ABSTRACT
Foot-and-mouth disease virus (FMDV) can result in economical destruction of cloven-hoofed animals. FMDV infection has been reported to induce macroautophagy/autophagy; however, the precise molecular mechanisms of autophagy induction and effect of FMDV capsid protein on autophagy remain unknown. In the present study, we report that FMDV infection induced a complete autophagy process in the natural host cells of FMDV, and inhibition of autophagy significantly decreased FMDV production, suggesting that FMDV-induced autophagy facilitates viral replication. We found that the EIF2S1-ATF4 pathway was activated and the AKT-MTOR signaling pathway was inhibited by FMDV infection. We also observed that ultraviolet (UV)-inactivated FMDV can induce autophagy. Importantly, our work provides the first piece of evidence that expression of FMDV capsid protein VP2 can induce autophagy through the EIF2S1-ATF4-AKT-MTOR cascade, and we found that VP2 interacted with HSPB1 (heat shock protein family B [small] member 1) and activated the EIF2S1-ATF4 pathway, resulting in autophagy and enhanced FMDV replication. In addition, we show that VP2 induced autophagy in a variety of mammalian cell lines and decreased aggregates of a model mutant HTT (huntingtin) polyglutamine expansion protein (HTT103Q). Overall, our results demonstrate that FMDV capsid protein VP2 induces autophagy through interaction with HSPB1 and activation of the EIF2S1-ATF4 pathway.
KEYWORDS: AKT, ATF4, autophagy, EIF2S1, FMDV, HSPB1, MTOR, replication, VP2
Abbreviations
- 3-MA
3-methyladenine
- AMPK
AMP-activated protein kinase
- ATF4
activating transcription factor 4
- Baf-A1
bafilomycin A1
- DAPI
4’,6-diamidino-2-phenylindole
- EIF2S1
eukaryotic translation initiation factor 2 subunit alpha
- ER
endoplasmic reticulum
- FMD
foot-and-mouth disease
- FMDV
foot-and-mouth disease virus
- hpi
hours post infection
- HSPB1
heat shock protein family B (small) member 1
- KD
knockdown
- MAP1LC3B/LC3B
microtubule associated protein 1 light chain 3 beta
- MOI
multiplicity of infection
- MTOR
mechanistic target of rapamycin
- shRNA
short hairpin RNA
- TCID50
50% tissue culture infectious doses
- ULK1
unc-51 like autophagy activating kinase 1.
Introduction
Autophagy is a catabolic process to maintain cellular homeostasis.1,2 During autophagy, transient double-membrane sequestering compartments called phagophores are formed, which sequester damaged organelles and misfolded proteins, or portions of the cytoplasm.2 The sequestered components are selected through a series of cargo-specific autophagy receptors, such as SQSTM1/p62 and NBR1.2–4 At late phases of autophagy, the phagophores mature into closed autophagosomes that fuse with lysosomes in which the cargos are degraded and metabolic byproducts, such as amino acids, are released.2,5,6 Autophagy is induced by a variety of cellular and environmental stresses, such as nutrient and energy deficiency, pathogen infection, endoplasmic reticulum (ER) stress, hypoxia, and so on.7,8 The numerous ATG (autophagy-related) proteins make up several core molecular components of autophagy, which are essential for the formation of autophagosomes.1,7,9 The autophagic ULK1 (unc-51 like autophagy activating kinase 1) complex is commonly considered as an initiator in the autophagic cascade, and consists of ULK1, ATG13, ATG101 and RB1CC1/FIP200.9,10 Both MTOR (mechanistic target of rapamycin) and AMP-activated protein kinase (AMPK) are key regulators of the ULK1 complex.9,11 AMPK promotes autophagy by directly activating ULK1 through phosphorylation of Ser317 and Ser777 under conditions of glucose starvation.11 Conversely, MTOR negatively regulates autophagy by inhibiting the activity of ULK1 through direct phosphorylation of Ser757.11 AMPK also indirectly activates ULK1 by phosphorylating TSC2 and RPTOR to inactivate MTOR.9,12 Apart from AMPK, AKT/PKB plays an important role in regulating activity of MTOR through the tuberous sclerosis complex.13,14
Foot-and-mouth disease virus (FMDV), a single-stranded positive-sense RNA virus, is the prototypic member of the Aphthovirus genus within the family Picornaviridae.15–17 FMDV is the causative agent of foot-and-mouth disease, a highly contagious and economically devastating disease of domestic and wild cloven-hoofed animals.17,18 Foot-and-mouth disease is the first disease on the World Organisation for Animal Health listed for which the agency established an official list of free countries and zones. A wide range of DNA and RNA viruses were found to regulate the autophagy pathway.19–23 For example, porcine circovirus type 2 induces autophagy via the AMPK-MAPK/ERK-TSC2-MTOR signaling pathway.23 As opposed to porcine circovirus type 2, murine γ-herpesvirus 68 encodes M11 (a viral BCL2 protein) which interacts with BECN1/Beclin 1, leading to anti-autophagic activity.19
A number of studies had demonstrated that viral infection can result in an array of diverse stress stimuli.24–26 The phosphorylation of EIF2S1 (eukaryotic translation initiation factor 2 subunit alpha) is a response to several forms of stress, such as ER stress, viral infection and amino acid starvation.26 Phosphorylation of EIF2S1 leads to a rapid reduction of RNA translation and the selective translation of the mRNA encoding ATF4 (activating transcription factor 4), which is essential for amino acid metabolism, protein folding and autophagy.25–27 The EIF2S1-ATF4 pathway could activate transcription of numerous autophagy genes in response to stresses, and ATF4 plays a critical role in bortezomib-induced autophagy.26,28,29 HSPB1 knockdown activates the EIF2S1-ATF4 pathway and induces autophagy in PC-3 cells.30 Although studies have shown that FMDV infection can induce autophagy31,32, detailed mechanisms and the effect of autophagy on FMDV remain elusive. In the present study, we investigated molecular mechanisms of FMDV-induced autophagy in its natural host cells. The results provide the first piece of evidence that FMDV could activate the EIF2S1-ATF4 pathway in infected cells. FMDV infection inhibits the AKT-MTOR pathway through the EIF2S1-ATF4 pathway, leading to autophagy. Ultraviolet (UV)-inactivated FMDV can induce autophagy. The FMDV structural protein VP2 is responsible for the induction of autophagy. Besides, expression of VP2 alone is enough to induce autophagy in a variety of cells. We also found that VP2 interaction with HSPB1 activated the EIF2S1-ATF4 pathway, leading to autophagy.
Results
FMDV-induced autophagy plays an important role in viral replication
Although the phenomenon that FMDV can induce autophagy had been reported before, those studies were mainly performed in non-natural host-cells of FMDV31,32, such as mouse embryonic fibroblasts, and MCF-10A. The effect of virus on autophagy might be a cell type-specific effect. In our system, porcine kidney 15 (PK-15) cells were used from a natural host of FMDV. To explore whether FMDV induces cellular autophagy, the levels of MAP1LC3B/LC3B (microtubule associated protein 1 light chain 3 beta) was examined in FMDV-infected PK-15 cells. We found that LC3B-II expression was significantly upregulated at 1.5, 2 and 3 h post-infection (hpi), as compared with mock cells (Fig. 1A). FMDV induction of autophagy was also determined by visualizing the fluorescent puncta of LC3B. In agreement with immunoblotting, there were few fluorescent puncta in uninfected PK-15 cells, whereas FMDV-infected cells displayed a significant accumulation of fluorescent puncta (Fig. 1B). The ATG5 protein, with ATG12 and ATG16L1, forms a conjugation system that is involved in the phagophore elongation process. ATG5 knockdown significantly decreased levels of endogenous ATG5 protein and mRNA (Fig. 1C and S1). As shown in Fig. 1C, ATG5 knockdown significantly inhibited FMDV-induced autophagy. These results show that FMDV infection can induce autophagy through an ATG5-dependent pathway in PK-15 cells.
