Abstract
The ocular onchocercosis is caused by the zoonotic parasite Onchocerca lupi (Spirurida: Onchocercidae). A major hindrance to scientific progress is the absence of a reliable diagnostic test in affected individuals. Microscopic examination of skin snip sediments and the identification of adults embedded in ocular nodules are seldom performed and labour-intensive. A quantitative real-time PCR (qPCR) assay was herein standardized for the detection of O. lupi DNA and the results compared with microscopic examination and conventional PCR (cPCR). The specificity of qPCR and cPCR was assessed by processing the most common filarial nematodes infecting dogs, skin samples from O. lupi infected (n = 35 dogs) or uninfected animals (n = 21 dogs; n = 152 cats) and specimens of potential insect vector (n = 93 blackflies; n = 59 mosquitoes/midges). The analytical sensitivity of both assays was assessed using 10-fold serial dilutions of DNA from adult specimen and from a pool of microfilariae. The qPCR on skin samples revealed an analytical specificity of 100% and a sensitivity up to 8 x 10−1 fg/2μl O. lupi adult-DNA and up to 3.6 x 10−1 pg/2μl of mfs-DNA (corresponding to 1 x 10−2 mfs/2μl). Only 9.5% O. lupi-infected skin samples were positive for cPCR with a sensitivity of 8 x 10−1 pg/2μl of DNA. Out of 152 blackflies and mosquitoes/midges, eight specimens experimentally infected (n = 1 S. erythrocephalum; n = 1 S. ornatum; n = 6 Simulium sp.) were positive by qPCR. The qPCR assay herein standardized represents an important step forward in the diagnosis of zoonotic onchocercosis caused by O. lupi, especially for the detection and quantification of low number of mfs. This assay provides a fundamental contribution for the establishment of surveillance strategies aiming at assessing the presence of O. lupi in carnivores and in insect species acting as potential intermediate hosts. The O. lupi qPCR assay will enable disease progress monitoring as well as the diagnosis of apparently clinical healthy dogs and cats.
Author summary
The diagnosis of zoonotic ocular onchocercosis caused by Onchocerca lupi (Spirurida: Onchocercidae) is currently based on microscopic examination of skin snip sediments and on the identification of adults embedded in ocular nodules. These methods are labour-intensive and require multiple steps to achieve the diagnosis. In this context, a novel quantitative real-time PCR assay (qPCR) has been herein standardized and analytical specificity and sensitivity assessed. The results indicate that the qPCR assay could represent an important step forward in the diagnosis of onchocercosis, in carnivores and in insect species acting as potential intermediate hosts.
Introduction
Within the genus Onchocerca (Spirurida: Onchocercidae), Onchocerca volvulus and Onchocerca lupi parasitize humans and carnivores, respectively [1–5], the latter being a zoonotic agent [6,7]. While O. volvulus is a well-known parasite of humans transmitted by blackflies (Simulium spp.) [8,9], the epidemiology of O. lupi is far from being understood, particularly because the information about insect species acting as vectors is lacking. Only Simulium tribulatum was suggested as the putative vector of this filarial worm in California (USA), but proof of its intermediate host competence is currently absent [10]. Onchocerca lupi belongs to the spirurids in the Nematode clade III [11] was first detected from a Caucasian wolf (Canis lupus) in Georgia [12], and, only recently, diagnosed in dogs and cats from Europe (Greece, Portugal, Spain, Germany, Hungary) and USA [13–20]. The reports of O. lupi infection are mainly based on the presence of ocular nodules on the eyelids, conjunctiva, and sclera [3,21,22], though the localization of adult worms in the retrobulbar area of the canine patients may impair the assessment of its distribution in endemic areas [23]. The detection of microfilariae (mfs) in skin snip sediments is the only available tool for the diagnosis of the infection when nodules are not apparent in the eyes. The retrieval and identification of mfs in skin snip samples is a rather invasive and time-consuming method, highly dependent on the anatomical location of skin biopsy and mfs density [24]. Again, the detection of mfs may depend upon the prepatent period, previous microfilaricidal treatments, and on the operator’s skills in examining skin sediments, as described for O. volvulus [25,26].
Conventional PCR (cPCR) amplification and sequencing of mitochondrial NADH dehydrogenase subunit 5 (ND5) and cytochrome c oxidase subunit 1 (cox1) genes are available for the molecular identification of O. lupi adults and mfs [7,27,28]. The cPCR, however, may be relatively labour-intensive and exhibit low sensitivity, mainly for mfs detection, limiting the establishment of large-scale epidemiological studies in vertebrate hosts and putative vectors.
Here, we developed a quantitative real-time PCR (qPCR) assay based on the hybridization probe to detect O. lupi DNA in host and putative vector samples. The diagnostic validity of qPCR assay was compared with microscopic examination and cPCR methods.
Methods
Ethics statement
All dogs’ and cats’ skin samples were collected in previous studies [17,29] and approved by the ethical committee of the Department of Veterinary Medicine of the University of Bari (Prot. Uniba 1/16) and by the ethical committee of the Faculty of Veterinary Medicine, Universidade Lusófona de Humanidades e Tecnologias.
