Summary
Genome editing in human pluripotent stem cells (hPSCs) enables the generation of reporter lines and knockout cell lines. Zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and CRISPR/Cas9 technology has recently increased the efficiency of proper gene editing by creating double strand breaks (DSB) at defined sequences in the human genome. These systems typically use plasmids to transiently transcribe nucleases within the cell. Here, we describe the process for preparing hPSCs for transient expression of nucleases via electroporation and subsequent analysis to create genetically-modified stem cell lines.
Keywords: genome editing, genomics, CRISPR/Cas9, electroporation
Introduction
Nuclease based gene-editing relies on endogenous DNA damage repair processes to edit desired sequences in the genome. Double strand breaks (DSBs) generated by nucleases are typically repaired via one of two processes: homology-directed recombination (HDR) and non-homologous end joining (NHEJ) [1]. HDR using donor DNA templates or single-stranded oligos (ssODNs) results in error-free incorporation of foreign DNA sequences. NHEJ, in contrast, generates a variety of insertions and deletions at the DSB site, which in some cases results in frameshifts causing premature stop codons to generate knockout cell lines.
Currently, the most common method to generate gene-edited hPSCs uses a clustered regularly interspaced palindromic repeat (CRISPR)/CRISPR associated protein (Cas9) system [2]. Engineering of the type II bacterial CRISPR associated (Cas) system into human cells [2] has dramatically increased the rate of which genetic engineering has progressed. Cas9 is an RNA-guided endonuclease that uses short guide RNAs (gRNAs) to selectively create double strand breaks (DSBs) at any point in the genome that is flanked by a 3’ protospacer adjacent motif (PAM). When DSBs are created in cells, endogenous repair mechanisms are activated, resulting in repair from a donor via HDR or NHEJ. This method of gene editing was previously shown using zinc-finger nucleases [3] and TALENs [4] in hPSCs. HDR mediated insertion of transgenes occurs naturally in hPSCs [5] but occurs at a much higher rate when DSBs are introduced. Transgenes can be stably inserted into the genome by HDR if a donor is supplied that contains the gene surrounded by regions homologous to the area where it should be inserted. Single nucleotide polymorphisms (SNP) can also be introduced by transfecting with a ssODN that is homologous to the region around the cut site but for the point mutation of interest [3] [6].
There are currently three methods in which the necessary components can be introduced into the cell: (i) transient transfection of two plasmids, one containing the coding sequence for Cas9 and the other expressing the gRNA [2] [6], (ii) viral integration of the Cas9 coding sequence and transient expression of the gRNA [7] and, (iii) viral integration of both Cas9 and gRNA sequences [8] [9] [10]. Delivery of nucleic acids to hPSCs is typically limited using standard lipofection reagents, so most studies utilize electroporation or nucleofection [11]. Efficiency of the two plasmid system has been increased by utilizing a Cas9-2A-GFP plasmid [12] which can be used to enrich the cell population expressing Cas9 via fluorescence-activated cell sorting (FACS) of GFP-expressing cells. Using these strategies, we have been able to insert transgenes (Fig. 1) [13] and detect frame-shifting indels causing a knockout phenotype in over 25% of cultured hPSCs.
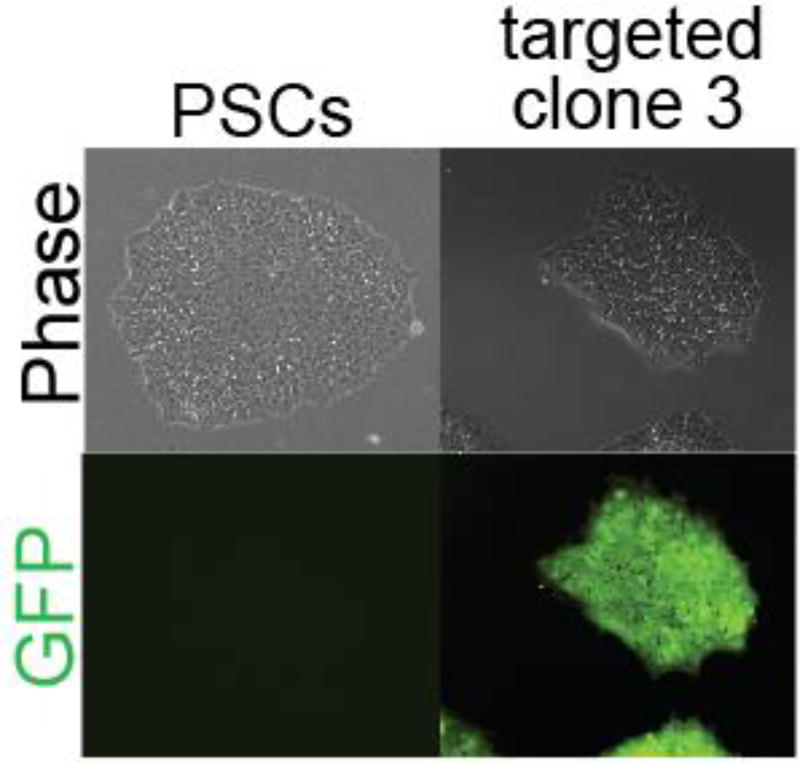
Fig. 1.
