Summary
Bacteria of the genus Pseudomonas are widespread in nature. In the last decades, members of this genus, especially Pseudomonas aeruginosa and Pseudomonas putida, have acquired great interest because of their interactions with higher organisms. Pseudomonas aeruginosa is an opportunistic pathogen that colonizes the lung of cystic fibrosis patients, while P. putida is a soil bacterium able to establish a positive interaction with the plant rhizosphere. Members of Pseudomonas genus have a robust metabolism for amino acids and organic acids as well as aromatic compounds; however, these microbes metabolize a very limited number of sugars. Interestingly, they have three‐pronged metabolic system to generate 6‐phosphogluconate from glucose suggesting an adaptation to efficiently consume this sugar. This review focuses on the description of the regulatory network of glucose utilization in Pseudomonas, highlighting the differences between P. putida and P. aeruginosa. Most interestingly, It is highlighted a functional link between glucose assimilation and exotoxin A production in P. aeruginosa. The physiological relevance of this connection remains unclear, and it needs to be established whether a similar relationship is also found in other bacteria.
Introduction
Bacteria of the genus Pseudomonas are ubiquitous inhabitants of soil, water, plant surfaces, animal and human tissue and have a robust metabolism for amino acids and organic acids as well as aromatic compounds (Jiménez et al., 2002; Puchalka et al., 2008; Valerie et al., 2013; Daniels et al., 2010), and the deciphering of the complete genomes of a number of Pseudomonas strains from different species has revealed that these microbes metabolize a very limited number of sugars (Buell et al., 2003; Feil et al., 2005; Joardar et al., 2005), which are mainly glucose, glucuronic acid and fructose (Daniels et al., 2010). This metabolic pattern has been associated with their lifestyle as they inhabit environmental niches characterized by a limited presence of sugars (Silby et al., 2011).
Studies in Pseudomonas putida have shown that there is a three‐pronged metabolic system to generate 6‐phosphogluconate from glucose (Del Castillo et al., 2007; Fig. 1). Glucose enters through the OprB porin into the periplasmic space. Once in the periplasm, glucose can be either transported to the cytosol or converted into gluconate and 2‐ketogluconate (KG) to be subsequently transferred to the cytosol via different transporters (GntP and KguT respectively). Gluconate can be phosphorylated to 6‐phosphogluconate by gluconokinase (GnuK), whereas the conversion of 2‐ketogluconate into 6‐phosphogluconate requires two enzymatic reactions mediated by KguK and KguD (Fig. 1); whereby, it starts the Entner–Doudoroff pathway (Nikel et al., 2015).
Figure 1.
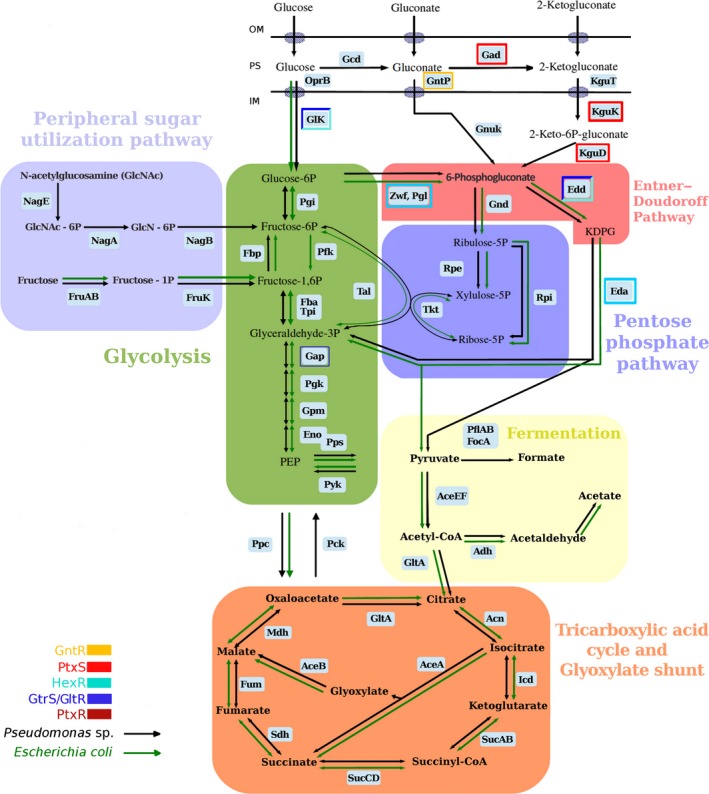
Schematic representation of the glucose metabolism in Pseudomonas and Escherichia coli as deduced from gene annotations and functional analysis in the wild‐type strain. Genes whose expression is controlled by the regulators described in this review are boxed in different colours.
In addition, the growing number of complete genome sequences of Pseudomonas strains in public databases (Jayal et al., 2017; Nesme et al., 2017; Wilson et al., 2017) along with the increasing sophistication of the techniques used in metabolomics and transcriptomics (La Rosa et al., 2015; Nikel et al., 2015) has provided us with key information for a further understanding of the complex regulation processes and functionality of the enzymes that participate in carbohydrate metabolism in Pseudomonas. The genes encoding the carbohydrate catabolic pathways are organized in operons (Fig. 2), which are under the control of different regulators that respond differentially to distinct pathway intermediates, suggesting a hierarchy in the control of glucose metabolism related to a tight gene expression (Rojo, 2010; Daddaoua et al., 2014; Figs 2 and 3).
Figure 2.
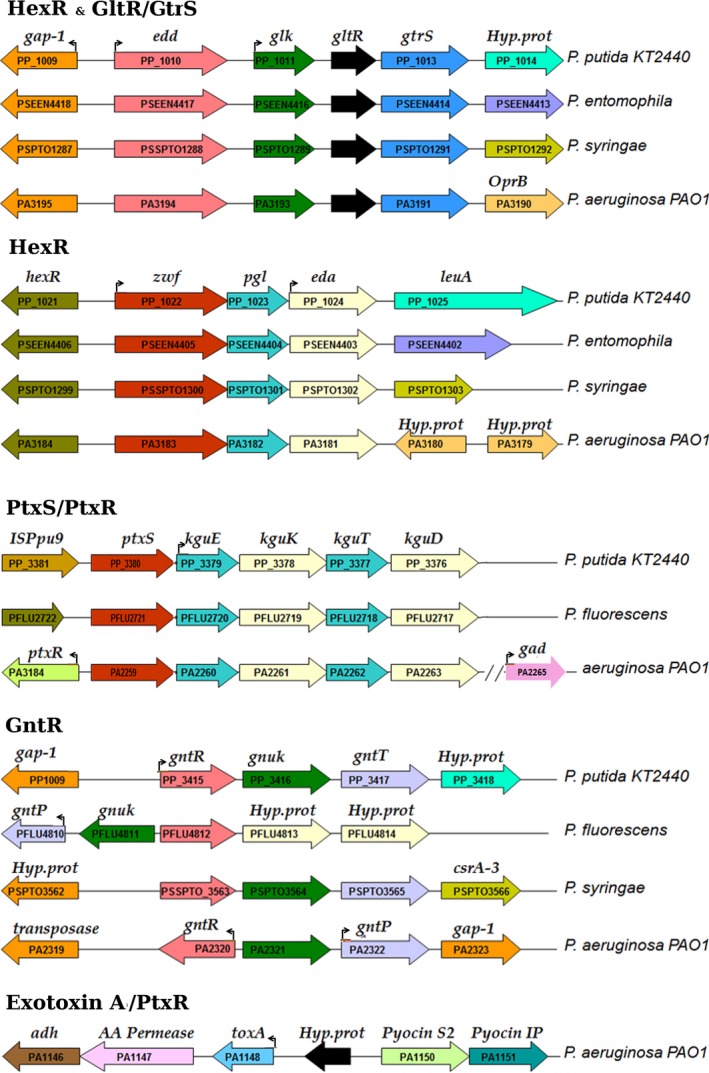
Genetic organization of genes encoding enzymes of carbohydrate degradation pathways and exotoxin A in different Pseudomonas strains. The genes that were found to be regulated are boxed, and the corresponding regulator y system is provided over each block of genes.
