Summary
Gut microbiota contributes positively to the physiology of their host. Some feed additives have been suggested to improve livestock health and stimulate growth performance by modulating gut bacteria species. Here, we fed grass carp with 0 (control), 8% (Treat1), 10% (Treat2), 12% (Treat3) and 16% (Treat4) of yeast culture (YC) for 10 weeks. The gut microbiota was analysed by 16S rRNA gene V3‐4 region via an Illumina MiSeq platform. PCoA test showed that gut bacterial communities in the control and Treat3 formed distinctly separate clusters. Although all the groups shared a large size of OTUs as a core microbiota community, a strong distinction existed at genus level. Treat3 contained the highest proportion of the beneficial bacteria and obviously enhanced the capacity of amino acid, lipid metabolism and digestive system. In addition, Treat3 significantly improved the fish growth and increased the liver and serum T‐SOD activities while dramatically decreased the liver GPT and GOT. Collectively, these findings demonstrate the beneficial effects of YC feeding on gut microbiota, growth and biochemical parameters and Treat3 might be the optimal supplementation amount for grass carp, which opens up the possibility that a new feed additive can be developed for healthy aquaculture.
Introduction
Aquaculture has emerged as one of the most promising and fastest growing industries, and healthy aquaculture technology and management is a major concern in aquaculture to provide high‐quality products for human consumption. Grass carp, Ctenopharyngodon idellus is one of the most importantly economic aquaculture species. It represents the largest freshwater aquaculture product and has great commercial value in the world. According to the latest statistics of Food and Agriculture Organization (FAO), global production of cultured or farmed grass carp is approximately 5.5 million tons in 2014, which account for 7.5% of global freshwater aquaculture production of this year (FAO, 2016).
In the past, antibiotics were used to prevent the spread of disease in commercial aquaculture. However, overuse of antibiotics in aquaculture led to an increasing concern over spread of antibiotic‐resistant bacterial genes in the environment and suppression of the aquatic animal's immune system (Cabello, 2006; Baquero et al., 2008). The restriction for use of antibiotics in aquaculture may increase fish disease rates and influence the total production of the world aquaculture. Therefore, along with the increasing demand for aquatic product, it is imperative to find some effective alternatives to improve the fish health and maintain efficiency of aquatic production. Recently, to replace antibiotics in aquaculture, some probiotics, prebiotics and other feed additives, as novel dietary supplements have increased a great deal of attention to improve fish health and growth (Nayak, 2010; Giri et al., 2013; Huang et al., 2015).
It is commonly known that vertebrate gut microbiome provide a number of benefits to their host health including improvement of growth performance, nutrient digestion, immune function and protection from invasive pathogens (Viaud et al., 2013; Blanton et al., 2016; Stanley et al., 2016). Accumulating evidence suggested that probiotics and prebiotics products could modulate the gut microbiota of both human beings and animals, with consequences for improvement of their physiology and health (Li et al., 2014; Johnson et al., 2015; Pourabedin et al., 2015). In particular, the prebiotics products, such as galactooligosaccharides, fructooligosaccharides (FOS), mannan‐oligosaccharides (MOS) and andxylo‐oligosaccharides (XOS), were widely used to modulate the gut microbiota diversity, selectively stimulate the growth of more beneficial bacteria in human (Tuohy et al., 2005), livestock (Li et al., 2014; De Maesschalck et al., 2015; Pourabedin et al., 2015) and fish species (Dimitroglou et al., 2010; Carda‐Diéguez et al., 2014; Guerreiro et al., 2016), and inhibit colonization of pathogenic bacteria by producing antimicrobial substances. For example, treatments with certain XOS modified the relative abundance of chicken microbial genera and increased the probiotic bacteria such as Bifidobacteria (Pourabedin et al., 2015). Dimitroglou et al. (2010) demonstrated that dietary MOS on gilthead sea bream affected the intestinal microbial species richness and diversity (Dimitroglou et al., 2010).
Recently, the yeast culture (YC), as one of the promising feed additives candidates with many benefits, has also been used to modulate the animal gut microbiota. For instance, dietary YC supplementation at 5 g kg−1 had a positive effect on growth performance of nursery pigs by modulating gut immune response (Shen et al., 2009). In human beings, dried yeast modulates both the luminal and mucosal gut microbiota and protects against inflammation (Possemiers et al., 2013). In addition, consistent supplement with yeast cell wall prebiotics significantly increases the proportion of Proteobacteria phyla and Faecalibacterium genus in chicken (Park et al., 2016). Similarly, yeast supplementation also exhibited a sensitive response of the hindgut microbial ecosystem in horses (Grimm et al., 2016). In fish species, the feed intake and weight gain were promoted by dietary supplementation of yeast extract in Nile tilapia and a more efficient defence response to disease was shown (Berto et al., 2016). Essa et al. (2011) evaluated the effects of different additive levels of yeast on the Egyptian African catfish (Clarias gariepinus), which indicated that adding high level of yeast recorded higher final body weight and growth rate. Although yeast supplementation could promote growth performances and immune response in several fish species, whether the YC could improve the structure and composition of gut microbiota and generate the beneficial bacteria in cyprinid grass carp is not clear.