Samples
Genomic DNA of adult specimens of O. lupi (n = 3), as well as DNA from single (n = 7) or pooled mfs (n = 10), collected from dogs in different geographical locations (Table 1) were used as control. All specimens were previously identified based on morphological and molecular analyses [18,30].
Table 1. Filarial nematodes used to assess the analytical specificity of the qPCR assay.
| Species | Host | Collection locality | Source | ID sample |
|---|---|---|---|---|
| Onchocerca lupi | Canis lupus familiaris | USA (Minnesota) | Adult | 100–14 |
| Canis lupus familiaris | USA (New Mexico) | Adult | 132–14 | |
| Canis lupus familiaris | USA (Colorado) | Adult | 478–15 | |
| Canis lupus familiaris | Portugal | Microfilariae | 63–12 | |
| Canis lupus familiaris | Greece | Microfilariae | 62–12 | |
| Canis lupus familiaris | Portugal | Skin | 537–15 | |
| Felis catus | Portugal | Skin | 61–15 | |
| Onchocerca armillata | Bos taurus | Cameroon | DNA | 54FKA2 |
| Onchocerca bohemi | Equus caballus | Italy | Adult | 409 |
| Onchocerca fasciata | Camelus dromedaries | Iraq | Adult | 200 |
| Onchocerca gutturosa | Bos taurus | Cameroon | DNA | 54FKG1 |
| Onchocerca ochengi | Bos taurus | Cameroon | DNA | 54FKO2 |
| Dirofilaria immitis | Vulpes vulpes | Italy | Adult | 377 |
| Dirofilaria repens | Canis lupus familiaris | Italy | Adult | 379 |
| Cercopithifilaria grasii | Canis lupus familiaris | Portugal | Skin | 81–16 |
| Cercopithifilaria bainae | Canis lupus familiaris | Portugal | Skin | 81–16 |
| Cercopithifilaria sp. II | Canis lupus familiaris | Portugal | Skin | 81–16 |
| Acanthocheilonema reconditum | Canis lupus familiaris | Italy | Blood | 496 |
| Brugia malayi | Meriones unguiculatus (experimental cycle) | FR3 strain | DNA | 8YT1 |
| Brugia pahangi | Meriones unguiculatus (experimental cycle) | FR3 strain | DNA | 46YT |
| Wuchereria bancrofti | Homo sapiens | Singapore | DNA | 82YT FIL13/01 |
Primers and probe design and qPCR run protocol
Primers (O.l.F 5′-GGAGGTGGTCCTGGTAGTAG-3′; O.l.R 5′- GCAAACCCAAAACTATAGTATCC-3′) and a TaqMan-MGB hydrolysis probe (FAM-5’-CTTAGAGTAGAGGGTCAGCC-3’-non-fluorescent quencher-MGB; Applied Biosystems; Foster City, CA, USA), targeting partial cox1 gene (90bp), were designed by alignment of sequences from a wide range of closely related filarial nematodes available from GenBank database (Table 2), using Primer Express 2.0 (Applied Biosystems, Foster City, CA). Specificity of the primers and probe for O. lupi were confirmed in silico using the basic local alignment search tool (BLAST, GenBank, NCBI).
Table 2. GenBank accession numbers (AN) of mitochondrial cytochrome c oxidase subunit 1 sequences of filarial nematodes used for primers and TaqMan-probe design.
| Parasite | AN | Host | Collection locality |
|---|---|---|---|
| Onchocerca lupi | KC686702 | Canis lupus familiaris | Greece |
| KC686701 | Canis lupus familiaris | Portugal | |
| EF521408 | Canis lupus familiaris | Hungary | |
| Onchocerca armillata | KP760200 | Bos taurus | Cameroon |
| Onchocerca boehmi | KX898458 | Equus caballus | Italy |
| Onchocerca dewittei japonica | AM749267 | Sus scrofa leucomystax | Japan: Oita |
| Onchocerca eberhardi | AM749268 | Cervus nippon | Japan: Oita |
| Onchocerca ochengi | KC167350 | Simulium damnosum sensu lato | Cameroon: northern |
| Onchocerca gibsoni | AJ271616 | Bos taurus | Australia: Queensland |
| Onchocerca gutturosa | KP760201 | Bos taurus | Cameroon |
| Onchocerca lienalis | KX853326 | Bos taurus | United Kingdom: Wales |
| Onchocerca ramachandrini | KC167356 | Simulium damnosum sensu lato | Cameroon: northern |
| Onchocerca suzukii | AM749277 | Nemorhaedus crispus | Japan: Yamagata |
| Onchocerca skrjabini | AM749269 | Cervus nippon | Japan: Oita |
| Onchocerca sp. ‘Siisa’ | KC167354 | Simulium damnosum sensu lato | Cameroon: northern |
| Onchocerca volvulus | KC167355 | Simulium damnosum sensu lato | Cameroon: northern |
| Brugia malayi | KP760171 | Meriones unguiculatus | FR3 strain |
| Brugia pahangi | EF406112 | Homo sapiens | Malaysia |
| Wuchereria bancrofti | AM749235 | Homo sapiens | Italy |
| Cercopithifilaria bainae | JF461457 | Canis lupus familiaris | Italy: Sicily |
| Cercopithifilaria bulboidea | AB178834 | Cervus nippon | Japan |
