Abstract
In this study, we investigated the mechanism of miR-200c-3p and SLC6A1 in regulating cell activity of clear cell renal cell carcinoma (CCRCC). The mRNA and miRNA expressions of tissue specimens were analyzed by CapitalBio Corporation (Beijing, China). The expression of SLC6A1 in CCRCC cells was examined through qRT-PCR and western blot. The migration and invasion ability of 786-O cells was testified by transwell assay after transfected. 786-O cell proliferation ability was detected by MTT assay. Dual luciferase reporter assay verified the association between SLC6A1 and miR-200c-3p. SLC6A1 was high expressed and miR-200c-3p was low expressed in CCRCC tissues and cells. Besides, lower SLC6A1 expression indicated longer survival time and higher survival rate. MiR-200c-3p could directly target at SLC6A1 and reduce its expression. MiR-200c-3p inhibited the proliferation, migration and invasion in 786-O cells by down-regulating SLC6A1 expression. The results suggested that the miR-200c-3p served as a suppressor for CCRCC via down-regulating SLC6A1.
KEYWORDS: clear cell renal cell carcinoma, miR-200c-3p, SLC6A1, migration, invasion, survival rate
Introduction
Renal cell carcinoma (RCC), has the highest mortality rate among patients with urological malignancies for its high rates of recurrence, metastases and resistance to radiotherapy and chemotherapy,1 which has been enjoying its increasing incidences around the world for years.2 Generally, the standard treatment for RCC involves radical chemotherapy,3 tumor nephrectomy, and nephron surgery.4 However, owing to obscure incipient symptoms and high-frequent metastases of CCRCC, there is a very poor clinical outcome for patients.5 Approximately 30% to 40% of patients generate metastases after radical nephrectomy, let alone 30% of CCRCC patients present with disseminated tumor stage at diagnosis.6 Hence, there is an urgent need to further study the development of CCRCC and find more effective therapeutic methods for people with CCRCC.
MicroRNAs (miRNAs), a class of small endogenous noncoding RNA molecules, play a vital role in gene expression regulation via interacting with specific transcripts. Increasing studies have shown that miRNAs take part in several processes of tumorigenesis, involving proliferation, migration, invasion and metastasis,7,8 which suggests potential values of miRNAs for disease biomarkers and new targets. Unsurprisingly, several kinds of miRNAs, which had unusual expression level in carcinoma cells, were observed to take a role in CCRCC prognosis. For example, Gao Y et al. reported that miR-155 promoted the proliferative and invasive ability of CCRCC cells via suppressing E2F2 expression.9 Wotschofsky Z et al. found three miRNAs (miR-146a-5p, miR-128a-3p, miR-17-5p) whose upregulation were in accordance with CCRCC metastasis.10 With further exploration of miRNAs involvement in human cancers, researchers have uncovered strong connections between miR-200c-3p and carcinoma cells. MiR-200c-3p is in part responsible for the survival rate of patients with high-grade serous ovarian carcinoma.11 Through miRNA target prediction analysis, Chang et al. suggested that miR-200c-3p targeted breast oncogenes, which contributed to patient survival.12 A previous studies showed that miR-200c-3p decreased the metastatic ability of CCRCC by regulating E-cadherin through ZEB1.13 Hence, above mentioned discoveries provided strong indication that miR-200c-3p might contribute to CCRCC prognosis and become valuable therapeutic targets.
SLC6A1 encodes the voltage-dependent γ-aminobutyric acid (GABA) transporter (GAT-1) that undertakes to recycle GABA in the synaptic structure.14 Previous studies concerning SLC6A1 and GAT-1 mainly concentrate on their neurological functions and their involvement in nervous disorders. Mutation or impaired expression of SLC6A1 has been found playing a vital part in various neurological diseases, such as epilepsy,15 hippocampal sclerosis,16 and schizophrenia.17 Apart from neurological disorders, SLC6A1 was also found ectopically expressed in various human cancers such as ovarian cancer and gastric cancer.18,19
In present study, we discovered that miR-200c-3p impeded the CCRCC cells from proliferating, migrating and invading via interacting with SLC6A1. Our study would contribute to provide new treatment target for patients with CCRCC.
Results
SLC6A1 was high expressed in CCRCC tissues
Differentially expressed mRNAs were screened out by the TCGA series (Figure 1A). 20 most-aberrant expressed miRNAs were illustrated (10 up-regulated and 10 down-regulated) in CCRCC tissues compared to in adjacent normal tissues (Figure 1B). SLC6A1 was one of the top 10 up-regulated miRNAs and the expression level was averagely 2.118242907 times higher in the CCRCC tissues than in the adjacent normal tissues (adj.P.Val = 3.25E-20). The results of qRT-PCR and western blot showed that the expression level of SLC6A1 of tumor tissues was higher than that of adjacent tissues (P<0.05,Figure 1C-1D). Log-rank test results showed that the survival rate of patients with high SLC6A1 expression was significantly lower than patients with low SLC6A1 expression (P<0.05,Figure 1E). According to pathological characteristics of 82 patients, SLC6A1 expression was notably related with clinical stage (P = 0.0138), tumor stage (P = 0.0380), lymph node metastasis (P = 0.0016) and distant metastasis (P = 0.0277), while not related with gender, age, or tumor size (all P>0.05,Table 1). Furthermore, SLC6A1 could provide a high accuracy on CCRCC tissue identification estimated by ROC curve analysis (Figure 1F), suggesting the prognostic value of SLC6A1 in clinical application for CCRCC. To further investigate whether the upregulated SLC6A1 correlated with the survivals of the CCRCC patients, we performed Kaplan-Meier and Cox's proportional hazards regression model analysis and found that high SLC6A1 level was significantly correlated with poor overall survivals of CCRCC patients (Table 2), indicating that the association between SCL6A1 and survival was independent of other clinical variables.
Figure 1.
