SUMMARY
Cancer progression involves the gradual loss of a differentiated phenotype and acquisition of progenitor and stem cell-like features. Here, we provide novel stemness indices for assessing the degree of oncogenic dedifferentiation. We used an innovative one-class logistic regression machine learning algorithm (OCLR) to extract transcriptomic and epigenetic feature sets derived from non-transformed pluripotent stem cells and their differentiated progeny. Using OCLR, we were able to identify previously undiscovered biological mechanisms associated with the dedifferentiated oncogenic state. Analyses of the tumor microenvironment revealed unanticipated correlation of cancer stemness with immune checkpoint expression and infiltrating immune system cells. We found that the dedifferentiated oncogenic phenotype was generally most prominent in metastatic tumors. Application of our stemness indices to single cell data revealed patterns of intra-tumor molecular heterogeneity. Finally, the indices allowed for the identification of novel targets and possible targeted therapies aimed at tumor differentiation.
ETOC
Stemness features extracted from transcriptomic and epigenetic data from TCGA tumors reveals new drug targets for anti-cancer therapies
INTRODUCTION
Stemness, defined as the potential for self-renewal and differentiation from the cell-of-origin, was originally attributed to normal stem cells that possess the ability to give rise to all cell types in the adult organism. Cancer progression involves gradual loss of differentiated phenotype and acquisition of progenitor-like, stem cell-like features. Undifferentiated primary tumors are more likely to result in cancer cell spread to distant organs, causing disease progression and poor prognosis, particularly because metastases are usually resistant to available therapies (Friedmann-Morvinski and Verma, 2014; Ge et al., 2017; Shibue and Weinberg, 2017; Visvader and Lindeman, 2012).
An increasing number of genomic, epigenomic, transcriptomic, and proteomic signatures have been associated with cancer stemness. Those molecular features are causally connected to particular oncogenic signaling pathways that regulate transcriptional networks that sustain the growth and proliferation of cancer cells (Ben-Porath et al., 2008; Eppert et al., 2011; Kim et al., 2010). Transcriptional and epigenetic dysregulation of cancer cells frequently leads to oncogenic de-differentiation and acquisition of stemness features by altering core signaling pathways that regulate the phenotypes of normal stem cells (Bradner et al., 2017; Young, 2011). Cell-extrinsic mechanisms can also affect maintenance of the undifferentiated state, largely through epigenetic mechanisms. Tumors comprise a complex, diverse, integrated ecosystem of relatively differentiated cancer cells, cancer stem cells, endothelial cells, tumor-associated fibroblasts, and infiltrating immune cells, among other cell types. The microenvironment of a tumor, considered as a pathologically formed “organ” is frequently characterized by hypoxia, as well as by abnormal levels of various cytokines, growth factors, and metabolites (Lyssiotis and Kimmelman, 2017). It provides numerous opportunities for cell-cell signals to modulate the epigenome and expression of stem cell-like programs in cancer cells, frequently independent of their genetic backgrounds (Gingold et al., 2016).
Over the last decade, The Cancer Genome Atlas (TCGA) has illuminated the landscapes of primary tumors by generating comprehensive molecular profiles composed of genomic, epigenomic, transcriptomic, and (post-translational) proteomic characteristics (Hoadley et al., 2014; Tomczak et al., 2015), along with histopathological and clinical annotations. The resulting resource enabled us to analyze cancer stemness quite extensively in almost 12,000 samples of 33 tumor types.
First, we defined signatures to quantify stemness, using publicly available molecular profiles from normal cell types that exhibit various degrees of stemness. By multi-platform analyses of their transcriptome, methylome, and transcription factor binding sites using an innovative one-class logistic regression machine-learning algorithm (OCLR) (Sokolov et al., 2016), we obtained two independent stemness indices. One (mDNAsi) was reflective of epigenetic features, the other (mRNAsi) was reflective of gene expression. We then identified associations between the two stemness indices and novel oncogenic pathways, somatic alterations, and microRNA and transcriptional regulatory networks. Those features correlated with, and perhaps govern, cancer stemness in particular molecular subtypes of TCGA tumors. Importantly, higher values for stemness indices were associated with biological processes active in cancer stem cells and with greater tumor dedifferentiation, as reflected in histopathological grade.
Metastatic tumor cells appeared more dedifferentiated phenotypically, probably contributing to their aggressiveness. We also found tumor heterogeneity at the single-cell level by measuring stemness in transcriptome profiles obtained from individual cancer cells. Using CIBERSORT to profile immune cell types in TCGA tumors, we obtained insight into the interface of the immune system with stemness. Finally, we identified compounds specific to selected molecular targets and mechanisms that may eventually lead to novel treatments that trigger differentiation and exhaust the stemness potential of highly aggressive neoplasms.
RESULTS
DNA methylation- and mRNA expression-based stemness classifiers
We analyzed publicly available non-tumor and tumor datasets for which transcriptomic and epigenomic molecular profiles were available (Figure 1A). We derived stemness indices using an OCLR algorithm trained on stem cells (SC; ESC/iPSC) classes and their differentiated ecto-, meso- and endoderm progenitors. We chose OCLR because it does not penalize misclassification of stem cell-derived progenitors at different stages of differentiation, which still carry some of the undifferentiated features in their molecular profiles (its output was also validated against Random Forests in Figure S1A). OCLR-based transcriptomic and epigenetic signatures were applied to TCGA datasets to calculate the mRNAsi and mDNAsi. Each stemness index (si) ranges from low (zero) to high (one) stemness (Table S1). The tumor samples stratified by the indices were used for the integrative analyses.
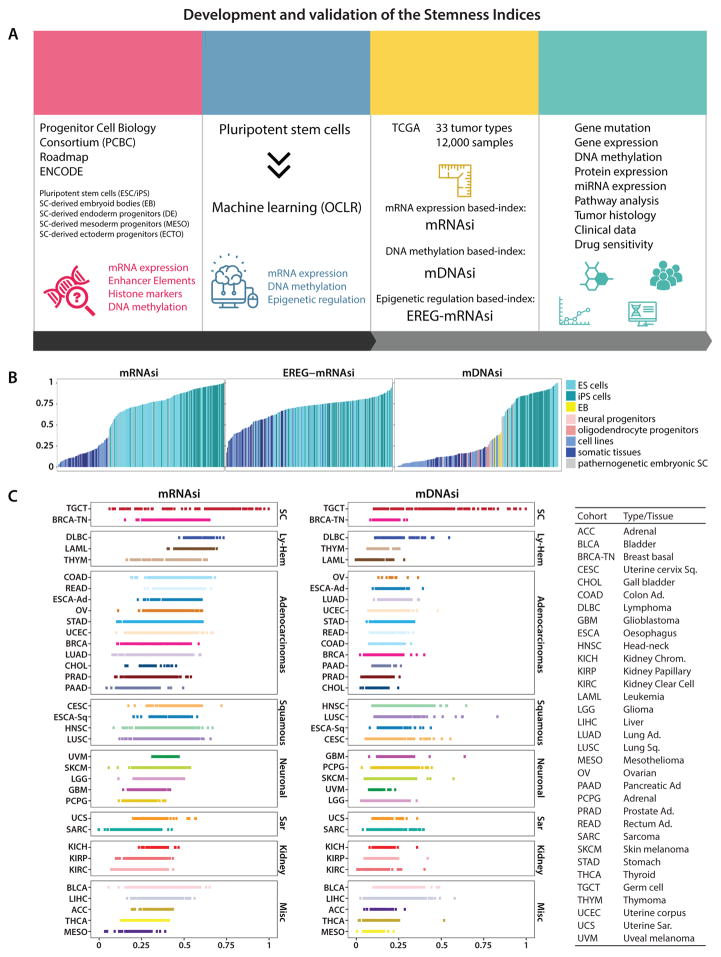
Figure 1. Development and validation of the Stemness Indices.
(A) Overall methodology. Highlighted are data sources Progenitor Cell Biology Consortium (PCBC), Roadmap and ENCODE databases, OCLR machine learning algorithm, and the resulting stemness indices mRNAsi, mDNAsi and EREG-mRNAsi. The indices for each TCGA tumor sample were correlated with known cancer biology, tumor pathology, clinical information, and drug sensitivity.
(B) Stemness indices of the validation set derived using our stemness signature.
(C) TCGA tumor types sorted by the stemness indices obtained from transcriptomic (mRNAsi) and epigenetic features (mDNAsi); indices were scaled from 0 (low) to 1 (high). The TCGA tumor types were grouped based on their histology and cell-of-origin into stem cell-like (SC), lympho-hematopoietic (Ly-Hem), Adenocarcinomas, Squamous Cell Carcinomas (Squamous), Neuronal lineage (Neuronal), Sarcomas (Sar), Kidney tumors (Kidney), and not belonging to any of the above (Misc) (Table S2).
See also Figures S1 and S2; and Tables S1 and S2.
mRNA expression-based stemness index
We validated the mRNAsi by applying it to an external dataset composed of both stem cells and somatic differentiated cells (Nazor et al., 2012) (Figure 1B), and by scoring molecular subtypes of breast cancers and gliomas that are characterized by different degrees of oncogenic dedifferentiation associated with pathology and clinical outcome (Figures S1B and S1C). All stem cell samples attained higher si values than samples from differentiated cells. TCGA tumors display various degrees of cancer stemness as revealed by mRNAsi (Figure 1C, left) and mDNAsi (Figure 1C, right). Germ-cell tumors, basal breast cancer, and Ly-Hem cancers displayed highly dedifferentiated phenotypes in comparison to other tumor types.
Using GSEA, we compared our signature to 16 gene sets that were associated with stemness in cancer and healthy cells in previous studies (Ben-Porath et al., 2008; Ivanova et al., 2006; Kim and Orkin, 2011; Mathur et al., 2008; Palmer et al., 2012; Sato et al., 2003; Venezia et al., 2004; Yan et al., 2011). These sets spanned 2,564 unique genes, with no two sets overlapping by more than 134 genes. In all cases, the published stemness gene sets were significantly enriched in mRNAsi (Figure 2A). We found that “Cancer Hallmark” gene sets were significantly enriched, as were MYC targets which significantly contributed to the positive side of the signature (Hanahan and Weinberg, 2011). This is consistent with MYC being one of the transcription factors that drive pluripotency in ESC (Young, 2011).
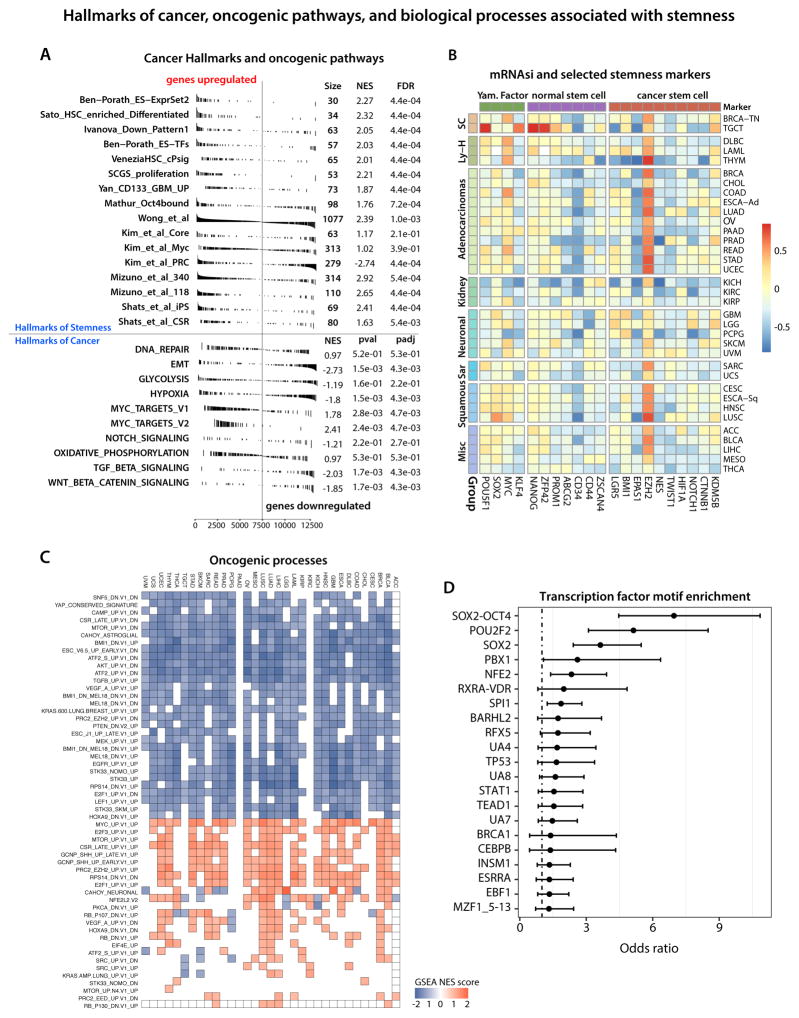
Figure 2. Biological processes associated with cancer stemness.
(A) Gene Set Enrichment Analysis showing RNAseq-based stemness signature evaluated in the context of gene sets representative for Hallmarks of Stemness and Cancer.