| Cercopithifilaria crassa | AB178840 | Cervus nippon | Japan |
| Cercopithifilaria grassii | JQ837810 | Canis lupus familiaris | Italy |
| Cercopithifilaria japonica | AM749263 | Ursus thibetanus | Japan: Gifu |
| Cercopithifilaria longa | AB178843 | Cervus nippon | Japan |
| Cercopithifilaria minuta | AB178846 | Cervus nippon | Japan |
| Cercopithifilaria multicauda | AB178848 | Cervus nippon | Japan |
| Cercopithifilaria rugosicauda | KF479370 | Capreolus capreolus | Italy |
| Cercopithifilaria sp. II | JQ837809 | Canis lupus familiaris | Italy |
| Cercopithifilaria shohoi | AB178850 | Cervus nippon | Japan |
| Cercopithifilaria tumidicervicata | AB178852 | Cervus nippon | Japan |
| Acanthocheilonema delicata | JQ289993 | Meles anakuma | Japan |
| Acanthocheilonema odendhali | KF038145 | Callorhinus ursinus | USA: Alaska |
| Acanthocheilonema reconditum | JF461456 | Canis lupus familiaris | Italy: Sicily |
| Acanthocheilonema spirocauda | KF038155 | Erignathus barbatus | USA: Alaska |
| Acanthocheilonema vitaea | KP760169 | Meriones unguiculatus | FR3 strain |
| Dirofilaria immitis | EU169124 | Ailurus fulgens | China |
| Dirofilaria repens | AM749230 | Canis lupus familiaris | Italy |
qPCR reactions were carried out in a final volume of 20μl, consisting of 10μl of IQ Supermix (Bio-Rad Laboratories, Hercules CA, USA), 7.1μl of Di-Ethyl Pyro-Carbonate (DEPC) treated pyrogen-free DNase/RNase-free water (Invitrogen, Carlsbad, CA, USA), 2μl of template DNA (except no-template controls), 5 pmol and 0.5 pmol for primers and probe, respectively.
The run protocol consisted of a hot-start at 95°C for 3 min, and 40 cycles of denaturation (95°C for 10 sec) and annealing-extension (64°C for 30 sec). All assays were carried out in duplicate and a no-template control was included in each run. The qPCR was performed in a CFX96 Real-Time System (Bio-Rad Laboratories, Inc., Hercules CA, USA) and the increase in the fluorescent signal was registered during the extension step of the reaction and analysed by the CFX Manager Software Version 3.1 (Bio-Rad).
Specificity and sensitivity of qPCR
To investigate the analytical specificity of the assay, genomic samples of Onchocerca spp. and of the most common filarial nematodes infesting dogs (Table 1) were used. The specificity of the assay was tested by using DNA from skin samples of naturally infected dogs, which were positive for O. lupi (n = 35) at microscopic examination [29]. Skin samples were divided in five groups (G1-G5) according to their mfs load (Table 3), being 14 also co-infected with Cercopithifilaria bainae and Cercopithifilaria sp. II. Skin samples (dogs n = 21; cats n = 152), which did not test positive to any mfs [17,29], were used as negative control.
Table 3. Skin samples tested for Onchocerca lupi by qPCR, divided (Groups 1–5) according to the parasitic load (mfs) microscopically detected.
The mean, minimum, maximum and standard deviation (sd) values of the threshold cycle (Cq), parasite load (Starting Quantity (SQ) value, expressed as ng/μl of DNA for reaction) and microfilariae concentration, assessed by qPCR is reported.
| Parasitic load (mfs) | Skin (n) | Cq | Mfs | DNA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SQ | SQ | ||||||||||
| Mean | Min-Max | SD | Mean | Min/Max | SD | Mean | Min/Max | SD | |||
| G1 | 1 < 5 | 16 | 33.49 | 32.12–35.89 | 1.2 | 1.9 | 2.6 x 10−1–3.8 | 1.2 | 6.1 x 10−2 | 9.5 x 10−3–1.3 x 10−1 | 4.3 x 10−2 |
| G2 | 6 < 10 | 7 | 31.24 | 30.24–31.75 | 0.5 | 6.9 | 4.9–10 | 1.8 | 2.5 x 10−1 | 1.8 x 10−1–3.8 x 10−1 | 6.5 x 10−2 |
| G3 | 11 < 25 | 8 | 29.92 | 29.06–31.3 | 0.8 | 19.1 | 6.7–29.8 | 8.6 | 6.9 x 10−1 | 3.1 x 10−1–1.1 | 3 x 10−1 |
| G4 | 26 < 40 | 2 | 28.65 | 28.37–28.93 | 0.4 | 38.4 | 3.5 x 101–4.1 x 101 | 4 | 1.4 | 1.3–1.5 | 1.5 x 10−1 |
| G5 | > 40 | 2 | 27.52 | 27.41–27.63 | 0.1 | 96 | 1 x 102–8.9 x 101 | 10.2 | 3.4 | 3.2–3.7 | 3.7 x 10−1 |
Specimens of blackflies (n = 66) and mosquitoes/midges (n = 39) collected from 2011 to 2014 in Greece [31], and 27 blackflies and 20 Aedes albopictus (colony specimens) experimentally infected by intrathoracic microinjection with mfs of O. lupi (parasitic load of 20mfs/μl) were analyzed after death (i.e., from one to 10 days post infection) (Table 4).