Figure 1.
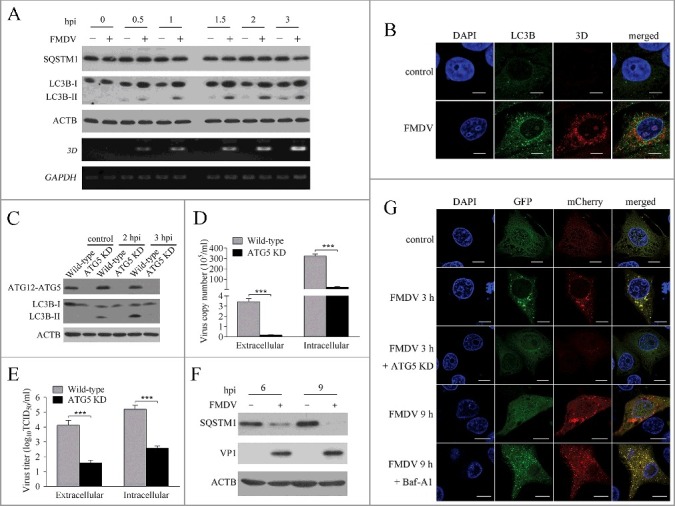
FMDV infection-induced autophagy plays an important role in viral replication. (A) PK-15 cells were mock infected or infected with FMDV (MOI = 1) for 0, 0.5, 1, 1.5, 2 and 3 h. The expression of LC3B and SQSTM1 was analyzed by western blot. The level of 3D was analyzed by RT-PCR. ACTB and GAPDH were used as a sample loading control. (B) The cells were then fixed and processed for indirect immunofluorescence using antibodies against LC3B and the 3D protein, followed by the corresponding secondary antibodies. The fluorescence signals were visualized by confocal immunofluorescence microscopy. (C) ATG5 knockdown (KD) and wild-type cells were infected with FMDV (MOI = 1) for 2 and 3 h. The expression of ATG5 and LC3B was analyzed by western blot. (D and E) ATG5 KD and wild-type cells were infected with FMDV (MOI = 1). At 3 hpi, both the extracellular and intracellular copy numbers of FMDV were measured by qRT-PCR; both the extracellular and intracellular virus titers were measured by TCID50. The data represent the mean ± SD of 3 independent experiments. ***P < 0.001. (F) PK-15 cells were mock infected or infected with FMDV (MOI = 1) for 6 and 9 h. The expression of SQSTM1 and VP1 were analyzed by western blot. (G) Cells were transfected with pmCherry-GFP-LC3B for 24 h, followed by FMDV infection (MOI = 1) and treatment with Baf-A1. The fluorescence signals were visualized by confocal immunofluorescence microscopy. Scale bars: 10 μm.
Next, we analyzed the effect of autophagy on FMDV replication. As shown in Fig. 1D and E, the copy number of FMDV RNA and viral titer significantly decreased in ATG5 knockdown PK-15 cells. 3-MA, an inhibitor of autophagy,33 also decreased FMDV production, and rapamycin, an inducer of autophagy, significantly increased FMDV yield (Fig. S2 and S3). These findings reveal that autophagy plays an important role in the replication of FMDV. Compared with control, the treatment (rapamycin and 3-MA) groups showed no differences in cell viability (Fig. S4).
FMDV infection enhanced autophagic flux
The accumulation of autophagosomes may be due to autophagy induction or a block in autophagosomal maturation.4 To confirm whether FMDV-induced autophagy is a complete process, we measured the degradation of SQSTM1, a marker for the autophagy-mediated protein degradation pathway.34 As shown in Fig. 1A, SQSTM1 was not significantly degraded at early stages of infection, but the level of SQSTM1 was significantly decreased at later stages of infection (6 and 9 hpi) (Fig. 1F), suggesting that the FMDV induced complete autophagy. To further confirm the observation, PK-15 cells were transfected with an mCherry-GFP-LC3B plasmid. This plasmid is the basis of a useful assay to monitor autophagic flux.4,35 The signal of green (GFP) is quenched by the low pH inside the lysosome lumen, whereas the red signal (mCherry) exhibits more stable fluorescence in acidic conditions.4,34 As shown in Fig. 1G, almost all of the green and red fluorescent puncta colocalized in the FMDV-infected PK-15 cells at 3 hpi. In contrast, numerous red fluorescent puncta were observed and numerous green fluorescent puncta were quenched at 9 hpi (Fig. 1G). Subsequently, PK-15 cells were treated with bafilomycin A1 (Baf-A1), a specific inhibitor of fusion between autophagosomes and lysosomes.36 Baf-A1 treatment dramatically recovered green fluorescent puncta and increased yellow puncta in FMDV-infected PK-15 cells (Fig. 1G). These data show that FMDV infection not only increases autophagosome formation, but also enhances autophagic flux.
FMDV triggered autophagy through the EIF2S1-ATF4-AKT-MTOR pathway
As MTOR and AMPK are key regulators of autophagy initiation11 the activity of MTOR and AMPK was analyzed in FMDV-infected PK-15 cells. As shown in Fig. 2A, the phosphorylation of MTOR S2448 was significantly inhibited by FMDV infection and became undetectable from 2 hpi, while the phosphorylation of T172 of AMPK was downregulated less than 1.5 fold in FMDV-infected PK-15 cells at 1.5 and 2 hpi (Fig. 2A). Further study revealed that phosphorylation of ULK1 S757 paralleled MTOR phosphorylation (Fig. 2A), suggesting that the activity of MTOR was inhibited by FMDV infection and low MTORC1 activity could not phosphorylate ULK1 S757, which resulted in autophagy initiation. It was known that AKT plays a key part in the maintenance of the activity of MTOR through the AKT-MTOR pathway13, so the activity of AKT was measured during FMDV infection. As shown in Fig. 2A, the phosphorylation of AKT S473 did significantly decrease in the FMDV-infected PK-15 cells. These data indicate that FMDV infection inhibited the AKT-MTOR pathway, leading to autophagy at early stages of infection.
Figure 2.