Figure 3.
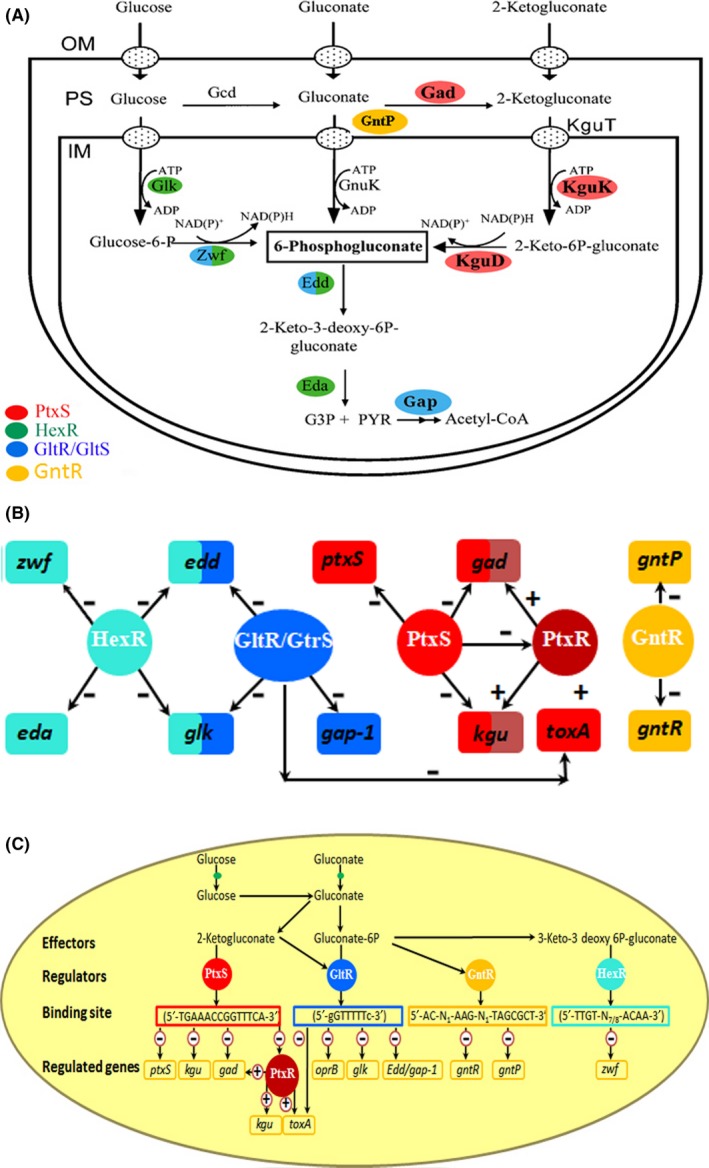
Schematic view of the concerted regulation of gene expression involved in glucose metabolism.
A. Schematic view of the three internalization routes.
B. The functional interconnectivity of the five regulatory mechanisms.
C. Summary of information flow from effectors to regulated genes. The − and + symbols indicate repression or activation of gene expression respectively.
Transcriptional regulation is the primary mechanism to control gene expression in prokaryotic cells (Ishihama, 2000). Typically, transcriptional regulators sense certain environmental cues and the resulting molecular stimulus modulates their interactions with RNA polymerases or DNA. In one‐component regulatory system (OCS), the input (i.e. sensing) and output functions are united in a single protein. In two‐component system (TCS), a membrane‐bound histidine kinase is dedicated to signal sensing, whereas the response regulator protein mediates a transcriptional response (Mitrophanov and Groisman, 2008). TCS represents the major regulatory mechanism in bacteria and archaea and is responsible for the transformation of external and internal stimuli into adaptive responses, including regulation of gene expression and methylation of target proteins (Skerker et al., 2005; Mitrophanov and Groisman, 2008). In a typical ligand‐induced TCS, changes in the autokinase activity of the sensor kinase modulate the rate of transphosphorylation to its cognate response regulator, which in turn defines the system output. In addition, there is evidence for TCS that contains additional signalling proteins (Mitchell et al., 2015; Popella et al., 2016).
Data so far available indicate that the regulation of the glucose catabolic pathways in Pseudomonas is controlled by the concerted action of the one‐component systems (OCSs) HexR, PtxS, PtxR and GntR as well as the two‐component system (TCS) GltR/GtrS (Daddaoua et al., 2009, 2012, 2014, 2017). This review focuses on the description of this regulatory network and highlights a number of differences in regulation between strains of the soil bacterium P. putida and the opportunistic human pathogen Pseudomonas aeruginosa. Glucose metabolism in Pseudomonads is fundamentally different to that in Escherichia coli (Lendenmann et al., 1996; Fuhrer et al., 2005; Fig. 1), and the current knowledge on its regulation is reviewed here.
Carbohydrate catabolic pathways in Pseudomonas
Metabolic flux analysis of wild‐type Pseudomonas and different mutant strains has permitted to estimate the carbon flow through the individual three peripheral uptake routes. These data revealed that the flow through the gluconate kinase (GnuK) route is minor, whereas the remaining flux appears to be similarly distributed to the routes that involve the phosphorylation of either glucose or 2‐ketogluconate (Del Castillo et al., 2007). Carbohydrates enter the periplasmic space through porins located in the outer membrane (OprB1/OprB2) (Figs 1 and 3A). Once in the periplasm, glucose can be oxidized to gluconate through the action of a glucose dehydrogenase (Gcd), and gluconate is transported to the cytoplasm and phosphorylated to 6‐phosphogluconate (6PG) by gluconate kinase (GnuK). Alternatively, gluconate, still in the periplasm, can be further oxidized to 2‐ketogluconate (2KG) by the action of gluconate dehydrogenase (Gad); then, 2KG enters the cytoplasm and is converted into 6PG via 2‐keto‐6‐phosphogluconate by the action of the 2‐ketogluconate kinase (KguK) and 2‐ketogluconate‐6‐phosphate reductase (KguD). Glucose can also be transported directly to the cytoplasm through an ABC uptake system, and the first acting enzyme is glucokinase (Glk) that phosphorylates glucose to give glucose 6‐phosphate (G6P). Next, the combined action of glucose 6‐phosphate dehydrogenase (Zwf) and 6‐phosphogluconolactonase (Pgl) convert G6P into 6‐phosphogluconate (6PG) (Del Castillo et al., 2007). The produced 6PG enters to the Entner–Doudoroff route and is converted into 2‐keto‐3‐deoxy‐6‐phosphogluconate (KDPG) by the action of the 6‐phosphogluconate dehydratase (Edd) and then hydrolysed to produce glyceraldehyde‐3‐phosphate and pyruvate by the action of the 2‐keto‐3‐deoxy‐6‐phosphogluconate aldolase (Eda) (Figs 1 and 3A; Braga et al., 2004). Glyceraldehyde‐3‐phosphate is further metabolized by the glyceraldehyde‐3‐phosphate dehydrogenase (Gap‐1) to d‐glycerate 1,3‐bisphosphate, while pyruvate is decarboxylated to acetyl‐coenzyme A (Acetyl‐CoA) and enters the Krebs cycle (Fig. 1).
