ABSTRACT
Macroautophagy/autophagy is a highly conserved process for degrading cytoplasmic contents, determines cell survival or death, and regulates the cellular homeostasis. Besides ATG proteins, numerous regulators together with various post-translational modifications (PTMs) are also involved in autophagy. In this work, we collected 4,237 experimentally identified proteins regulated in autophagy and cell death pathways from the literature. Then we computationally identified potential orthologs of known proteins, and developed a comprehensive database of The Autophagy, Necrosis, ApopTosis OrchestratorS (THANATOS, http://thanatos.biocuckoo.org), containing 191,543 proteins potentially associated with autophagy and cell death pathways in 164 eukaryotes. We performed an evolutionary analysis of ATG genes, and observed that ATGs required for the autophagosome formation are highly conserved across eukaryotes. Further analyses revealed that known cancer genes and drug targets were overrepresented in human autophagy proteins, which were significantly associated in a number of signaling pathways and human diseases. By reconstructing a human kinase-substrate phosphorylation network for ATG proteins, our results confirmed that phosphorylation play a critical role in regulating autophagy. In total, we mapped 65,015 known sites of 11 types of PTMs to collected proteins, and revealed that all types of PTM substrates were enriched in human autophagy. In addition, we observed multiple types of PTM regulators such as protein kinases and ubiquitin E3 ligases or adaptors were significantly associated with human autophagy, and again the results emphasized the importance of PTM regulations in autophagy. We anticipated THANATOS can be a useful resource for further studies.
KEYWORDS: ATG, autophagy, phosphorylation, post-translational modification, ubiquitin
Abbreviations
- AGC
protein kinase A, G, and C
- ARN
autophagy regulatory network
- ATG
autophagy related
- BECN1
Beclin-1
- Cdc
cell-division cycle
- CDP
cell death proteomics
- COSMIC
Catalogue Of Somatic Mutations In Cancer
- Cvt
cytoplasm-to-vacuole targeting
- DES
diethylstilbestrol
- DMPK
dystrophia myotonica protein kinase
- DUB
deubiquitinating enzyme
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin-protein ligase
- EKPD
Eukaryotic Kinase and Phosphatase Database
- E-ratio
enrichment ratio
- FDA
food and drug administration
- GPS
group-based prediction system
- HADb
human autophagy database
- HGNC
HUGO Gene Nomenclature Committee
- I2D
Interologous Interaction Database
- ICGC
International Cancer Genome Consortium
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KSPN
kinase-substrate phosphorylation network
- MAPT
microtubule-associated protein TAU
- ncRNA
noncoding RNA
- PCD
programmed cell death
- PPI
protein-protein interaction
- PTM
post-translational modification
- RB1CC1
RB1-inducible coiled-coil protein 1
- RBH
reciprocal best hit
- SKP2
S-phase kinase-associated protein 2
- ssKSR
site specific kinase-substrate relationship
- TF
transcription factor
- THANATOS
The Autophagy, Necrosis, ApopTosis OrchestratorS
- ULK1
Unc-51-like kinase 1.
Introduction
Autophagy is a highly conserved “self-eating” process that controls the degradation of cytoplasmic contents within the lysosome and vacuole, and ensures the cellular homeostasis and the recycling of macromolecular constituents.1-5 Although Christian de Duve coined the term “autophagy” at the Ciba Foundation symposium on lysosomes in 1963, the upsurge of research on autophagy emerged only after the discovery of autophagy-related (ATG) genes by using the yeast S. cerevisiae as a wonderful model organism for genetic screening.3,6 To date, 41 ATG genes have been identified, and nearly half of them are well conserved from yeast to human.2,4,7 ATG genes and the autophagy process are extensively controlled by post-translational modifications (PTMs), transcriptional regulations, post-transcriptional regulations, and protein-protein interactions (PPIs), whereas hundreds of small chemicals can either induce or inhibit autophagy.4,7-9 Recent findings have clearly proved that autophagy play critical roles in the regulation of metabolism and membrane transport,2,4 and has diverse physiological and pathophysiological roles in starvation adaptation, antiaging, immunity and various human diseases.1,10,11
Although autophagy mainly serves as a cell survival mechanism during nutrient starvation, in certain states it may also induce programmed cell death (PCD) by excessively degrading the cellular contents.12,13 Besides autophagic cell death, apoptosis and necrosis can also trigger cell suicide, as the other two types of PCDs.12-14 Autophagy, apoptosis, and necrosis have a complicated crosstalk to determine cell survival or suicide.12,13 The induction of apoptosis is inhibited by autophagy and apoptosis-associated caspase activation can diminish the autophagy process.13 Under certain conditions, autophagy suppresses apoptosis to avoid cell death, whereas in other special cases, autophagy serves as an alternative cell death pathway or promotes apoptosis or necrosis to initiate cell death together.12,13 Previous studies suggest that autophagy, apoptosis and necrosis processes share common pathways in certain circumstances, and maintaining the relationship or balance among them is important for normal pathophysiological functions of organisms.13,15
Numerous experimental studies have identified a large number of genes and proteins involved in autophagy, apoptosis, and/or necrosis, while the collection, integration, and annotation of the data have emerged to be a great challenge.2,4,5,13 In 2003, Doctor et al. has first developed an apoptosis database, containing proteins with apoptotic domains, although these proteins may also be functional in nonapoptotic processes.16 Later, Díez et al. mainly focuses on apoptosis, and constructs the DeathBase by collecting 213 PCD proteins from 5 model species.17 More specifically, yApoptosis has been designated for the annotation of 51 apoptosis-associated proteins in S. cerevisiae.18 Due to the rapid progresses in quantitative proteomics, a great number of proteins differentially expressed under various PCD conditions have been detected and maintained in ApoptoProteomics,19 which has been further integrated into the cell death proteomics (CDP) database, containing 3,667 proteins potentially involved in cell death.20 In 2010, Homma et al. developed the first autophagy database, containing 133 experimentally identified autophagy genes or regulators in S. cerevisiae, H. sapiens and M. musculus. They further predict 499 orthologs and 1,531 homologs across 41 eukaryotic species.21 Then Moussay et al. collect nearly 222 human genes directly or indirectly involved in autophagy, and construct a human autophagy database (HADb).22 More recently, 739 autophagy-modulating proteins and 385 chemical inducers or inhibitors have carefully been curated, although a public database has not been released.9 Besides the data collection and integration, computational analysis of autophagy and its crosstalk with cell death pathways has also become an attractive topic. For example, by collecting 416 human and murine genes with functions in autophagy, Jegga et al. have systematically modeled a transcriptional regulatory network and demonstrate a strong relation between the autophagy-lysosomal pathway and neurodegenerative diseases.23 Moreover, Wu et al. have developed a database of ncRDeathDB, containing more than 4,600 noncoding RNA (ncRNA)-mediated PCD-associated entries, and further analyze the ncRNA-regulated cell death systems.24 The same authors also implement the miRDeathDB database for maintaining the miRNA-target relations in PCD network.8,25 In 2015, a highly useful autophagy resource termed the Autophagy Regulatory Network (ARN) was reported, containing 2,240 proteins and 386 miRNAs, including 38 ATG genes.7 Multiple levels of regulations, such as PPIs, transcriptional regulations by transcription factors (TFs) and post-transcriptional regulations by miRNAs, are comprehensively considered and integrated.7 However, the PTM regulations are not included, and the PTM-mediated crosstalk of autophagy with cell death pathways still remains to be dissected.
Recently, the importance of PTM regulations for ATG proteins has been well documented.4 Here we further analyzed proteins and PTMs involved in autophagy and cell death pathways using Xie's review4 as a starting point. In this work, we first collected 4,237 experimentally identified proteins associated with autophagy, apoptosis and necrosis from the literature, and observed that a considerable proportion of proteins were involved in multiple processes. For simplicity, these proteins were hereafter referred to as AT, AP or NE proteins, respectively. Then we computationally identified potential orthologs of 3,882 known proteins of 8 model organisms, and developed an integrative database of The Autophagy, Necrosis, ApopTosis OrchestratorS (THANATOS), containing 191,543 AT, AP and NE proteins in 164 eukaryotes. By analyzing the evolutionary conservation of 41 ATG genes, our results demonstrated that the key machinery of autophagy is highly conserved across eukaryotes. The statistical results demonstrated that human AT proteins were highly enriched in known cancer genes and drug targets, whereas a functional enrichment analysis revealed that human AT proteins were significantly associated with a number of signaling and disease pathways. By analyzing cancer mutations, we found up to 854 and 54 human AT genes are frequently mutated in pancreatic adenocarcinoma and prostate cancer. The analysis of drug-target relations demonstrated that a considerable number of mutated AT proteins can be potential targets for drugs in the treatment of the 2 cancers. Furthermore, we mapped human AT proteins to known protein kinases and phosphatases, and observed that both kinases and phosphatases were highly over-represented in autophagy. By predicting potential site-specific kinase-substrate relations (ssKSRs) of known phosphorylation sites, we reconstructed a human kinase-substrate phosphorylation network (KSPN) among key ATG proteins and upstream kinases for multiple eukaryotes, and found that mammalian BECN1/Beclin 1 has the most potential kinase-substrate relations. Moreover, we mapped known sites of 11 types of PTMs to AT proteins, and observed that all PTMs are statistically enriched in human AT proteins. By mapping ubiquitin and ubiquitin-like regulators to human AT proteins, we found that multitypes of regulators were significantly associated with autophagy. Finally, the online service of THANATOS was implemented in PHP + MySQL + JavaScript, while known PTMs, PPIs, primary references and other annotations were also present.