Table 4. Blackflies and mosquitoes/midges specimens used to test the analytical specificity of qPCR assay.
| Geographical origin | Blackflies species | Number (pos/tot) |
Mosquitoes/ midges species | Number (pos/tot) |
|---|---|---|---|---|
| Greece | Simulium balcanicum | 0/14 | Culex pipiens pipiens | 0/10 |
| Simulium erythrocephalum | 0/6 | Ochlerotatus caspius | 0/10 | |
| Simulium pseudequinum | 0/10 | Anopheles maculipennis | 0/4 | |
| Simulium reptans | 0/23 | Coquilletidia richiardii | 0/2 | |
| Simulium ornatum | 0/4 | Culiseta annulata | 0/3 | |
| Simulium velutium | 0/9 | Culicoides spp. | 0/1 | |
| Ceratopogonidae | 0/5 | |||
| Psychodidae | 0/4 | |||
| Italy | ||||
| Basilicata region | Simulium erythrocephalum | 1/1* | ||
| Simulium linatum | 0/5* | |||
| Simulium ornatum | 1/4* | |||
| Simulium pseudequinum | 0/10* | |||
| Simulium sp. | 6/7* | |||
| Reggio Emilia | Aedes albopictus | 0/20* | ||
| Total | 8/93 | 0/59 |
* = Specimens experimentally infected by intrathoracic microinjection with microfilariae of Onchocerca lupi.
The analytical sensitivity of the qPCR assay was assessed using 10-fold serial dilutions of DNA from adult specimen (i.e., ranging from 8 × 104 to 8 × 10−3 fg/2μl of reaction) and from a pool of 10 mfs (i.e., ranging from 10 to 10 × 10−3 microfilariae/2μl of reaction, corresponding to 3.6 ×10−1 ng/2μl to 3.6 ×101 fg/2μl of DNA). Ten replicates of each serial dilution were submitted to the same run for assessment of intra-assay reproducibility.
Genomic DNA was isolated from all skin samples and from O. lupi adults and mfs, blackflies, mosquitoes and midges specimens using the commercial kits DNeasy Blood & Tissue Kit (Qiagen, GmbH, Hilden, Germany), respectively, following the manufacturers’ instructions. The amounts of purified DNA were determined spectrophotometrically using the Qubit (Applied Biosystems, Foster City, CA, USA).
Specificity and sensitivity of cPCR
The analytical specificity and sensitivity of the cPCR for the specific amplification of cox1 gene fragment (∼689bp; [32]) was assessed by testing genomic DNA of: i) skin samples with different parasitic load of O. lupi (Table 3), ii) serial dilution of O. lupi mfs DNA (i.e., from 3.6 ×101 pg/2μl to 3.6 ×10−3 pg/2μl) and iii) DNA of adult specimens (i.e., from 8 ×101 ng/2μl to 8 x 10−3 fg/2μl).
All cPCR products were resolved in 0.5x GelRed stained (Biotium, CA, USA) agarose gels (2%), purified using enzymatic purification (Exo I-FastAP; Thermo Fisher Scientific, MA, USA) and sequenced in an automated sequencer (3130 Genetic Analyzer). All sequences generated were compared with those available in GenBank using Basic Local Alignment Search Tool (BLAST) [33].
Results
All O. lupi naturally-infected dog positive at skin samples examination by microscopy, considered the gold standard method as true positives, were positive by the O. lupi qPCR herein assessed (specificity of 100%). Out of 21 skin samples microscopically and qPCR positive for O. lupi, two were positive by cPCR (parasite load of 8 and 25 mfs), revealing a low analytical cPCR specificity (i.e., 9.5%). None of cat’s skin samples were positive by qPCR.
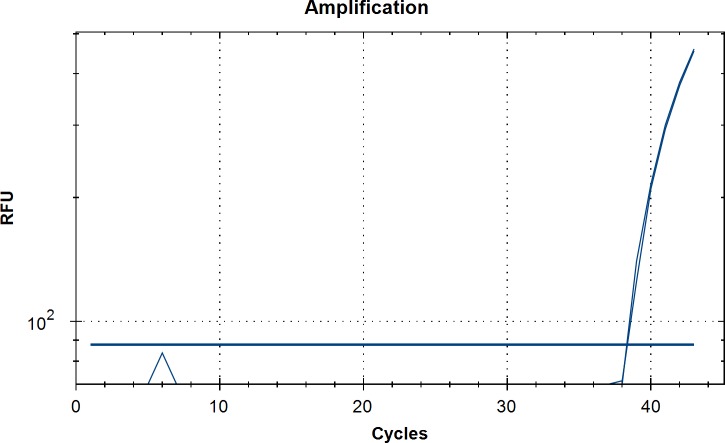
A specific fluorescent signal was recorded for all O. lupi adult and mfs positive controls tested (Fig 1). No fluorescence was obtained for all other Onchocerca species or filarial nematodes examined as well as for skin samples used as negative control.