(B) Correlation between mRNAsi and mRNA expression for published hallmarks of stemness.
(C) Correlation between mRNAsi and selected oncogenic processes.
(D) Association between the epigenomic-based stemness signature (EREG-mDNAsi and EREG-mRNAsi) and enrichment in the transcription factor binding sites.
Wnt/β-catenin and TGFβ signaling pathways were significantly enriched on the negative side of the stemness signature. This negative enrichment does not imply absence of specific signals in cancer stem cells, but rather that this signaling is lower relative to stem cell-derived progenitors, as captured by the signature weights. This is again consistent with other GSEA results, as both signaling pathways are known mediators of the EMT mechanism (Gonzalez and Medici, 2014). We also computed the correlation of mRNAsi against mRNA expression of published pan-cancer EMT markers (Mak et al., 2016), which revealed significant correlations with for most tumors (Figures S2C). This is consistent with the biology of ESCs, which grow as epithelioid, polygonal cells in vitro and epithelial cancer precursors having stem-like properties. Importantly, most TCGA samples are primary tumors of an epithelial phenotype. Most skin melanoma cases come from lymph nodes and this tumor type shows higher expression of vimentin, a key marker of mesenchymal phenotype. mRNAsi is positively correlated with other core stem cell factors: EZH2, OCT4, and SOX2 (Figure 2B and Table S2). Finally Moonlight analysis of the oncogenic signatures from MSigDB further validated our gene expression based index and confirmed engagement of MYC, EZH2, along with E2F3, MTOR, SHH in driving oncogenic dedifferentiation (Figure 2C) (Colaprico et al., 2018).
DNA methylation-based stemness index
We defined the mDNAsi using OCLR by combining: 1) supervised classification between ESC/iPSC and their progenies; 2) stem cell signatures associated with pluripotency-specific genomic enhancer elements based on ChromHMM from Roadmap, and 3) ELMER, which uses DNA methylation to identify enhancer elements and correlates their state with the expression of nearby genes. 219 CpG probes (Figure S2A) were selected in training OCLR using the PCBC datasets. By selecting probes previously defined to be active stemness-specific enhancers, we confirmed the ability of our approach to derive an mDNAsi. Since we focused exclusively on hypomethylated, functionally important CpG probes associated with stem cells, we further explored cis-activated genes.
We scored each TCGA sample using the mDNAsi and used an external dataset to confirm that stem cells had higher mDNAsi than differentiated samples (Figure 1B, left plot). TCGA tumor types show different degrees of inferred dedifferentiated phenotype (Figure 1C, right). Within these, Individual tumor samples show variation for cancer stemness. As anticipated, TCGA samples derived from the primary tumors show higher cancer stemness indices compared to non-tumor samples obtained from adjacent normal tissue-of-origin (Figure S1E, bottom).
Most of our selected probes fell within non-promoter elements, yet the SOX2-OCT4 transcription factor binding motif is one of the most highly enriched signatures within these regions. The SOX2-OCT4 complex is a critical master regulator of pluripotency and stemness, and is highly enriched in tumor samples with high mDNAsi (Figure 2D).
Correlations of mRNAsi and mDNAsi
Since the inputs for mDNAsi and mRNAsi are not necessarily complementary, we explored stratification of glioma samples by the epigenetically regulated-mRNAsi (EREG-mRNAsi), a stemness index generated using a set of stemness-related epigenetically regulated genes. The EREG-mRNAsi, based on both RNA expression and epigenetics, elucidates the discrepancy between mDNAsi and mRNAsi and shows a positive correlation with both indices (Figure S1F). Both mRNAsi and mDNAsi show good correspondence for a majority of tumors (Figures S1F and S2B). We observed major discrepancies in the case of LGG, THCA, and THYM. For gliomas, mDNAsi is correlated positively with tumor pathology and clinical features, while mRNAsi shows a negative correlation. This result could arise from a high frequency of IDH1/2 mutations and resulting DNA hypermethylation.
Stemness index can stratify recognized undifferentiated cancers
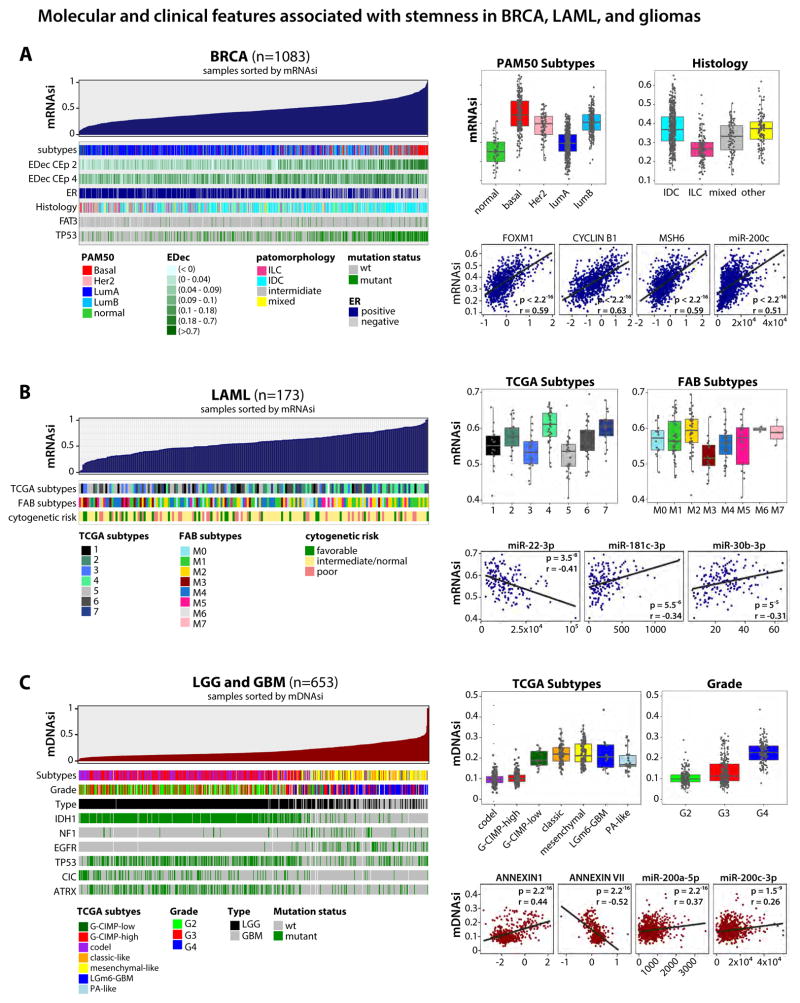
We examined BRCA, AML, and gliomas to study if the mRNAsi/mDNAsi predict stemness in poorly differentiated tumors. In BRCA, we found a strong association between the stemness index and known clinical and molecular features (Figure 3A, left). The mRNAsi was highest in the basal subtype, known to exhibit an aggressive phenotype associated with an undifferentiated state. BRCA samples with high mRNAsi were more likely to be ER-negative, and enriched for FAT3 and TP53 mutations. We noted that high mRNAsi was associated with higher protein expression of FOXM1, CYCLINB1 and MSH6 as well as higher microRNA-200 family expression (Figure 3A, right). Invasive lobular type of BRCA (ILC), characterized by better prognosis in comparison to invasive ductal carcinoma (IDC), has a lower mRNAsi (Figure 3A, right). We also applied our indices to non-TCGA BRCA samples (Reyngold et al., 2014), and found a similar correlation between mRNAsi and mDNAsi in those samples. Moreover, mRNAsi also stratified BRCA samples with distinct histology in this dataset (Figure S1B). Using datasets with estimated tumor cell type composition provided by the epigenetic deconvolution method (Onuchic et al., 2016) we found that both mRNAsi and mDNAsi were more highly correlated with malignant epithelial cells than with normal epithelial cells suggesting that our indices identify distinct cancerous epithelial cell populations characterised by different features or degrees of stemness (Figure S1D).
Figure 3. Molecular and clinical features associated with stemness in breast cancer, acute myeloid leukemia, and gliomas.
(A) An overview of the association between known molecular and biological processes and stemness in BRCA (Left). Columns represent samples sorted by mRNAsi from low to high (top row). Rows represent molecular and biological processes associated with mRNAsi. Rows named “EDec CEp 2 and 4” represent estimated cell type proportions. Top right, boxplots of mRNAsi in individual samples, stratified by molecular subtype and histology. Bottom right, correlation of mRNAsi and representative protein expression and microRNA.
(B) Similar to A, association of mRNAsi in AML. Top right, mRNAsi by mRNA-based molecular subtype and by FAB classification. Bottom right, correlation scores of mRNAsi and representative microRNA.
(C) As in A and B, GBM and LGG sorted by mDNAsi. Top right, mDNAsi by molecular subtype and grade. Bottom right, correlation scores of mDNAsi and representative protein expression and microRNA. All molecular and clinical features shown are statistically significant.
See also Figures S1, S3, S4, and S5.
We found an association between the mRNAsi, RNA expression subtypes previously defined by TCGA, and the French-American-British (FAB) classification of AML (Figure 3B). The mRNAsi showed the strongest correlation with the stage of myeloid differentiation of the AML samples. FAB subtypes M0 (undifferentiated), M1 (with minimal maturation), and M2 (with maturation) were characterized by high mRNAsi. In contrast, M3 well-matured promyelocytic subtype, which is associated with benign chromosomal abnormalities and favorable clinical outcome had low mRNAsi (Figure 3B, right upper). High mRNAsi was associated with higher expression of miR-181c-3p, miR-22-3p, and miR-30b-3p (Figure 3B, right bottom).
We found a strong association between high mDNAsi, high pathologic grade and recently published molecular subtypes of glioma (Figure 3C). mDNAsi was low in less aggressive gliomas that are characterized by codel and G-CIMP-high features and was highest in highly aggressive GBMs characterized by IDH mutations (G-CIMP-low) and poor clinical outcome. Also, high mDNAsi is strongly associated with more aggressive classical and mesenchymal subtypes of GBM, suggesting that it can stratify tumors with distinct clinical outcomes. We also found that high mDNAsi was associated with mutations in NF1 and EGFR and infrequent mutations in IDH1, TP53, CIC, and ATRX (Figure 3C, left), with higher expression of ANNEXIN-A1 protein and lower expression of ANNEXIN-A7, and with expression of the miR-200 family (Figure 3C, right bottom).
We obtained similar results on non-TCGA glioma samples for which both mRNA expression and DNA methylation data were available (Turcan et al., 2012) (Figure S1C). The negative correlation between mDNAsi and mRNAsi was restricted to LGG samples, specifically the IDH mutant subtypes (G-CIMP high and codel). IDH1 mutations are known to reduce cell differentiation, and high values of the mRNAsi in a subset of IDH mutant gliomas might capture this phenomenon (Lu et al., 2012).
Pan-cancer stemness landscape
Next, we tested the ability of our indices to identify previously unexplored features of cancer stemness across all TCGA tumors. First, we performed an enrichment analysis by sorting all TCGA samples by stemness index for each tumor type and looking for associations with mutations, molecular and clinical features. The most salient associations of mRNAsi and mDNAsi are presented in Figure 4, while the following results of the comprehensive analyses are shown in the supplementary material: associations with mutations (Figure S3), associations with miRNA expression and protein abundance (Figure S4), associations with the tumor grading and clinical outcome (Figure S5).
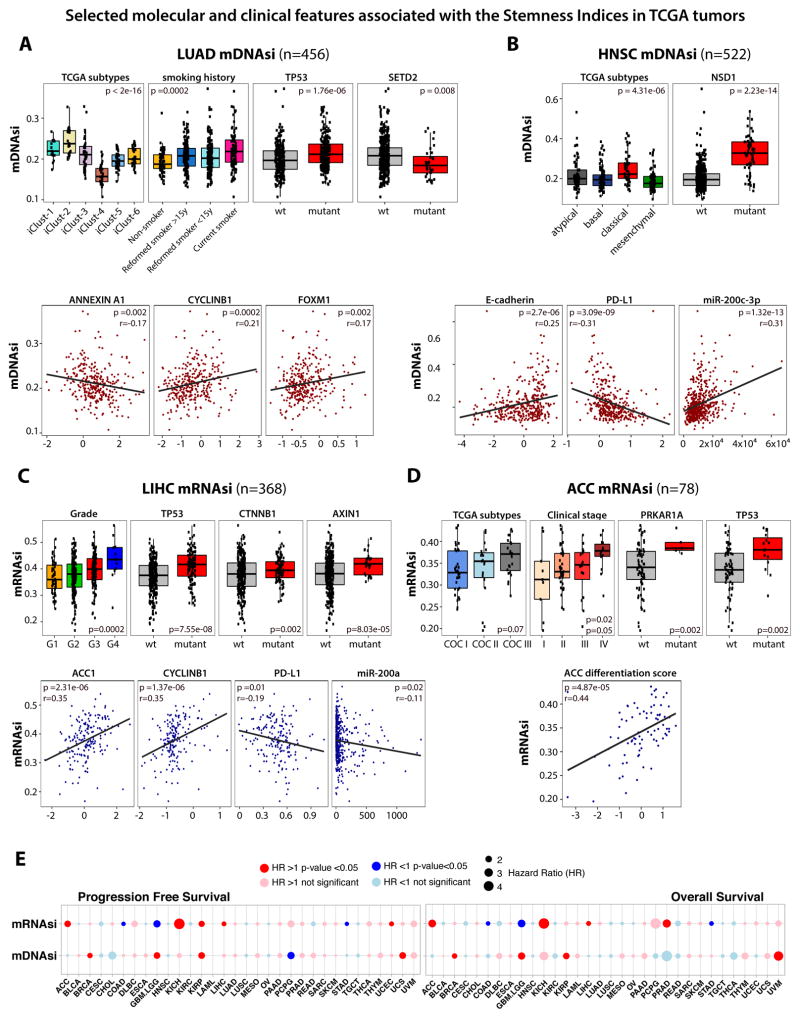
Figure 4. Selected molecular and clinical features associated with the Stemness Indices in TCGA tumors.