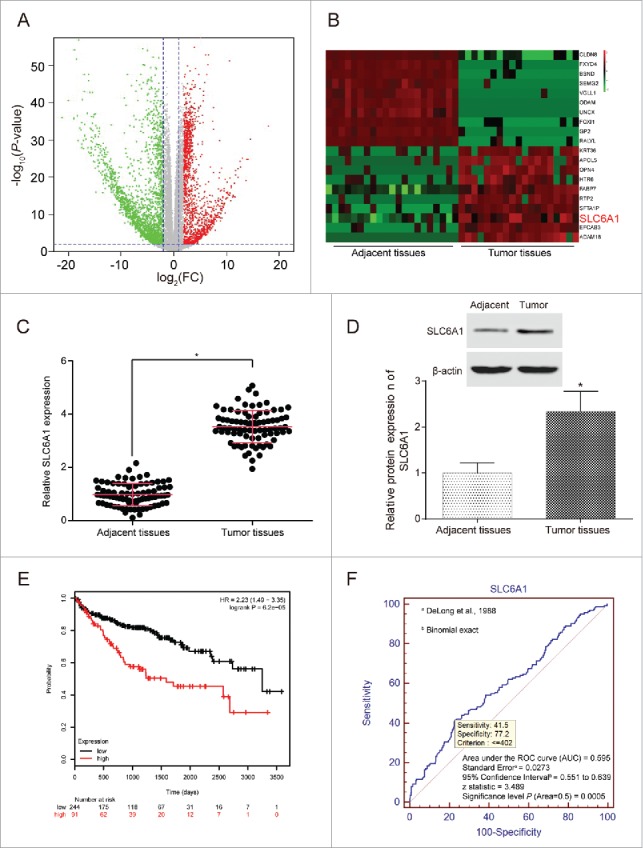
SLC6A1 was high expressed in CCRCC tissues. (A) Abnormal expression mRNAs in CCRCC tissues were reflected by the volcano plot. (B) The heat map showed SLC6A1 was a higher-expression mRNA in CCRCC tissues than in adjacent tissues. (C) SLC6A1 was high expressed in tumor tissues according to qRT-PCR. *P<0.05, compared with adjacent tissues. (D) SLC6A1 was high expressed in tumor tissues according to western blot. *P<0.05, compared with adjacent tissues. (E) Patients with lower SLC6A1 expression had higher overall survival rate and longer survival time shown by Kaplan-Meier curve. (F) An ROC curve built on a univariate classification model based on SLC6A1 expression across independent TCGA dataset for predicting CCRCC.
Table 1.
Clinicopathologic characteristics of patients in the study.
| The expression of SLC6A1 |
||||
|---|---|---|---|---|
| Clinicopathologic Characteristics | High expression(n = 56) | Low expression(n = 26) | P value | |
| Gender | Male | 42 | 19 | 0.8527 |
| Female | 14 | 7 | ||
| Age(years) | <60 | 26 | 14 | 0.5318 |
| ≥60 | 30 | 12 | ||
| Tumor size(cm) | <4 | 21 | 15 | 0.0864 |
| ≥4 | 35 | 11 | ||
| Clinical stage | I-II | 8 | 10 | 0.0138* |
| III-IV | 48 | 16 | ||
| Tumor stage | T1-T2 | 31 | 8 | 0.0380* |
| T3-T4 | 25 | 18 | ||
| Lymph node metastasis | Absence | 36 | 7 | 0.0016** |
| Presence | 20 | 19 | ||
| Distant metastasis | Absence | 34 | 9 | 0.0277** |
| Presence | 22 | 17 | ||
P < 0.5,
P < 0.01 determined by Chi-Square test.
Table 2.
Univariate and multivariate analyses of clinicopathological characteristics.
| Univariate analysisa |
Multivariate analysisb |
|||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| laterality | 1.454(1.056-2.003) | 0.021 | 1.390 (0.962-2.009) | 0.08 |
| gender | 0.950 (0.684-1.321) | 0.762 | ||
| age | 0.823(0.458–1.478) | 0.514 | ||
| person neoplasm cancer status | 6.080(4.241-8.717) | <0.001 | 3.540(2.322-5.397) | <0.001 |
| lactate dehydrogenase | 1.488 (0.507-4.363) | 0.467 | ||
| serum calcium | 0.756 (0.530-1.079) | 0.122 | ||
| hemoglobin | 2.109(1.450-3.068) | <0.001 | 1.364 (0.886-2.102) | 0.159 |
| platelet qualitative | 2.415(1.690-3.450) | <0.001 | 1.680(1.121-2.518) | 0.012 |
| white cell count | 0.696 (0.486-0.998) | 0.047 | 0.998 (0.667-1.494) | 0.992 |
| stage event system version | 0.698 (0.252-1.932) | 0.486 | ||
| stage event pathologic stage | 4.591(3.242-6.502) | <0.001 | 2.406(1.540-3.757) | <0.001 |
| Neoplasm histologic grade | 2.838(1.950-4.129) | <0.001 | 1.168 (0.757-1.801) | 0.484 |
| T stage | 5.149(2.691-9.852) | <0.001 | 1.975 (0.889-4.388) | 0.095 |
| N stage | 1.559 (0.354-6.876) | 0.555 | ||
| M stage | 0.253 (0.062-1.027) | 0.038 | 0.367 (0.080-1.677) | 0.196 |
| Has-miR-200c | 0.580 (0.344-0.977) | 0.038 | 0.571 (0.325-1.002) | 0.051 |
| SLC6A1 | 1.498(1.084-2.072) | 0.014 | 1.454(1.056-2.003) | 0.021 |
The data were subjected to Cox's proportional hazards regression model. Bold italics indicate statistically significant values (P < 0.05).
Multivariate analysis used stepwise addition and removal of clinical covariates found to be associated with survival in univariate models (P < 0.05) and final models, include only those covariates that were significantly associated with survival (Wald statistic, P < 0.05). Bold italics indicate statistically significant values (P < 0.05).
SLC6A1 knockdown inhibited CCRCC cells to proliferate, migrate and invade
SLC6A1 siRNA transfection significantly downregulated the expression of SLC6A1 (P<0.05, Figure 2A), and pcDNA3.1-SLC6A1 transfection significantly upregulated the expression of SLC6A1 (P < 0.05), indicating that the transfection was conducted successfully. At 48h, cell number in SLC6A1 siRNA group showed a significant decrease and continued the trend in the following hours, indicating that SLC6A1 siRNA could inhibit cell proliferation (P<0.05, Figure 2B). The cell proliferation activity in SLC6A1 group had the opposite trend occurred. SLC6A1 siRNA could inhibit 786-O cells migration and invasion as well, while SLC6A1 could promote the migration and invasion ability of 786-O cells (both P<0.05, Figure 2C-2D). Therefore, it came to a conclusion that SLC6A1 overexpression accelerated cell proliferation, migration and invasion, whereas SLC6A1 knockdown inhibited CCRCC cells to proliferate, migrate and invade.
Figure 2.