Fig 1. Assessment of the specificity of qPCR assay in the detection of Onchocerca lupi DNA.
The amplification plot is represented by the fluorescent signal accordingly to relative fluorescence units (RFU) and threshold cycle.
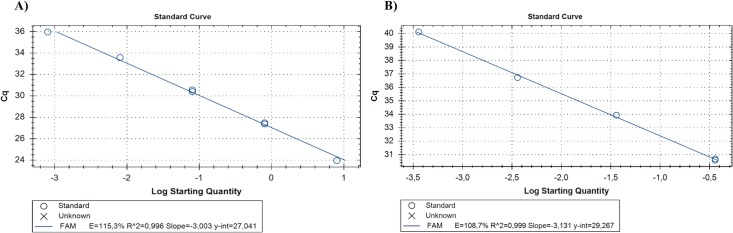
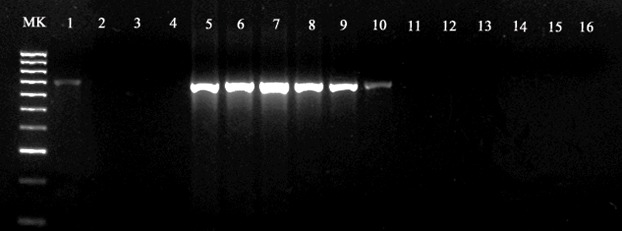
The analytical sensitivity of qPCR was confirmed by detection of up to 8 x 10−1 fg/2μl and 3.6 x 10−1 pg/2μl of DNA (i.e., corresponding to 1 x 10−2 mfs/2μl) of O. lupi adult worm and mfs, respectively (Fig 2A and 2B). qPCR efficiencies ranged from 108.7 to 115.3% with an R2 from 0.996 to 0.999 and Slope from -3.003 to -3.131, for both adult and mfs (Fig 2A and 2B). The mean parasite load detected for the positive skin samples, ranged from 1.9 to 96 mfs/2μl of reaction, corresponding to 6.1 x 10−2 ng/2μl (mean cycle threshold of 33.49) and to 3.4 ng/2μl DNA (mean cycle threshold of 27.52), respectively (Table 3). The results of mfs detection by qPCR overlapped the values obtained by the microscopic examination. The detection limit registered for cPCR was up to 8 x 10−1 pg/2μl for adult worms and up to 3.6 x 101 pg/2μl for mfs DNA (i.e., corresponding to 1 mf/2μl), respectively (Fig 3).
Fig 2.
Standard curves generated from serial dilutions of (A) genomic DNA from adult (from 8 × 104 to 8 × 10−3 fg/2μl of reaction) and microfilariae (B) (from 3.6 ×10−1 ng/2μl to 3.6 ×101 fg/2μl of reaction) of Onchocerca lupi. Each point was tested in triplicate. Slope, efficacy and R2 are reported on the bottom.
Fig 3. Detection limit of the conventional PCR assay determined by 10-fold serial dilution of genomic DNA of microfilariae and adult of Onchocerca lupi.
Lanes 1–4, from 3.6 ×101 pg/2μl to 3.6 ×10−3 pg/2μl of O. lupi mfs DNA (i.e., from 1 to 1×10−4 mfs); Lanes 5–15, from 8 ×101 ng/2μl to 8 x 10−3 fg/2μl of O. lupi adult DNA; Line 16, no-DNA control; M, 100 bp DNA marker.
Out of 152 blackflies, mosquitoes and midges, eight Simulim spp. (n = 1 S. erythrocephalum; n = 1 S. ornatum; n = 6 Simulium sp.), experimentally infected and died from 1 to three days post infection, returned positive signal for O. lupi DNA (Table 4). All field-collected blackflies and mosquitoes were negative for O. lupi DNA using qPCR (Table 4). All blackflies positive for qPCR scored positive also for cPCR.
Sequences derived from all amplicons of cPCR matched with 99–100% nucleotide identity appropriate reference sequences of O. lupi available from GenBank (accession numbers KC686702, KC686701).
