ABSTRACT
Bleomycin is a clinically potent anticancer drug used for the treatment of germ-cell tumors, lymphomas and squamous-cell carcinomas. Unfortunately, the therapeutic efficacy of bleomycin is severely hampered by the development of pulmonary fibrosis. However, the mechanisms underlying bleomycin-induced pulmonary fibrosis, particularly the molecular target of bleomycin, remains unknown. Here, using a chemical proteomics approach, we identify ANXA2 (annexin A2) as a direct binding target of bleomycin. The interaction of bleomycin with ANXA2 was corroborated both in vitro and in vivo. Genetic depletion of anxa2 in mice mitigates bleomycin-induced pulmonary fibrosis. We further demonstrate that Glu139 (E139) of ANXA2 is required for bleomycin binding in lung epithelial cells. A CRISPR-Cas9-engineered ANXA2E139A mutation in lung epithelial cells ablates bleomycin binding and activates TFEB (transcription factor EB), a master regulator of macroautophagy/autophagy, resulting in substantial acceleration of autophagic flux. Pharmacological activation of TFEB elevates bleomycin-initiated autophagic flux, inhibits apoptosis and proliferation of epithelial cells, and ameliorates pulmonary fibrosis in bleomycin-treated mice. Notably, we observe lowered TFEB and LC3B levels in human pulmonary fibrosis tissues compared to normal controls, suggesting a critical role of TFEB-mediated autophagy in pulmonary fibrosis. Collectively, our data demonstrate that ANXA2 is a specific bleomycin target, and bleomycin binding with ANXA2 impedes TFEB-induced autophagic flux, leading to induction of pulmonary fibrosis. Our findings provide insight into the mechanisms of bleomycin-induced fibrosis and may facilitate development of optimized bleomycin therapeutics devoid of lung toxicity.
KEYWORDS: Annexin A2, autophagy, bleomycin, chemical proteomics, pulmonary fibrosis, transcription factor EB
Abbreviations
- ANXA2
annexin A2
- ATII
alveolar epithelial type II
- BALF
bronchoalveolar lavage fluid
- F-Bleo
fluorescein-labeled bleomycin
- H&E
hematoxylin and eosin
- His
hexahistidine
- IPF
idiopathic pulmonary fibrosis
- MAP1LC3B/LC3B
microtubule associated protein 1 light chain 3 beta
- MTOR
mechanistic target of rapamycin
- RNA-seq
RNA sequencing
- SQSTM1/p62
sequestosome 1
- TFEB
transcription factor EB
- TGFB1
transforming growth factor beta 1
- WT
wild type
Introduction
Bleomycin is a glycopeptide antibiotic originally isolated from Streptomyces verticillus in 1966.1 For about half a century, bleomycin has been used clinically in combination with chemotherapy for the treatment of testicular cancer, malignant lymphomas and squamous cell carcinomas with attractive therapeutic efficacy due to minimal myelosuppression and immunosuppression.2,3 In addition, bleomycin also exhibits potent and broad anticancer activities against other types of cancer, including ovarian cancer, cervical cancer, sarcoma and melanoma.2,4 However, the clinical use and therapeutic efficacy of bleomycin are severely limited by interstitial pneumonitis with an incidence rate of up to 46%, which can frequently develop into pulmonary fibrosis.2–6 Given that the toxic side effects are major clinical problems encountered for chemotherapy,7 deciphering the molecular mechanisms underlying this side effect of bleomycin will contribute to optimizing bleomycin therapeutics.
During the past decades, many studies have sought to decipher the mechanism of pulmonary fibrosis induced by bleomycin. Previous studies have suggested that bleomycin-induced oxidative stress and inflammatory response contribute to the development of pulmonary fibrosis.2,8–10 However, the use of the antioxidant N-acetylcysteine or antiinflammatory agents (such as corticosteroids) to protect against lung toxicity of bleomycin remains equivocal.9–13 These findings imply that in addition to oxidative stress and inflammatory response, additional undefined mechanisms might be involved. Although recent transcriptomics and proteomics studies have made much progress in deciphering the cellular consequences during bleomycin-induced pulmonary fibrosis in mice,14,15 the mechanisms of bleomycin-induced pulmonary fibrosis still merits further investigation. In this regard, identifying the binding targets of bleomycin will gain new perspectives for the understanding of bleomycin-induced pulmonary fibrosis.
Here, using a chemical proteomics strategy, we have identified ANXA2 (annexin A2) as a direct binding target of bleomycin in lung epithelial cells. Binding of bleomycin to ANXA2 impedes TFEB (transcription factor EB)-induced autophagic flux, leading to induction of pulmonary fibrosis. Our study reveals a novel cellular outcome in bleomycin-induced pulmonary fibrosis and may help optimize the clinical use of bleomycin in cancer therapy.
Results
ANXA2 directly binds to bleomycin in lung epithelial cells
To identify the cellular targets of bleomycin, we performed compound-centric chemical proteomics to purify and characterize bleomycin-binding proteins (Fig. S1).16–19 Bleomycin A5 was covalently conjugated to NHS-activated Sepharose beads with high coupling efficacy (90.26 ± 7.54%) (Fig. S2A-C). The immobilized bleomycin A5 was then incubated with lung homogenates from normal C57BL/6J mice (Fig. 1A) or cell lysates of human A549 lung epithelial cells (Fig. S3A). High-affinity binders were competitively eluted with free bleomycin A5 and then subjected to SDS-PAGE and mass spectrometry analysis. We found that only ANXA2 could be affinity-purified from both mouse lung homogenates (Fig. 1A, lane 3 and Table S1) and A549 cell lysates (Fig. S3A, lane 2 and Table S2). As expected, ANXA2 was not identified when free bleomycin A5 was added to mouse lung homogenates before incubation with the beads (Fig. 1A, lane 4), suggesting that bleomycin A5 selectively bound to ANXA2. We then confirmed the binding of bleomycin A5 with ANXA2 using immunoblotting both in mouse lung homogenates (Fig. 1A, bottom) and A549 cell lysates (Fig. S3A, bottom).
Figure 1.
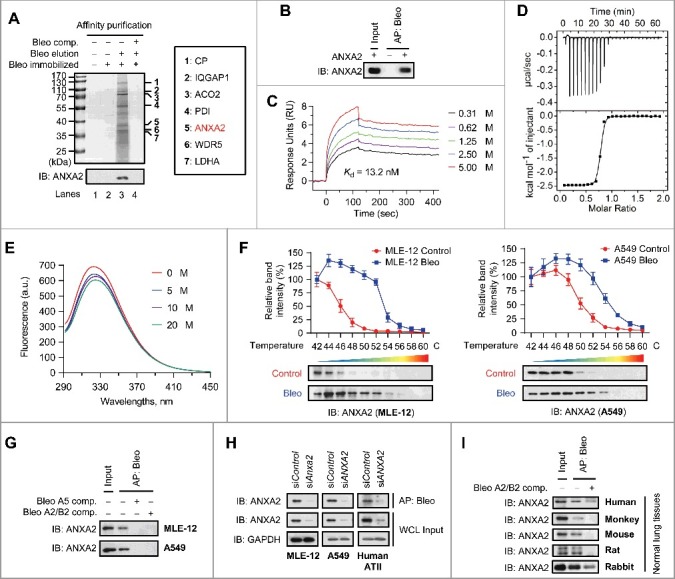
ANXA2 is a binding target of bleomycin in vitro and in vivo. (A) Bleomycin (Bleo)-binding proteins were affinity-purified from lung homogenates of normal mice using bleomycin-immobilized NHS-activated Sepharose beads. Bound proteins were eluted with free bleomycin (lane 3). The SDS-PAGE gel was stained with Coomassie Brilliant Blue (top) or immunoblotted with anti-ANXA2 antibody (bottom). Candidate binding targets are listed (right). (B) Recombinant human ANXA2 (non-His tagged) was incubated with bleomycin-immobilized beads for affinity purification and then immunoblotted with anti-ANXA2 antibody. (C) Surface plasmon resonance analysis of the binding of bleomycin with increasing concentrations of recombinant human ANXA2 (non-His tagged). (D) Isothermal titration calorimetry analysis of bleomycin binding with recombinant human ANXA2 (non-His tagged). (E) Fluorescence quenching assay to examine the conformational changes of ANXA2 (non-His tagged) caused by bleomycin binding. (F) Cellular thermal shift assay showing ANXA2 target engagement by bleomycin A2 (Bleo) in MLE-12 cells (left) and A549 cells (right). The thermal stability of ANXA2 was quantified as indicated at the top of each immunoblot (n = 3). Data are shown as mean ± s.d. (G) Target proteins were affinity-purified using bleomycin-immobilized beads from MLE-12 or A549 cells and then immunoblotted with anti-ANXA2 antibody. (H) Target proteins were affinity-purified using bleomycin-immobilized beads from MLE-12, A549 or primary human ATII cells treated with ANXA2 siRNA (siANXA2) or control siRNA (siControl) and then immunoblotted with anti-ANXA2 antibody. GAPDH, loading control. (I) Target proteins was affinity-purified using bleomycin-immobilized beads from normal lung homogenates of human, monkey, mouse, rat or rabbit and then immunoblotted with anti-ANXA2 antibody. In the competition experiments (comp.) in (A, G and I), 5-fold excess bleomycin was added to protein extracts before incubation with the beads. Results are representative of 3 independent experiments unless otherwise stated.
Bleomycin A5 was conjugated to NHS-activated Sepharose beads via the active amino group at the end of its bithiazole tail (Fig. S2A and B). Because the bleomycin A2/B2 complex (the main component of Blenoxane), which shares the same core structure with bleomycin A5 but differs by the bithiazole tail, is the clinically administered form of this drug,2 we used bleomycin A2/B2 (hereafter called bleomycin) for subsequent experiments.
