
In addition to these two common conserved motifs shared by the 95 peach bHLH proteins, the eight peach bHLH proteins (from Prupe.5G130300.1 to Prupe.2G195300.1) of subfamily III e from the phylogenetic tree harbor another highly conserved motif with a length of 50 amino acids. This motif is indicated by the green block and is composed of logo 3 (Fig 3 and S3 Table). This result is consistent with previous reports that members of a given subfamily of the bHLH superfamily exhibit another conserved motif (conserved non-bHLH motif) in addition to the common bHLH motif of plants [6]. In contrast, the existence of a conserved non-bHLH motif in addition to the bHLH domain strongly illustrates the correction of the subfamily classification based on the phylogenetic tree generated in this study. The conservation of motif 3 during peach evolution suggests that this motif must be essential for the function of the eight bHLH proteins in this subfamily. In addition, among the 95 bHLH proteins, Prupe.7G040500.1 and Prupe.1G213700.1 exhibited incomplete bHLH domains, whereas the remaining 93 bHLH proteins all presented complete bHLH domains. Similar observations have been made in other plant species, such as blueberry [11].
Additionally, to analyze the sequence features of the bHLH domains at the amino acid level, we performed multiple sequence alignment using the sequences of the 95 bHLH proteins (Fig 4). There were two conserved regions in the sequences of the bHLH domain: the basic region plus helix 1 and the loop region plus helix 2. The results of multiple sequence alignment also confirmed the correction of the result obtained using the MEME tool described above. For example, a long amino acid sequence was found between the two highly conserved regions (Fig 4), which is consistent with the long distance between the two motifs in Prupe.2G136300.1, as shown in Fig 2. Additionally, the empty spaces in the second conserved region due to the absence of certain amino acids in the protein sequences of Prupe.7G040500.1 and Prupe.1G213700.1 (Fig 4) is consistent with the fact that Prupe.7G040500.1 and Prupe.1G213700.1 do not contain motif 2, as shown in Fig 2. Generally, the sequences of the basic region and the two helix regions were more conserved than the sequence of the loop region. These results obtained using MEME and sequence alignment tools further confirm that the results obtained from the HMM are highly reliable. These results also illustrate that the HMM is able to make highly accurate predictions for a given superfamily.
Fig 4. Multiple sequence alignment of the peach bHLH proteins.
Amino acids with more than 50% identity are labeled with colored boxes. The ID order of 95 peach bHLH proteins is the same as in Fig 3.
Characteristics and structures of bHLH genes
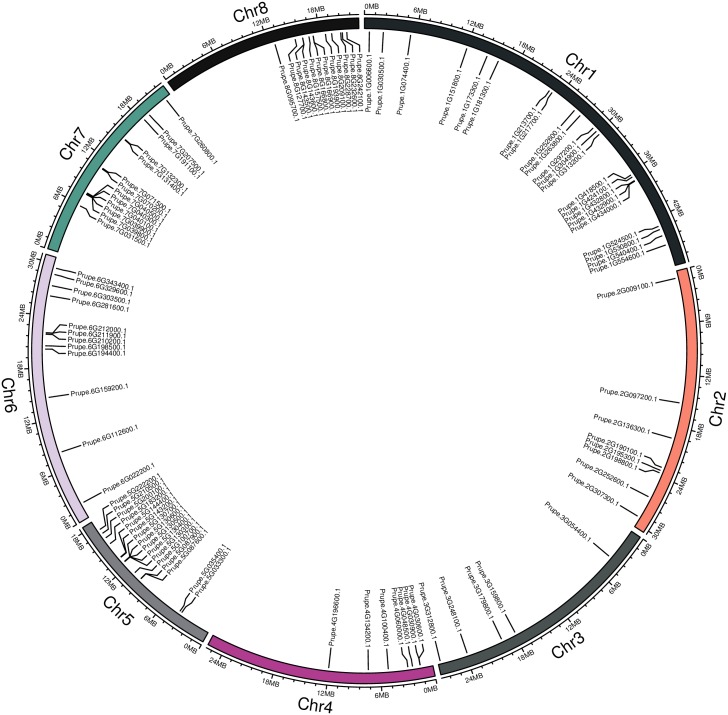
Peach has eight chromosomes. Based on the data (S2 Table) regarding the distribution of peach bHLH genes on the chromosomes and the length of each chromosome obtained from the database, the chromosomal location of each bHLH gene was mapped to each chromosome using software to display the position intuitively. The 95 bHLH genes were found to be unevenly distributed on chromosomes 1 to 8 of the peach genome. The numbers of bHLH genes distributed on chromosomes 1 to 8 were 22,8,5,7,16,12,13, and 12, respectively (S2 Table and Fig 5). Chromosome 1 is the longest among the eight chromosomes in peach and exhibited the greatest number of bHLH genes. The 22 and 12 bHLH genes on chromosomes 1 and 6, respectively, were relatively well distributed. In contrast, the 12 and seven bHLH genes on chromosomes 8 and 4, respectively, were clustered near one of the two termini. Prupe.5G130300.1, Prupe.5G130400.1, Prupe.5G130500.1, Prupe.5G130600.1, and Prupe.5G130700.1 were not only clustered together, as shown in Fig 1, but were also located next to each other on chromosome 5 (Fig 5).