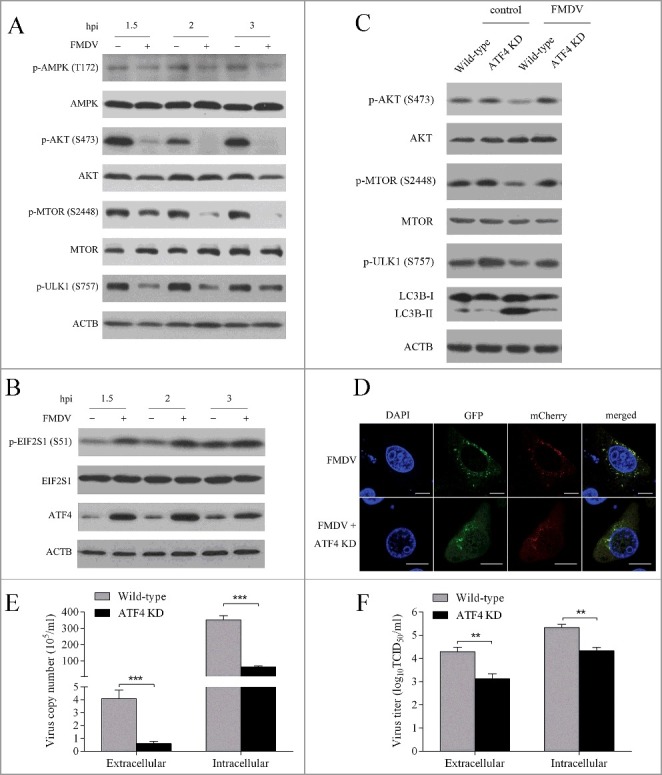
FMDV infection induces autophagy through the EIF2S1-ATF4-AKT-MTOR cascade. (A) PK-15 cells were mock infected or infected with FMDV (MOI = 1) for 1.5, 2 and 3 h. The phosphorylation of AKT, AMPK, MTOR and ULK1 were analyzed by western blot. ACTB was used as a sample loading control. (B) PK-15 cells were mock infected or infected with FMDV (MOI = 1) for 1.5, 2 and 3 h. ATF4 and phosphorylation of EIF2S1 were analyzed by western blot. (C) ATF4 KD and wild-type cells cells were infected with FMDV (MOI = 1). ATF4, LC3B and phosphorylation of AKT, MTOR and ULK1 were analyzed by western blot. (D) ATF4 and scrambled knockdown cells were transfected with pmCherry-GFP-LC3B for 24 h, followed by FMDV infection (MOI = 1) for 3 h. The fluorescence signals were visualized by confocal immunofluorescence microscopy. Scale bars: 10 μm. (E and F) ATF4 KD and wil-type cells cells were infected with FMDV (MOI = 1) for 3 h. At 3 hpi, both the extracellular and intracellular copy numbers of FMDV were measured by qRT-PCR; both the extracellular and intracellular virus titers were measured by TCID50. The data represent the mean ± SD of 3 independent experiments. **P < 0.01, ***P < 0.001.
A number of studies had demonstrated that viral infection can result in diverse stress.27,37,38 EIF2S1 responds to the stress of viral infection and interesting functional links have been revealed between the EIF2S1-ATF4 pathway and autophagy.28,39–41 We determined whether the EIF2S1-ATF4 pathway was activated by FMDV infection. As shown in Fig. 2B, the phosphorylation of EIF2S1 dramatically increased in the FMDV-infected cells. Consistent with results of EIF2S1, the level of ATF4 protein also significantly improved. These findings reveal that FMDV infection led to activation of the EIF2S1-ATF4 pathway during early stages of infection. To address whether the EIF2S1-ATF4 pathway and autophagy are linked during early stages of FMDV infection, ATF4 knockdown cells were used. As shown in Figure S5G and S5H, the levels of endogenous ATF4 protein and mRNA were significantly decreased compared with the control group. We found that suppression of ATF4 expression significantly decreased the accumulation of LC3B-II, suggesting that the EIF2S1-ATF4 pathway was involved in FMDV-induced autophagy (Fig. 2C). Consistent with immunoblotting data, ATF4 knockdown indeed inhibited the accumulation of fluorescent puncta induced by FMDV infection (Fig. 2D).
Given that the AKT-MTOR pathway was also involved in FMDV-induced autophagy, the relationship between the EIF2S1-ATF4 pathway and the AKT-MTOR pathway is unclear. The activity of AKT and MTOR was therefore examined in ATF4 knockdown cells. Immunoblotting revealed that ATF4 knockdown partly recovered the phosphorylation of AKT S473 and MTOR S2448 that were inhibited by FMDV infection (Fig. 2C). Similarly, the phosphorylation of ULK1 S757 was also partly rescued (Fig. 2C). These findings reveal that the FMDV-activated EIF2S1-ATF4 pathway inhibited the AKT-MTOR pathway, resulting in autophagy.
To evaluate the impact of the EIF2S1-ATF4 pathway on FMDV replication, the FMDV RNA copy number and virus titer were measured in ATF4 knockdown cells. As shown in Fig. 2E and F, ATF4 knockdown decreased FMDV production, suggesting that the inhibition of the EIF2S1-ATF4 pathway may decrease FMDV replication.
UV-inactivated FMDV could induce autophagy
To investigate whether FMDV induced-autophagy required viral replication, UV inactivation of FMDV was performed. Loss of infectivity was confirmed by detecting virus titers (data not shown). As shown in Fig. 3A, UV-FMDV infection increased the expression of LC3B-II in PK-15 cells, indicating that viral replication was not required for FMDV-induced autophagy. Next we examined whether the mechanism of UV-FMDV-induced autophagy agreed with FMDV-induced autophagy. Consistent with FMDV, UV-FMDV also activated the EIF2S1-ATF4 pathway and inhibited the AKT-MTOR pathway (Fig. 3A). To confirm UV-FMDV induced autophagy through the EIF2S1-ATF4 and AKT-MTOR pathways, the AKT- MTOR pathway and LC3B were examined in ATF4 knockdown cells. As shown in Fig. 3B, suppression of ATF4 expression partly recovered the activity of AKT and MTOR inhibited by UV-FMDV. The expression of LC3B-II was also decreased in ATF4 knockdown cells (Fig. 3B). These data reveal that the UV-FMDV-activated EIF2S1-ATF4 pathway leads to autophagy through the inhibition of the AKT-MTOR pathway.
Figure 3.
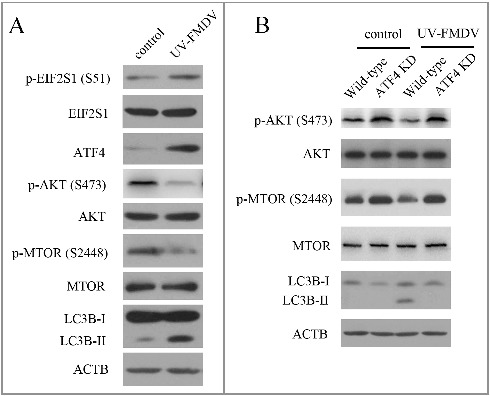
UV-FMDV infection induces autophagy. (A) PK-15 cells were mock infected or infected with UV-FMDV for 3 h (MOI = 10). LC3B and phosphorylation of EIF2S1, AKT and MTOR were analyzed by western blot. ACTB was used as a sample loading control. (B) ATF4 KD and wild-type cells were infected as described in (A).
The FMDV capsid protein VP2 induced autophagy
The FMDV capsid consists of 4 structural proteins, and 3 (VP1, VP2 and VP3) are exposed on the surface of the virus.42 VP1, VP2 and VP3 are able to interact with host receptors and are important to viral antigenicity.42 To further determine the effect of viral capsid proteins on autophagy, we transfected the cells with a capsid protein (VP1, VP2 or VP3) expression plasmid. When VP1, VP2 and VP3 were expressed in PK-15 cells, VP2 increased the level of LC3B-II, but VP1 and VP3 did not (Fig. 4A). Consistent with immunoblotting data, expression of VP2 increased the accumulation of fluorescent puncta (Fig. 4B). FMDV capsid protein VP2 could induce autophagy, suggesting that VP2 plays an important role in FMDV-induced autophagy. Compared with the empty vector control, VP2 activated the EIF2S1-ATF4 pathway and inhibited the AKT-MTOR pathway (Fig. 4C). To confirm if the mechanism of VP2-induced autophagy was consistent with FMDV, we examined the AKT- MTOR pathway and LC3B in ATF4 knockdown cells. When ATF4 was suppressed, the activity of AKT and MTOR inhibited by VP2 was partly recovered; and the level of LC3B-II was decreased (Fig. 4D). These data show that VP2 induces autophagy through the activation of the EIF2S1-ATF4 pathway and the inhibition of the AKT-MTOR pathway. Previous studies showed that amino acid changes in VP2 affect the FMDV replicative ability and virulence.42 Therefore the effect of a VP2 mutant on autophagy was detected in PK-15 cells. We found that expression of the VP2 mutant reduced the level of LC3B-II, compared with VP2 WT (Fig. 5B). Given that autophagy plays an important role in the replication of FMDV, VP2-induced autophagy may be a mechanism of FMDV infection.