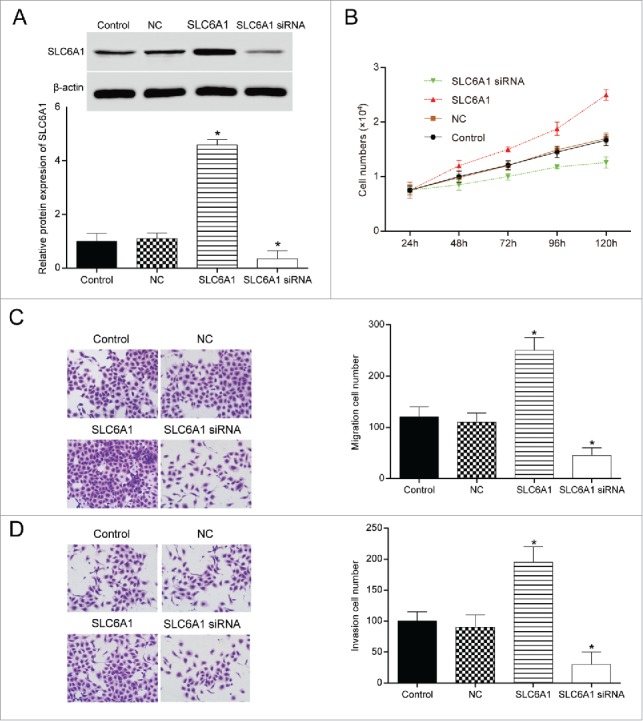
. SLC6A1 knockdown inhibited CCRCC cells to proliferate, migrate and invade. (A) The transfection efficiency was observed by RT-qPCR. *P < 0.05, compared with control group. (B) SLC6A1 siRNA transfection inhibited the proliferation of 786-O cells. *P<0.05, compared with control group. (C-D) SLC6A1 siRNA transfection inhibited the migration and invasion of 786-O cells (200 ×). *P < 0.05, compared with control group.
MiR-200c-3p targeted at SLC6A1 directly
MiR-200c-3p was significantly low expressed in tumor tissues (P < 0.05, Figure 3C, Supplementary Table S1), and it could target at SLC6A1 3’UTR as shown in Figure 3A. After co-transfection with miR-200c-3p mimics and SLC6A1 3’UTR-wt into HEK-293T cells, luciferase activity in 786-O cells decreased significantly (P<0.05, Figure 3B), while little changed in co-transfection group with SLC6A1 3’UTR-mut and miR-200c-3p mimics (P>0.05, Figure 3B). Hence, we confirmed that miR-200c-3p directly targeted at SLC6A1. Furthermore, the Pearson Correlation Analysis demonstrated that miR-200c-3p was negatively correlated with SLC6A1 with P<0.01 and R2 = 0.524 (Figure 3D).
Figure 3.

. MiR-200c-3p directly targeted at SLC6A1. (A) MiR-200c-3p was predicted to target SLC6A1 and the targeting scheme was illustrated. (B) MiR-200c-3p could directly targeted at SLC6A1 3’UTR. *P < 0.05, compared with NC mimics group. (C) MiR-200c-3p was low expressed in tumor tissues by qRT-PCR detection. *P < 0.05, compared with adjacent tissues. (D) MiR-200c-3p was negatively related with SLC6A1 expression with P < 0.01 and R2 = 0.524.
MiR-200c-3p reversed the effects of SLC6A1
MiR-200c-3p mimics and SLC6A1 cDNA could inhibit and promote SLC6A1 expression respectively, but their combination failed to affect SLC6A1 expression (both P<0.05,Figure 4A). Compared with control group, cell proliferation decreased in miR-200c-3p mimics group but increased in SLC6A1 group (both P < 0.05). However, joint expression of miR-200c-3p and SLC6A1 failed to affect 786-O cell proliferation (P > 0.05, Figure 4B). MiR-200c-3p mimics could effectively inhibit cell invasion and migration, which was opposite to the synergist effect of SLC6A1 cDNA (both P<0.05). Similarly, joint expression of miR-200c-3p and SLC6A1 displayed no significant difference (P > 0.05, Figure 4C-4D). Therefore, miR-200c-3p could reverse effects of SLC6A1.
Figure 4.
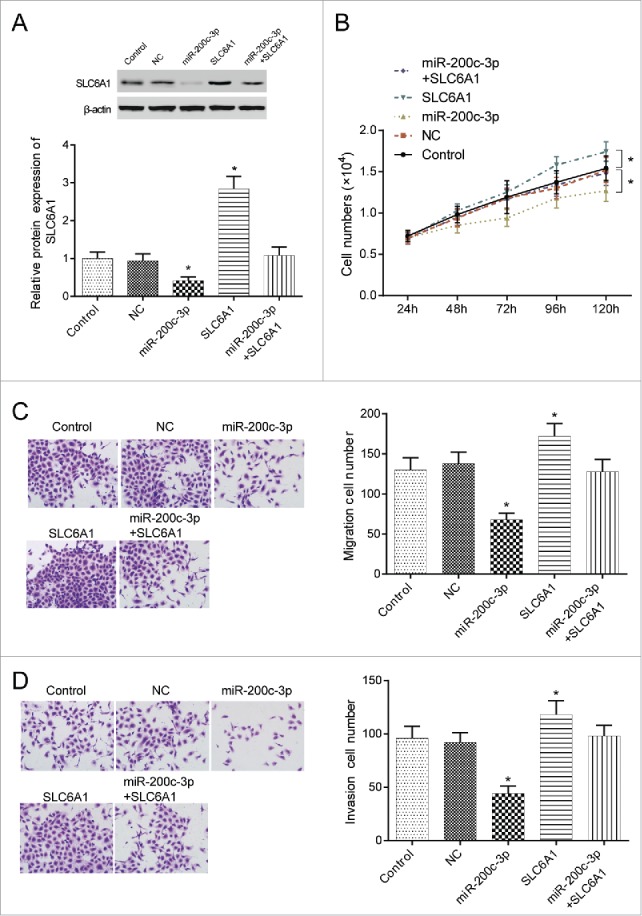
. MiR-200c-3p reversed the progression of SLC6A1. (A) MiR-200c-3p mimics transfection downregulated SLC6A1 expression while SLC6A1 cDNA upregulated SLC6A1 expression. *P < 0.05, compared with control group. (B) MiR-200c-3p mimics transfection decreased proliferation but SLC6A1 cDNA transfection increased proliferation in 786-O cells. Meanwhile, there was no significant difference between miR-200c-3p+SLC6A1 group and control group. *P < 0.05, compared with control group. (C-D) MiR-200c-3p mimics transfection inhibited but SLC6A1 cDNA transfection increased the migration and invasion in 786-O cells. There was no significant difference between miR-200c-3p+SLC6A1 group and control group despite of migration or invasion (200 ×). *P < 0.05, compared with control group.