(A) Association of molecular and clinical features with stemness in LUAD. Top, mDNAsi by integrative molecular subtypes, smoking history, and mutations of TP53 and SETD2. Bottom, correlation scores of mDNAsi and representative protein expression.
(B) Stemness in HNSC. Top, mDNAsi stratified by molecular subtypes and mutation of NSD1. Bottom, correlation scores of mDNAsi and representative protein and microRNA expression.
(C) Stemness in LIHC. Top, mRNAsi stratified by grade and mutations of TP53, CTNNB1, and AXIN1. Bottom, correlation scores of mRNAsi and representative protein and microRNA expression.
(D) Stemness in ACC. Top, mRNAsi stratified by mRNA molecular subtypes, clinical stage, and mutations of PRKAR1A and TP53. Bottom, correlation scores of mRNAsi and adrenal differentiation score.
(E) Cox proportional hazards model analysis. Left, progression-free survival; right, overall survival. Hazard ratio greater than one denotes a trend toward higher stemness index with worse outcome.
See also Figures S3, S4, and S5.
Correlations of mRNAsi and mDNAsi with mutations in genes, miRNA and expression of proteins
We found a strong association between mDNAsi and known molecular subtypes, somatic mutations in SETD2 and TP53 genes, and with tobacco smoking status in LUAD (Figures 4A and S3). Current smokers and recently reformed smokers have higher mDNAsi than non-smokers or long-term reformed smokers. This suggests that the stemness of LUAD tumors might be activated in response to environmental stimuli such as smoking, and might influence te aggressiveness of the tumor. We also found an association between mDNAsi and higher protein expression of CYCLINB1 and FOXM1, which is a pro-stemness transcription factor upstream of CYCLINB1 (Figure 4A, lower plots). FOXM1 has been associated with dedifferentiation in pancreatic cancer cells (Bao et al., 2011) as well as tumor proliferation in the kidney (Xue et al., 2012) and ovarian (Wen et al., 2014) cancers. Our result suggests that it could be a driver of dedifferentiation and proliferation in breast and lung cancers. Stemness of LUAD tumors is also associated with lower expression of ANNEXIN-A1 (Figure 4A). ANNEXIN-A1 has been indicated as a differentiation marker in pancreatic (Bai et al., 2004) and urothelial cancers (Kang et al., 2012), therefore we suspect that the relationship between ANNEXIN and FOXM1 expression and tumor differentiation may extend to other tumor types (Figure S4C).
Analyses of HNSC samples revealed that high indices are correlated with NSD1 mutation, E-cadherin protein expression, miR-200-3p, and previously identified classical molecular subtypes (Figure 4B). NSD1 mutation was recently linked in HNSC tumors to blockade of cellular differentiation and promotion of oncogenesis (Papillon-Cavanagh et al., 2017). Interestingly, miR-200 family members have been implicated in cancer initiation and metastasis, as well as self-renewal of healthy stem cells (Gregory et al., 2008; Tellez et al., 2011). HNSC tumors with high mDNAsi have reduced programmed death ligand 1 (PD-L1) protein level (Figure 4B).
In LIHC samples, we found an association between mRNAsi and high pathological grade (Figure 4C). Negative associations between mRNAsi and the probability of OS or PFS were detected (Figures 4E and S5C). In contrast to the majority of tumor types, LIHC samples with high mRNAsi have low expression of miR-200 family members (Figure 4C). The miR-200 family is known to be associated with progression of hepatocellular carcinoma (Tsai et al., 2017; Wong et al., 2015), and the miR-200b-ZEB1 circuit has been suggested as a master regulator of stemness in these cancers (Tsai et al., 2017). We found associations of mRNAsi with higher CYCLINB1 and ACC1 and with lower PD-L1 and ANNEXIN A1 protein expression in LIHC (Figure 4C). ACC1 was associated with pathomorphological markers of LIHC aggressiveness (vascular invasion and poor differentiation) and its upregulation was correlated with poor OS and disease recurrence in hepatocellular carcinoma patients (Wang et al., 2016). LIHC samples with high mRNAsi were associated with specific genomic alterations (e.g., TP53, CTNNB1, AXIN1, among others).
Detailed analyses of ACC samples revealed an association between high mRNAsi and defined molecular subtypes (Zheng et al., 2016), clinical stage, and mutations in PRKAR1A and TP53 genes (Figure 4D). We found a positive correlation between mRNAsi and adrenal differentiation score, that is based on expression of 25 genes that are important for adrenal function (Zheng et al., 2016) (Figure 4D).
Stemness indices are correlated with tumor pathology and predictive of clinical outcome
We observed a positive correlation between tumor histology and pathology grading and both stemness indices for the majority of the TCGA cases (Figures S5A, S5B, and Figures 3A, 3C, 4C, 4D, and S1B). For mRNAsi, the most significant correlations were found for BRCA (IDC and ILC), CESC, LIHC, PAAD, UCEC (Figure S5A). Interestingly, mRNAsi shows low values in GBM and STAD. On the other hand, mDNAsi strongly stratifies glioma by the pathology grade culminating with the highest value for GBM (Figure S5B). The reversed values of mDNAsi and mRNAsi in case of gliomas were also evident in the clinical data analyses. An adverse association between the mRNAsi and survival was detected (Figure 4E), which was significant for OS and PFS after adjusting for clinical factors (Figures S5C). In contrast, the mDNAsi had no significant association with OS and PFS after correcting for clinical factors. We found a positive correlation between previously published glioma subtypes and mDNAsi suggesting that mDNAsi might recapitulate prognostic molecular subtypes (Figure 3C). The discordance between the mRNAsi and the mDNAsi for gliomas may be explained in part by the dominant genomic alteration associated with the LGG tumor type. Roughly 80% of LGG tumors carry an IDH1/2 mutation and, as demonstrated by our group and others, tconfers a genome-wide hypermethylator phenotype (G-CIMP) (Noushmehr et al., 2010; Turcan et al., 2012). Given that the mDNAsi is driven primarily by low methylation levels associated with the stemness phenotype, the LGG tumors might resemble non-stem like phenotypes, which are predominantly hypermethylated. The subgroup of G-CIMP with the lowest overall DNA methylation levels (G-CIMP-low) is associated with the worst outcomes. Compared to G-CIMP-high tumors, G-CIMP-low tumors are known to be more proliferative, express cell-cycle-related genes, and have various stem cell-like genomic features (Ceccarelli et al., 2016).
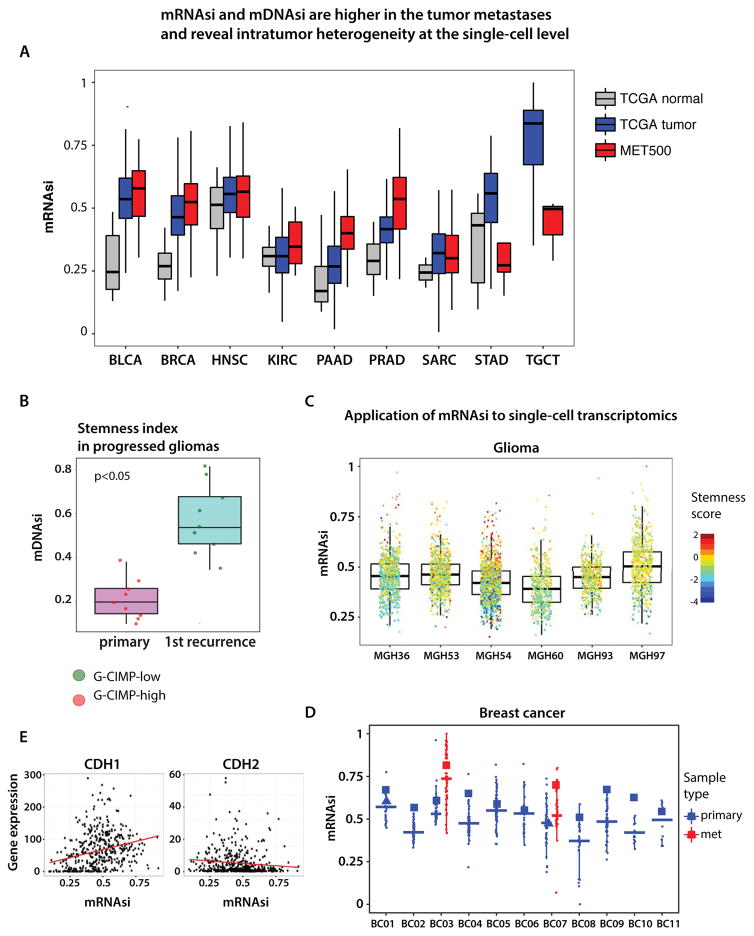
Cancer stemness indices are higher in tumor metastases and reveal intratumor heterogeneity
The TCGA samples are derived mostly from primary tumors except for skin melanoma for which tissues are mostly metastatic lymph nodes. We used the mRNAsi to interrogate the MET500 dataset comprising expression profiles from 500 metastatic samples obtained from 22 different organs (Robinson et al., 2017). In most cases, mRNAsi was significantly higher in e metastatic samples compared to primary TCGA tumors (Figure 5A). Prostate and pancreatic adenocarcinomas metastases had the most dedifferentiated phenotypes, and are also more aggressive and resistant to therapies in contrast to primary tumors. Weaker association with the mRNAsi was due to a small number of available samples (n<20). Interestingly, TGCT presents the less differentiated phenotype in primary tumors when compared to distant metastases. Primary TGCT tuor cells have high mRNAsi and may differentiate when metastasizing to distant organs. A similar trend was observed for STAD.
Figure 5. Analysis of cancer stemness in the context of metastatic state and intratumor heterogeneity.
(A) mRNAsi is higher in cancer metastases in comparison to the TCGA primary tumors.
(B) mDNAsi is higher in recurrent glioma samples compared to the primary glioma occurrence from the same patient. G-CIMP - glioma CpG methylator phenotype.
(C) and (D) Application of mRNAsi to single-cell transcriptome of gliomas and breast cancer reveal intratumor heterogeneity and various degrees of the oncogenic dedifferentiation. (E) Correlation of mRNAsi and mRNA expression of CDH1 (epithelial marker) and CDH2 (mesenchymal marker) in the cancer metastases.
Using another dataset, we found that mDNAsi was significantly higher in glioma samples obtained at first recurrence in contrast to primary gliomas (Figure 5B). Our results reveal significant dedifferentiation of glioma cancer cells that contribute to glioma recurrence which is frequently associated with poor prognosis and resistance to treatment (de Souza et al., 2017).
By taking advantage of single-cell transcriptome datasets, we used mRNAsi to probe tumor heterogeneity for oncogenic dedifferentiation of individual cancer cells (Chung et al., 2017; Tirosh et al., 2016). We revealed high variation of stemness in the glioma and breast primary tumors. Individual glioma cells showed higher variegation of oncogenic dedifferentiation in comparison to breast cancer cells (Figure 5C). Single cells from metastases had higher stemness index in breast cancer (Figure 5D). Interestingly, the negative correlation of EMT signature and stemness that we observed in TCGA primary tumors was also found in metastatic samples (Figure 5E).