Fig 5. Distribution of bHLH superfamily genes on the eight chromosomes of peach.
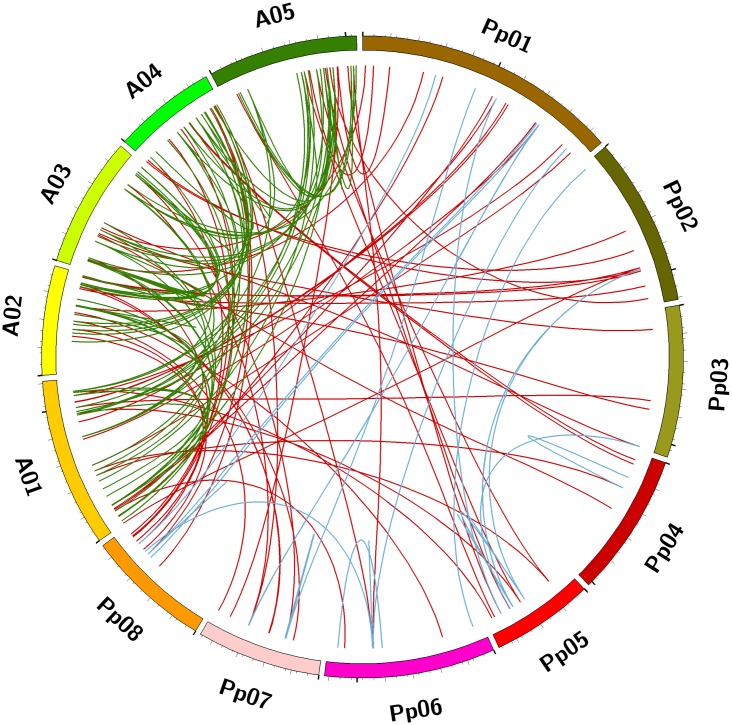
To further intuitively display and illustrate the genetic divergence and gene duplication within the bHLH superfamily, homology comparisons and syntenic relationships were analyzed to identify orthologous bHLH genes between peach and Arabidopsis as well as paralogous bHLH genes in peach or Arabidopsis using the OrthoMcl and Circos tools. As a result, 57 pairs of orthologous bHLH genes between peach and Arabidopsis were identified among the 95 bHLH genes of peach and 161 bHLH genes of Arabidopsis (Fig 6 (red line) and S4 Table). Among the orthologous gene pairs between peach and Arabidopsis, we found that each Arabidopsis bHLH gene shared only one orthologous gene with peach. This observation is not consistent with the findings reported in cabbage, where each Arabidopsis bHLH gene presents one to three orthologous genes in Chinese cabbage [13]. Nine of the 13 bHLH genes on chromosome 1 of peach were orthologous to nine bHLH genes on chromosome 1 of Arabidopsis, and the remaining four peach bHLH genes were orthologous to two bHLH genes on each of chromosomes 3 and 5 of Arabidopsis (A03 and A05). Four of the eight bHLH genes on chromosome 2 of peach were orthologous to four bHLH genes on chromosome 2 of Arabidopsis, and the remaining four peach bHLH genes were orthologous to four bHLH genes on chromosomes 1, 4, and 5 of Arabidopsis. Among the bHLH orthologous genes on chromosome 3 of peach, 50% exhibited orthologs on chromosome 3 of Arabidopsis. The same situation was observed for peach chromosome 4. Five of the eight bHLH genes on chromosome 5 of peach were orthologous to bHLH genes on chromosome 5 of Arabidopsis, and the remaining three were orthologous to bHLH genes on chromosome 1 of Arabidopsis (Fig 6 (red line) and S4 Table). Four orthologous bHLH genes on chromosome 6 of peach were orthologous to corresponding bHLH genes on chromosome 1, 2, 4, and 5 of Arabidopsis. Half of the orthologous bHLH genes on chromosome 7 of peach were orthologous to bHLH genes on chromosome 2 of Arabidopsis. Five of the eight bHLH genes on chromosome 8 of peach were orthologous to five bHLH genes on chromosome 4 of Arabidopsis. These results indicated that the bHLH genes on chromosome 1 of peach are more homologous to bHLH genes on the corresponding chromosome 1 of Arabidopsis than genes on the other Arabidopsis chromosomes. The same tendency was observed for the bHLH genes on chromosomes 2 to 5 of peach. These findings indicated that orthologous genes are evolutionarily conserved in land plants. Most orthologous genes of one plant species might evolve and originate from the corresponding chromosome of the ancestor plant. Additionally, bHLH