Effects of the miR-200c-3p and SLC6A1 on CCRCC in vivo
The orthotopically implanted tumors in nude mice of the five groups with 786-O cells transfected with Control, NC, miR-200c-3p, SLC6A1 and miR-200c-3p+SLC6A1 were taken and measured respectively (Figure 5A). SLC6A1 group had the heaviest tumor (P<0.05). The tumor weight of miR-200c-3p+SLC6A1 group were equivalent to that of NC group (P > 0.05). The tumor weight in miR-200c-3p group were minimal (P < 0.05, Figure 5B). The lungs of mice injected cells in miR-200c-3p group had almost no visible metastatic nodules, while that of mice injected cells in SLC6A1 group was fully covered with nodules. The number of lung nodules in the mice lungs injected with SLC6A1 and miR-200c-3p+SLC6A1 group was comparable to that of NC group. The counting results under dissecting microscopy were shown in Figure 5C. The weight of the lungs in nude mice was weighed after they were sacrificed. After being transfected with miR-200c-3p, the lung weight significantly decreased (P < 0.05, Figure 5D), while the SLC6A1 group increased sharply (P < 0.01). Additionally, miR-200c-3p+SLC6A1 group and NC group showed no significant difference (P > 0.05). The results from the H&E staining assay (Figure 5E) were similar to the above.
Figure 5.
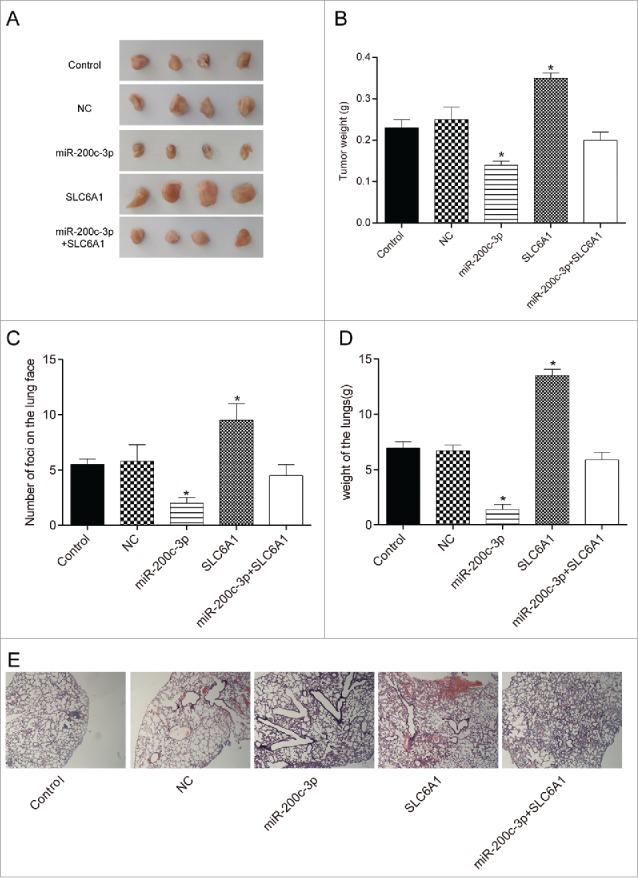
. Effects of the miR-200c-3p and SLC6A1 on CCRCC in vivo. (A) Tumor specimens from nude mice divided into five groups. (B) Tumor weights from nude mice divided into five groups. *P < 0.05, compared with control group. (C) The number of murine pulmonary nodules determined by dissecting microscope. *P < 0.05, compared with control group. (D) The weight of lungs in nude mice from 5 respective groups. *P < 0.05, compared with control group. (E) H&E staining was conducted to detect the number of lung metastatic nodules (× 40). *P < 0.05, compared with control group.
Discussion
The present study demonstrated that SLC6A1 was high expressed whereas miR-200-3p was low expressed in CCRCC tissues. SLC6A1 knockdown significantly inhibited CCRCC cells to proliferate, migrate and invade. There was a negative correlation between miR-200-3p level and SLC6A1 level. We also confirmed that miR-200c-3p directly targeted at SLC6A1 3’UTR. SLC6A1 overexpression could promote CCRCC cell growth and metastasis, whereas increased miR-200c-3p expression could reverse the effect.
SLC6A1 encodes a gamma-aminobutyric acid (GABA) transporter-1 (GAT-1), whose mutations lead to dysfunction of transporting GABA from extracellular space to the presynaptic terminal.20 GAT transporter has long been studied in psychiatric disorders. Recently, SLC6A1 has been speculated to participate in cancer development. For instance, Januchowski et al. found that the expression of SLC6A1 was upregulated in drug-resistant ovarian cancer cell lines.18 It was also found upregulated by over 10-fold in mucosa of atrophic gastritis and intestinal metaplasia. Thus, SLC6A1 could possibly be a participant in gastric cancer.19 In addition, other GAT transporter family members have also been found to be involved in cancer development. For example, low expression of SLC6A10P might indicate poor conditions of prognosis for ovarian cancer patients.21 SLC6A14 was found upregulated in cancers and its blockade in cancer cell lines with alpha-methyltryptophan might lead to cell cycle arrest.22 In our experiments, we also found the ectopic overexpression of SLC6A1 in CCRCC, and the modulation of SLC6A1 expression in CCRCC cell line resulted in the alteration of cell proliferation and metastasis, suggesting its tumor facilitator role.
By comparing the cancer tissues and its corresponding normal tissues, scientists have identified numerous kinds of miRNAs that are upregulated or downregulated during the tumor development.23,24 As for miR-200c-3p, its upregulated expression level has been observed in various cancers, and high miR-200c-3p level frequently indicates a poor survival outcome. For instance, Jonasch et al. found that miR-200c-3p expression was significantly enhanced in early stage of colorectal cancer compared with normal colorectal mucosa.25 Zhang et al. discovered that gastric cancer patients with high level of miR-220c-3p showed poor disease-free survival (DFS), which provided further comprehension for miR-220c-3p with cancer prognostic relevance.26 Moreover, pathway enrichment analysis and expression correlation analysis suggested that miR-200c-3p might account for epithelial ovarian cancer progression by affecting cellular adhesion process.27 Previous studies reached the agreement that miR-200c-3p promoted the tumorigenesis and metastases, though, via different ways. However, Wang et al. found that miR-200c was significantly downregulated in RCC tissues and its restoration suppressed the metastasis of 786-0 cells by regulating epithelial-to-mesenchymal transition (EMT) in human RCC.28 Similarly, Yoshino et al. discovered that EMT-related miR-200 was significantly suppressed in RCC, and miR-200c could affect RCC oncogenesis and metastasis via focal adhesion pathway.29 A miRNA can be both an oncogenic agent and a tumor suppressor dependent on cancer type. In our study, we observed miR-200c-3p was underexpressed in CCRCC tissues, possibly indicating its tumor suppressor role. Supportingly, we also found that enforced expression of miR-200c-3p impeded CCRCC cell 786-O proliferation, migration and invasion. Recent studies have proposed several targets for miR-200c-3p, such as solute carrier family 1 (GLA1)30 and epithelial-mesenchymal transition (EMT).11 No study has been done to discover the target relationship between miR-200c-3p and SLC6A1. In this study, we illustrated new correlation between miR-200c-3p and SLC6A1. Taken together, we thus speculated that miR-20c-3p could inhibit CCRCC progression by suppressing SLC6A1. In vitro experiments had been thoroughly conducted in the study, meanwhile, in vivo experiments were also performed to validate the tumor suppressor role of miR-200c-3p/SLC6A1 in CCRCC.