In the present study, we used high‐throughput sequencing of the V3‐V4 region of 16S rRNA gene to assess the effects of the different YC dietary supplementation on the gut microbiota composition, diversity and metabolic capacity in grass carp. In addition, the growth performance and biochemical parameters were also determined. The main objective of this work was to clarify how the YC dietary treatment influences the fish gut microbiota by evaluating the variability among different YC dietary treatments and confirm which additive amount is optimal for grass carp.
Results
The influence of YC treatment on gut microbial diversity
In total, 1.5 million quality‐controlled reads were generated from 16S rRNA gene V3 + V4 amplicons with an average of 34 774 reads per subject (ranging from 26 203 to 43 032) (Table S1). The assembled sequences had an average length of 437 bp. Operational taxonomic units (OTUs) clustering at 97% cutoff yielded a total of 6916 OTUs for the entire data set. The microbial complexity within and between samples was estimated on the basis of alpha‐diversity and beta‐diversity respectively. Tukey's HSD test for multiple comparisons found that there were no significant differences (P > 0.05) in terms of alpha‐diversity based on the observed richness (OTUs, ACE, Chao1) and Shannon's or Simpson's diversity indices between the control and YC‐treated groups (Table 1). From the rarefaction curves, we found similar trend in the microbial diversity among the 43 specimens, approaching the saturation plateau (Fig. S1). In terms of beta‐diversity analysis based on the overall community composition revealed that the extent of similarity between gut microbiota clustered according to YC dietary treatments (Fig. 1). Our UniFrac principal coordinate analysis (PCoA) of 6,916 OTUs (grouped at 97% sequence identity) indicated a clear separation between the control and Treat3 group using both weighted (Fig. 1A) and unweighted (Fig. 1B) analysis. However, within the Treat1 and Treat2, samples of the same treatment have higher dispersion and did not show separate clustering to the control. It should also be noted that distinct clusters were observed in Treat4, but obviously clustered together to the control.
Table 1.
Diversity of the gut microbiome was evaluated using OTUs defined at 97% sequence similarity
| Groups | YC% | Sequenced library No. | Total filtered quality sequences | Richness estimates | Diversity estimates | |||
|---|---|---|---|---|---|---|---|---|
| OTUs | ACE | Chao1 | Shannon | Simpson | ||||
| Control | 0 | 9 | 295510 | 148 ± 9.63 | 172 ± 8.44 | 171 ± 10.76 | 2.16 ± 0.084 | 0.23 ± 0.015 |
| Treat1 | 8 | 9 | 314307 | 153 ± 6.89 | 174 ± 8.46 | 179 ± 9.67 | 2.47 ± 0.15 | 0.20 ± 0.032 |
| Treat2 | 10 | 8 | 256851 | 166 ± 9.34 | 193 ± 9.78 | 197 ± 10.11 | 2.46 ± 0.55 | 0.21 ± 0.052 |
| Treat3 | 12 | 8 | 316645 | 158 ± 4.65 | 180 ± 5.96 | 183 ± 4.91 | 2.66 ± 0.16 | 0.18 ± 0.037 |
| Treat4 | 16 | 9 | 311973 | 179 ± 7.59 | 200 ± 9.33 | 199 ± 8.12 | 2.66 ± 0.21 | 0.17 ± 0.030 |
Figure 1.
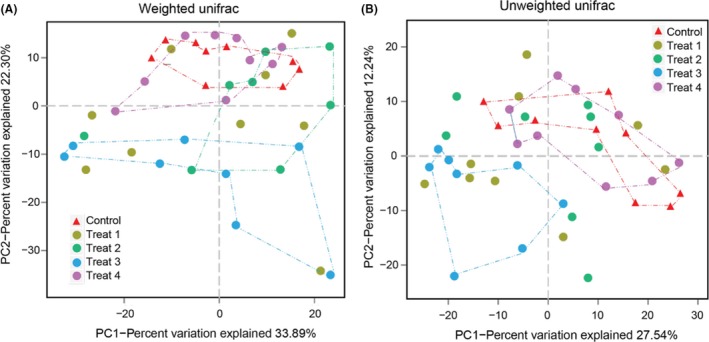
YC dietary treatments differently shift the gut microbial community structure of grass carp. Principle coordinates analyses (PCoA) of weighted (A) and unweighted (B) UniFrac distances show that gut communities cluster by different YC dietary treatments.