To determine whether bleomycin binds to ANXA2 directly, we prepared recombinant human ANXA2 protein and found that recombinant ANXA2 could be affinity-purified by bleomycin (Fig. 1B). This direct interaction was further confirmed to have high affinity by surface plasmon resonance (Fig. 1C) and isothermal titration calorimetry analysis (Fig. 1D). In addition, fluorescence titration against bleomycin with recombinant ANXA2 resulted in fluorescence quenching, revealing that bleomycin binding causes conformational changes of ANXA2 (Fig. 1E).
Because injury in lung epithelium and activation of lung epithelial cells are associated with both the initiation and progression of pulmonary fibrosis,20–23 we considered the possibility that the binding of bleomycin to ANXA2 might occur in lung epithelial cells. To this end, we intratracheally injected fluorescein-labelled bleomycin (F-Bleo) in normal C57BL/6J mice and found that F-Bleo colocalized with ANXA2 in mouse lung epithelial cells in lung sections (Fig. S3B). This colocalization was also observed in cultured primary human alveolar epithelial type II (ATII) cells (Fig. S3C). We then used 2 lung epithelial cell lines (mouse MLE-12 and human A549 cells) to validate the interaction of bleomycin with ANXA2. In living cells, thermal stabilization of a cellular protein will be enhanced when binding with a chemical agent.24 Using a cellular thermal shift assay, we found that ANXA2 was physically engaged and stabilized against thermal changes in bleomycin-treated MLE-12 (Fig. 1F, left) and A549 (Fig. 1F, right) cells. In addition, we found that ANXA2 could be affinity-purified with bleomycin-immobilized beads from both MLE-12 and A549 cells, which could be reversed when bleomycin A5 or bleomycin A2/B2 was added into lysates prior to incubation with the beads (Fig. 1G). Consistent with these results, siRNA knockdown of ANXA2 clearly diminished bleomycin binding in MLE-12, A549 and primary human ATII cells (Fig. 1H). The selective binding of bleomycin with ANXA2 was also observed in a number of species (including human, monkey, mouse, rat and rabbit) that can develop pulmonary fibrosis after bleomycin exposure (Fig. 1I).25 Collectively, these findings demonstrate that ANXA2 is a specific target of bleomycin in lung epithelial cells both in vitro and in vivo.
ANXA2 facilitates bleomycin-induced pulmonary fibrosis
To determine the role of ANXA2 in bleomycin-induced pulmonary fibrosis, we intratracheally injected bleomycin or saline (control) into wild-type (WT) and anxa2−/- mice and analyzed the lung tissues, bronchoalveolar lavage fluid (BALF) and body weights. We showed that bleomycin elicited alveolar septal thickening, alveolar structure distortion and fibrotic foci in wild-type (WT) mice compared to anxa2−/− mice (Fig. 2A). We then histopathologically evaluated the severity of fibrosis in hematoxylin and eosin (H&E)-stained lung sections using the Ashcroft score, an established continuous numerical scale for determining the degree of pulmonary fibrosis.26 Bleomycin-challenged WT mice exhibited a great extent of fibrosis as evidenced by elevated fibrotic score, which was considerably alleviated in bleomycin-treated anxa2−/− mice (Fig. 2B). Next, we assessed abnormal collagen deposition (a hallmark of pulmonary fibrosis) in mouse lungs by trichrome staining, Sircol assay and hydroxyproline (a marker of collagen) quantification. Notably, bleomycin-treated WT mice exhibited prominent collagen accumulation and increased hydroxyproline content compared with saline controls, whereas bleomycin-injected anxa2−/− mice showed attenuated collagen deposition and hydroxyproline levels (Fig. 2C-E). We then prepared BALF, and found increased total protein contents and TGFB1 (transforming growth factor beta 1) levels in bleomycin-treated WT mice compared to bleomycin-challenged anxa2−/− mice (Fig. 2F-G). In addition, body weight loss induced by bleomycin was more pronounced in WT mice than that in anxa2−/− mice (Fig. 2H). Overall, these data indicate that ANXA2 is required for bleomycin-induced pulmonary fibrosis.
Figure 2.
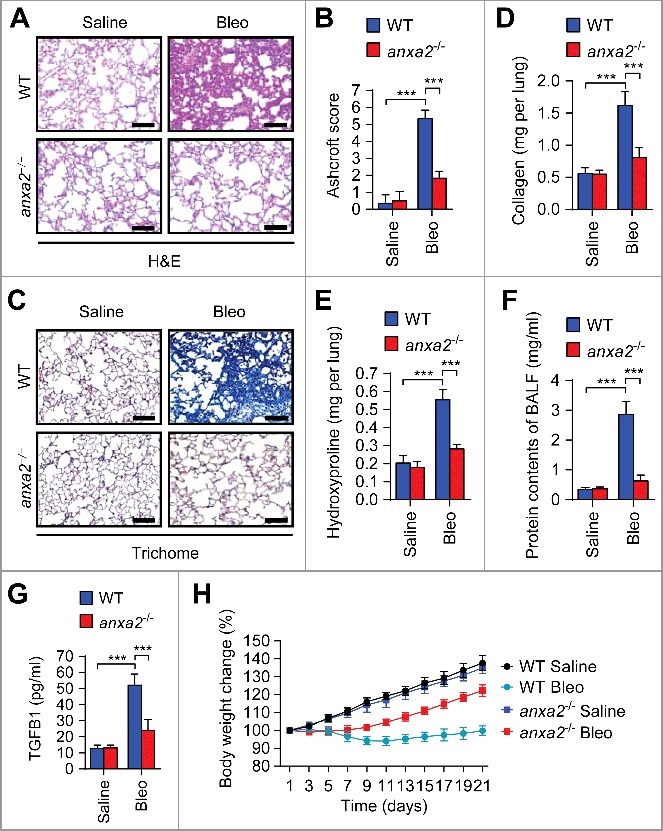
ANXA2 deficiency ameliorates bleomycin-induced pulmonary fibrosis. Wild-type (WT) and anxa2−/− mice were treated intratracheally with bleomycin (Bleo) or saline for 21 days (n = 5 per group). (A) Representative images of hematoxylin and eosin (H&E) staining of lung tissue sections. Scale bars: 100 μm. (B) Ashcroft fibrotic scores were determined by H&E staining of lung sections. (C) Representative images of Masson's trichrome staining of lung tissue sections. Scale bars: 100 μm. (D and E) Whole lung homogenates were analyzed to examine collagen contents using the Sircol assay (D) and hydroxyproline contents (E). (F and G) Total protein contents (F) and TGFB1 levels (G) were determined in bronchoalveolar lavage fluid (BALF). (H) Body weights were measured over time. Data are means ± s.d. ***P < 0.001 (Student t test).
Glu139 of ANXA2 is required for bleomycin binding in lung epithelial cells
To investigate the mechanism by which ANXA2 contributes to bleomycin-induced pulmonary fibrosis, we first examined ANXA2 expression upon bleomycin treatment. Intriguingly, bleomycin has no obvious effect on ANXA2 expression both in vivo and in vitro (Fig. S4A-D). In this regard, we next set out to identify the binding site of ANXA2 with bleomycin. We generated a series of ANXA2 truncation mutants and mapped a central 82-amino acid region (105-186) of ANXA2 as a bleomycin-binding region (Fig. 3A and B). Using docking analysis, we identified Glu139 (E139) and Asp182 (D182) in this 82-amino acid region as possible binding sites (Fig. 3C, D and S5A). We then constructed plasmids containing these 2 point mutants of ANXA2 (E139A and D182A), and purified recombinant proteins of these 2 ANXA2 mutants. We found that recombinant ANXA2E139A barely interacted with bleomycin, whereas ANXA2D182A retained bleomycin binding activity comparable to ANXA2WT (Fig. 3E). A fluorescence quenching assay revealed no obvious conformational change of ANXA2E139A compared with ANXA2WT protein (Fig. 3F). These data suggest that Glu139 of ANXA2 may be required for bleomycin binding. In addition, the docking analysis also showed that bleomycin may bind to ANXA2 through the amide group on its sugar moiety (Fig. S5A). We thus presumed that bleomycin lacking this sugar moiety may disrupt ANXA2 binding. Indeed, both recombinant ANXA2 and endogenous ANXA2 in lung epithelial cells failed to be affinity-purified by deglycosylated bleomycin (Fig. S5B and C), indicating that the sugar moiety of bleomycin is required for ANXA2 binding.
Figure 3.