genes on chromosomes 7 and 8 of peach shared higher homology with bHLH genes on chromosomes 2 and 4 of Arabidopsis, respectively, than genes on other Arabidopsis chromosomes. However, the orthologous bHLH genes on chromosome 6 of peach did not exhibit this characteristic but rather presented orthologs distributed on four of the five chromosomes of Arabidopsis. Hence, the mechanism underlying the evolution of these syntenic pairs on different or corresponding chromosomes between Arabidopsis and peach remains unclear. These genes may be subjected to several types of evolutionary selection. Because integration of the multiple aspects of gene evolution is very complex, it is impossible to summarize based on a single factor.
Fig 6. Syntenic relationships of bHLH superfamily genes in peach and Arabidopsis.
Eight chromosomes of peach (Pp01–Pp08) and five chromosomes of Arabidopsis (A01–A05) are mapped in different colors. The red lines connect the orthologous bHLH genes between peach and Arabidopsis. The blue and green lines connect the paralogous bHLH genes in peach and Arabidopsis, respectively.
In addition, 48 pairs of paralogous bHLH genes were identified on eight chromosomes of peach (linked by blue lines in Fig 6; S5 Table). Each gene exhibited one to seven paralogous bHLH genes in peach; for example, Prupe.5G087600.1 and Prupe.2G195300.1 presented six and seven paralogous bHLH genes, respectively. In fact, the 48 pairs of paralogous bHLH genes in peach were found to be composed of 40 peach bHLH genes. Regarding paralogous bHLH genes in peach (blue line in Fig 6), no paralogous bHLH genes were found to exist on the two adjacent peach chromosomes. In contrast, we observed that among the 161 bHLH genes of Arabidopsis, 81 pairs of paralogous bHLH genes were present on five chromosomes (shown with green lines in Fig 6; S5 Table). Similar to peach, each gene exhibited one to six paralogous bHLH genes in Arabidopsis; for example, AT5G54680 and AT4G25410 presented six and five paralogous bHLH genes, respectively. This feature has also been observed in other studies [13,27]. Similar to peach, the 81 pairs of paralogous bHLH genes in Arabidopsis are composed of 88 Arabidopsis bHLH genes. Consistent with a previous study in Brachypodium distachyon [2], most of the paralogous bHLH genes in peach were grouped within the same subfamily as the original genes. For example, the seven bHLH genes paralogous to Prupe.2G195300.1 were clustered in subfamily III d+e (Fig 1 and S2 Table). Based on these results, we deduced that the genes of the bHLH superfamily likely shared a common origin and gradually expanded under intense selective pressure approximately 4–14 Mya, leading to the functional conservation and divergence of the bHLH superfamily observed in peach.
