Abstract
The level of indole-3-acetic acid (IAA) was locally modified in cambial tissues of transgenic aspen (Populus tremula L. × Populus tremuloides Michx.). We also demonstrate the use of a linked reporter gene to visualize the expression of the iaa genes. The rate-limiting bacterial IAA-biosynthetic gene iaaM and the reporter gene for β-glucuronidase (GUS), uidA, were each fused to the cambial-region-specific Agrobacterium rhizogenes rolC promoter and linked on the same T-DNA. In situ hybridization of the iaaM gene confirmed that histochemical analysis of GUS activity could be used to predict iaaM gene expression. Moreover, quantitative fluorometric analysis of GUS activity allowed estimation of the level of de novo production of IAA in transgenic lines carrying a single-copy insert of the iaaM, uidA T-DNA. Microscale analysis of the IAA concentration across the cambial region tissues showed an increase in IAA concentration of about 35% to 40% in the two transgenic lines, but no changes in the radial distribution pattern of IAA compared with wild-type plants. This increase did not result in any changes in the developmental pattern of cambial derivatives or the cambial growth rate, which emphasizes the importance of the radial distribution pattern of IAA in controlling the development of secondary xylem, and suggests that a moderate increase in IAA concentration does not necessarily stimulate growth.
It is well established that exogenous indole-3-acetic acid (IAA) affects several aspects of secondary growth of the stem, in particular cambial cell division and radial enlargement of xylem elements (Little and Savidge, 1981; Little and Pharis, 1995; Sundberg et al., 2000). Therefore, to study the regulation of these processes, it is of interest to modify the endogenous level of IAA in stem tissues. This can be accomplished by transforming plants with Agrobacterium tumefaciens T-DNA IAA-biosynthetic iaaM and iaaH genes, which encodes enzymes that convert Trp to IAA via indole-3-acetamid (Klee and Lanahan, 1995). In transgenic petunia (Klee et al., 1987) and tobacco (Sitbon et al., 1992a), ectopic expression of these genes under the control of the strong cauliflower mosaic virus 19S or 35S promoters caused a several-fold increase in the concentration of IAA and alterations in xylem formation. More recently, this approach was applied to hybrid aspen, Populus tremula L. × Populus tremuloides Michx., by expressing the iaa genes from the weaker mannopine synthase 1′2′ promoter (Tuominen et al., 1995). This resulted in a change in the xylem structure of the transgenic trees. A detailed analysis of these trees revealed that the alterations in cambial growth were related to alterations in the concentration and radial distribution pattern of IAA across the cambial meristem and its differentiating derivatives (Tuominen et al., 1997). As in previous studies on transgenic petunia and tobacco, the overall growth pattern of these transgenic hybrid aspen trees was altered, which complicated comparison with the corresponding wild-type plants. Such unwanted effects on growth may be minimized by using promoters with a known, tissue-specific expression pattern (Rotino et al., 1997).
Even though the expression pattern from a particular promoter can be predicted from reporter gene studies, the precise localization and level of transgene expression from the promoter could vary considerably between independent transformants due to positional effects (Stam et al., 1997; Matzke and Matzke, 1998). Such variation could be minimized by fusing the transgene and a reporter gene to the same promoter and linking them into the same T-DNA. In previous studies, however, such linkage of two genes has often resulted in poor coordination of gene expression. This was attributed to the use of a selectable marker as one of the genes, which would be preferentially expressed in the regenerated transgenic plants (Nagy et al., 1985; An, 1986; Sanger et al., 1990). In addition, different promoters were often used to direct the expression of the linked genes, which might have resulted in poor coordination of gene expression due to a differential response of the promoters to surrounding DNA, or to environmental and/or developmental factors (Nagy et al., 1985; Jones et al., 1987; Sanger et al., 1990; Peach and Velten, 1991). The use of identical promoter sequences in front of a reporter gene and the transgene of interest should prevent the above-mentioned problems.
The aim of this study was to investigate the effect of localized expression of the IAA-biosynthetic genes on cambial growth of hybrid aspen. The rolC promoter from Agrobacterium rhizogenes was chosen because, in addition to the previously reported location of rolC expression in phloem tissues (Nilsson et al., 1996, 1997), the promoter is also expressed in the cambial meristem and its expanding derivatives (Regan et al., 1999). The rolC promoter was fused to the IAA-biosynthetic gene iaaM, since it has been previously shown that it is the expression from this first IAA-biosynthetic gene that determines the level of de novo production of IAA in the transgenic plants (Klee et al., 1987; Sitbon et al., 1992b; Rotino et al., 1997). In addition, applicability of a reporter gene-based analysis of transgene expression was studied by also fusing the rolC promoter to the GUS reporter gene uidA and by linking the chimeric rolC:iaaM and rolC:uidA genes into the same T-DNA of a plant gene expression vector.
RESULTS
Southern-Blot Analysis of the Hybrid Aspen Lines
Seventeen independent lines were regenerated after transformation of iaaH-expressing hybrid aspen with the plant expression vector p812C1C carrying the chimeric rolC:iaaM and rolC:uidA genes in tandem. Southern-blot analysis was performed on 13 of these lines to verify proper insertion and to determine the copy number of the inserted T-DNA from p812C1C.
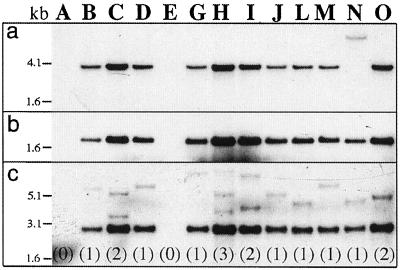
Insertion of the XbaI fragment containing rolC:iaaM:ocspA was demonstrated by digestion of the genomic DNA with the restriction enzyme XbaI and hybridization with the probe from the iaaM gene. A band of the expected size (3,773 bp) was observed in 10 out of 13 lines (Fig. 1A). A band of a larger Mr was observed in line N, which indicated a partial deletion or a rearrangement close to the T-DNA left border of this line. Two lines, A and E, showed no hybridization to the iaaM-specific probe.
Figure 1.