Figure 4.
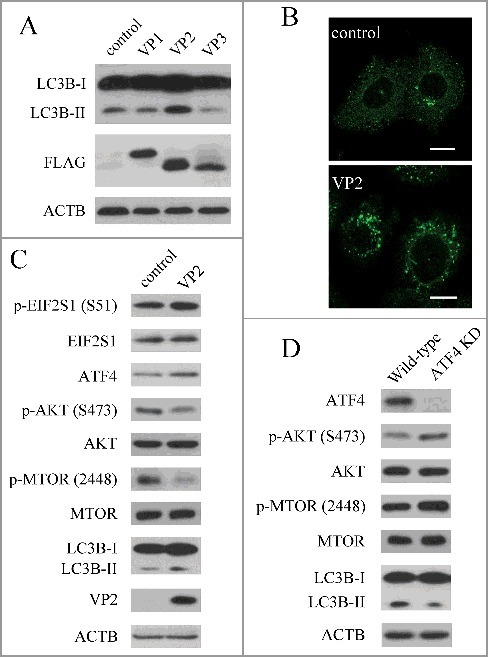
FMDV capsid protein VP2 induced autophagy. (A) PK-15 cells were transfected with empty vectors or various plasmids expressing FLAG-tagged VP1, VP2and VP3 proteins for 24 h. LC3B and FMDV capsid proteins were analyzed by western blot. ACTB was used as a sample loading control. (B) Cells were transfected with empty vectors or pCMV-Flag-VP2 for 24 h. Cells were fixed and analyzed by immunofluorescence using anti-LC3B antibodies. The fluorescence signals were visualized by confocal immunofluorescence microscopy. Scale bars: 10 μm. (C) PK-15 cells were transfected with empty vectors or pCMV-Flag-VP2 for 24 h. LC3B and phosphorylation of EIF2S1, AKT and MTOR were analyzed by western blot. ACTB was used as a sample loading control. (D) ATF4 and scrambled knockdown cells were transfected with pCMV-Flag-VP2 for 24 h. LC3B and phosphorylation of AKT and MTOR were analyzed by western blot. ACTB was used as a sample loading control.
Figure 5.
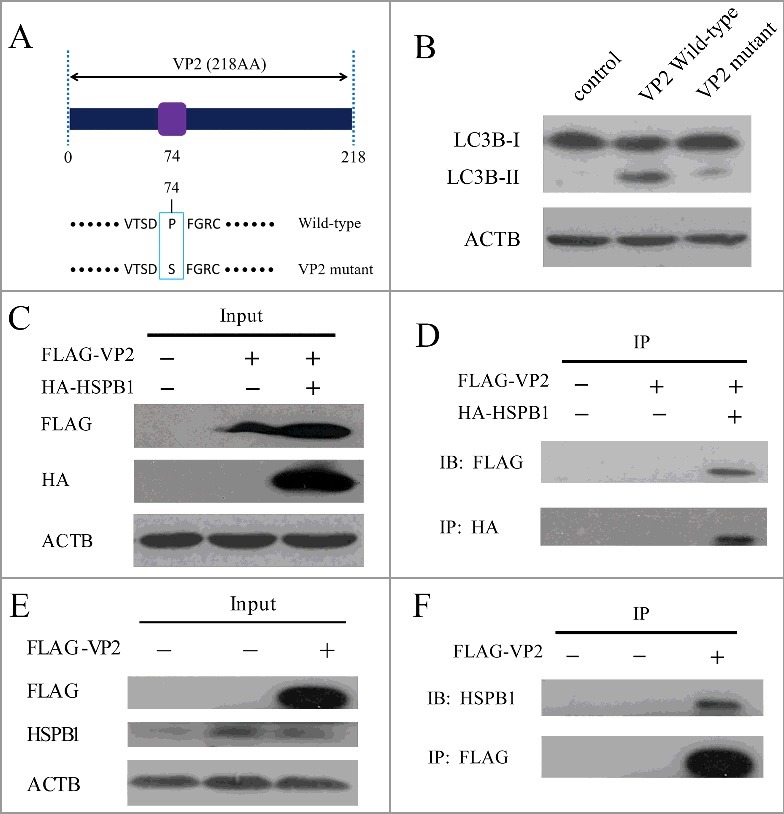
VP2 mutant and interaction between VP2 and HSPB1 in PK-15 cells. (A) A scheme of the VP2 muant. (B) PK-15 cells were transfected with empty vectors, pCMV-Flag-VP2, or VP2 mutant for 24 h. LC3B and ACTB were analyzed by western blot. (C and D) The interaction between FMDV VP2 and HSPB1 in PK-15 cells was verified. The PK-15 cells were co-transfected with 10 μg pCMV-Flag-VP2 plasmid and 10 μg pEGFP-HA-HSPB1 plasmid or transfected with just 10 μg pCMV-Flag-VP2 plasmid, and immunoprecipitation was performed with anti-HA antibody. Immunoblotting analysis was performed with anti-HA antibody and anti-FLAG antibody. (E and F) The PK-15 cells were transfected with 10 μg pCMV-Flag-VP2 plasmid or 10 μg empty vector pCMV-Flag plasmid, and immunoprecipitation was performed with anti-FLAG antibody. Immunoblotting analysis was performed with anti-FLAG antibody and anti-HSPB1 antibody.
VP2 interaction with HSPB1 activated the EIF2S1-ATF4 pathway
We performed immunoprecipitation to identify cellular proteins that interact with the VP2 protein. The protein that interacts with FLAG-VP2, but not the FLAG tag, was identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). HSPB1 (heat shock protein family B [small] member 1), a stress-activated multifunctional chaperone that inhibits treatment-induced apoptosis, was one of the top hits.30 Recent studies indicate that HSPB1 plays an important role in ER stress and activation of the unfolded protein response. HSPB1 silencing induced autophagy through induction of ER stress.30,43 To further examine the physical association between VP2 and HSPB1, 2 immunoprecipitation assays were performed. First, when overexpressed in PK-15 cells, FLAG-VP2 coprecipitated with HA-HSPB1 as shown in Fig. 5C and D. In addition, the endogenous HSPB1 that coprecipitated with the overexpressed FLAG-VP2 could be detected by the HSPB1 antibody (Fig. 5E and F). These results suggest that VP2 is able to interact with HSPB1. VP2 protein binding to HSPB1 may block the functionality of HSPB1, leading to activation of the EIF2S1-ATF4 pathway.
VP2-induced autophagy decreased aggregates of HTT103Q
Next VP2 was expressed in a variety of mammalian cell lines including HEK293T, and Vero. As shown in Fig. 6A and B, VP2 could efficiently induce autophagy in these cells. The expansion of a polyglutamine repeat in HTT (huntingtin) causes Huntington disease (HD). Mutant HTT (mHTT) forms aggregates and disrupts various cellular functions.44 Studies demonstrate that autophagy plays an important role in reduction of mHTT aggregate formation.45–47 A model of mutant HTT polyglutamine expansion protein (HTT103Q) was expressed in PK-15 cells. We found that VP2 decreased aggregates of HTT103Q in PK-15 cells, but not in ATG5 knockdown cells, suggesting that VP2-induced degradation of HTT103Q depends on the autophagy process (Fig. 6C, D and E).