The schematic structure of peach bHLH genes was analyzed using the GSDS tool (Fig 7). Prupe.5G130400.1, Prupe.5G130300.1, Prupe.5G130500.1, Prupe.5G130700.1, Prupe.5G130600.1, Prupe.5G087600.1, and Prupe.5G087900.1 of subfamily III d+e were observed to exhibit two exons and one intron and to be clustered within a single branch of the phylogenetic tree (Fig 1). Prupe.6G212000.1, Prupe.6G211900.1, and Prupe.8G157500.1 of subfamily IV exhibited four exons and three introns and were also clustered within a single branch of the phylogenetic tree (Fig 1). Prupe.6G210200.1, Prupe.1G540400.1, Prupe.5G143200.1, and Prupe.3G312800.1 of subfamily III b presented four exons and three introns and were clustered within a single clade in the phylogenetic tree (Fig 1). Members of subfamily I also exhibited three exons and two introns. The length and location of each exon in the genome sequence of these four genes were also highly conserved among the four genes (Fig 7). The members of subfamilies III a, V a, VII a+b, IV d, IX, X, XI, and XII presented relatively greater numbers of exons than the other subfamilies in this study. Overall, the numbers, lengths, and positions of the exons were relatively well conserved in each subfamily of peach bHLH genes. This finding is consistent with previous reports about both bHLH genes in other plant species [2] and other TF families [38,39]. Exon/intron organization is considered to play an important role in the evolution of the genes of a given family [40].
Fig 7. Locations and lengths of the exons and introns of peach bHLH superfamily genes.
Exons and introns are presented as filled yellow sticks and thin gray single lines, respectively. UTRs are represented by dark blue bars at the ends. The red dotted line separates each subfamily and clearly presents the member conservation of each subfamily. The phylogenetic tree on the right was constructed using only the sequences of bHLH proteins in peach.
Expression analysis of peach bHLH superfamily genes
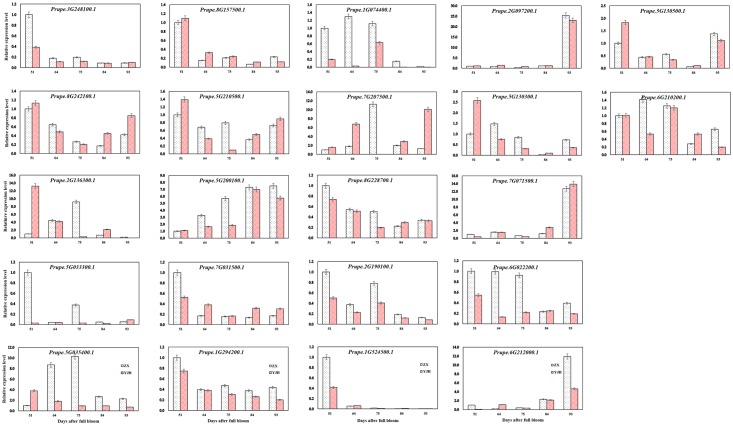
To assess the potential regulatory role of bHLH superfamily genes in peach fruits, the expression levels of 22 bHLH superfamily genes in the white-fleshed fruit of ZX and red-fleshed fruit of YJH sampled 51, 64, 75, 84, and 93 DAFB were analyzed in this study (Fig 8). The expression of Prupe.1G524500.1 in the fruits of ZX and YJH decreased significantly, by approximately 1- and 0.4-fold compared with the initial level, and was almost undetectable at 93 DAFB in the fruits of both cultivars. At 51 DAFB, the expression level of Prupe.1G524500.1 in the fruit of ZX was lower than that in the fruit of the YJH cultivar. The expression of Prupe.1G294200.1 in the fruit of YJH decreased gradually with development from young to harvested fruit. In contrast, in the ZX cultivar, no significant difference in the expression level of Prupe.1G294200.1 was observed from 64 to 93 DAFB. However, from 51 to 64 DAFB, there was a sharp decrease in the expression level of Prupe.1G294200.1. A similar expression pattern to that found for Prupe.1G294200.1 was observed for Prupe.8G228700.1 in both cultivars. The expression of Prupe.5G035400.1 in the fruit of YJH decreased gradually as the fruit matured, whereas the expression level of Prupe.5G035400.1 in the fruit of ZX increased from 51 to 75 DAFB and then decreased until harvest. A similar expression pattern to that of Prupe.5G035400.1 was observed for Prupe.2G136300.1 in both cultivars. A sharp decrease in the expression of Prupe.7G031500.1 from 51 to 64 DAFB followed by relatively stable expression from 64 to 93 DAFB was observed in the ZX cultivar. The expression level of Prupe.3G248100.1 decreased gradually from 51 to 93 DAFB in both cultivars. The expression level of Prupe.3G248100.1 in ZX was relatively higher than that in YJH from 51 to 75 DAFB. The expression level of