Southern-blot analysis of hybrid aspen lines regenerated after transformation with the vector p812C1C. Ten micrograms of genomic DNA, digested with various restriction enzymes, was loaded on each lane of a agarose gel, transferred to a Hybond-N membrane, and probed with 32P-labeled DNA from either the iaaM or the uidA gene. a, Digestion with XbaI and hybridization with an iaaM-specific probe. b, Digestion with SacI and hybridization with an uidA-specific probe. c, Digestion with EcoRV and hybridization with an uidA-specific probe. Letters on the top of the figure (A–O) indicate the different lines. The copy number of the inserted T-DNA(s) is shown in parentheses.
Hybridization with a probe from the uidA gene revealed proper insertion of the rolC:uidA fragment in 11 of the transgenic lines. DNA digested with SacI and hybridized with the uidA-specific probe revealed a band of the expected size (1,947 bp) corresponding to the uidA gene together with the 3′ end of the rolC promoter (Fig. 1B). Furthermore, DNA digested with EcoRV and hybridized with the uidA-specific probe revealed a band of expected size (2,786 bp) from the fragment containing the rolC promoter, the 3′ end of the iaaM gene and the 5′ end of the uidA gene in the same 11 lines (Fig. 1C). Thus, the rearrangement or deletion in the line N, which was detected in connection to the iaaM gene, did not affect the sequences between the EcoRV site of the iaaM gene and the SacI site at the 3′ end of the uidA gene. Lines A and E did not show any hybridization to the uidA-specific probe.
The copy number of the inserted T-DNA in each line was deduced from the number of bands exceeding 3,088 bp when DNA was digested with EcoRV and hybridized to the uidA-specific probe (Fig. 1C). These bands corresponded to DNA fragments between the second internal EcoRV site of the uidA gene and the closest EcoRV site of the flanking genomic DNA outside of the right border right. There was one T-DNA insert in seven lines, two inserts in three lines, and three inserts in one line. This result, and the absence of tandem repeats of T-DNA, was verified by digestion of DNA with EcoRV and hybridization with the iaaM-specific probe (data not shown). This latter hybridization also verified the presence of two T-DNA copies in line O. As shown in Figure 1C, this line seems to have only one DNA fragment flanking the right border, but a strong hybridization signal is indicative of the presence of two T-DNA copies with similar lengths of the fragments flanking the right border. Lines A and E, which did not show hybridization to either the iaaM or the uidA gene, and line N, which displayed a non-intact T-DNA on Southern-blot analysis, were not studied further. Of the remaining 10 lines, eight lines were randomly chosen for detailed characterization.
Phenotypic Characterization
Compared to the wild type, the different transgenic hybrid aspen lines displayed relatively modest alterations in their phenotype (Fig. 2, A and B). Internode length was significantly increased and the number of axillary buds released after decapitation significantly decreased in all lines (Table I). Leaf width and length, stem diameter, and height of the trees were significantly reduced in a few transgenic lines; leaf width in lines C and G, leaf length in lines C and H, stem diameter in lines B and C, and the height of the trees in line C. In addition, the trees of the lines C, G, and H displayed slightly hyponastic leaves (Fig. 2, A and B). No phenotypic alterations in root growth were detected (data not shown).
Figure 2.

Phenotype of the wild type and selected transgenic hybrid aspen lines, and the expression of the reporter gene GUS and iaaM gene in transgenic line G trees. A, Trees from the wild type and the transgenic lines C, G, H, and O with an approximate height of 1 m. B, Close-up image of the phenotype of a wild-type and a line G tree. Note the vigorous bursting of the axillary buds in the wild type, and the hyponastic leaves in line G. C, GUS activity in a transverse section from the base of the stem. D, In situ hybridization of the iaaM gene in a transverse section from the base of the stem. Co, Cortex; P, phloem; CZ, cambial zone; X, xylem.
Table I.
Phenotypic characters of transgenic and wild-type hybrid aspen trees
| Line | n | Tree Height | Stem Diameter | Internode Length | Leaf Width | Leaf Length | Secondary Shoots |
|---|---|---|---|---|---|---|---|
| cm | mm | no. | |||||
| Wild type | 9 | 128 | 10.6 | 29.1 | 145 | 192 | 13.7 |
| B | 4 | 122 | 9.2* | 33.1* | 133 | 173 | 5.8** |
| C | 5 | 99*** | 9.1* | 34.1** | 115** | 152** | 3.2*** |
| D | 5 | 131 | 11.3 | 33.8** | 132 | 179 | 5.8*** |
| G | 6 | 127 | 9.4 | 35.0*** | 125* | 173 | 6.2*** |
| H | 5 | 125 | 9.7 | 36.5** | 128 | 166* | 3.6*** |
| J | 5 | 131 | 11.2 | 36.1*** | 143 | 184 | 4.2*** |
| L | 4 | 127 | 10.2 | 34.3** | 133 | 177 | 5.0*** |
| O | 4 | 124 | 10.9 | 33.1* | 144 | 181 | 5.5*** |
Stem diameter was measured 7 cm from the base. Internode length was measured for 10 internodes subjacent to the uppermost fully expanded leaf for each tree. Leaf width and length was measured in three full-sized mature leaves for each tree. The no. of secondary shoots from axillary buds was counted 30 d after decapitation. The asterisks indicate statistical significance of difference between the means of the wild type and the transgenic line (Student's t test); *, 0.05 > P > 0.01; **, 0.01 > P > 0.001; and ***, P < 0.001.
The phenotype of the trees in lines A, E, and N, as well as untransformed regenerants were indistinguishable from the wild type (data not shown), confirming that the changes observed in the other lines resulted from the expression of the iaa genes and not from the transformation procedure or somaclonal variation. Expression of the iaaM gene was sufficient to produce the above-mentioned phenotypes, since transgenic lines expressing only the iaaM gene (wild type transformed with the construct p812C1C) had similar phenotypes as the lines expressing both the iaaM and iaaH genes (data not shown).