Discussion
A qPCR assay has been developed for the detection of O. lupi in animal skin snip samples and potential vectors and proved to be a sensitive and specific tool for the diagnosis of this parasite, with a mean detection limit as low as 1.9 mfs per reaction. In addition, the high sensitivity of the qPCR protocol has been demonstrated by detecting a small amount of DNA (up to 8 x 10−1 fg/2μl for adult and up to 3.6 x 10−1 pg/2μl for mfs), by the slope value of standard curve (−3.131), the efficiency (115.3%) and the coefficient of determination (R2 = 0.999). These features of the assay are due to the selection of a stable hydrolysis probe designed (100% specific for O. lupi DNA), as well as to the choice of the target gene used. Indeed, cox1 gene of the mitochondrial DNA has been well recognized as a “barcode” for filarial nematodes [34], with a high amplification efficiency, also due to the large copy numbers enabling the detection of minimum amounts of DNA [35–37]. Though few Onchocerca species DNA were herein tested, which may represent a limitation of the qPCR assay, this new tool provides an alternative to the labor intensive microscopic examination of skin snip samples and to cPCR for the diagnosis of O. lupi [38]. The qPCR assay was highly specific in revealing O. lupi DNA both in co-infected samples from dogs as well as in potential vector species, avoiding the sequencing confirmation needed using cPCR with filarioid generic primers [32]. Overall, the positive fluorescent signal from samples of O. lupi, from different geographical areas (i.e., Europe and USA), which displayed genetic intraspecific variability [18], indicates the usefulness of the qPCR also for the surveillance of O. lupi where the parasite has been reported [13,14,16,17,19,39–41]. Similarly, even if the qPCR cannot discriminate between viable and nonviable parasites or immature and infective larvae, the assay could be useful for detecting O. lupi in blackfly, mosquito and/or midge species, potentially involved in the transmission of this parasite. Indeed, the specificity of the qPCR to amplify exclusively the DNA of the pathogen in potential insect vectors herein tested, may ultimately assist in the quest to identify the elusive vector of O. lupi.
The newly designed assay represents an improvement in the diagnosis of onchocercosis, by the detection and quantification of low mf densities from tissue samples and could provide a contribution to disease progress monitoring and to the surveillance of O. lupi-infected dogs, avoiding the introduction and/or spread of this life-threatening parasitic nematode, as well as to the identification of apparently healthy animals [29, 42].
The qPCR may speed-up time of diagnosis and prompt treatments of infected animals, which may avoid the appearance of nodular lesions in the eyes or in other anatomical localizations [43].
A TaqMan-based specific and sensitive assay without sequencing is expected to assist high-throughput analysis of samples, eventually leading to improve disease monitoring under the frame of a Public Health perspective. This would be particularly relevant considering that, since its first description of its zoonotic potential [7], cases of zoonotic onchocercosis are being detected increasingly in people from Europe, Iran and the USA [44–47].
Data Availability
All relevant data are within the paper.
Funding Statement
CM has the support of the Portuguese Ministry of Education and Science (via Fundação para a Ciência e a Tecnologia), through an Investigator Starting Grant (IF/01302/2015). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Eberhard ML, Ortega Y, Dial S, Schiller CA, Sears AW, Greiner E. Ocular Onchocerca infections in two dogs in western United States. Vet Parasitol. 2000;90: 333–338. [DOI] [PubMed] [Google Scholar]
- 2.Bain O. Evolutionary relationships among filarial nematodes In: Klei TR, Rajan TV, editors. The Filaria. Boston, USA: Kluwer Academic Publishers; 2002. pp. 21–29. [Google Scholar]
- 3.Sreter T, Szell Z. Onchocercosis: a newly recognized disease in dogs. Vet Parasitol 2008;15: 1–13. [DOI] [PubMed] [Google Scholar]
- 4.Bain O, Mutafchiev Y, Junker K. Order Spirurida In: Schmidt-Rhaesa A, editor. Handbook of Zoology. Nematoda. De Gruyter, Berlin, Germany; 2013. pp. 661–732. [Google Scholar]
- 5.Lefoulon E, Giannelli A, Makepeace BL, Mutafchiev Y, Townson S, Uni S, et al. Whence river blindness? The domestication of mammals and host-parasite co-evolution in the nematode genus Onchocerca. Int J Parasitol. 2017;47: 457–470. doi: 10.1016/j.ijpara.2016.12.009 [DOI] [PubMed] [Google Scholar]
- 6.Sréter T, Széll Z, Egyed Z, Varga I. Subconjunctival zoonotic onchocerciasis in man: aberrant infection with Onchocerca lupi? Ann Trop Med Parasitol. 2002;96: 497–502. doi: 10.1179/000349802125001267 [DOI] [PubMed] [Google Scholar]