Prupe.3G248100.1 was almost equal in ZX and YJH from 84 to 93 DAFB. A trend consisting of a gradual decrease in expression followed by a gradual increase was observed for Prupe.8G242100.1 in both cultivars. Prupe.5G210500.1 exhibited a similar expression pattern to Prupe.5G033300.1, except at 75 DAFB in ZX. Prupe.5G033300.1 presented very low and relatively stable expression throughout the development of YJH, whereas in the young fruit of ZX, Prupe.5G033300.1 exhibited relatively high levels of expression. The expression of Prupe.5G200100.1 increased gradually with fruit development from 51 to 93 DAFB in both cultivars, except at 93 DAFB in YJH. Overall, the expression level of Prupe.5G200100.1 in ZX was higher than that in YJH, and the expression level of Prupe.5G200100.1 remained stable in ZX from 84 to 93 DAFB. Surprisingly, the variable expression trend of Prupe.2G190100.1 was the same in both cultivars, and the expression level of Prupe.2G190100.1 in ZX was always higher than that in YJH. Prupe.5G033300.1 and Prupe.1G524500.1, both of which belong to subfamily I a, exhibited relatively similar expression patterns in this study. This finding illustrates that members of the same bHLH subfamily might have redundant functions, although this hypothesis requires further verification. These proteins probably participate in the same network to regulate certain biological or biochemical processes simultaneously. Prupe.7G071500.1, Prupe.2G097200.1, and Prupe.6G212000.1 presented relatively low expression levels from 51 to 84 DAFB but displayed high expression levels in the harvested mature fruits of both cultivars.
Fig 8. Relative expression levels of selected bHLH genes in fruits of the two peach cultivars.
Error bars represent standard errors from three independent replicates.
The different expression patterns involving gradual increases and decreases observed in this study illustrate that some bHLH superfamily genes may be associated with fruit development. This finding was consistent with previous findings that bHLH proteins play a regulatory role in fruit ripening in peach and other fruit species [41]. Additionally, the findings of this study are in agreement with transcriptomic results [42], which revealed that the transition from developmental stage 3 to stage 4 is accompanied by up-regulated expression of some bHLH genes in peach [36]. Similarly, it has been previously reported that bHLH3 together with MYB and WD40 can form a regulatory complex to regulate flavonoid biosynthesis by activating the transcription of downstream genes in nectarine [43]. Similar results have been observed for anthocyanin accumulation in apple [44] and peach [45] fruits, where MYB TFs use bHLH proteins as a partner to activate the transcription of anthocyanin pathway genes. Montefiori et al. [46] reported that bHLH TFs are involved in the regulation of the flavonoid pathway in a variety of species. Liu et al. showed that significantly increased anthocyanin accumulation in peels is accompanied by enhanced expression of bHLH3 [47]. In rice, it has been reported that bHLH proteins are involved in controlling grain length and weight [48]. The results of the present study indicated that Prupe.8G242100.1, Prupe.7G031500.1, and Prupe.7G207500.1 may be associated with anthocyanin biosynthesis, which needs to be further confirmed experimentally. Gene expression is one of the vital regulatory mechanisms utilized by plant cells to carry out their functions. During fruit development and ripening, fleshy fruits undergo a series of complex biochemical reactions, in addition to physiological and dynamic changes. These internal and environmental factors collectively affect the phenotypes of fruits, including their acidity or sweetness, coloration, hardness or softness, flavor, and aroma, increasing the nutritional value and attractiveness of the fruits [36].