Genomic distribution of genes involved in Pseudomonas carbohydrate catabolism
The analysis of the genomic localization of genes involved in glucose catabolism (Fig. 2) revealed that genes are arranged in operons that encode for different sets of catabolic enzymes, some transcriptional regulators as well as specific porins. Interestingly, the two genes that encode the key enzymes of the Entner–Doudoroff pathway, edd and eda, are located in different operons. The edd and glk genes (phosphorylative branch) form part of the same operon together with the genes encoding the regulatory proteins GltR, GtrS and the gap‐1 gene (Fig. 2). The eda gene forms an operon with the zwf and pgl genes (phosphorylative branch) as well as the gene encoding the regulatory protein HexR (Fig. 2). This genomic organization (edd/glk and zwf/pgl/eda containing operons) suggests a complex regulation, because the main route of glucose metabolism in Pseudomonads occurs through the 2KG degradation pathway, which, however, are encoded by kguT, kguK and kguD that form another operon with ptxS, encoding a transcriptional regulator (Fig. 2). The fact that the genes that control the expression of Edd and Eda proteins are located in the same operons as the genes involved in the glucose phosphorylative pathway, which is not the main glucose degradative pathway in Pseudomonas, can be a reminiscence of an ancestral organism and explains why this pathway is still active in Pseudomonas; both edd and eda genes are necessary for the 6PG conversion into tricarboxylic acid (TCA) intermediates, regardless of the peripheral pathway by which glucose has been converted into 6PG.
Transcriptional regulators involved in Pseudomonas carbohydrate degradation pathway
In E. coli, 300 types of transcription factors have been identified and about 10% of them are TCSs (Yamamoto et al., 2005; Ishihama et al., 2014). In bacteria, extracellular signals are recognized predominantly by TCSs (Hoch, 2000; Stock et al., 2000; Mascher, 2006; Gao et al., 2007; Lacal et al., 2010) which typically recognize signals at extracytoplasmic ligand binding domains.
Usually, promoters that control the expression of genes that encode proteins implicated in the synthesis of cell structures, such as flagella, pili and fimbriae, or those involved in complex cellular processes, that is, virulence or biofilm formation, are often controlled by multiple environmental signals that are recognized by multiple transcription factors (Dalebroux et al., 2010; Rasamiravaka et al., 2015; ).
Data currently available indicate that the control of glucose metabolism in Pseudomonas is controlled by the concerted action of the HexR, PtxS, PtxR and GntR, which are OCSs, whereas and the GltR/GtrS is TCS. The transcriptional regulation of glucose degradation has been studied in Pseudomonas putida KT2440 and P. aeruginosa PAO1, and the current knowledge is summarized in Figure 1.
OCS involved in carbohydrate catabolism pathways
HexR
The HexR regulator belongs to the RpiR family of transcriptional regulators whose members typically act as transcriptional regulators in sugar catabolism and have been identified as both repressors and activators in Gram‐negative and Gram‐positive bacteria (Yamamoto et al., 2001). In E. coli, RpiR negatively regulates the expression of the rpiB gene that encodes a ribose 5‐phosphate isomerase, an enzyme that catalyses the reversible reaction of ribose 5‐phosphate to ribulose 5‐phosphate and forms part of the pentose phosphate pathway (Fig. 1; Sorensen and Hove‐Jensen, 1996). Sugar‐responsive RpiR proteins form dimers in solution and have an N‐terminal helix–turn–helix (HTH) DNA‐binding motif and a sugar isomerase‐like binding (SIS) domain at their C‐terminal extension (Bateman, 1999).
In Pseudomonas, HexR regulator is divergently transcribed from the zwf/pgl/eda operon, and the physical organization of these genes is highly conserved within Pseudomonas, which suggests the conservation of the identified regulatory mechanism (Fig. 2). HexR regulator controls the zwf/pgl/eda and edd/glk/gltR‐2 operons as well as the gap‐1 gene (Daddaoua et al., 2009; Fig. 3 and Table 2) and is also required for the metabolism of other sugars such as fructose and gluconate (Fig. 1). The HexR regulator exerts its regulatory action by binding to specific sequences in the target promoters (Figs 3 and 4 and Table 1), which in turn prevents the progress of RNA polymerase. HexR recognizes 2‐keto‐3‐deoxy‐6‐phosphogluconate (KDPG), an intermediate of the Entner–Doudoroff pathway (Table 2), and is required not only for glucose catabolism but also for the metabolism of other sugars such as fructose and gluconate (Fig. 1). HexR acts as a transcriptional repressor in the absence of a specific effector; but binding of KDPG to DNA‐bound HexR causes protein dissociation and transcriptional activation (Fig. 4). Furthermore, it has been speculated that this is not a ‘random genetic organization’ as KDPG plays a relevant role as a signalling molecule in catabolite repression and in the response to oxidative stress in Pseudomonas (Daddaoua et al., 2009; Rojo, 2010).
Table 2.
Specific regulator binding site involved in Pseudomonas carbohydrate catabolic pathways
| Promoter | Regulator | Effector | Sequence of operator site | Position of operator | K D | Reference |
|---|---|---|---|---|---|---|
| zwf | HexR | KDPG | 5′‐TTGT‐N7/8‐ACAA‐3′ | +30; +1 | 780 ± 40 nM | Daddaoua et al. (2009) |
| edd | HexR | KDPG | 5′‐TTGT‐N7/8‐ACAA‐3′ | +16; +41 | 774 ± 80 nM | Daddaoua et al. (2009) |
| gap‐1 | HexR | KDPG | 5′‐TTGT‐N7/8‐ACAA‐3′ | −6; −18 | 480 ± 50 nM | Daddaoua et al. (2009) |
| ptxS | PtxS | 2 ketogluconate | 5′‐TGAAACCGGTTTCA‐3′ | +5; −9 | ND | Daddaoua et al. (2010, 2012) |
| kgu | PtxS | 2 ketogluconate | 5′‐TGAAACCGGTTTCA‐3′ | +5; +18 | ND | Daddaoua et al. (2010, 2012) |
| gad | PtxS | 2 ketogluconate | 5′‐TGAAACCGGTTTCA‐3′ | +10; +23 | ND | Daddaoua et al. (2010, 2012) |
| toxA | PtxS | 2 ketogluconate | 5′‐TGAAACCGGTTTCA‐3′ | ND | ND | Daddaoua et al. (2010, 2012) |
| gad | PtxR | Unknown | 5′‐CGGCGCGCCCG‐3′ | −32; −41 | ND | Daddaoua et al. (2010, 2012, 2013) |
| kgu | PtxR | Unknown | 5′‐CGGCGCGCCCG‐3′ | −34; −43 | ND | Daddaoua et al. (2010, 2012, 2013) |
| toxA | PtxR | Unknown | 5′‐CGGCGCGCCCG‐3′ | −42; −51 | ND | Daddaoua et al. (2010, 2012, 2013) |
| oprB | GltR/GtrS | 2‐ketogluconate and Gluconate | 5′‐gGTTTTTc ‐3′ | −10; −21 | ND | Daddaoua et al. (2014) |
| glk | GltR/GtrS | 2‐ketogluconate and Gluconate | 5′‐gGTTTTTc ‐3′ | +22; +33 | ND | Daddaoua et al. (2014) |
| edd | GltR/GtrS | 2‐ketogluconate and Gluconate | 5′‐gGTTTTTc ‐3′ | −4; +9 | ND | Daddaoua et al. (2014) |
| gap‐1 | GltR/GtrS | 2‐ketogluconate and Gluconate | 5′‐gGTTTTTc ‐3′ | −12; +2 | ND | Daddaoua et al. (2014) |
| toxA | GltR/GtrS | 2‐ketogluconate and Gluconate | 5′‐gGTTTTTc ‐3′ | +177; +190 | ND | Daddaoua et al. (2014) |
| gntR | GntR | Gluconate and 6‐phosphogluconate | 5′‐AC‐N1‐AAG‐N1‐TAGCGCT‐3′ | −4, −17 | ≈1 mM | Daddaoua et al. (2017) |
| gntP | GntR | Gluconate and 6‐phosphogluconate | 5′‐AC‐N1‐AAG‐N1‐TAGCGCT‐3′ | −4, −17 | ≈1 mM | Daddaoua et al. (2017) |
Figure 4.