- 7.Otranto D, Sakru N, Testini G, Gürlü VP, Yakar K, Lia RP, et al. Case report: First evidence of human zoonotic infection by Onchocerca lupi (Spirurida, Onchocercidae). Am J Trop Med Hyg. 2011;84: 55–58. doi: 10.4269/ajtmh.2011.10-0465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Post RJ, Cheke RA, Boakye DA, Wilson MD, Osei-Atweneboana MY, Tetteh-Kumah A, et al. Stability and change in the distribution of cytospecies of the Simulium damnosum complex (Diptera: Simuliidae) in southern Ghana from 1971 to 2011. Parasit Vectors. 2013;6: 205 doi: 10.1186/1756-3305-6-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamberton PH, Cheke RA, Walker M, Winskill P, Osei-Atweneboana MY, Tirados I, et al. Onchocerciasis transmission in Ghana: biting and parous rates of host-seeking sibling species of the Simulium damnosum complex. Parasit Vectors. 2014;7: 511 doi: 10.1186/s13071-014-0511-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan HK, Bolcen S, Kubofcik J, Nutman TB, Eberhard ML, Middleton K, et al. Isolation of Onchocerca lupi in dogs and black flies, California, USA. Emerg Infect Dis. 2015;5: 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JK, Sultana T, Lee SH, Kang S, Kim HK, Min GS, et al. Monophyly of clade III nematodes is not supported by phylogenetic analysis of complete mitochondrial genome sequences. BMC Genomics. 2011;12: 392 doi: 10.1186/1471-2164-12-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodonaja TE. A new species of nematode, Onchocerca lupi n. sp., from Canis lupus cubanensis. Bulletin of the Academic of Science of Georgian SSR. 1967;45: 715–719. [Google Scholar]
- 13.Hermosilla C, Hetzel U, Bausch M, Grübl J, Bauer C. First autochthonous case of canine ocular onchocercosis in Germany. Vet Rec. 2005;156: 450–452. [DOI] [PubMed] [Google Scholar]
- 14.Szell Z, Erdelyi I, Sreter T, Albert M, Varga I. Canine ocular onchocercosis in Hungary. Vet Parasitol. 2001;97: 243–249. [DOI] [PubMed] [Google Scholar]
- 15.Labelle AL, Daniels JB, Dix M, Labelle P. Onchocerca lupi causing ocular disease in two cats. Vet Ophthalmol. 2011;14: 105–110. doi: 10.1111/j.1463-5224.2011.00911.x [DOI] [PubMed] [Google Scholar]
- 16.Labelle AL, Maddox CW, Daniels JB, Lanka S, Eggett TE, Dubielzig RR, et al. Canine ocular onchocercosis in the United States is associated with Onchocerca lupi. Vet Parasitol. 2013;193: 297–301. doi: 10.1016/j.vetpar.2012.12.002 [DOI] [PubMed] [Google Scholar]
- 17.Maia C, Annoscia G, Latrofa MS, Pereira A, Giannelli A, Pedroso L, et al. Onchocerca lupi nematode in cat, Portugal. Emerg Infect Dis. 2015;21: 2252–2253. doi: 10.3201/eid2112.150061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otranto D, Dantas-Torres F, Giannelli A, Latrofa MS, Papadopoulos E, Cardoso L, et al. Zoonotic Onchocerca lupi in dogs from Greece and Portugal. Emerg Infect Dis. 2013;19: 2000–2003. doi: 10.3201/eid1912.130264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otranto D, Giannelli A, Trumble SN, Chavkin M, Kennard G, Latrofa MS, et al. Clinical case presentation and a review of the literature of canine onchocercosis by Onchocerca lupi in the United States. Parasit Vectors. 2015;8: 89 doi: 10.1186/s13071-015-0699-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miró G, Montoya A, Checa R, Gálvez R, Mínguez JJ, Marino V, et al. First detection of Onchocerca lupi infection in dogs in southern Spain. Parasit Vectors. 2016;9: 290 doi: 10.1186/s13071-016-1587-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarfoss MK, Dubielzig RR, Eberhard ML, Schmidt KS. Canine ocular onchocerciasis in the United States: two new cases and a review of the literature. Vet Ophthalmol. 2005;8: 51–57. doi: 10.1111/j.1463-5224.2005.00348.x [DOI] [PubMed] [Google Scholar]
- 22.Komnenou AT, Thomas ALN, Papadopoulos E, Koutinas AF. Intraocular localization of Onchocerca lupi adult worm in a dog with anterior uveitis: a case report. Vet Ophthalmol. 2015;19: 245–249. doi: 10.1111/vop.12277 [DOI] [PubMed] [Google Scholar]
- 23.Franchini D, Giannelli A, Di Paola G, Cortes H, Cardoso L, Lia RP, et al. Image diagnosis of zoonotic onchocercosis by Onchocerca lupi. Vet Parasitol. 2014; 203: 91–95. doi: 10.1016/j.vetpar.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 24.Otranto D, Dantas-Torres F, Giannelli A, Abramo F, Ignjatović Ćupina A, Petrić D, et al. Cutaneous distribution and circadian rhythm of Onchocerca lupi microfilariae in dogs. PLoS Negl Trop Dis. 2013;7: e2585 doi: 10.1371/journal.pntd.0002585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawking F. The 24-hour periodicity of microfilariae: biological mechanisms responsible for its production and control. Proc R Soc B. 1967;169: 59–67. [Google Scholar]
- 26.Sasa M, Tanaka H. Studies on the methods for statistical analysis of the microfilarial periodicity survey data. Southeast Asian J Trop Med Public Health. 1972;3: 518–536. [Google Scholar]