Figure 6.
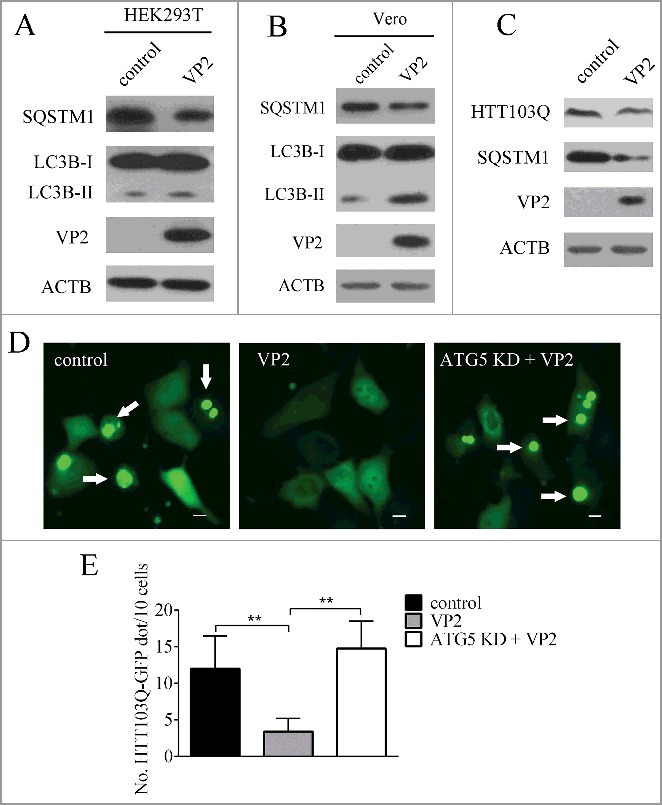
VP2 decreases aggregation of HTT103Q. (A) HEK293T cells and (B) Vero cells were transfected with empty vectors or pCMV-N-VP2 for 24 h. LC3B and SQSTM1 by were analyzed by western blot. ACTB was used as a sample loading control. (C) PK-15 cells were transfected with empty vectors or pCMV-EGFP-HTT103Q for 24 h. HTT103Q and SQSTM1 were analyzed by western blot. ACTB was used as a sample loading control. (D) PK-15 cells and ATG5 KD cells were co-transfected with pCMV-Flag-VP2 and pCMV-EGFP-HTT103Q for 24 h. The fluorescence signals were visualized by confocal immunofluorescence microscopy. Scale bars: 10 μm. (E) The number of HTT103Q dot was counted. The data represent the mean ± SD of 3 independent experiments. **P < 0.01.
Discussion
Autophagy plays an important role in intracellular homeostasis through degradation.2,5 This process not only degrades damaged organelles and misfolded or long-lived proteins, but also is a recycling pathway to provide nutrients and energy under various stresses.2 Our results show that FMDV infection can induce complete autophagy in PK-15 cells. Treatment of FMDV-infected PK-15 cells with rapamycin increased viral yield, and 3-MA treatment decreased viral replication. ATG5 knockdown further demonstrated the important role of autophagy in replication of FMDV. Autophagy has been proposed to provide a membrane platform for viral replication complexes or to mediate virus assembly and release.25 Consistent with Berryman's report31, FMDV nonstructural protein 3D, which is associated with viral replication, did not colocalize with autophagosomes, indicating that FMDV replication might not occur in autophagosomes. Type I IFN (interferon) production is a central component of the innate immune response to FMDV.48–50 Autophagy plays an important role in antiviral innate immunity and the autophagic machinery can regulate type I IFN signaling.51–54 Previous research shows that the ATG12–ATG5 conjugate negatively regulates type I IFN production pathway during VSV infection.53 And in the case of hepatitis C virus, several unfolded protein response-autophagy molecules (such as DDIT3/CHOP, ATG5 and LC3B) also play a negative role in regulation of innate immune responses.54 Autophagy could facilitate virus replication by suppressing innate antiviral immunity. We speculated that FMDV-induced autophagy may enhance FMDV replication through blocking type I IFN signaling. However, the relationship of FMDV-induced autophagy and host antivirus immunity needs further work.
As an important autophagy regulator, MTOR integrates signals from nutrients, growth factors and stress stimuli, with AMPK and AKT transducing them to the downstream autophagy pathway.9 We found that FMDV infection significantly inhibited phosphorylation of MTOR S2448. The phosphorylation of ULK1 S757 and AKT S473 was also inhibited in FMDV-infected PK-15 cells, indicating that FMDV infection inhibited the AKT-MTOR signaling pathway, leading to autophagy. It is well known that MTOR can activate RPS6KB/p70S6 kinase by phosphorylating Thr389 and RPS6KB can also phosphorylate MTOR at Ser2448.13,55,56 Therefore MTOR and RPS6KB form a positive-feedback loop that enforces both kinases’ activity downstream of AKT. The detailed relationship between MTOR and RPS6KB was not investigated further due to the lack of a porcine antibody to RPS6KB. The evidence that phosphorylation of EIF2S1 and ATF4 significantly increased in FMDV-infected cells indicates that FMDV infection could activate the EIF2S1-ATF4 pathway at the early stages of infection. ATF4 knockdown not only decreased FMDV-induced autophagy, but also partly recovered the activity of the AKT-MTOR pathway inhibited by FMDV. These results indicate that the activation of the EIF2S1-ATF4 pathway by FMDV infection inhibits the AKT-MTOR pathway and leads to autophagy.
The crosstalk between ATF4 and AKT-MTOR is complicated. In pancreatic islet cells, ATF4-DDIT3/CHOP stimulates TRIB3 expression, and TRIB3 reduces AKT phosphorylation through an undefined mechanism.57 Cancer cells treated with an ER stress-inducing agent show reduced MTOR activity and associated increases in the expression levels of ATF4. The reduction of MTOR activity is at least partially caused by ATF4-mediated MTOR inhibitor SQSN2 (sestrin 2) expression.58 Collectively, the phenomenon that ATF4 and ER stress can inhibit the AKT-MTOR pathway had been observed in many cell types, but the mechanism had not been carefully studied; it may be complicated and diverse in different cell types. UV-FMDV can also induce autophagy through the EIF2S1-ATF4 and AKT-MTOR pathways, suggesting that FMDV-induced autophagy is independent of viral replication. However, there are also other mechanisms of autophagy involved in the EIF2S1-ATF4 pathway. The EIF2S1-ATF4 pathway could activate transcription of numerous autophagy genes in response to stresses and ATF4 plays a critical role in bortezomib-induced autophagy.28,29
We found that FMDV-induced autophagy plays a key role in viral replication and the FMDV structural protein VP2 could induce autophagy, suggesting that VP2-induced autophagy may favor viral infection and replication in host cells. Consistent with a previous study that amino acid changes in VP2 affect FMDV replication and virulence42, we found that expression of a VP2 mutant cannot induce autophagy in PK-15 cells. In particular, the VP2 protein interacted with HSPB1. Recent reports showed that HSPB1 inhibition induces ER stress and autophagy in PC-3 cells.30 These data link the FMDV structural protein VP2 to activation of the EIF2S1-ATF4 pathway. During FMDV infection in PK-15 cells, VP2-blockade against HSPB1 resulted in activation of the EIF2S1-ATF4 pathway. Vaccination is currently a major strategy to control FMDV infection, and peptide vaccines against FMDV are based on capsid protein VP1 and a nonstructural protein.59–63 However, the role of VP2 in peptide vaccines has been largely ignored. Now we found that VP2 may be used as a new therapeutic target against FMDV. Studies demonstrated that inhibition of MTOR induces autophagy and decreases aggregate formation in a model of Huntington disease, and rapamycin or CCI-779 might be used to treat Huntington disease.44 Consistent with rapamycin, FMDV capsid protein VP2 induces autophagy through inhibition of MTOR. VP2 may be used as a potential therapeutic molecule against Huntington disease. The full-length VP2 maybe induce host immune responses; therefore, it is necessary to identify a short core peptide of VP2 that is sufficient for autophagy induction.
