ABSTRACT
Ubiquitin chains are formed with 8 structurally and functionally distinct polymers. However, the functions of each polyubiquitin remain poorly understood. We developed a polyubiquitin-mediated fluorescence complementation (PolyUb-FC) assay using Kusabira Green (KG) as a split fluorescent protein. The PolyUb-FC assay has the advantage that monoubiquitination is nonfluorescent and chain-specific polyubiquitination can be directly visualized in living cells without using antibodies. We applied the PolyUb-FC assay to examine K33-linked polyubiquitin. We demonstrated that SQSTM1/p62 puncta colocalized with K33-linked polyubiquitin and this interaction was modulated by the ZRANB1/TRABID-K29 and -K33 linkage-specific deubiquitinase (DUB). We further showed that the colocalization of K33-linked polyubiquitin and MAP1LC3/LC3 (microtubule associated protein 1 light chain 3) puncta was impaired by SQSTM1/p62 deficiency. Taken together, these findings provide novel insights into how atypical polyubiquitin is recruited by SQSTM1/p62. Finally, we developed an inducible-PolyUb-FC system for visualizing chain-specific polyubiquitin. The PolyUb-FC will be a useful tool for analyzing the dynamics of atypical polyubiquitin chain generation.
KEYWORDS: autophagy, BiFC, K33, SQSTM1/p62, ubiquitin
Abbreviations
- ACTB
actin beta
- BiFC
bimolecular fluorescence complementation
- DUB
deubiquitinase
- GFP
green fluorescent protein
- iPolyUb-FC
inducible polyubiquitin-mediated fluorescence complementation
- KG/Mkg
Kusabira Green, monomeric
- LAMP2
lysosomal associated membrane protein 2
- LLOMe
L-leucyl-L-leucine methyl ester
- MAP1LC3/LC3
microtubule associated protein 1 light chain 3
- MEFs
mouse embryonic fibroblasts
- MW
molecular weight
- NCS
neocarzinostatin
- PBS
phosphate-buffered saline
- PolyUb-FC
polyubiquitin-mediated fluorescence complementation
- SQSTM1/p62
sequestosome 1
- Ub
ubiquitin
- UBA
ubiquitin-associated
- ZRANB1/TRABID
zinc finger RANBP2-type containing 1
Introduction
Ubiquitination is one of the most abundant post-translational modifications found in cells. Ubiquitin chains can be linked through one of 7 ubiquitinated Lys residues (which are K6, K11, K27, K29, K33, K48, and K63) or through the ubiquitin amino terminal Met1 residue (which generates linear chains). Whereas the roles of K48- and K63-linked polyubiquitin are well characterized, the function of the remaining 6 atypical ubiquitin chain types (linked via K6, K11, K27, K29, K33, and Met1) remain poorly defined.1 Polyubiquitin can contain a single type of linkage or multiple linkage types (termed heterotypic chains). Heterotypic chains are either branched (also termed ‘forked’) or non-branched (also termed ‘mixed’).1-4 Unanchored polyubiquitin chains can also be found in cells.4,5
To monitor the dynamics of ubiquitin chain formation in vivo, chain-specific antibodies,6,7 ubiquitin sensor proteins,8-11 tandem ubiquitin-binding entities technology,12 and genetic approaches (inducible knock-in system to replace endogenous ubiquitin)13 were developed. Nevertheless, it is still not possible to visualize all types of atypical ubiquitin chains. Therefore, we examined bimolecular fluorescence complementation (BiFC) methods for polyubiquitin chain analysis. BiFC has the advantage that a complex can be directly visualized in living cells without the need for staining with exogenous molecules.14 To develop the polyubiquitin-mediated fluorescence complementation (PolyUb-FC) assay, we used monomeric Kusabira Green (mKG) as a split fluorescent protein.15 mKG has sharper emission spectra than blue fluorescent protein or cyan fluorescent protein, and it is smaller than green fluorescent protein (GFP). N-terminal mKG (mKG[N]) is 507 base pairs with an estimated molecular mass of 19.0 kDa. C-terminal mKG (mKG[C]) is156 base pairs with an estimated molecular mass of 5.7 kDa.
Atypical ubiquitin chains remain poorly characterized. In Jurkat T cells, the E3 ubiquitin ligases CBLB and ITCH cooperate to induce T-cell receptor z-chain K33-ubiquitination, which inhibits ZAP70 phosphorylation.16-18 In addition, K33-polyubiquitination is involved in adenosine monophosphate-activated protein kinase signaling19 and protein trafficking.17 Recently, quantitative proteomics revealed that K33-ubiquitination occurs in response to UV radiation.20 Furthermore, ZRANB1/TRABID (zinc finger RANBP2-type containing 1) has been identified as a K29- and K33-specific deubiquitinase (DUB).21-23 Nevertheless, K33-linked polyubiquitin remains largely unexplained, and it has not been directly visualized on proteins in living cells. Therefore, we implemented the PolyUb-FC assay to detect K33-linked polyubiquitin by mutating potential ubiquitin Lys residues.
SQSTM1/p62 (sequestosome 1) regulates various signal transduction pathways,24,25 including T-cell receptor signaling.26,27 It is also a selective autophagy receptor for ubiquitinated cargo, including ubiquitinated aggregates, damaged mitochondria, and ubiquitinated microbes.28-30 SQSTM1/p62 binds to polyubiquitin (K48, K63) via its C-terminal ubiquitin-associated (UBA) domain.31,32 We demonstrated using PolyUb-FC that K33-linked polyubiquitin is also recruited to the phagophore, the autophagosome precursor, via the SQSTM1/p62 UBA domain.
Results
Visualization of polyubiquitin by fluorescence complementation in living cells
We constructed ubiquitin expression plasmids that were N- or C-terminally fused with truncated mKG proteins (Fig. 1A). First, to determine which combination of ubiquitin-fused truncated mKG expression vector could produce a fluorescence signal by complementation, we cotransfected 4 different complementary pairs of expression plasmids. Cotransfection of mKG(N)-ubiquitin (mKG[N]-Ub) and mKG(C)-ubiquitin (mKG[C]-Ub) induced fluorescence in a high percentage of cells as shown by microscopy and flow cytometry analysis. In this study, for detecting polyubiquitin chains, we chose the highest percentages of fluorescence-positive sets of mKG-fused ubiquitin vectors: mKG(N)-Ub and mKG(C)-Ub. To analyze polyubiquitin chain specificity, we generated ubiquitin mutants (Fig. 1B). mKG(N)-K63Ub and mKG(C)-K63Ub linked through K63 only to form polyubiquitin. These mutants showed a higher percentage of fluorescence-positive cells than truncated mKG-only expression vector. We also generated K48 and K33 vectors, which also showed similar results of higher percentages of fluorescence-positive cells than the truncated mKG-only expression vector. Thus, we could analyze chain-specific polyubiquitin chains by fluorescence complementation.
Figure 1.