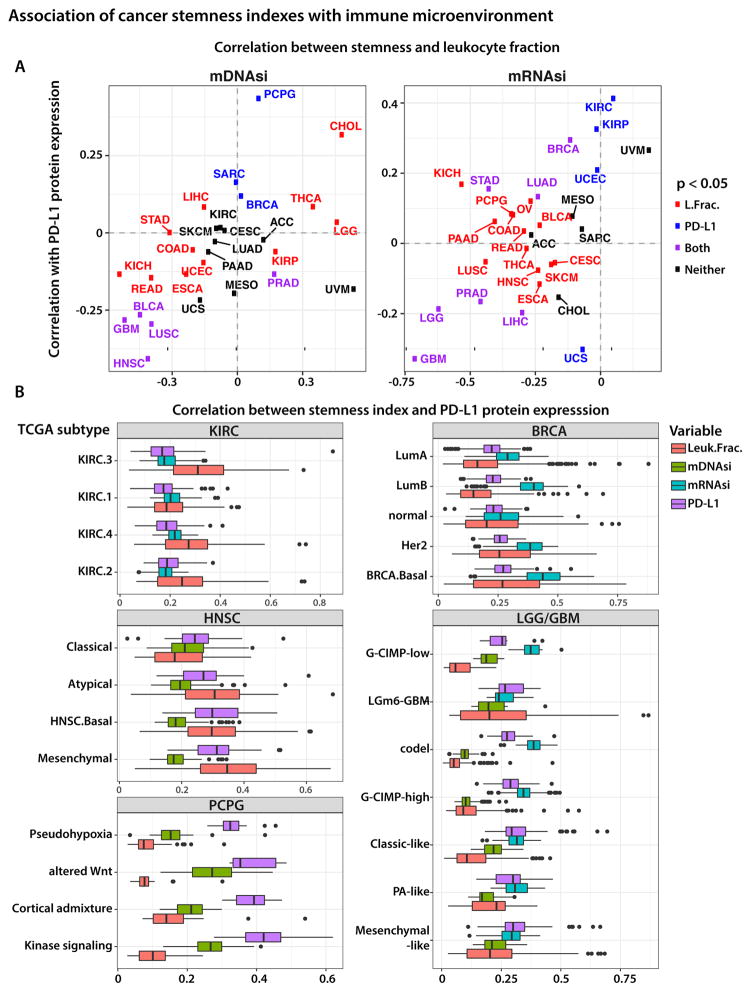
Stemness index evaluated in the context of immune response
We found that, for many tumors, higher stemness indices are associated with a reduced leukocyte fraction and lower PD-L1 expression (Figure 6A). For mDNAsi, the most distinctive negative correlations were found in the PanCan-12 squamous cluster (LUSC, HNSC, BLCA) (Hoadley et al., 2014) and in GBM (Figures 6A [left panel] and S6B). For the mRNAsi, the highest negative correlation values were seen in GBM/LGG, prostate adenocarcinoma (PRAD), LICH, and UCS tumors (Figures 6A [right panel] and S6A). We expect that such tumors will be less susceptible to immune checkpoint blockade treatments, due to insufficient immune cell infiltration or e pre-existing downregulation of the PD-L1 pathway, which makes further inhibition ineffective. Our findings are consistent with previous reports showing a strong correlation between PD-L1 protein expression and infiltration of CD8+ cytotoxic lymphocytes (Zaretsky et al., 2016).
Figure 6. Association of stemness index with immune microenvironment.
(A) mDNAsi and mRNAsi in the context of immune microenvironment. Each panel shows the Spearman correlation between the stemness index and PD-L1 protein expression plotted against Spearman correlation between the same stemness index and total leukocyte fraction, as estimated from DNA methylation data.
(B) Highlight of tumor types that exhibit strong correlation between stemness and PD-L1 expression or total leukocyte fraction. See also Figure S6.
We further explored correlations between stemness and immune microenvironment variables in the context of molecular subtypes of tumors. Figure 6B highlights several tumor types with the strongest (positive or negative) correlations. Except for KIRC, the association between stemness and PD-L1 expression and leukocyte fraction is readily apparent from the increasing and decreasing trends of individual variables across the molecular subtypes. For example, we found mesenchymal tumors to have the highest PD-L1 expression levels, the most significant leukocyte fractions, and lowest mDNAsi compared to other HNSC subtypes, suggesting potential susceptibility to checkpoint blockade inhibitors. The use of immunotherapy for HNSC tumors is under active investigation (Economopoulou et al., 2016; Fuereder, 2016), with the recent FDA approval of pembrolizumab; however, whether the effectiveness of therapy is limited to specific HNSC molecular subtypes is not clear from those reports.
To assess other relationships between stemness and tumor microenvironment, we computed correlations between stemness indices and individual types of immune cells. By applying CIBERSORT, we scored 22 immune cell types for their relative abundance in TCGA tumor samples. These cell types included NK cells, monocytes, macrophages, dendritic and mast cells, eosinophils, and neutrophils. We also obtained absolute estimates by scaling their relative abundance by overall leukocyte infiltration in each tumor, as determined by ESTIMATE applied to DNA methylation data. For any given TCGA sample, we calculated the correlation between mDNAsi/mRNAsi and the estimated fraction of individual immune cell types. In addition to individual immune subpopulation fractions, we considered the functional activation of distinct cells by measuring the difference between activated and resting fractions of NK cells, CD4+ T cells, and macrophages. This approach was motivated by recent observations that activation of peripheral CD4+ T cells triggered by immunotherapy is responsible for the specific killing of tumor cells (Spitzer et al., 2017).
Although the squamous cluster tumors had a negative correlation between stemness and the fraction of CD4+ T cell populations, the activation state of the CD4+ T cells was higher in dedifferentiated tumors. This finding is consistent with our observation that PD-L1 protein expression is lower in these tumors, suggesting again that immune checkpoint blockade might be ineffective and an additional mechanism of immune evasion may be operative. The opposite trend is present in thymomas, where PD-L1 protein expression and the fraction of the CD4+ T cell population are positively correlated with tumor dedifferentiation. Likewise, the activation state of CD4+ T cells is lower in dedifferentiated tumors, suggesting that they might be more susceptible to immunotherapy treatments (Figures S6AB).
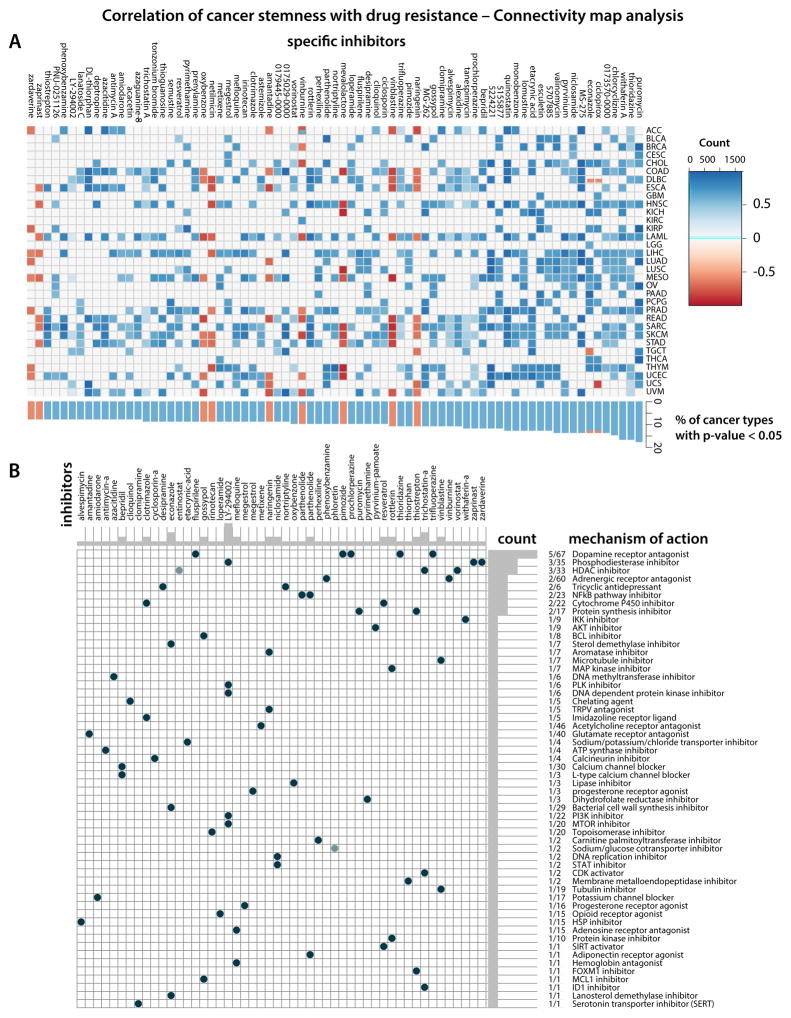
Connectivity map (CMap) analysis identifies potential compounds/inhibitors capable of targeting the stemness signature
We employed CMap, a data-driven, systematic approach for discovering associations among genes, chemicals, and biological conditions, to search for candidate compounds that might target pathways associated with stemness. We found enrichment for compounds associated with stemness in at least three cancer types Figure 7A. Five compounds are significantly enriched in more than ten cancer types and have been reported to inhibit stemness-related tumorigenicity: the dopamine receptor antagonists thioridazine and prochlorperazine (Cheng et al., 2015; Lu et al., 2015, (Dolma et al., 2016)), the WNT signaling inhibitor pyrvinium (Xu et al., 2016), the HSP90 inhibitor tanespimycin and the protein synthesis inhibitor puromycin. Further, telomerase inhibitor gossypol induced apoptosis and growth inhibition of CSCs (Volate et al., 2010), and histone deacetylase inhibitors such as trichostatin A (SAHA) reduced glioblastoma stem cell growth (Chiao et al., 2013). According to our analysis, pyrvinium and puromycin could inhibit stemness in LUAD. We found several candidates with recognized anti-CSC activity for HNSCC, including the aforementioned compounds. For LIHC, thioridazine, a prospective inhibitor of lung cancer stem cells (Yue et al., 2016), pyrvinium, puromycin, prochlorperazine, and others are potential compounds targeting undifferentiated tumors (Figure 7).
Figure 7. Correlation of cancer stemness with drug resistance – Connectivity map analysis.
(A) Heatmap showing enrichment score (positive in blue, negative in red) of each compound from the CMap for each cancer type. Compounds sorted from right to left by descending number of cancer type significantly enriched.
(B) Heatmap showing each compound (perturbagen) from the CMap that share Mechanism of actions (rows). Sorted by descending number of compound with shared mechanism of actions.
CMap Mode-of-action (MOA) analysis of the 74 compounds revealed 56 mechanisms of action shared by the above compounds (Figure 7B and Table S4B). Five compounds (fluspirilene, pimozide, prochlorperazine, thioridazine, and trifluoperazine) shared the MoA of Dopamine receptor antagonist. We observed that entinostat, trichostatin-a, vorinostat shared MoA as HDAC inhibitors, and LY-294002, zaprinast, zardaverine as Phosphodiesterase inhibitors.
CMap Target analysis revealed 212 distinct drug-target genes shared by the mentioned compounds (Figure S7 and Table S4C). Eight genes are targets of five different compounds, namely DRD2 (8 drugs), HTR2A (7 drugs), HRH1 (6), ADRA1A (5), CALM1 (5), CHRM3 (5), HTR1A (5), HTR2C (5).
Recent polypharmacology studies suggest the need to design compounds that act on multiple genes or molecular pathways. In this study, we observed similar mechanisms of action among different compounds suggesting selective therapies can target the undifferentiated phenotypes for selected cancer types.
DISCUSSION
This study is based on integrated analysis of cancer stemness in almost 12,000 primary human tumors of 33 different cancer types. We interrogated TCGA data for mutations, DNA methylation, expression of mRNA and miRNA, expression and post-translational modification of proteins, histopathological grade, and clinical outcome. Applying CIBERSORT, we gained insight into the tumor microenvironment and composition of immune cell infiltrates. By applying a machine-learning algorithm to molecular datasets from normal stem cells and their progeny, we developed two different molecular metrics of stemness and then used them to assess epigenomic and transcriptomic features of TCGA cancers according to their grade of oncogenic dedifferentiation. Ultimately, the analyses led us to potentially actionable targets (and their modes-of-action), as candidates for possible differentiation therapy of solid tumors and metastases. Our approach could be applied to longitudinal study of samples from primary, recurrent, and metastatic cancers and gene expression signatures identified in the tumor samples can be used to interrogate CMap to suggest actionable targets and inhibitors for further analysis.
To the best of our knowledge, this is the first study in which molecular PCBC datasets comprised of stem cells and defined populations of their differentiated progeny have been leveraged to develop a classification tool and machine-learning algorithm for analysis of a spectrum of human malignancies. A number of cancer stemness scores, based on genes that are differentially expressed between CSCs and non-CSCs, have been published and are relevant to clinical outcomes in AML (Eppert et al., 2011; Gentles et al., 2010; Ng et al., 2016). In those studies, gene sets enriched in ESCs (e.g., targets of NANOG, OCT4, SOX2, and c-MYC) were frequently overexpressed in poorly differentiated tumors compared with well-differentiated ones. In breast cancers, those gene sets were associated with high-grade estrogen receptor-negative, basal-like tumors and poor clinical outcome (Ben-Porath et al., 2008). Another web-based tool, StemChecker, uses a curated set of 49 published stemness signatures defined by gene expression, RNAi screens, transcription factor binding sites, text-mining of the literature, and other computational approaches. But it has been tested only for pancreatic ductal adenocarcinoma. In that case, high expression of stemness genes correlated with poor prognosis (Pinto et al., 2015). All previous studies were transcriptome-based and limited to a narrow set of genes and a small number of tumor types.
In the present study, we found oncogenic dedifferentiation to be associated with several characteristics: mutations in genes that encode oncogenes and epigenetic modifiers, perturbations in specific mRNA/miRNA transcriptional networks, and deregulation of signaling pathways. Cancer stemness also appeared to involve core expression of Myc, Oct4, Sox2, and other genes involved in the regulatory circuitry that underlies normal and malignant self-renewal potential. Our indices derived from mRNA expression and DNA methylation signatures reliably stratified tumors of known stemness phenotype. High mRNAsi was associated with basal breast carcinomas but also Her2 and lumB subtypes that are more aggressive than the hormone-dependent lumA group. In contrast, high mDNAsi was strongly associated with high-grade glioblastomas, poor overall and progression-free survival. The association between stemness signatures and adverse outcome for some tumor types, including gliomas, may reflect malignant cell origins or the impact of their microenvironment.