Conclusions
This study constitutes the first comprehensive and systematic analysis of bHLH superfamily genes in peach based on the whole-genome sequence. As a result, a total of 95 bHLH superfamily genes were identified from the peach genome and were clustered into 19 subfamilies. The characterization of the genes and proteins, including their motif compositions and exon/intron organizations, indicated that the members within each subfamily are highly conserved. Additionally, some common bHLH subfamilies that exist in other plant species were also identified in peach, indicating that the highly conserved bHLH subfamilies among plant species may play fundamental roles in the growth and development of these plant species. The 95 bHLH genes of peach were found to be unevenly distributed on chromosomes 1 to 8 of the peach genome. Among these genes, there are 48 pairs of paralogous bHLH genes on the eight chromosomes of the peach genome. Additionally, 57 pairs of bHLH members were found to be orthologous between peach and Arabidopsis. The identification of peach bHLH genes homologous to Arabidopsis genes enriches the genome annotation and facilitates the prediction of the functions of bHLH superfamily genes in peach. The expression patterns observed in both white-fleshed and red-fleshed fruits in this study provide preliminary data for further analysis of bHLH genes associated with fruit development. This study lays the foundation for further functional verification of bHLH superfamily genes in peach and enriches the knowledge of bHLH superfamily genes in plant species.
Supporting information
(XLSX)
(XLSX)
(XLSX)
Each pair of orthologous bHLH genes between peach and Arabidopsis is listed on the same line.
(XLSX)
Each pair of paralogous bHLH genes in peach or Arabidopsis is listed on the same line. The order number is used for displaying repeated gene IDs.
(XLSX)
Acknowledgments
The authors wish to thank the National Natural Science Foundation of China (No. 31471848 and No. 31401851) and the earmarked fund of the China Agriculture Research System (No. CARS-30) for providing financial support.
Abbreviations
- bHLH
basic helix-loop-helix
- CDSs
coding sequences
- DAFB
days after full bloom
- GDR
Genome Database for Rosaceae
- GSDS
Gene Structure Display Server
- HMM
hidden markov model
- MEME
Multiple EM for Motif Elicitation
- ORF
the open reading frame
- qRT-PCR
quantitative real-time polymerase chain reaction
- RP II
RNA polymerase II
- TAIR
The Arabidopsis Information Resource
- TEF2
translation elongation factor 2
- TFs
transcription factors
- YJH
Yejihong
- ZX
Zhaoxia
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Natural Science Foundation of China (No. 31471848), https://isisn.nsfc.gov.cn/egrantweb/, ML Yu received the funding; National Natural Science Foundation of China (No. 31401851), https://isisn.nsfc.gov.cn/egrantweb/, CH Zhang received the funding; and China Agriculture Research System (No. CARS-30), http://123.127.160.231/, ML Yu received the funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000; 20:429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niu X, Guan YX, Chen SK, Li HF. Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in Brachypodium distachyon. BMC Genomics. 2017; 18:619–639. doi: 10.1186/s12864-017-4044-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. The Plant Cell. 2003; 15:1749–1770. doi: 10.1105/tpc.013839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Duan XP, Jiang HX, Sun YJ, Tang YP, Yuan Z, et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiology. 2006; 141:1167–1184. doi: 10.1104/pp.106.080580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal. 2011; 66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x [DOI] [PubMed] [Google Scholar]
- 6.Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010; 27(4):862–874. doi: 10.1093/molbev/msp288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck MJ, Atchley WR. Phylogenetic analysis of plant basic helix-loop-helix proteins. Journal of Molecular Evolution. 2003; 56(6):742–750. doi: 10.1007/s00239-002-2449-3 [DOI] [PubMed] [Google Scholar]