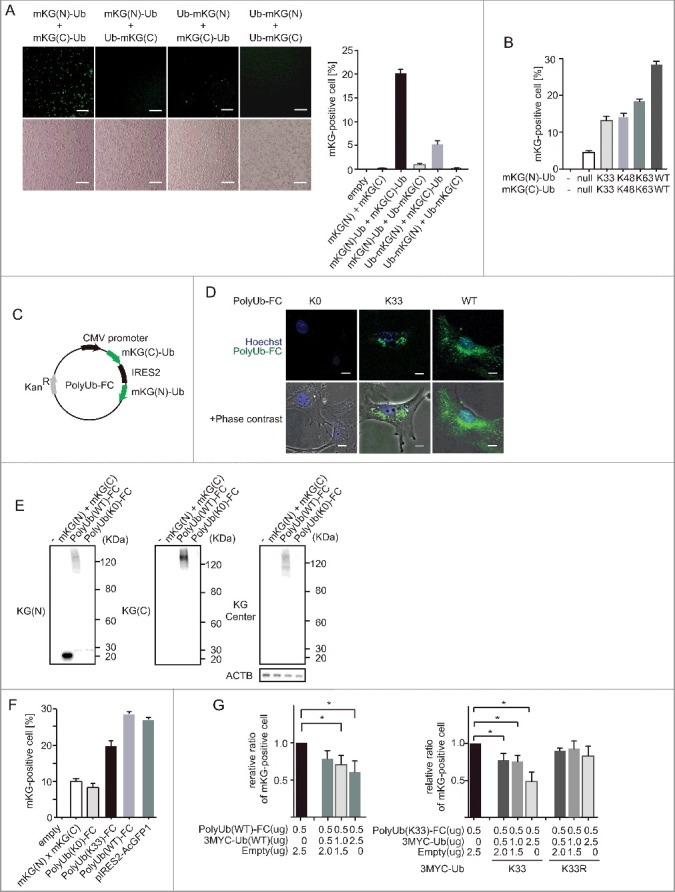
Visualization of polyubiquitin by fluorescence complementation in living cells (A) BiFC assay for polyubiquitin. Complementary pairs of N- or C-terminal mKG (mKG[N] or mKG[C])-fused ubiquitin expression plasmids (mKG[N]-Ub, Ub-mKG[N], mKG[C]-Ub and Ub-mKG[C]) were cotransfected into HEK 293T cells. After 22 h, the mKG-positive cells were counted using a conventional epifluorescence microscope or flow cytometry. Plasmids expressing N- or C-terminal mKG(N) and mKG(C) were used as negative controls. Scale bars: 200 µm. Assays were performed in triplicate. Data are representative of 2 independent experiments. Error bars indicate standard deviations. (B) mKG-fused ubiquitin expression plasmids were mutated for chain-specific analysis. Complementary pairs of N- or C-terminally mKG-fused ubiquitin expression plasmids were cotransfected into HEK 293T cells. After 20 h, mKG-positive cells were counted using a conventional epifluorescence microscope or flow cytometry. Data are representative of 2 independent experiments. Error bars indicate standard deviations. (C) Generation of the PolyUb-FC vector. mKG(N)-Ub and mKG(C)-Ub were cloned into the pIRES2-AcGFP1 vector, and AcGFP1 was excised. Schematic representation of PolyUb-FC vector plasmids encoding mKG(C)-Ub-IRES-mKG(N)-Ub. (D) Visualization of polyubiqutin K33 chains by using PolyUb(K33)-FC. PolyUb(K33)-FC vector, which contains ubiquitin with K33 alone; other Lys residues were mutated to Arg. MEFs were electroporated with PolyUb(K0)-FC, PolyUb(K33)-FC and PolyUb(WT)-FC. After 22 h, images were acquired using an FV10i confocal laser microscope. Scale bars: 20 µm. Data are representative of 3 independent experiments. (E) Polyubiquitination of PolyUb-FC. HEK 293T cells were transfected with mKG(N), mKG(C), PolyUb(WT)-FC, and PolyUb(K0)-FC. PolyUb(WT)-FC is a wild-type ubiquitin, and PolyUb(K0)-FC is a mutant Lys-less form of ubiquitin. After 20 h, cells were lysed and immunoblotted with mKG antibodies for the N-terminal, C-terminal, and central (center) domains. Data are representative of 3 independent experiments. (F) PolyUb-FC fluorescence by flow cytometry. PolyUb-FC expression plasmids were transfected in HEK 293T cells. pIRES2-AcGFP1 was transfected to demonstrate transfection efficiency. After 20 h, mKG-positive the cells were counted using flow cytometry. Data are representative of 2 independent experiments. Error bars indicate standard deviations. (G) Competition assay of PolyUb-FC and MYC-Ub vectors. HEK 293T cells were cotransfected with PolyUb-FC and MYC-Ub vectors. All samples were transfected with 0.5 µg PolyUb-FC DNA. Empty vector and MYC-Ub DNA were added to reach 3 µg DNA/sample. mKG-positive cells were counted using flow cytometry. Data represent the average of 4 experiments. *, p < 0.05 in Student t test. Error bars indicate standard deviations.
To efficiently analyze the dynamics of the ubiquitin chains, we expressed complementary ubiquitin-fused truncated mKG in one vector (mKG[C]-Ub-IRES-mKG[N]-Ub) and we called this vector PolyUb-FC (Fig. 1C). K0 ubiquitin (a Lys-less mutant) was fused with truncated mKG, and it could not generate polyubiquitin chains through Lys residues. Wild-type ubiquitin (PolyUb[WT]-FC) induced fluorescence in cells. To understand the localization of PolyUb-FC puncta, we examined PolyUb (WT)-FC vector-transfected cells by confocal microscopy (Fig. 1D). PolyUb-FC puncta were mainly detected in the cytoplasm, and a few were localized in the nucleus without stimulation. These puncta were consistent with a previous report using ubiquitin antibodies.33 Naturally, PolyUb-FC were not diffuse throughout the cytoplasm, which differed from ubiquitin antibody and GFP-fused ubiquitin results.34 These data indicated that fluorescence was generated through Lys residues.
Next, we examined the formation of atypical ubiquitin chains using our PolyUb-FC system. We generated PolyUb(K33)-FC, which could produce polyubiquitin chains only through K33. In a previous report, FLAG-Ub (K33 only) vectors displayed puncta formation.21 We found that not only PolyUb (WT)-FC ubiquitin but also PolyUb (K33)-FC showed puncta formation in mouse embryonic fibroblasts (MEFs; Fig. 1D). Next, to further confirm polyubiquitination, we analyzed transfected cells by immunoblotting with 3 mKG antibodies (Fig. 1E). mKG N-terminal antibody recognized mono Ub-mKG(N), mKG(N) and high molecular weight (MW) smears in PolyUb (WT)-FC vector-transfected cells. mKG C-terminal antibody recognized high-MW smears in PolyUb (WT)-FC vector-transfected cells. It was difficult to detect mono Ub-mKG(C) and mKG(C) because C-terminal mKG is very small. These data indicated that Poly Ub(WT)-FC linked together via the Lys residue (similar to endogenous ubiquitin) and that mKG alone were not polyubiquitinated under these conditions.