Dedifferentiated cells can arise from different sources: from long-lived stem or progenitor cells that accumulate mutations in oncogenic pathways, or via dedifferentiation from non-stem cancer cells that convert to CSCs through deregulation of developmental and/or non-developmental pathways. It is important to distinguish between the inherent stemness of CSCs and dedifferentiation induced by the tumor microenvironment. However, addressing that issue would require further validation beyond the scope of this study using other genomic datasets and/or laboratory experiments.
Both stemness indices were lowest in normal cells, increased in primary tumors, and highest in metastases, consistent with the idea that tumor progression generally involves oncogenic dedifferentiation. Interestingly, we observed negative associations between stemness and EMT gene signatures. The relationship between EMT and stemness remains a hotly debated topic, with several studies showing that EMT is necessarily associated with stemness (Fabregat et al., 2016). However, most TCGA data are obtained from primary tumors, which exist in a pre-EMT state, since EMT is strongly associated with tumor progression and with metastasis for many tumor types. Cancer cells in many primary solid tumors are basically epithelial regardless of their degrees of dedifferentiation, but some cells in such contexts could acquire mesenchymal characteristics, either by accumulating additional mutations or by undergoing epigenetic changes shaped by the tumor microenvironment. Those mesenchymal cells can traverse the underlying tissue, enter the bloodstream and seed distant organs where they reacquire an epithelial phenotype to form metastatic tumors.
We observed epithelial phenotype and increased stemness index in molecular profiles of tumor type-matched metastatic samples in the MET500 cohort. This portends an association between dedifferentiation and spread of tumor cells to distant organs. The observation is further supported by high mDNAsi in samples from recurrent gliomas. It appears that tumor growth de novo, or at recurrence/metastasis, is associated with an increased stemness phenotype. Decreased mRNAsi levels seen in TGCT suggest its possible differentiation as a germ cell tumor type induced by the microenvironment of liver or lung parenchyma, the organs it most often colonises. Clinically, in general, tumor progression is associated with greater aggressiveness and resistance to therapy of almost all types.
The mRNAsi was high for individual primary glioma and breast cancer cells. Interestingly, when applied to transcriptomic profiles obtained from analysis of single cancer cells in bulk tumors, stemness indices revealed a high degree of intratumor heterogeneity with respect to dedifferentiation phenotype. The heterogeneity was greater in gliomas than in breast cancer, suggesting that intratumor environment, including stromal cells, hypoxia, and infiltration of immune cells, may play a role in shaping CSC niches, and affect cancer cell developmental plasticity. Further molecular analyses of cancer cells stratified by the stemness phenotype would provide novel insights into the biology of primary tumors.
We found that, for a number of tumor types (GBM, LUSC, HNSC, and BLCA), higher mDNAsi was associated with reduced leukocyte fraction and/or lower PD-L1 expression. Such tumors are expected to be less susceptible to immune checkpoint blockade, due either to insufficient immune cell infiltration of tumors or to inherent downregulation of the PD-L1 pathway. Both factors can render immune checkpoint immunotherapy ineffective. The interaction between PD-L1 on cancer cells and PD1 receptor on T-cells helps cancer cells elude the immune system by preventing activation of cytotoxic T cells in lymph nodes and subsequent recruitment of other immune cell types to the tumor site (Chen and Mellman, 2013). The presence of tumor-infiltrating lymphocytes and/or PD-L1 expression correlates with aggressiveness in gastrointestinal stromal tumors (Bertucci et al., 2015) and breast carcinomas (Polónia et al., 2017).
Common features shared between cancer cells and stem cells in the context of the immune response are being highlighted by a growing number of studies showing that vaccination with ESC or iPSC can raise specific immune response against cancer cells (Kooreman et al., 2018). That finding may indicate that both cell populations use protein networks that, in tumors, result in uncontrolled self-renewal and de-differentiated phenotypes histopathologically defined by loss of architecture specific to the tissue of origin. We speculate that the indices described here may help predict the efficacy of stem-cell based immunotherapies and contribute to the identification of patients who will respond to such therapies.
We interrogated CMap using the gene expression signatures from tumor samples with the highest and lowest mRNAsi levels. Surprisingly perhaps, the Cmap analysis, which is based on only a limited number of treated cell lines, very precisely selected drugs that have been shown to affect cancer stem cells with specificity. These translational analyses may ultimately pave the way for implementation of differentiation therapies for solid tumors.
Here, we have also shown that cancer hallmarks can be extracted from datasets on cells with defined phenotypes and used to train machine-learning methods applicable to index molecular profiles of cancer. Our mRNAsi and mDNAsi can be translated into stemness scores (e.g., STEM50) that stratify tumors based on their dedifferentiation features, thus providing biomarkers for prediction of patient outcomes and response to to differentiation therapies.
By defining new metrics of cancer stemness and using them to interrogate TCGA datasets, our results provide a comprehensive characterization of dedifferentiation as new and significant hallmarks of cancer. The strengths of the approach are that it leverages features of dedifferentiated cells across a spectrum of tumor types that reflect tumor pathology and, in some cases, clinical outcome. This study also provides strategies for integrated analysis of cancer genomics based on machine-learning methods trained on molecular profiles obtained from cells with defined phenotypes. The findings based on those methods may advance the development of objective diagnostics tools for quantitating cancer stemness in clinical tumors, perhaps leading eventually to new biomarkers that predict tumor recurrence, guide treatment selection, or improve responses to therapy.
STAR★Methods
Detailed methods are provided in the online version of this paper and include the following:
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by Maciej Wiznerowicz (maciej.wiznerowicz@iimo.pl).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Clinical and molecular data were collected from the NIH Genomic Data Commons (GDC) of 11,392 participants from The Cancer Genome Atlas PanCancer Atlas cohort (https://gdc.cancer.gov/about-data/publications/PanCanStemness-2018).
METHODS DETAILS
DNA Methylation Data
A total of 9,627 PanCancer TCGA samples across 33 different tumor types were available for DNA methylation using the robust Illumina HumanMethylation 450 (HM450) platform. TCGA samples included primary (8,471), recurrent (41), and metastatic tumor (394) tissues and a set of 721 non-tumor tissues.
Level 3 data were downloaded from TCGA Data Portal using TCGAbiolinks functions GDCquery, GDCdownload and GDCprepare importing into R (http://www.r-project.org) for further analysis (Colaprico et al., 2016).
DNA methylation level 3 data are β-values that were calculated from pre-processed raw data using the methylumi Bioconductor package (Davis et al., 2015). Pre-processing steps included background correction, dye-bias normalization, and calculation of β-values and detection p-values. β-values range from zero to one, with zero indicating no DNA methylation and one indicating complete DNA methylation. A detection p-value compares the signal intensity difference between the analytical probes and a set of negative control probes on the array. Any data point with a corresponding p-value greater than 0.01 is deemed not statistically significantly different from background and is thus masked as “NA” in TCGA level 3 data. The data levels and the files contained in each data level package are on the NIH Genomic Data Commons (GDC).
In addition to TCGA data, we used a dataset of 99 human stem/progenitor cells from the Progenitor Cell Biology Consortium (PCBC) (https://www.synapse.org/pcbc) to define stem cell signatures (Daily et al., 2017; Salomonis et al., 2016). PCBC samples were profiled using the Illumina HumanMethylation 450 (HM450) platform and consisted of 4 embryonic stem cells (ESC), 40 induced pluripotent stem cells (iPSC), 22 stem cell (SC)-derived embryoid bodies (EB), 11 SC-derived mesoderm day 5 (MESO), 11 SC-derived ectoderm (ECTO), and 11 SC-derived definitive endoderm (DE). We downloaded raw IDAT files from PCBC Genomic Data Commons and processed the data according to the TCGA standard level 3 protocol described above.
RNA Expression Data
PanCancer TCGA RNA sequence level 3 normalized data were downloaded from the GDC Data Portal using TCGAbiolinks functions GDCquery, GDCdownload and GDCprepare importing into R (http://www.r-project.org) for further analysis (Colaprico et al., 2016). A total of 10,852 samples across 33 tumor types were available, including primary (9,702), recurrent (45) and metastatic tumor (395) tissues and a set of 710 non-tumor tissues.
We also downloaded PCBC RNA sequence data from the PCBC Synapse Portal (https://www.synapse.org/pcbc), consisting 16 ESC, 77 iPSC, 66 SC-derived EB, 29 SC-derived MESO, 29 SC-derived ECTO, and 36 SC-derived DE (Daily et al., 2017; Salomonis et al., 2016).
Stemness Index Derived Using OCLR
To calculate a stemness index (si) based on mRNA expression or DNA methylation, we built a predictive model using one-class logistic regression (OCLR) (Sokolov et al., 2016) on the pluripotent stem cell samples (ESC and iPSC) from the PCBC dataset (Daily et al., 2017; Salomonis et al., 2016).
For mRNA expression-based signatures, to ensure compatibility with the TCGA PanCancer cohort, we first mapped the gene names from Ensembl IDs to Human Genome Organisation (HUGO), dropping any genes that had no such mapping. The resulting training matrix contained 12,945 mRNA expression values measured across all available PCBC samples. For DNA methylation-based signatures, we used each of the signatures (probe set) described below.
We mean-centered the data, then applied OCLR to just the samples labeled SC (which included both ESC and iPSC). We chose to use the one-class framework because of its robustness in the absence of the a “negative” class. The PCBC data does not have data for fully differentiated cells, and progenitor cell types might exhibit some of the stemness signals.
Once the signature is obtained, it can be applied to score new samples. For RNA expression data, we computed Spearman correlations between the model’s weight vector and the new sample’s expression profile. We advocate for the use of Spearman correlation over the more traditional dot product operation because it is more robust with respect to potential cross-dataset batch effects that may arise. For DNA methylation data, which follow the beta distribution, the samples were scored using the standard application of a linear model: f(x) = w^T x + b.
We validated our approach using leave-one-out cross-validation by withholding each SC sample in turn. A separate signature was then trained on all other SC samples and used to score the withheld sample as well as all the non-SC samples. The performance was measured using the area under the curve (AUC) metric, which can be interpreted as the probability that the model correctly ranks a positive sample above a negative (Agarwal et al., 2005). In our cross-validation experiment, every withheld SC sample was scored higher than all the non-SC samples, yielding an overall AUC of 1.0.
We performed additional validation of the stemness signature by applying it to an external dataset composed of pluripotent stem cells (ESC and iPSC), somatic cells (17 distinct tissue types and several primary cell lines of diverse origin), and hydatidiform mole samples (Nazor et al., 2012). The mRNA expression data for the study were downloaded from GEO (GSE30652) as were DNA methylation data (GSE30654). We observed that all of the SC samples were correctly scored above all of the somatic samples by both platforms (Figure 1B). This is particularly striking for mRNA expression, because mRNA expression in study by the Nazor et al. was measured using microarrays, whereas the signature was trained using RNA-seq data.
Having validated the signature by using cross-validation and external SC data, we then applied it to score the TCGA PanCancer cohort using the same Spearman correlation (RNA expression) or linear model (DNA methylation) operators. The indices were subsequently mapped to the [0,1] range by using a linear transformation that subtracted the minimum and divided by the maximum. The mapping was done to assist with interpretation as well as integration with the stemness indices derived from other data platforms (i.e., DNA methylation and mRNA expression).
Additionally, we downloaded independent, non-TCGA datasets of gliomas [(Sturm et al., 2012) (GSE36245, GSE36278) and (Turcan et al., 2012) (GSE30339)] and BRCA samples (Reyngold et al., 2014) (GSE59000) and applied our metrics to measure the stemness in the validation data. For mRNA expression, the preprocessing consisted of mapping the Illumina probe IDs (Illumina HumanHT-12 V3.0 platform) to HUGO symbols, and then reducing the signature and the external dataset to a common set of genes. We then computed the Spearman correlation between the signature and the external samples. For DNA methylation, we applied the linear model.
DNA Methylation Stemness Signatures
Due to the magnitude of the available DNA methylation platform Infinium HumanMethylation450 (HM450), we defined DNA methylation-based stemness signatures as a reduced input to the OCLR machine learning algorithm. For the DNA methylation-based stemness indices, three signatures were utilized, each defining a distinct, biologically relevant, molecular phenotype of stemness. First, we performed a supervised analysis between human pluripotent stem cells (ESC and iPSC) and stem cell-derived progenitors (embryoid bodies [EB], mesoderm [MESO], ectoderm [ECTO], and definitive endoderm [DE]) (β value mean difference < −0.4 and false discovery rate [FDR] < 10e-22; β value mean difference > 0.3 and false discovery rate [FDR] < 10e-17).All ‘rs’ and ‘ch’ probes were removed prior to analyses. To eliminate somatic tissue-specific probes, we removed probes that were consistently methylated (standard deviation β value > 0.05) in non-tumor adult tissues available through TCGA. This resulted in a set of 62 pluripotent cell-specific and differentially methylated regions, which was then used as input for the OCLR to determine the stemness index for each TCGA tumor sample, named “differentially methylated probes-based stemness index” (DMPsi). Interestingly, most of these probes (85%) were positioned within intergenic regions known as open seas (Figure S2A).