Northern-Blot Analysis of the Introduced Genes
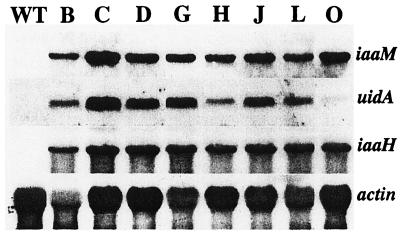
Sequential transformation of hybrid aspen with the plant expression vectors pPUV7022 and p812C1C was expected to result in the constitutive expression of the iaaH gene and coordinate expression of the iaaM and uidA genes in the transgenic lines. After visual compensation for differences in sample loading, the iaaH gene was found to be equally expressed in all lines, whereas the expression levels of the iaaM and uidA genes varied significantly between the different lines (Fig. 3). The expression of the linked iaaM and uidA genes seemed to be coordinated in lines B, C, D, G, J, and L, less coordinated in line H, and not at all coordinated in line O.
Figure 3.
RNA-blot analysis of iaaM, uidA, iaaH, and actin genes in wild-type (WT) and transgenic hybrid aspen lines B through O. Total RNA was obtained from the cambial region tissues of the stem. Approximately 15 μg of total RNA was loaded on each lane. The expression level of the actin gene was used to estimate differences in the loading of the samples.
Localization of the uidA and iaaM Gene Expression
The pattern of uidA gene expression, as studied by histochemical staining for GUS activity, was identical in mature stems of all transgenic lines except line O, which had a low level and an indistinct pattern of expression (data not shown). GUS activity was localized to the cambial meristem, its expanding derivatives, and mature phloem (Fig. 2C). In situ hybridization was performed to detect iaaM gene expression in line G. Similar to the expression revealed by the histochemical GUS assay, the expression of the iaaM gene was detected in the cambial meristem, its expanding derivatives, and in the mature phloem. (Fig. 2D). The results confirm that the expression of these two genes is co-localized in the hybrid aspen stem.
Correlation between the Levels of Free IAA and GUS Activity in the Transgenic Lines
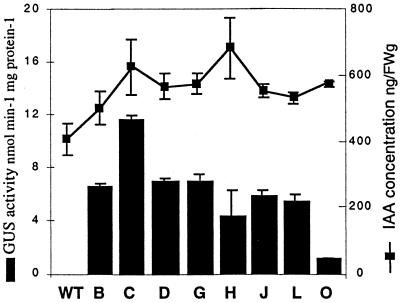
Co-expression of the IAA-biosynthetic genes and the reporter gene GUS was expected to raise the levels of both IAA and GUS activity in a coordinated manner. The concentration of free IAA and GUS activity were increased in the different transgenic lines compared with the wild type (Fig. 4). When the levels of free IAA and GUS activity were compared in the different transgenic lines, a poor correlation was obtained when all transgenic lines were included. However, a good correlation was found (Spearman rank correlation coefficient = 0.81) when excluding lines H and O, which did not show a coordinate expression of the iaaM and uidA genes as determined by the northern-blot analysis.
Figure 4.
GUS activity and IAA concentration in the cambial region of the wild type (WT) and the transgenic hybrid aspen lines B through O at the base of the stem. GUS activity was determined in the extraxylary tissues of four to six trees in each line. IAA was determined in six trees of the wild type and in three trees of all the other lines. The vertical bars indicate se.
The level of IAA conjugates was characterized in wild-type plants and transgenic lines C and G, which displayed the greatest phenotypic deviation from the wild type. Conjugated IAA could not be detected in the wild-type plants. In line C, three out of four analyzed plants had detectable levels of IAA conjugates, with an average of 33 ng/g fresh weight (±7 sd, n = 3), and in line G the average level was 59 ng/g fresh weight (±14 sd, n = 5). Thus, the pool of IAA conjugates induced in the transgenes is minor in relation to the pool of free IAA.
Correlation between Localization of IAA Overproduction and GUS Expression Pattern
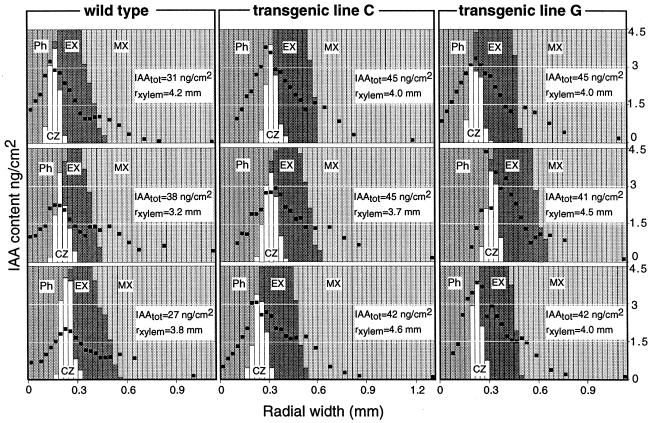
Localization of de novo production of IAA in the stem was studied by characterization of the radial distribution of free IAA in the wild type and in the transgenic lines C and G. IAA was shown to be distributed along a steep gradient across developing cambial derivatives in both transgenic and wild-type lines (Fig. 5). The level of IAA was highest in the cambial meristem and decreased toward both xylem and phloem. In the transition zone between expanding and maturing xylem, the level of IAA was raised locally. No major differences were found in the radial pattern of IAA distribution between the wild type and the transgenic lines. However, both the total amount of IAA per square centimeter of stem area and the peak concentration of IAA in the cambial zone cells were higher in the two transgenic lines than in the wild type (Fig. 5). Comparison of the radial distribution pattern of IAA and the GUS expression pattern (Fig. 2C) of the transgenic lines revealed that the co-localized expression of the iaaM and uidA genes also resulted in co-localized increase in the levels of IAA and GUS activity.
Figure 5.
Radial distribution pattern of IAA across the cambial region in the wild type and the transgenic lines C and G. Three replicate trees were analyzed in each line. Each column represents a single tangential 30-μm-thick section, and its relative composition of different tissue types. The amount of IAA per square centimeter of a section is indicated with a black dot. The radial width of the xylem (rxylem) is indicated for each tree. The total amount of IAA per square centimeter stem area (IAAtot), indicated for each tree, was approximated from the integrated area under the gradient. Ph, Phloem; CZ, cambial zone; EX, expanding xylem; MX, maturing and mature xylem.