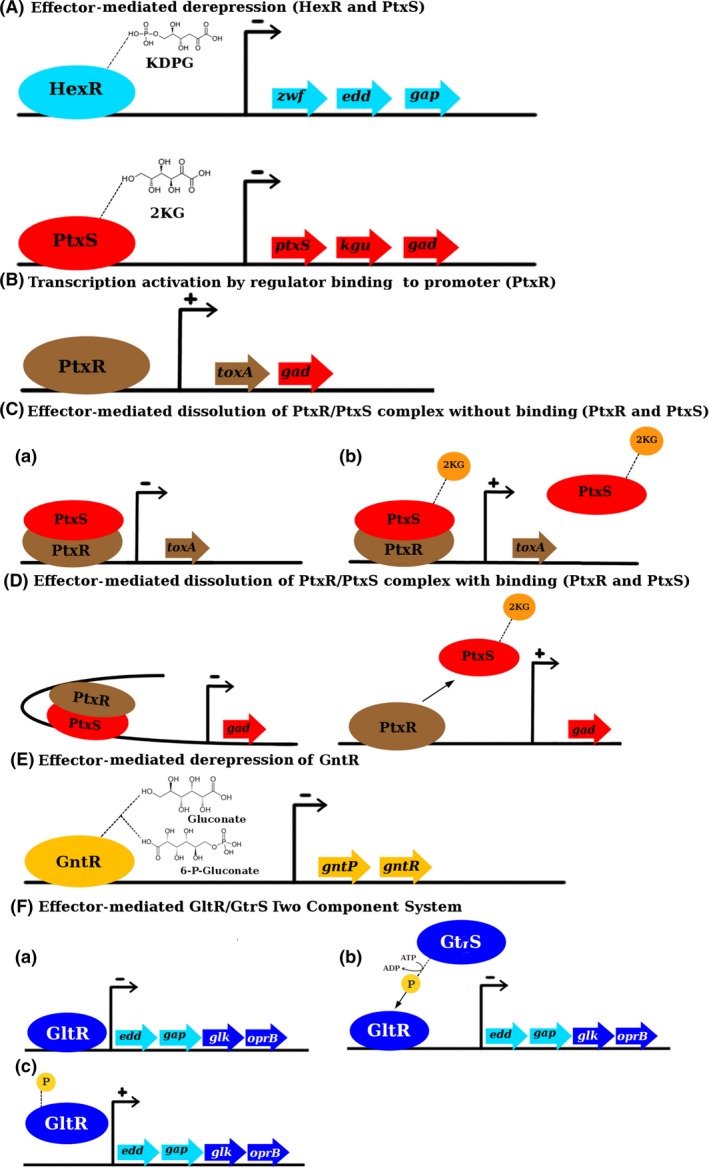
The mechanisms’ action of transcriptional regulators involved in the regulation of carbohydrate catabolism pathways. The − and + symbols indicate repression or activation of gene expression respectively.
A. Effector‐mediated derepression (HexR and PtxS).
B. Transcription activation by regulator binding to promotor (PtxR).
C. Effector‐mediated dissolution PtxR/PtxS complex without binding (PtxR and PtxS).
D. Effector‐mediated dissolution of PtxR/PtxS complex with binding (PtxR and PtxS).
E. Effector‐mediated derepression of GntR.
F. Effector‐mediated GltR/GtrS two component system.
Table 1.
Regulatory proteins involved in the transcriptional control of Pseudomonas carbohydrate catabolic pathways
| Regulator | Activation of genes | Repression of genes | Effector | Reference |
|---|---|---|---|---|
| HexR | None | edd, gap‐1, zwf | KDPG | Daddaoua et al. (2009) and Del Castillo et al. (2007) |
| PtxS | None | ptxS, kgu, gad, ptxR, toxA | 2‐ketogluconate | Hamood et al. (1996) and Daddaoua et al. (2010, 2012) |
| PtxR | toxA | None | Unknown | Hammod et al. (1996) and Daddaoua et al. (2012, 2013) |
| GltR/GtrS | edd, gap‐1, glk, oprB, toxA | None | 6‐phosphogluconate and 2‐ketogluconate | Daddaoua et al. (2014) |
| GntR | None | gntR, gntP | Gluconate and 6‐phosphogluconate | Daddaoua et al. (2017), Izu et al. (1997), and Peekhaus and Conway (1998) |
Homology modelling of HexR using the structure of the YbbH transcriptional regulator from Bacillus subtilis (PDB: 2O3F) as a template (36% sequence identity) revealed that HexR is composed of two distinct domains, namely an N‐terminal HTH containing DNA‐binding domain (residues 20‐126) and a C‐terminal domain (residues 117‐256) that is homologous to the phosphosugar isomerase domain of the RpiR family. Furthermore, the inspection of this model suggested that the amino acids involved in DNA recognition are Gln‐43, Lys‐46, Glu‐49, Arg‐54 and Arg‐57, and the amino acids Arg‐54 and Arg‐57 may be directly interacting with DNA (Fig. 5; Daddaoua et al., 2009).
Figure 5.
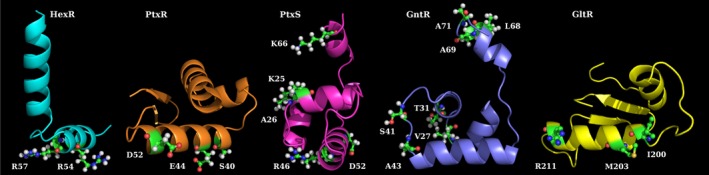
Homology models of NH 2 or COOH‐terminal extensions corresponding to the DNA‐binding domains of regulators involved in carbohydrate catabolism pathways. The amino acids involved in the interaction of the regulator with DNA are highlighted.