Second, we defined a stem cell signature associated with genomic enhancer elements. Enhancers have been shown to be a critically relevant functional element for defining gene target expression and chromatin organization. For this, we downloaded Chromatin State data (ChromHMM) from the NIH Roadmap Epigenomics Consortium (http://www.roadmapepigenomics.org), which defined 18 chromatin states (based on 6 different histone marks: H3K4me3, H3K27ac, H3K4me1, H3K36me3, H3K9me3, and H3K27me3) across 98 different cell types (Roadmap Epigenomics Consortium et al., 2015). Briefly, by using ChromHMM data we mapped the HM450 probes to the chromatin states in each individual cell type; then we identified genomic regions corresponding to active enhancers that are specific to pluripotent stem cell states (ESC and iPSC), meaning that each region was defined as active enhancers (according to their states: 9-EnhA1 and 10-EnhA2 (Roadmap Epigenomics Consortium et al., 2015)) in all pluripotent stem cells (n=9) whereas not enhancer (enhancer in less than 25% of non-pluripotent stem cells (n= 89)) in non-pluripotent stem cells. We identified 82 DNA methylation probes of the HM450 platform that mapped to enhancer elements and considered them to be a DNA methylation-based pluripotent stem cell enhancer signature, which was then used as input for the OCLR to evaluate stemness signatures for TCGA samples, named “enhancer-based stemness index” (ENHsi) (Figure S2A).
Third, we applied ELMER (Enhancer Linking by Methylation/Expression Relationships), an R/Bioconductor package (Yao et al., 2015) that uses DNA methylation to identify enhancer elements and correlates enhancer state with expression of nearby genes to identify putative transcriptional targets. Using ELMER, we compared pluripotent stem cells (ESC and iPSC) to stem cell-derived progenitors (EB, MESO, ECTO, DE) from PCBC and identified 87 CpGs that were hypomethylated in the pluripotent state (ESC and iPSC) compared to stem cell-derived progenitors and that potentially regulate 103 genes. We confirmed the importance of these probe-gene pair targets by identifying that the SOX2-OCT4 transcription factor binding motif was among the most highly enriched signatures within these elements (+/−250 bp from the center). The SOX2-OCT4 complex is an important master regulator of pluripotency and stemness. We then derived a new set of signatures using the OCLR and defined TCGA samples’ stemness as “epigenetically regulated stemness indices” for each molecular feature (RNA expression-based Epigenetically regulated-mRNAsi [EREG-mRNAsi] and DNA methylation-based [EREG-mDNAsi]).
Because there was high concordance among the three DNA methylation-based indices (DMPsi, ENHsi, and EREG-mDNAsi) (not shown) and each contributes important and complementary biological relevance to stemness, we combined the three stemness signatures (total of 219 probes) and derived a comprehensive DNA methylation index, named mDNAsi (Figure S2A). The lists of probes and genes used to derive the stemenss indices are provided on the publication portal accompanying this publication (https://gdc.cancer.gov/about-data/publications/PanCanStemness-2018).
Stemness vs Molecular and Clinical Features
To evaluate the performance of our stemness indices across the entire TCGA cohort, we performed an enrichment analysis by sorting TCGA samples by stemness index for each tumor type and looked for associations with all available genomic features (by using comprehensive mutation data [MC3]), molecular features (previously published TCGA molecular subtypes available at TCGAbiolinks package (http://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html) (Colaprico et al., 2016; Silva et al., 2017), through the function “PanCancerAtlas_subtypes()”, which provides full access to the curated matrix used for this study), and clinical features (more than 10,000 features). We used the fgsea R/Bioconductor package to compute the enrichment scores (Sergushichev, 2016). Briefly, for each tumor type we ranked the TCGA samples according to their stemness index (from -low to -high stemness index) and tested if any particular genomic/molecular/clinical feature was associated with either -low or -high stemness index in a non-random behavior. We performed 10,000 permutations for each parameter analyzed to calculated our enrichment score. We then normalized the enrichment scores to mean enrichment of random samples of the same size (NES - normalized enrichment score). Tables containing all the results can be accessed at https://gdc.cancer.gov/about-data/publications/PanCanStemness-2018. In addition, an interactive portal with the results across all tumor samples/types vs. mDNAsi and mRNAsi can be accessed at https://bioinformaticsfmrp.github.io/PanCanStem_Web/where the user can search for any gene or molecular/clinical feature of interest.
Stemness versus Clinical Predictors
The associations between the three stemness indices and overall survival (OS) and progression free survival (PFS) in different tumors were evaluated in two stages. First, the proportional hazard (PH) model with the index as a single continuous covariate was used to test whether there was a statistically significant effect on OS or PFS. Given that, for each outcome, the effects of the three indices were tested for 33 cancer types. The significance level of the tests was adjusted for multiple testing to control the overall type I error probability at 5%. In the next stage, the cancer types for which at least one index showed a statistically significant association with either OS or PFS were analyzed in more detail by using a multivariable PH model that included relevant clinical factors. Moreover, the model included a functional form of the index obtained by using degree-2 fractional polynomials (Royston and Altman, 1994). The plausibility of the PH assumption was checked by using the test based on the scaled Schoenfeld residuals (Therneau and Grambsch, 2000). The analyses were conducted using STATA v13 software.
To select the clinical factors for inclusion in the PH model used in the second stage of the OS/PFS analysis for selected cancer types, a detailed analysis of the association between the stemness indices and demographic and clinical features (such as sex, age, race, stage, grade, etc.) was carried out by using linear models. mRNAsi and EREG-mRNAsi were analyzed on the original scale, while mDNAsi was transformed logarithmically to make its distribution more symmetric. The fit of the constructed models was assessed by using residual plots. The analyses were conducted using STATA v13 software.
The screening of the association between the stemness indices and OS (Figure 4E) by using univariable proportional hazard (PH) models indicated a statistically significant (using p values adjusted for multiple testing) effect of mRNAsi on OS for LGG (p < 0.0001) and STAD (p = 0.005) and on PFS for GBM (p = 0.04), LGG (p < 0.0001), LIHC (p = 0.05), STAD (p = 0.04), and UCEC (p = 0.03). For mDNAsi, an effect on OS was found for LGG (p < 0.0001) and on PFS for KIRP (p = 0.04) and LGG (p < 0.0001). Finally, for EREG-mRNAsi, a statistically significant effect on OS was found for ACC (p = 0.005), KIRC (p = 0.008), and LGG (p = 0.03), and on PFS for ACC (p = 0.03), LGG (p = 0.03), and UCEC (p = 0.04). In these selected cases, multivariable analyses were conducted (using STATA v13 software), which took into account the effect of clinical factors. The analyses confirmed (by using unadjusted p values) the effect of mRNAsi on OS for STAD (p = 0.0001) and for GBM/LGG (p = 0.002) and the effect on PFS for GBM/LGG (p = 0.008) and LIHC (p = 0.002). For mDNAsi, the effect on PFS in KIRP was confirmed (p = 0.0001), while for EREG-mRNAsi, the effect on PFS in UCEC was confirmed (p = 0.05). These confirmed results indicate that the indices have a potential role as novel, independent prognostic factors for the indicated tumor types.
Compounds Targeting with Cancer Stemness
To determine which target drugs might be useful against cancer stem cells, we used the Broad Institute’s Connectivity Map build 02 (CM) (Lamb et al., 2006), a public online tool (https://portals.broadinstitute.org/cmap/) (with registration) that allows users to predict compounds that can activate or inhibit based on a gene expression signature.
To further investigate about mechanism of actions (MoA) and drug-target we performed specific analysis within Connectivity Map tools (https://clue.io/) (Subramanian et al., 2017).
Using Connectivity Map (Query) in May 2017 having data available from a collection of cell lines (MCF, PC3, HL60 and SKMEL5) and 164 compounds as small molecules perturbagens. We obtained 33 mRNA expression signatures (one for each cancer type) by applying a differential expression analysis to samples with high mRNAsi and low mRNAsi, using the function TCGAanalyze_DEA from the the R/Bioconductor package TCGAbiolinks version 2.5.9 (Colaprico et al., 2016), carrying edgeR pipeline. The table with differentially expressed genes is reported as Table S3. Due to a limitation of the Connectivity Map tool that matches gene symbol and HG-U133A probe set (eg 200800_s_at) GPL96 platform ID, we had to remove duplicate IDs after sorting by decreasing |logFC|. We selected the top 1000 genes (500 up regulated and 500 downregulated) where the number of differentially expressed genes was enough or considering the aggregation of up-regulated or down-regulated genes.
Connectivity MAP is a method similar to GSEA analysis and follows a 4 step approach: (i) looking for similarity between a query signature (diff.expr. genes) and expression profiles present in the dataset using pattern-matching strategy based on Kolmogorov-Smirnov test (ii) rank-ordering the list of genes according their diff.expr. relative to the control from the above expression profiles with significantly similarity (iii) comparison of each rank-ordered list with a query signature to specify when up-regulated query genes appear in the proximity of the top of the list or near the bottom (“positive connectivity”) or vice versa (“negative connectivity”) producing an Enrichment Score (ES) from −1 to 1. (iv) All instances in the database are then ranked according to their connectivity scores; those at the top are most strongly correlated to the query signature, and those at the bottom are most strongly anticorrelated.
For each cancer type we obtained two tables that applied the Connectivity Map’s findings to stemness mRNA expression signatures, namely, “detailed results” and “permuted results”. We used the permuted results and filter (with p < 0.05), to identify an average of 74 compounds per tumor type that are predicted to repress or activate the stemness signature (Table S4A).
Connectivity Map (CMap) was recently updated (September 2017) (Subramanian et al., 2017), providing the end-users new functionalities and new graphical interface as web-server, previous registration (https://clue.io/) allowing easily the extraction of drug-interaction knowledge using as input a signature of genes or compounds.
The new interface (https://clue.io/), provided 7 different analysis (query, touchstone, proteomics query, command, data library, repurposing, morpheus).
In particular CMap Query it is a tool for perturbagens that give rise to similar (or opposing) expression signatures, for a technical limit, the CMap Query 2017 allows only to upload 150 genes max for up-regulated genes and 150 genes for down-regulated genes. For this reason we considered the results analysed in May 2017 using 500 genes for up-down regulated genes.
STATISTICAL ANALYSIS
R version 3.3.1 was used for all statistical analyses, unless specified otherwise. The statistical details of all experiments are reported in the figure legends and figures, including statistical analysis performed, statistical significance and exact n values.
To identify differentially methylated DNA methylation probes, we used the Wilcoxon test followed by multiple testing using the Benjamini-Hochberg (BH) method to estimate false discovery rate (Benjamini and Hochberg, 1995).
To identify proteins and microRNAs differentially expressed between tumors with low vs. high stemness index, we used a t-test followed by multiple testing using BH.
P values for the association between stemness index and continuous clinical data were also computed using a t-test followed by multiple testing using BH.
DATA AND SOFTWARE AVAILABILITY
All data are available on the NIH Genomic Data Commons (GDC), https://gdc.cancer.gov/about-data/publications/PanCanStemness-2018.
The workflow to reproduce the stemness index, including downloading PCBC and TCGA PanCan33 datasets, training a stemness signature, and applying it to score TCGA samples can be accessed at https://bioinformaticsfmrp.github.io/PanCanStem_Web/.
An interactive portal with the results for enrichment of molecular and clinical features and Stemness Indices across all tumor samples/types can be accessed at https://bioinformaticsfmrp.github.io/PanCanStem_Web/ where the user can search for any gene or molecular/clinical feature of interest.
Supplementary Material
(A) Comparison of mRNAsi and mDNAsi values, as scored by signatures learned from PCBC data by OCLR (x axis) and Random Forest (y axis).
(B) Validation of mRNAsi in non-TCGA breast cancer (BRCA) samples (Reyngold et al., 2014) to define stemness status. Stratification of mRNAsi according to histology of TCGA samples (left) and of the validation cohort (right). IDC - invasive ductal carcinoma; ILC - invasive lobular carcinoma.
(C) Validation of stemness indices (mDNAsi and mRNAsi) in non-TCGA glioma samples (Sturm et al., 2012; Turcan et al., 2012) using our metric to define stemness status. The validation sets were previously classified into molecular subtypes of TCGA gliomas. Stratification of stemness indices according to molecular subtypes in the TCGA (left) and in the validation cohort (right).
(D) Correlation of stemness indices and estimated cell composition derived from deconvolution of tumor epigenomic features in TCGA breast cancer samples (Onuchic et al., 2016).
(E) Stemness indices in tumor vs. non-tumor samples across tissue type. Top plots show mRNAsi and bottom plots show mDNAsi. See list of Abbreviations of the TCGA Tumor Types.