Results
The collection and integration of experimentally identified proteins in the regulation of autophagy and cell death pathways
The flowchart of the study was shown in Fig. 1A. First, we searched PubMed to find experimentally identified AT, AP and/or NE proteins, and annotated each collected protein with a “+” or “-” to distinguish the positive or negative regulation in autophagy or PCDs (Fig. 1A). For example, it was demonstrated that the loss of ATG3 results in the deficiency of autophagosome formation in mice.26 Thus, mouse ATG3 positively regulates autophagy, and has been annotated as AT+. Also, the inhibition of human SKP2, an important component of the SCF-SKP2 E3 ligase complex, can arrest the cell cycle progression and activate autophagy in myeloma cells.27 In this regard, human SKP2 is negatively associated with autophagic activation, and was annotated as AT-. In addition, mouse ATG5 is essential for autophagic clearance of apoptotic cells during embryonic development, while autophagy is abolished in atg5−/− embryoid bodies.28 However, the phosphorylation of mouse ATG5 at Thr75 by MAPK14 inhibits starvation-induced autophagy.29 Thus, mouse ATG5 was annotated as both AT+ and AT- (Fig. 1A).
Figure 1.
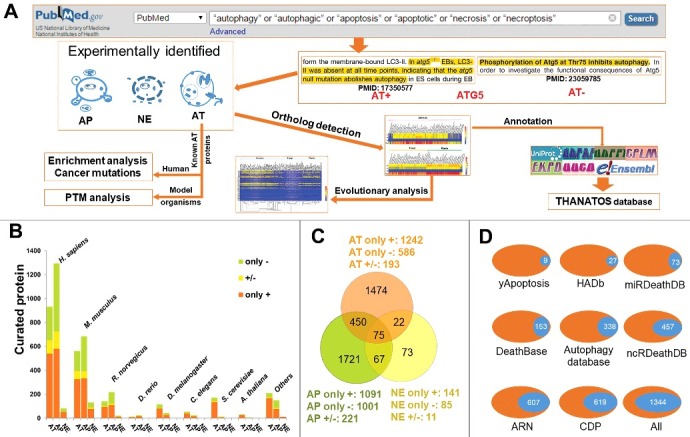
The collection and curation of proteins that were experimentally identified to be associated with autophagy and cell death pathways from the literature. (A) In this study, we used multiple keywords to search the PubMed search engine, and obtained a total of 4,237 known AT, AP and NE proteins. Using 3,882 known proteins from 8 model organisms, we computationally detected their potential orthologs in 164 eukaryotes, and further performed an evolutionary analysis of ATG genes. Also, we carried out the enrichment analysis and the cancer mutation analysis for known human AT proteins, while the PTM analysis was conducted for known AT proteins in model species. Finally, we combined both known and computationally identified AT, AP and NE proteins together and developed the THANATOS database. (B) Based on experimental evidence, we annotated each known protein with a “+” or “-” to distinguish the positive or negative regulation in autophagy or PCDs. For 3,882 known AT, AP and NE proteins of 8 organisms, the proteins annotated only with “+” (only +) or “-” (only -), and with both “+” and “-” (+/-) were separately present. (C) The overlap of different types of known proteins for 8 model species. (D) The comparison of curated proteins from the literature between THANATOS and other existing resources. i. All, the number of nonredundant proteins in 8 public databases.
In this study, in total we obtained 4,237 experimentally characterized proteins in eukaryotes, with 3,882 proteins from 8 model organisms, including H. sapiens, M. musculus, R. norvegicus, D. rerio, D. melanogaster, C. elegans, S. cerevisiae and A. thaliana (Fig. 1B and Table S1). For each process, the proteins annotated only with “+” (only +) or “-” (only -), and with both “+” and “-” (+/-) were counted and shown, separately (Fig. 1B). From the results, we observed that the numbers of known AP proteins are considerably higher than AT proteins in H. sapiens, M. musculus, R. norvegicus, and D. rerio (Fig. 1B and Table S1). However, there were 115, 54 and 176 AT proteins identified in D. melanogaster, C. elegans and S. cerevisiae, whereas only 42, 25 and 14 AP proteins were reported in the 3 species, respectively. Thus, our analysis emphasized the importance of less complex model organisms in autophagy research, for their convenient usage in genetic screening.3,6 Also, we observed that there were 1,909 proteins from H. sapiens implicated in autophagy and cell death pathways, while only 189 autophagy and PCD proteins were reported in S. cerevisiae (Fig. 1B and Table S1). Thus, different species might contain considerably different numbers of proteins that participate in autophagy and PCDs. Furthermore, we found that a considerable proportion of proteins were involved in multiple processes (Fig. 1C), and the overlap of known AT, AP and NE proteins for each organism was present (Fig. S1). For example, there was 26.18% (528/2017) of total AT proteins also implicated in apoptosis, while 22.83% of 2,313 AP proteins also participated in the regulation of autophagy. In particular, up to 69.20% (164/237) of NE proteins played roles in other types of processes. These multifunctional proteins might be important for mediating the crosstalk of autophagy and cell death pathways. In addition, we revealed that 9.52% (192/2017), 9.56% (221/2313), and 4.64% (11/237) of AT, AP and NE proteins can both positively and negatively regulate corresponding processes, respectively (Fig. 1C). These bifunctional regulators might be essential to balance and ensure the fidelity of autophagy, apoptosis or necrosis.
To identify how many entries came entirely from the literature and were not yet accumulated in any of the previously established databases, we downloaded the data sets from 8 public databases for autophagy and/or PCDs, including DeathBase,17 yApoptosis,18 CDP,19,20 Autophagy database,21 HADb,22 miRDeathDB,8,25 ncRDeathDB,24 and ARN.7 In total, there were 6,550 nonredundant proteins contained in the 8 databases, whereas 85.88% of the proteins (5,625) were integrated in only one database (Fig. S2A). Only 2 proteins were collected in up to 6 databases, whereas no proteins were curated in ≥ 7 resources (Fig. S2A). Also, we compared our data set with the 8 databases, and found that only 31.72% (1,344/4,237) of our proteins to be included in at least one public database (Fig. 1D and Fig. S2B). In this regard, our curated data set is much larger than previous data resources, mainly due to the rapid progress in the study of autophagy.
Development of THANATOS for eukaryotic proteins and PTMs involved in autophagy, apoptosis and necrosis
The orthologous information of known AT, AP and NE proteins will be potentially useful for discovering new regulators, since orthologs across different species might have conserved functions in autophagy and cell death pathways. Then we computationally identified 191,543 potential orthologs of 3,882 experimentally identified proteins across the 164 eukaryotic species (Table S2). Combined with both known and computationally identified proteins, the distribution of the number of identified proteins for each organism is shown in Fig. 2. In our results, there were only 1,134, 340, 39, 153, 77 and 37 known AT, AP and/or NE proteins experimentally identified in M. musculus, R. norvegicus, D. rerio, D. melanogaster, C. elegans and A. thaliana respectively, whereas the integrative data set in total contained 9,208 proteins for the 6 species, with a >4-fold increase (Table S2). Even for the 2 most studied organisms, H. sapiens and S. cerevisiae, the experimentally identified proteins were only 1,909 and 189, while the final data set contained 2,498 and 516 for the 2 species, respectively (Table S2). In this regard, our analyses greatly expanded the reservoir of candidates for further experimental studies.
Figure 2.
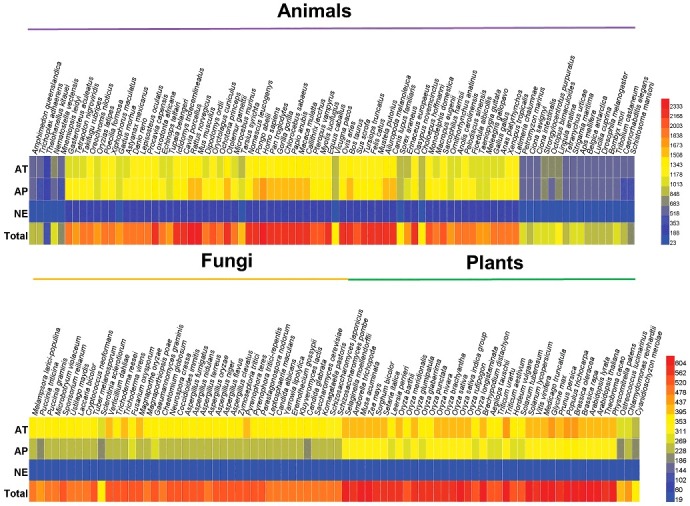
The distribution of experimentally and computationally identified AT, AP and NE proteins across 164 eukaryotes in THANATOS database. In our results, there were only 1,909, 1,134, 340, 39, 153, 77, 189 and 37 known AT, AP and/or NE proteins experimentally identified in H. sapiens, M. musculus, R. norvegicus, D. rerio, D. melanogaster, C. elegans, S. cerevisiae and A. thaliana respectively, whereas the integrative data set in total contained 12,222 proteins for the 8 species, with a >2-fold increase (Table S2). In total, THANATOS contains 191,543 proteins potentially associated with autophagy cell death pathways.