In addition, we generated an mKG center antibody that predominantly recognized full-length mKG and very weakly recognized C-terminal mKG (Fig. S1). PolyUb (WT)-FC vector-transfected cells also displayed high-MW smears with mKG center antibody. Faint smears were found in PolyUb (K0)-FC vector-transfected cells; these smears are likely to be mKG center antibody recognizing the end of a polyubiquitin chain with C-terminal mKG-K0, and also we could not exclude the possibility of K33-linked mixed chains. Thus, these data indicated that the PolyUb-FC fluorescence was generated by polyubiquitination. Next, we analyzed PolyUb-FC fluorescence by flow cytometry (Fig. 1F). PolyUb(WT)-FC generated fluorescence in a percentage similar to that in positive cells with GFP vector, indicating that almost all vector-transfected cells generated PolyUb(WT)-FC fluorescence. In contrast, PolyUb(K33)-FC generated a lower proportion of positive cells than PolyUb(WT)-FC, but it was still much higher than the negative control. Therefore, PolyUb(K33)-FC generated fluorescence. However, endogenous wild-type ubiquitin is abundant, and it forms polyubiquitin chains through internal lysine residues. We confirmed similar levels of expression of the mRNA corresponding to N-terminal and C-terminal mKG using real-time PCR (Fig. S2). To further confirm the specificity of PolyUb-FC, we performed a competition assay (Fig. 1G). PolyUb-FC fluorescence was reduced by the addition of non-mKG-tagged Ub expression vector in a dose-dependent manner. These data indicated that PolyUb-FC fluorescence is generated through ubiquitin. To rule out the possibility of fluorescent artifacts, we transfected MYC-K33 or MYC-K33R expression vectors for a competition assay (Fig. 1G). MYC-K33 vectors, but not MYC-K33R vectors, decreased PolyUb(K33)-FC fluorescence. These findings indicated that PolyUb(K33)-FC generated through K33-linked polyubiquitin may contain K33-linked mixed and forked polyubiquitin. Thus, by using mKG as a split fluorescent protein, we have established the PolyUb(K33)-FC assay as a useful method for studying K33-linked polyubiquitination.
PolyUb(WT)-FC puncta were visualized after neocarzinostatin (NCS) and L-leucyl-L-leucine methyl ester (LLOMe) treatments
To test the ability of PolyUb (WT)-FC to monitor polyubiquitination in live cells, we analyzed the generation of PolyUb (WT)-FC foci after DNA damage (Fig. 2A). The E3 ubiquitin ligase RNF8 initiates DNA damage-dependent polyubiquitination.35 We treated cells with NCS to induce DNA damage. To analyze polyubiquitin chain specificity, we generated ubiquitin mutants that could only forms links with other ubiquitin monomers through K63 to generate polyubiquitin (PolyUb[K63]-FC). K63-linked polyubiquitin was observed following DNA damage, as confirmed by the detection of PolyUb(K63)-FC puncta with a K63-specific antibody. Although PolyUb (WT)-FC and PolyUb (K63)-FC puncta were observed in the cytoplasm 180 min after stimulation, both foci also localized in nuclei after stimulation. PolyUb (K63)-FC puncta were colocalized with K63-specific antibody. PolyUb (WT)-FC foci exhibited a broader localization than those detected with the K63-specific antibody. It is possible that wild-type ubiquitin generated polyubiquitin other than K63-linked. Without NCS stimulation, the number of PolyUb(WT)-FC and PolyUb (K63)-FC foci in the nuclei remained unchanged. PolyUb(K0)-FC fluorescence was not detected with or without NCS stimulation. These data indicated that PolyUb (WT)-FC can be used for nuclear analysis and detection of polyubiquitination induction in living cells.
Figure 2.

PolyUb(WT)-FC puncta were visualized after NCS and LLOMe treatment. (A) Visualization of PolyUb-FC puncta after NCS stimuli. HEK 293T cells were transfected with a plasmid encoding PolyUb(WT)-FC, PolyUb(K63)-FC and PolyUb(K0)-FC. After 20 h, cells were stained with anti-K63 polyubiquitin antibodies and analyzed by immunofluorescence microscopy. Images were acquired using a confocal laser microscope (FV10i, Olympus). Cells were treated with 5 ng/mL NCS for the last 180 min. Scale bars: 20 µm. We counted cells with more than 10 foci in the nuclear area. In all, 100 cells from each indicated strain were analyzed. Data are representative of 3 independent experiments. Error bars indicate standard deviations. (B) Dynamic visualization of PolyUb-FC puncta after LLOMe stimuli in living cells. HEK 293T cells were transfected with plasmids encoding PolyUb(K0)-FC and PolyUb(WT)-FC. After 17 h, cells were treated with 500 µg/mL LLOMe. Images were acquired using an FV10i confocal laser microscope every 3 min. Data are representative of 4 independent experiments. Scale bars: 10 µm. Data are representative of 2 independent experiments.
The lysosomotropic reagent LLOMe stimulates the formation of polyubiquitin puncta in damaged lysosomes.36 After transfection with PolyUb(WT)-FC, we treated cells with LLOMe (Fig. 2B). Consistent with previous results, PolyUb (WT)-FC puncta were generated after treatment. In addition, we detected the induction of polyubiquitination by live imaging. PolyUb(K0)-FC fluorescence was not observed with or without LLOMe stimulation. Collectively, these data showed that our novel PolyUb-FC vector displays fluorescence complementation upon polyubiquitination. Furthermore, PolyUb-FC did not show baseline or monoubiquitin intensity, which gives it an advantage over sensor proteins and GFP-fused ubiquitin.
SQSTM1/p62 recognized K33-linked polyubiquitination
Ubiquitinated protein aggregates can be recognized and targeted for degradation by autophagy.37,38 To examine the localization of K33-linked polyubiquitination, we tested aggresome staining with Poly-Ub(K33)-FC (Fig. 3A, S3A). Some parts of PolyUb(K33)-FC puncta colocalize with aggresomes. SQSTM1/p62 is required for the concentration of ubiquitinated proteins forming aggregates and is crucial for their clearance.39 Previous studies have shown that the UBA domain of SQSTM1/p62 preferentially binds K63 and K48 polyubiquitin.32,40-42 However, SQSTM1/p62 may also recognize K33-linked polyubiquitin, and it may have specific functions. Therefore, we examined additional roles of SQSTM1/p62 using the PolyUb (K33)-FC assay. First, we tested whether endogenous SQSTM1/p62 colocalizes with PolyUb (K33)-FC (Fig. 3B). Additionally, we cotransfected PolyUb(K33)-FC into wild-type MEFs with AsRED-mouse SQSTM1/p62 (Fig. S3B). PolyUb(K33)-FC was mainly localized in the cytoplasm, and some puncta colocalized with SQSTM1/p62 puncta. In contrast, PolyUb(K0)-FC puncta were not detected.
Figure 3.