Electroporation of Cas9 plasmid and AAVS1 T2 gRNA plasmid (Addgene #41818), and GFP donor plasmid (Addgene #28014) yielded target GFP cell lines. This example uses HDR targeting techniques.
Materials
Dulbecco’s Phosphate-Buffered Saline, without calcium, without magnesium (DPBS, Life Technologies)
mTeSR1 media (STEMCELL Technologies)
Y-27632 (ROCK Inhibitor, STEMCELL Technologies)
Cas9-2A-GFP Plasmid (Addgene #44719)
gRNA vector (Addgene #41824)
DMEM/F12 (Life Technologies)
Accutase (STEMCELL Technologies)
Microcentrifuge Tubes (Fischer Scientific or other source)
15mL Conical Tubes (BD Biosciences or other source)
Standard micropipettes with P2.5, P20, P200, P1000 + tips
Hemocytometer (Hausser Scientific or other source)
Bio-Rad cuvette, 0.4 cm (1 per transfection)
Matrigel-coated tissue culture plates (Corning)
Electroporator (Bio-Rad or equivalent for mammalian cells)
QuickExtract (Epicenter)
AccuPrime Taq High Fidelity (Life Technologies)
Forward and reverse primers for genomic locus (IDT or other source)
Transfection Protocol
Set Up
Seed hPSCs at a density such that they are about 70–80% confluent in 2–3 days (see Note 1).
24 hours prior to transfection, change media to include 10 µM ROCK Inhibitor (see Note 2).
Prepare Matrigel plates for subsequent culture of electroporated cells. Cells may be plated at a density of 104–105 cells per cm2.
Prepare plasmid stocks for electroporation (see Note 3).
Warm Accutase and cell culture reagents to 37°C prior to starting experiments.
Experimental
- Prepare electroporation solution containing plasmids and media:
- Determine plasmid volume for each plasmid. 2.5 µg of Cas9 plasmid and 1.0 µg of gRNA plasmid are needed for every 1×106 cells to be transfected (see Note 4). If using a donor DNA template, add 2.0 µg per 1×106 cells. If using a ssODN template, add 1µL of a 10 µM stock (see Note 5) [6]. For protocol modifications for ZFN and TALEN strategies, see Note 6.
- Pipette 400 µL mTeSR1 media into a to microcentrifuge tube.
- Thaw and gently vortex plasmid stocks.
- Add plasmids to media.
- Plasmid solution should be kept on ice.
Aspirate media from the hPSC wells and wash with 1–2 mL of DPBS.
- Harvest cells via Accutase enzymatic detachment:
- Add enough enzyme to cover well and place in 37°C incubator for 5–7 minutes.
- Dislodge cells by tapping the culture plate, then collect cells from each well using a P1000 pipette and add cell suspension to a 15 mL conical tube.
- Gently pipette up and down to dissociate any remaining clumps (see Note 7).
- Immediately rinse each well with an additional 0.5 volumes of culture media or DMEM/F12 and add to the conical tube to collect any remaining cells and neutralize Accutase by dilution.
Obtain cell count via hemocytometer or flow cytometry-based methods.
Remove sufficient volume to obtain the desired cell count per transfection (0.5–5×106 per transfection has been shown to be effective) and place into a 15 mL conical tube.
Pellet cells by centrifuging 5 min at 200 × g.
Aspirate supernatant and resuspend pellet in the cold electroporation solution created in step 1 (see Note 8).
Transfer solution into a 0.4 cm cuvette (see Notes 9, 10).
Pulse cuvette in electroporator. High efficiency has been observed using an exponential decay waveform, 250V, 750 µF capacitance, and infinite resistance (see Notes 11, 12, 13).
Using a P200 pipette remove electroporation solution from cuvette and transfer into a 15 mL conical tube, containing sufficient volume of mTeSR + 10 µM ROCK Inhibitor medium for desired number of wells (see Note 14).