In this study, we report that the EIF2S1-ATF4 pathway regulates autophagy through the AKT-MTOR signaling pathway during FMDV infection. We show that FMDV-induced autophagy facilitates viral replication. We also found that UV-inactivated FMDV could induce autophagy. Moreover, we identified for the first time that the FMDV capsid protein VP2 induced autophagy via interaction with HSPB1.
Materials and methods
Antibodies, reagents and plasmids
The antibodies used and their sources are as follows: Anti-LC3B (2775), anti-AKT (9272), anti-phospho-AKT (4060), anti-phospho-MTOR (5536), anti-phospho-ULK1 (6888), anti-EIF2S1 (2103) and anti-phospho-AMPK (2535) antibodies were obtained from Cell Signaling Technology. Anti-SQSTM1/p62 (18420-1-AP), anti-ATF4 (10835-1-AP), anti-HA (66006) and anti-ACTB (66009) antibodies were obtained from Proteintech Group. Anti-MTOR (ab2731), anti-HSPB1 (ab2790) and anti-phospho-EIF2S1 (ab32157) antibodies were obtained from Abcam. Anti-ATG5 (NB110-53818) antibody was obtained from Novus Biologicals. Horseradish peroxidase-labeled goat anti-rabbit (170-6515) and anti-mouse (170-6516) secondary antibodies were obtained from Bio-Rad. 3-methyladenine (3-MA, M9281) was purchased from Sigma-Aldrich. Bafilomycin A1 (Baf-A1, B-1080) and rapamycin (R-5000) were purchased from LC Laboratories. pCMV-N-Flag (D2722) and pCMV-C-EGFP (D2626) were obtained from Beyotime. pmCherry-GFP-LC3B was obtained from MiaoLingBio. HTT103Q was provided by Dr. Ye-Guang Chen (School of Life Sciences, Tsinghua University).
Cell cultures, transfection, and virus infection
The swine kidney cell line PK-15 (ATCC, CCL-33), HEK293T (ATCC, CRL-11268) and Vero (ATCC, CCL-81) cells were maintained in complete Dulbecco's modified Eagle's medium (Gibco, C11995500BT) supplemented with 10% fetal bovine serum and 1% antibiotics. The cells were incubated at 37°C with 5% CO2. Cells grown to approximately 80% confluence in a petri dish with a glass bottom (NEST, GBD-35-20) were transfected with plasmid by using Lipofectamine® 2000 Reagent (Life Technologies, 11668-019).
The FMDV type O strain used in the study was prepared in our laboratory. The FMDV type O strain was propagated in PK-15 cells cultured with 2% fetal bovine serum. Viral stocks were prepared at a multiplicity of infection (MOI) of 1 for autophagy induction. Virus titers were calculated according to Kaerber64 and expressed as 50% tissue culture infectious doses (TCID50) per milliliter. PK-15 cells were grown to approximately 80% confluence in cell culture plates and were infected with FMDV at 1 MOI. After 1 h, the inoculum was removed by aspiration. The cells were then washed twice with phosphate-buffered saline (Gibco, 10010023) and incubated in complete medium at 37°C for different times until harvesting. UV-inactivated FMDV was generated by exposing to a UV light source for 15 min. Loss of infectivity was confirmed by detecting virus titers.
Immunoblotting
Cell monolayers were incubated on ice with RIPA lysis buffer (Beyotime, P0013B) containing a protease inhibitor cocktail (Roche, 4693132001) and a phosphatase inhibitor cocktail (Roche, 4906845001). The lysates were then clarified by centrifugation at 16,000 × g for 5 min at 4°C, and the protein concentration was quantified using the BCA protein assay kit (Thermo Scientific, 23225). Equal amounts of protein samples were denatured for 5 min in 2 × SDS-PAGE loading buffer (Sigma-Aldrich, S3401). Proteins were separated on SDS-PAGE gels and then electrotransferred onto nitrocellulose membrane (Bio-Rad, 162-0115) or polyvinylidene fluoride membranes (Millipore, ISEQ00010), which were then blocked for 1 h at room temperature in Tris-buffered saline (Sigma-Aldrich, V900838-4L) containing 5% nonfat milk powder and 0.1% Tween 20 (Sigma-Aldrich, P7949). Next, the membranes were incubated with primary antibodies at 4°C overnight and then with the corresponding secondary antibodies conjugated to horseradish peroxidase at room temperature for 1 h. The protein bands were detected using the ECL kit (Thermo, 34080).
Immunoprecipitation
PK-15 cells were transfected for 24 h and were incubated on ice with RIPA lysis buffer containing a protease inhibitor cocktail, a phosphatase inhibitor cocktail, 1 mM PMSF and 1% Trion X-100. For each sample, 0.6 ml of lysate was incubated with 1 μg of antibody and 600 μl protein G-Sepharose beads (GE Healthcare, 17-0618-01) overnight. The Sepharose beads were washed 3 times with 1 ml of lysis buffer containing 1% NP-40 (Beyotime, ST366). The precipitates were detected by immunoblotting.
Lentiviral shRNA packaging, infection and selection
For packaging lentiviruses, 1 μg pLKO.1 shRNA plasmid (Addgene, 24150), 750 ng psPAX2 packaging plasmid (Addgene, 12260), and 250 ng pMD2.G envelope plasmid (Addgene, 12259) were cotransfected into 7 × 104 HEK293 cells with 6 μl FuGENE® HD Transfection Reagent (Promega, E2311). The supernatants containing lentiviruses were collected, filtered and stored at -80°C as aliquots. For infection, PK-15 cells were incubated with viral stocks supplemented with 8 μg/ml polybrene (Solarbio, H8761) for 10 h, and then supplied with fresh medium. Cells were selected with hygromycin (Invitrogen, 10687010) at 24 h postinfection. The following primers were used:
ATG5 shRNA: GCACACCACTGAAATGGCATT;
ATF4 shRNA: GGAAATCTCGGAAGGAGATAG
Reverse transcription PCR and quantitative real-time PCR
Total RNA was isolated using a RNeasy Mini Kit (Qiagen, 74104) according to the provided handbook and 1 μg of the total RNA was reverse-transcribed using a Transcriptor First Strand cDNA Synthesis Kit (Roche, 04897030001). The real-time RT-PCR was performed using the SuperScript® III Platinum®One-Step qRT-PCR Kit (Thermo Fisher Scientific, 11732-020) and DyNAmo Flash SYBR Green qPCR Kit (Thermo Fisher Scientific, F-415S).