We also compiled an integrative data set containing both known and precalculated PPIs for 6 organisms, including H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, and S. cerevisiae (Table S3). For each species, we mapped the PPIs to AT, AP and/or NE proteins in the THANATOS database (Table S3). In addition, we mapped known PTM sites to all integrated proteins for 7 eukaryotes, including H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, A. thaliana and S. cerevisiae (Table S4). Most of the PTM sites were identified from mass spectrometry-based proteomic profiling, while whether and how many PTM sites directly linked to autophagy and PCDs still remain to be dissected. Finally, we developed the comprehensive THANATOS database for known and predicted proteins, while primary references for known proteins and other annotations from the UniProt database30 were also present.
The online service of the THANATOS database was developed in an easy-to-use manner. The database contained 4 search options, including “Simple search” (Fig. 3A), “Advanced search” (Fig. 3B), “Batch search” (Fig. 3C), and “BLAST search” (Fig. 3D). For example, if a keyword “ulk1” in “Gene/Protein Name” was directly submitted for a simple search (Fig. 3A), all related proteins across eukaryotes, will be shown. Also, the option of “Advanced Search” allows a more accurate query that 2 terms combined with operators of “and”, “or” and “exclude” can be specified in 2 different fields (Fig. 3B). For example, searching the database with “Homo sapiens” in “Species” and “ulk1” in “Gene/Protein Name” will return the information of human ULK1 in a tabular format with accession, evidence, species, and protein or gene names (Fig. 3B). By clicking the accession “ANA-HSA-111744”, the detailed annotations of human ULK1 can be shown. Moreover, users can submit a list of keywords for a batch search (Fig. 3C). In addition, users can submit a protein sequence in FASTA format in “BLAST Search” to find identical or homologous proteins in THANATOS (Fig. 3D). THANATOS can also be browsed by multiple options (Fig. S3).
Figure 3.
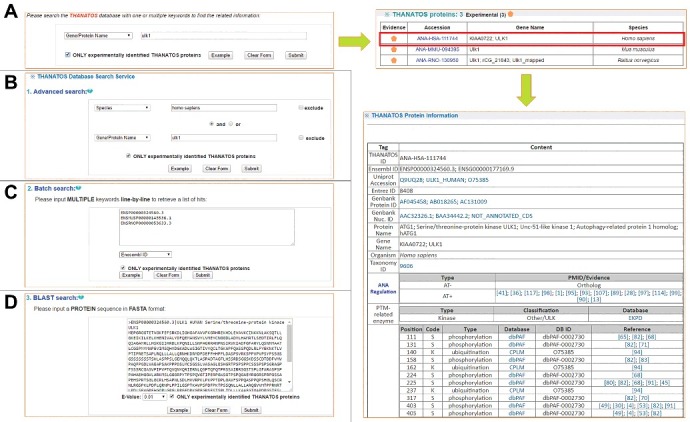
The search options of THANATOS database. (A) Simple search. The THANATOS database can be queried with one or multiple keywords. (B) Advanced search. This option allows a more precise search that 2 terms combined with operators of “and”, “or” and “exclude” can be specified in 2 different fields. (C) Batch search. The option permits users to input multiple keywords such as accession numbers or gene names in a line-by-line format for querying the database. (D) BLAST search. The option was designed for searching the database with a protein sequence in FASTA format.
An evolutionary analysis of ATG genes
Previously, an evolutionary analysis was performed for 17 ATG genes across 17 photosynthetic eukaryotes, and demonstrated that the autophagy pathway is conserved in green algae and chromalveolates, but not in red algae.31 With the orthologous information, here we performed a comprehensive analysis of the evolution of 41 ATG genes (Fig. 4), and the known ATG genes in the 8 model organisms were also shown (Table S5). In eukaryotes, only orthologs of the known ATG genes were adopted for further analyses. It should be noted that ATG39, ATG40 and ATG41 were only found in the yeast S. cerevisiae, and currently no orthologs were detected in other eukaryotes (Fig. 4). Also, 5 ATG genes including ATG25, ATG28, ATG30, ATG35 and ATG37 are encoded in the yeast Komagataella pastoris (also called Pichia pastoris) but not in S. cerevisiae (Table S5). Although ATG25 and ATG30 were only found in K. pastoris, the orthologs of ATG28, ATG35 and ATG37 were detected in a number of other eukaryotes (Fig. 4).
Figure 4.
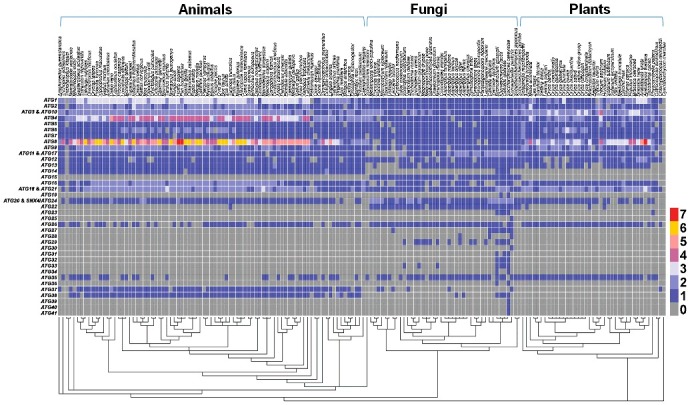
The evolutionary conservation of 41 ATG genes across 164 species. ATG39, ATG40 and ATG41 were exclusively found in the yeast S. cerevisiae, whereas ATG25 and ATG30, ATG35 and ATG37 are only encoded in the K. pastoris. We classified ATG11 and ATG17 into a single group due to the sequence similarity in more complex eukaryotes and the functional similarity in autophagy. Based on the sequence and functional similarity, we also classified ATG3 and ATG10, ATG18 and ATG21, as well as ATG20 and SNX4/ATG24 into 3 groups, respectively. Clearly, there were 18 highly conserved ATG genes including ATG1 to ATG10, ATG11 and ATG17, ATG13, ATG16, ATG18 and ATG21, ATG20 and SNX4/ATG24. Their orthologs were detected in more than 85% (140) of 164 eukaryotes, and most of these ATG genes are involved in autophagosome formation.
In S. cerevisiae, both Atg11 and Atg17 interact with Atg1 and Atg9 as scaffold/adaptor proteins, and mainly participate in the cytoplasm-to-vacuole targeting (Cvt) and macroautophagy pathways, respectively.32,33 It has been demonstrated that Drosophila Atg17/RB1CC1 and mammalian RB1CC1/FIP200 are functional equivalents of yeast ATG17.32,34 However, Lin et al. have found that the C termini of nematode ATG-11/EPG-7, mammalian RB1CC1 and yeast Atg11 are considerably similar and contain the Atg11 motif (Pfam motif PF10377).35 Indeed, nematode ATG-11 was computationally identified as an ortholog of human RB1CC1 in this study. Thus, although there was no significant similarity in protein sequences between yeast Atg11 and Atg17, we classified ATG11 and ATG17 into a single group due to the sequence similarity in more complex eukaryotes and the functional similarity in autophagy (Fig. 4, Table S5). In protein sequences, yeast Atg3 and Atg10 possess the same consensus domain as Autophagy_act_C (Pfam domain PF03987), and both were annotated as ubiquitin-like conjugating enzymes in UniProt. Also, yeast Atg18 is highly similar with Atg21 in sequences, and both proteins contain WD repeats. Although Atg20 and Snx4/Atg24 only exhibit considerable sequence similarity in S. cerevisiae, the 2 proteins were annotated with a conserved PX domain (Pfam domain PF00787). Due to the sequence and functional similarity, we classified ATG3 and ATG10, ATG18 and ATG21, as well as ATG20 and SNX4/ATG24 into 3 groups, respectively (Fig. 4, Table S5). In addition, although yeast Atg38 and mammalian NRBF2 are not significantly similar in amino acid sequences, experimental studies demonstrate that NRBF2 is a functional ortholog of Atg38.36 We manually added the information for further analyses (Table S5).
One ATG gene can be a singleton in one species but have multiple paralogs in other organisms. For example, yeast ATG1 has 5 human orthologs including ULK1, ULK2, ULK3, ULK4 and STK36.37 Besides ATG1, we observed that ATG2, ATG4, ATG6, ATG8, ATG9 and ATG16 are singleton genes in S. cerevisiae but have multiple copies in animals or plants (Fig. 4). In particular, although ATG12 has only one copy in most of species, it has a few duplications in Brassicales, which underwent large-scale duplication events that occurred at 100 to 200 million years ago.38 In total, there were 15 ATG genes including ATG15, ATG19, ATG25, ATG27 to ATG34, ATG36, and ATG39 to ATG41 to be fungus specific, whereas their orthologs in animals or plants were not detected (Fig. 4). Furthermore, 18 ATG genes were highly conserved (ATG1 to ATG10, ATG11 and ATG17, ATG13, ATG16, ATG18 and ATG21, ATG20 and SNX4/ATG24), and their orthologs can be readily found in over 85% (140) of 164 eukaryotes (Fig. 4). Most of these ATG genes are involved in autophagosome formation, and our analysis supported the idea that the machinery of the autophagy pathway is highly conserved in eukaryotes.39 In addition, for human ATG genes, there is 1 representative for ATG5, ATG7, ATG11 and ATG17, ATG12 to ATG14, ATG37, ATG38, 2 representatives for ATG2, ATG3 and ATG10, VPS30/ATG6, ATG9, ATG16, ATG18 and ATG21, ATG20 and SNX4/ATG24, 3 representatives for ATG1, 4 representatives for ATG4, 7 representatives for ATG8 and 0 for ATG15, ATG19, ATG22, ATG23, ATG25 to ATG36, and ATG39 to ATG41, respectively (Fig. 4 and Table S5).
