Abstract
Non-alcoholic fatty liver disease (NAFLD) is the most common liver disorder in developed countries [1]. A subset of individuals with NAFLD progress to non-alcoholic steatohepatitis (NASH), an advanced form of NAFLD which predisposes individuals to cirrhosis, liver failure and hepatocellular carcinoma. The current gold standard for NASH diagnosis and staging is based on histological evaluation, which is largely semi-quantitative and subjective. To address the need for an automated and objective approach to NASH detection, we combined Raman micro-spectroscopy and machine learning techniques to develop a classification model based on a well-established NASH mouse model, using spectrum pre-processing, biochemical component analysis (BCA) and logistic regression. By employing a selected pool of biochemical components, we identified biochemical changes specific to NASH and show that the classification model is capable of accurately detecting NASH (AUC=0.85–0.87) in mice. The unique biochemical fingerprint generated in this study may serve as a useful criterion to be leveraged for further validation in clinical samples.
Keywords: non-alcoholic fatty liver disease, non-alcoholic steatohepatitis, Raman micro-spectroscopic imaging, biochemical component analysis, model fitting
Graphical abstract
1. Introduction
As one of the leading causes of chronic liver disease worldwide, non-alcoholic fatty liver disease (NAFLD) is estimated to afflict 20–30 % of the general population in Western countries [2]. While most patients are histologically categorized to have relatively benign simple steatosis, a subset of NAFLD patients eventually progress to non-alcoholic steato-hepatitis (NASH), histologically characterized by the presence of hepatocellular steatosis [3] and necro-inflammatory reactions [4]. In many instances, NASH is an indication of a systemic disorder, such as type 2 diabetes, metabolic syndrome or hyperlipidemia [5]. With disease progression in patients with advanced NASH, extensive liver fibrosis is associated with an increased risk for developing cirrhosis, liver failure, and/or hepatocellular carcinoma (HCC) [6].
Clinically, the histopathological evaluation of liver biopsies is currently the gold standard for NASH diagnosis. However, significant variation exists amongst pathologists in the definition of NASH and an unequivocal agreement has not been reached. Additionally, there is also inter-observer variation in the diagnosis [7]. Besides NASH diagnosis, efforts have also been directed towards the development of a NAFLD grading system that would be sensitive to underlying changes in the disease process, such as those occurring during the course of natural disease progression to NASH or during therapeutic interventions to treat NASH. While histological scoring systems that enable the staging of NAFLD have been developed [8], their value in routine practice remains to be determined and are therefore, primarily used only in treatment trials. In sum, whether for diagnosing NASH or for NAFLD staging, the traditional histological approach is fundamentally semi-quantitative, observer-dependent, and includes only a very limited set of pathological features.
As many diseases lead to changes in tissue composition, Raman micro-spectroscopy has emerged as a promising diagnostic tool in recent years [9], particularly for the diagnosis of cancer [10]. Besides being a label-free approach that enables multiplexing, Raman micro-spectroscopy provides a biochemical map of the tissue of interest that potentially enables the identification of spatial-temporal changes in tissue composition. Indeed, several groups have reported the use of Raman micro-spectroscopy-based techniques such as coherent Anti-Stokes Raman scattering and confocal Raman imaging for the detection of NAFLD and NASH [11–15]. These label-free approaches enable objective and quantitative measurements of lipid accumulation within the liver without the need for exogenous contrast agents. However, the intrinsic molecular vibration-based imaging techniques employed in these systems are limited to the detection of microvesicular or macro-vesicular steatosis. What has not been shown, is whether these techniques can similarly be used to detect spatio-temporal changes in other biochemical moieties in the liver during disease progression. Furthermore, while it has been shown that these systems are capable of detecting lipid droplets and characterizing the response of hepatocytes to metabolic disease, a fully automated model that can distinguish NAFLD/NASH tissue from normal tissue has not been developed; such a generalized model is needed for an improved understanding of fatty liver pathogenesis.
To address this need for a fully quantitative and observer-independent approach to NASH diagnosis (and ultimately NAFLD staging), we combined spontaneous Raman micro-spectroscopy and machine learning techniques to identify spectral signatures that are specific to NASH using the STAM™ NASH mouse model [16], a well-established murine model that recapitulates NASH progression in humans [17,18]. Using this approach, we report the development of a classification model that may be further leveraged for the development of a quantitative and objective scoring system for NASH in patients.