Anatomical Characterization of the Stem Tissues
Stem anatomy was characterized at the base of the wild type and transgenic lines C and G by measuring radial widths of the different tissue types of the stem and morphological parameters of xylem fibers, vessel elements, cambial zone cells, sieve tubes, and companion cells (Table II). No statistically significant differences were found in any of the measured parameters between the transgenic lines and the wild type.
Table II.
Stem anatomy in the wild type and in the transgenic lines C and G
| Anatomical Characteristics | Unit | Wild Type | Transgenic C | Transgenic G |
|---|---|---|---|---|
| Radial width of stem tissues | ||||
| Cortex | μm | 336 ± 29 | 316 ± 21 | 335 ± 16 |
| Phloem | μm | 351 ± 41 | 369 ± 16 | 433 ± 27 |
| Xylem | mm | 4.14 ± 0.31 | 4.16 ± 0.17 | 3.90 ± 0.14 |
| Phloem/xylem ratio | 0.085 ± 0.010 | 0.089 ± 0.004 | 0.111 ± 0.009 | |
| Cell morphology | ||||
| Cambial zone cells, no. | 12.2 ± 1.1 | 12.3 ± 0.41 | 9.6 ± 0.40 | |
| Sieve tubes, radial diameter | μm | 18.8 ± 1.1 | 16.9 ± 0.67 | 18.1 ± 0.45 |
| Companion cells, length | μm | 44.2 ± 2.1 | 41.5 ± 0.65 | 43.5 ± 1.43 |
| Xylem fibers, length | μm | 519 ± 10 | 516 ± 8.8 | 521 ± 17 |
| Xylem fibers, lumen area | μm2 | 116 ± 2.0 | 112 ± 6.5 | 120 ± 7.3 |
| Xylem vessel elements, length | μm | 326 ± 7.3 | 318 ± 6.3 | 320 ± 7.4 |
| Xylem vessel elements, lumen area | μm2 | 1,274 ± 62 | 1,216 ± 32 | 1,219 ± 41 |
All measurements, except for fiber and vessel element length, were done on microscopic sections taken from the base of the stem in one section per tree. Cortex, phloem, sieve tubes, and companion cells were measured in radial sections, while the other measurements were done in transverse sections. The lengths of the fibers and the vessel elements were measured in macerate samples from the outermost xylem of the basal stem sample. Radial widths of different stem tissues and the number of cambial zone cells were measured in 10 cell files. Sieve tubes and companion cells and the lumen area of xylem fibers and vessels were measured in images from the mature phloem and mature xylem with a computer-assisted image analysis system. All parameters were measured in five trees per line, except the lengths of the xylem fibers and vessel elements, which were measured in four trees per line. Values are means ± se.
DISCUSSION
Localized expression of transgenes is necessary when studying their role in specific processes of plant growth and development. We have expressed the bacterial iaa genes under the control of the cambial-region-specific A. rhizogenes rolC promoter. Expression of the rate-limiting IAA-biosynthetic iaaM gene was detected by using a linked reference gene, uidA, as a coordinately expressed control. The results show that a reporter gene can be utilized to localize expression of a transgene that is linked on the same T-DNA. In situ hybridization of the iaaM gene and histochemical staining for GUS activity confirmed that the expression of the linked genes iaaM and uidA was co-localized in the cambial region tissues of the mature stem (Fig. 2, C and D). Therefore, when linked to the uidA gene, the expression pattern of the iaaM gene, and thus the de novo production of IAA, could be accurately predicted by the GUS expression pattern. In contrast to expression analysis by promoter-GUS fusions in a separate set of transgenic plants, this approach takes into consideration possible effects that the expression of the actual transgene might have on the activity of the promoter used, and is clearly preferred when studying tissue-specific expression and function of a transgene.
Linkage to a reporter gene also allowed estimation of the level of expression from the iaaM gene by means of the quantitative GUS assay. Six out of eight transgenic hybrid aspen lines showed a coordinate expression of the iaaM and uidA genes (Fig. 3) and a good correlation between GUS activity and IAA level in the cambial region tissues (Fig. 4). However, no coordination was observed in line O. This could reflect epigenetic silencing of the uidA gene due to, for example, methylation of the promoter or to a local modification of the chromatin structure (Assaad et al., 1993; Vaucheret et al., 1998). Similar effects might also have induced the minor deviation from coordinate expression found in line H (Fig. 3). However, deviations from the coordinate expression in these lines coincided with the presence of more than one insert of T-DNA from the vector p812C1C (Fig. 1). Even though an increase in the copy number of T-DNA has occasionally been reported to elevate the level of transgene expression (Gendloff et al., 1990; Hobbs et al., 1993; Voelker et al., 1996), it more often results in an increased frequency of transgene silencing (Jones et al., 1987; Linn et al., 1990; Mittelsten Scheid et al., 1991; Hobbs et al., 1993). Therefore, we suggest that the deviation from coordinate expression of the iaaM and uidA genes in lines O and H was due to the presence of several T-DNA inserts. These results indicate that reporter-gene activity can predict the level of expression of a linked transgene when the T-DNA containing the two genes is present as a single copy. This approach is particularly attractive when studying genes such as the IAA-biosynthetic genes, in which the product is not easily detected.
Based on the expression pattern of both iaaM and uidA genes, de novo production of IAA from the IAA-biosynthetic genes was expected in the cambial meristem, its expanding derivatives, and the phloem of transgenic hybrid aspen stems (Fig. 2, C and D). This was confirmed by using high-sensitivity mass spectrometry to visualize the radial distribution pattern of IAA in transgenic lines C and G. The level of IAA was increased about 35% throughout the cambial region tissues of the stem, and there were no obvious differences in the radial distribution pattern of IAA between the different lines (Fig. 5). De novo production of IAA in stem tissues of transgenic trees was also demonstrated in decapitated plants, which displayed an inhibition of axillary bud release typically observed in response to elevated auxin levels (Table I). Earlier work in tobacco had demonstrated the importance of conjugation in controlling homeostasis of IAA in IAA-overexpressing transgenics (Sitbon et al., 1993). In our case, the increase in IAA conjugates constituted only a small portion of the total IAA pool (see “Results”). This is consistent with earlier findings of insignificant IAA-conjugate levels in stem tissues of hybrid aspen (Tuominen et al., 1995) and Scots pine (Sundberg et al., 1990). Other pathways of IAA metabolism (Tuominen et al., 1994) may be involved in controlling homeostasis of IAA in cambial region tissues of these transgenic hybrid aspen trees.