YC treatments affect the taxonomic composition of gut microbiome
In order to determine how the YC dietary treatments affect gut bacterial communities of grass carp, and how much of the YC supplement is optimal, the gut microbiomes of five groups with different YC treatments were compared at different taxonomic scales. Figure S2 illustrated approximately 99% of the total bacteria abundance was classified and the most abundant taxa (top 12) of bacteria were showed in all fish gut samples. Fusobacteria, Bacteroidetes, Firmicutes and Proteobacteria constituted the four most dominant phyla of bacterial communities in grass carp with different YC treatments, followed by CKC4 and Verrucomicrobia (Fig. 2A). Metastats‐based analysis of differential abundances among these phyla found that Fusobacteria (21.47 ±3.11%, P = 0.050) and CKC4 (3.19 ± 0.71%, P = 0.016) were significantly decreased in Treat3, whereas the Proteobacteria (27.80 ± 5.51%, P = 0.048) and Actinobacteria (2.37 ± 0.45%, P = 0.001) distinctively increased as compared with the control (Fig. S3). There is no significant difference of above‐mentioned bacterium in other YC treatments except CKC4 decreasing in Treat1 and Actinobacteria increasing in Treat1 and Treat2 (Fig. S3).
Figure 2.
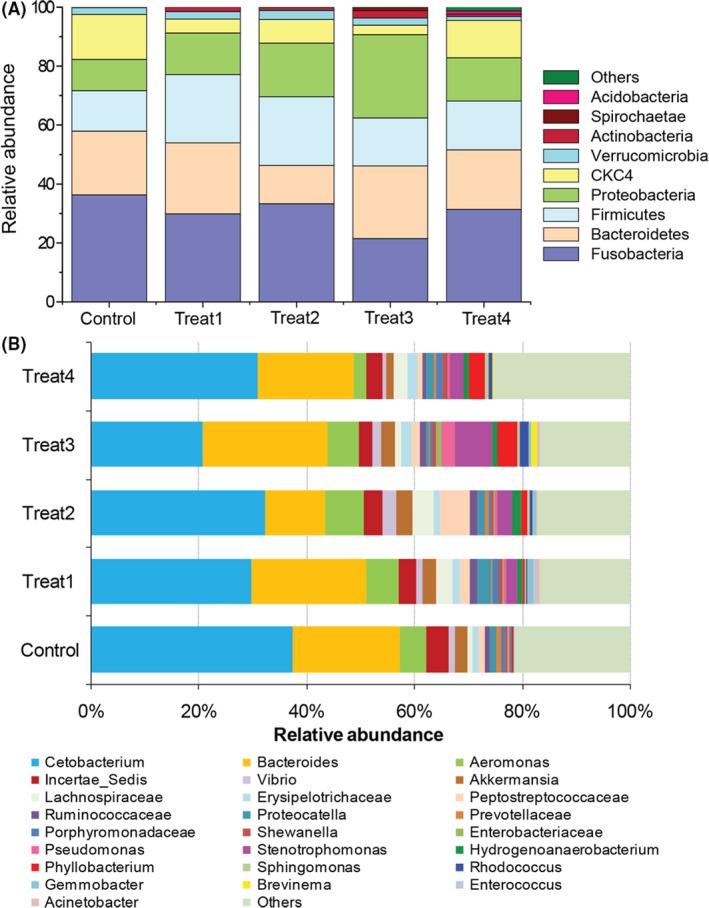
Taxonomic compositions of gut bacterial communities with different YC dietary treatments in grass carp. (A) The relative abundance of bacterial phyla. (B) Their relative abundance of each bacterial taxon (top 25 taxa) within a group at genus level. Control, n = 9; Treat1, n = 9; Treat2, n = 8; Treat3, n = 8; Treat4, n = 9.
Inspection of taxonomic profiles at genus level for all samples, the most abundant taxa (top 25) of bacteria were shown in Fig. 2B, while the rest of the less frequent taxa were categorized as ‘others’. Three prominent members Cetobacterium, Bacteroides and Aeromonas were detected (Fig. 2B). Interestingly, the most distinct changes at the genus level included a considerable reduction of Cetobacterium (20.60 ± 4.04%, P = 0.050) in Treat3 compared with the control and accompanied by a prominent increase of Stenotrophomonas (6.51 ± 1.83%, P = 0.038), Pseudomonas (2.67 ± 0.91%, P = 0.010), Phyllobacterium (3.61 ± 1.02%, P = 0.027) and Rhodococcus (1.45 ± 0.62%, P = 0.008) (Fig. S4). In contrast, no distinct differences in the relative abundance of the above‐mentioned bacteria were found in Treat1, Treat2 and Treat4 compared with the control, except the Rhodococcus in Treat2.
To further discriminate as many taxa as possible for meaningful comparisons, we also performed LEfSe to detect differential abundance of bacterial taxa among different YC treatments. As shown in Fig. 3A, the phylogenetic composition of gut microbiota was noticeably different among YC‐treated samples. The results showed that a total of 42 bacterial biomarkers at five different taxonomic levels were differentially abundant among the five groups (Fig. 3B). In comparison, Treat1 and Treat3 account for the majority of the 42 bacterial clades. Lactobacillales, Flavobacteriales and Rhodobacterales were the dominant orders in the Treat1 while Legionellales were mostly in the control. It is necessary to note that some important cellulose‐degrading bacteria including Streptococcus, Leptotrichia and Pseudomonas were differentially enriched in different YC treatments (Fig. 3B). Bacteria with differential abundance between control and Treat3 were also detected using LefSe (Fig. S5A). For example, the most differentially over represented taxa (LDA score > 3.0) in Treat3 were the genera Rhodococcus, Chryseobacterium, Brevundimonas, Bradyrhizobium, Phyllobacterium, Serratia, Acinetobacter and Pseudomonas, while in the control only the genera Epulopiscium and Erysipelothrix were the most differentially abundant taxa (Fig. S5B).