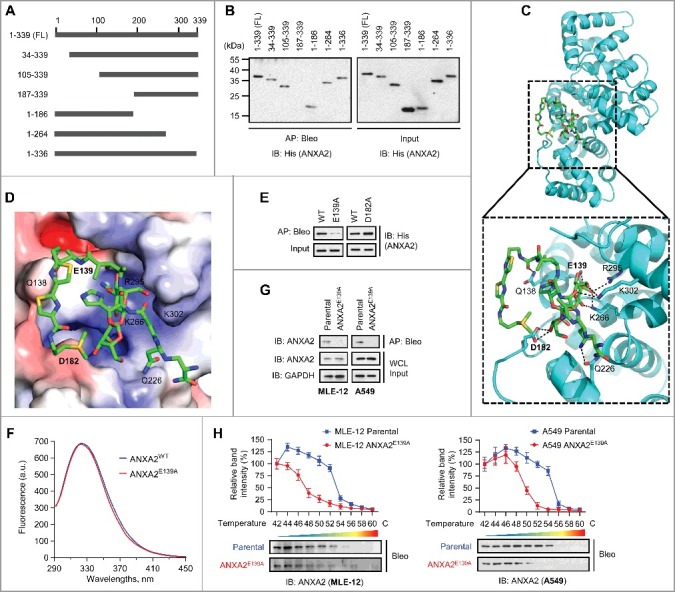
Glu139 of ANXA2 is required for bleomycin binding. (A) Schematic representation of full-length (FL) ANXA2 and its truncation mutants. (B) Recombinant His-ANXA2-FL and its truncation mutants were affinity-purified using bleomycin-immobilized NHS-activated Sepharose beads and then immunoblotted with anti-His antibody. (C and D) Cartoon representation (C) and electrostatic surface representation (D) of the docking model of bleomycin binding to H. sapiens ANXA2. (E) Recombinant His-ANXA2E139A or His-ANXA2D182A was affinity-purified using bleomycin-immobilized NHS-activated Sepharose beads and then immunoblotted with anti-His antibody. (F) Fluorescence quenching assay to examine the conformational changes of recombinant ANXA2 and ANXA2E139A (non-His tagged). (G) Target proteins were affinity-purified with bleomycin-immobilized beads from MLE-12 parental or MLE-12 ANXA2E139A cells and A549 parental or A549 ANXA2E139A cells, and then immunoblotted with anti-ANXA2 antibody. GAPDH, loading control. (H) Cellular thermal shift assay showing ANXA2 or ANXA2E139A target engagement by bleomycin (Bleo) in MLE-12 parental or MLE-12 ANXA2E139A cells (left) and A549 parental or A549 ANXA2E139A cells (right). The thermal stability of ANXA2 was quantified as indicated at the top of each immunoblot. Results are representative of 3 independent experiments unless stated otherwise. Data in (H) are means ± s.d.
To further determine whether Glu139 is required for bleomycin-ANXA2 binding, we used CRISPR-Cas9-based genome editing to engineer the E319A mutation in lung epithelial MLE-12 and A549 cells (Fig. S6A and B). The E139A mutation in living cells had no apparent effect on cell morphology and ANXA2 localization (Fig. S6C and D). In addition, ANXA2 expression was marginally affected in response to E139A mutation (Fig. S6E). As expected, CRISPR-Cas9-engineered ANXA2E139A-mutant cells (MLE-12 ANXA2E139A and A549 ANXA2E139A cells) revealed no obvious bleomycin binding in contrast to MLE-12 parental and A549 parental cells (Fig. 3G). Consistently, the thermal stabilization of ANXA2 in bleomycin-treated cells was markedly decreased when the E139A mutation was introduced (Fig. 3H). These findings demonstrate that Glu139 is required for the binding of ANXA2 with bleomycin. These data also suggest that the paired parental and ANXA2E139A lung epithelial cells are suitable models to dissect the molecular events downstream of bleomycin-ANXA2 binding that may play a role in the development of pulmonary fibrosis.
Bleomycin-ANXA2 interaction impedes autophagic flux in lung epithelial cells
Bleomycin induces autophagy in lung epithelial cells, and autophagy deficiency exacerbates bleomycin-induced pulmonary fibrosis in atg4−/− mice.27,28 We presumed that the contribution of ANXA2 binding to bleomycin-induced pulmonary fibrosis might be due to the regulation of autophagy. Bleomycin treatment increased LC3B-II abundance and GFP-LC3B puncta formation, and decreased autophagic substrate SQSTM1/p62 levels in MLE-12 and A549 parental cells (Figs. 4A-E). LC3B-II levels were further elevated after lysosomal inhibition with chloroquine or bafilomycin A1 (Fig. 4A). These data indicate that bleomycin induces autophagic flux in lung epithelial cells, which is consistent with a previous report.28 Notably, we observed markedly reduced LC3B-II abundance and GFP-LC3B puncta formation, as well as decreased SQSTM1 levels in bleomycin-treated ANXA2E139A cells compared to bleomycin-treated parental cells (Figs. 4A-E). In addition, chloroquine or bafilomycin A1 significantly increased LC3B-II levels in bleomycin-treated ANXA2E139A cells in comparison with bleomycin-treated parental cells (Fig. 4A and B). These data imply that the decrease of the bleomycin-ANXA2 interaction may dramatically accelerate autophagic flux.
Figure 4.
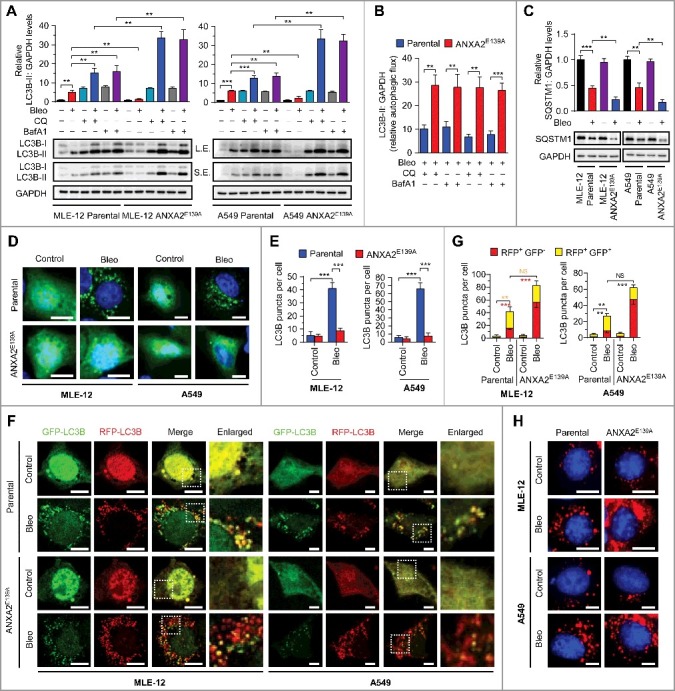
Disruption of bleomycin-ANXA2 interaction accelerates autophagic flux in lung epithelial cells. (A) Immunoblots for LC3B in parental and ANXA2E139A cells treated with bleomycin (50 μM) or cotreated with chloroquine (CQ, 20 μM) or bafilomycin A1 (BafA1, 100 nM) for 24 h. GAPDH, loading control. S.E., short exposure; L.E., long exposure. Quantification of LC3B levels in parental and ANXA2E139A cells treated with or without bleomycin is shown. (B) LC3B-II:GAPDH levels in parental and ANXA2E139A cells treated with bleomycin in the presence of CQ or BafA1 shown in (A). LC3B-II:GAPDH was calculated as: (LC3B-II level treated with bleomycin combined with CQ or BafA1 − LC3B-II level treated with bleomycin alone) ÷ LC3B-II basal levels. (C) Immunoblots for SQSTM1 in parental and ANXA2E139A cells treated with or without bleomycin (50 μM). GAPDH, loading control. Quantification of SQSTM1 levels is shown. (D) Representative images of parental or ANXA2E139A cells transiently expressing GFP-LC3B plasmids followed by bleomycin (Bleo, 50 μΜ) treatment for 24 h. DNA was counterstained with Hoechst 33342 (blue). Scale bars: 10 μm. (E) Quantification of LC3B puncta shown in (D). (F) Representative images of parental or ANXA2E139A cells transiently expressing mRFP-GFP-LC3B plasmids followed by treatment with bleomycin (50 μM) for 24 h. Scale bars: 10 μm. (G) Quantification of LC3B puncta shown in (F). (H) Representative images of parental or ANXA2E139A cells incubated with BODIPY-conjugated bovine serum (DQ-BSA, red) followed by bleomycin treatment (50 μΜ) for 24 h. DNA was counterstained with Hoechst 33342 (blue). Scale bars: 10 μm. Data in (B, D, F and H) are means ± s.d., Results are representative of 3 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (Student t test). NS, nonsignificant.
To corroborate these findings, we used LC3B tandemly tagged with acid-resistant monomeric RFP and acid-sensitive GFP (mRFP-GFP-LC3B) to measure the formation of autophagosomes (RFP+ GFP+ signal) and autolysosomes (RFP+ GFP− signal).29 As expected, the number of RFP+ GFP− puncta (autolysosomes) was increased in bleomycin-treated ANXA2E139A cells compared with bleomycin-treated parental cells (Fig. 4E and F), suggesting increased formation of autolysosomes caused by disrupting the bleomycin-ANXA2 interaction. To determine whether this autolysosome increase led to enhanced autophagic degradation, we used a highly self-quenched BODIPY-conjugated bovine serum albumin (DQ-BSA) that displays bright red fluorescence only when proteolytic degradation occurs.29,30 Indeed, the increase of degradation-induced DQ-BSA fluorescent signal in bleomycin-treated parental cells was further elevated in bleomycin-treated ANXA2E139A cells (Fig. 4G). In agreement with these observations, bleomycin induced LC3B accumulation in the lungs of wild-type (WT) mice, whereas with much reduced LC3B level in the lungs of anxa2−/− mice, which might be attributed to accelerated autophagic flux in bleomycin-treated anxa2−/− mice (Fig. S7). Collectively, these data suggest that ANXA2 binding impedes bleomycin-induced autophagic flux.
Bleomycin binding with ANXA2 impedes autophagic flux by inactivating TFEB in lung epithelial cells
To interrogate how ANXA2 binding attenuates bleomycin-induced autophagic flux, we profiled the global gene expression by RNA sequencing (RNA-seq) analysis in MLE-12 parental and MLE-12 ANXA2E139A cells in response to bleomycin treatment (Fig. 5A and Table S3). The RNA-seq data identified a set of autophagy-lysosome genes that were upregulated in bleomycin-treated ANXA2E139A cells in comparison with bleomycin-treated parental cells. The upregulation of these genes was further confirmed by qPCR analysis (Fig. 5B). These autophagy-lysosome genes have been previously reported to be regulated by TFEB, a master regulator of autophagy.31–33 Indeed, bleomycin treatment markedly induced nuclear translocation of TFEB in ANXA2E139A cells compared to parental cells (Fig. 5C and S8).
Figure 5.