The tissue-specific overproduction of IAA resulted in plants with alterations in IAA levels that were physiologically relevant. Interestingly, despite an increase in IAA concentration of 35% to 40% in the cambial meristem and the expanding cells, and despite the general phenotypic effects, no effect could be observed in the rate of xylem cell production or xylem morphology. Although the stimulating effect of exogenous IAA on cambial growth processes has repeatedly been demonstrated, an understanding of the relationship between endogenous IAA concentration and xylem growth has been lacking. In Scots pine, however, the radial distribution pattern of IAA across cambial tissues, rather than its actual concentration in the meristem, was recently found to be important in growth control (Uggla et al., 1998). This observation supports the hypothesis that IAA functions as a positional signal and stimulates growth by controlling the size of the cambial meristem (Uggla et al., 1996).
Further evidence for a role of IAA in positional signaling during xylem development comes from our previous study on hybrid aspen, where the Agrobacterium iaa genes were expressed under the control of the mannopine synthase 1′2′ promoter (Tuominen et al., 1997). In these trees, the IAA content of the stem tissues was not altered, but a change in the radial distribution of IAA was related to alterations in the pattern of xylem development and xylem cell morphology. Earlier results from transgenic tobacco and petunia expressing the iaa genes are more difficult to interpret because the IAA concentration and distribution pattern across the cambial region are not known. However, a moderate overproduction of IAA in tobacco, as measured in whole internodes, did not increase xylem production, whereas a several-fold increase in IAA resulted in an overall reduction in growth rate, including xylem production (Sitbon et al., 1992a). In contrast, transgenic petunia with a 10-fold increase in IAA concentration, as measured in leaves, was claimed to increase xylem production (Klee et al., 1987). Both in tobacco and petunia, high non-specific overproduction of IAA also resulted in aberrant phenotypes indicating a distorted IAA balance.
The localized expression of IAA-biosynthetic transgenes in this study maintained the IAA distribution across cambial tissues and did not alter the developmental pattern or morphology of the cambial derivatives, which is consistent with a function of IAA in pattern formation. The increase in IAA concentration observed here, coincident with a conserved pattern of distribution, did not affect the cell cycling rate or xylem cell expansion, but does not exclude a role of absolute IAA concentration in cambial growth processes.
MATERIALS AND METHODS
Construction of the Vector p812C1C
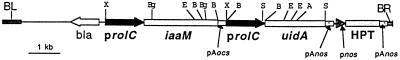
The IAA-biosynthetic iaaM gene from Agrobacterium tumefaciens T-DNA, and the uidA gene were linked in tandem, both under the control of the Agrobacterium rhizogenes rolC promoter to produce the plant gene expression vector p812C1C (Fig. 6).
Figure 6.
Schematic drawing of the T-DNA of the binary vector p812C1C. prolC, rolC promoter; iaaM, Trp mono-oxygenase gene; uidA, β-glucuronidase gene; HPT, hygromycin phosphotransferase gene; bla, ampicillin resistance gene; pAnos and pAocs, polyadenylation signals from A. tumefaciens T-DNA nopaline synthase and octopine synthase; pnos, nopaline synthase promoter; BL, border left; BR, border right. Selected restriction enzyme sites are shown for XbaI (X), SacI (S), BglII (Bg), EcoRV (E), BamHI (B), and AseI (A).
The iaaM fragment (Sitbon et al., 1992a) was linked with a BclI-linker and subcloned into a modified pUC19 containing a BclI site instead of BamHI site in the multiple cloning site. The 1,053-bp-long untranslated 3′ region of the iaaM gene fragment was removed by partial digestion with BamHI. An octopine synthase (ocs) polyadenylation signal from the plasmid pPCV6NF (Koncz et al., 1989) was ligated as a BamHI-XbaI fragment at the 3′ end of the iaaM gene, and the rolC promoter from the plasmid p812rolC (Nilsson et al., 1996) was ligated as a BamHI fragment at the 5′ end of the iaaM gene. The plasmid was thereafter digested with EcoRI, an XbaI linker was added, and the rolC:iaaM:pAocs fragment was excised by digestion with XbaI. The excised fragment was inserted into the XbaI site next to the rolC:uidA:pAnos construct of the disarmed binary vector p812rolC (Nilsson et al., 1996), resulting in the vector p812C1C. This was transformed into A. tumefaciens strain GV3101 (pMP90RK; Koncz and Schell, 1986) by electroporation (Nilsson et al., 1992).
Plant Transformation and Cultivation
Hybrid aspen (Populus tremula L. × Populus tremuloides Michx.) was transformed and regenerated according to the method of Nilsson et al. (1992). As a first step, hybrid aspen was transformed with an A. tumefaciens strain carrying the plant expression vector pPUV7022 harboring the second IAA-biosynthetic gene from A. tumefaciens T-DNA, indoleacetamid hydrolase (iaaH), under the control of cauliflower mosaic virus 35S promoter (Tuominen et al., 1997). After regeneration, one line with a single T-DNA insert from the vector pPUV7022 and a strong expression of the iaaH gene was selected for transformation with the vector p812C1C. Regeneration of these double transformants was achieved by selection for both kanamycin (resistance conferred by vector pPUV7022) and hygromycin (resistance conferred by vector p812C1C). The vector p812C1C was also transformed into wild-type hybrid aspen as a control.
The transgenic hybrid aspen lines were propagated in sterile culture in parallel with wild-type plants. Rooted plantlets were potted in mineral wool, acclimatized, and transferred to a greenhouse with an 18-h photoperiod, a temperature of 22°C:17°C (day:night), and a relative humidity of at least 70%. The natural daylight was supplemented with light from metal halogen lamps (HQI-TS 400-W/DH, Osram, Munich). Ample watering with a complete nutrient solution (Ingestad, 1970) was done daily.