Finally, the HexR operator sites have been identified in the zwf, edd and gap‐1 promoters of P. putida KT2440, what permitted the definition of the consensus sequence 5′‐TTGT‐N7/8‐ACAA‐3′, which likely corresponds to the specific HexR binding motif (Daddaoua et al., 2009; Table 2 and Fig. 3).
PtxS and PtxR
In Pseudomonas, the ptxS gene is located next to the kguE/kguK/kguT/and/kguD genes (Fig. 2). Similarly, to other members of the well‐characterized LacI family, PtxS has an N‐terminal DNA‐binding domain and a C‐terminal effector binding domain. In several species of the genus Pseudomonas, the role of PtxS in the control of the gluconate degradation pathway has been elucidated (Daddaoua et al., 2012; Suresh et al., 2014). In P. aeruginosa PAO1, PtxS binds to the palindromic sequence 5′‐TGAAACCGGTTTCA‐3′ (Fig. 3 and Table 2) located near the ‐10 region repressing the expression of the corresponding gene (Table 1; Daddaoua et al., 2010). The three‐dimensional model of the PtxS N‐terminal domain (PF00356) was generated with I‐TASSER server (C‐score: 0.74) based on the transcriptional regulator CcpA (Schumacher et al., 2006, 2011; PDB: 1ZVV and 3OQMA) that shares 26% sequence identity with PtxS. The model suggested that amino acids Lys‐25, Ala‐26, Arg‐46, Asp‐52 and Lys‐66 are important for DNA recognition (Yang et al., 2015; Fig. 5).
In P. putida, KT2440 PtxS controls its own expression as well as that of the operons gad and kgu (Daddaoua et al., 2012). However, in P. aeruginosa PAO1, PtxS regulates, in addition (Figs 1 and 3), the expression of the toxA gene that encodes the exotoxin A, a primary virulence factor (Daddaoua et al., 2012; Fig. 3). This protein is an ADP‐ribosyl transferase that irreversibly inhibits protein synthesis in eukaryotic cells causing cell death.
In P. aeruginosa, but not in other Pseudomonas, an additional transcriptional regulator ptxR is located within the kgu cluster and transcribed divergently from the ptxS gene (Fig. 2). PtxR belongs to the LysR‐type of transcriptional regulators and does not share any significant sequence similarities with PtxS (13% sequence identity in an alignment with seven gaps). PtxR is also involved in the regulation of toxA expression (Hamood et al., 1996) which encodes an ADP‐ribosyl transferase that irreversibly inhibits protein synthesis in eukaryotic cells causing cell death. It has been shown that endotoxin A production is triggered by certain environmental conditions (such as cation concentration, iron and oxygen levels or temperature) and is controlled by different regulators, and the involvement of RegA, PtxR and the iron‐starvation alternative sigma factor PvdS in this complex regulatory process has been documented (Wick et al., 1990; Hamood et al., 2004).
PtxR is also involved in the regulation of several genes of the glucose degradation pathway (Daddaoua et al., 2012) and recognizes a pseudopalindrome with a consensus sequence of 5′‐ GGC‐N4‐6‐GCC ‐3′ (Fig. 3 and Table 2) which overlapped with the RNA polymerase binding site of PtoxA, Pkgu and Pgad promoters. PtxR has a DNA‐binding HTH domain (in the N‐terminal region) and has a signal receptor domain at its C‐terminal extension that is composed of two subdomains: one is responsible for inducer recognition, whereas the other is involved in the response (Maddocks and Oyston, 2008). A three‐dimensional homology model of PtxR was built using, the LysR family protein member, CrgA of Neisseria meningitidis (PDB: 3 hhg) (Sainsbury et al., 2009) as a template. The in silico analysis of the model together with isothermal titration calorimetry (ITC) assays using PtxR mutants revealed that residues Ser‐40, Glu‐44 and Asp‐52 in the HTH are involved in interaction with the target DNA (Fig. 5).
In P. aeruginosa, PtxS regulates the expression of ptxR; therefore, it is involved in the indirect control of the production of exotoxin A (Fig. 3). PtxS does not directly bind to the toxA promoter; instead, ITC analyses demonstrated that PtxS forms a tight complex with PtxR, either in its free form or when bound to the PtoxA DNA (Colmer and Hamood, 1998; Daddaoua et al., 2012; ). The binding of PtxS to DNA‐bound PtxR prevents PtxR from activating transcription (Fig. 4). The binding of 2‐ketogluconate to PtxS causes its dissociation from PtxR allowing the activation of transcription (Daddaoua et al., 2012). It could be speculated that PtxR might be responsible for the recruitment of RNA polymerase allowing transcription.
In contrast to the mechanism by which PtxR and PtxS control the expression of PtoxA , both regulators bind to Pkgu and Pgad. As both proteins interact with each other and as their operator sites are separated by around 50 bp, it has been shown that the interaction between these two DNA‐bound provokes the formation of a DNA loop (Daddaoua et al., 2013). It has been suggested that this loop structure prevents the RNA polymerase to access the promoter (Huo et al., 2009).
The binding of 2‐ketogluconate to PtxS breaks the loop permitting RNA polymerase recruitment for the transcription of the genes involved in 2‐ketogluconate catabolism (Fig. 4; Daddaoua et al., 2012, 2013). Therefore, it seems that there are two different control mechanisms exerted by the same regulator, in the PtoxA promoter via PtxS/PtxR/DNA complex formation and in the case of Pkgu and Pgad promoters by forming a DNA/PtxS/PtxR/DNA loop structure (Fig. 4).
GntR
As has been mentioned by Daddaoua et al. (2017), the inspection of the genetic context of genes involved in glucose metabolism in P. aeruginosa resulted in the detection of a GntR‐like transcriptional regulator (Jain, 2015), that is predicted to possess an N‐terminal HTH DNA‐binding motif and a periplasmic binding protein‐like domain for effector binding (Daddaoua et al., 2017). In P. aeruginosa, it was found that the gntR gene is transcribed divergently to the gnuK gene (gluconokinase), which is located adjacent to those of the gluconate transporter (GntP) and a glyceraldehyde‐3‐phosphate dehydrogenase (GapN) (Fig. 2) It has been proposed that GntR regulates its own expression, as well as that of a gluconokinase (GnuK), gluconate permease (GntP) and gluconate 6‐phosphate dehydrogenase (GntZ) gene (Daddaoua et al., 2017; Del Castillo et al., 2007).
However, the GntR homologue of P. aeruginosa shared only modest sequence identities (11–37%) with characterized paralogues in Corynebacterium glutamicum (Frunzke et al., 2008), Sinorhizobium meliloti (Steele et al., 2009) and Vibrio cholerae (Roy et al., 2016). Recent data confirm that GntR represses its own expression as well as that of the GntP gluconate permease (Table 1). In contrast to PtxS and GtrS/GltR, GntR did not modulate expression of the toxA gene encoding the P. aeruginosa exotoxin A virulence factor. GntR bound to promoters PgntR and PgntP, and the consensus sequence of its operator was defined as 5′‐AC‐N1‐AAG‐N1‐TAGCGCT‐3′ (Table 2). Both operator sites overlapped with the RNA polymerase binding site. GntR employs an effector‐mediated derepression mechanism (Fig. 4) because the release of promoter‐bound GntR is induced by gluconate and 6‐phosphogluconate that bind with similar apparent affinities to the GntR/DNA complex (Table 2). Surprisingly, GntR and PtxS are paralogous which may have evolved from a common ancestor (Daddaoua et al., 2017).