Activation of the epithelial-mesenchymal transition (EMT) is a prior condition for tumor metastasis because it makes it possible for the dissemination of oncocytes to other organs via blood stream.31 EMT has been observed in several human cancers previously. For example, EMT was induced by SET in pancreatic cancer, lung cancer,32 prostatic carcinoma and cervical cancer.33 As for CCRCC, it was revealed that EMT was attenuated by miR-138 in CCRCC cells, indicating the tumor suppression function of miR-138.34 on the contrary, IL-6 induces EMT and stimulates metastasis in CCRCC.35 Furthermore, Jiang J et al. Has reported that increased expression of miR-200c inhibits the cell viability and EMT of clear cell renal cell carcinoma.36 Therefore, we summarized the above points and scheduled the following plan of experiments that the highly related signaling pathway EMT should be taken into consideration for researches on CCRCC. What's more, one of the major limitations in this study was that all the mechanistic studies such as SLC6A1 knockdown and miR-200c-3p overexpression experiments were performed in only one cell line. It would be the next focus of our study to find out whether the consistent biological effect is a cell-line dependent.
In summary, we explored how miR-200c-3p mediated CCRCC tumorigenesis and progression by targeting SLC6A1. We demonstrated that miR-200c-3p suppressed while SLC6A1 promoted the propagation and metastasis of CCRCC cells. The target relationship between miR-200c-3p and SLC6A1 was validated in HEK-293T cell line. Moreover, when we overexpressed miR-200c-3p and SLC6A1 at the same time, the propagation, migration and invasiveness of CCRCC cells had no significant change, confirming the antagonism between SLC6A1 and miR-200c-3p. Our study may become a crucial reference for developing new drug target for CCRCC treatment.
Materials and methods
Tissue specimens
Human CCRCC tissues and normal adjacent tissues were obtained from 82 CCRCC patients who had not undergone chemotherapy or radiotherapy before the study. Patients of either gender were recruited by random and the study population characteristics were listed in Supplementary Table S2. The mRNA and miRNA expressions of tissue specimens was analyzed by CapitalBio Corporation (Beijing). The final diagnosis results were determined through the routine observation for clinicopathologic characteristics of patients. These specimens were snap-frozen in liquid nitrogen at -80°C and embedded in paraffin for the follow-up assays. This study had gotten approval from the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University and obtained informal written consents from each participant prior to the research.
Microarray analysis
The data for analysis of gene differential expression was obtained from The Cancer Genome Atlas (TCGA) Datasets. R programming language, Bayesian test and fold change were used to select differentially expressed miRNAs and mRNAs (P<0.05 and |log(fold change)|>1). Prognosis data was analyzed and survival rate as well as ROC curve analysis correlating with SLC6A1 in CCRCC was plotted by R studio (TIBCO, USA).
Cell culture and transfection
CCRCC 786-O cell line were acquired from Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in serum-free Dulbecco's modified Eagle medium (DMEM) (Gibco BRL, Grand Island, NJ, USA) plus 10% FBS (Invitrogen) at 37°C. PCR primers, miR-200c-3p mimics, SLC6A1 siRNA and SLC6A1 cDNA (plasmid vector pcDNA3.1) (Invitrogen, Carlsbad, CA, USA) used in the experiment were synthesized by Sangon Biotech (Shanghai, China). 4 × 105/ml cells were maintained in 6-well plates overnight. The plasmids were put into the 200 μl opti-MEM (HyClone, South Logan, UT, USA) and cultured in an incubator overnight. The cells were transfected with miR-200c-3p mimics, pcDNA3.1-SLC6A1 or SLC6A1 siRNA at the concentration of 50 nM and 100 nM respectively through Lipofectamine 2000 (Life Technologies) according to the producer's manual. 24 h after transfection, the cell transfection efficiency was detected for further analysis. The grouping of 786-O cells was as following: (1) Control group (non-transfection); (2) NC group (transfected with negative control siRNA); (3) miR-200c-3p group (transfected with miR-200c-3p mimics); (4) SLC6A1 group (transfected with SLC6A1); (5) miR-200c-3p+SLC6A1 group (co-transfected with miR-200c-3p mimics and SLC6A1).
Dual-luciferase reporter gene assay
HEK-293T cells purchased from the stem cell bank of Chinese Academy of Sciences were used for transfection in the dual-luciferase reporter gene assay. Passive lysis buffer (PLB), luciferase assay reagent II and Stop&Glo®Reagent (PLB, Promega, Madison, WI, USA) were prepared in accordance with the kit instructions. 3’UTR wild-type (WT) and 3’UTR-mutation (Mut) of SLC6A1 were constructed by Sangon Biotech (Shanghai, China) and were cloned into pGL3 vector. The cells were incubated the 24-well plates. The transfection was performed when the cells grew to 80% confluence. The 786-O cells were co-transfected with miR-200c-3p mimics or negative control mimics (NC mimics) with WT- or Mut 3’-UTR of SLC6A1 3’UTR using Lipofectamine 2000 reagent (Life Technologies). 48 h post-transfection, the cells were lysed using PLB and the luciferase activity was detected using dual-luciferase reporter assay system (E1910; Promega, USA) following the manufacturer's guidelines.
RNA extraction and qRT-PCR
Total RNA of 786-O cells was extracted with Trizol reagent (Invitrogen) according to the producer's protocols. The reverse transcription of total RNA (2 μg) was performed by means of Superscript II First-Strand Synthesis System for RT-PCR (Invitrogen). According to the mature sequence of miR-200c-3p (5’-UAAUACUGCCGGGUAAUGAUGCA-3’), the reverse transcription primer was designed (5’-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTG AGTCCATCAT-3’) and synthesized by Sangon Biotech, Shanghai. PCR was conducted through a 7300 real-time PCR system (Applied Biosystems, USA) using SYBR-Green mix (Applied Biosystems, USA). The amplification profile was initiated by 15 min of incubation at 95°C, followed by amplification of 30 sec at 95°C, 30 sec at 72°C and 30 sec at 55°C for 40 cycles. The expression levels of miR-200c-3p or SLC6A1 were determined by the 2−ΔΔCt method. U6 was an internal control for miRNA, whereas GAPDH was an reference gene for SLC6A1. The primers were generated by Sangon Biotech (Shanghai, China). The primers designed for RT-PCR were displayed in Table 3.