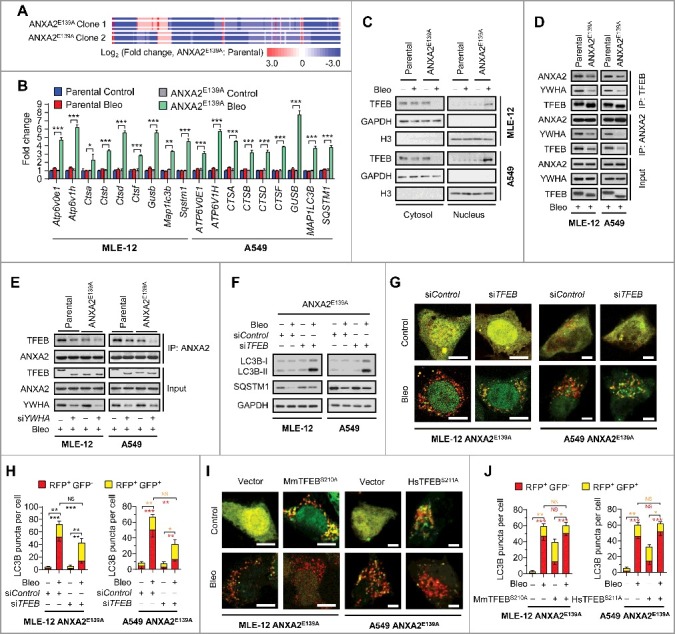
Disrupting bleomycin-ANXA2 interaction accelerates autophagic flux by activating TFEB in lung epithelial cells. (A) RNA-seq analysis of the differential expression profiles in 2 different clones of MLE-12 ANXA2E139A cells compared with MLE-12 parental cells following bleomycin treatment for 24 h. (B and C) Parental and ANXA2E139A cells were treated with or without bleomycin (Bleo, 50 μM) for 24 h. mRNA levels of autophagy-lysosome genes controlled by TFEB were measured by qPCR analysis (B). TFEB levels in cytosolic and nuclear fractions were examined by immunoblotting. GAPDH and H3F3/H3 are loading controls for the cytosol and nucleus, respectively (C). (D) Parental and ANXA2E139A cells were treated with bleomycin (Bleo, 50 μM) for 24 h. Immunoprecipitation of TFEB with ANXA2 or YHWA was detected by immunoblotting analysis. (E) Parental and ANXA2E139A cells were transfected with control siRNA or YWHA siRNA followed by bleomycin (50 μM) treatment for 24 h. Immunoprecipitation of ANXA2 with TFEB was detected by immunoblotting analysis. (F) ANXA2E139A cells transfected with siControl or siTFEB were treated with or without bleomycin (50 μM) for 24 h and immunoblotted for LC3B and SQSTM1. GAPDH, loading control. (G) Representative images of ANXA2E139A cells transiently expressing mRFP-GFP-LC3B plasmids transfected with control siRNA or siTFEB, followed by treatment with bleomycin (50 μM) for 24 h. Scale bars: 10 μm. (H) Quantification of LC3B puncta shown in (G). (I) Representative images of ANXA2E139A cells transiently expressing mRFP-GFP-LC3B plasmids transfected with vector or TFEB mutants (MmTFEBS210A in MLE-12 cells and HsTFEBS211A in A549 cells, respectively), followed by treatment with bleomycin (50 μM) for 24 h. Scale bars: 10 μm. (J) Quantification of LC3B puncta shown in (I). Data in (B, H and J) are means ± s.d. Results are representative of 3 independent experiments unless stated otherwise. *P < 0.05, **P < 0.01, ***P < 0.001 (Student t test). NS, nonsignificant.
It has been reported that YWHA/14-3-3 interacts with TFEB to inhibit the translocation of TFEB into the nucleus,34,35 we therefore performed an immunoprecipitation assay to determine whether YWHA was involved in bleomycin-induced nuclear translocation of TFEB in ANXA2E139A cells. As expected, ANXA2, TFEB and YWHA exhibited strong interactions with each other in bleomycin-treated parental cells, while these interactions were markedly reduced in bleomycin-treated ANXA2E139A cells (Fig. 5D). YWHA knockdown using siRNA significantly attenuated the interaction of TFEB with ANXA2 in bleomycin-treated parental cells, to a similar level observed in bleomycin-treated ANXA2E139A cells (Fig. 5E). These results indicate that ANXA2, YWHA and TFEB form a triplex resulting in TFEB cytoplasmic retention and inactivation, and that YWHA mediates ANXA2 and TFEB interaction.
We further presumed that TFEB might contribute to the accelerated autophagic flux in bleomycin-treated ANXA2E139A cells. As expected, TFEB knockdown in bleomycin-treated ANXA2E139A cells increased LC3B-II levels and decreased RFP+ GFP− puncta (Fig. 5F-H and S9A), akin to what were observed in bleomycin-treated parental cells (Fig. 4A, B, G and H). Consistently, constitutive activation of TFEB in ANXA2E139A cells using TFEB-mutants (MmTFEBS210A in MLE-12 parental cells and HsTFEBS211A in A549 parental cells)33,34 did not further accelerate bleomycin-induced autophagic flux (Fig. 5I and J). In addition, constitutive activation of TFEB in bleomycin-treated parental cells using TFEB mutants decreased LC3B-II and increased RFP+ GFP− puncta (Fig. S9B-D), to a similar extent to that in bleomycin-treated ANXA2E139A cells (Fig. 4A, B, G and H). Collectively, these results demonstrate that the bleomycin-ANXA2 interaction impedes autophagic flux by inducing cytoplasmic retention of TFEB in lung epithelial cells.
Pharmacological activation of TFEB-induced autophagy ameliorates bleomycin-induced pulmonary fibrosis
Given the TFEB inactivation and autophagy impediment caused by bleomycin-ANXA2 binding, we next investigated whether stimulation of TFEB-mediated autophagy by Torin 1 (treated from day 1), a TFEB activator that inhibits its phosphorylation and induces its nuclear translocation,32,35,36 could prevent the occurrence of pulmonary fibrosis under bleomycin treatment conditions. As expected, Torin 1 treatment from d 1 significantly reduced LC3B expression levels in lungs from bleomycin-treated mice (Fig. 6A and B), which might be attributed to TFEB-accelerated autophagic flux. In addition, Torin 1 reduced bleomycin-induced alveolar septal thickening, fibrotic foci, alveolar structure distortion and fibrotic scores (Fig. 6C and D). Moreover, Torin 1 alleviated collagen accumulation and hydroxyproline content in bleomycin-induced mice (Fig. 6E-G). Total protein contents and TGFB1 levels in BALF were also decreased by Torin 1 treatment in bleomycin-injected mice (Fig. 6H and I). As expected, body weight loss induced by bleomycin was restored by Torin 1 treatment (Fig. 6J). These results indicate that the TFEB activator Torin 1 induces autophagic flux and ameliorates bleomycin-induced pulmonary fibrosis in mice.
Figure 6.
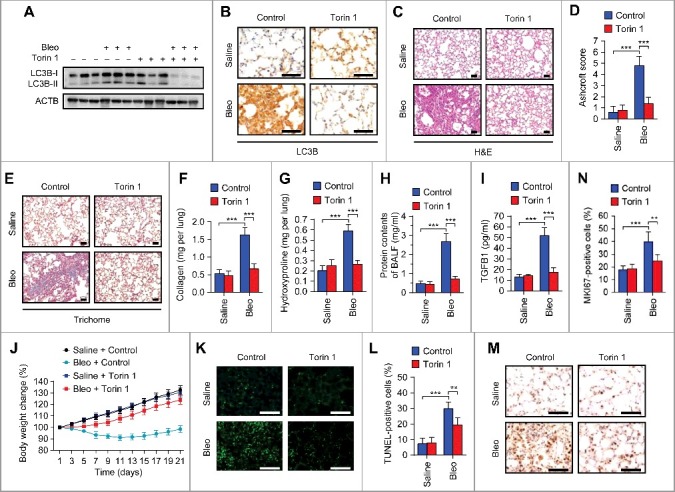
Pharmacological activation of TFEB by Torin 1 elevates autophagic flux and ameliorates pulmonary fibrosis in bleomycin-treated mice. Mice were injected intratracheally with bleomycin or saline, and treated with Torin 1 (20 mg/kg) every other day for 21 d (n = 5 per group). (A) Lung tissues were homogenized and immunoblotted for LC3B. ACTB, loading control. (B) Representative images of LC3B immunohistochemical staining of lung tissue sections. Scale bars: 50 μm. (C) Representative images of hematoxylin and eosin (H&E) staining of lung tissue sections. Scale bars: 50 μm. (D) Ashcroft fibrotic scores were determined by H&E staining of lung sections. (E) Representative images of Masson's trichrome staining of lung tissue sections. Scale bars: 50 μm. (F and G) Whole lung homogenates were analyzed to examine collagen contents using the Sircol assay (F) and hydroxyproline contents (G). (H and I) Total protein contents (H) and TGFB1 levels (I) were determined in bronchoalveolar lavage fluid (BALF). (J) Body weights were measured over time. (K) Representative images of TUNEL staining of apoptotic cells in lung tissue sections. Scale bars: 50 μm. (L) Quantification of apoptotic cells shown in (A). (M) Representative images of MKI67 immunohistochemical staining in lung tissue sections. Scale bars: 50 μm. (N) Percentage of MKI67-positive cells shown in (M). Data are means ± s.d. **P < 0.01, ***P < 0.001 (Student t test).