Autophagy proteins and regulators are preferentially associated with human diseases
To investigate whether and how AT proteins are preferentially involved in human diseases, we first obtained 2,247 known human drug targets from the DrugBank database,40 and 559 well-curated cancer genes from the Cancer Gene Census in the Catalogue Of Somatic Mutations In Cancer (COSMIC).41 Then we mapped human AT proteins to the 2 datasets, and found that 261 and 95 AT proteins were annotated as known drug targets and cancer genes, with the enrichment ratios of 2.85- and 4.17-fold, respectively (Fig. 5A). Obviously, our results proposed that known drug targets and cancer genes were enriched in autophagy against the human proteome (Fig. 5A). To further understand the functional distribution of known human AT proteins beyond the regulation of autophagy, we performed an enrichment analysis based on pathway annotations from the database of Kyoto Encyclopedia of Genes and Genomes (KEGG),42 using the hypergeometric distribution (P value < 1E-12). From the results, we observed that AT proteins were significantly overrepresented in several signaling pathways, such as the TNF signaling pathway (KEGG ID: hsa04668), the NOD-like receptor signaling pathway (hsa04621), the TLR signaling pathway (hsa04620), and the FOXO signaling pathway (hsa04068) (Fig. 5B and Table S6). In particular, a number of disease pathways are enriched in human AT proteins (Fig. 5B and Table S6). For example, a considerable proportion of AT proteins were enriched in the pathway of hepatitis B (KEGG ID: hsa05161). Previously, Shin et al. have reported that the HBx protein induces the degradation of human TNFRSF10B through the autophagy pathway to promote the survival of hepatocytes infected by HBV.43 Also, Tian et al. have proved that autophagy is required for HBV replication, whereas the HBV DNA level in sera is greatly reduced by more than 90% in atg5−/− transgenic mice.44 Thus, the statistical results are consistent with previous experiments. Besides the KEGG term of pathways in cancer (hsa05200), human AT proteins were also enriched in pancreatic cancer (hsa05212) and prostate cancer (hsa05215) (Fig. 5B). For pancreatic cancer, 42 out of 112 (37.50%) KEGG annotated proteins were identified as AT proteins, whereas 44 out of 139 (31.65%) annotated proteins were detected as AT proteins for prostate cancer (Table S6).
Figure 5.
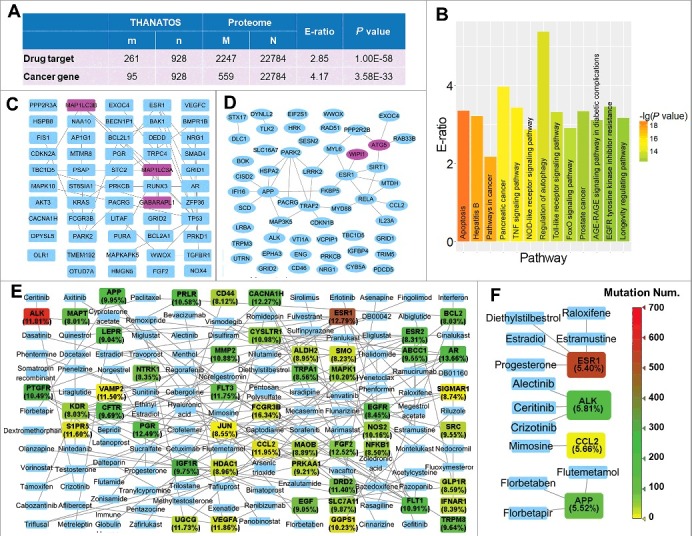
The statistical enrichment analyses revealed that AT proteins are preferentially associated with human diseases, using the hypergeometric distribution. (A) AT proteins are significantly enriched in drug targets and cancer genes. (B) The KEGG-based enrichment analysis found that AT proteins are statistically over-represented in a number of cellular signaling and disease pathways. (C) There were 54 AT genes with a mutation frequency of ≥ 12% visualized for pancreatic adenocarcinoma. The PPI relations among these proteins are also present if available, 3 ATG genes, MAP1LC3A/LC3A, GABARAPL1 and MAP1LC3B/LC3B, are shown in pink. (D) The 54 mutated AT genes with a frequency of ≥ 5% were present for prostate cancer. Two ATG genes, ATG5 and WIPI1, are shown in pink. (E) A network of mutated AT proteins with at least one approved drug in pancreatic adenocarcinoma. The mutation frequency was shown in parentheses for each gene. The color indicates the mutation number of each gene detected in pancreatic adenocarcinoma samples from ICGC database. Genes with mutation frequency ≥ 8% were shown. (F) A drug-target network of mutated AT genes in prostate cancer. Genes with mutation frequency ≥ 5% were shown.
Currently, KEGG annotations are still limited and the numbers of proteins involved in biological pathways are far from fully annotated. For example, KEGG only annotated 53 human proteins as “regulation of autophagy” (hsa04140) (Fig. 5B and Table S6). To avoid any bias, we performed a systematic analysis of cancer mutations of AT genes, by using the cancer genomic data. First, we downloaded all nonsynonymous somatic mutations together with mutated gene lists of pancreatic adenocarcinoma and prostate cancer from the International Cancer Genome Consortium (ICGC) database,45 respectively. We mapped 928 known human AT proteins to mutated genes in the 2 cancers, and found that there were 854 and 54 AT genes with a mutation frequency of ≥ 5% in pancreatic adenocarcinoma and prostate cancer, respectively (Table S7). For simplicity, 54 AT genes with a mutation frequency of ≥ 12% were visualized for pancreatic adenocarcinoma (Fig. 5C), and all mutated AT genes with a frequency of ≥ 5% were present for prostate cancer (Fig. 5D). The known and highly potential PPIs among these AT genes were also shown, if available (Fig. 5C and D). From the results, we observed that up to 30 ATG genes were frequently mutated in pancreatic adenocarcinoma (Table S7). For example, MAP1LC3A/LC3A, GABARAPL1 and MAP1LC3B/LC3B, 3 mammalian ortholog members of the yeast ATG8 family, were highly mutated with the frequency values of 13.58%, 12.83% and 12.50%, respectively (Fig. 5C and Table S7). In contrast, there were only 2 ATG genes, ATG5 and WIPI1, the latter being an ortholog of yeast ATG18, that are frequently mutated in prostate cancer (Fig. 5D and Table S7).
To probe how many mutated AT proteins can be potentially therapeutic drug targets for the 2 cancers, we obtained human drug-target relations from DrugBank,40 and then mapped all mutated AT genes to the data set. From the results, we found that 79 and 4 proteins might be targeted pharmacologically in pancreatic adenocarcinoma and prostate cancer, with at least one applicable drug approved by Food and Drug Administration (FDA), respectively (Table S7). For convenience, the relations between available drugs and 50 AT genes with a mutation frequency of ≥ 8% were shown for pancreatic adenocarcinoma (Fig. 5E), while we also presented the drug-target relations for all mutated AT genes in prostate cancer (Fig. 5F). Although no ATG proteins were observed to be targeted pharmacologically in the current stage, we found that a number of autophagy regulators can be potentially targeted in the 2 cancers (Fig. 5E, F, and Table S7). For example, microtubule-associated protein TAU (MAPT) has been detected to be highly mutated only in pancreatic adenocarcinoma, while 2 taxanes, docetaxel and paclitaxel, can stabilize microtubule dynamics and target MAPT (Fig. 5E).46 Both agents are effective anticancer drugs, and nanoparticle albumin-bound paclitaxel (nab-paclitaxel) plus gemcitabine was approved by the FDA in 2013 for the treatment of advanced pancreatic adenocarcinoma.47 Interestingly, autophagy promotes chemoresistance of cancer cells to docetaxel and paclitaxel, whereas targeting autophagy enhances the anti-tumor effects of the 2 drugs.48,49 Also, we observed that ESR1 (estrogen receptor 1) is highly mutated in both cancers, and up to 31 drugs were approved to target this protein (Fig. 5E and F). Although none of the drugs have been approved for the prevention of advanced pancreatic adenocarcinoma, a number of them were widely used for the therapy of prostate cancer, such as diethylstilbestrol (DES) and estramustine.50 In addition, a tyrosine kinase gene, ALK, is frequently mutated in both cancers (Fig. 5E and F). At least 3 small-molecule drugs, including crizotinib, ceritinib and alectinib, have been approved for the therapy of non-small-cell lung cancer.51 In particular, crizotinib activates autophagy in multiple lung cancer cell lines, and the inhibition of autophagy enhances its efficacy for the induction of cell death.52 Although the effectiveness of most drugs for the 2 cancers remains to be characterized, our multiple analyses demonstrated that AT proteins are highly associated with human diseases, indicating autophagy can be a promising target in disease therapy.