Wash cuvette with a 200µL aliquot of mTeSR media to remove any remaining cells, add wash to the 15 mL conical tube.
Plate cells into culture dish.
At 24 hours post electroporation change media with mTeSR1 + 10 µM ROCK Inhibitor.
At 48 hours post electroporation, ROCK Inhibitor may be removed and selection agents (if any) may be added.
To Assay Efficiency of NHEJ-mediated Indels
Allow cells to grow to ~80% confluency.
Passage wells at a 1:2 split ratio.
Allow to grow until all wells are again ~80% confluent with daily media changes of mTeSR without ROCK inhibitor.
-
Harvest genomic DNA using QuickExtract solution or equivalent extraction method (see Note 15).
Run one of the following assays for detection of NHEJ: T7 Endonuclease (Fig. 2) or SURVEYOR [14] (see Note 16), PAGE gel [15], MiSeq [16], HRMA [17]. Mutational frequencies can vary between assays, as do the lower limits of resolution for each assay. T7 Endonuclease assays require ~5% NHEJ to be resolved whereas MiSeq can detect a single cell in which a mutation has occurred. The sensitivity of other assays fall between these two extremes.
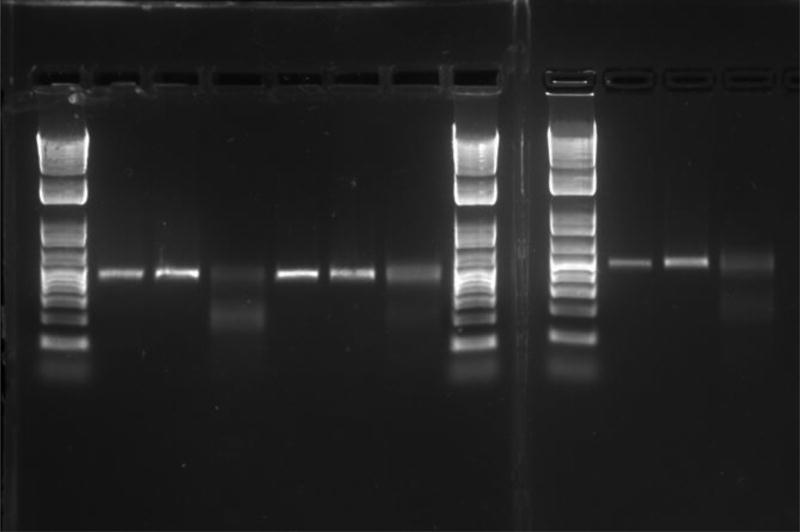
Fig. 2.
Representative image from 3 T7 assays done in parallel. Each sample is from a different gRNA targeted against a mCherry transgene. 3 lanes of each genomic PCR product are run: lanes 2, 5, and 10 are purified genomic PCR product, lanes 3, 6, and 11 are PCR product after hybridization to form heterodimers and in lanes 4, 7, and 12 the heterodimers have been cleaved with T7 endonuclease. Primers: Forward: AAGGGCGAGGAGGATAACATGG; Reverse: TTGTACAGCTCGTCCATGCCG.
To Isolate Clonal Populations via FACS sorting
Schedule a time to use the FACS sorting facility 2–3 days after electroporation.
Follow electroporation protocol. In step 1 of the electroporation protocol, substitute Cas9 plasmid with Cas9-2A-GFP plasmid. Alternatively, in step 1 of the electroporation protocol, add 250 ng of GFP plasmid per 1×106 cells (Addgene #13031).
At 24–48 hours post electroporation, confirm transfection success using fluorescent microscopy.
Add 10 µM ROCK Inhibitor to transfected wells one hour prior to dissociation (see Note 17).
Obtain a single cell dissociation using Accutase as described in step 3 of electroporation protocol.
Centrifuge for 5 min at 200 × g.
Resuspend in 2–3 mL of mTeSR media + 10 µM ROCK inhibitor and pass through a 40 µm cell strainer cap into a FACS tube.
Keep cells and collection tubes on ice (see Note 18).
Using FACS, collect cells that are expressing GFP. Ideally, use cells that were mock electroporated (no plasmid) to set up gates for fluorescence (Fig. 3).
Transfer cells into fresh wells of a 6-well plate at a density of ~1000 cells/cm2 in mTeSR1 with 10uM ROCK inhibitor.
Allow cells to grow for 7–10 days while performing daily media changes with mTeSR1 with 10uM ROCK inhibitor (see Note 19). Other cloning methods can also be successful (see Note 20)
When colonies begin to form, use a P200 pipette to lightly scrape at colonies of interest while slowly releasing the plunger (see Note 21).