2. Materials and methods
2.1 Animal model and preparation of sample tissue
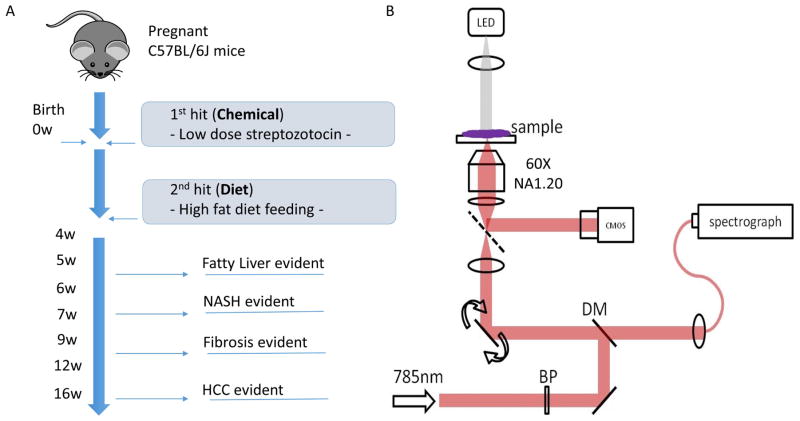
Liver tissue was obtained from the STAM™ NASH mouse model (Stelic Institute & Co., Inc.) (Figure 1). This model is generated by administering a single low dose of Streptozotocin at 2-days of age to induce insulin resistance, followed by a high-fat diet from 4-weeks of age. The entire lifespan for the mice is approximately 16 weeks during which steatohepatitis, NASH, fibrosis and HCC is expected to develop progressively over time. In this study, 42 STAM™ mice were generated and sacrificed at 6 time points with Raman data recorded with 1 section per mice. Among them, 14 were non-NASH (control) group and 28 were NASH group according to the NASH score (mathematically adding up of steatosis, ballooning and inflammation scores) given from the Supporting Figure 1. Liver tissues from both the non-NASH (control) and NASH groups were collected and snap-frozen with liquid nitrogen. The tissues were then cryo-sectioned into 20 μm-thick tissue slices and placed on quartz coverslips (ALFA43210.KJ, VWR) for Raman imaging. Every consecutive three sections were stained with H&E, Oil Red and Sirius Red.
Figure 1.
STAM mice model and imaging apparatus set up. a) Figure illustration of STAM mice model. The model was created by using both the effect of chemical toxic STZ (0 week) and high fat dietary (4 weeks) on non-genetic C57BL/6 mice with measurements conducted on the 5 different time points. A total of 42 cryo-preserved tissues were harvested from the mice. b) Schematic illustration of Raman micro-spectroscope set-up. Excitation laser was a continuous wave (CW) tunable Ti: Sapphire laser (wavelength set at 785 nm) with a frequency-doubles Nd: YAG laser (wavelength set at 532 nm) used as he pump source. The collimated beam passed through a laser line bandpass filter (BP), reflected by a dichroic mirror (DM) and focused onto the sample by a water-immersion objective lens (60X, NA=1.2). The dual-axis galvanometer mirrors were used to scan the beam over the sample. The inelastically scattered Raman signal was then delivered to the imaging spectrograph which covers Raman shift of −34 to 1894 cm−1 and the spectral resolution of 2.0 cm−1/pixel. Spectra are captured by a thermo electrically cooled charge-coupled device (CCD) and the bright field images were captured using an intensity controlled white light-emitting diode (LED) and the complementary metal-oxide semiconductor (CMOS) camera.
2.2 Evaluation of non–alcoholic steatohepatitis (NASH)
For each sample, histological slides stained separately with H&E, Oil Red and Sirius Red were reviewed by experienced pathologists with blind reading to minimize bias. The histological criteria employed to detect NASH included the presence of steatosis, inflammation and hepatocellular ballooning [7], as previously developed by Brunt et al. [19]. Scores for steatosis, inflammation and hepatocellular ballooning were recorded separately for subsequent analysis.
2.3 System setup and image acquisition
The Raman spectroscope employed in this study was based on a home-built inverted microscope [20]. A frequency-doubled Nd: YAG laser with wavelength of 532 nm (Millennia 5sJ, Spectra-physics) was used as the pump source of a CW tunable Ti: sapphire laser with wavelength of 785 nm (3900S, Spectra-Physics). The collimated beam was firstly filtered by the laser line clean-up filter (BPF, LL01-785-12.5, Semrock) and directed by a dichroic mirror (LPD01-785RU-25, Semrock) to the dual-axis galvanometer mirrors (CT-6210, Cambridge Technology). Subsequently, the beam size was adjusted by a telescope and focused at the sample plane by the water immersion objective lens (Olympus UPLSA-PO60XWIR 60X/1.20) and the laser power at the sample location was approximately 60 mW. The XY position was achieved by a micrometer-controlled stage and the z-focus was controlled by combining a piezo actuator with a differential micrometer (DRV517, Thorlabs). Collected Raman signal was delivered to the imaging spectrograph (HoloSpec f/1.8i, Kaiser Optical Systems), which was featured by high throughput and low aperture ratio. The Raman grating (HSG-785-LF, Kaiser Optical Systems) enabled a spectra shift coverage of −34 to 1894 cm−1 and spectra resolution of 2.0 cm−1/pixel. Spectra was captured using a TE-cooled CCD. An intensity controlled white LED was used as the illumination source for bright field imaging and images were captured using the CMOS camera (BCN–B050-U, MightTex). The Labview 8.2 software (National Instruments) accompanied with a data acquisition board (PCI-6251, National Instruments) was used to control the devices.