To achieve a more rational strategy for clinical intervention, we evaluated the anti-fibrotic effect of Torin 1 (treated from d 7). As expected, Torin 1 treatment from d 7 significantly ameliorated bleomycin-induced pulmonary fibrosis (Fig. S10A-G), which is consistent with that of Torin 1 treatment from d 1. However, in addition to TFEB activation, Torin 1 was also reported to induce autophagy as an MTOR (mechanistic target of rapamycin) inhibitor.37 Thus, we further investigated the anti-fibrotic effect of the curcumin analog compound C1, an MTOR-independent activator of TFEB.38 As shown in Fig. S11A-G, compound C1 significantly attenuated bleomycin-induced pulmonary fibrosis. These results strongly support the idea that TFEB activation plays an important role in ameliorating bleomycin-induced pulmonary fibrosis.
It has been recently reported that apoptosis and hyperproliferation of lung epithelial cells contribute to bleomycin-induced lung injury and fibrosis.39–41 Consistently, we found that bleomycin increased TUNEL- and MKI67-positive cells in mice, whereas the increase of apoptotic and proliferative cells was attenuated by Torin 1 treatment (Fig. 6K-N). Together, these findings indicate that pharmacological activation of TFEB-induced autophagy ameliorates bleomycin-induced pulmonary fibrosis in mice.
TFEB-induced autophagy is negatively associated with human pulmonary fibrosis
To evaluate the clinical relevance of TFEB, we examined the expression of TFEB in 41 paired human idiopathic pulmonary fibrosis (IPF) tissues and adjacent normal lung tissues (Table S4). Compared with normal lung tissues, TFEB expression in lung epithelial cells was downregulated from pulmonary fibrosis tissues (Fig. 7A and B). Consistently, we noted that in contrast to fibrosis tissues, TFEB was preferentially expressed in the nuclei of lung epithelial cells in normal tissues (Fig. 7C). In addition, we found that LC3B was also downregulated in fibrotic tissue (Fig. 7D and E), and TFEB expression positively correlated with LC3B expression (Fig. 7F). These data suggest a critical role of TFEB-mediated autophagy in IPF. Although we have not directly evaluated the role of bleomycin-ANXA2 binding in bleomycin-induced fibrosis samples from patients due to unavailability, the potential signaling pathways including TFEB-induced autophagy in IPF human samples could partially address the clinical relevance of our study.
Figure 7.
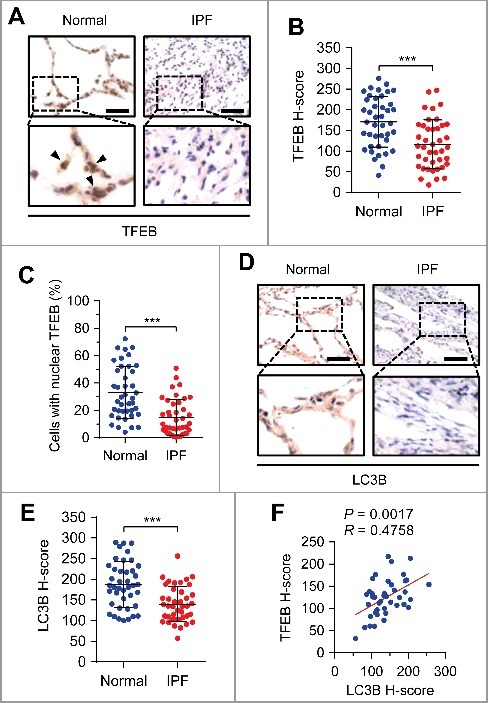
Expressions of TFEB and LC3B are negatively associated with human pulmonary fibrosis. (A) Representative images of TFEB immunohistochemical staining in pulmonary fibrosis (PF) tissues or adjacent normal lung tissues from patients. Arrows represent lung epithelial cells with nuclear TFEB staining. Scale bars: 50 μm. (B and C) TFEB staining intensity (B) and percentage of nuclear TFEB-positive cells (C) of lung epithelial cells in 41 PF tissues and paired adjacent normal lung tissues from patients. (D) Representative images of LC3B immunohistochemical staining in pulmonary fibrosis (PF) tissues or adjacent normal lung tissues from patients. Scale bars: 50 μm. (E) LC3B staining intensity in PF tissues and paired adjacent normal lung tissues from patients (n = 41). (F) Association of the staining intensity of TFEB and LC3B in PF tissues (n = 41). Concordance was determined by Pearson correlation and linear regression. Data in (B, C and E) are means ± s.d. Results are representative of 3 independent experiments unless stated otherwise. ***P < 0.001 (Student t test).
Discussion
Despite being a potent anticancer chemotherapeutic drug, bleomycin has a main limitation due to causing severe pulmonary fibrosis that shortens patient life span. Studies have thus far focused on investigating cellular and physiological details of bleomycin-induced pulmonary fibrosis. However, virtually no attempt has been made to identify bleomycin's direct molecular targets in eukaryotes. Here, we identify and demonstrate that ANXA2 is a binding target of bleomycin in causing pulmonary fibrosis. Blocking this binding by introducing the E139A mutant into ANXA2 or modifying the chemical structure of bleomycin may represent feasible strategies to prevent pulmonary fibrosis during anticancer therapy with bleomycin.
In our experiments, we show that bleomycin is anchored to E139 of ANXA2 through the amide group on the sugar moiety. Deletion of this sugar moiety of bleomycin disrupts ANXA2 binding and should therefore reduce pulmonary toxicity. In accordance with this concept, it has been recently reported that deglycosylated bleomycin, a bleomycin-derived drug in which the sugar residue located in the glycosylated portion is removed, shows similar anticancer activity to bleomycin, but does not induce pulmonary fibrosis.42 Our findings together with this previous study suggest that deglycosylated bleomycin may represent a potential alternative to bleomycin for cancer treatment.
ANXA2, an evolutionarily conserved calcium-dependent protein, has been reported to be involved in initiating the autophagy process either by promoting phagophore assembly via actin in physiological contexts or enabling ATG9A trafficking to phagophores under starvation conditions.43,44 In addition, our previous study suggests that ANXA2 regulates autophagy through the AKT-MTOR pathway in alveolar macrophages in a mouse infection model.45 These studies address the important role of ANXA2 in autophagy, but the regulatory mechanisms seem to be context dependent. In the current study, we characterize an additional mechanism of ANXA2-regulated autophagy in the context of bleomycin treatment. ANXA2 is selectively targeted by bleomycin, leading to recruitment of TFEB to form a bleomycin-ANXA2-YWHA-TFEB complex in the cytoplasm. The cytoplasmic retention of TFEB blunts its transcriptional activity and impedes autophagic flux (Fig. S12).
Recently, increasing evidence has suggested that autophagy plays a role in pulmonary fibrosis. For instance, insufficient autophagy has been observed in this condition.46 Induction of autophagy in epithelial cells by blocking IL17A enhances collagen degradation and decreases pulmonary fibrosis.47 In addition, it has been found that bleomycin induced more severe pulmonary fibrosis in atg4b−/− mice.28 These findings suggest that autophagy plays a protective role in the development of pulmonary fibrosis. In this study, we found that bleomycin-ANXA2 binding impedes autophagic flux by TFEB inactivation. This autophagic event occurs in lung epithelial cells, the injury and activation of which are required for the initiation and progression of pulmonary fibrosis.20–22 Consistent with our data, it has been reported that rapamycin (an MTOR inhibitor promoting autophagic flux) partially reversed bleomycin-induced pulmonary fibrosis, and this effect could be inhibited by chloroquine (an inhibitor of autolysosome formation blocking autophagic flux).48 Our data together with other studies reveal a protective role of autophagic flux in bleomycin-induced pulmonary fibrosis, suggesting that promotion of autophagic flux, such as via TFEB activators, may represent a possible intervention strategy against pulmonary fibrosis.
It is well known that bleomycin-induced lung toxicity involves oxidative stress and DNA damage that causes an inflammatory response.2 These events occur in the nucleus whereas autophagy takes place in the cytoplasm, implying that both cytoplasmic and nuclear events play essential roles in bleomycin-induced pulmonary fibrosis. Autophagy has been reported to be involved in regulating oxidative stress, DNA damage and inflammatory response49–52; whether autophagy affects bleomycin-induced oxidative stress, DNA damage or inflammatory response requires further investigation. Moreover, although TFEB-induced autophagy is negatively associated with IPF in human samples, it should be noted that ANXA2, the molecular target of bleomycin, has only been demonstrated to be involved in bleomycin-induced pulmonary fibrosis in the current study. Whether ANXA2 is relevant to IPF is currently not clear.
In summary, we demonstrate that bleomycin binds directly to ANXA2, impedes TFEB-induced autophagic flux and thereby contributes to the development of pulmonary fibrosis. Our study offers insight into the mechanistic basis of bleomycin-induced pulmonary fibrosis, and opens new perspectives for developing potential strategies to minimize the pulmonary toxicity of bleomycin.
Materials and methods
Cell culture
MLE-12, A549 and 293T cells were obtained from the American Type Culture Collection (CRL-2110, CCL-185 and CRL-3216). A549 and 293T cells were cultured in Dulbecco's modified Eagle's medium (Thermo Fisher Scientific, 12800017) supplemented with 10% fetal bovine serum (FBS; HyClone, SH30088.03). MLE-12 cells were maintained in DMEM/F12 containing 2% FBS. Short tandem-repeat profiling was performed for all cell lines. Primary human alveolar epithelial type II (ATII) cells were isolated from distal normal lung tissue of lung cancer as previously described and cultured in DMEM supplemented with 10% FBS.53 All cell lines and primary cells were free of mycoplasma contamination as tested using the LookOut Mycoplasma PCR Detection Kit (Sigma-Aldrich, MP0035).