3D sense Primer: 5′-ACTGGGTTTTAYAAACCTGTGATG-3′,
3D anti-sense Primer: 5′-TCAACTTCTCCTGKATGGTCCCA-3′,
Probe: ATCCTCTCCTTTGCACGC;
ATG5 sense: 5′-GGGAGGCAGAACCGTATTATTT-3′,
ATG5 anti-sense: 5′-TCAGTGGTGTGCCTTCATATTC-3′;
ATF4 sense: 5′-GTGGAAATCTCGGAAGGAGATAG-3′,
ATF4 anti-sense: 5′-AGGAGTCAGGGCTCATACA-3′;
GAPDH sense: 5′-TGGAGTCCACTGGTGTCTTCAC-3′,
GAPDH anti-sense: 5′-TTCACGCCCATCACAAACA-3′.
Immunofluorescence and confocal microscopy
PK-15 cells grown to approximately 80% confluence in a petri dish with a glass bottom (NEST, GBD-35-20) were infected with FMDV at 1 MOI. After 3 hpi, cells were fixed using methanol for 15 min at -20°C. Cells were blocked with phosphate-buffered saline containing 5% goat serum (Life Technologies, 16210064) for 1 h at room temperature. Next, cells were incubated overnight at 4°C in the presence of primary antibody (anti-LC3B and anti-3D) (1:200), followed by incubation in fluorochrome-conjugated secondary antibody (Alexa Fluor® 488 and 546, 1:200; Thermo, A11070 and A11018) diluted in antibody dilution buffer for 1 h at room temperature in the dark. Then cells were incubated with DAPI (Roche, 10236276001) for nuclear staining. The fluorescence signals were visualized with a TCS SP8 confocal fluorescence microscope (Leica).
Cell viability assay
Cell viability was determined by the MTT assay as described previously.64
Statistical analysis
The Student t test was used for statistical analysis. The results are expressed as mean ± SD.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary Material
Funding Statement
This work was supported by National Natural Science Foundation of China (NSFC) [grant number 31572522, 31201943]; DH | National Institute for Health Research (NHS) [grant number AI103807, AI099625]; Fundamental Research Funds of the Chinese Academy of Agricultural Sciences [grant number 1610312016026].
Acknowledgments
This work was supported by National Natural Science Foundation of China (No.31572522 and No. 31201943), Fundamental Research Funds of the Chinese Academy of Agricultural Sciences (1610312016026) and the National Institutes of Health research support to P.W. (AI103807 and AI099625). We thank Dr. Pu Wang (Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences) for suggestions.
References
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. PMID:20811353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–20. doi: 10.1038/ncb2788. PMID:23817233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katsuragi Y, Ichimura Y, Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282(24):4672–8. doi: 10.1111/febs.13540. PMID:26432171 [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Abdalla FC, Abeliovich H, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. PMID:22966490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hale AN, Ledbetter DJ, Gawriluk TR, et al. . Autophagy: regulation and role in development. Autophagy. 2013;9:951–72. doi: 10.4161/auto.24273. PMID:24121596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–72. doi: 10.1038/nrm4024. PMID:26177004 [DOI] [PubMed] [Google Scholar]
- 7.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. PMID:20965422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King JS, Veltman DM, Insall RH. The induction of autophagy by mechanical stress. Autophagy. 2011;7:1490–9. doi: 10.4161/auto.7.12.17924. PMID:22024750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong PM, Puente C, Ganley IG, et al. . The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–37. doi: 10.4161/auto.23323. PMID:23295650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nazarko VY, Zhong Q. ULK1 targets Beclin-1 in autophagy. Nat Cell Biol. 2013;15:727–8. doi: 10.1038/ncb2797. PMID:23817237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Kundu M, Viollet B, et al. . AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011;13:132–41. doi: 10.1038/ncb2152. PMID:21258367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ha J, Guan KL, Kim J. AMPK and autophagy in glucose/glycogen metabolism. Mol Aspects Med. 2015;46:46–6. doi: 10.1016/j.mam.2015.08.002. PMID:26297963 [DOI] [PubMed] [Google Scholar]
- 13.Buchkovich NJ, Yu Y, Zampieri CA, et al. . The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat Rev Microbiol. 2008;6:266–75. doi: 10.1038/nrmicro1855. PMID:18311165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, et al. . The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014;26:2694–701. doi: 10.1016/j.cellsig.2014.08.019. PMID:25173700 [DOI] [PubMed] [Google Scholar]
- 15.Mason PW, Grubman MJ, Baxt B. Molecular basis of pathogenesis of FMDV. Virus Res. 2003;91:9–32. doi: 10.1016/S0168-1702(02)00257-5. PMID:12527435 [DOI] [PubMed] [Google Scholar]
- 16.Tulloch F, Pathania U, Luke GA, et al. . FMDV replicons encoding green fluorescent protein are replication competent. J Virol Methods. 2014;209:35–40. doi: 10.1016/j.jviromet.2014.08.020. PMID:25194890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein J. Understanding the molecular epidemiology of foot-and-mouth-disease virus. Infect Genetics Evolution: J Mol Epidemiol Evolutionary Genetics Infect Dis. 2009;9:153–61. doi: 10.1016/j.meegid.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrillo C, Tulman ER, Delhon G, et al. . Comparative genomics of foot-and-mouth disease virus. J Virol. 2005;79:6487–504. doi: 10.1128/JVI.79.10.6487-6504.2005. PMID:15858032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ku B, Woo JS, Liang C, et al. . Structural and biochemical bases for the inhibition of autophagy and apoptosis by viral BCL-2 of murine gamma-herpesvirus 68. PLoS Pathogens. 2008;4:e25. doi: 10.1371/journal.ppat.0040025. PMID:18248095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dreux M, Gastaminza P, Wieland SF, et al. . The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106:14046–51. doi: 10.1073/pnas.0907344106. PMID:19666601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor MP, Kirkegaard K. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy. 2008;4:286–9. doi: 10.4161/auto.5377. PMID:18094610 [DOI] [PubMed] [Google Scholar]
- 22.Orvedahl A, MacPherson S, Sumpter R Jr, et al. . Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–27. doi: 10.1016/j.chom.2010.01.007. PMID:20159618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu B, Zhou Y, Xu F, et al. . Porcine circovirus type 2 induces autophagy via the AMPK/ERK/TSC2/mTOR signaling pathway in PK-15 cells. J Virol. 2012;86:12003–12. doi: 10.1128/JVI.01434-12. PMID:22915817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Gassart A, Bujisic B, Zaffalon L, et al. . An inhibitor of HIV-1 protease modulates constitutive eIF2alpha dephosphorylation to trigger a specific integrated stress response. Proc Natl Acad Sci U S A. 2016;113:E117–26. doi: 10.1073/pnas.1514076113. PMID:26715744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiramel AI, Brady NR, Bartenschlager R. Divergent roles of autophagy in virus infection. Cells. 2013;2:83–104. doi: 10.3390/cells2010083. PMID:24709646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donnelly N, Gorman AM, Gupta S, et al. . The eIF2alpha kinases: their structures and functions. Cell Mol Life Sci. 2013;70:3493–511. doi: 10.1007/s00018-012-1252-6. PMID:23354059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fung TS, Torres J, Liu DX. The emerging roles of viroporins in ER stress response and autophagy induction during virus infection. Viruses. 2015;7:2834–57. doi: 10.3390/v7062749. PMID:26053926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.B'Chir W, Maurin AC, Carraro V, et al. . The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–99. doi: 10.1093/nar/gkt563. PMID:23804767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milani M, Rzymski T, Mellor HR, et al. . The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–23. doi: 10.1158/0008-5472.CAN-08-2839. PMID:19417138 [DOI] [PubMed] [Google Scholar]