Table 3.
Primer sequences used in qRT-PCR.
| Primers | |||
|---|---|---|---|
| miR-200c | F | 5’- TCGTCTTACCCAGCAGTG-3’ | |
| R | 5’- CGGCAGTATTAGAGACTCC-3’ | ||
| U6 | F | 5’-CTCGCTTCGGCAGCACATA-3’ | |
| R | 5’-AACGATTCACGAATTTGCGT-3’ | ||
| SLC6A1 | F | 5’-GTGAGGCAACTCCAAGGT-3’ | |
| R | 5’-CAGATGGACGTGCGATGT-3’ | ||
| GAPDH | F | 5’-GAAGGTGAAGGTCGGAGTC-3’ | |
| R | 5’-GAAGATGGTGATGGGATTTC-3’ | ||
F: forward primers; R: reverse primers.
Western blot
Cells in logarithmic phase were harvested. RIPA lysis buffer (Sigma) was applied to the extraction of total protein. Protein concentration was measured through BCA protein kit (Bio-Rad). Afterwards, the proteins were segregated using 10% SDS-PAGE (Bio-Rad, USA) for 2 h and placed into the PVDF membranes (Invitrogen) in accordance with the producer's manual. 5% nonfat milk was used to block the membranes for 1 h, which were then incubated with primary antibodies against SLC6A1 (ab177483, 1:1000, Abcam, USA) and β-actin (ab8226, 1:000, Abcam, USA) at 4°C overnight. The membranes were subsequently rinsed using tris buffer saline Tween-20 (TBST) three times and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2000) secondary antibody at 37°C. After incubation for 1 h, the membranes were rinsed three times for 10 min. The electrochemiluminescent (ECL) detection system (Thermo Scientific) was utilized to detect the immunoreactive proteins. The immunoblot strips were analyzed using ImageJ software.
MTT assay
The cell viability or growth of the transfected 786-O cells was confirmed by MTT assay. The cells were seeded in 96-well plate (1 × 104 cells/well) overnight and maintained in DMEM, supplemented with FBS. After that, the cell growth was measured at different time points (24 h, 48 h, 72 h, 96 h and 120 h) after adding 20 μL of MTT (0.5 mg/ml, Sigma) at 37°C for 4 h. After the medium was removed, 150 μL dimethylsulfoxide (DMSO) was added to solubilize the crystals. The optical density (OD) was measured at 490 nm by Automatic Microplate Reader (Bio-Tec, Minneapolis, MN, USA).
Transwell assay
The cell migration and invasion abilities were determined by transwell assay. Matrigel (BD Biosciences, MA, USA) was first diluted with 100 μL serum-free medium and added to the upper chamber of transwell, which then transferred to 24-well chambers. 1 × 104 cells were seeded into the upper chamber 24 h post-transfection. 600 μl DMEM (10% FBS) was put into the lower chamber. After incubation at 37°C for 24 h, non-invading cells were gently removed with cotton swab. For the invading or migrating cells in the lower chamber, methyl alcohol and 0.1% crystal violet were respectively applied to the fixing and staining of the cells. The migration or invasion cells were counted by a microscope (× 200).
Models of human CCRCC in nude mice by orthotopic transplantation
Nude mice (SPF grade, 4–5 weeks, 17 ∼ 20 g) were provided by Animal research center of the First Affiliated Hospital of Xinjiang Medical University, half male and half female. 786-O Cells in logarithmic phase cultured in vitro were collected and subcutaneously injected into the right thigh of each nude mouse (0.2 ml, 1 × 107/ml). After 7 d incubation period, subcutaneously transplanted tumors began to grow to the diameter of 2 ∼ 3 cm. Then 1 mm3 size tumors were removed and transplanted in the subcutis of nude mice (2 per generation). The success rate of subcutaneous transplantation was l00%, the average incubation period 6 d, and each passage interval was 18 d or so. The experiment did not appear no cause of death and tumor spontaneous regression phenomenon. The subcutaneously transplanted tumors were taken out after passaged successfully 5 generations, removing the necrotic tissues, and transplanted under the renal capsule. After tumor inoculation, the left kidney area of nude mice was observed twice a day to understand the growth of transplanted tumors. All mice were sacrificed after renal failure and then tumor growth status, the renal hilum or hilar lymph node metastasis and pulmonary metastasis focuses were observed. And the weight of lungs was tested. At last, 4% paraformaldehyde was used to fix tissues for H&E staining and histological examination. All the experimental procedures were approved by Animal Ethics Committee in the First Affiliated Hospital of Xinjiang Medical University.
H&E staining
The tissues embedded in paraffin were dewaxed using xylene, and soaked in alcohol with gradient concentration (100%, 95%, 90%, 80%, 70% ethyl alcohol) for 5 min. After being washed with distilled water for 2 min, hematoxylin was added dropwise to stain the tissues for 15 min. The stained tissues were disposed in 1% hydrochloric acid ethanol for color separation for 10s, and then soaked in warm water (50°C) for 5 min. The 0.25% eosin dye solution was added for staining again for 2 min, and the stained tissues were dehydrated by being soaked in gradient concentration of alcohol (70%, 80%, 90%, 95%, 100% ethyl alcohol) for 5 min. At last, the optical microscope was used to observe and photograph (40 ×).
Statistical analysis
Statistical analyses were carried out using R and GraphPad Prism 6.0 (GraphPad Software, California, USA) and all data were presented as means ± standard deviation. Differences between groups were evaluated by Student's t-test and one-way ANOVA. Spearman correlation analysis was applied for linear correlation relationship. Clinical outcomes were estimated by means of Kaplan-Meier analysis and log-rank test. A regression analysis was conducted to prove that the association between SCL6A1 and survival was independent of other clinical variables. CCRCC clinical data were obtained from TCGA database (https://cancergenome.nih.gov/). Log2 (Foldchange) was set at >8 or <-8 with P<0.05, and 18 genes with aberrant expressions were screened out. P < 0.05 signified a statistically significance.
Ethic approval
This study had gotten approval from the Ethics Committee of the First Affiliated Hospital of Xinjiang Medical University and obtained informal written consents from each participant prior to the research.
Supplementary Material
Disclosure of interest
No potential conflict of interest was reported by the authors.