Reagents
Antibodies for ANXA2 (annexin A2) (PA5-14318), GAPDH (MA5-15738), 6 × His Epitope Tag (MA1-21315) and Flag Epitope Tag (MA1-91878) were from Thermo Fisher Scientific. Antibody for LC3B (NB100-233) was from Novus Biologicals. Antibody for SQSTM1 (PM045) was from MBL International Corporation. Antibodies for TFEB (sc-48784) and H3F3/histone H3 (sc-10809) were from Santa Cruz Biotechnology. Antibody for YWHA (8312) was from Cell Signaling Technology. Antibodies for goat anti-mouse IgG-HRP (sc-2005) and goat anti-rabbit IgG-HRP (sc-2004) were from Santa Cruz Biotechnology.
ANXA2 siRNA (Hs) (sc-270151), Anxa2 siRNA (Mm) (sc-29683), TFEB siRNA (Hs) (sc-38509) and Tfeb siRNA (Mm) (sc-38509) were purchased from Santa Cruz Biotechnology. Bleomycin A5 (MB1038) and bleomycin A2/B2 (MB1039) were from Melone Pharmaceutical Co. Chloroquine (1327000) and bafilomycin A1 (B1793) were purchased from Sigma-Aldrich. Torin 1 (A8312) was from ApexBio Technology. Lipofectamine 3000 (L3000015) and Hoechst 33342 (H3570) were purchased from Thermo Fisher Scientific.
Affinity purification with bleomycin-immobilized beads
The scheme for coupling bleomycin A5 to NHS-activated Sepharose beads is shown in Fig. S2B. NHS-Activated Sepharose 4 Fast Flow agarose matrix (GE Healthcare Life Sciences, 17-0906-01) was washed with 1 mM cold HCl and incubated with bleomycin A5 in coupling buffer (0.2 M NaHCO3, 0.5 M NaCl, pH 8.3) overnight at 4ºC. Bleomycin A5 was immobilized on beads via its amino reactivity. Unreacted residues were blocked with 0.5 M ethanolamine, pH 8.3. Beads were washed and stored at 4ºC for use. HPLC analysis of bleomycin A5 solutions before or after bleomycin conjugation was performed to determine the coupling efficacy.
Bleomycin-immobilized beads were equilibrated with 1% NP-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40 (Sigma-Aldrich, 74385) and then incubated with protein lysates prepared from mouse MLE-12, human A549 cells or lung homogenates of human, mouse, rabbit, rat and monkey for 2 h at 4ºC. The beads were washed 3 times with 1% NP-40 lysis buffer. Bound proteins were eluted with 5 mM free bleomycin A5. In the competition experiment, 1 mM free bleomycin A5 was added to protein extracts before incubation with the beads. Bound proteins were separated by SDS-PAGE and subjected to mass spectrometry or immunoblotting analysis.
Expression of recombinant human ANXA2 (rHsANXA2)
Human ANXA2 cDNA (provided by Dr. Jiahuai Han, Xiamen University) was cloned into a pET28a vector with an N-terminal hexahistidine (His) tag (Provided by Dr. Yamei Yu, Sichuan University) between restriction sites BamHI and XhoI. cDNAs encoding residues 34–399, 105–339, 187–339, 1–186, 1–264, 1–336 of human ANXA2 were individually amplified by PCR and inserted into the pET28a vector as above. Mutations of E139A and D182A were created by site directed mutagenesis. All primers are shown in Table S5. pET28a His-ANXA2 plasmids were transformed into Rosetta (DE3) E. coli cells (EMD Millipore, 70954). Cells were inoculated in Luria-Bertani medium (10 g NaCl, 10 g tryptone and 5 g yeast extract in 1 L ddH2O) containing kanamycin and induced with 400 μM IPTG (EMD Millipore, 420322). Cells were harvested and lysed by sonication. His-tag ANXA2 was purified using a Ni2+-charged HiTrap Chelating column (GE Healthcare Life Sciences, 17-0409-05) on an AKTA Explorer Purification Unit (GE Healthcare Life Sciences, Piscataway, NJ, USA). Where necessary, the His tag was removed by TEV protease (Sigma-Aldrich, T4455) cleavage. The purified human ANXA2 was characterized by SDS-PAGE, Immunoblotting and ESI-Q-TOF-MS analysis.
Surface plasmon resonance analysis
Surface plasmon resonance experiments were performed on a BIAcore T200 biosensor system (GE Healthcare Life Sciences, Piscataway, NJ, USA) at 25ºC. Bleomycin A5 was dissolved in 10 mM sodium borate, pH 8.5 and immobilized on an activated CM5 dextran chip using amine-coupling chemistry (GE Healthcare Life Sciences, 29104988). The remaining active sites were blocked with 1 M ethanolamine, pH 8.3. The binding of rHsANXA2 at different concentrations was performed in 1 × PBS buffer (Beyotime, C0221A) containing 0.05% Tween-20 (Sigma-Aldrich, P1379), pH 7.4 at a flow rate of 30 μl min−1 for 120 s. For chip regeneration, bound rHsANXA2 was removed by injection of 5 mM NaOH for 30 sec. The binding kinetics was analyzed by BIAevaluation software 2.0 using the 1:1 Langmuir binding model.
Isothermal titration calorimetry
Isothermal titration calorimetry experiments were carried out on a MicroCal iTC200 calorimeter (Malvern Instruments, Malvern, UK) at 25ºC in 1 × PBS buffer, pH 7.4. Bleomycin (1 mM) was titrated into 50 μM rHsANXA2. The data were analyzed with the Origin calorimetry software package using a single site-binding model.
Fluorescence quenching assay
Fluorescence titrations were performed using a Lumina Fluorescence Spectrometer (Thermo Fisher Scientific, MA, USA) at 25ºC with 1-nm excitation and 10-nm emission bandwidths. The excitation wavelength was 280 nm, and the emission spectra were measured between 290 and 450 nm. Different concentrations of bleomycin were titrated into 200 μM rHsANXA2 in 1 × PBS buffer, pH 7.4. During titrations, the additives were not allowed to exceed 2% of the total solution volume.
Cellular thermal shift assay
The stabilization of targets in cells by compound interaction was evaluated by cellular thermal shift assay as described previously.24 Briefly, cells grown in 100-mm dishes to 80% confluency were treated with 50 μM bleomycin or PBS for 6 h. Cells were harvested by trypsin and subsequently resuspended in PBS. The cell suspension was then aliquoted into 10 PCR tubes, heated for 5 min to 42, 44, 46, 48, 50, 52, 54, 56, 58 or 60ºC followed by 3 cycles of freeze-thawing with liquid nitrogen and centrifugation at 17,000 g for 15 min. The soluble fractions were analyzed by SDS-PAGE and immunoblotted with anti-ANXA2 antibody.
Preparation of fluorescein-labeled bleomycin
Fluorescein-labeled bleomycin (F-Bleo) was prepared as described by Aouida et al.54 Briefly, 2.1 mM 5-(and-6)-carboxyfluorescein, succinimidyl ester (5(6)-FAM, SE (Thermo Fisher Scientific, C1311) was incubated with 0.6 mM bleomycin A5 in 0.2 M NaHCO3, pH 8.3, at 4ºC overnight. Unreacted residues were blocked with 1.0 M hydroxylamine, pH 8.3. The products were then separated on a 1% agarose gel with 40 mM MES buffer, pH 6.5. The F-Bleo product, which slowly migrated towards the cathode, was visualized by UV and excised followed by 3 cycles of freeze-thawing at -80ºC. Excised gels were centrifuged at 12,000 g for 10 min, freeze dried and dissolved in sterile deionized water. F-Bleo products were then purified and quantified by RP-HPLC using an Acquity BEH C18 column (100 mm × 2.1 mm × 1.8 μm; Waters, 186008755).
Immunofluorescence
Cells were seeded onto glass cover slips in 24-well culture plates and incubated overnight. After treatment, cells were fixed with 4% paraformaldehyde at room temperature for 30 min. The slips were rinsed with PBS and cells were permeabilized with 0.04% Triton X-100 for 10 min and blocked with 5% goat serum (EMD Millipore, S26). Cells were incubated with primary antibody at 4ºC overnight, washed in PBS, incubated with appropriate secondary antibody (Thermo Fisher Scientific, 35560 for DyLight 594-conjugated goat anti-rabbit IgG, and 35502 for DyLight 488-conjugated goat anti-mouse IgG) and then counterstained with Hoechst 33342 (Sigma-Aldrich, 14533). For the DQ-BSA assay, cells were incubated with 10 µg/ml DQ-BSA Red (Thermo Fisher Scientific, D-12051) for 1 h and then treated with or without bleomycin (50 µM) for 24 h. Cells were washed, fixed and counterstained with Hoechst 33342. The red fluorescent signal indicating degradation capacity was measured.
Immunoblotting
Total protein lysates were prepared in RIPA buffer (50 mM Tris base, 150 mM NaCl, 0.1% SDS, 1% NP-40, pH 7.6) and quantified using the BCA Protein Assay (Thermo Fisher Scientific, 23250). Nuclear and cytoplasmic fractionation was performed using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, 78833). Proteins were separated by SDS-PAGE gels, transferred onto PVDF membranes (EMD Millipore, ISEQ00010) and blocked with 5% skimmed milk in TBST (20 mM Tris base, 137 mM NaCl, 0.1% Tween-20, pH 7.6) for 2 h. The membranes were probed with primary antibodies at 4ºC overnight, and then washed with TBST followed by incubation with HRP secondary antibodies (Thermo Fisher Scientific, 31430 for goat anti-mouse IgG (H+L) secondary antibody, and 31460 for goat anti-rabbit IgG (H+L) secondary antibody) for 1 h at room temperature. Immunoreactivity was visualized by enhanced chemiluminescence reagents (EMD Millipore, WBKLS0500).