Phenotypic Characterization and Sampling Procedure
The following characteristics were measured in each tree: height, basal stem diameter, length of internodes adjacent to fully expanded leaves, and length and width of fully expanded leaves. The main stem was decapitated 20 cm from the base and divided into segments for further analysis according to the following: 20 to 30 cm from the base for histochemical GUS analysis and anatomical analysis; 30 to 40 cm from the base for RNA analysis; 40 to 41 cm from the base for quantitative GUS analysis; and 41 to 54 cm from the base for IAA analysis. The samples intended for microscopy were trimmed and fixed in 10% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol, while the other stem segments were quickly frozen in liquid N2 and kept at −70°C. Young leaves were also sampled and frozen in liquid N2. The decapitated plants were cultivated further in the greenhouse under the same conditions as before. After 30 d, the number of released axillary buds was recorded for each tree.
Southern- and Northern-Blot Analysis
Chromosomal DNA for the Southern-blot analysis was isolated from young leaves (Doyle and Doyle, 1990). Ten micrograms of DNA was digested with the restriction enzymes XbaI, EcoRV, or SacI, separated on a 0.8% (w/v) agarose gel, and blotted onto a Hybond-N membrane (Amersham, Little Chalfont, UK) according to the manufacturer's instructions.
Total RNA for the northern-blot analysis was isolated from cambial region tissues of the stem using a plant total RNA kit (Rneasy, Qiagen, Hilden, Germany). The tissues were obtained by peeling the bark and scraping the exposed surfaces with a scalpel. Microscopic investigation showed that the sample contained the cambial meristem, radially expanding xylem elements, some xylem elements undergoing secondary wall thickening, developing phloem, plus part of the mature phloem. For each tissue sample, 15 μg of RNA was separated on a formaldehyde agarose gel (Sambrook et al., 1989), and blotted onto a Hybond-N membrane.
Hybridization of the Southern and northern blots were performed as described by Church and Gilbert (1984). An internal 1,697-bp BglII-fragment from the iaaM gene, a 2,093-bp BamHI-fragment from the iaaH gene spanning the whole coding sequence, or an internal 968-bp BamHI-AseI fragment from the uidA gene were labeled with [32P]dCTP in a random-primed reaction to a high specific activity and used as probes. A 186-bp fragment of an actin gene from Populus trichocarpa cv Trichobel was also labeled and used as a heterologous probe to estimate differences in sample loading. Hybridization signals were detected by autoradiography.
IAA Measurements
IAA was quantified in cambial region tissues obtained as described above. The tissues were scraped directly into liquid N2 and homogenized in conical 10-mL test tubes with a metal pestle connected to an electrical drill. The samples were extracted at 4°C for 1 h in 3 mL of sodium phosphate buffer, pH 7.0, containing 0.02% (w/v) of sodium diethyldithiocarbamate as an antioxidant and 1 μg of [13C6]IAA (Cambridge Isotope Laboratories, Woburn, MA) per gram fresh weight as an internal standard. After extraction, the pH was adjusted to approximately 2.7 with 120 μL of 1 m HCl, and the samples were slurried with 60 mg of XAD-7 (Serva, Heidelberg). The XAD-7 was washed with 2 × 3 mL of 1% (v/v) acetic acid and eluted with 2× 2 mL of dichloromethane. The samples were evaporated to dryness, derivatized, and analyzed by the isotope dilution technique and gas chromatography-selected reaction monitoring-mass spectrometry as described in Edlund et al. (1995). For quantification of IAA conjugates, the extract was divided into two portions and the concentration of IAA conjugates (hydrolyzable IAA) was calculated as the difference between total and free IAA. From one portion, free IAA was determined as described above. Total IAA was measured in the other portion by subjecting the extract to hydrolysis in 7 n NaOH at 100°C for 3 h in a N2 atmosphere, neutralizing with HCl, and quantifying as described above.
The radial distribution pattern of IAA across the cambial region tissues was determined as previously described (Uggla et al., 1996). Stem pieces containing part of the mature xylem and all of the extraxylary tissues were trimmed to approximately 1.3 (tangentially) × 10 (vertically) × 10 mm (radially). Thirty-micrometer-thick tangential cryosections were obtained across the cambial region tissues. IAA was not measured in tissues from the later stages of phloem differentiation due to the formation of phloem fibers that interfered with the collection of intact tangential sections. The radial position of the tangential sections was determined in cross-sections sampled after every third tangential section. Endogenous IAA was measured in each tangential section. For each sample, 100 pg of [13C6]IAA was included as an internal standard, and the samples were extracted and analyzed for IAA (Edlund et al., 1995).
Anatomical Investigation
The fixed samples were dehydrated in an ascending series of acetone and embedded in a methacrylate resin (S. Fink, unpublished results). Transverse and radial sections were obtained at 3 μm with a microtome (model 2065, Leica, Nussloch, Germany) using a diamond knife. The sections were stained polychromatically with successive incubations in 0.1% (w/v) acriflavin/3% (w/v) safranin O, 1% (w/v) auramin O, and 2% (w/v) methylene blue. The sections were mounted in Eukitt (Thoma, Freiburg, Germany) and observed under a light microscope (Axiophot, Carl Zeiss, Oberkochen, Germany).
The radial widths of cortex, phloem and xylem and the number of cambial zone cells were measured under the microscope in 10 radial cell files on one section per tree. Radial width was measured with an ocular measuring scale. The cortex and phloem width was measured on radial sections, and the xylem width and number of cambial zone cells was measured on transverse sections. Transition from cortex to phloem was defined by the appearance of sieve tubes. Cambial zone cells were defined as cells that had thin cell walls and were not radially expanded.
The cross-sectional lumen area of xylem fibers and vessels, the radial diameter of phloem sieve tubes, and the length of phloem companion cells were measured using a computer-assisted image analysis system described previously (Tuominen et al., 1997). Xylem fibers and vessels were measured automatically in three images taken from the most recently matured xylem of one transverse section per tree. The radial diameter of sieve tubes and the length of companion cells were measured manually in images taken from the mature phloem of one radial section per tree. On average, 2,800 fibers, 150 vessels, 35 sieve tubes, and 50 companion cells were measured in each section.