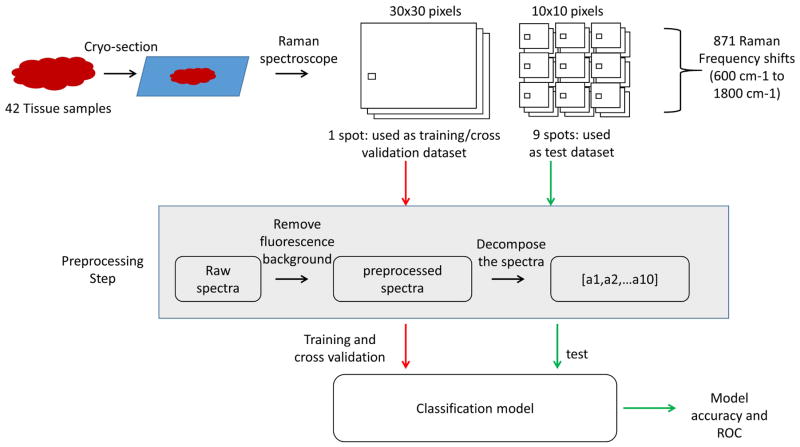
To perform tissue sample imaging, two types of data were acquired from the slides. The first type was 30×30 pixels’ high-resolution spectral images –one image was acquired from each tissue sample and used for image-based analysis. The second type was 10×10 pixels’ low-resolution spectral images –nine images were acquired from each tissue sample and used for spectral – based analysis from wide tissue area and minimize the tissue heterogeneity effect. The signal acquisition interval time was 1 second, therefore each 30×30 pixels’ spectral image and 10×10 pixels’ spectral image was acquired every 20 min and 2 min, with the dimension being 45×45 μm and 15×15 μm, respectively. The spectra of different selected chemicals were acquired when a small amount of chemical powder was put on the quartz cover slip (spectra image resolution was set as 3×3 and the signal acquisition integral time was 10 s). The nine measurements were taken from the same sample spot and then averaged into the spectra of the selected chemicals.
2.4 Spectrum processing and image analysis
The initial spectra were collected over the range of −175 cm−1 to 1843 cm−1. Since the signal from the quartz substrate is primarily in the range of 250 cm−1 to 580 cm−1 [21], spectra within the range of 600 cm−1 to 1800 cm−1 were specifically selected for analysis. Spectrum processing was then performed using MATLAB (Mathworks, Massachusetts) where the narrow spikes induced by cosmic rays were first removed. Then, as the Raman spectra of tissues typically contains Raman scattering, intrinsic tissue fluorescence and noise, background fluorescence from the tissue was estimated and removed using the adaptive minimax method [22]. Based on the method developed by Cao et al. [23], polynomial fits [24] (based on the fluorescence-to-signal (F/S) ratio) were used to minimize the residual mean square (RMS) error. The F/S ratio is defined as the maximum fluorescence divided by the maximum Raman scattering signal, where the minimum intensity value was set as zero. The proposed method, ‘adaptive minimax method’, consists of two steps. The first step involved the use of constrained and unconstrained polynomial fit for two consecutive orders, in which the order was determined by the adaptive part of the algorithm based on the F/S ratio; in the second step, the maximum value among the initial fits were used as the points for the final fit. The acquired spectrum had a better result compared with other methods using minimized RMS error and was followed by smoothing before further analysis was performed.
2.5 Chemical component decomposition and model fitting
The biochemical components analysis (BCA) is commonly used in Raman spectra analysis [25]. The assumption in this method is that the overall tissue spectra is the linear combination of the spectra belonging to each biochemical component. The BCA method estimates the contribution of each component’s whole spectrum using least square regression and this approach has been investigated at both the tissue level (for breast cancer diagnosis [26]) and cell level (study of necrotic cells [27]). To perform the BCA analysis, prior knowledge of the constituents of the tissue is required. Biochemical components linked to liver structure, including saturated lipid acid, unsaturated lipid acid, collagen, glycogen, phenylalanine, reduced glutathione, DNA, retinyl acetate and 3-nitro-L-tyrosine were purchased from Sigma Aldrich (USA) and used directly for Raman microscopy analysis. To further decompose the spectra of the sample tissue, the least square regression method was adapted [28], using a linear curve fitting function lsqnonneg in MATLAB. This function finds the non-negative coefficients of all component spectra that best fits the tissue spectra. To also generate the Raman images, the fitting coefficients of each compound were used to reconstruct the pixel intensity for both non-NASH (control) and NASH groups. A leave-one-out cross-validation method was also adopted where one sample was used as the test set to calculate its probability of being diagnosed as a NASH sample while all other samples were used for training [29]. The diagnostic model was established by using the logistic regression while its performance was evaluated based on the area under the receiver operating characteristic curve (AUC) [30] for both all tissues (with HCC) and less diseased tissues (without HCC).
2.6 K-means cluster analysis (KCA)
Using full range of the Raman spectrum as input, KCA was performed using MATLAB based program. Briefly, in this method spectra with similar profiles are grouped together as part of one cluster in an unsupervised manner. A pseudo color is assigned to a cluster and spectra are believed to have similar biochemical and molecular composition. The procedure is repeated until a stable solution is obtained. This method has been utilized and explained in several Raman mapping studies [31–33].
2.7 Statistical analysis
Data in this report were presented as average ± standard error of the mean (SEM). To compare the fitting coefficient between different groups, the initial test of difference across groups was analyzed using one-way ANOVA, The HSD correction method was then used as a post-hoc test to identify the significant differences among the different conditions using adjusted p values. p values <0.05 (*), p <0.01 (**) were considered statistically significant.