Animal studies
anxa2−/− mice were kindly provided by Dr. Katherine A. Hajjar (Cornell University).55 C57BL/6J female mice of 6–8 wk of age were used. Bleomycin (2.5 U/kg) in 50 μl saline was intratracheally administered in anxa2−/− and wild-type (WT) mice. Where indicated, Torin 1 (20 mg/kg) in 5% DMSO:50% PEG-400 (Sigma-Aldrich, 202398) in PBS was injected intraperitoneally every other day from after bleomycin administration (d 1 or d 7). Curcumin analog compound C1 (20 mg/kg; Sigma-Aldrich, L157139) in 1% sodium carbonyl methylcellulose (Sigma-Aldrich, C5678) was orally administered by gavage every other day. The body weights were measured on each day of treatment. Lungs were removed and bronchoalveolar lavage fluid (BALF) was obtained from anesthetized mice on d 21. The measurement of collagen (Sicrol Collagen Assay; Biocolor, S2000) and hydroxyproline (Sigma-Aldrich, MAK008) contents of whole lungs were performed according to the manufacturer's instructions. Protein contents of BALF were measured with a BCA Protein Assay Kit (Thermo Fisher Scientific, 23250). TGFB1 levels in BALF were determined with the ELISA Ready-SET-Go! Kit (Thermo Fisher Scientific, 88-8350-22). For histopathological examination, mouse lungs were fixed in 4% paraformaldehyde and embedded in paraffin. The lung sections were stained with H&E or Masson's trichrome reagent. The severity of fibrosis was evaluated by Ashcroft score in H&E-stained sections.26 All animal studies were reviewed and approved by the Institutional Animal Care and Treatment Committee of Sichuan University, and the University of North Dakota Institutional Animal Care and Use Committee.
CRISPR-Cas9 genome editing
Homology directed repair-mediated E139A mutation of ANXA2 in A549 or MLE-12 cells was performed by CRISPR-Cas9 genome editing. The single guide RNA sequence for targeting human ANXA2 in A549 cells (5’-CCTGCTGATGTTCGTGCTCCT-3’) or mouse ANXA2 in MLE-12 cells (5’-ACGCATGCAAGAGTGCGCACGG-3’), respectively, was designed using the CRISPR design web interface (http://crispr.genome-engineering.org/). The guide sequence was cloned into the pX330 vector as described previously.56 The E139A repair template was amplified from genomic DNA in A549 or MLE-12 cells and inserted into the pcDNA3.1 vector (Thermo Fisher Scientific, V79020). Then the E139A mutation and silent mutation (preventing re-cutting of the targeted locus following homologous recombination) were introduced into the repair template by site directed mutagenesis. The proper pX330/guide RNA and pcDNA3.1/repair template plasmids were cotransfected into A549 or MLE-12 cells using Lipofectamine 3000 (Thermo Fisher Scientific, L3000001). Cells were then expanded as single clones, of which the genomic DNA was amplified by PCR and sequenced to examine the E139A mutation.
Quantitative RT–PCR and RNA-seq
Total cellular RNAs were extracted using Trizol (Thermo Fisher Scientific, 15596018). Total RNA (1 µg) was subjected to reverse transcription using the SuperScript III FirstStrand Synthesis System (Thermo Fisher Scientific, 18080051). Quantitative RT-PCR was performed with iTaq Universal SYBR Green Supermix (Bio-Rad, 1725120) in a CFX Connect Real-Time PCR Detection System (Bio-Rad). The relative transcript levels were normalized to endogenous GAPDH and expressed as 2−ΔΔCt values. Primer sequences are described in Table S5. RNA-Seq was performed with Illumina HiSeq 2000 by Beijing Genomics Institute (BGI, Shenzhen, China).
Human samples
A total of 41 idiopathic pulmonary fibrosis tissues and paired adjacent normal lung tissues were obtained under informed consent from patients who underwent surgical resection at either West China Hospital or Sichuan Provincial People's Hospital (Chengdu, China). Table S4 shows patient demographics and clinical characteristics. Ethics approval was obtained from the Institutional Ethics Committee of Sichuan University. Excised Lungs were fixed with 4% paraformaldehyde and embedded with paraffin for immunohistochemical examination.
Immunohistochemistry
Immunohistochemical staining was performed as described previously.57 Briefly, the tissue sections were deparaffinized, rehydrated and blocked with 3% H2O2 followed by microwave antigen retrieval in citrate buffer (pH 6.0). Sections were then treated with 10% normal serum, incubated with primary antibody at 4ºC overnight. The EnVision Detection System (Agilent Technologies, K5007) was used to detect antigen expression. Staining intensity was graded as: negative (0), weak (1), moderate (2), and strong (3). H-Score (0 – 300) = (1 × % of weak staining) + (2 × % moderate staining) + (3 × % strong staining). Protein staining was evaluated independently by 2 experienced investigators.
Statistics
Significant differences between 2 groups were evaluated using a 2-tailed Student t test. Concordance was determined by Pearson correlation and linear regression. GraphPad Prism 6.0 software was used for plotting graphs and statistical analysis. Data were expressed as means ± s.d. and significance was designated as follows: *P < 0.05, **P < 0.01, *** P < 0.001.
Supplementary Material
Funding Statement
Chinese NSFC [grant numbers 81602194, 81225015, 81430071, 81773143 and 81401951]; National 973 Basic Research Program of China [grant number 2013CB911300]; US National Institute of Health [grant number AI101973-01]; US National Institute of Health [grant number AI109317-01A1].
Acknowledgements
This work was supported by National 973 Basic Research Program of China (2013CB911300), the Chinese NSFC (81602194, 81225015, 81430071, 81773143 and 81401951), and US National Institute of Health (AI109317-01A1 and AI101973-01).
Disclosure of interest
The authors report no conflict of interest.
References
- 1.Umezawa H, Maeda K, Takeuchi T, Okami Y. New antibiotics, bleomycin A and B. J Antibiot (Tokyo) 1966;19:200–209. PMID:5953301 [PubMed] [Google Scholar]
- 2.Chen J, Stubbe J. Bleomycins: towards better therapeutics. Nat Rev Cancer 2005;5:102–112. doi: 10.1038/nrc1547. PMID:15685195 [DOI] [PubMed] [Google Scholar]
- 3.Galm U, Hager MH, Van Lanen SG Ju J, Thorson JS, Shen B. Antitumor antibiotics: bleomycin, enediynes, and mitomycin. Chem Rev 2005;105:739–758. doi: 10.1021/cr030117g. PMID:15700963 [DOI] [PubMed] [Google Scholar]
- 4.Lazo JS, Hoyt DG, Sebti SM, Pitt BR. Bleomycin: a pharmacologic tool in the study of the pathogenesis of interstitial pulmonary fibrosis. Pharmacol Ther 1990;47:347–358. doi: 10.1016/0163-7258(90)90061-6. PMID:1705351 [DOI] [PubMed] [Google Scholar]
- 5.Sleijfer S. Bleomycin-induced pneumonitis. Chest 2001;120:617–624. doi: 10.1378/chest.120.2.617. PMID:11502668 [DOI] [PubMed] [Google Scholar]
- 6.Della Latta V, Cecchettini A, Del Ry S, Morales MA. Bleomycin in the setting of lung fibrosis induction: From biological mechanisms to counteractions. Pharmacol Res 2015;97:122–130. doi: 10.1016/j.phrs.2015.04.012. PMID:25959210 [DOI] [PubMed] [Google Scholar]
- 7.Bouwman P, Jonkers J. The effects of deregulated DNA damage signalling on cancer chemotherapy response and resistance. Nat Rev Cancer 2012;12:587–598. doi: 10.1038/nrc3342. PMID:22918414 [DOI] [PubMed] [Google Scholar]
- 8.Bernard K, Hecker L, Luckhardt TR, Cheng G, Thannickal VJ. NADPH oxidases in lung health and disease. Antioxid Redox Signal 2014;20:2838–2853. doi: 10.1089/ars.2013.5608. PMID:24093231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oga T, Matsuoka T, Yao C, Nonomura K, Kitaoka S, Sakata D, Kita Y, Tanizawa K, Taguchi Y, Chin K, et al. Prostaglandin F(2alpha) receptor signaling facilitates bleomycin-induced pulmonary fibrosis independently of transforming growth factor-beta. Nat Med 2009;15:1426–1430. doi: 10.1038/nm.2066. PMID:19966781 [DOI] [PubMed] [Google Scholar]
- 10.Hay J, Shahzeidi S, Laurent G. Mechanisms of bleomycin-induced lung damage. Arch Toxicol 1991;65:81–94. doi: 10.1007/BF02034932. PMID:1711838 [DOI] [PubMed] [Google Scholar]
- 11.Hagiwara SI, Ishii Y, Kitamura S. Aerosolized administration of N-acetylcysteine attenuates lung fibrosis induced by bleomycin in mice. Am J Respir Crit Care Med 2000;162:225–231. doi: 10.1164/ajrccm.162.1.9903129. PMID:10903246 [DOI] [PubMed] [Google Scholar]
- 12.Shahzeidi S, Sarnstrand B, Jeffery PK, McAnulty RJ, Laurent GJ. Oral N-acetylcysteine reduces bleomycin-induced collagen deposition in the lungs of mice. Eur Respir J 1991;4:845–852. PMID:1720100 [PubMed] [Google Scholar]
- 13.Ward HE, Nicholson A, Berend N. Failure of systemic N-acetyl cysteine to protect the rat lung against bleomycin toxicity. Pathology 1987;19:358–360. doi: 10.3109/00313028709103883. PMID:2451202 [DOI] [PubMed] [Google Scholar]
- 14.Konigshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, et al. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest 2009;119:772–787. PMID:19287097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decaris ML, Gatmaitan M, FlorCruz S, Luo F, Li K, Holmes WE, Hellerstein MK, Turner SM, Emson CL. Proteomic analysis of altered extracellular matrix turnover in bleomycin-induced pulmonary fibrosis. Mol Cell Proteomics 2014;13:1741–1752. doi: 10.1074/mcp.M113.037267. PMID:24741116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raida M. Drug target deconvolution by chemical proteomics. Curr Opin Chem Biol 2011;15:570–575. doi: 10.1016/j.cbpa.2011.06.016. PMID:21763176 [DOI] [PubMed] [Google Scholar]