- 27.Egyed Z, Sréter T, Széll Z, Beszteri B, Oravecz O, Márialigeti K, et al. Morphologic and genetic characterization of Onchocerca lupi infecting dogs. Vet Parasitol. 2001;102: 309–319. [DOI] [PubMed] [Google Scholar]
- 28.Sréter-Lancz Z, Széll Z, Sréter T. Molecular genetic comparison of Onchocerca sp. Infecting dogs in Europe with other spirurid nematodes including Onchocerca lienalis. Vet Parasitol. 2007;148: 365–370. doi: 10.1016/j.vetpar.2007.06.021 [DOI] [PubMed] [Google Scholar]
- 29.Colella V, Maia C, Pereira A, Gonçalves N, Caruso M, Martin C, et al. Evaluation of oxfendazole in the treatment of zoonotic Onchocerca lupi infection in dogs. PLos Neglected Tropical Diseases. 2018;1: e0006218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otranto D, Giannelli A, Latrofa MS, Dantas-Torres F, Trumble NS, Chavkin M, et al. Canine infections with Onchocerca lupi nematodes, United States, 2011–2014. Emerging Infect Dis. 2015;21: 868–871. doi: 10.3201/eid2105.141812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Otranto D, Dantas-Torres F, Papadopoulos E, Petrić D, Ćupina AI, Bain O. Tracking the vector of Onchocerca lupi in a rural area of Greece. Emerg Infect Dis. 2012;18: 1196–1200. doi: 10.3201/eid1807.AD1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casiraghi M, Bazzocchi C, Mortasino M, Ottina E, Genchi C. A simple molecular method for discrimination common filarial nematodes of dogs (Canis familiaris). Vet Parasitol. 2006;141: 368–372. doi: 10.1016/j.vetpar.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferri E, Barbuto M, Bain O, Galimberti A, Uni S, Guerriero R, et al. Integrated taxonomy: traditional approach and DNA barcoding for the identification of filarioid worms and related parasites (Nematoda). Front Zool. 2009;6: 1 doi: 10.1186/1742-9994-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnheim N. Concerted evolution of multigne families In: Nei M, Koehn RK, editors. Evolution of Genes and Proteins. Sunderland, Massachusetts: Sinauer Associates; 1983. pp. 38–61. [Google Scholar]
- 36.Hillis DM, Dixon MT. Ribosomal DNA: molecular evolution and phylogenetic inference. Q Rev Biol. 1991;66; 411–453. [DOI] [PubMed] [Google Scholar]
- 37.Mishra K, Raj DK, Hazra RK, Dash AP, Supakar PC. The development and evaluation of a single step multiplex PCR method for simultaneous detection of Brugia malayi and Wuchereria bancrofti. Mol Cell Probes. 2007;21: 355–362. doi: 10.1016/j.mcp.2007.05.001 [DOI] [PubMed] [Google Scholar]
- 38.Otranto D, Brianti E, Latrofa MS, Annoscia G, Weigl S, Lia RP, et al. On a Cercopithifilaria sp. transmitted by Rhipicephalus sanguineus: a neglected, but widespread filarioid of dogs. Parasit Vectors. 2012;5: 1 doi: 10.1186/1756-3305-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sréter T, Széll Z, Egyed Z, Varga I. Ocular onchocercosis in dogs: a review. The Veterinary Record. 2002;151: 176–180. [DOI] [PubMed] [Google Scholar]
- 40.Faísca P, Morales-Hojas R, Alves M, Gomes J, Botelho M, Melo M, et al. A case of canine ocular onchocercosis in Portugal. Vet Ophthalmol. 2010;13: 117–121. doi: 10.1111/j.1463-5224.2010.00763.x [DOI] [PubMed] [Google Scholar]
- 41.Tudor P, Turcitu M, Mateescu C, Dantas-Torres F, Tudor N, Bărbuceanu F, et al. Zoonotic ocular onchocercosis caused by Onchocerca lupi in dogs in Romania. Parasitol Res. 2016;115: 859–862. doi: 10.1007/s00436-015-4816-1 [DOI] [PubMed] [Google Scholar]
- 42.Colella V, Lia RP, Di Paola G, Cortes H, Cardoso L, Otranto D. International dog travelling and risk for zoonotic Onchocerca lupi. Transbound Emerg Dis. 2018;00: 1–3. [DOI] [PubMed] [Google Scholar]
- 43.Alho AM, Cruz L, Coelho A, Martinho F, Mansinho M, Annoscia G, et al. Aberrant laryngeal location of Onchocerca lupi in a dog. Parasitol Int. 2016;65: 218–220. doi: 10.1016/j.parint.2015.12.010 [DOI] [PubMed] [Google Scholar]
- 44.Eberhard ML, Ostovar GA, Chundu K, Hobohm D, Feiz-Erfan I, Mathison BA, et al. Zoonotic Onchocerca lupi infection in a 22-month-old child in Arizona: first report in the United States and a review of the literature. Am J Trop Med Hyg. 2013;88: 601–605. doi: 10.4269/ajtmh.12-0733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mowlavi G, Farzbod F, Kheirkhah A, Mobedi I, Bowman DD, Naddaf SR. Human ocular onchocerciasis caused by Onchocerca lupi (Spirurida, Onchocercidae) in Iran . J Helminthol. 2014;88: 250–255. doi: 10.1017/S0022149X13000060 [DOI] [PubMed] [Google Scholar]
- 46.Dudley RW, Smith C, Dishop M, Mirsky D, Handler MH, Rao S. A cervical spine mass caused by Onchocerca lupi. Lancet. 2015;386: 1372 doi: 10.1016/S0140-6736(14)62255-8 [DOI] [PubMed] [Google Scholar]
- 47.Cantey PT, Eberhard M, Weeks J, Swoboda S, Ostovar GA. Letter to the Editor: Onchocerca lupi infection. J Neurosurg Pediatr. 2016;17: 118–119. doi: 10.3171/2015.6.PEDS15344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.