3. Results and discussion
3.1 Raman micro-spectroscopy reflects spatial distribution of biochemical moieties in tissues
Signals in the range of 600 cm−1 to 1800 cm−1 were retained and spike signals (defined as signal intensities that are beyond the recording range of the spectrograph) were excluded for further analysis. Following background subtraction and smoothing, the mean Raman spectrum of the NASH and control groups were plotted (Figure 3A). The overall spectrum of each group was obtained by averaging over all samples. Qualitatively, the signal intensity and peak location of certain Raman peaks differed between control and NASH samples. This difference was due to a change in lipid droplets and fat accumulation and subsequent cellular change within the tissue as most shifts could be attributed to peaks that were generated from non-saturated fat. There was almost no signal arising from non-saturated fat in the control group, signals were obtained for the later stage samples, suggesting that Raman micro-spectroscopy is able to detect at the least, macro-steatosis with variation in both its size and distribution during disease progression, as a characteristic feature of NASH [34,35].
Figure 3.

Raman spectrum and signal decomposition. a) The averaged Raman spectrum of non-NASH (control) and NASH groups across 871 Raman shifts. b) The Raman spectra of the selected nine compounds selected for the Raman signal decomposition. c) The name of the nine compounds. d) and e) The spectrum fitting result of the non-NASH (control) and NASH groups. The blue line represents the original spectrum and the green one represents the reconstructed spectrum using the nine selected compounds after spectrum decomposition while the red line gives the residual.
3.2 Raman spectra of NASH samples can be approximated using known biochemical components
Next, hypothesizing that the original Raman spectra can be approximated by summing up the Raman spectra from a set of known biochemical components, we selected 9 compounds that are NASH-related to recapitulate the biochemical signature characteristic of the disease. The spectrum of each individual biochemical component was vertically segregated for clarity (Figure 3B and C) [25]. The 9 compounds were selected to recapitulate the biochemical signature characteristic of the disease as they are indicative of inflammatory reactions, oxidative stress, cell viability and unsaturated fatty acids of NASH [36,37].
3-nitro-L-tyrosine is a marker of peroxynitrite formation and is a known indicator of inflammation-induced tissue damage [38]. Phospholipids, a key component of the cell membrane, has direct association with a high fat diet [39]. Representative peaks for arachidic acid, a saturated fatty acid with a 20-carbon chain, were those at 1302 cm−1 and 1446 cm−1 [40]. These two peaks were attributed to the CH2 stretching mode. The representative peaks of linoleic acid, a polyunsaturated acid, were those at 1266 cm−1 and 1655 cm−1, with abundant double bonds (C=C). Both play an important role during NASH disease progression as fat accumulates. Reduced glutathione (GSH) is an important anti-oxidant that prevents damage to important cellular components caused by reactive oxygen species and are converted into oxidized glutathione (GSSH) [41]. An increase in the GSSG-to-GSH ratio is considered an indicator of oxidative stress [42]. We also included DNA and phenylalanine as increase in these molecules may be indicative of malignancy. Cancer cells exhibit an increased uptake of amino acids such as phenylalanine, which serve as an energy source [43]. Of note, the STAM™ NASH model is known to progress to HCC over time. Indeed, tumor cells were detected in 20 % of the analyzed NASH samples. The peak at 1004 cm−1 corresponds to C–C stretching in phenylalanine [44]. The peak at 1600 cm−1 was generated from retinyl acetate, a natural form of vitamin. The Raman signals from actin, collagen and DNA were also used for compensating the spectrum from the liver tissue as they exhibit various characteristic spectra in the Raman signal [45].
By using the spectra from the selected compounds, the total Raman spectrum was further decomposed into the contributions of each biochemical component based on the original and fitted Raman spectrum of the liver tissue from both the non-NASH (control) and NASH groups (Figure 3D and E). Comparable to previous studies which used the same curve fitting approach [46], the fitting residue fluctuated between −0.08 to 0.09 with most spectral features of the original spectrum preserved. Each fitting coefficient was divided by the sum of all coefficients and converted to a percentage [25] to represent the relative contributions of the selected biochemical components spectra to the whole tissue spectrum and further analyzed.
3.3 Differences in biochemical composition between control and NASH samples
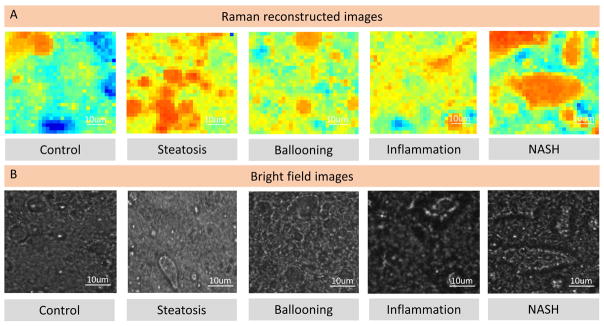
Using the fitting coefficients calculated from Raman spectrum decomposition via BCA method, the Raman reconstructed images were generated for lipid content in comparison with bright field images (Figure 4A and B), suggesting that Raman micro-spectroscopy can detect at the least, macro-steatosis with variation in both its size and distribution during disease progression, as a characteristic feature of NASH [34,35]. More compounds were mapped using such method in the Sup Figure 2 to show the bio-molecular variation for both non-NASH (control) and NASH groups. These results were further evaluated by performing K-means cluster analysis (KCA) using full range of the Raman spectrum. Three (3) cluster images were generated for both non-NASH (control) and NASH groups and compared with BCA images, Supporting Figure 2. Similar image profile of the cluster images suggest that Raman micro-spectroscopy in combination with statistical tools can be applied to facilitate objective identification of molecular changes with high spatial resolution with the onset of NASH.