- 17.Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol 2009;5:616–624. doi: 10.1038/nchembio.216. PMID:19690537 [DOI] [PubMed] [Google Scholar]
- 18.Wang K, Yang T, Wu Q, Zhao X, Nice EC, Huang C. Chemistry-based functional proteomics for drug target deconvolution. Expert Rev Proteomics 2012;9:293–310. doi: 10.1586/epr.12.19. PMID:22809208 [DOI] [PubMed] [Google Scholar]
- 19.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science 2010;327:1345–1350. doi: 10.1126/science.1177319. PMID:20223979 [DOI] [PubMed] [Google Scholar]
- 20.Kulkarni T, de Andrade J, Zhou Y, Luckhardt T, Thannickal VJ. Alveolar epithelial disintegrity in pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2016;311:L185–L191. PMID:27233996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thannickal VJ, Toews GB, White ES, Lynch JP, 3rd, Martinez FJ. Mechanisms of pulmonary fibrosis. Annu Rev Med 2004;55:395–417. doi: 10.1146/annurev.med.55.091902.103810. PMID:14746528 [DOI] [PubMed] [Google Scholar]
- 22.Wynn TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med 2011;208:1339–1350. doi: 10.1084/jem.20110551. PMID:21727191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest 2012;122:2756–2762. doi: 10.1172/JCI60323. PMID:22850886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 2013;341:84–87. doi: 10.1126/science.1233606. PMID:23828940 [DOI] [PubMed] [Google Scholar]
- 25.Chua F, Gauldie J, Laurent GJ. Pulmonary fibrosis: searching for model answers. Am J Respir Cell Mol Biol 2005;33:9–13. doi: 10.1165/rcmb.2005-0062TR. PMID:15964990 [DOI] [PubMed] [Google Scholar]
- 26.Ashcroft T, Simpson JM, Timbrell V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J Clin Pathol 1988;41:467–470. doi: 10.1136/jcp.41.4.467. PMID:3366935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakahira K, Cloonan SM, Mizumura K, Choi AM, Ryter SW. Autophagy: a crucial moderator of redox balance, inflammation, and apoptosis in lung disease. Antioxid Redox Signal 2014;20:474–494. doi: 10.1089/ars.2013.5373. PMID:23879400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabrera S, Maciel M, Herrera I, Nava T, Vergara F, Gaxiola M, López-Otín C, Selman M, Pardo A. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy 2015;11:670–684. doi: 10.1080/15548627.2015.1034409. PMID:25906080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. PMID:26799652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reis RC, Sorgine MH, Coelho-Sampaio T. A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur J Cell Biol 1998;75:192–197. doi: 10.1016/S0171-9335(98)80061-7. PMID:9548376 [DOI] [PubMed] [Google Scholar]
- 31.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science 2009;325:473–477. PMID:19556463 [DOI] [PubMed] [Google Scholar]
- 32.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. TFEB links autophagy to lysosomal biogenesis. Science 2011;332:1429–1433. doi: 10.1126/science.1204592. PMID:21617040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol 2014;15:65–74. doi: 10.1038/nrm3716. PMID:24326622 [DOI] [PubMed] [Google Scholar]
- 34.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012;8:903–914. doi: 10.4161/auto.19653. PMID:22576015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM.. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012;31:1095–1108. doi: 10.1038/emboj.2012.32. PMID:22343943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest 2015;125:25–32. doi: 10.1172/JCI73939. PMID:25654547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song JX, Sun YR, Peluso I, Zeng Y, Yu X, Lu JH, Xu Z, Wang MZ, Liu LF, Huang YY, et al. A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of MTOR inhibition. Autophagy 2016;12:1372–1389. doi: 10.1080/15548627.2016.1179404. PMID:27172265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang D, Liang J, Fan J, Yu S, Chen S, Luo Y, Prestwich GD, Mascarenhas MM, Garg HG, Quinn DA, et al. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat Med 2005;11:1173–1179. doi: 10.1038/nm1315. PMID:16244651 [DOI] [PubMed] [Google Scholar]
- 40.Pullamsetti SS, Savai R, Dumitrascu R, Dahal BK, Wilhelm J, Konigshoff M, Zakrzewicz D, Ghofrani HA, Weissmann N, Eickelberg O, et al. The role of dimethylarginine dimethylaminohydrolase in idiopathic pulmonary fibrosis. Sci Transl Med 2011;3:87ra53. doi: 10.1126/scitranslmed.3001725. PMID:21677199 [DOI] [PubMed] [Google Scholar]
- 41.Weng T, Poth JM, Karmouty-Quintana H, Garcia-Morales LJ, Melicoff E, Luo F, Chen N-Y, Evans CM, Bunge RR, Bruckner BA, et al. Hypoxia-induced deoxycytidine kinase contributes to epithelial proliferation in pulmonary fibrosis. Am J Respir Crit Care Med 2014;190:1402–1412. doi: 10.1164/rccm.201404-0744OC. PMID:25358054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burgy O, Wettstein G, Bellaye PS, Decologne N, Racoeur C, Goirand F, Beltramo G, Hernandez JF, Kenani A, Camus P, et al. Deglycosylated bleomycin has the antitumor activity of bleomycin without pulmonary toxicity. Sci Transl Med 2016;8:326ra20. doi: 10.1126/scitranslmed.aad7785. PMID:26888428 [DOI] [PubMed] [Google Scholar]
- 43.Morozova K, Sridhar S, Zolla V, Clement CC, Scharf B, Verzani Z, Diaz A, Larocca JN, Hajjar KA, Cuervo AM, et al. Annexin A2 promotes phagophore assembly by enhancing Atg16L(+) vesicle biogenesis and homotypic fusion. Nat Commun 2015;6:5856. doi: 10.1038/ncomms6856. PMID:25597631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreau K, Ghislat G, Hochfeld W, Renna M, Zavodszky E, Runwal G, Puri C, Lee S, Siddiqi F, Menzies FM, et al. Transcriptional regulation of Annexin A2 promotes starvation-induced autophagy. Nat Commun 2015;6:8045. doi: 10.1038/ncomms9045. PMID:26289944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li R, Tan S, Yu M, Jundt MC, Zhang S, Wu M. Annexin A2 Regulates Autophagy in Pseudomonas aeruginosa Infection through the Akt1-mTOR-ULK1/2 Signaling Pathway. J Immunol 2015;195:3901–3911. doi: 10.4049/jimmunol.1500967. PMID:26371245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Araya J, Kojima J, Takasaka N, Ito S, Fujii S, Hara H, Yanagisawa H, Kobayashi K, Tsurushige C, Kawaishi M, et al. Insufficient autophagy in idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2013;304:L56–L69. doi: 10.1152/ajplung.00213.2012. PMID:23087019 [DOI] [PubMed] [Google Scholar]
- 47.Mi S, Li Z, Yang HZ, Liu H, Wang JP, Ma YG, Wang X-X, Liu H-Z, Sun W, Hu Z-W. Blocking IL-17A promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1-dependent and -independent mechanisms. J Immunol 2011;187:3003–3014. doi: 10.4049/jimmunol.1004081. PMID:21841134 [DOI] [PubMed] [Google Scholar]
- 48.Gui YS, Wang L, Tian X, Li X, Ma A, Zhou W, Zeng N, Zhang J, Cai B, Zhang H, et al. mTOR Overactivation and Compromised Autophagy in the Pathogenesis of Pulmonary Fibrosis. PLoS One 2015;10:e0138625. doi: 10.1371/journal.pone.0138625. PMID:26382847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013;13:722–737. doi: 10.1038/nri3532. PMID:24064518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scherz-Shouval R, Elazar Z. Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci 2011;36:30–38. doi: 10.1016/j.tibs.2010.07.007. PMID:20728362 [DOI] [PubMed] [Google Scholar]
- 51.Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy 2010;6:838–854. doi: 10.4161/auto.6.7.12113. PMID:20505317 [DOI] [PubMed] [Google Scholar]
- 52.Feng Y, Klionsky DJ. Autophagy regulates DNA repair through SQSTM1/p62. Autophagy 2017;13:995–996. doi: 10.1080/15548627.2017.1317427. PMID:28650265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bove PF, Grubb BR, Okada SF, Ribeiro CM, Rogers TD, Randell SH, O'Neal WK, Boucher RC. Human alveolar type II cells secrete and absorb liquid in response to local nucleotide signaling. J Biol Chem 2010;285:34939–34949. doi: 10.1074/jbc.M110.162933. PMID:20801871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aouida M, Poulin R, Ramotar D. The human carnitine transporter SLC22A16 mediates high affinity uptake of the anticancer polyamine analogue bleomycin-A5. J Biol Chem 2010;285:6275–6284. doi: 10.1074/jbc.M109.046151. PMID:20037140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ling Q, Jacovina AT, Deora A, Febbraio M, Simantov R, Silverstein RL, Hempstead B, Mark WH, Hajjar KA. Annexin II regulates fibrin homeostasis and neoangiogenesis in vivo. J Clin Invest 2004;113:38–48. doi: 10.1172/JCI19684. PMID:14702107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819–823. doi: 10.1126/science.1231143. PMID:23287718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K, Liu R, Li J, Mao J, Lei Y, Wu J, Zeng J, Zhang T, Wu H, Chen L, et al. Quercetin induces protective autophagy in gastric cancer cells: involvement of Akt-mTOR- and hypoxia-induced factor 1alpha-mediated signaling. Autophagy 2011;7:966–978. doi: 10.4161/auto.7.9.15863. PMID:21610320 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