The lengths of xylem fibers and vessel elements were measured in a sample from the outermost mature xylem of the basal stem sample. The xylem sample was macerated by boiling in 50% (v/v) acetic acid and 4.5% (v/v) hydrogen peroxide for 16 h. A minimum of 100 fibers and 50 vessel elements were measured with a computer-assisted image analysis system (Visor, Rimbo, Sweden) in each sample. The length of the vessel elements was defined as the distance between the midpoint of the perforation plate at each end.
Histochemical and Quantitative GUS Analysis
GUS activity was localized histochemically in 2-mm-thick hand-cut sections excised from the stem of selected transgenic lines and the wild type as a control. After cutting, the pieces were immersed in cold 90% (w/v) acetone to facilitate penetration of 5-bromo-4-chloro-3-indolyl glucuronide (X-gluc), the substrate for GUS (Hawkins et al., 1997; Regan et al., 1999). Samples were then rinsed with water and incubated from 2 h to overnight at 37°C in a medium containing 1 mm X-gluc, 1 mm K3Fe(CN)6, 1 mm K4Fe(CN)6, 50 mm sodium phosphate buffer (pH 7.0), and 0.1% (v/v) Triton X-100. The samples were then rinsed with water, dehydrated to 50% (v/v) ethanol, fixed for 10 min in 5% (v/v) formaldehyde, 5% (v/v) acetic acid, and 50% (v/v) ethanol, and cleared in 100% (v/v) ethanol. Once cleared, the samples were rehydrated, frozen, and transverse sections were cut at 30 to 50 μm with a cryomicrotome (model HM 505E, Microm Laborgeräte, Walldorf, Germany). The sections were mounted in water for microscopic evaluation.
Quantitative analysis for GUS activity was done in extraxylary tissues obtained from a 0.5-cm-long piece of the stem. The samples were prepared and analyzed according to the fluorescent method (Jefferson et al., 1987), using a fluorometer (model TKO 100, Hoefer Scientific Instruments, San Francisco). Enzyme activity in each sample was determined as a mean of three replicate measurements, and expressed against the protein content of the extract (Bradford, 1976).
In Situ Hybridization
The expression pattern of the iaaM gene was localized according to the method of Regan et al. (1999). mRNA was cryo-immobilized by fast freezing and freeze substitution, and the tissue was embedded in a methacrylate resin for high resolution analysis. Two-micrometer sections were hybridized with an antisense iaaM digoxigenin-labeled riboprobe prepared according to manufacturer's instructions (Boehringer Mannheim, Basel).
ACKNOWLEDGMENTS
The authors wish to thank Karin Waldmann and Kjell Olofsson for technical assistance, Dr. Antje Rohde for the gift of the P. trichocarpa actin fragment, Dr. Nigel Chaffey for providing the method for maceration of the xylem, and Anneli Stenberg for initial help with the vector construction.
Footnotes
This work was supported by the Swedish Council for Forestry and Agricultural Research, the Swedish Natural Sciences Research Council, Foundation for Strategic Research, the Academy of Finland (to H.T.), and European Commission (COST E6, to L.P.).
LITERATURE CITED
- An G. Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol. 1986;81:86–91. doi: 10.1104/pp.81.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Tucker KL, Signer ER. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol. 1993;22:1067–1085. doi: 10.1007/BF00028978. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- Edlund A, Eklöf S, Sundberg B, Moritz T, Sandberg G. A microscale technique for gas chromatography-mass spectrometry measurements of picogram amounts of indole-3-acetic acid in plant tissues. Plant Physiol. 1995;108:1043–1047. doi: 10.1104/pp.108.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendloff EH, Bowen B, Buchholz WG. Quantitation of chloramphenicol acetyl transferase in transgenic tobacco plants by ELISA and correlation with gene copy number. Plant Mol Biol. 1990;14:575–583. doi: 10.1007/BF00027503. [DOI] [PubMed] [Google Scholar]
- Hawkins S, Samaj J, Lauvergeat V, Boudet A, Grima-Pettenati J. Cinnamyl alcohol dehydrogenase: identification of new sites of promoter activity in transgenic poplar. Plant Physiol. 1997;113:321–325. doi: 10.1104/pp.113.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs SLA, Warkentin TD, DeLong CMO. Transgene copy number can be positively or negatively associated with transgene expression. Plant Mol Biol. 1993;21:17–26. doi: 10.1007/BF00039614. [DOI] [PubMed] [Google Scholar]
- Ingestad T. A definition of optimum nutrient requirements in birch seedlings. Physiol Plant. 1970;23:1127–1138. [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Gilbert DE, Grady KL, Jorgensen RA. T-DNA structure and gene expression in petunia plants transformed by Agrobacterium tumefaciens C58 derivatives. Mol Gen Genet. 1987;207:478–485. [Google Scholar]
- Klee HJ, Horsch RB, Hinchee MA, Hein MB, Hoffman NL. The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Genes Dev. 1987;1:86–96. [Google Scholar]
- Klee HJ, Lanahan MB. Transgenic plants in hormone biology. In: Davies PJ, editor. Plant Hormones Physiology, Biochemistry, and Molecular Biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 340–353. [Google Scholar]
- Koncz C, Martini N, Mayerhofer R, Koncz-Kalman Z, Körber H, Redei GP, Schell J. High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci USA. 1989;86:8467–8471. doi: 10.1073/pnas.86.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Linn F, Heidmann I, Saedler H, Meyer P. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol Gen Genet. 1990;222:329–336. doi: 10.1007/BF00633837. [DOI] [PubMed] [Google Scholar]
- Little CHA, Pharis RP. Hormonal control of radial and longitudinal growth in the tree stem. In: Gartner BL, editor. Plant Stems: Physiology and Functional Morphology. San Diego: Academic Press; 1995. pp. 281–319. [Google Scholar]