Transfer each clone to a single well of a 24-well plate.
When each clone is individually confluent (typically 4–5 days after step 13), passage at a 1:3 ratio into 3 wells of a 12 well plate.
From one well, harvest genomic DNA using QuickExtract solution (see Note 15) or equivalent extraction method.
Individually freeze other two wells (see Chapter 12 in this book) in liquid nitrogen.
Prepare harvested genomic DNA for Sanger sequencing [18]. After receiving sequencing results, thaw clones that contain desired mutations (see Chapter 11 in this book).
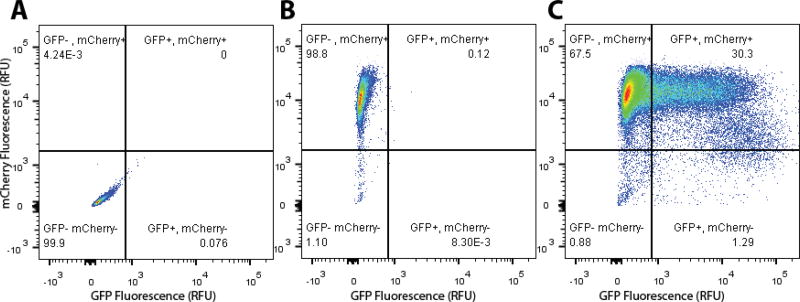
Fig. 3. Representative flow cytometry plot for sorting Cas9- and GFP- expressing hPSCs.
(A) Non-fluorescent hPSC negative control (B) Mock electroporated H9-H2B-mCherry expressing cells. (C) Cas9, eGFP-N1 co-electroporated hPSCs. This method shows >30% efficiency of introduction into hPSCs.
NOTES
Cells must be in an actively dividing state
The longer that the cells are in ROCK Inhibitor, the greater the cell attachment following electroporation (18–24 hours is recommended). The minimum amount of time for cells to be in ROCK inhibitor that has an appreciable effect is 1 hour.
Concentration will depend on efficiency of plasmid isolation protocol. Ideal values will be greater than 1 µg/mL as determined by NanoDrop or equivalent method. Plasmid purification protocol must contain an endotoxin removal step.
An alternative to transfecting plasmids is to directly transfect the RNA and Cas9 protein required. The advantage of this alternative is that there is less of a lag time until results can be collected. In this alternative, generate gRNAs via in vitro transcription [7] [19] off of the created gRNA plasmid using the MEGAshortscript T7 kit (Ambion). Cas9 coding sequence can be cloned into a bacterial expression vector and expressed in a bacterial host before being purified via an attached His tag [20].
Optimal ssODN design is necessary for successful HDR. Some of the considerations that must be taken into account are the mutation of the PAM sequence via codon wobble to produce a silent mutation [21]. Without this modification, the Cas9-gRNA complex will cleave the ssODN. Secondly, design intended point mutations in the center of ssODN with ~40 bp flanking homology on either side [22]. It is also advisable to design your ssODN to either introduce or remove a restriction enzyme site [23]. This consideration will allow for facile analysis of clones, without having to perform one of the NHEJ assays mentioned in step 4.
Both zinc finger nucleases (ZFNs) and TALENs remain viable options for gene editing. If using a either method, electroporate using 0.5 µg of nuclease encoding plasmid and 4 µg of donor DNA per 1×106 cells [4] [24]. All other steps of the protocol remain the same.
Single cell dissociations are important as electroporation of cell clumps decreases efficiency.
It is important to remove as much supernatant as possible as excess volume during electroporation will decrease efficiency.
Bubbles must be avoided, as they will cause increased cell death during electroporation.
Avoid touching the metal sides of cuvette as oils may transfer and cause waveform to be disrupted.
Individual machines vary, so these values will have to be optimized on a per-machine basis. High voltage electroporations will increase efficiency while also increasing cell death. We recommend starting at our settings at changing the voltage in increments of ±25 V while holding the capacitance constant. Then increment capacitance ±50 µF at optimum voltage.
Make a note of time constant (TC) values. Highly variable values may indicate a malfunctioning machine.
Variability between stem cell lines, especially between embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), can have a drastic effect of cell viability. If you are experiencing high amounts of cell death go back and reoptimize electroporation conditions for each cell line. This is especially necessary if transitioning conditions from an ESC line to an iPSC line.
We generally electroporate enough cells to provide 6 replicates in a full 6 well plate. Using this setup 12 mL of mTeSR would be needed per electroporation.