Phosphorylation is essential in regulating the autophagy pathway
As one of the most important and well-studied PTMs, phosphorylation participates almost all of biological processes and reversibly determines cellular dynamics and plasticity. Recent studies demonstrated that phosphorylation plays an important role in the regulation of autophagy.4 Previously, we developed an integrative resource of Eukaryotic Kinase and Phosphatase Database (EKPD),53 containing 516 protein kinases and 160 protein phosphatases in H. sapiens. Here, we mapped 1,909 curated human proteins (Table S1) to EKPD database, and identified 92 kinases and 11 phosphatases to be involved in autophagy (Fig. 6A and Table S8). Using the hypergeometric distribution (P value < 0.05),54 statistical analyses demonstrated that kinases were more significantly over-represented in AT than phosphatases (Fig. 6A and Table S8). Interestingly, the results can be analogous to a previous study in D. melanogaster, which in total identified 80 of 228 fly kinases to be essential for cell cycle progression, including 34 mitosis-associated kinases.55 Because it is well documented that mitosis and cell cycle processes are tightly regulated by phosphorylation, our analyses suggested that phosphorylation might play a similar role in the regulation of autophagy. We further mapped 512,059 known phosphorylation sites of 63,151 substrates for the 7 model organisms, including H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, S. cerevisiae and A. thaliana to our dataset (Fig. 6B and Table S4).
Figure 6.
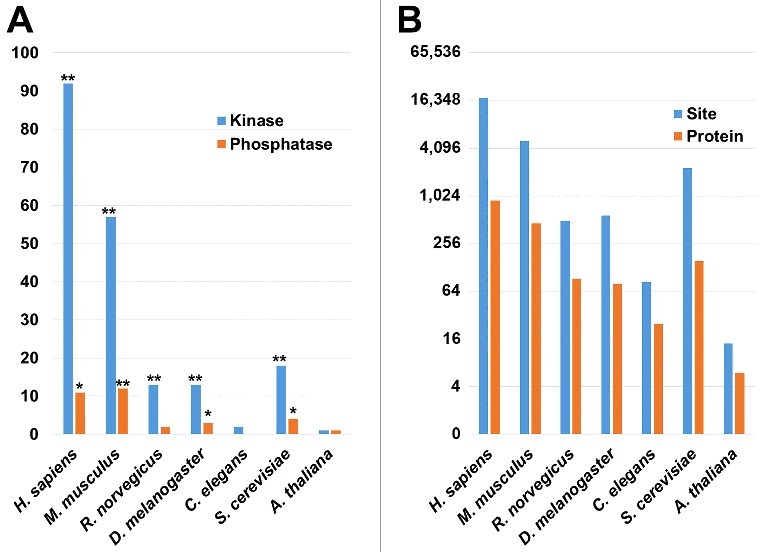
The phospho-regulation of human AT proteins. (A) The kinases and phosphatases were mapped from EKPD to AT proteins in H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, S. cerevisiae and A. thaliana, respectively. The enrichment analysis was performed for each of the 7 species. Protein kinases were significantly enriched in most of the species, Except in C. elegans and A. thaliana, mainly due to the data limitation of known AT proteins in the 2 organisms. (B) The distribution of phosphorylated AT proteins and sites in 7 species. i. *, P value < 0.05; ii. **, P value < 0.01.
For a better understanding of phosphoregulations in the machinery of the autophagy pathway, we used a previously developed tool of in vivo group-based prediction system (iGPS)56 for the reconstruction of the KSPNs among ATGs and their regulatory kinases for H. sapiens (Fig. 7A), M. musculus (Fig. 7B) and S. cerevisiae (Fig. 7C), respectively. In both human and mouse networks, BECN1/Vps30/Atg6 has the most kinase-substrate relations, and can be phosphorylated by 92 and 71 kinases in H. sapiens (Fig. 7A) and M. musculus (Fig. 7B), respectively. However, no kinase was found to phosphorylate its ortholog in yeast, Vps30/Atg6/BECN1, whereas Atg1 was mostly phosphorylated by 21 kinases in S. cerevisiae (Fig. 7C). For regulatory protein kinases, the serine/threonine kinase AKT1 phosphorylates the most ATG proteins, with 9 and 7 substrates in H. sapiens (Fig. 7A) and M. musculus (Fig. 7B), respectively. Interestingly, a member of AKT family, Ypk2, also phosphorylates the most ATG proteins with 21 substrates in S. cerevisiae (Fig. 7C). Although human ULK1 (one of the human orthologs of yeast Atg1) was predicted to be regulated by 40 kinases, it was the mostly hyperphosphorylated protein among all ATGs with up to 61 phosphorylation sites. The phosphorylation sites with at least one predicted kinase were shown for human ULK1, and nearly half of the sites were modified by the kinase activity of MTOR (Fig. 7D). Most of these ssKSRs have not been reported previously, and our predictions can be useful for further experimental consideration. Taken together, our results suggested that phosphorylation plays an important role in the regulation of autophagy.
Figure 7.
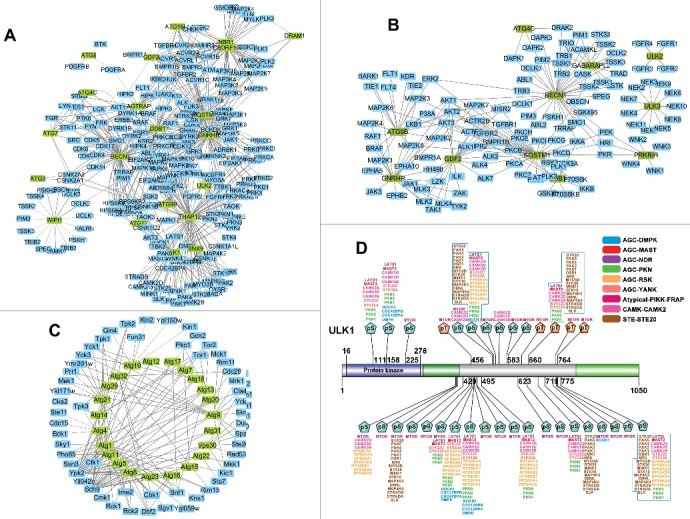
The phosphorylation networks among ATG proteins and their regulatory kinases for (A) H. sapiens, (B) M. musculus, and (C) S. cerevisiae. (D) The phosphorylation sites predicted kinases of human ULK1. The protein kinase family was shown as “group-family”. For example, AGC-DMPK refers to the family of dystrophia myotonica protein kinases (DMPKs) in the protein kinase A, G, and C (AGC) group. The detailed classifications of eukaryotic protein kinases can be accessed at EKPD (http://ekpd.biocuckoo.org/). pS, phospho-serine; pT, phospho-threonine.
Various PTMs are highly associated with autophagy
Next, we extended the PTM analysis by mapping known PTM sites of ten types of protein lysine modifications to all integrated proteins in H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, S. cerevisiae and A. thaliana. For known AT proteins in the 7 organisms, in total we obtained 35,420 PTM sites of 3,396 substrates, including 26,371 phosphorylation sites of 1,724 proteins, 6,377 ubiquitination sites of 932 proteins, 2,067 acetylation sites of 532 proteins, and 269 sumoylation sites of 79 proteins, respectively (Fig. 8A and Table S4). From the results, we observed a complex overlap existed among different types of PTM substrates, and a considerable proportion of proteins can be regulated by multiple PTMs (Fig. 8B). Using the human proteome as the background, the statistical analyses demonstrated that all types of PTMs were statistically enriched in human AT proteins (Fig. 8C and Table S9). Our analyses are consistent with a previous summarization, which emphasized the importance of phosphorylation, ubiquitination and acetylation in autophagy.4 Again, the results suggested that other types of PTMs might also be important in autophagy (Fig. 8B).
Figure 8.
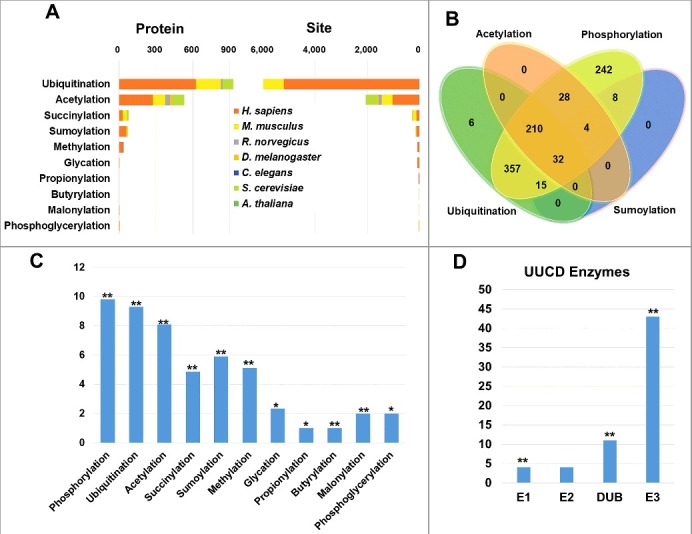
Multiple PTMs are significantly associated with AT proteins. (A) The distribution of numbers of mapped substrates and sites of ubiquitination, acetylation, succinylation, sumoylation, methylation, glycation, propionylation, butyrylation, malonylation and phosphoglycerylation in 7 species. (B) The overlap of 4 major types of PTMs including phosphorylation, ubiquitination, acetylation and sumoylation for AT proteins. (C) The distribution and enrichment analysis of 11 PTMs mapped to human AT proteins. (D) The distribution of ubiquitin and ubiquitin-like enzymes mapped to human AT proteins. i. *, P value < 0.05; ii. **, P value < 0.01.