Author contribution
Naibijiang·Maolakuerban and Baihetiya·Azhati contributed to research conception and design as well as manuscript drafting; Hamulati·Tusong analyzed and interpreted data; Asimujiang·Abula and Anniwaer·Yasheng made statistical analysis; Ayiding·Xireyazidan revised the manuscript. In addition, all authors approved final manuscript.
References
- 1.Jonasch E, Futreal PA, Davis IJ, Bailey ST, Kim WY, Brugarolas J, Giaccia AJ, Kurban G, Pause A, Frydman J, et al. . State of the science: an update on renal cell carcinoma. Mol Cancer Res 2012; 10(7): 859–80; PMID:22638109; https://doi.org/10.1158/1541-7786.MCR-12-0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin 2013; 63(1): 11–30; PMID:23335087; https://doi.org/10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 3.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ 2014; 349: g4797; PMID:25385470; https://doi.org/10.1136/bmj.g4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Poppel H Da Pozzo L, Albrecht W, Matveev V, Bono A, Borkowski A, Marechal JM, Klotz L, Skinner E, Keane T, et al. . A prospective randomized EORTC intergroup phase 3 study comparing the complications of elective nephron-sparing surgery and radical nephrectomy for low-stage renal cell carcinoma. Eur Urol 2007; 51(6): 1606–15; PMID:17140723; https://doi.org/10.1016/j.eururo.2006.11.013 [DOI] [PubMed] [Google Scholar]
- 5.Thomas JS, Kabbinavar F. Metastatic clear cell renal cell carcinoma: A review of current therapies and novel immunotherapies. Crit Rev Oncol Hematol 2015; 96(3): 527–33; PMID:26299335; https://doi.org/10.1016/j.critrevonc.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 6.Skolarikos A, Alivizatos G, Laguna P, de la Rosette J. A review on follow-up strategies for renal cell carcinoma after nephrectomy. Eur Urol 2007; 51(6): 1490–500; discussion 501; PMID:17229521; https://doi.org/10.1016/j.eururo.2006.12.031 [DOI] [PubMed] [Google Scholar]
- 7.Fendler A, Jung K. MicroRNAs as new diagnostic and prognostic biomarkers in urological tumors. Crit Rev Oncog 2013; 18(4): 289–302; PMID:23614616; [DOI] [PubMed] [Google Scholar]
- 8.Fendler A, Stephan C, Yousef GM, Jung K. MicroRNAs as regulators of signal transduction in urological tumors. Clin Chem 2011; 57(7): 954–68; PMID:21632885; https://doi.org/10.1373/clinchem.2010.157727 [DOI] [PubMed] [Google Scholar]
- 9.Gao Y, Ma X, Yao Y, Li H, Fan Y, Zhang Y, Zhao C, Wang L, Ma M, Lei Z, et al. . miR-155 regulates the proliferation and invasion of clear cell renal cell carcinoma cells by targeting E2F2. Oncotarget 2016; 7(15): 20324–37; PMID:26967247; https://doi.org/10.18632/oncotarget.7951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wotschofsky Z, Gummlich L, Liep J, Stephan C, Kilic E, Jung K, Billaud JN, Meyer HA. Integrated microRNA and mRNA Signature Associated with the Transition from the Locally Confined to the Metastasized Clear Cell Renal Cell Carcinoma Exemplified by miR-146-5p. PLoS One 2016; 11(2): e0148746; PMID:26859141; https://doi.org/10.1371/journal.pone.0148746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilming Elgaaen B, Olstad OK, Haug KB, Brusletto B, Sandvik L, Staff AC, Gautvik KM, Davidson B. Global miRNA expression analysis of serous and clear cell ovarian carcinomas identifies differentially expressed miRNAs including miR-200c-3p as a prognostic marker. BMC Cancer 2014; 14: 80; PMID:24512620; https://doi.org/10.1186/1471-2407-14-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang JT, Wang F, Chapin W, Huang RS. Identification of MicroRNAs as Breast Cancer Prognosis Markers through the Cancer Genome Atlas. PLoS One 2016; 11(12): e0168284; PMID:27959953; https://doi.org/10.1371/journal.pone.0168284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butz H, Szabo PM, Khella HW, Nofech-Mozes R, Patocs A, Yousef GM. miRNA-target network reveals miR-124as a key miRNA contributing to clear cell renal cell carcinoma aggressive behaviour by targeting CAV1 and FLOT1. Oncotarget 2015; 6(14): 12543–57; PMID:26002553; https://doi.org/10.18632/oncotarget.3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Burgos G. GABA transporter GAT1: a crucial determinant of GABAB receptor activation in cortical circuits? Adv Pharmacol 2010; 58: 175–204; PMID:20655483; https://doi.org/10.1016/S1054-3589(10)58008-6 [DOI] [PubMed] [Google Scholar]
- 15.Carvill GL, McMahon JM, Schneider A, Zemel M, Myers CT, Saykally J, Nguyen J, Robbiano A, Zara F, Specchio N, et al. . Mutations in the GABA Transporter SLC6A1 Cause Epilepsy with Myoclonic-Atonic Seizures. Am J Hum Genet 2015; 96(5): 808–15; PMID:25865495; https://doi.org/10.1016/j.ajhg.2015.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schijns O, Karaca U, Andrade P, de Nijs L, Kusters B, Peeters A, Dings J, Pannek H, Ebner A, Rijkers K, et al. . Hippocampal GABA transporter distribution in patients with temporal lobe epilepsy and hippocampal sclerosis. J Chem Neuroanat 2015; 68: 39–44; PMID:26212582; https://doi.org/10.1016/j.jchemneu.2015.07.004 [DOI] [PubMed] [Google Scholar]
- 17.Curley AA, Eggan SM, Lazarus MS, Huang ZJ, Volk DW, Lewis DA. Role of glutamic acid decarboxylase 67 in regulating cortical parvalbumin and GABA membrane transporter 1 expression: implications for schizophrenia. Neurobiol Dis 2013; 50: 179–86; PMID:23103418; https://doi.org/10.1016/j.nbd.2012.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Januchowski R, Zawierucha P, Andrzejewska M, Rucinski M, Zabel M. Microarray-based detection and expression analysis of ABC and SLC transporters in drug-resistant ovarian cancer cell lines. Biomed Pharmacother 2013; 67(3): 240–5; PMID:23462296; https://doi.org/10.1016/j.biopha.2012.11.011 [DOI] [PubMed] [Google Scholar]