To perform an extraction using QuickExtract cells are first trypsinized, pelleted, and resuspended in 500 µL of QuickExtract before being transferred in to a microcentrifuge tube. Then place microcentrifuge tube in a heat block at 65°C for 15min, 68°C for 15min, and 98°C for 10 min.
SURVEYOR from Transgenomic is similar to the T7 Endonuclease assay. We have found that reagents can be exchanged between the two kits.
ROCK inhibitor may be left in culture from pre-electroporation through FACS without decrease in viability or proliferation rate.
Sorting cells into collection tubes may be beneficial to sorting directly into culture plates. Sorting into plates causes cell death due to slamming into the plastic on bottom of plate. Sorting parameters need also to be modified for hPSCs. For a BD Aria, we use a nozzle size of 100 µm and a pressure of 20 psi.
Do not allow colonies to grow too large or merge.
Single cell cloning can also be done in a high-throughput method if mutations happen at a frequency of <1%. This method, known as sib-cloning, combines a TaqMan PCR assay with fluorescent probes that distinguish between mutant and wild type products. Mutant frequencies as low as 0.1% can be detected and enriched by selection and propagation to a further round of cloning [25].
-
Be sure to remove entire colony from culture plate. If cells are detached they may float through media and contaminate other clones being isolated. To avoid this contamination, change media between each clone being selected.
FACS sorting at clonal density culture can survive without differentiation 48–72 hours without changing media. This simulates the use of conditioned media and decreases stress on single cells. After this period change media daily. When colonies form after 7–10 days use a P200 pipette set to 200 µL and, with the plunger depressed, gently scrape at the colony edge causing it to lift off the plate. Simultaneously, slowly release the plunger to suck the entire colony into pipette tip. Transfer into new well and gently pipette up and down 3–4 times to dissociate the colony. Empirically, this dissociation dramatically speeds up the rate of proliferation. Change media in new well every 24 hours. Wait until cells are ~80% confluent, usually about four days, and then passage 1:3 into 3 wells. Wait another 4–5 days or until wells are again ~80% confluent before freezing 2 wells. 2 wells are frozen to provide a backup in the event one sample does not survive the freeze/thaw cycle. Collected genomic DNA from final well should be PCR amplified using AccuPrime Taq with primers for the locus of interest and then Sanger sequenced using the same forward primer as used for amplification.
References
- 1.Sonoda E, Hochegger H, Saberi A, et al. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 2.Mali P, Yang L, Esvelt KM, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen F, Pruett-Miller SM, Huang Y, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8:753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hockemeyer D, Wang H, Kiani S, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 6.Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.González F, Zhu Z, Shi Z-D, et al. An iCRISPR Platform for Rapid, Multiplexable, and Inducible Genome Editing in Human Pluripotent Stem Cells. Cell Stem Cell. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalem O, Sanjana NE, Hartenian E, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lakshmipathy U, Pelacho B, Sudo K, et al. Efficient Transfection of Embryonic and Adult Stem Cells. STEM CELLS. 2004;22:531–543. doi: 10.1634/stemcells.22-4-531. [DOI] [PubMed] [Google Scholar]
- 12.Ding Q, Regan S, Xia Y, et al. Enhanced Efficiency of Human Pluripotent Stem Cell Genome Editing through Replacing TALENs with CRISPRs. Cell Stem Cell. 2013;12:393–394. doi: 10.1016/j.stem.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cordie T, Harkness T, Jing X, et al. Nanofibrous Electrospun Polymers for Reprogramming Human Cells. Cell Mol Bioeng. 2014;7:379–393. doi: 10.1007/s12195-014-0341-z. [DOI] [Google Scholar]
- 14.Qiu P. Mutation detection using Surveyor™ nuclease. BioTechniques. 2004;36:702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 15.Zhu X, Xu Y, Yu S, et al. An Efficient Genotyping Method for Genome-modified Animals and Human Cells Generated with CRISPR/Cas9 System. Sci Rep. 2014 doi: 10.1038/srep06420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wittwer CT, Reed GH, Gundry CN, et al. High-Resolution Genotyping by Amplicon Melting Analysis Using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 18.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013 doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 20.Jinek M, Chylinski K, Fonfara I, et al. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013 doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L, Guell M, Byrne S, et al. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu Y, Sander JD, Reyon D, et al. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hockemeyer D, Soldner F, Beard C, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyaoka Y, Chan AH, Judge LM, et al. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods. 2014 doi: 10.1038/nmeth.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]