Additionally, we obtained 886 annotated human ubiquitin and ubiquitin-like conjugation regulators including 10 ubiquitin-activating enzymes (E1s), 43 ubiquitin-conjugating enzymes (E2s), 700 ubiquitin-protein ligases (E3s) and 120 deubiquitination enzymes (DUBs) from a previously constructed database of UUCD.57 We mapped curated human AT proteins to the data set, and observed that E1s, E3s and DUBs but not E2s were significantly enriched against the human proteome (Fig. 8D and Table S8). Taken together, our analyses suggested that various PTMs are important in regulating autophagy pathways, while the results can be useful for further experimental manipulation.
Discussion
Recently, autophagy has emerged to be an intriguing biological process, and has attracted much attention for extensive research.1-6 It is demonstrated that autophagy plays essential roles in a broad spectrum of physiological, developmental and pathophysiological processes, and has been associated with human diseases such as neurodegenerative diseases, inflammatory diseases, and cancers.1,10,11 Besides 41 ATG proteins, a large number of regulators have also been discovered to regulate autophagy in multilevels.2,4,7-9 In particular, numerous PTMs, such as phosphorylation, ubiquitination, acetylation and sumoylation, are important in the regulation of autophagy.4 Thus, the collection, curation and integration of experimentally identified regulators and PTMs will be helpful for understanding the molecular mechanisms of autophagy at a systems-level, and provide highly useful information for further experimental consideration. Although a number of data sets or resources for autophagy and cell death pathways have been developed,7-9,16-25 the corresponding known PTM information still remains to be integrated.4
In this study, we manually collected and curated 4,237 known AT, AP and NE proteins from the literature, and this number was much greater than previous efforts (Fig. 1D and Fig. S2). Using 3,882 known proteins of 8 model species, we carried out a computational detection of potential orthologs in 164 eukaryotes, and annotated the orthologous proteins with potential regulations in autophagy and PCDs by using known information. To test the reliability of such a transfer of the data from species to species, we compared the experimental evidence of 352 known human AT proteins with their orthologs in other species, if available (Table S10). Our results demonstrated that the regulations of most of known human AT proteins were consistent with their orthologs (93.75%, 330/352), and there were only 22 human proteins with inconsistently annotated orthologs (Table S10). For these inconsistent annotations, we carefully traced the original literature and found the inconsistency was generated mainly 2 reasons. First, one protein can play different roles in different types of cells or tissues. For example, a tumor suppressor RB1 activates autophagy in human tumor cells,58 but inhibits autophagy in mouse primary cells.59 Second, the regulatory functions of orthologous proteins might be different in distinct species. For example, it is reported that a temperature sensitive mutant of SEC17 blocks autophagy in S. cerevisiae,60 however, the depletion of its mammalian ortholog NAPA/αSNAP stimulates the autophagic flux in human epithelial cells.61 Although not all annotations were consistent between human proteins and their ortholgs in other organisms, the high consistency of the experimental evidences suggested that the orthologous transfer of annotations across species is much reliable.
By mapping protein kinases to human AT proteins, we observed that phosphorylation play a similar role in autophagy and mitosis and cell cycle pathways (Fig. 6). For example, Dr. Leland H. Hartwell firstly established the yeast S. cerevisiae as an excellent model for genetic screening, and identified cell-division cycle (Cdc) mutants that regulate mitosis and cell cycle.62 Similarly, a number of ATG genes were also screened and discovered in yeast.6 Second, both types of processes are highly conserved across eukaryotes with conserved genes.2,3,5,6,62 Third, besides phosphorylation,4 both autophagy, and mitosis and cell cycle were dynamically but precisely regulated in multiple levels, such as transcriptional and post-translational regulations.4,7-9 Fourth, both autophagy and mitosis are multistage process. For example, a typical autophagic process contains 3 steps, including phagophore formation, autophagosome generation, and its fusion with lysosomes for degrading the contents.4,5 Analogously, mitosis also contains several steps, including prophase, metaphase, anaphase and telophase, to orchestrate the proper segregation of sister chromatids.54 Fifth, various protein complexes will be formed during either autophagy or mitosis at distinct cellular compartment. For example, the ATG1/ULK complex, ATG9 and its cycling system, and the PtdIns3K complex are part of the machinery of autophagy,2,4,39 whereas hundreds of proteins form different complexes at midbody, centrosome and kinetochore during mitosis.54,63 Sixth, both autophagy and mitosis and the cell cycle are highly associated with human diseases.1,10,11 Finally, 2 processes can crosstalk with each other mediated by specific regulators.27,64,65 For example, although MAPK1 and MAPK3 regulate the autophagic process,65 they also play a role in determining mitotic spindle angle during early lung development.64 The numerous analogies of autophagy, and mitosis and cell cycle emphasized the importance of scientific researches in autophagy.
Taken together, we manually collected 4,237 known proteins and further computationally characterized 191,543 potentials AT, AP and NE proteins in 164 eukaryotes. For our future plans, a number of efforts should be taken. First, more species will be considered and included in the database. Second, proteins may have different regulatory functions in different types of cells and tissues.58,59 Thus, the tissue-specific information will be carefully curated from the primary literature and integrated into the database. Also, the PTM sites were mainly identified from high-throughput experiments, and the exact functions of most of the sites were unknown. Besides the curation of autophagy-associated PTM events from the literature, we will perform experiments to discover new PTM regulators or substrates involved in autophagy. Moreover, since over 380 autophagy inducers or inhibitors together with their targeting proteins have been reported, the information is highly useful for further research, and will be integrated into our database. In addition, the multilayer data beyond proteins and PTMs, such as mRNA expression data, ncRNAs and cancer mutations, will be collected and maintain THANATOS. The database will be continuously maintained and updated, and we believe such a data resource can provide helpful information for both experimental and computational analyses.
Materials and methods
Data collection and curation
From the scientific literature, we manually collected experimentally identified proteins that participate in autophagy and cell death pathways. Multiple keywords, such as “autophagy”, “autophagic”, “apoptosis”, “apoptotic”, “necrosis” and “necroptosis”, were used to query the PubMed search engine. We also considered the information from several well annotated data resources, such as ARN,7 DeathBase,17 and the autophagy census.9 Each protein entry in these databases was rechecked by searching PubMed to ensure the data quality. The obtained abstracts or full papers were carefully read, and proteins with unambiguously experimental evidence were preserved. As previously described,2,4,7 41 ATG genes were denoted as “autophagy proteins”, whereas other proteins that also contribute in regulating autophagy were denoted as “autophagy regulators”. For each protein entry, the “+” or “-” was used to distinguish the positive or negative regulation for autophagy, apoptosis or necrosis, as previously described.9 All protein sequences were retrieved from the Ensembl database.66
Orthologous detection
To identify potential orthologs of known AT, AP and NE proteins, we downloaded the complete proteome sets of 164 eukaryotes including 84 animals, 39 plants and 41 fungi, from Ensembl66 (release version 84, http://www.ensembl.org/, under the directory of “/pub/release-84/fasta”), Ensembl Metazoa (release version 35, http://metazoa.ensembl.org/), EnsemblPlants (release version 31, http://plants.ensembl.org/) and EnsemblFungi (release version 31, http://fungi.ensembl.org/), respectively. Besides C. elegans and D. melanogaster, we also included 16 additional metazoan species, including Amphimedon queenslandica (Sponge), Apis mellifera (Honeybee), Belgica antarctica (Antarctic midge), Lucilia cuprina (Green bottle fly), Bombyx mori (Silk moth), Lingula anatina, Mnemiopsis leidyi (Sea walnut), Nematostella vectensis (Starlet sea anemone), Thelohanellus kitauei (Myxosporean), Octopus bimaculoides (California two-spotted octopus), Schistosoma mansoni (Blood fluke), Strigamia maritima (European centipede), Strongylocentrotus purpuratus (Purple sea urchin), Tetranychus urticae (Two-spotted spider mite), Tribolium castaneum (Red flour beetle) and Trichoplax adhaerens (Trichoplax reptans). Because multiple variant nucleotide sequences or peptides can be originated from a single gene, we used Ensembl Gene ID as the unique accession to eliminate the redundancy. For multiple alternatively splicing isoforms of a single gene, only the longest one was reserved. As previously described,63 the strategy of reciprocal best hits (RBHs) was chosen, and the blastall program in the BLAST package was utilized.67
The data set of PPIs
We obtained 322,043 experimental and computational PPI pairs of 6 model organisms from the Interologous Interaction Database (I2D),68 containing 296,008, 264,554, 205,384, 123,713, 55,236, and 334,197 pairwise PPIs of H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, and S. cerevisiae, respectively. Furthermore, PPIs in 2 public databases, IntAct69 and MINT,70 were also integrated. From IntAct, we integrated 135,570, 17,691, 2,564, 38,646, 12,159 and 78,512 PPIs of H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, and S. cerevisiae, while 17,538, 9,408, 942, 568, 476, 28,274 PPIs were retrieved from MINT for these 6 organisms respectively. We mapped the PPI data set to all integrated proteins in the 6 species, and found their interacting partners if available. Finally, 38,088, 24,457, 14,317, 3,809, 3,267, 9,284 PPIs were compiled to THANATOS for H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, and S. cerevisiae respectively.