- 8.Gao C, Sun J, Wang C, Dong Y, Xiao S, Wang X, et al. Genome-wide analysis of basic/helix-loop-helix gene family in peanut and assessment of its roles in pod development. PLoS ONE. 2017; 12 (7): e0181843 doi: 10.1371/journal.pone.0181843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun H, Fan HJ, Ling HQ. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics. 2015; 16:9–21. doi: 10.1186/s12864-014-1209-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao K, Dong Q, Li C, Liu C, Ma F. Genome wide identification and characterization of apple bHLH transcription factors and expression analysis in response to drought and salt stress. Front Plant Sci. 2017; 8:480–495. doi: 10.3389/fpls.2017.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Y, Liu HD, Zhou Q, Zhang HJ, Zhang ZD, Li YD, et al. High-throughput sequencing of highbush blueberry transcriptome and analysis of basic helix-loop-helix transcription factors. Journal of Integrative Agriculture. 2017; 16 (3):591–604. [Google Scholar]
- 12.Carretero-Paulet L, Galstyan A, Roig-Villanova I, Martinez-Garcia JF, Bilbao-Castro JR, Robertson DL. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010; 153:1398–1412. doi: 10.1104/pp.110.153593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song XM, Huang ZN, Duan WK, Ren J, Liu TK, Li Y, et al. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol Genet Genomics. 2014; 289:77–91. doi: 10.1007/s00438-013-0791-3 [DOI] [PubMed] [Google Scholar]
- 14.Ogo Y, Itai RN, Nakanishi H, Kobayashi T, Takahashi M, Mori S, et al. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe deficient conditions. Plant J. 2007; 51:366–377. doi: 10.1111/j.1365-313X.2007.03149.x [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Luo H, Xu Z, Zhu Y, Ji A, Song J, et al. Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci Rep. 2015; 5:11244–11254. doi: 10.1038/srep11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szecsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, et al. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J. 2006; 25:3912–3920. doi: 10.1038/sj.emboj.7601270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kavas M, Baloğlu MC, Atabay ES, Ziplar UT, Daşgan HY, Ünver T. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Molecular Genetics and Genomics. 2016; 291:129–143. doi: 10.1007/s00438-015-1095-6 [DOI] [PubMed] [Google Scholar]
- 18.Nakatsuka T, Haruta KS, Pitaksutheepong C, Abe Y, Kakizaki Y, Yamamoto K, et al. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 2008; 49:1818–1829. doi: 10.1093/pcp/pcn163 [DOI] [PubMed] [Google Scholar]
- 19.Xiang LL, Liu XF, Li X, Yin XR, Grierson D, Li F, et al. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS ONE. 2015; 10 (11):e0143892 doi: 10.1371/journal.pone.0143892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Ren LP, Fan XY, Tang KJ, Li B. Identification of putative flavonoid-biosynthetic genes through transcriptome analysis of Taihe Toona sinensis bud. Acta Physiol Plant. 2017; 39:122–135. [Google Scholar]
- 21.Xu HF, Wang N, Liu JX, Qu CZ, Wang YC, Jiang SH, et al. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol Biol. 2017; 94:149–165. doi: 10.1007/s11103-017-0601-0 [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, Xia R, Zhao B, An YQ, Dardick CD, Callahan AM, et al. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC plant biology. 2012; 12:149–167. doi: 10.1186/1471-2229-12-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2016; 44:279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28:2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001; 25: 402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 26.Wang RQ, Zhao P, Kong NN, Lu RZ, Pei Y, Huang CX, et al. Genome-wide identification and characterization of the potato bHLH transcription factor family. Genes. 2018; 9:54–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo XJ, Wang JR. Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Biology. 2017; 17:90–102. doi: 10.1186/s12870-017-1038-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen YY, Li MY, Wu XJ, Huang Y, Ma J, Xiong AS. Genome-wide analysis of basic helix-loop-helix family transcription factors and their role in responses to abiotic stress in carrot. Mol Breeding. 2015; 35:125–137. [Google Scholar]
- 29.Verde I, Jenkins J, Dondini L, Micali S, Pagliarani G, Vendramin E, et al. The peach v2.0 release: high-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics. 2017; 18:225–243. doi: 10.1186/s12864-017-3606-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Ohashi-Ito K, Bergmann DC. Orthologs of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development. 2009; 136: 2265–2276. doi: 10.1242/dev.032938 [DOI] [PubMed] [Google Scholar]