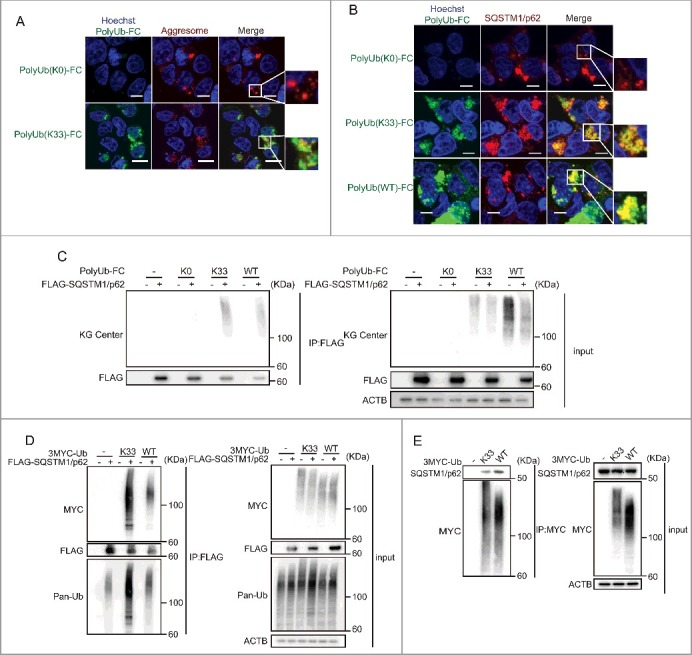
SQSTM1/p62 recognizes K33-linked polyubiquitination. (A) Colocalization of PolyUb-FC puncta with aggresomes. HEK 293T cells were transfected with PolyUb-FC vectors. After 20 h, aggresomes in the cells were stained. Images were acquired using an FV10i confocal laser microscope every 3 min. Data are representative of 4 independent experiments. Scale bars: 10 μm. Data are representative of 2 independent experiments. (B) Colocalization of Poly-Ub(K33)-FC with endogenous SQSTM1/p62. HEK 293T cells were cotransfected with PolyUb-FC vectors. After 20 h, cells were stained with anti-SQSTM1/p62 antibodies. Images were acquired using an FV10i confocal laser microscope. Scale bars: 10 µm. Data are representative of 2 independent experiments. (C) Recruitment of PolyUb(K33)-FC to FLAG-SQSTM1/p62. HEK 293T cells were cotransfected with PolyUb-FC vectors and a plasmid encoding FLAG-SQSTM1/p62. After 20 h, cells were lysed and protein extracts were immunoprecipitated (IP) with FLAG antibody and immunoblotted (IB) for the indicated proteins. Data are representative of 3 independent experiments. (D) Interaction of MYC-K33Ub and FLAG-SQSTM1/p62. HEK 293T cells were cotransfected with plasmids encoding MYC-K33Ub and FLAG-SQSTM1/p62. Twenty h later, protein extracts were immunoprecipitated (IP) with FLAG antibody and immunoblotted for the indicated proteins. Data are representative of 2 independent experiments. (E) Recruitment of MYC-K33Ub to endogenous SQSTM1/p62. HEK 293T cells were transfected with a plasmid encoding MYC-K33Ub. After 20 h, cells were lysed and protein extracts were immunoprecipitated with MYC antibody and immunoblotted (IB) for the indicated proteins. Data are representative of 3 independent experiments.
We confirmed these observations using immunoprecipitation. PolyUb(K33)-FC and FLAG-mouse-SQSTM1/p62 were cotransfected into MEFs and we affinity isolated the SQSTM1/p62 complex with anti-FLAG antibody (Fig. 3C). FLAG-SQSTM1/p62 bound to polyubiquitinated ubiquitin-fused mKG (wild type and K33). However, it was possible that these observations are because of the nonspecificity of our new PolyUb-FC system. It is difficult to generate K33 polyubiquitin in vitro23; therefore, we cotransfected a MYC-K33-only vector (which generated polyubiquitin with K33 alone) with a plasmid expressing FLAG-mouse SQSTM1/p62 and affinity isolated the SQSTM1/p62 complex with anti-FLAG antibody (Fig. 3D). These experiments showed that SQSTM1/p62 bound to K33-linked polyubiquitin. Next, we examined whether K33 polyubiquitin bound to endogenous SQSTM1/p62 (Fig. 3E). We transfected HEK-293T cells with a MYC-K33-only vector and pulled this down with anti-MYC antibody. This experiment supported our observation that K33-linked polyubiquitin interacts with SQSTM1/p62. Collectively, these data illustrated that SQSTM1/p62 forms complexes with K33-linked polyubiquitin.
SQSTM1/p62 regulates colocalization of K33-linked polyubiquitinated cargo to MAP1LC3/LC3 (microtubule associated protein 1 light chain 3 puncta
We investigated the mechanisms used for SQSTM1/p62 binding to K33-linked polyubiquitin. To examine whether SQSTM1/p62 interacts with K33 polyubiquitin via the UBA domain (which is critical for K63 binding), we cotransfected PolyUb(K33)-FC with SQSTM1/p62 lacking the UBA domain (Fig. 4A). Confocal microscopy revealed that PolyUb (K33)-FC puncta colocalized with SQSTM1/p62 puncta; however, PolyUb(K33)-FC did not colocalize with the SQSTM1/p62 UBA mutant. In addition, immunoprecipitation assay revealed reduced MYC smearing with the SQSTM1/p62 UBA mutant (Fig. S3C). These results demonstrated that SQSTM1/p62 interacts with K33 polyubiquitin via the UBA domain. To determine the ability of K33-linked ubiquitin to bind SQSTM1/p62, we performed a cell-free assay (Fig. 4B) and found that tetra K33 bound to SQSTM1/p62. In contrast, K63-linked polyubiquitin strongly bound SQSTM1/p62 as a positive control.
Figure 4.

SQSTM1/p62 regulates colocalization of K33-linked polyubiquitinated protein to LC3 puncta. (A) Disappearing colocalization of PolyUb(K33)-FC with AsRED-SQSTM1/p62ΔUBA. MEFs were electroporated with PolyUb(K33)-FC vector and AsRED-SQSTM1/p62 (or SQSTM1/p62ΔUBA) plasmid. After 24 h, images were acquired using an FV10i confocal laser microscope. Scale bars: 20 µm. Data are representative of 2 independent experiments. (B) Affinity isolation assay showing ubiquitin chains binding to SQSTM1/p62. Ubiquitin chains bound to negative samples or GST-SQSTM1/p62. Ubiquitin chain input is shown as a control in the left lane of each GST affinity isolation experiment. Input of negative sample and GST-tagged SQSTM1/p62 is shown as a control in the left lanes. Data are representative of 3 independent experiments. (C) Modification of the interaction between K33-linked polyubiquitin chains and SQSTM1/p62 by ZRANB1. HEK293T cells were cotransfected with plasmids encoding MYC-K33Ub, FLAG-SQSTM1/p62, and siRNA against ZRANB1. After 24 h, cells were lysed and protein extracts were immunoprecipitated (IP) with FLAG antibody and immunoblotted (IB) for the indicated proteins. To confirm the knockdown efficiency of ZRANB1, cells were harvested for RT PCR 24 h after transfection. Data are representative of 2 independent experiments. Error bars indicate standard deviations. (D) Quantification of the number of punctate structures positive for PolyUb(K33)-FC and LC3 per cell. HEK 293T cells were cotransfected with PolyUb(K33)-FC vector and siRNA for SQSTM1/p62. After 20 h, cells were stained with anti-LC3 antibodies and analyzed by immunofluorescence microscopy. Immunoblots indicate the knockdown efficiency of SQSTM1/p62. In all, 30 cells from each indicated strain were analyzed. Data are representative of 3 independent experiments. *p < 0.05 in student's t-test. Error bars indicate standard deviations. (E) Schematic showing that SQSTM1/p62 puncta also colocalize with K33-linked polyubiquitin the same as K48- and K63-linked polyubiquitin. The interaction between SQSTM1/p62 and K33-linked polyubiquitin is modulated by ZRANB1. K33-linked polyubiquitin also colocalizes with LC3 puncta.