The data set of PTM sites
Previously, we have collected 565,176 known phosphorylation sites of 85,160 proteins from 27 eukaryotes.71,72 Here, we obtained 512,059 known phosphorylation sites of 63,151 substrates for the 7 model organisms, including H. sapiens, M. musculus, R. norvegicus, D. melanogaster, C. elegans, S. cerevisiae and A. thaliana. Furthermore, we used the data set of a previously developed database of CPLM, which contained 189,919 sites in 45,748 proteins for 12 types of protein lysine modifications, including acetylation, butyrylation, crotonylation, glycation, malonylation, methylation, phosphoglycerylation, propionylation, pupylation, succinylation, sumoylation, and ubiquitination.73 The pupylation sites were not used because pupylation only occurs in prokaryotes. Also, because crotonylation sites were mainly identified in histones, this PTM was not considered. All PTM sites were mapped to all identified proteins of the 7 species, to pinpoint the exact modification sites.
The data set of cancer genes, drug targets and cancer mutations
We obtained 559 well-curated cancer genes from the Cancer Gene Census in COSMIC,41 and 2,247 human drug targets together with corresponding FDA-approved drugs from the DrugBank database (version 5.0).40 We downloaded all nonsynonymous somatic mutations together with mutated gene lists of prostate cancer (Project ID: EOPC-DE, PRAD-CA, PRAD-UK, PRAD-US) and pancreatic adenocarcinoma (Project ID: PACA-CA, PACA-AU, PAAD-US) from the ICGC database (Data Release 22, August 23rd, 2016),45 and acquired 845,198 mutations on 53,882 genes, antisenses and noncoding RNAs for prostate cancer, while 2,966,546 mutations on 55,220 genes, antisenses and noncoding RNAs were obtained for pancreatic adenocarcinoma.
The statistical enrichment analysis
To analyze the preferentially distributed pathways of known human AT proteins, we purchased a KEGG FTP subscription for personal use,42 and mapped all human proteins to KEGG pathways if available. In total, there were 6,178 human proteins annotated with at least one KEGG entry, while 564, 783 and 62 collected AT, AP and NE proteins were annotated with at least one KEGG entry, respectively. Here we defined:
N = number of proteins in human proteome annotated by at least one KEGG pathway
n = number of proteins in human proteome annotated by the KEGG pathway t
M = number of proteins in human AT proteins annotated by at least one KEGG pathway
m = number of proteins in human AT proteins annotated by the KEGG pathway t
Then the enrichment ratio (E-ratio) of the KEGG pathway t was calculated, and the P value was calculated with the hypergeometric distribution as below:
In this work, we only considered the over-represented KEGG pathways with E-ratio ≥ 1.
Reconstruction of human kinase-substrate phosphorylation network
Previously, we developed the iGPS software packages (http://igps.biocuckoo.org) for the prediction of in vivo ssKSRs of 408 human kinases from the phosphoproteomic data.56 A sequence-based algorithm of Group-based Prediction System (GPS) was adopted, while the PPI information between protein kinases and substrates was used as an additional filter to greatly reduce false positive predictions. Using iGPS with the default threshold values, we predicted potential ssKSRs for experimentally identified phosphorylation sites in known human AT proteins. For the reconstruction of human KSPN, the orientation was defined as Kinase -> Substrate. Because a proportion of substrates can be kinases, the orientation can also be Kinase A -> Kinase B (A phosphorylates B) or Kinase A <-> Kinase B (A and B mutually phosphorylate with each other).
Supplementary Material
Funding Statement
This work was supported by grants from the Special Project on Precision Medicine under the National Key R&D Program (2017YFC0906600), the National Basic Research Program (973 project) (2013CB933900), Natural Science Foundation of China (31671360), and International Science & Technology Cooperation Program of China (2014DFB30020).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors are grateful to Prof. Li Yu (Tsinghua Univ.), who kindly encouraged us to transfer from pure bioinformatics to autophagy. We also thank Dr. Cong Yi, Dr. Wenzhi Feng, and Dr. Jingjing Tong in Prof. Yu's lab, for their great helps. We are thankful for Prof. Hong Zhang (IBP, CAS) for his suggestive discussion during this work. We thank Prof. Dong Wang (HRBMU) for his helpful advice during the manuscript preparation. The authors also thank Dr. Min Li (SYSU), Dr. Yan Zhao (IBP), and Dr. Qingqiu Gong (Nankai Univ.) for their helpful comments on ATG11 and ATG17.
References
- 1.Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69–79; doi: 10.1038/cr.2013.161. PMID:24323045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24–41; doi: 10.1038/cr.2013.168. PMID:24366339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822; doi: 10.1038/ncb0910-814. PMID:20811353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11:28–45; doi: 10.4161/15548627.2014.984267. PMID:25484070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klionsky DJ. Coming soon to a journal near you – the updated guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2014;10:1691; doi: 10.4161/auto.36187. PMID:25208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9–23; doi: 10.1038/cr.2013.169. PMID:24366340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turei D, Foldvari-Nagy L, Fazekas D, Modos D, Kubisch J, Kadlecsik T, Demeter A, Lenti K, Csermely P, Vellai T, et al. . Autophagy Regulatory Network – a systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy. 2015;11:155–165; doi: 10.4161/15548627.2014.994346. PMID:25635527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Zhuang L, Wang Y, Hu Y, Wu Y, Wang D, Xu J. Connect the dots: a systems level approach for analyzing the miRNA-mediated cell death network. Autophagy. 2013;9:436–439; doi: 10.4161/auto.23096. PMID:23322033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenzi PL, Claerhout S, Mills GB, Weinstein JN. A curated census of autophagy-modulating proteins and small molecules: candidate targets for cancer therapy. Autophagy. 2014;10:1316–1326; doi: 10.4161/auto.28773. PMID:24906121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695; doi: 10.1016/j.cell.2011.07.030. PMID:21884931 [DOI] [PubMed] [Google Scholar]
- 11.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335; doi: 10.1038/nature09782. PMID:21248839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758; doi: 10.1016/j.cell.2011.10.033. PMID:22078876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94; doi: 10.1038/nrm3735. PMID:24401948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linkermann A, Green DR. Necroptosis. N Engl J Med. 2014;370:455–465; doi: 10.1056/NEJMra1310050. PMID:24476434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323; doi: 10.1016/j.cell.2008.10.044. PMID:19109899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doctor KS, Reed JC, Godzik A, Bourne PE. The apoptosis database. Cell Death Differ. 2003;10:621–633; doi: 10.1038/sj.cdd.4401230. PMID:12761571 [DOI] [PubMed] [Google Scholar]
- 17.Diez J, Walter D, Munoz-Pinedo C, Gabaldon T. DeathBase: a database on structure, evolution and function of proteins involved in apoptosis and other forms of cell death. Cell Death Differ. 2010;17:735–736; doi: 10.1038/cdd.2009.215. PMID:20383157 [DOI] [PubMed] [Google Scholar]
- 18.Wanichthanarak K, Cvijovic M, Molt A, Petranovic D. yApoptosis: yeast apoptosis database. Database (Oxford). 2013;2013:bat068; doi: 10.1093/database/bat068. PMID:24082050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arntzen MO, Thiede B. ApoptoProteomics, an integrated database for analysis of proteomics data obtained from apoptotic cells. Mol Cell Proteomics. 2012;11:M111 010447; doi: 10.1074/mcp.M111.010447. PMID:22067098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arntzen MO, Bull VH, Thiede B. Cell death proteomics database: consolidating proteomics data on cell death. J Proteome Res. 2013;12:2206–2213; doi: 10.1021/pr4000703. PMID:23537399 [DOI] [PubMed] [Google Scholar]
- 21.Homma K, Suzuki K, Sugawara H. The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res. 2011;39:D986–D990; doi: 10.1093/nar/gkq995. PMID:20972215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moussay E, Kaoma T, Baginska J, Muller A, Van Moer K, Nicot N, Nazarov PV, Vallar L, Chouaib S, Berchem G, et al. . The acquisition of resistance to TNFalpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy. 2011;7:760–770; doi: 10.4161/auto.7.7.15454. PMID:21490427 [DOI] [PubMed] [Google Scholar]
- 23.Jegga AG, Schneider L, Ouyang X, Zhang J. Systems biology of the autophagy-lysosomal pathway. Autophagy. 2011;7:477–489; doi: 10.4161/auto.7.5.14811. PMID:21293178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Huang Y, Kang J, Li K, Bi X, Zhang T, Jin N, Hu Y, Tan P, Zhang L, et al. . ncRDeathDB: A comprehensive bioinformatics resource for deciphering network organization of the ncRNA-mediated cell death system. Autophagy. 2015;11:1917–1926; doi: 10.1080/15548627.2015.1089375. PMID:26431463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, Li YH. miRDeathDB: a database bridging microRNAs and the programmed cell death. Cell Death Differ. 2012;19:1571; doi: 10.1038/cdd.2012.87. PMID:22743998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, Hara T, Sawada N, Yamada A, Mizushima N, Uchiyama Y, et al. . The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775; doi: 10.1091/mbc.E08-03-0309. PMID:18768753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, et al. . Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699; doi: 10.1182/blood-2007-09-112904. PMID:18305219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946; doi: 10.1016/j.cell.2006.12.044. PMID:17350577 [DOI] [PubMed] [Google Scholar]