Figure 4.
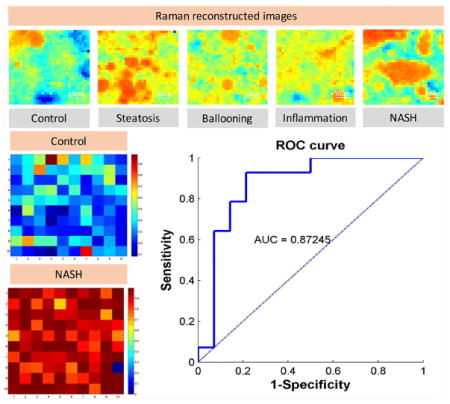
Raman reconstructed images using calculated fitting coefficients. a) Selected Raman reconstructed images using calculated fitting coefficients and b) bright field images of lipid content for non-NASH (control), steatosis, ballooning, inflammation and NASH stages.
Figure 2.
Data acquisition and processing flow. The pre-processed averaged tissue spectrums were collected from both training and testing groups for selected 871 Raman frequency shifts before being decomposed to the selected compounds’ spectrums to build a multinomial logistic regression model, which was then evaluated by Receiver operating characteristics curve (ROC) analysis.
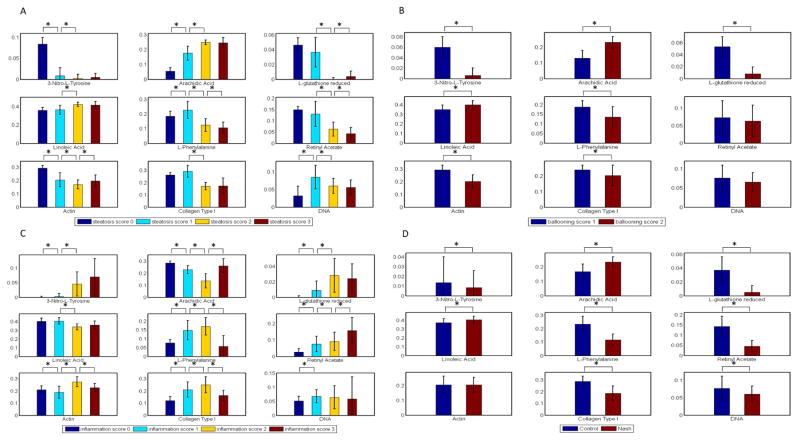
The fitting coefficients of the 9 selected biochemical components derived from spectrum decomposition for the different disease patterns, including steatosis, ballooning, inflammation and NASH detection, are shown in Figure 5A–D. For steatosis and ballooning, the increase in lipid content in the NASH samples as compared to the control samples was significant – the fitting coefficients for arachidic acid (saturated fat) and linoleic acid (unsaturated fat) increased from 8 to 25 % and 30 to 40 %, respectively. These findings corroborate with previous studies that reported similar findings, that fat accumulates during NAFLD progression towards NASH [47]. We also observed an increase in the amount of 3-nitro-L-tyrosine with increasing severity of inflammation (overall assessment of all inflammatory foci) suggesting that 3-nitro-L-tryosine may be a potential indicator for cell damage, inflammation as well as NO (nitric oxide) production in NASH.
Figure 5.
Fitting result for diseases using bar chart. a) The fitting result for the selected compounds by steatosis scoring. Figure 5b) The fitting result for the selected compounds by ballooning scoring. Figure 5c) The fitting result for the selected compounds by inflammation scoring. Figure 5d) The fitting result for the selected compounds by NASH scoring.
Comparing the NASH samples against the controls, the decrease in 3-nitro-L-tyrosine in the NASH as compared to the control samples is contradictory to what was observed based on inflammation scoring; this difference may be due to a small sample size as most of the NASH tissues were classified as mildly inflammatory during pathological evaluation. The decrease in the amount of reduced L-glutathione in the NASH samples as compared to the control group indicates the presence of increased oxidative damage in NASH [48], which has been associated with the onset of HCC [49]. Retinyl acetate is a natural form of vitamin A and a reduction in this compound typically is indicative of decreased cell turnover [50]. The decrease in this compound in the NASH samples suggests that there is rapid acceleration of cell death and decreased cell proliferation triggered by inflammation and degeneration [51]. While an increase in actin content was also observed, this difference was small and remained unchanged at later time-points. While we are unable to explain the decrease in DNA, one possibility is that the onset of NASH is associated with extensive cell death, which was the case in most of the analyzed NASH samples.
3.4 Classification model for NASH
The diagnostic plot for true negative (control) and true positive (NASH) samples with the resolution being 10×10 pixels’ size using the developed classification model is shown in Figure 6, where the different pixels represent the probability of a certain region being categorized as normal or diseased (Figure 6A and B) using logistic regression for both all tissues (with HCC) and less diseased tissues (with HCC). To further determine whether the classification model can accurately identify NASH, the receiver operating characteristic (ROC) curve was generated. The area under the ROC (AUROC) curve was found to be 0.87, suggesting that the selected biochemical compounds are indeed useful for the development of a NASH classification model. As expected, accuracy of the model from less diseased tissue is slightly lower compared to the model with HCC (end-stage disease) included. This is because an increase in the number of histological patterns incorporated or stored in the database for the learning algorithm typically increases the diagnostic power of a model. This data could potentially be used together with the multiple classifiers under the statistical framework of supervised learning, as proposed by Sowa et al. [52].