To better understand the molecular mechanism by which SQSTM1/p62 binds to K33-linked polyubiquitin, we considered ZRANB1 to be a K29- and K33-specific DUB. To evaluate this linkage, we knocked down ZRANB1 via small interfering (si) RNA and analyzed polyubiquitin using western blot (Fig. 4C). The interaction of SQSTM1/p62 with K33-linked polyubiquitin was enhanced by ZRANB1 knockdown. These data indicated that SQSTM1/p62-K33 polyubiquitin chain interaction can be modulated by ZRANB1, and that K33-linked polyubiquitin may have physiological functions. Finally, to understand the functions of SQSTM1/p62 mediated by K33-linked polyubiquitin, we considered that SQSTM1/p62 localizes to phagophores.43 We analyzed the localization of PolyUb(K33)-FC to LC3 and showed that they colocalize (Fig. 4D). Colocalization between PolyUb(K33)-FC and LC3 was impaired by SQSTM1/p62 deficiency. To further investigate the function of K33 polyubiquitin, we analyzed the lysosomal marker LAMP2 (lysosome associated membrane protein 2) (Fig. S4A). Some PolyUb(K33)-FC puncta colocalized with LAMP2. These data indicated that the PolyUb(K33)-FC localized to late endosomes or lysosomes. Furthermore, PolyUb(K33)-FC localization with LC3 was modified with bafilomycin A1 and ZRANB1 knockdown (Fig. S4B and S4C). These data indicated that this interaction is modulated by ZRANB1. Taken together, these findings provide novel insights into not only K48- and K63-linked polyubiquitination, but also indicate that K33-linked ubiquitin is recruited by SQSTM1/p62 (Fig. 4E).
Generation of iPolyUb-FC: a useful tool for atypical polyubiquitin chain analysis
To analyze polyubiquitin more extensively and efficiently, we modified PolyUb-FC. We transferred mKG(N)-Ub-IRES-mKG(C)-Ub into tetracycline inducible vectors, and we generated inducible PolyUb-FC (iPloyUb-FC) (Fig. 5A). iPloyUb-FC can be used to control the expression levels of mKG by time and tetracycline concentration. We were able to confirm the colocalization of iPoly-Ub-FC puncta with endogenous SQSTM1/p62 (Fig. 5B and S5) at any tetracycline concentration. Furthermore, colocalization of iPolyUb(K33)-FC and LC3 was also impaired by SQSTM1/p62 knockdown (Fig. 5C). We could assess iPolyUb(K33)-FC at low concentrations of tetracycline and also confirm the K33-linked polyubiquitin. Taken together, iPolyUb-FC is a useful tool for analyzing atypical ubiquitin chain generation.
Figure 5.
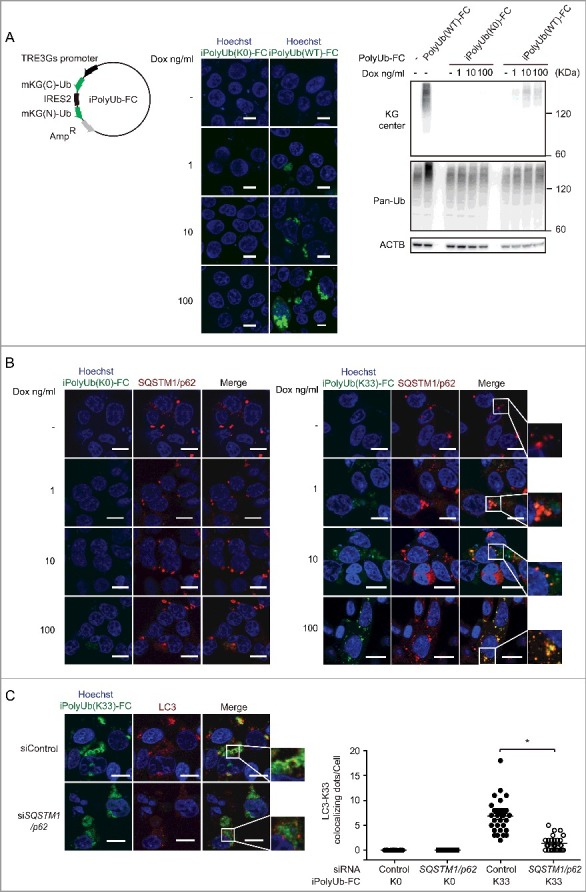
Generation of iPolyUb-FC plasmids and validation of their function. (A) Generation of iPolyUb-FC vector. Schematic represents iPolyUb-FC vector plasmids encoding mKG(C)-Ub-IRES-mKG(N)-Ub. mKG(N)-Ub-IRES-mKG(C)-Ub coding sequences were cloned into the pTet-One vector from PolyUb(FC). HEK 293T cells were transfected with iPolyUb(K0)-FC and iPolyUb(WT)-FC. After 24 h, images were acquired using an FV10i confocal laser microscope. Scale bars: 10 µm. HEK 293T cells were transfected with PolyUb(WT)-FC and iPolyUb-FC vectors. After 24 h, cells were lysed and immunoblotted. The Dox concentrations used are indicated. Induction was for 24 h. Data are representative of 2 independent experiments. (B) Colocalization of iPoly-Ub-FC vectors with endogenous SQSTM1/p62. HEK 293T cells were cotransfected with iPolyUb-FC vectors. After 24 h, cells were stained with anti-SQSTM1/p62 antibodies. The Dox concentrations used are indicated. Induction was for 24 h. Images were acquired using an FV10i confocal laser microscope. Scale bars: 10 µm. Data are representative of 2 independent experiments. (C) Quantification of the number of punctate structures positive for iPolyUb(K33)-FC and LC3 per cell. HEK 293T cells were cotransfected with iPolyUb(K33)-FC vector and siRNA for SQSTM1/p62. After 24 h, cells were stained with anti-LC3 antibodies and analyzed by immunofluorescence microscopy. The Dox concentration used was 100 ng/ml. Induction was for 24 h. In all, 30 cells from each indicated strain were analyzed. Data are representative of 2 independent experiments. *p < 0.05 in Student t test. Error bars indicate standard deviations.