- 19.Kim KR, Oh SY, Park UC, Wang JH, Lee JD, Kweon HJ, Kim SY, Park SH, Choi DK, Kim CG, et al. . [Gene expression profiling using oligonucleotide microarray in atrophic gastritis and intestinal metaplasia]. Korean J Gastroenterol 2007; 49(4): 209–24; PMID:17464166; [PubMed] [Google Scholar]
- 20.Sipila S, Huttu K, Voipio J, Kaila K. GABA uptake via GABA transporter-1 modulates GABAergic transmission in the immature hippocampus. J Neurosci 2004; 24(26): 5877–80; PMID:15229234; https://doi.org/10.1523/JNEUROSCI.1287-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganapathi MK, Jones WD, Sehouli J, Michener CM, Braicu IE, Norris EJ, Biscotti CV, Vaziri SA, Ganapathi RN. Expression profile of COL2A1 and the pseudogene SLC6A10P predicts tumor recurrence in high-grade serous ovarian cancer. Int J Cancer 2016; 138(3): 679–88; PMID:26311224; https://doi.org/10.1002/ijc.29815 [DOI] [PubMed] [Google Scholar]
- 22.Karunakaran S, Umapathy NS, Thangaraju M, Hatanaka T, Itagaki S, Munn DH, Prasad PD, Ganapathy V. Interaction of tryptophan derivatives with SLC6A14 (ATB0,+) reveals the potential of the transporter as a drug target for cancer chemotherapy. Biochem J 2008; 414(3): 343–55; PMID:18522536; https://doi.org/10.1042/BJ20080622 [DOI] [PubMed] [Google Scholar]
- 23.Jung M, Mollenkopf HJ, Grimm C, Wagner I, Albrecht M, Waller T, Pilarsky C, Johannsen M, Stephan C, Lehrach H, et al. . MicroRNA profiling of clear cell renal cell cancer identifies a robust signature to define renal malignancy. J Cell Mol Med 2009; 13(9B): 3918–28; PMID:19228262; https://doi.org/10.1111/j.1582-4934.2009.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakada C, Matsuura K, Tsukamoto Y, Tanigawa M, Yoshimoto T, Narimatsu T, Nguyen LT, Hijiya N, Uchida T, Sato F, et al. . Genome-wide microRNA expression profiling in renal cell carcinoma: significant down-regulation of miR-141 and miR-200c. J Pathol 2008; 216(4): 418–27; PMID:18925646; https://doi.org/10.1002/path.2437 [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Chen L, Jin H, Wang S, Zhang Y, Tang X, Tang G. Screening miRNAs for early diagnosis of colorectal cancer by small RNA deep sequencing and evaluation in a Chinese patient population. Onco Targets Ther 2016; 9: 1159–66; PMID:27022275; https://doi.org/10.2147/OTT.S100427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang L, Huang Z, Zhang H, Zhu M, Zhu W, Zhou X, Liu P. Prognostic value of candidate microRNAs in gastric cancer: A validation study. Cancer Biomark 2017; 18(3): 221–30; PMID:27983528; https://doi.org/10.3233/CBM-160091 [DOI] [PubMed] [Google Scholar]
- 27.Teng Y, Su X, Zhang X, Zhang Y, Li C, Niu W, Liu C, Qu K. miRNA-200a/c as potential biomarker in epithelial ovarian cancer (EOC): evidence based on miRNA meta-signature and clinical investigations. Oncotarget 2016; 7(49): 81621–33; PMID:27835595; https://doi.org/10.18632/oncotarget.13154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Chen X, Wang R, Xiao P, Xu Z, Chen L, Hang W, Ruan A, Yang H, Zhang X. microRNA-200c modulates the epithelial-to-mesenchymal transition in human renal cell carcinoma metastasis. Oncol Rep 2013; 30(2): 643–50; PMID:23754305; https://doi.org/10.3892/or.2013.2530 [DOI] [PubMed] [Google Scholar]
- 29.Yoshino H, Enokida H, Itesako T, Tatarano S, Kinoshita T, Fuse M, Kojima S, Nakagawa M, Seki N. Epithelial-mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Genet 2013; 58(8): 508–16; PMID:23635949; https://doi.org/10.1038/jhg.2013.31 [DOI] [PubMed] [Google Scholar]
- 30.Capri M, Olivieri F, Lanzarini C, Remondini D, Borelli V, Lazzarini R, Graciotti L, Albertini MC, Bellavista E, Santoro A, et al. . Identification of miR-31-5p, miR-141-3p, miR-200c-3p, and GLT1 as human liver aging markers sensitive to donor-recipient age-mismatch in transplants. Aging Cell 2017; 16(2): 262–72; PMID:27995756; https://doi.org/10.1111/acel.12549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mody HR, Hung SW, Naidu K, Lee H, Gilbert CA, Hoang TT, Pathak RK, Manoharan R, Muruganandan S, Govindarajan R. SET contributes to the epithelial-mesenchymal transition of pancreatic cancer. Oncotarget 2017; 8(40): 67966–79; PMID:28978088; https://doi.org/10.18632/oncotarget.19067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu WW, Chen PC, Chen JM, Wu YM, Liu PY, Lu CH, Lin YF, Tang CH, Chao CC. Periostin promotes epithelial-mesenchymal transition via the MAPK/miR-381 axis in lung cancer. Oncotarget 2017; 8(37): 62248–60; PMID:28977942; https://doi.org/10.18632/oncotarget.19273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin S, Im HJ, Kwon YJ, Ye DJ, Baek HS, Kim D, Choi HK, Chun YJ. Human steroid sulfatase induces Wnt/beta-catenin signaling and epithelial-mesenchymal transition by upregulating Twist1 and HIF-1alpha in human prostate and cervical cancer cells. Oncotarget 2017; 8(37): 61604–17; PMID:28977889; https://doi.org/10.18632/oncotarget.18645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F, Wu L, Wang A, Xu Y, Luo X, Liu X, Hua Y, Zhang D, Wu S, Lin T, et al. . MicroRNA-138 attenuates epithelial-to-mesenchymal transition by targeting SOX4 in clear cell renal cell carcinoma. Am J Transl Res 2017; 9(8): 3611–22; PMID:28861152; [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Q, Yang D, Zong H, Zhu L, Wang L, Wang X, Zhu X, Song X, Wang J. Growth-induced stress enhances epithelial-mesenchymal transition induced by IL-6 in clear cell renal cell carcinoma via the Akt/GSK-3beta/beta-catenin signaling pathway. Oncogenesis 2017; 6(8): e375; PMID:28846080; https://doi.org/10.1038/oncsis.2017.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang J, Yi BO, Ding S, Sun J, Cao W, Liu M. Demethylation drug 5-Aza-2'-deoxycytidine-induced upregulation of miR-200c inhibits the migration, invasion and epithelial-mesenchymal transition of clear cell renal cell carcinoma in vitro. Oncol Lett 2016; 11(5): 3167–72; PMID:27123083; https://doi.org/10.3892/ol.2016.4364 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