(F) Correlation of stemness indices across TCGA tumor types. Tumor types on the x-axis are sorted by low-to-high correlation between mDNAsi and mRNAsi. See list of Abbreviations of the TCGA Tumor Types. mRNAsi, RNA-based stemness index. mDNAsi, DNA methylation-based stemness index. EREG-mRNAsi, epigenetically regulated-mRNAsi.
Table S1: Stemness indices derived for all PanCancer 33 TCGA cohort, Related to Figure 1.
Table S2: List of published stem cell markers (healthy and cancer) used to correlate to mRNAsi, Related to Figures 1 and 2.
(A) Heatmaps of DNA methylation levels of probe signature used to derive the mDNAsi in: Progenitor Cell Biology Consortium (PCBC) dataset (left), TCGA normal adjacent tissue (middle), and TCGA tumors (right). Columns represent samples. TCGA tumor samples are sorted from lowest to highest mDNAsi. Rows represent the DNA methylation probe signature (219 probes), which is a combination of three DNA methylation-based signatures: differentially methylated probes stemness index (DMPsi), enhancer stemness index (ENHsi), and epigenetically regulated-mDNAsi (EREG-mDNAsi) (see STAR Methods for detail). Annotation of probes are provided in the left of the heatmaps. First column (mDNAsi) indicates to which signature the probe belong. Second column (genomic location) indicates the genomic location annotation of each probe from the UCSC genome browser. Chromatin State columns indicates the chromatin state of pluripotent stem cells (iPSC and ESC) from Roadmap (n=9) (Roadmap Epigenomics Consortium et al., 2015).
(B) Scatter plots showing correlation of mDNAsi (x axis) and mRNAsi (y axis) by tumor type. Samples are colored according to TCGA classification in: primary, recurrent, metastatic, and non-tumors.
(C) Correlation between mRNAsi and mRNA expression of epithelial-to-mesenchymal transition (EMT) markers. The markers are grouped according to whether they are associated with epithelial (pink) or mesenchymal (teal blue) phenotype.
(A) (Left Panel) Comparison of normalized enrichment scores (NES) in mDNAsi (x-axis) and mRNAsi (y-axis) computed by tumor type in the mutation enrichment analysis. Positive and negative NES entail enrichment and depletion of a given mutation being associated with a high stemness index, respectively. The quadrants and colors represent genes with agreement or disagreement between the two distinct indices. (Right Panel) Stemness indices stratified by representative mutation status and tumor types.
(B) Relationship of mutations in epigenetic modifier genes and stemness indices mRNAsi (top) and mDNAsi (bottom). NES, normalized enrichment scores.
(A) (Left Panel) Comparison of miRNA expression associated with mDNAsi and mRNAsi. Log10-normalized FDR-adjusted P value is plotted for mDNAsi (x-axis) and mRNAsi (y-axis) for each miRNA probe for each tumor type. If a particular miRNA is upregulated in samples with a high stemness index, the values are multiplied by −1. The quadrants and colors represent proteins with agreement or disagreement between the two distinct indices. (Right Panel) Correlation of stemness indices and representative miRNAs for selected tumor types.
(B) Comparison of protein expression associated with mDNAsi and mRNAsi (left panel). Log10 (FDR-adjusted P value) is plotted for mDNAsi (x-axis) and mRNAsi (y-axis) for each protein for each tumor type. If the protein is upregulated in high stemness index, −1 is multiplied to log10 providing positive values. The quadrants and colors represent proteins with agreement or disagreement between the two distinct indices. Right, correlation scores of stemness indices and representative proteins and tumor types.
(C) Correlation of ANNEXIN1 and FOXM1 with mDNAsi (top) and mRNAsi (bottom) across tumor types.
(A and B) Tumor pathology grade associations with mRNAsi (A) and mDNAsi (B) by TCGA tumors types.
(C) OS and PFS curves for four (quartile-based) categories of the relevant indices. The figure also includes the estimated (solid line) functional forms of the dependence of the logarithm of hazard ratio (log-HR) of OS or PFS on the indices. The forms in general correspond to the ordering of the survival curves. For selected indices and outcomes the estimated functional form suggests a non-monotonic relationship between log-HR and the index.
(A) Plots show correlations between the mRNAsi (top) and mDNAsi (bottom) and the estimated fraction of cells for a selected panel of immune cell types. Additional correlations of stemness with estimated immune cell activity, computed as the difference between activated and resting population fractions, are included for NK cells, CD4+ T cells and macrophages. Correlation with PD-L1 protein expression is included for reference and the tumor types are sorted according to it.
(B) Moonlight dynamic recognition analysis to identify associations with the mRNAsi across tumor types.
Heatmap showing each compound (perturbagen) from the Connectivity Map that share gene targets in (rows). Sorted by descending number of targets.
Table S3: Differentially expressed genes for each tumor using mRNAsi score, Related to Figures 7A, 7B, S6C, and S7.
Table S4A: Output from the Connectivity Map, Related to Figure 7A.
Table S4B: Output from the Connectivity Map, Related to Figure 7B.
Table S4C: Output from the Connectivity Map, Related to Figure S7.
HIGHLIGHTS.
Epigenetic and expression-based stemness indices measure oncogenic dedifferentiation
Immune microenvironment content and PD-L1 levels associate with stemness indices
Stemness index is increased in metastatic tumors and reveals intratumor heterogeneity
Applying stemness indices reveals potential drug targets for anti-cancer therapies
Acknowledgments
We thank Marcin Cieślik from Michigan Center for Translational Pathology at University of Michigan for providing the MTE500 dataset. This work was supported by the following grants: NIH grants: U54 HG003273, U54 HG003067, U54 HG003079, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845, U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025, P30 CA016672; NCI grants: 5R01CA180778, 3U24CA143858, 1U24CA210990, 5U54HG006097; NIGMS grant 5R01GM109031; Henry Ford Cancer Institute’s Early Career Investigator Award; Sao Paulo Research Foundation (FAPESP) grants: 2014/02245-3, 2014/08321-3, 2015/07925-5, 2016/01389-7, 2016/10436-9, 2016/06488-3, 2016/12329-5, 2016/01975-3, 2016/15485-8; institutional grants from Henry Ford Hospital; Spanish Institute of Health Carlos III: CP14/00229; Mary K. Chapman Foundation gift to JNW; CA143883; CPRIT# RP13039; the Michael & Susan Dell Foundation; CA016672; Polish Science Foundation Welcome grant 2010/3-3 to MW.
ADDITIONAL RESOURCES - Abbreviations of the TCGA Tumor Types
- ACC
Adrenocortical carcinoma
- AML
Acute myeloid leukemia
- BLCA
Bladder urothelial carcinoma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangiocarcinoma
- COAD
Colon adenocarcinoma
- DLBC
Lymphoid neoplasm diffuse large B-cell lymphoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HNSC
Head and neck squamous cell carcinoma
- KICH
Kidney chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LGG
Brain lower grade glioma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin cutaneous melanoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular germ cell tumors
- THCA
Thyroid carcinoma
- THYM
Thymoma
- UCEC
Uterine corpus endometrial carcinoma
- UCS
Uterine carcinosarcoma
- UVM
Uveal melanoma
Footnotes
AUTHOR CONTRIBUTIONS
The TCGA Research Network contributed collectively to this study. The contributions of other authors are as follows: epigenetic-derived stemness index, T.M.M., P.W.L., and H.N.; mRNA expression-derived stemness index, A.S.; methodology, T.M.M., A.S., and H.N.; integrative analysis, T.M.M., A.S., and H.N.; clinical analysis, T.B. and L.P; drug analysis, A.C.; data interpretation, T.M.M., A.S., J.H., B.K., H.H., A.J.G., A.C., L.M., P.W.L., H.N., and M.W.; data curation, L.O.; writing, T.M.M., A.S., J.W., B.K., J.H., A.C., J.N.W., H.N., and M.W.; visualization, T.M.M., A.S., H.N., and M.W.; overall concept and coordination, T.M.M., A.S., P.W.L., H.N., and M.W.
Supplemental Information includes seven figures and five tables and can be found with this article online at: TBD
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal Graepel S, Herbrich T, Har-Peled R, Roth S, Dan Generalization Bounds for the Area Under the ROC Curve. The Journal of Machine Learning Research 2005 [Google Scholar]
- Bai X-F, Ni X-G, Zhao P, Liu S-M, Wang H-X, Guo B, Zhou L-P, Liu F, Zhang J-S, Wang K, et al. Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J Gastroenterol. 2004;10:1466–1470. doi: 10.3748/wjg.v10.i10.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao B, Wang Z, Ali S, Kong D, Banerjee S, Ahmad A, Li Y, Azmi AS, Miele L, Sarkar FH. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J Cell Biochem. 2011;112:2296–2306. doi: 10.1002/jcb.23150. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertucci F, Finetti P, Mamessier E, Pantaleo MA, Astolfi A, Ostrowski J, Birnbaum D. PDL1 expression is an independent prognostic factor in localized GIST. Oncoimmunology. 2015;4:e1002729. doi: 10.1080/2162402X.2014.1002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh A, Pagnotta SM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T, Ryu HS, Kim S, Lee JE, Park YH, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44:e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Olsen C, Cava C, Terkelsen T, Silva TC, Olsen A, Cantini L, Bertoli G, Zinovyev A, Barillot E, et al. Moonlight: a tool for biological interpretation and driver genes discovery. BioRxiv. 2018:265322. [Google Scholar]
- Daily K, Ho Sui SJ, Schriml LM, Dexheimer PJ, Salomonis N, Schroll R, Bush S, Keddache M, Mayhew C, Lotia S, et al. Molecular, phenotypic, and sample-associated data to describe pluripotent stem cell lines and derivatives. Sci Data. 2017;4:170030. doi: 10.1038/sdata.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Bilke S, Triche T, Jr, Bootwalla M. R Package Version 2.18.0. 2015. methylumi: Handle Illumina methylation data. [Google Scholar]
- Dolma S, Selvadurai HJ, Lan X, Lee L, Kushida M, Voisin V, Whetstone H, So M, Aviv T, Park N, et al. Inhibition of dopamine receptor D4 impedes autophagic flux, proliferation, and survival of glioblastoma stem cells. Cancer Cell. 2016;29:859–873. doi: 10.1016/j.ccell.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economopoulou P, Perisanidis C, Giotakis EI, Psyrri A. The emerging role of immunotherapy in head and neck squamous cell carcinoma (HNSCC): anti-tumor immunity and clinical applications. Ann Transl Med. 2016;4:173. doi: 10.21037/atm.2016.03.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- Fabregat I, Malfettone A, Soukupova J. New Insights into the Crossroads between EMT and Stemness in the Context of Cancer. J Clin Med. 2016:5. doi: 10.3390/jcm5030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15:244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuereder T. Immunotherapy for head and neck squamous cell carcinoma. Memo. 2016;9:66–69. doi: 10.1007/s12254-016-0270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Gomez NC, Adam RC, Nikolova M, Yang H, Verma A, Lu CPJ, Polak L, Yuan S, Elemento O, et al. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169:636–650.e14. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304:2706–2715. doi: 10.1001/jama.2010.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O, Villalobos V, Sikic BI, Plevritis SK. Identification of ovarian cancer driver genes by using module network integration of multi-omics data. Interface Focus. 2013;3:20130013. doi: 10.1098/rsfs.2013.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold J, Zhou R, Lemischka IR, Lee DF. Modeling Cancer with Pluripotent Stem Cells. Trends Cancer. 2016;2:485–494. doi: 10.1016/j.trecan.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Kang W-Y, Chen W-T, Huang Y-C, Su Y-C, Chai C-Y. Overexpression of annexin 1 in the development and differentiation of urothelial carcinoma. Kaohsiung J Med Sci. 2012;28:145–150. doi: 10.1016/j.kjms.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Orkin SH. Embryonic stem cell-specific signatures in cancer: insights into genomic regulatory networks and implications for medicine. Genome Med. 2011;3:75. doi: 10.1186/gm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooreman NG, Kim Y, de Almeida PE, Termglinchan V, Diecke S, Shao N-Y, Wei T-T, Yi H, Dey D, Nelakanti R, et al. Autologous iPSC-Based Vaccines Elicit Anti-tumor Responses In Vivo. Cell Stem Cell. 2018 doi: 10.1016/j.stem.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The Connectivity Map: using gene-expression signatures to connect small molecules genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Kimmelman AC. Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 2017;27:863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, Skoulidis F, Parra ER, Rodriguez-Canales J, Wistuba II, et al. A Patient-Derived, Pan-Cancer EMT Signature Identifies Global Molecular Alterations and Immune Target Enrichment Following Epithelial-to-Mesenchymal Transition. Clin Cancer Res. 2016;22:609–620. doi: 10.1158/1078-0432.CCR-15-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur D, Danford TW, Boyer LA, Young RA, Gifford DK, Jaenisch R. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazor KL, Altun G, Lynch C, Tran H, Harness JV, Slavin I, Garitaonandia I, Müller FJ, Wang YC, Boscolo FS, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–634. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SWK, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540:433–437. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuchic V, Hartmaier RJ, Boone DN, Samuels ML, Patel RY, White WM, Garovic VD, Oesterreich S, Roth ME, Lee AV, et al. Epigenomic Deconvolution of Breast Tumors Reveals Metabolic Coupling between Constituent Cell Types. Cell Rep. 2016;17:2075–2086. doi: 10.1016/j.celrep.2016.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer NP, Schmid PR, Berger B, Kohane IS. A gene expression profile of stem cell pluripotentiality and differentiation is conserved across diverse solid and hematopoietic cancers. Genome Biol. 2012;13:R71. doi: 10.1186/gb-2012-13-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papillon-Cavanagh S, Lu C, Gayden T, Mikael LG, Bechet D, Karamboulas C, Ailles L, Karamchandani J, Marchione DM, Garcia BA, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet. 2017;49:180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto JP, Kalathur RK, Oliveira DV, Barata T, Machado RSR, Machado S, Pacheco-Leyva I, Duarte I, Futschik ME. StemChecker: a web-based tool to discover and explore stemness signatures in gene sets. Nucleic Acids Res. 2015;43:W72–7. doi: 10.1093/nar/gkv529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polónia A, Pinto R, Cameselle-Teijeiro JF, Schmitt FC, Paredes J. Prognostic value of stromal tumour infiltrating lymphocytes and programmed cell death-ligand 1 expression in breast cancer. J Clin Pathol. 2017 doi: 10.1136/jclinpath-2016-203990. [DOI] [PubMed] [Google Scholar]