The three‐dimensional model of the GntR N‐terminal domain was generated with the I‐TASSER server (C‐score: 0.48) based on the transcriptional regulator from Bacillus subtilis (PDB: 1ZVV) which had 25% of identity with GntR (Schumacher et al., 2006). The analysis of the model suggested that the amino acids Val‐27, Tyr‐31, Ser‐41, Ala‐43, Leu‐68, Ala‐69 and Ala‐71 were important for the recognition of its target DNA (Fig. 5).
Role of the GltR/GtrS (TCS) in the regulation of the carbohydrate catabolism
The TCS GtrS/GltR was also found to participate in the transcriptional regulation of glucose catabolism. Different research groups have provided initial information on the role GtrS and GltR, but they did not release that they do form a TCS. Whereas GltR was identified to be essential for efficient glucose transport (Sage et al., 1996), GtrS was found to be important for optimal host colonization and dissemination in a mouse infection model by modulating type III secretion in response to host cells (O'Callaghan et al., 2012). However, Daddaoua et al. (2014) demonstrated that the GltR and GtrS form indeed a TCS. GtrS is a transmembrane sensor kinase that contains a periplasmic ligand binding domain. Efficient GtrS autophosphorylation and transphosphorylation to the GltR response regulator have been observed. GtrS recognizes specifically 2‐ketogluconate and 6‐phosphogluconate (Table 1), causing a modulation of its autokinase activity, leading in turn to changes in GltR transphosphorylation activity (Fig. 4).
GltR interacts with different promoters regulating the expression of the oprB, glk, edd and gap‐1 genes (Fig. 3). Most interestingly, GltR also binds to the PtoxA promoter regulating toxA expression, underlining the interconnectivity of regulatory mechanisms for glucose metabolism and exotoxin A expression in P. aeruginosa (Daddaoua et al., 2014). GltR acts as a transcriptional repressor that is released from DNA upon phosphorylation and the consensus sequence for GltR was determined to be 5′‐tgGTTTTTc‐3′ (Table 1 and 2; Daddaoua et al., 2014).
The inspection of the homology model generated using OMPR/phoB type DNA‐binding domain (amino acids 134‐234) PDB: 2OQR) (King‐Scott et al., 2007) as template (Fig. 6) suggests that residues Ile‐200, Met‐203 and Arg‐211 are involved in recognition of its DNA‐binding site (Fig. 5).
Figure 6.
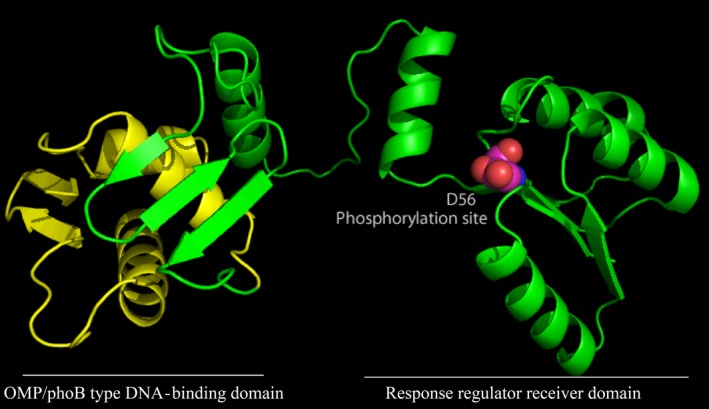
Homology model of the GltR regulator involved in the regulation of carbohydrate catabolism pathways and exotoxin A expression. The model has been generated using the I‐TASSER server software using the RegX3 from Mycobacterium tuberculosis structure as a template (PDB: 2OQR). The phosphoryl group accepting aspartate residue (D56) in the receiver domain is highlighted. The green part of the structure indicates the response regulator receiver domain, and the yellow part indicates the OMP/phoB type DNA‐binding domain.
This review summarizes the current knowledge on the specific regulatory circuits that govern glucose metabolism in Pseudomonads. However, there is also evidence for global regulation and interconnection with other processes, although the precise mechanisms have not been elucidated yet. Available information suggests the ketogluconate branch plays a role in the control virulence; and An and Moe (2016) showed that Gcd levels varied significantly with the carbon source used by Pseudomonas, showing that expression in glucose was higher than in glycerol, LB and citrate, which was consistent with the requirements in glucose dehydrogenase activity. These authors also showed that gcd expression was downregulated by inorganic phosphate, a demonstration of interconnection among metabolism of different nutrients.
Concluding remarks
Electrophoresis mobility shift assay, footprinting, isothermal titration calorimetry, classical expression assays and computer modelling analysis allowed to gain insight into the molecular mechanisms that govern carbohydrate degradation pathways in Pseudomonas. These analyses have answered a number of questions that emerged from the early biochemical studies. This review has focussed in the latest advances in the regulatory mechanism rather than in the carbohydrate metabolism by itself. The complexity of the glucose degradation in Pseudomonas is given by the fine regulation of glucose fluxes among three different convergent pathways and how these pathways are coordinately expressed.
Pseudomonas aeruginosa is among the most feared human pathogens. Importantly, the transcriptional regulation of glucose metabolism in P. aeruginosa is intimately linked to bacterial virulence. So far, five specific regulatory systems have been shown to modulate glucose metabolism and transport (HexR, PtxS, PtxR, GtrS/GltR and GntR), of which two, PtxS and GtrS/GltR, were found to regulate, directly or through PtxR, the expression of toxA, encoding the primary virulence factor; exotoxin A. There is thus a functional link between glucose assimilation and exotoxin A production in P. aeruginosa. The physiological relevance of this connection remains unclear, and it needs to be established whether a similar relationship is also found in other bacteria.
In general, the effector molecules of most signal transduction systems are unknown. However, the effectors for four of the five systems have been established. In all cases, these effectors were intermediates of the glucose metabolism, whereas glucose itself is not recognized by any of the sensor proteins. 2KG and 6‐phosphogluconate play central roles as they are recognized by two different sensor proteins. In the case of 6‐phosphogluconate, this role may be related to the fact that all three glucose metabolism pathways converge in this metabolite. The importance of 2KG as an effector molecule may suggest that the corresponding metabolic route is of particular relevance. The structural basis for their recognition is different as, for example, 2KG is recognized by a periplasmic binding protein type of sensor domain (Pfam00532) at PtxS, whereas the GtrS sensor domain remains unannotated.
Another interesting feature is the cellular compartment at which the signals are of sensed. Whereas 2KG and 6‐phosphogluconate are sensed in the cytosol by PtxS and GntR, respectively, both ligands are sensed by GtrS which contains a periplasmic sensor domain. TCS represents a genetic and metabolic burden as compared to OCS, but its capacity to sense ligands in the extracytosolic space is considered as a major advantage over OCSs. In the present case, a system based on the dual sensing of the same effector in different cellular compartments has evolved, which may suggest that sampling information on intermediate concentrations in both cell compartments is advantageous.
Taken together, the molecular mechanism of transcriptional regulation of carbohydrate metabolism in P. aeruginosa and P. putida can be considered a model system to understand complex regulatory processes in bacteria.
Conflict of interest
None declared.