Figure 3.
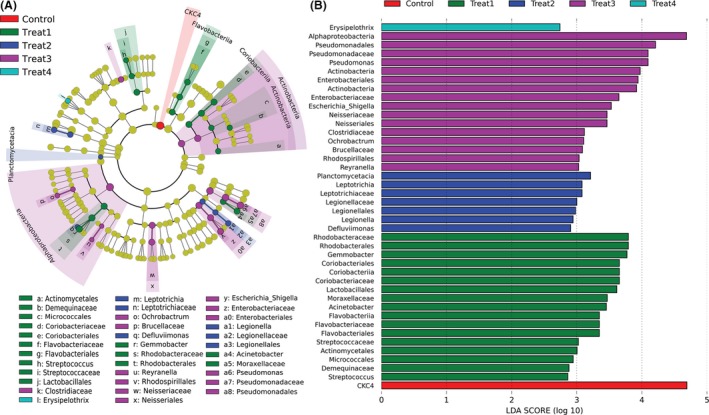
Differences in the gut microbiota of grass carp among the different YC dietary treatments. (A) Circular cladogram reporting LEfSe results presenting the identified OTUs distributed according to phylogenetic characteristics around the circle. Biomarker taxa are highlighted by coloured and shaded circles. Each circle's diameter is relative to abundance of taxa in community. (B) Linear discriminant analysis (LDA) effect size (LEfSe) results show that bacteria were significantly different in abundance between control and YC‐treated groups.
Shared and unique microbial populations
A heatmap showed the abundance with large variation of the 50 most abundant bacterial taxa at family level among five groups (Fig. 4A). Interestingly, among these groups, the pattern of bacterial abundance in Treat4 was much similar to the control, which indicated that the high YC supplement might be with minor effect on gut microbiota compared with other groups. To further investigate the microbial community in different YC dietary treatments, the shared and unique OTUs were analysed through a Venn diagram. We identified 241 taxa present in all samples as the shared OTUs (Fig. S6). The most abundant shared OTUs at the phylum level were Bacteroidetes, Firmicutes, Proteobacteria, followed by Actinobacteria and CKC4 (Fig. 4B). Treat3 showed the largest difference compare to the control. Therefore, the unique and shared OTUs in control and Treat3 were analysed (Fig. 4C) and the five most abundant genera of the unique OTUs in Treat3 were Incertae_Sedis, Acinetobacter, Clostridium, Desulfovibrionaceae and Nocardia (Fig. 4D).
Figure 4.
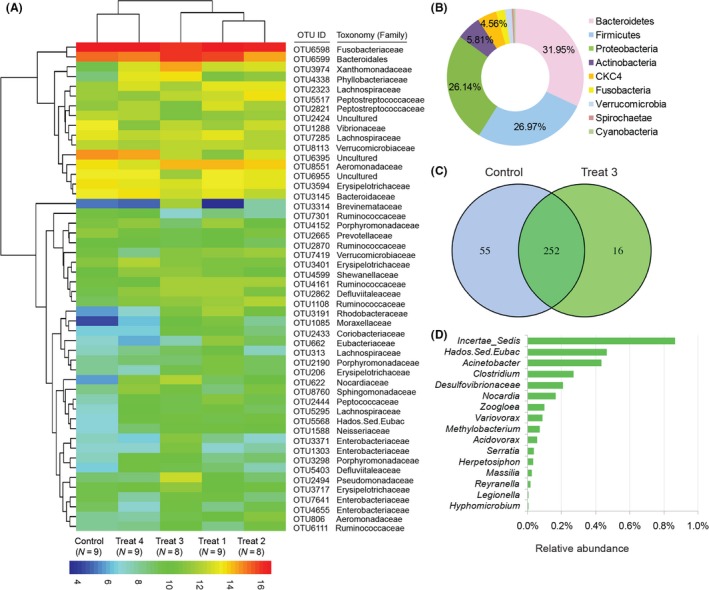
Shared and unique gut bacterial community composition of grass carp with different YC dietary treatments. (A) A heat map of changes in the relative abundances of the 50 most abundant OTUs summarized at the family level in gut microbial communities collected from YC dietary‐treated grass carp gut. (B) The composition of shared gut microbiota among five groups at the phylum level. (C) Number of shared OTUs between the Control and Treat3 and unique taxa. (D) The relative abundance of gut microbiota uniquely present in Treat3 at genus level.