Discussion
We have developed a PolyUb-FC assay to study atypical polyubiquitination (Fig. 1C and 5A). The PolyUb-FC assay has 3 advantages over existing techniques. First, compared with sensor proteins and GFP-fused ubiquitin, PolyUb-FC does not produce baseline and monoubiquitin fluorescence (Fig. 1D). Second, PolyUb-FC revealed the dynamics of polyubiquitin generation in living cells (Fig. 2A and 2B). Conventional methods, such as GFP-fused ubiquitin sensor proteins, have their own fluorescence intensity without binding to polyubiquitin,31 making it very difficult to detect polyubiquitination directly after stimulation. Third, PolyUb-FC can easily recruit atypical polyubiquitin chains assayed through mutation of the Lys residue(s) (Fig. 1B and 1D). Although some linkage-specific polyubiquitin antibodies are commercially available, they do not recognize all types of polyubiquitin chains. Moreover, antibodies recognize only small epitopes; therefore, it may be difficult to generate antibodies to mixed polyubiquitin chains such as ubiquitin-Lys 29-ubiquitin-K33 and to noncanonical branched polyubiquitin chains.44 KG tags may also block associations or generate artificial associations. Furthermore, PolyUb-FC may misrepresent the localization, because of endogenous ubiquitin. It is also possible to visualize homotypic and heterotypic K33 polyubiquitin chains.
Using this method, PolyUb-FC can be used to analyze the dynamics of polyubiquitin chain generation and explore new targets of atypical polyubiquitination. Among atypical ubiqutin chains, most functions of the K33-linked polyubiquitin chain are unknown.1 ZRANB1 was identified as a K29- and K33-specific DUB21 (Fig. 4C and S4C). In addition, zranb1 homozygous knockout mice show viviparous lethality, whereas heterozygous knockout mice display deficiencies in T-cell functions. In other words, K33-linked polyubiquitination (or conjugated cargo) may regulate T-cell immune function. The PolyUb(K33)-FC assay can be used to further elucidate roles of T-cell immune function. In this study, we successfully imaged polyubiquitination in real time. By using PolyUb-FC, we showed that K33-linked polyubiquitin was colocalized with SQSTM1/p62 and that colocalization with LC3 was dependent on SQSTM1/p62 (Fig. 4A, 4B, 4D and 4E). SQSTM1/p62 is thought to bind mainly through K63-linked and K48-linked polyubiquitin, and these functions have been analyzed. However, the full physiological significance of K33-linked ubiquitin is not completely understood. In future studies, we plan to use PolyUb-FC to identify the stimuli of K33-linked polyubiquitin and LC3 colocalization. In addition, we will identify cargos that display K33-linked polyubiquitination. Further research may elucidate a novel function for K33-linked polyubiquitin in autophagy.
PolyUb-FC and iPolyUb-FC are good tools for ubiquitin analysis. It is possible to analyze the functions of polyubiquitin, not only K63 and K48 chains but also atypical chains. As our data showed, these tools are good candidates for screening the unknown substrates in the ubiquitin and autophagy networks.
Materials and methods
Cell culture and reagents
Human embryonic kidney (HEK) 293T cells and MEFs were cultured in Dulbecco's Modified Eagle Medium (SIGMA, D5796) with fetal bovine serum (BioWest, S1820) and penicillin/streptomycin (Nacalai Tesque, 26253–84) at 37°C. HEK 293T cells were transiently transfected with plasmids using Lipofectamine 2000 (Invitrogen, 11668019) as indicated by the supplier. MEFs were transiently transfected using 4D-Nucleofector X Unit (Lonza, AAF-1002B and AAF 1002X) and P4 Primary Cell 4D Nucleofector X Kit L (Lonza, V4XP-4012) as recommended by the supplier. To induce DNA damage, HEK 293T cells were treated for 180 min with 5 ng/mL NCS (Sigma-Aldrich, N9162). To disrupt the lysosomal membrane and induce autophagy, HEK 293T cells were exposed for 1 h to 500 µM LLOMe (Sigma-Aldrich, L7393). Bafilomycin A1 (Merck Millipore; 196000) was dissolved in dimethyl sulfoxide at a concentration of 100 μM and was used at a final concentration of 100 nM. Doxycycline (Clontech, 631311) was dissolved in double-distilled H2O at a concentration of 1, 10 and 100 µg/ml and was used at a final concentration of 1, 10 and 100 ng/ml. Tetracycline-free fetal bovine serum were purchased from Clontech (631105).
Plasmids and siRNA oligonucleotides
Expression plasmids of ubiquitin fused with N- or C-terminally truncated mKG were constructed by inserting PCR-amplified fragments encoding ubiquitin, into fragmented mKG vector (Coral Hue Fluo-Chase Kit; MBL) using BamHI and ApaI.
Cells were transfected with a complementary pair of mKG fusion plasmids. Mouse Sqstm1/p62 complementary DNA (cDNA) was kindly provided by Prof. Masaaki Komatsu (Niigata University). Mouse Sqstm1/p62 cDNA was cloned into the p3XFLAG-CMV-10 expression vector (Sigma-Aldrich, E7658). FLAG-Sqstm1/p62ΔUBA was constructed by TAA (Stop) insertion in the sequence position 1156–1158 of Sqstm1/p62 cDNA using the PrimeSTAR mutagenesis basal kit (TaKaRa, R046A). Mouse Sqstm1/p62 cDNA and Sqstm1/p62ΔUBA cDNA were cloned into pAsRED2-N1 plasmids (Clontech, 632449). Human ubiquitin cDNA was cloned into pCMV-3Tag-2C plasmids (Agilent Technologies, 240196). K0, K33, K48 and K63 were prepared using the PrimeSTAR mutagenesis basal kit. iPoly-Ub (K0, K33 and WT)-FC was constructed by inserting fragments encoding KG(C)-ubiquitin-IRES-KG(N)-ubiquitin into fragmented pTet-One vector (Clontech, 634301), which was constructed at the SacI site insertion in regions 4206–42011, using SacI and NotI. ON-TARGETplus SMARTpool siRNA oligonucleotides specific for human SQSTM1/p62, human ZRANB1, and nontargeting siRNA (control siRNA) were purchased from ThermoFisher Scientific. The pIRES2-AcGFP1 vector (Clontech, 632435) was purchased from Clontech.
Flow cytometry
The mKG fluorescent signal was acquired using a FACS CantoII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star). DAPI (D3571) was purchased from Invitrogen.