- 29.Keil E, Hocker R, Schuster M, Essmann F, Ueffing N, Hoffman B, Liebermann DA, Pfeffer K, Schulze-Osthoff K, Schmitz I. Phosphorylation of Atg5 by the Gadd45beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ. 2013;20:321–332; doi: 10.1038/cdd.2012.129. PMID:23059785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The UniProt Consortium Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014;42:D191–D198; doi: 10.1093/nar/gkt1140. PMID:24253303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shemi A, Ben-Dor S, Vardi A. Elucidating the composition and conservation of the autophagy pathway in photosynthetic eukaryotes. Autophagy. 2015;11:701–715; doi: 10.1080/15548627.2015.1034407. PMID:25915714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5:85–87; doi: 10.4161/auto.5.1.7180. PMID:18981720 [DOI] [PubMed] [Google Scholar]
- 33.Liu X, Klionsky DJ. The Atg17-Atg31-Atg29 complex and Atg11 regulate autophagosome-vacuole fusion. Autophagy. 2016;12:894–895; doi: 10.1080/15548627.2016.1162364. PMID:26986547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy P, Karpati M, Varga A, Pircs K, Venkei Z, Takats S, et al. . Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy. 2014;10:453–467; doi: 10.4161/auto.27442. PMID:24419107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin L, Yang P, Huang X, Zhang H, Lu Q, Zhang H. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J Cell Biol. 2013;201:113–129; doi: 10.1083/jcb.201209098. PMID:23530068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, et al. . Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013;203:299–313; doi: 10.1083/jcb.201304123. PMID:24165940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12:1–222; doi: 10.1080/15548627.2015.1100356. PMID:26799652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290:2114–2117; doi: 10.1126/science.290.5499.2114. PMID:11118139 [DOI] [PubMed] [Google Scholar]
- 39.Noda NN, Inagaki F. Mechanisms of Autophagy. Annu Rev Biophys. 2015;44:101–122; doi: 10.1146/annurev-biophys-060414-034248. PMID:25747593 [DOI] [PubMed] [Google Scholar]
- 40.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–D672; doi: 10.1093/nar/gkj067. PMID:16381955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. . COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811; doi: 10.1093/nar/gku1075. PMID:25355519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280; doi: 10.1093/nar/gkh063. PMID:14681412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin GC, Kang HS, Lee AR, Kim KH. Hepatitis B virus-triggered autophagy targets TNFRSF10B/death receptor 5 for degradation to limit TNFSF10/TRAIL response. Autophagy. 2016:1–16; doi: 10.1080/15548627.2016.1239002. PMID:27740879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol. 2011;85:13453–13456; doi: 10.1128/JVI.06064-11. PMID:21957292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.International Cancer Genome C, Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR, Bhan MK, Calvo F, Eerola I, et al. . International network of cancer genome projects. Nature. 2010;464:993–998; doi: 10.1038/nature08987. PMID:20393554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGrogan BT, Gilmartin B, Carney DN, McCann A. Taxanes, microtubules and chemoresistant breast cancer. Biochim Biophys Acta. 2008;1785:96–132; doi: 10.1016/j.bbcan.2007.10.004. PMID:18068131 [DOI] [PubMed] [Google Scholar]
- 47.Ojima I, Lichtenthal B, Lee S, Wang C, Wang X. Taxane anticancer agents: a patent perspective. Expert Opin Ther Pat. 2016;26:1–20; doi: 10.1517/13543776.2016.1111872. PMID:26651178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng X, Gong F, Chen Y, Jiang Y, Liu J, Yu M, et al. . Autophagy promotes paclitaxel resistance of cervical cancer cells: involvement of Warburg effect activated hypoxia-induced factor 1-alpha-mediated signaling. Cell Death Dis. 2014;5:e1367; doi: 10.1038/cddis.2014.297. PMID:25118927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan B, Chen D, Huang J, Wang R, Feng B, Song H, et al. . HMGB1-mediated autophagy promotes docetaxel resistance in human lung adenocarcinoma. Mol Cancer. 2014;13:165; doi: 10.1186/1476-4598-13-165. PMID:24996221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benson RC, Jr., Gill GM. Estramustine phosphate compared with diethylstilbestrol. A randomized, double-blind, crossover trial for stage D prostate cancer. Am J Clin Oncol. 1986;9:341–351; doi: 10.1097/00000421-198608000-00014. PMID:3529921 [DOI] [PubMed] [Google Scholar]
- 51.Holla VR, Elamin YY, Bailey AM, Johnson AM, Litzenburger BC, Khotskaya YB, Sanchez NS, Zeng J, Shufean MA, Shaw KR, et al. . ALK: a tyrosine kinase target for cancer therapy. Cold Spring Harb Mol Case Stud. 2017;3:a001115; doi: 10.1101/mcs.a001115. PMID:28050598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.You L, Shou J, Deng D, Jiang L, Jing Z, Yao J, Li H, Xie J, Wang Z, Pan Q, et al. . Crizotinib induces autophagy through inhibition of the STAT3 pathway in multiple lung cancer cell lines. Oncotarget. 2015;6:40268–40282; doi: 10.18632/oncotarget.5592. PMID:26384345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Liu Z, Cheng H, Gao T, Pan Z, Yang Q, Guo A, Xue Y. EKPD: a hierarchical database of eukaryotic protein kinases and protein phosphatases. Nucleic Acids Res. 2014;42:D496–D502; doi: 10.1093/nar/gkt1121. PMID:24214991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z, Ren J, Cao J, He J, Yao X, Jin C, Xue Y. Systematic analysis of the Plk-mediated phosphoregulation in eukaryotes. Brief Bioinform. 2013;14:344–360; doi: 10.1093/bib/bbs041. PMID:22851512 [DOI] [PubMed] [Google Scholar]
- 55.Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. . Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987; doi: 10.1038/nature03160. PMID:15616552 [DOI] [PubMed] [Google Scholar]
- 56.Song C, Ye M, Liu Z, Cheng H, Jiang X, Han G, Songyang Z, Tan Y, Wang H, Ren J, et al. . Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol Cell Proteomics. 2012;11:1070–1083; doi: 10.1074/mcp.M111.012625. PMID:22798277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao T, Liu Z, Wang Y, Cheng H, Yang Q, Guo A, Ren J, Xue Y. UUCD: a family-based database of ubiquitin and ubiquitin-like conjugation. Nucleic Acids Res. 2013;41:D445–D451; doi: 10.1093/nar/gks1103. PMID:23172288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang H, Martin V, Gomez-Manzano C, Johnson DG, Alonso M, White E, Xu J, McDonnell TJ, Shinojima N, Fueyo J. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010;70:7882–7893; doi: 10.1158/0008-5472.CAN-10-1604. PMID:20807803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ciavarra G, Zacksenhaus E. Multiple pathways counteract cell death induced by RB1 loss: implications for cancer. Cell Cycle. 2011;10:1533–1539; doi: 10.4161/cc.10.10.15520. PMID:21540641 [DOI] [PubMed] [Google Scholar]
- 60.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al. . SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302; doi: 10.1016/j.cell.2011.06.022. PMID:21784249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naydenov NG, Harris G, Morales V, Ivanov AI. Loss of a membrane trafficking protein alphaSNAP induces non-canonical autophagy in human epithelia. Cell Cycle. 2012;11:4613–4625; doi: 10.4161/cc.22885. PMID:23187805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hartwell LH, Culotti J, Reid B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc Natl Acad Sci U S A. 1970;66:352–359; doi: 10.1073/pnas.66.2.352. PMID:5271168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Z, Ma L, Wang Y, Pan Z, Ren J, Liu Z, Xue Y. MiCroKiTS 4.0: a database of midbody, centrosome, kinetochore, telomere and spindle. Nucleic Acids Res. 2015;43:D328–D334; doi: 10.1093/nar/gku1125. PMID:25392421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tang N, Marshall WF, McMahon M, Metzger RJ, Martin GR. Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science. 2011;333:342–345; doi: 10.1126/science.1204831. PMID:21764747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Y, Li X, Ma K, Yang J, Zhou J, Fu W, Wei F, Wang L, Zhu WG. The axis of MAPK1/3-XBP1u-FOXO1 controls autophagic dynamics in cancer cells. Autophagy. 2013;9:794–796; doi: 10.4161/auto.23918. PMID:23426330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biasoli D, Kahn SA, Cornelio TA, Furtado M, Campanati L, Chneiweiss H, Moura-Neto V, Borges HL. Retinoblastoma protein regulates the crosstalk between autophagy and apoptosis, and favors glioblastoma resistance to etoposide. Cell Death Dis. 2013;4:e767; doi: 10.1038/cddis.2013.283. PMID:23949216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, et al. . BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 2013;41:W29–W33; doi: 10.1093/nar/gkt282. PMID:23609542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:R95; doi: 10.1186/gb-2007-8-5-r95. PMID:17535438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, et al. . The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363; doi: 10.1093/nar/gkt1115. PMID:24234451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. . MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861; doi: 10.1093/nar/gkr930. PMID:22096227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ullah S, Lin S, Xu Y, Deng W, Ma L, Zhang Y, Liu Z, Xue Y. dbPAF: an integrative database of protein phosphorylation in animals and fungi. Sci Rep. 2016;6:23534; doi: 10.1038/srep23534. PMID:27010073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheng H, Deng W, Wang Y, Ren J, Liu Z, Xue Y. dbPPT: a comprehensive database of protein phosphorylation in plants. Database (Oxford). 2014;2014:bau121; doi: 10.1093/database/bau121. PMID:25534750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Z, Wang Y, Gao T, Pan Z, Cheng H, Yang Q, Cheng Z, Guo A, Ren J, Xue Y. CPLM: a database of protein lysine modifications. Nucleic Acids Res. 2014;42:D531–D536; doi: 10.1093/nar/gkt1093. PMID:24214993 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