Acknowledgements
This work was supported by a grant from the Spanish Ministry for Economy and Competitiveness (BIO2016‐76779‐P).
Microbial Biotechnology (2018) 11(3), 442–454
Funding Information
This work was supported by a grant from the Spanish Ministry for Economy and Competitiveness (BIO2016‐76779‐P).
References
- An, R. , and Moe, L.E. (2016) Regulation of pyrroloquinoline quinone‐dependent glucose dehydrogenase activity in the model rhizosphere‐dwelling bacterium Pseudomonas putida KT2440. Appl Environ Microbiol 82: 4955–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A. (1999) The SIS domain: a phosphosugar‐binding domain. Trends Biochem Sci 24: 94–95. [DOI] [PubMed] [Google Scholar]
- Braga, R. , Hecquet, L. , and Blonski, C. (2004) Slow‐binding inhibition of 2‐keto‐3 deoxy‐6phosphogluconate (KDPG) aldolase. Bioorg Med Chem 12: 2965–2972. [DOI] [PubMed] [Google Scholar]
- Buell, C.R.V. , Joardar, M. , Lindeberg, J. , Selengut, I. , Paulsen, T. , et al (2003) The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. Tomato DC3000. Proc Natl Acad Sci USA 100: 10181–10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer, J.A. , and Hamood, A.N. (1998) Characterization of ptxS, a Pseudomonas aeruginosa gene which interferes with the effect of the exotoxin A positive regulatory gene, ptxR . Mol Gen Genet 258: 250–259. [DOI] [PubMed] [Google Scholar]
- Daddaoua, A. , Krell, T. , and Ramos, J.L. (2009) Regulation of glucose metabolism in Pseudomonas: the phosphorylative branch and Entner‐Doudoroff enzymes are regulated by a repressor containing a sugar isomerise domain. J Biol Chem 284: 21360–21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua, A. , Krell, T. , Alfonso, C. , Morel, B. , and Ramos, J.L. (2010) Compartmentalized glucose metabolism in Pseudomonas putida is controlled by the PtxS repressor. J Bacteriol 192: 4357–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua, A. , Fillet, S. , Fernandez, M. , Udaondo, Z. , Krell, T. , and Ramos, J.L. (2012) Genes for carbon metabolism and the ToxA virulence factor in Pseudomonas aeruginosa are regulated through molecular interactions of PtxR and PtxS. PLoS ONE 7: e39390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua, A. , Krell, T. , and Ramos, J.L. (2013) Transcriptional control by two interacting regulatory proteins: identification of the PtxS binding site at PtxR. Nucleic Acids Res 41: 10150–10156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua, A. , Molina‐Santiago, C. , de la Torre, J. , Krell, T. , and Ramos, J.L. (2014) GtrS and GltR form a two‐component system: the central role of 2‐ketogluconate in the expression of exotoxin A and glucose catabolic enzymes in Pseudomonas aeruginosa . Nucleic Acids Res 42: 7654–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daddaoua, A. , Corral‐Lugo, A. , Ramos, J.L. , and Krell, T. (2017) Identification of GntR as regulator of the glucose metabolism in Pseudomonas aeruginosa . Environ Microbiol 19: 3721–3733. [DOI] [PubMed] [Google Scholar]
- Dalebroux, Z.D. , Svensson, S.L. , Gaynor, E.C. , and Swanson, M.S. (2010) ppGpp Conjures bacterial virulence. Microbiol Mol Biol Rev 74: 171–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, C. , Godoy, P. , Duque, E. , Molina‐Henares, M.A. , de la Torre, J. , Del Arco, J.M. , et al (2010) Global regulation of food supply by Pseudomonas putida DOT‐T1E. J Bacteriol 192: 2169–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Castillo, T. , Ramos, J.L. , Rodríguez‐Hervá, J.J. , Fuhrer, T. , Sauer, U. , and Duque, E. (2007) Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: genomic and flux analysis. J Bacteriol 189: 5142–5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil, H. , Feil, W.S. , Chain, P. , Larimer, F. , DiBartolo, G. , et al (2005) Comparison of the complete genome sequences of Pseudomonas syringae pv.syringae B728a and pv. tomato DC3000. Proc Natl Acad Sci USA 102: 11064–11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frunzke, J. , Engels, V. , Hasenbein, S. , Gatgens, C. , and Bott, M. (2008) Co‐ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol 67: 305–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer, T. , Fischer, E. , and Sauer, U. (2005) Experimental identification and quantification of glucose metabolism in seven bacterial species. J Bacteriol 187: 1581–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, R. , Mack, T.R. , and Stock, A.M. (2007) Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 32: 225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamood, A.N. , Colmer, J.A. , Ochsner, U.A. , and Vasil, M.L. (1996) Isolation and characterization of a Pseudomonas aeruginosa gene, ptxR, which positively regulates exotoxin A production. Mol Microbiol 21: 97–110. [DOI] [PubMed] [Google Scholar]
- Hamood, A.N. , Colmer‐Hamood, J.A. , and Carty, N.L. (2004) Regulation of Pseudomonas aeruginosa exotoxin A synthesis In Pseudomonas: Virulence and Gene Regulation. Ramos J.‐L. (ed). New York, NY: Kluwer Academic/Plenum Publishers, pp. 389–423. [Google Scholar]
- Hoch, J.A. (2000) Two‐component and phosphorelay signal transduction. Curr Opin Microbiol 3: 165–170. [DOI] [PubMed] [Google Scholar]
- Huo, Y.X. , Zhang, Y.T. , Xiao, Y. , Zhang, X. , Buck, M. , Kolb, A. , and Wang, Y.P. (2009) IHF binding sites inhibit DNA loop formation and transcription initiation. Nucleic Acids Res 37: 3878–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama, A. (2000) Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol 54: 499–518. [DOI] [PubMed] [Google Scholar]
- Ishihama, A. , Kori, A. , Koshio, E. , Yamada, K. , Maeda, H. , Shimada, T. , et al (2014) Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli . J Bacteriol 196: 2718–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izu, H. , Adachi, O. , and Yamada, M. (1997) Gene organization and transcriptional regulation of the gntRKU operon involved in gluconate uptake and catabolism of Escherichia coli . J Mol Biol 267: 778–793. [DOI] [PubMed] [Google Scholar]
- Jain, D. (2015) Allosteric control of transcription in GntR family of transcription regulators: a structural overview. IUBMB Life 67: 556–563. [DOI] [PubMed] [Google Scholar]
- Jayal, A. , Johns, B.E. , Purdy, K.J. , and Maddocks, S.E. (2017) Draft genome sequence of Pseudomonas aeruginosa ATCC 9027, originally isolated from an outer ear infection. Genome Announc 5: e01397‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez, J.I. , Miñambres, B. , García, J.L. , and Díaz, E. (2002) Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol 4: 824–841. [DOI] [PubMed] [Google Scholar]
- Joardar, V. , Lindeberg, M. , Jackson, R.W. , Selengut, J. , Dodson, R. , Brinkac, L.M. , et al (2005) Whole‐genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J Bacteriol 187: 6488–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King‐Scott, J. , Nowak, E. , Mylonas, E. , Panjikar, S. , Roessle, M. , Svergun, D.I. , and Tucker, P.A. (2007) The structure of the response regulator RegX3 from Mycobacterium tuberculosis . J Biol Chem 282: 37717–37729. [DOI] [PubMed] [Google Scholar]