Microscopy analysis
Cells were washed with phosphate-buffered saline (PBS; Nacalai Tesuque, 14249–24) and fixed in 4% paraformaldehyde (Nacalai Tesque, 09154–85) for 10 min at 4°C. Fixed cells were permeabilized with 50 µg/mL digitonin (Invitrogen, BN2006) in PBS for 5 min, blocked with 3% bovine serum albumin (Nacalai Tesque, 01863–48) in PBS for 15 min, and incubated with anti-LC3A/B antibody (MBL, M152-3), anti-LAMP2 antibody (H4B4; abcam, ab25631), anti-SQSTM1/p62 (MBL, PM045) or anti-polyubiqutin (linkage-specific K63) antibody (ab179434) for 1 h. After washing, cells were incubated with Alexa Fluor 594-conjugated anti-mouse secondary antibody (Invitrogen, A11032) or anti-rabbit antibody (Invitrogen, A21207) for 30 min. The nucleolus was stained using VECTASHIELD® Mounting Medium with 4',6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, H-1200). For live-cell fluorescence imaging, the nucleolus was counterstained using Hoechst 33342 (Nacalai Tesque, 04915–81). To stain aggregesomes, the ProteoStat Aggregesome Detection kit (Enzo, ENZ-51035) was used according to the manufacturer's instructions. Images were acquired using a conventional epifluorescence microscope (BZ-8000, KEYENCE, Tokyo, Japan) or a confocal laser microscope (FV10i, Olympus) with a 60 × oil-immersion objective lens.
Quantitative real-time (RT) PCR
Total RNA was isolated using the RNeasy mini kit (QIAGEN, 74106). cDNA was synthesized using the QuantiTect reverse transcription kit (QIAGEN, 205313). qPCR was performed using QuantiTect SYBR Green Master Mix (QIAGEN, 204243) and the StepOnePlus RT PCR system (Applied Biosystems, 4376600). Gene expression was normalized to the expression of the housekeeping gene ACTB. Gene expression was determined using the following primers: ZRANB1, 5'-CAATGCTTGTGTGGGGGTTG-3' and 5'-GCAAGCGTACTTCATCTGCG-3'; ACTB, 5'-GACAGGATGCAGAAGGAGA-3' and 5'-GTACTTGCGCTCAGGAGGAG-3'; mKG(N), 5'- ATCAAGCCCGAGATGAAGAT-3' and 5'-GTACTTGGTAAACACCCTGT-3'; and mKG(C) 5'- CGGCAACCACAAGTGCCA-3' and 5'-TGATGTTGCCCTCGGTCTT -3'.
Immunoblotting
Cells were incubated in lysis buffer (either 20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5% CHAPS [Nacalai Tesuque, 07957-64], 10% glycerol, 2 mM N-ethylmaleimide [Sigma Aldrich, 04259-5G], and Halt protease and phosphatase inhibitor cocktail [Pierce, 1861280], or 20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.2% NP-40 [Nacalai Tesuque, 23640–65], 10% glycerol, 2 mM N-ethylmaleimide, and protease and phosphatase inhibitor cocktail) on ice for 20 min and centrifuged at 14,000 × g for 20 min. For FLAG or MYC immunoprecipitation, cell lysates were incubated with anti-DDDDK-tag monoclonal antibody-magnetic beads (MBL, M185-9) or anti-MYC-tag monoclonal antibody-magnetic beads (MBL, M047-9). Cell lysates were incubated with the pre-coupled beads for 1 h at 4°C. Samples were resolved on NuPage precast 4–12% Bis-Tris gels (Invitrogen, NP0323) and transferred to polyvinylidene difluoride membranes. The following antibodies and reagents were used for immunoprecipitation and immunoblotting studies: anti-ACTB (Sigma-Aldrich, A5441), anti-FLAG (Sigma-Aldrich, F7425), anti-MYC (A-14; Santa Cruz Biotechnology, sc-789), KG (N; MBL, M148-3), KG (C; MBL, M149-3), anti-Ub (P4D1; Santa Cruz Biotechnology, sc-8017), anti-SQSTM1/p62 (MBL, PM045) and anti-GST (MBL, PM013). Secondary antibodies (mouse anti-rabbit and goat anti-mouse; 211-032-171 and 115-035-174, respectively) were purchased from Jackson Laboratories. Anti-mKG rabbit polyclonal antibody (Center, 015) was generated against full-length mKG recombinant protein.
Affinity isolation assay
GST-tagged human recombinant SQSTM1/p62 (ENZ-PRT120-0050) was purchased from Enzo Life Sciences. A ubiqutin mutant with K33-only (UM-K330), ubiquitin mutant with K63-only (UM-K630), tetra-ubiquitin/Ub4 WT chains (K33-linked) (UC-103) and tetra-ubiquitin/Ub4 WT chains (K63-linked) (UB-310B) were purchased from Boston Biochem. Ubiquitin-binding assays with SQSTM1/p62 proteins were performed with the following steps: (i) Incubation of Dynabeads protein G (DB10007, Veritas) with anti-GST tag polyclonal antibody (MBL, PM013). (ii) Incubation of Dynabeads bound to anti-GST tag polyclonal antibody with GST-SQSTM1/p62 protein for 10 min at room temperature in ubiquitin binding buffer (25 mM HEPES, pH 7.5, 150 mM KCl, 2 mM MgCl2, 0.5% Triton X-100 [Sigma Aldrich, 30-5140-5], 1 mM EGTA, 1 mg/ml BSA [GIBCO, 10437-028]). (iii) After serial washing with ubiquitin binding buffer, SQSTM1/p62-bound Dynabeads were incubated with free ubiquitin overnight at 4℃ in ubiquitin-binding buffer. (iv) After serial washing with binding buffer flow-through samples were analyzed by immunoblotting for ubiquitin and GST using commercial antibodies.
Competition assay
HEK 293T cells were cotransfected with PolyUb(K33)-FC and MYC-K33Ub or MYC-K33RUb. All samples were transfected with 0.5 µg PolyUb(K33)-FC DNA, supplemented with an empty vector and MYC-K33Ub or MYC-K33RUb to reach 3 µg of total DNA/sample. As a control experiment, HEK 293T cells were cotransfected with PolyUb(WT)-FC and MYC-WT Ub. All samples were transfected with 0.5 µg of PolyUb(WT)-FC DNA/sample, supplemented with an empty vector and MYC-WT Ub to reach 3 µg of total DNA. The number of mKG-positive cells were counted using flow cytometry 20 h after transfection.
Statistical analysis
Data are expressed as the mean ± standard deviation. Statistical significance was calculated using unpaired Student t test. Statistical analysis was performed using Prism 5.0 (GraphPad).
Disclosure of potential conflict of interest
The authors declare that they have no conflicts of interest.
Supplementary Material
Funding Statement
This work was supported by the Japan Agency for Medical Research and Development [grant number 15AeK0109047h0002]; Japan Society for the Promotion of Science (JSPS) [grant number 16K15423], [grant number 25460946].
Acknowledgements
We thank Eisuke Itakura and Averil Ma for critically reading the manuscript. This work was supported by the Takeda Science Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, and Daiichi Sankyo Foundation of Life Science. This study was also supported by MEXT/JSPS KAKENHI (Grant Number 25460946, 26221307, 15H04808, 16K15423), the Research Centre Network Program for Realization of Regenerative Medicine from the Japan Science and Technology Agency (JST) and the Japan Agency for Medical Research and Development (AMED), the Practical Research Project for Rare/Intractable Diseases from AMED, the Practical Research Project for Rare/Intractable Diseases (15AeK0109047h0002) from AMED, and the Practical Research Project for Innovative Cancer Control (15Ack0106017h0002) from AMED.