Figure 6.
Diagnostic plot and ROC graph. The diagnostic plot for a true positive and true negative sample where the pixels’ intensity represents the probability of being diagnosed as normal or diseased tissue with receiver operating characteristics curve (ROC) analysis of the classification model for a) all tissues (with HCC) and b) less diseased tissue (without HCC).
Recently, nonlinear optical microscopy imaging techniques such as stimulated Raman spectroscopy (SRS) and coherent anti-Stokes Raman scattering (CARS), have become powerful tools for label-free imaging in cells and tissues as they enable deep tissue imaging that is non-destructive, has biochemical specificity, and scalable resolution [53–55]. Previous studies have demonstrated the potential of this approach to identify patients with gradually progressive fibrotic processes such as NAFLD [56], fatty liver disease (FLD) and cirrhosis [57]. However, the translation of Raman imaging into potential clinical applications or in vivo studies requires further innovation of minimally invasive diagnostic tools [29] and more information on the spatio-temporal distribution. In this study, we further demonstrate that Raman micro-spectroscopy can similarly be used for the detection of NAFLD/NASH through quantification of the biochemical and biological changes at the cell and tissue level with spatio-temporal resolution. Using such approach, it would enable us to discover the disease pattern such as compounds’ variation and cellular change. On the other hand, since the features used in the proposed classification system are currently limited to content-based level, the model could potentially be retrained and reinforced by incorporating morphological and textural features of compound distribution at the cellular level to increase diagnostic capabilities. Further studies are ongoing in our laboratory to improve the classification methods with advanced algorithms for spectrum decomposition and different field-of-view or spatial resolution.
4. Conclusions
Being a label-free approach that enables the quantitative evaluation of spatiotemporal variations in tissue chemical composition with disease progression, Raman micro-spectroscopy is an untapped imaging modality that has the potential to greatly support the study of fatty liver disease. Elucidation of the spatio-temporal changes in chemical components comprising the Raman spectrum of liver tissue with progressing NAFLD/NASH is critically important for understanding the disease. As this study aims to create a classification model for NASH investigation in the mice sample given that the well-recognized histopathological features of NASH include hepatocellular steatosis and ballooning, mixed acute and chronic lobular inflammation, and zone 3 peri-sinusoidal fibrosis, we employed those features as part of required components to assess the diseased tissue and developed a quantitative approach to NASH detection using Raman micro-spectroscopy, coupled with least square regression and logistic regression. We showed that Raman micro-spectroscopy can accurately reveal the spatial-temporal distribution of biochemical in tissues. Subsequent analysis of the decomposed spectrum showed that the progression of NAFLD is associated mainly with an accumulation of fatty acids. We also identified 3-nitro-L-tryosine and reduced L-glutathione as potential biomarkers for inflammation and oxidative damage, respectively, by employing a machine learning-based algorithm to differentiate normal from NASH samples. This animal study also aims to identify possible markers in terms of chemical identities and/or signatures in Raman shifts. With such classification model and algorithms developed, we will adapt these on human biopsy samples and eventually correlate with non-invasive imaging or blood test markers. Though we realize that the model developed here would not be compatible with the in vivo fiber probes, such model would still be potential diagnostic tools for NASH in clinical settings.
Acknowledgments
This research was supported by the National Research Foundation (NRF), Prime Minister’s Office, Singapore, under its CREATE programme, Singapore-MIT Alliance for Research and Technology (SMART) BioSystems and Micromechanics (BioSyM) IRG, the Institute of Bioengineering and Nanotechnology, Biomedical Research Council, Agency for Science, Technology and Research (A*STAR), A*STAR, (Project Number 1334i00051); NMRC (R-185-000-294-511) and Mechanobiology Institute of Singapore (R-714-001-003-271) funding to HYU. JWK and PTCS acknowledge support from NIH 9P41EB015871-28 and Samsung Advanced Institute of Technology. This work was also supported by the Singapore Ministry of Health’s National Medical Research Council (NMRC) under its Open Fund Individual Research Grant scheme (OFIRG15nov062).
Footnotes
Supporting information for this article is available on the WWWunder https://doi.org/10.1002/jbio.201600303
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher’s website. Sup Figure 1. The properties of selected compounds for Raman reconstructed images. Processed data for the mice samples (No Raman data for red labels). Sup Figure 2a) and b) Raman reconstructed image using the fitting coefficient calculated from spectrum decomposition using BCA in comparison with multivariate K-means cluster analysis.