Comparison of the potential beneficial bacteria abundance
In our present study, distinct and diverse putative beneficial candidates and taxonomic groups were identified in different dietary YC‐treated groups. Interestingly, the diversity of potential beneficial bacteria was relatively high, and a number of OTUs also had a high sequence similarity to these bacteria (Fig. 5). The relative abundance of these bacteria in Treat3 (13.50%) is twofold higher than that in the control, and close to Treat2 (11.33%), while a relatively low proportion was found in Treat4 (4.71%) (Fig. 5A). Among these favourable bacteria, the three most abundant were Aeromonas, Vibrio and Pseudomonas in all fish samples, but more in Treat2 and Treat3. To better visualize the OTUs diversity of the beneficial bacteria with a broader evolutionary context in different YC‐treated groups, a maximum likelihood phylogeny was constructed. As shown in Fig. 5B, a total of 39 OTUs were identified as 17 different favourable bacteria species. Among these OTUs, 24 were classified as Proteobacteria, seven as Firmicutes, five as Bacterioidetes and three as Actinobacteria, which differently distributed in the five groups.
Figure 5.
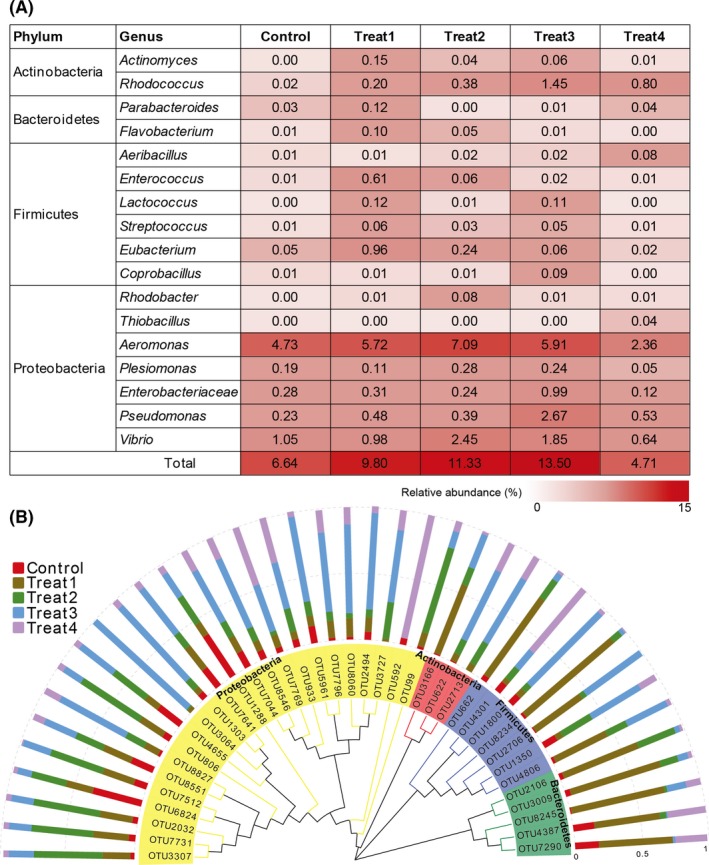
Relative abundance (%) and phylogenetic relationship of potential beneficial bacteria in gut samples from different YC dietary treatments. (A) A heat map of changes in the relative abundance of putative beneficial candidates in five trials. (B) Dendrogram of potential beneficial bacteria represented OTUs and their host occurrence patterns. Semicircle indicates the phylogenetic relationship of 17 above‐mentioned putative favourable species. Bars show the proportion of fish samples from different YC treatments in which the given OTUs is present.
Predicted the functions of gut microbiota
Based on the functionality prediction, a distinct difference in the KEGG Orthologues (KO) composition among different YC‐treated groups was detected (Fig. 6A), especially between the control and Treat1, Treat3 (Fig. 6B), respectively. In contrast, no obvious difference was detected between the control and Treat4 and they closely clustered each other. Pathways involving cell processes and signalling, digestive system, energy metabolism and immune system diseases were found over represented in each group (Fig. S7). Deeper analysis of the KEGG between control and Treat3 revealed that 17 of the 234 categories at levels II were shown to achieve a statistical significant difference at P < 0.05 (Fig. 6C). Notably, significant elevation in cell motility, cellular processes and signalling, digestive system, amino acid metabolism, metabolism of terpenoids and polyketides and lipid metabolism pathways was observed in Treat3 whereas pathways related to the energy metabolism, nucleotide metabolism, translation, replication and repair, genetic information processing and folding, sorting and degradation were more detected in control.
Figure 6.