- 30.Kumano M, Furukawa J, Shiota M, et al. . Cotargeting stress-activated Hsp27 and autophagy as a combinatorial strategy to amplify endoplasmic reticular stress in prostate cancer. Mol Cancer Therapeutics. 2012;11:1661–71. doi: 10.1158/1535-7163.MCT-12-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berryman S, Brooks E, Burman A, et al. . Foot-and-mouth disease virus induces autophagosomes during cell entry via a class III phosphatidylinositol 3-kinase-independent pathway. J Virol. 2012;86:12940–53. doi: 10.1128/JVI.00846-12. PMID:22993157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Donnell V, Pacheco JM, LaRocco M, et al. . Foot-and-mouth disease virus utilizes an autophagic pathway during viral replication. Virology. 2011;410:142–50. doi: 10.1016/j.virol.2010.10.042. PMID:21112602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–92. doi: 10.1073/pnas.79.6.1889. PMID:6952238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. PMID:20144757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klionsky DJ, Abdelmohsen K, Abe A, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. PMID:26799652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto A, Tagawa Y, Yoshimori T, et al. . Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. PMID:9639028 [DOI] [PubMed] [Google Scholar]
- 37.Benali-Furet NL, Chami M, Houel L, et al. . Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921–33. doi: 10.1038/sj.onc.1208673. PMID:15897896 [DOI] [PubMed] [Google Scholar]
- 38.Blazquez AB, Escribano-Romero E, Merino-Ramos T, et al. . Stress responses in flavivirus-infected cells: activation of unfolded protein response and autophagy. Frontiers Microbiol. 2014;5:266. doi: 10.3389/fmicb.2014.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yorimitsu T, Klionsky DJ. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy. 2007;3:160–2. doi: 10.4161/auto.3653. PMID:17204854 [DOI] [PubMed] [Google Scholar]
- 40.Yorimitsu T, Nair U, Yang Z, et al. . Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006;281:30299–304. doi: 10.1074/jbc.M607007200. PMID:16901900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kouroku Y, Fujita E, Tanida I, et al. . ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differentiation. 2007;14:230–9. doi: 10.1038/sj.cdd.4401984. PMID:16794605 [DOI] [PubMed] [Google Scholar]
- 42.Xue M, Wang H, Li W, et al. . Effects of amino acid substitutions in the VP2 B-C loop on antigenicity and pathogenicity of serotype Asia1 foot-and-mouth disease virus. Virol J. 2012;9:191. doi: 10.1186/1743-422X-9-191. PMID:22963009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee JJ, Seah JB, Chow VT, et al. . Comparative proteome analyses of host protein expression in response to Enterovirus 71 and Coxsackievirus A16 infections. J Proteomics. 2011;74:2018–24. doi: 10.1016/j.jprot.2011.05.022. PMID:21621020 [DOI] [PubMed] [Google Scholar]
- 44.Walter C, Clemens LE, Muller AJ, et al. . Activation of AMPK-induced autophagy ameliorates Huntington disease pathology in vitro. Neuropharmacol. 2016;108:24–38. doi: 10.1016/j.neuropharm.2016.04.041. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006;172:719–31. doi: 10.1083/jcb.200510065. PMID:16505167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar S, Ravikumar B, Floto RA, et al. . Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differentiation. 2009;16:46–56. doi: 10.1038/cdd.2008.110. PMID:18636076 [DOI] [PubMed] [Google Scholar]
- 47.Sarkar S, Rubinsztein DC. Huntington's disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–70. doi: 10.1111/j.1742-4658.2008.06562.x. PMID:18637946 [DOI] [PubMed] [Google Scholar]
- 48.Chinsangaram J, Moraes MP, Koster M, et al. . Novel viral disease control strategy: adenovirus expressing alpha interferon rapidly protects swine from foot-and-mouth disease. J Virol. 2003;77:1621–5. doi: 10.1128/JVI.77.2.1621-1625.2003. PMID:12502879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Summerfield A, Guzylack-Piriou L, Harwood L, et al. . Innate immune responses against foot-and-mouth disease virus: current understanding and future directions. Veterinary Immunol Immunopathol. 2009;128:205–10. doi: 10.1016/j.vetimm.2008.10.296. [DOI] [PubMed] [Google Scholar]
- 50.Chinsangaram J, Koster M, Grubman MJ. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J Virol. 2001;75:5498–503. doi: 10.1128/JVI.75.12.5498-5503.2001. PMID:11356957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sumpter R, Jr, Levine B. Autophagy and innate immunity: triggering, targeting and tuning. Seminars Cell Dev Biol. 2010;21:699–711. doi: 10.1016/j.semcdb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richetta C, Faure M. Autophagy in antiviral innate immunity. Cell Microbiol. 2013;15:368–76. doi: 10.1111/cmi.12043. PMID:23051682 [DOI] [PubMed] [Google Scholar]
- 53.Jounai N, Takeshita F, Kobiyama K, et al. . The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–5. doi: 10.1073/pnas.0704014104. PMID:17709747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ke PY, Chen SS. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. PMID:21135505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiang GG, Abraham RT. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J Biol Chem. 2005;280:25485–90. doi: 10.1074/jbc.M501707200. PMID:15899889 [DOI] [PubMed] [Google Scholar]
- 56.Holz MK, Blenis J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J Biol Chem. 2005;280:26089–93. doi: 10.1074/jbc.M504045200. PMID:15905173 [DOI] [PubMed] [Google Scholar]
- 57.Bromati CR, Lellis-Santos C, Yamanaka TS, et al. . UPR induces transient burst of apoptosis in islets of early lactating rats through reduced AKT phosphorylation via ATF4/CHOP stimulation of TRB3 expression. Am J Physiol Regulatory, Integrative Comparative Physiol. 2011;300:R92–100. doi: 10.1152/ajpregu.00169.2010. [DOI] [PubMed] [Google Scholar]
- 58.Bruning A, Rahmeh M, Friese K. Nelfinavir and bortezomib inhibit mTOR activity via ATF4-mediated sestrin-2 regulation. Mol Oncol. 2013;7:1012–8. doi: 10.1016/j.molonc.2013.07.010. PMID:23916134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Z, Pan L, Ding Y, et al. . Efficacy of synthetic peptide candidate vaccines against serotype-A foot-and-mouth disease virus in cattle. Applied Microbiol Biotechnol. 2015;99:1389–98. doi: 10.1007/s00253-014-6129-1. [DOI] [PubMed] [Google Scholar]
- 60.Cao Y, Lu Z, Liu Z. Foot-and-mouth disease vaccines: progress and problems. Exp Rev Vaccines. 2016;15:783–9. doi: 10.1586/14760584.2016.1140042. [DOI] [PubMed] [Google Scholar]
- 61.Ludi A, Rodriguez L. Novel approaches to foot-and-mouth disease vaccine development. Dev Biol. 2013;135:107–16. [DOI] [PubMed] [Google Scholar]
- 62.Rodriguez LL, Gay CG. Development of vaccines toward the global control and eradication of foot-and-mouth disease. Exp Rev Vaccines. 2011;10:377–87. doi: 10.1586/erv.11.4. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez LL, Grubman MJ. Foot and mouth disease virus vaccines. Vaccine. 2009;27(Suppl 4):D90–4. doi: 10.1016/j.vaccine.2009.08.039. PMID:19837296 [DOI] [PubMed] [Google Scholar]
- 64.Pei J, Zhao M, Ye Z, et al. . Autophagy enhances the replication of classical swine fever virus in vitro. Autophagy. 2014;10:93–110. doi: 10.4161/auto.26843. PMID:24262968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