References
- 1.Shaker M, Tabbaa A, Albeldawi M, Alkhouri N. World Jf Gastroenterol. 2014;20:5320–5330. doi: 10.3748/wjg.v20.i18.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masarone M, Federico A, Abenavoli L, Loguercio C, Persico M. Rev Recent Clin Trials. 2014;9:126–133. doi: 10.2174/1574887109666141216111143. [DOI] [PubMed] [Google Scholar]
- 3.White DL, Kanwal F, El-Serag HB. Clin Gastroenterol Hepatol. 2012;10:1342–1359. e1342. doi: 10.1016/j.cgh.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinella ME. Jama. 2015;313:2263–2273. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 5.Cusi K. Curr Opin Endocrinol Diabetes Obes. 2009;16:141–149. doi: 10.1097/MED.0b013e3283293015. [DOI] [PubMed] [Google Scholar]
- 6.Malhotra N, Beaton MD. World J Hepatol. 2015;7:2962–2967. doi: 10.4254/wjh.v7.i30.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi Y, Fukusato T. World J Gastroenterol. 2014;20:15539–15548. doi: 10.3748/wjg.v20.i42.15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunt EM, Tiniakos DG. World J Gastroenterol. 2010;16:5286–5296. doi: 10.3748/wjg.v16.i42.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kochan K, Maslak E, Krafft C, Kostogrys R, Chlopicki S, Baranska M. J Biophotonics. 2015;8:597–609. doi: 10.1002/jbio.201400077. [DOI] [PubMed] [Google Scholar]
- 10.Kourkoumelis N, Balatsoukas I, Moulia V, Elka A, Gaitanis G, Bassukas ID. Int J Mol Sci. 2015;16:14554–14570. doi: 10.3390/ijms160714554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schie IW, Wu J, Zern M, Rutledge JC, Huser T. J Biophotonics. 2011;4:425–434. doi: 10.1002/jbio.201000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le TT, Ziemba A, Urasaki Y, Brotman S, Pizzorno G. PLoS ONE. 2012;7:e51092. doi: 10.1371/journal.pone.0051092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kochan K, Maslak E, Kostogrys R, Chlopicki S, Baranska M. Biomed Spectrosc Imaging. 2013;2:331–337. [Google Scholar]
- 14.Pirhonen J, Arola J, Sädevirta S, Luukkonen P, Karppinen S-M, Pihlajaniemi T, Isomäki A, Hukkanen M, Yki-Järvinen H, Ikonen E. PLoS ONE. 2016;11:e0147804. doi: 10.1371/journal.pone.0147804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kochan K, Maslak E, Krafft C, Kostogrys R, Chlopicki S, Baranska M. J Biophotonics. 2015;8:597–609. doi: 10.1002/jbio.201400077. [DOI] [PubMed] [Google Scholar]
- 16.Saito K, Uebanso T, Maekawa K, Ishikawa M, Taguchi R, Nammo T, Nishimaki-Mogami T, Udagawa H, Fujii M, Shibazaki Y, Yoneyama H, Yasuda K, Saito Y. Sci Rep. 2015;5:12466. doi: 10.1038/srep12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi Y, Soejima Y, Fukusato T. World J Gastroenterol. 2012;18:2300–2308. doi: 10.3748/wjg.v18.i19.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anstee QM, Goldin RD. Int J Exp Pathol. 2006;87:1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunt EM, Janney CG, Di Bisceglie AM, Neuschwander-Tetri BA, Bacon BR. Am J Gastroenterol. 1999;94:2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 20.Kang JW, Lue N, Kong CR, Barman I, Dingari NC, Goldfless SJ, Niles JC, Dasari RR, Feld MS. Biomed Opt Express. 2011;2:2484–2492. doi: 10.1364/BOE.2.002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palonpon AF, Ando J, Yamakoshi H, Dodo K, Sodeoka M, Kawata S, Fujita K. Nat Protoc. 2013;8:677–692. doi: 10.1038/nprot.2013.030. [DOI] [PubMed] [Google Scholar]
- 22.Weakley AT, Griffiths PR, Aston DE. Appl Spectrosc. 2012;66:519–529. doi: 10.1366/110-06526. [DOI] [PubMed] [Google Scholar]
- 23.Cao A, Pandya AK, Serhatkulu GK, Weber RE, Dai H, Thakur JS, Naik VM, Naik R, Auner GW, Rabah R. J Raman Spectrosc. 2007;38:1199–1205. [Google Scholar]
- 24.Lieber CA, Mahadevan-Jansen A. Appl Spectrosc. 2003;57:1363–1367. doi: 10.1366/000370203322554518. [DOI] [PubMed] [Google Scholar]
- 25.Ong YH, Lim M, Liu Q. Opt Express. 2012;20:22158–22171. doi: 10.1364/OE.20.022158. [DOI] [PubMed] [Google Scholar]
- 26.Shafer-Peltier KE, Haka AS, Fitzmaurice M, Crowe J, Myles J, Dasari RR. M S Feld J Raman Spectrosc. 2002;33:552–563. [Google Scholar]
- 27.Kunapareddy N, Freyer JP. J R Mourant J Biomed Opt. 2008;13:054002. doi: 10.1117/1.2978061. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y. H Cao Sci World J. 2013;2013:306937. doi: 10.1155/2013/306937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu S, Kang CH, Gou X, Peng Q, Yan J, Zhuo S, Cheng CL, He Y, Kang Y, Xia W, So PT, Welsch R, Rajapakse JC, Yu H. J Biophotonics. 2016;9:351–363. doi: 10.1002/jbio.201500001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanley JA, McNeil BJ. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 31.Kang JW, Singh SP, Nguyen FT, Lue N, Sung Y, So PT, Dasari RR. Sensors. Vol. 16. Basel, Switzerland: 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G, Xu XX, Lu SH, Zhang CZ, Song ZF, Zhang CP. Guang Pu Xue Yu Guang Pu Fen Xi. 2006;26:869–873. [PubMed] [Google Scholar]