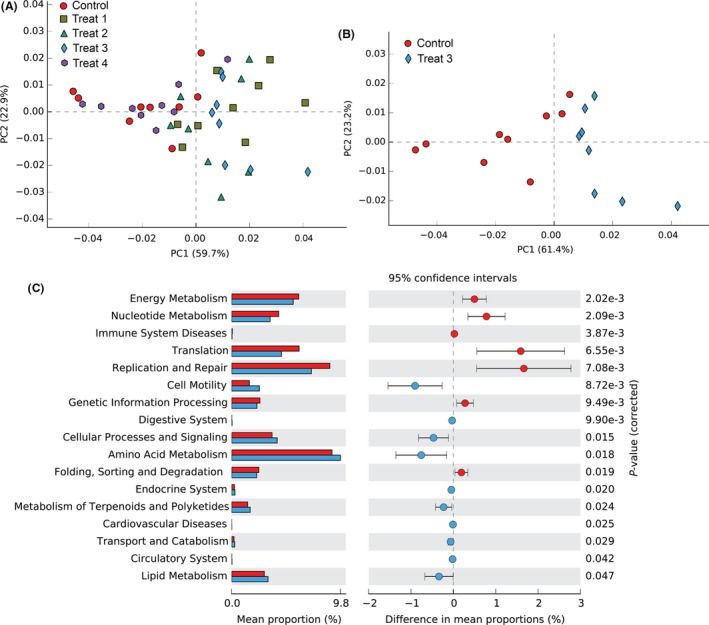
The predicted functional metagenomes of gut microbiota in grass carp with different YC dietary treatments. (A) Principal component analysis (PCA) predicted the functional metagenomes among control and YC dietary treatments. (B) PCA plot compared the taxonomic functional profiles between control and Treat3. (C) Extended error bar plot showing the normalized relative abundance of KEGG metabolic pathways (at level II) differing significantly between Control (red) and Treat3 (blue).
YC treatments improve the growth performances of grass carp
After 10 weeks of different concentrations of YC feeding, the YC dietary‐treated groups showed good growth performance as indicated by feed conversion ratio (FCR), condition factor (CF), gain rate (GR) and survival rate (SR) (Table 2). Although there were no significant differences between trials in SR and CF, significant differences in GR and FCR were detected (P < 0.05). It is worth noting that Treat3 (12% YC) was with the highest GR. For the whole trial period (day 0–70), the FCR was significantly (P < 0.05) more favourable for grass carp fed YC‐supplemented diet compared with the fish fed control diet, especially the Treat3 and Treat4. These results together with the increased GR show a biologically improved performance for fish fed YC.
Table 2.
Growth performances of grass carp with yeast culture dietary treatments for 10 weeks
| Control | Treat1 | Treat2 | Treat3 | Treat4 | |
|---|---|---|---|---|---|
| FCR | 1.95 ± 0.24a | 1.65 ± 0.05ab | 1.71 ± 0.10ab | 1. 60 ± 0.06b | 1.56 ± 0.02b |
| CF | 1.80 ± 0.02 | 1.80 ± 0.04 | 1.91 ± 0.05 | 1.77 ± 0.03 | 1.84 ± 0.05 |
| GR | 1.39 ± 0.05a | 1.65 ± 0.35b | 1.65 ± 0.01b | 1.80 ± 0.03c | 1.70 ± 0.02bc |
| SR (%) | 95.61 ± 2.26 | 96.72 ± 1.93 | 94.43 ± 2.54 | 97.70 ± 1.18 | 91.15 ± 5.93 |
All data represent means ± SEM. In the same row, means with different letters are significantly different (P < 0.05), means with the same letters are not significantly different (P > 0.05). Absence of letters indicates no significant difference between treatments.
FCR, Feed conversion ratio; CF, Condition factor; GR, Gain rate; SR, Survival rate.
Effects of YC treatments on liver and serum biochemical parameters
Despite the differences in the composition of gut microbiota and growth performance, the liver and serum T‐SOD, T‐AOC, GPT and GOT enzymes activities were differently influenced by YC dietary treatments in grass carp (Fig. 7) after 10 weeks of feeding. Specifically, 12% YC dietary treatment (Treat3) significantly increased (P < 0.05) T‐SOD enzyme activity in the liver (Fig. 7A) and serum (Fig. 7E) while there was no difference (P > 0.05) in T‐AOC enzyme activity in all YC‐treated groups (Fig. 7B and F). The liver GPT (Fig. 7C) and GOT (Fig. 7D) enzyme activities were dramatically decreased (P < 0.05) in Treat3 and Treat4 (16% YC), as compared with the control, while the serum GPT (Fig. 7G) and GOT (Fig. 7H) among the groups with YC dietary treatments were similar.
Figure 7.