- 31.Pillitteri LJ, Sloan DB, Bogenschutz NL, Torii KU. Termination of asymmetric cell division and differentiation of stomata. Nature. 2007; 445: 501–505. doi: 10.1038/nature05467 [DOI] [PubMed] [Google Scholar]
- 32.Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013; 25:371–386. doi: 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamaoka S, Nishihama R, Yoshitake Y, Ishida S, Inoue K, Saito M, et al. Generative cell specification requires transcription factors evolutionarily conserved in land plants. Current Biology. 2018; 28:479–486. doi: 10.1016/j.cub.2017.12.053 [DOI] [PubMed] [Google Scholar]
- 34.Yuan YX, Zhang J, Wang DW, Ling HQ. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 2005; 15:613–621. doi: 10.1038/sj.cr.7290331 [DOI] [PubMed] [Google Scholar]
- 35.Hou XJ, Li JM, Liu BL, Wei L. Co-expression of basic helix–loop–helix protein (bHLH) and transcriptional activator-Myb genes induced anthocyanin biosynthesis in hairy root culture of Nicotiana tabacum L and Ipomea tricolor. Acta Physiol Plant. 2017; 39:59–66. [Google Scholar]
- 36.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology.1994; 28–36. [PubMed]
- 37.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003; 20:735–747. doi: 10.1093/molbev/msg088 [DOI] [PubMed] [Google Scholar]
- 38.Hajiebrahimi A, Owji H, Hemmati S. Genome-wide identification, functional prediction, and evolutionary analysis of the R2R3-MYB superfamily in Brassica napus. Genome. 2017; 60:797–814. doi: 10.1139/gen-2017-0059 [DOI] [PubMed] [Google Scholar]
- 39.Zhang CH, Shangguan LF, Ma RJ, Sun X, Tao R, Guo L, et al. Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica). Genet Mol Res. 2012; 11(4):4789–4809. doi: 10.4238/2012.October.17.6 [DOI] [PubMed] [Google Scholar]
- 40.Xu G, Guo C, Shan H, Kong H. Divergence of duplicate genes in exon-intron structure. Proc Natl Acad Sci U S A. 2012; 109 (4):1187–1192. doi: 10.1073/pnas.1109047109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziliotto F, Begheldo M, Rasori A, Bonghi C, Ramina A, Tonutti P. Molecular and genetic aspects of ripening and qualitative traits in peach and nectarine fruits. Acta Hort. 2005; 682:237–246. [Google Scholar]
- 42.Trainotti L, Bonghi C, Ziliotto F, Zanin D, Rasori A, Casadoro G, et al. The use of microarray mPEACH1.0 to investigate transcriptome changes during transition from pre-climacteric to climacteric phase in peach fruit. Plant Science. 2006; 170:606–613. [Google Scholar]
- 43.Ravaglia D, Espley RV, Henry-Kirk RA, Andreotti C, Ziosi V, Hellens RP, et al. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biology. 2013; 13:68–82. doi: 10.1186/1471-2229-13-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL. Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc Natl Acad Sci U S A. 2000; 97:13579–13584. doi: 10.1073/pnas.250379897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou H, Lin-Wang K, Liao L, Gu C, Lu ZQ, Allan AC, et al. Peach MYB7 activates transcription of the proanthocyanidin pathway gene encoding leucoanthocyanidin reductase, but not anthocyanidin reductase. Frontiers in Plant Science. 2015; 6:908–921. doi: 10.3389/fpls.2015.00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montefiori M, Brendolise C, Dare AP, Lin-Wang K, Davies KM, Hellens RP, et al. In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. Journal of Experimental Botany. 2015; 66:1427–1436. doi: 10.1093/jxb/eru494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu T, Song S, Yuan YB, Wu DJ, Chen MJ, Sun QA, et al. Improved peach peel color development by fruit bagging. Enhanced expression of anthocyanin biosynthetic and regulatory genes using white non-woven polypropylene as replacement for yellow paper. Scientia Horticulturae. 2015; 184:142–148. [Google Scholar]
- 48.Heang D, Sassa H. An atypical bHLH protein encoded by positive regulator of grain length 2 is involved in controlling grain length and weight of rice through interaction with a typical bHLH protein APG. Breeding Science. 2012; 62:133–141. doi: 10.1270/jsbbs.62.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
Each pair of orthologous bHLH genes between peach and Arabidopsis is listed on the same line.
(XLSX)
Each pair of paralogous bHLH genes in peach or Arabidopsis is listed on the same line. The order number is used for displaying repeated gene IDs.
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.