References
- 1.Kulathu Y, Komander D. Atypical ubiquitylation – the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–523. doi: 10.1038/nrm3394. PMID:22820888 [DOI] [PubMed] [Google Scholar]
- 2.Ordureau A, Münch C, Harper JW. Quantifying ubiquitin signaling. Mol Cell. 2015;58:660–676. doi: 10.1016/j.molcel.2015.02.020. PMID:26000850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–921. doi: 10.1016/j.cell.2014.03.037. PMID:24813613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. PMID:22524316 [DOI] [PubMed] [Google Scholar]
- 5.Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. PMID:19675569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. PMID:18724939 [DOI] [PubMed] [Google Scholar]
- 7.Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. PMID:20655260 [DOI] [PubMed] [Google Scholar]
- 8.Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Yen B, Woo T, et al. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. PMID:19060883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Wijk SJ, Fiškin E, Dikic I. Selective monitoring of ubiquitin signals with genetically encoded ubiquitin chain-specific sensors. Nat Protoc. 2013;8:1449–1458. doi: 10.1038/nprot.2013.089. PMID:23807287 [DOI] [PubMed] [Google Scholar]
- 10.van Wijk SJ, Fiskin E, Putyrski M, Pampaloni F, Hou J, Wild P, Kensche T, Grecco HE, Bastiaens P, Dikic I. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol Cell. 2012;47:797–809. doi: 10.1016/j.molcel.2012.06.017. PMID:22819327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sims JJ, Scavone F, Cooper EM, Kane LA, Youle RJ, Boeke JD, Cohen RE. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat Methods. 2012;9:303–309. doi: 10.1038/nmeth.1888. PMID:22306808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. PMID:19798103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009;36:302–314. doi: 10.1016/j.molcel.2009.10.002. PMID:19854138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerppola TK. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. PMID:16625152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueyama T, Kusakabe T, Karasawa S, Kawasaki T, Shimizu A, Son J, Leto TL, Miyawaki A, Saito N. Sequential binding of cytosolic Phox complex to phagosomes through regulated adaptor proteins: evaluation using the novel monomeric Kusabira-Green System and live imaging of phagocytosis. J Immunol. 2008;181:629–640. doi: 10.4049/jimmunol.181.1.629. PMID:18566430 [DOI] [PubMed] [Google Scholar]
- 16.Huang H, Jeon MS, Liao L, Yang C, Elly C, Yates JR, Liu YC. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. PMID:20637659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan WC, Lee YR, Lin SY, Chang LY, Tan YP, Hung CC, Kuo JC, Liu CH, Lin MY, Xu M, et al. K33-Linked Polyubiquitination of Coronin 7 by Cul3-KLHL20 Ubiquitin E3 Ligase Regulates Protein Trafficking. Mol Cell. 2014;54:586–600. doi: 10.1016/j.molcel.2014.03.035. PMID:24768539 [DOI] [PubMed] [Google Scholar]
- 18.Yang M, Chen T, Li X, Yu Z, Tang S, Wang C, Gu Y, Liu Y, Xu S, Li W, et al. K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8(+) T cell activation. Nat Immunol. 2015;16:1253–1262. doi: 10.1038/ni.3258. PMID:26390156 [DOI] [PubMed] [Google Scholar]
- 19.Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR. Control of AMPK-related kinases by USP9X and atypical Lys(29)/Lys(33)-linked polyubiquitin chains. Biochem J. 2008;411:249–260. doi: 10.1042/BJ20080067. PMID:18254724 [DOI] [PubMed] [Google Scholar]
- 20.Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol Cell. 2015;59:867–881. doi: 10.1016/j.molcel.2015.05.006. PMID:26051181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licchesi JD, Mieszczanek J, Mevissen TE, Rutherford TJ, Akutsu M, Virdee S, El Oualid F, Chin JW, Ovaa H, Bienz M, et al. An ankyrin-repeat ubiquitin-binding domain determines TRABID's specificity for atypical ubiquitin chains. Nat Struct Mol Biol. 2012;19:62–71. doi: 10.1038/nsmb.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. PMID:23827681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michel MA, Elliott PR, Swatek KN, Simicek M, Pruneda JN, Wagstaff JL, Freund SM, Komander D. Assembly and specific recognition of k29- and k33-linked polyubiquitin. Mol Cell. 2015;58:95–109. doi: 10.1016/j.molcel.2015.01.042. PMID:25752577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komatsu M, Kageyama S, Ichimura Y. p62/SQSTM1/A170: physiology and pathology. Pharmacol Res. 2012;66:457–462. doi: 10.1016/j.phrs.2012.07.004. PMID:22841931 [DOI] [PubMed] [Google Scholar]
- 25.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. PMID:23911927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin P, Diaz-Meco MT, Moscat J. The signaling adapter p62 is an important mediator of T helper 2 cell function and allergic airway inflammation. EMBO J. 2006;25:3524–3533. doi: 10.1038/sj.emboj.7601250. PMID:16874300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. PMID:22658522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. PMID:16286508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. PMID:17580304 [DOI] [PubMed] [Google Scholar]
- 30.Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. PMID:24462201 [DOI] [PubMed] [Google Scholar]
- 31.Donaldson KM, Li W, Ching KA, Batalov S, Tsai CC, Joazeiro CA. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc Natl Acad Sci U S A. 2003;100:8892–8897. doi: 10.1073/pnas.1530212100. PMID:12857950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long J, Gallagher TR, Cavey JR, Sheppard PW, Ralston SH, Layfield R, Searle MS. Ubiquitin recognition by the ubiquitin-associated domain of p62 involves a novel conformational switch. J Biol Chem. 2008;283:5427–5440. doi: 10.1074/jbc.M704973200. PMID:18083707 [DOI] [PubMed] [Google Scholar]
- 33.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. PMID:22266820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dantuma NP, Groothuis TA, Salomons FA, Neefjes J. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol. 2006;173:19–26. doi: 10.1083/jcb.200510071. PMID:16606690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato K, Nakajima K, Ui A, Muto-Terao Y, Ogiwara H, Nakada S. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol Cell. 2014;53:617–630. doi: 10.1016/j.molcel.2014.01.030. PMID:24560272 [DOI] [PubMed] [Google Scholar]
- 36.Maejima I, Takahashi A, Omori H, Kimura T, Takabatake Y, Saitoh T, Yamamoto A, Hamasaki M, Noda T, Isaka Y, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. PMID:23921551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. PMID:20811356 [DOI] [PubMed] [Google Scholar]
- 38.Kirkin V, Lamark T, Sou YS, Bjørkøy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. PMID:19250911 [DOI] [PubMed] [Google Scholar]
- 39.Gamerdinger M, Hajieva P, Kaya AM, Wolfrum U, Hartl FU, Behl C. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009;28:889–901. doi: 10.1038/emboj.2009.29. PMID:19229298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. PMID:15340068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. PMID:18174161 [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. PMID:22017874 [DOI] [PubMed] [Google Scholar]
- 43.Itakura E, Mizushima N. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011;192:17–27. doi: 10.1083/jcb.201009067. PMID:21220506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. PMID:18516089 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