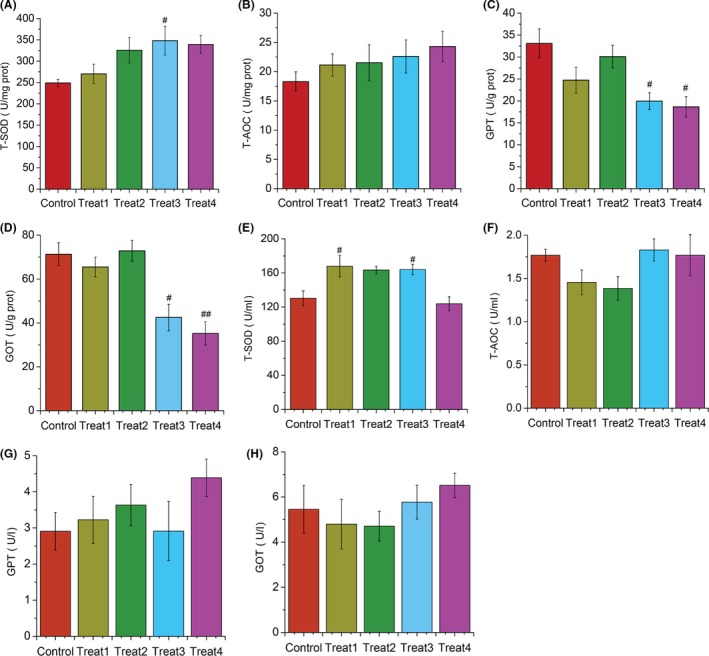
Effects of YC dietary treatment for 10 weeks on biochemical parameters in liver and serum of grass carp. (A) Liver T‐SOD. (B) Liver T‐AOC. (C) Liver GPT. (D) Liver GOT. (E) Serum T‐SOD. (F) Serum T‐AOC. (G) Serum GPT. (H) Serum GOT. Values are expressed as means ± S.E.M (n = 9). #P < 0.05, ##P < 0.01 compared with control group.
Discussion
More recently, high‐throughput sequencing of 16S rRNA gene amplicons has been used to explore the diversity and composition of gut microbiota in fish species, including three‐spined stickleback and Eurasian perch (Bolnick et al., 2014), fathead minnows (Narrowe et al., 2015), surgeonfishes (Miyake et al., 2015), Trinidadian guppies (Sullam et al., 2015), tilapia (Giatsis et al., 2014), Atlantic cod (Forberg et al., 2016), Atlantic salmon (Llewellyn et al., 2016) and some commercially viable cyprinids (Eichmiller et al., 2016; Liu et al., 2016). Most of these fishes were studied in the rearing conditions fed basic diet or in wild conditions, which well demonstrated the influence of basic dietary input and environmental locations on the gut microbiota. However, so far very little is known about the effects of dietary YC on the composition and functional diversity of fish gut microbiota.
There is increasing evidence that prebiotics as non‐digestible food ingredients beneficially affects the physiology of host to improve growth performance, resistance against pathogenic bacteria or by stimulating the growth of beneficial bacteria (Li et al., 2014; Johnson et al., 2015; Pourabedin et al., 2015). Fully fermented YC is a dried product and contains yeast and various metabolites of yeast fermentation (Shen et al., 2009), as one of the promising feed additives candidates with many benefits in many terrestrial animals, which has been also used in aquaculture (Essa et al., 2011; Berto et al., 2016). Our results clearly indicated that YC dietary treatments for 10 weeks affected the diversity and composition of gut microbiota in grass carp and increased the relative abundance of potential beneficial bacteria species, especially in Treat3. Furthermore, YC dietary Treat3 significantly influenced the metabolic capacity of gut microbiota, exhibited good growth performances (weight gain rate and FCR) and biochemical parameters of fish (liver and serum T‐SOD, T‐AOC, GPT and GOT) compared with control diet. These results provide an explicit understanding how YC supplement in diet influences the gut microbiota diversity of grass carp and how much of YC addition is optimal to the host's food digestion and growth.
Although no significant differences were found in terms of alpha‐diversity between the control and YC treatment groups (Table 1), beta‐diversity analysis based on PCoA indicated a clear separate cluster between the control and YC Treat3 (Fig. 1), suggesting that the microbial complexity and enrichments were affected by YC diet in Treat3. This observation is in accordance with the study of Dimitroglou et al. (2010) who demonstrated a clear shift of intestinal microbial profiles with prebiotic MOS treatments, clustered into distinct groups in gilthead sea bream (Dimitroglou et al., 2010). On the contrary, Ran et al. (2015) found that yeast supplementation on Nile Tilapia exerted a significant influence on the alpha‐diversity of the autochthonous microbiota, but no significant difference was in beta‐diversity (Ran et al., 2015). This discrepancy is likely due to the intestinal microbiota variably depending on the environmental factors and the variety of host species.