- R Core Team, R.C.T. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- Reyngold M, Turcan S, Giri D, Kannan K, Walsh LA, Viale A, Drobnjak M, Vahdat LT, Lee W, Chan TA. Remodeling of the methylation landscape in breast cancer metastasis. PLoS ONE. 2014;9:e103896. doi: 10.1371/journal.pone.0103896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Lonigro RJ, Vats P, Cobain E, Everett J, Cao X, Rabban E, Kumar-Sinha C, Raymond V, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royston P, Altman DG. Regression Using Fractional Polynomials of Continuous Covariates: Parsimonious Parametric Modelling. Appl Stat. 1994;43:429. [Google Scholar]
- Salomonis N, Dexheimer PJ, Omberg L, Schroll R, Bush S, Huo J, Schriml L, Ho Sui S, Keddache M, Mayhew C, et al. Integrated Genomic Analysis of Diverse Induced Pluripotent Stem Cells from the Progenitor Cell Biology Consortium. Stem Cell Reports. 2016;7:110–125. doi: 10.1016/j.stemcr.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Sergushichev A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv. 2016:060012. [Google Scholar]
- Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva TC, Colaprico A, Olsen C, Bontempi G, Ceccarelli M, Berman BP, Noushmehr H. TCGAbiolinksGUI: A graphical user interface to analyze cancer molecular and clinical data. BioRxiv. 2017:147496. [Google Scholar]
- Sokolov A, Paull EO, Stuart JM. One-class detection of cell states in tumor subtypes. Pac Symp Biocomput. 2016;21:405–416. [PMC free article] [PubMed] [Google Scholar]
- de Souza CF, Sabedot TS, Malta TM, Stetson L, Morozova O, Sokolov A, Laird PW, Wiznerowicz M, Iavarone A, Snyder J, et al. Distinct epigenetic shift in a subset of Glioma CpG island methylator phenotype (G-CIMP) during tumor recurrence. Cell Reports. 2017 doi: 10.1016/j.celrep.2018.03.107. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. Systemic immunity is required for effective cancer immunotherapy. Cell. 2017;168:487–502.e15. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- StataCorp. Stata Statistical Software: Release 13. College Station TX: StataCorp LP; 2013. [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DTW, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellez CS, Juri DE, Do K, Bernauer AM, Thomas CL, Damiani LA, Tessema M, Leng S, Belinsky SA. EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res. 2011;71:3087–3097. doi: 10.1158/0008-5472.CAN-10-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York, NY: Springer New York; 2000. Estimating the survival and hazard functions; pp. 7–37. [Google Scholar]
- Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19:A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S-C, Lin C-C, Shih T-C, Tseng R-J, Yu M-C, Lin Y-J, Hsieh S-Y. The miR-200b-ZEB1 circuit regulates diverse stemness of human hepatocellular carcinoma. Mol Carcinog. 2017 doi: 10.1002/mc.22657. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venezia TA, Merchant AA, Ramos CA, Whitehouse NL, Young AS, Shaw CA, Goodell MA. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- Wang MD, Wu H, Fu GB, Zhang HL, Zhou X, Tang L, Dong LW, Qin CJ, Huang S, Zhao LH, et al. Acetyl-coenzyme A carboxylase alpha promotion of glucose-mediated fatty acid synthesis enhances survival of hepatocellular carcinoma in mice and patients. Hepatology. 2016;63:1272–1286. doi: 10.1002/hep.28415. [DOI] [PubMed] [Google Scholar]
- Wen N, Wang Y, Wen L, Zhao S-H, Ai Z-H, Wang Y, Wu B, Lu H-X, Yang H, Liu W-C, et al. Overexpression of FOXM1 predicts poor prognosis and promotes cancer cell proliferation, migration and invasion in epithelial ovarian cancer. J Transl Med. 2014;12:134. doi: 10.1186/1479-5876-12-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. ggplot2 - Elegant Graphics for Data Analysis. New York, NY: Springer New York; 2009. [Google Scholar]
- Wong CM, Wei L, Au SLK, Fan DNY, Zhou Y, Tsang FHC, Law CT, Lee JMF, He X, Shi J, et al. MiR-200b/200c/429 subfamily negatively regulates Rho/ROCK signaling pathway to suppress hepatocellular carcinoma metastasis. Oncotarget. 2015;6:13658–13670. doi: 10.18632/oncotarget.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhang L, Hu C, Liang S, Fei X, Yan N, Zhang Y, Zhang F. WNT pathway inhibitor pyrvinium pamoate inhibits the self-renewal and metastasis of breast cancer stem cells. Int J Oncol. 2016;48:1175–1186. doi: 10.3892/ijo.2016.3337. [DOI] [PubMed] [Google Scholar]
- Yan X, Ma L, Yi D, Yoon J, Diercks A, Foltz G, Price ND, Hood LE, Tian Q. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci USA. 2011;108:1591–1596. doi: 10.1073/pnas.1018696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Shen H, Laird PW, Farnham PJ, Berman BP. Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 2015;16:105. doi: 10.1186/s13059-015-0668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Huang D, Qin L, Zheng Z, Hua L, Wang G, Huang J, Huang H. Targeting Lung Cancer Stem Cells with Antipsychological Drug Thioridazine. Biomed Res Int. 2016;2016:6709828. doi: 10.1155/2016/6709828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016;375:819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Cherniack AD, Dewal N, Moffitt RA, Danilova L, Murray BA, Lerario AM, Else T, Knijnenburg TA, Ciriello G, et al. Comprehensive Pan-Genomic Characterization of Adrenocortical Carcinoma. Cancer Cell. 2016;29:723–736. doi: 10.1016/j.ccell.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Comparison of mRNAsi and mDNAsi values, as scored by signatures learned from PCBC data by OCLR (x axis) and Random Forest (y axis).
(B) Validation of mRNAsi in non-TCGA breast cancer (BRCA) samples (Reyngold et al., 2014) to define stemness status. Stratification of mRNAsi according to histology of TCGA samples (left) and of the validation cohort (right). IDC - invasive ductal carcinoma; ILC - invasive lobular carcinoma.
(C) Validation of stemness indices (mDNAsi and mRNAsi) in non-TCGA glioma samples (Sturm et al., 2012; Turcan et al., 2012) using our metric to define stemness status. The validation sets were previously classified into molecular subtypes of TCGA gliomas. Stratification of stemness indices according to molecular subtypes in the TCGA (left) and in the validation cohort (right).
(D) Correlation of stemness indices and estimated cell composition derived from deconvolution of tumor epigenomic features in TCGA breast cancer samples (Onuchic et al., 2016).
(E) Stemness indices in tumor vs. non-tumor samples across tissue type. Top plots show mRNAsi and bottom plots show mDNAsi. See list of Abbreviations of the TCGA Tumor Types.
(F) Correlation of stemness indices across TCGA tumor types. Tumor types on the x-axis are sorted by low-to-high correlation between mDNAsi and mRNAsi. See list of Abbreviations of the TCGA Tumor Types. mRNAsi, RNA-based stemness index. mDNAsi, DNA methylation-based stemness index. EREG-mRNAsi, epigenetically regulated-mRNAsi.
Table S1: Stemness indices derived for all PanCancer 33 TCGA cohort, Related to Figure 1.
Table S2: List of published stem cell markers (healthy and cancer) used to correlate to mRNAsi, Related to Figures 1 and 2.
(A) Heatmaps of DNA methylation levels of probe signature used to derive the mDNAsi in: Progenitor Cell Biology Consortium (PCBC) dataset (left), TCGA normal adjacent tissue (middle), and TCGA tumors (right). Columns represent samples. TCGA tumor samples are sorted from lowest to highest mDNAsi. Rows represent the DNA methylation probe signature (219 probes), which is a combination of three DNA methylation-based signatures: differentially methylated probes stemness index (DMPsi), enhancer stemness index (ENHsi), and epigenetically regulated-mDNAsi (EREG-mDNAsi) (see STAR Methods for detail). Annotation of probes are provided in the left of the heatmaps. First column (mDNAsi) indicates to which signature the probe belong. Second column (genomic location) indicates the genomic location annotation of each probe from the UCSC genome browser. Chromatin State columns indicates the chromatin state of pluripotent stem cells (iPSC and ESC) from Roadmap (n=9) (Roadmap Epigenomics Consortium et al., 2015).
(B) Scatter plots showing correlation of mDNAsi (x axis) and mRNAsi (y axis) by tumor type. Samples are colored according to TCGA classification in: primary, recurrent, metastatic, and non-tumors.
(C) Correlation between mRNAsi and mRNA expression of epithelial-to-mesenchymal transition (EMT) markers. The markers are grouped according to whether they are associated with epithelial (pink) or mesenchymal (teal blue) phenotype.
(A) (Left Panel) Comparison of normalized enrichment scores (NES) in mDNAsi (x-axis) and mRNAsi (y-axis) computed by tumor type in the mutation enrichment analysis. Positive and negative NES entail enrichment and depletion of a given mutation being associated with a high stemness index, respectively. The quadrants and colors represent genes with agreement or disagreement between the two distinct indices. (Right Panel) Stemness indices stratified by representative mutation status and tumor types.
(B) Relationship of mutations in epigenetic modifier genes and stemness indices mRNAsi (top) and mDNAsi (bottom). NES, normalized enrichment scores.
(A) (Left Panel) Comparison of miRNA expression associated with mDNAsi and mRNAsi. Log10-normalized FDR-adjusted P value is plotted for mDNAsi (x-axis) and mRNAsi (y-axis) for each miRNA probe for each tumor type. If a particular miRNA is upregulated in samples with a high stemness index, the values are multiplied by −1. The quadrants and colors represent proteins with agreement or disagreement between the two distinct indices. (Right Panel) Correlation of stemness indices and representative miRNAs for selected tumor types.
(B) Comparison of protein expression associated with mDNAsi and mRNAsi (left panel). Log10 (FDR-adjusted P value) is plotted for mDNAsi (x-axis) and mRNAsi (y-axis) for each protein for each tumor type. If the protein is upregulated in high stemness index, −1 is multiplied to log10 providing positive values. The quadrants and colors represent proteins with agreement or disagreement between the two distinct indices. Right, correlation scores of stemness indices and representative proteins and tumor types.
(C) Correlation of ANNEXIN1 and FOXM1 with mDNAsi (top) and mRNAsi (bottom) across tumor types.
(A and B) Tumor pathology grade associations with mRNAsi (A) and mDNAsi (B) by TCGA tumors types.
(C) OS and PFS curves for four (quartile-based) categories of the relevant indices. The figure also includes the estimated (solid line) functional forms of the dependence of the logarithm of hazard ratio (log-HR) of OS or PFS on the indices. The forms in general correspond to the ordering of the survival curves. For selected indices and outcomes the estimated functional form suggests a non-monotonic relationship between log-HR and the index.
(A) Plots show correlations between the mRNAsi (top) and mDNAsi (bottom) and the estimated fraction of cells for a selected panel of immune cell types. Additional correlations of stemness with estimated immune cell activity, computed as the difference between activated and resting population fractions, are included for NK cells, CD4+ T cells and macrophages. Correlation with PD-L1 protein expression is included for reference and the tumor types are sorted according to it.
(B) Moonlight dynamic recognition analysis to identify associations with the mRNAsi across tumor types.
Heatmap showing each compound (perturbagen) from the Connectivity Map that share gene targets in (rows). Sorted by descending number of targets.
Table S3: Differentially expressed genes for each tumor using mRNAsi score, Related to Figures 7A, 7B, S6C, and S7.
Table S4A: Output from the Connectivity Map, Related to Figure 7A.
Table S4B: Output from the Connectivity Map, Related to Figure 7B.
Table S4C: Output from the Connectivity Map, Related to Figure S7.