- La Rosa, R. , Nogales, J. , and Rojo, F. (2015) The Crc/CrcZ‐CrcY global regulatory system helps the integration of gluconeogenic and glycolytic metabolism in Pseudomonas putida . Environ Microbiol 17: 3362–3378. [DOI] [PubMed] [Google Scholar]
- Lacal, J. , García‐Fontana, C. , Muñoz‐Martínez, F. , Ramos, J.L. , and Krell, T. (2010) Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol 12: 2873–2884. [DOI] [PubMed] [Google Scholar]
- Lendenmann, U. , Snozzi, M. , and Egli, T. (1996) Kinetics of the simultaneous utilization of sugar mixtures by Escherichia coli in continuous culture. Appl Environ Microbiol 62: 1493–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks, S.E. , and Oyston, P.C. (2008) Structure and function of the LysR‐type transcriptional regulator (LTTR) family proteins. Microbiology 154: 3609–3623. [DOI] [PubMed] [Google Scholar]
- Mascher, T. (2006) Intramembrane‐sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett 264: 133–144. [DOI] [PubMed] [Google Scholar]
- Mitchell, S.L. , Ismail, A.M. , Kenrick, S.A. , and Camilli, A. (2015) The VieB auxiliary protein negatively regulates the VieSA signal transduction system in Vibrio cholerae . BMC Microbiol 4: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov, A.Y. , and Groisman, E.A. (2008) Signal integration in bacterial two‐ component regulatory systems. Genes Dev 22: 2601–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesme, J. , Cania, B. , Zadel, U. , Schöler, A. , Płaza, G.A. , and Schloter, M. (2017) Complete genome sequences of two plant‐associated Pseudomonas putida isolates with increased heavy‐metal tolerance. Genome Announc 5: e01330‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikel, P.I. , Chavarría, M. , Fuhrer, T. , Sauer, U. , and de Lorenzo, V. (2015) Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the entner‐doudoroff, embden‐meyerhof‐parnas, and pentose phosphate pathways. J Biol Chem 290: 25920–25932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan, J. , Reen, F.J. , Adams, C. , Casey, P.G. , Gahan, C.G. , and O'Gara, F. (2012) A novel host‐responsive sensor mediates virulence and type III secretion during Pseudomonas aeruginosa‐host cell interactions. Microbiology 158: 1057–1070. [DOI] [PubMed] [Google Scholar]
- Peekhaus, N. , and Conway, T. (1998) Positive and negative transcriptional regulation of the Escherichia coli gluconate regulon gene gntT by GntR and the cyclic AMP (cAMP)‐cAMP receptor protein complex. J Bacteriol 180: 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popella, P. , Krauss, S. , Ebner, P. , Nega, M. , Deibert, J. , and Götz, F. (2016) VraH is the third component of the Staphylococcus aureus VraDEH system involved in gallidermin and daptomycin resistance and pathogenicity. Antimicrob Agents Chemother 60: 2391–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchalka, J. , Oberhardt, M.A. , Godinho, M. , Bielcka, A. , Regenhardt, D. , Timmis, K.N. , et al (2008) Genome‐scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol 4: e1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasamiravaka, T. , Labtani, Q. , Duez, P. , and El Jaziri, M. (2015) The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int 2015: 759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, F. (2010) Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34: 658–684. [DOI] [PubMed] [Google Scholar]
- Roy, S. , Patra, T. , Golder, T. , Chatterjee, S. , Koley, H. , and Nandy, R.K. (2016) Characterization of the gluconate utilization system of Vibrio cholerae with special reference to virulence modulation. Pathog Dis 74: ftw085. [DOI] [PubMed] [Google Scholar]
- Sage, A.E. , Proctor, W.D. , and Phibbs, P.V. Jr (1996) A two‐component response regulator, gltR, is required for glucose transport activity in Pseudomonas aeruginosa PAO1. J Bacteriol 178: 6064–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury, S. , Lane, L.A. , Ren, J. , Gilbert, R.J. , Saunders, N.J. , Robinson, C.V. , et al (2009) The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res 37: 4545–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M.A. , Seidel, G. , Hillen, W. , and Brennan, R.G. (2006) Phosphoprotein Crh‐Ser46‐P displays altered binding to CcpA to effect carbon catabolite regulation. J Biol Chem 281: 6793–6800. [DOI] [PubMed] [Google Scholar]
- Schumacher, M.A. , Sprehe, M. , Bartholomae, M. , Hillen, W. , and Brennan, R.G. (2011) Structure of ccpa‐hpr‐ser46p‐ackA2 complex. Nucleic Acids Res 39: 2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silby, M.W. , Winstanley, C. , Godfrey, S.A. , Levy, S.B. , and Jackson, R.W. (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35: 652–680. [DOI] [PubMed] [Google Scholar]
- Skerker, J.M. , Prasol, M.S. , Perchuk, B.S. , Biondi, E.G. , and Laub, M.T. (2005) Two‐component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system‐level analysis. PLoS Biol 3: e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, K.I. , and Hove‐Jensen, B. (1996) Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerise B and of the rpiR gene, which is involved in regulation of rpiB expression. J Bacteriol 178: 1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, T.T. , Fowler, C.W. , and Griffitts, J.S. (2009) Control of gluconate utilization in Sinorhizobium meliloti . J Bacteriol 191: 1355–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock, A.M. , Robinson, V.L. , and Goudreau, P.N. (2000) Two‐component signal transduction. Annu Rev Biochem 69: 183–215. [DOI] [PubMed] [Google Scholar]
- Suresh, S. , Sarah, D. , Lars, M.B. , Martin, S.H. , and Andreas, S. (2014) The functional structure of central carbon metabolism in Pseudomonas putida KT2440. Appl Environ Microbiol 80: 5292–5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valerie, V. , Klement, R. , and Pavel, F. (2013) Aliphatic, cyclic, and aromatic organic acids, vitamins, and carbohydrates in soil. Sci World J Rev 2013: 524239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick, M.J. , Frank, D.W. , Storey, D.G. , and Iglewski, B.H. (1990) Structure, function, and regulation of Pseudomonas aeruginosa exotoxin A. Annu Rev Microbiol 44: 335–363. [DOI] [PubMed] [Google Scholar]
- Wilson, A.K. , Watral, V.G. , Kent, M.L. , Sharpton, T.J. , and Gaulke, C.A. (2017) Draft genome sequence of Pseudomonas sp. strain DrBHI1 (Phylum Proteobacteria). Genome Announc 5: e01090‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, H. , Serizawa, M. , Thompson, J. , and Sekiguchi, J. (2001) Regulation of the glv operon in Bacillus subtilis: YfiA (GlvR) is a positive regulator of the operon that is repressed through CcpA and cre . J Bacteriol 183: 5110–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K. , Hirao, K. , Oshima, T. , Aiba, H. , Utsumi, R. , and Ishihama, A. (2005) Functional characterization in vitro of all two component signal transduction systems from Escherichia coli . J Biol Chem 280: 1448–1456. [DOI] [PubMed] [Google Scholar]
- Yang, J. , Yan, R. , Roy, A. , Xu, D. , Poisson, J. , and Zhang, Y. (2015) The I‐TASSER suite: protein structure and function prediction. Nat Methods 12: 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]