- 33.Hedegaard MA, Bergholt MS, Stevens MM. J Biophotonics. 2016;9:542–550. doi: 10.1002/jbio.201500238. [DOI] [PubMed] [Google Scholar]
- 34.Tandra S, Yeh MM, Brunt EM, Vuppalanchi R, Cummings OW, Ünalp-Arida A, Wilson LA, Chalasani N. J Hepatol. 2011;55:654–659. doi: 10.1016/j.jhep.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abd El-Kader SM, El-Den Ashmawy EMS. World J Hepatol. 2015;7:846–858. doi: 10.4254/wjh.v7.i6.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogliolo L, Murrone O, Di Emidio G, Piccinini M, Ariu F, Ledda S, Tatone C. J Assist Reprod Genet. 2013;30:877–882. doi: 10.1007/s10815-013-0046-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haka AS, Sue E, Zhang C, Bhardwaj P, Sterling J, Carpenter C, Leonard M, Manzoor M, Walker J, Aleman JO, Gareau D, Holt PR, Breslow JL, Zhou XK, Giri D, Morrow M, Iyengar N, Barman I, Hudis CA, Dannenberg AJ. Anal Chem. 2016;88:2140–2148. doi: 10.1021/acs.analchem.5b03696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn B, Han BS, Kim DJ, Ohshima H. Carcinogenesis. 1999;20:1337–1344. doi: 10.1093/carcin/20.7.1337. [DOI] [PubMed] [Google Scholar]
- 39.Kahle M, Schäfer A, Seelig A, Schultheiß J, Wu M, Aichler M, Leonhardt J, Rathkolb B, Rozman J, Sarioglu H, Hauck SM, Ueffing M, Wolf E, Kastenmueller G, Adamski J, Walch A, Hrabé de Angelis M, Neschen S. Mol Metab. 2015;4:39–50. doi: 10.1016/j.molmet.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beattie JR, Bell SE, Moss BW. Lipids. 2004;39:407– 419. doi: 10.1007/s11745-004-1245-z. [DOI] [PubMed] [Google Scholar]
- 41.Couto N, Malys N, Gaskell SJ, Barber J. J Proteome Res. 2013;12:2885–2894. doi: 10.1021/pr4001948. [DOI] [PubMed] [Google Scholar]
- 42.Zitka O, Skalickova S, Gumulec J, Masarik M, Adam V, Hubalek J, Trnkova L, Kruseova J, Eckschlager T, Kizek R. Oncol Lett. 2012;4:1247–1253. doi: 10.3892/ol.2012.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Qu W, Lieberman BP, Plossl K, Kung HF. Nucl Med Biol. 2011;38:53–62. doi: 10.1016/j.nucmedbio.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajkumar BJ, Ramakrishnan V. Spectrochim Acta A Mol Biomol Spectrosc. 2002;58:1923–1934. doi: 10.1016/s1386-1425(01)00641-2. [DOI] [PubMed] [Google Scholar]
- 45.Shetty G, Kendall C, Shepherd N, Stone N, Barr H. Br J Cancer. 2006;94:1460–1464. doi: 10.1038/sj.bjc.6603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulze HG, Atkins CG, Devine DV, Blades MW, Turner RF. Appl Spectrosc. 2015;69:26–36. doi: 10.1366/14-07510. [DOI] [PubMed] [Google Scholar]
- 47.Leamy AK, Egnatchik RA, Young JD. Prog Lipid Res. 2013;52 doi: 10.1016/j.plipres.2012.1010.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Physiol Rev. 2014;94:329–354. doi: 10.1152/physrev.00040.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao R, Cheng J, Fan C, Shi X, Cao Y, Sun B, Ding H, Hu C, Dong F, Yan X. Sci Rep. 2015;5:18175. doi: 10.1038/srep18175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mukherjee S, Date A, Patravale V, Korting HC, Roeder A, Weindl G. Clin Interv Aging. 2006;1:327–348. doi: 10.2147/ciia.2006.1.4.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bushue N, Wan YJY. Adv Drug Deliv Rev. 2010;62:1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sowa JP, Atmaca Ö, Kahraman A, Schlattjan M, Lindner M, Sydor S, Scherbaum N, Lackner K, Gerken G, Heider D, Arteel GE, Erim Y, Canbay A. PloS ONE. 2014;9:e101444. doi: 10.1371/journal.pone.0101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin J, Lu F, Zheng W, Xu S, Tai D, Yu H, Huang Z. J Biomed Opt. 2011;16:116024. doi: 10.1117/1.3655353. [DOI] [PubMed] [Google Scholar]
- 54.Lu F, Zheng W, Lin J, Huang Z. Appl Phys Lett. 2010;96:133701. [Google Scholar]
- 55.Hu CR, Slipchenko MN, Wang P, Wang P, Lin JD, Simpson G, Hu B, Cheng JX. Opt Lett. 2013;38:1479–1481. doi: 10.1364/OL.38.001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sowa JP, Heider D, Bechmann LP, Gerken G, Hoffmann D, Canbay A. PloS ONE. 2013;8:e62439. doi: 10.1371/journal.pone.0062439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nanyue W, Youhua Y, Dawei H, Bin X, Jia L, Tongda L, Liyuan X, Zengyu S, Yanping C, Jia W. Sci World J. 2015;2015:859192. doi: 10.1155/2015/859192. [DOI] [PMC free article] [PubMed] [Google Scholar]