- Little CHA, Savidge RA. The role of plant growth regulators in forest tree cambial growth. Plant Growth Regul. 1981;6:137–169. [Google Scholar]
- Matzke AJM, Matzke MA. Position effects and epigenetic silencing of plant transgenes. Curr Opin Plant Biol. 1998;1:142–148. doi: 10.1016/s1369-5266(98)80016-2. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O, Paszkowski J, Potrykus I. Reversible inactivation of a transgene in Arabidopsis thaliana. Mol Gen Genet. 1991;228:104–112. doi: 10.1007/BF00282454. [DOI] [PubMed] [Google Scholar]
- Nagy F, Morelli G, Fraley RT, Rogers SG, Chua N-H. Photoregulated expression of a pea rbcS gene in leaves of transgenic plants. EMBO J. 1985;12:3063–3068. doi: 10.1002/j.1460-2075.1985.tb04046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson O, Aldén T, Sitbon F, Little CHA, Chalupa V, Sandberg G, Olsson O. Spatial pattern of cauliflower mosaic virus 35S promoter-luciferase expression in transgenic hybrid aspen trees monitored by enzymatic assay and non-destructive imaging. Transgenic Res. 1992;1:209–220. [Google Scholar]
- Nilsson O, Little CHA, Sandberg G, Olsson O. Expression of two heterologous promoters, Agrobacterium rhizogenes rolC and cauliflower mosaic virus 35S, in the stem of transgenic hybrid aspen plants during the annual cycle of growth and dormancy. Plant Mol Biol. 1996;31:887–895. doi: 10.1007/BF00019475. [DOI] [PubMed] [Google Scholar]
- Nilsson O, Tuominen H, Sundberg B, Olsson O. The Agrobacterium rhizogenes rolB and rolC promoters are expressed in pericycle cells competent to serve as root initials in transgenic hybrid aspen. Physiol Plant. 1997;100:456–462. [Google Scholar]
- Peach C, Velten J. Transgene expression variability (position effect) of CAT and GUS reporter genes driven by linked divergent T-DNA promoters. Plant Mol Biol. 1991;17:49–60. doi: 10.1007/BF00036805. [DOI] [PubMed] [Google Scholar]
- Regan S, Bourquin V, Tuominen H, Sundberg B. Accurate and high resolution in situ hybridization analysis of gene expression in secondary stem tissues. Plant J. 1999;19:363–369. doi: 10.1046/j.1365-313x.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- Rotino GL, Perri E, Zottini M, Sommer H, Spena A. Genetic engineering of parthenocarpic plants. Nat Biotechnol. 1997;15:1398–1401. doi: 10.1038/nbt1297-1398. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger M, Daubert S, Goodman RM. Characteristics of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol. 1990;14:433–443. doi: 10.1007/BF00028779. [DOI] [PubMed] [Google Scholar]
- Sitbon F, Hennion S, Sundberg B, Little CHA, Olsson O, Sandberg G. Transgenic tobacco plants coexpressing the Agrobacterium tumefaciens iaaM and iaaH genes display altered growth and indoleacetic acid metabolism. Plant Physiol. 1992a;99:1062–1069. doi: 10.1104/pp.99.3.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitbon F, Little CHA, Olsson O, Sandberg G. Correlation between the expression of T-DNA IAA biosynthetic genes from developmentally regulated promoters and the distribution of IAA in different organs of transgenic tobacco. Physiol Plant. 1992b;85:679–688. [Google Scholar]
- Sitbon F, Östin A, Sundberg B, Olsson O, Sandberg G. Conjugation of indole-3-acetic acid (IAA) in wild-type and IAA-overproducing transgenic tobacco plants, and identification of the main conjugates by frit-fast atom bombardment liquid chromatography-mass spectrometry. Plant Physiol. 1993;101:313–320. doi: 10.1104/pp.101.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam M, Mol JNM, Kooter JM. The silence of genes in transgenic plants. Ann Bot. 1997;79:3–12. [Google Scholar]
- Sundberg B, Little CHA, Cui K. Distribution of indole-3-acetic acid and the occurrence of its alkali-labile conjugates in the extraxylary region of Pinus sylvestris stems. Plant Physiol. 1990;93:1295–1302. doi: 10.1104/pp.93.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg B, Uggla C, Tuominen H. Cambial growth and auxin gradients. In: Savidge R, Barnett J, Napier R, editors. Cell and Molecular Biology of Wood Formation. Oxford: BIOS Scientific Publishers; 2000. (in press) [Google Scholar]
- Tuominen H, Östin A, Sandberg G, Sundberg B. A novel metabolic pathway for indole-3-acetic acid in apical shoots of Populus tremula (L.) × Populus tremuloides (Michx.) Plant Physiol. 1994;106:1511–1520. doi: 10.1104/pp.106.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H, Puech L, Fink S, Sundberg B. A radial concentration gradient of indole-3-acetic acid is related to secondary xylem development in Populus. Plant Physiol. 1997;115:577–585. doi: 10.1104/pp.115.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuominen H, Sitbon F, Jakobsson C, Sandberg G, Olsson O, Sundberg B. Altered growth and wood characteristics in transgenic hybrid aspen expressing Agrobacterium tumefaciens T-DNA indoleacetic acid-biosynthetic genes. Plant Physiol. 1995;109:1179–1189. doi: 10.1104/pp.109.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla C, Mellerowicz E, Sundberg B. Indole-3-acetic acid controls cambial growth by positional signaling in Pinus sylvestris (L.) Plant Physiol. 1998;117:113–121. doi: 10.1104/pp.117.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uggla C, Moritz T, Sandberg G, Sundberg B. Auxin as a positional signal in pattern formation in plants. Proc Natl Acad Sci USA. 1996;93:9282–9286. doi: 10.1073/pnas.93.17.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Béclin C, Elmayan T, Feuerbach F, Godon C, Morel J-B, Mourrain P, Palaqui J-C, Vernhettes S. Transgene-induced gene silencing in plants. Plant J. 1998;16:651–659. doi: 10.1046/j.1365-313x.1998.00337.x. [DOI] [PubMed] [Google Scholar]
- Voelker TA, Hayes TR, Cranmer AM, Turner JC, Davies HM. Genetic engineering of a quantitative trait: metabolic and genetic parameters influencing the accumulation of laurate in rapeseed. Plant J. 1996;9:229–241. [Google Scholar]