In the current study, Fusobacteria, Bacteroidetes, Firmicutes and Proteobacteria constituted the most dominant phyla of bacterial communities in grass carp with different YC treatments for 10 weeks. This result is different from our previous study in gut microbiota of wild adult grass carp (Liu et al., 2016), which might be caused by diet composition (Bolnick et al., 2014; Liu et al., 2016) or fish age (Stephens et al., 2015) in addition to other environmental factors (Eichmiller et al., 2016). Our further metastats‐based analysis found that Actinobacteria significantly increased in Treat1, 2 and 3. It was known that Actinobacteria phylum includes Collinsella, Bifidobacterium (which contains some probiotic strains) (Tremaroli and Bäckhed, 2012) and Actinomyces (which was known as a cellulose‐degrading bacteria) (Ye et al., 2014), indicating that YC dietary treatments might be beneficial for potential favourable bacteria. Similar results have been previously reported in mice (Everard et al., 2011) and human (Geurts et al., 2014). Although fish in different YC dietary treatments shared a large size of OTUs comprising a core microbiota community at the genus level, a strong distinction existed. Some important cellulose‐degrading bacteria (Ye et al., 2014; Liu et al., 2016) including Streptococcus, Leptotrichia and Pseudomonas were differentially enriched in different YC treatments except Treat4. Intriguingly, some of the most abundant genera of the unique OTUs in Treat3 were also the important cellulose‐degrading candidates, such as Clostridium and Methylobacterium. It is well known that grass carp is a typical herbivorous fish species with a longer intestine, which need more cellulose‐degrading bacteria to effectively degrade the ingested food sources. This result is accordant with the previous study in ruminant that the presence of the live yeast resulted in a significant increase of cellulolytic bacterial species, indicating a beneficial effect of yeast on rumen fermentation (Mosoni et al., 2007).
There is increasing evidence that dietary supplementation of prebiotics on various terrestrial species beneficially affects the host to increase the number of health‐promoting bacteria such as Lactobacillus and Bifidobacterium in their intestinal tract, while decreasing potentially pathogenic bacteria (De Maesschalck et al., 2015; Johnson et al., 2015; Pourabedin et al., 2015). In the current study, distinct and diverse putative beneficial bacteria recognized as probiotics in aquaculture (Nayak, 2010; Lazado and Caipang, 2014; Dawood and Koshio, 2016) were identified in different YC dietary‐treated groups, especially in Treat3 (13.50%), approximately twofold higher than that in the control (Fig. 5). These microorganisms in the gastrointestinal tract could process indigestible carbohydrates and provide nutrients and vitamins to their host (Aron‐Wisnewsky and Clément, 2015; Hanning and Diaz‐Sanchez, 2015). However, the well‐known health beneficial bacteria Lactobacillus and Bifidobacterium were not enhanced after YC treatments, different from the studies on the effects of prebiotic XOS dietary in chinken reported by Pourabedin et al. (2015) and De Maesschalck et al. (2015). This difference could explain that the effects of prebiotics may vary depending on the concentration of the prebiotic in the diet, specific host and the length of feeding period. Another important factor should be also considered that the living environment for fish is more complicated and variable than that for terrestrial animals (De Schryver and Vadstein, 2014). The possibility with the presence of other putative favourable bacteria needs to be further confirmed.
Furthermore, we also found that certain concentration of YC (Treat3 and Treat4) feeding grass carp for 10 weeks showed good growth performances including the FCR and GR. Similar results have been previously reported that yeast supplementation could also promote growth performances and immune response of human (Possemiers et al., 2013), chicks (Gao et al., 2008), pigs (Shen et al., 2009) and fish species (Essa et al., 2011; Huang et al., 2015; Ran et al., 2015; Berto et al., 2016). For example, dietary supplemental YC at 2.5 g kg−1 on chicks distinctively improved the growth performances including the average daily gain and feed conversion (Gao et al., 2008). Also, Berto et al. (2016) and Ran et al. (2015) reported that dietary supplementation of yeast extract and live baker's yeast improved the growth, feed utilization and blood immunological indexes of Nile tilapia (Ran et al., 2015; Berto et al., 2016). It is known that antioxidant enzymes such as superoxide dismutase (SOD) and catalase (CAT) could protect against reactive oxygen species (ROS) that induced oxidative damage. We also found that YC dietary Treat3 significantly increased (P < 0.05) the liver (Fig. 7A) and serum (Fig. 7E) T‐SOD enzymatic activity and dramatically decreased liver GPT (Fig. 7C) and GOT (Fig. 7D) enzyme activities compared with the control, which suggested that the biochemical parameters of grass carp were stimulated and host antioxidant capacity was enhanced by dietary supplementation of certain concentration of YC. A study carried out by Dong and Wang (2013) suggested that red swamp cray fish fed dietary certain dose of prebiotics FOS significantly increased the activity of SOD and the expression of immune genes. To further explore their potential use in aquaculture, the effects of YC on systemic immunity and how YC activates the responses of the antioxidant system should be considered for future experiments.
In conclusion, this study describes for the first time the comprehensive, high‐throughput analyses of gut microbiota diversity and demonstrates clear beneficial effects of YC in fish species. Our results clearly showed that YC dietary treatment modulated the composition and relative abundance of gut microbiota without changing the overall microbial structure. Importantly, the fact that certain YC treatment significantly increased variety and proportion of the putative beneficial bacteria species in grass carp could be an intestinal health‐promoting attribute, which might contribute to promote the growth performance and enhance the host antioxidant defences. Thus, these results indicated that YC could be used as a potential new feed additive for healthy aquaculture.
Experimental procedures
Experimental diets
The YC used in the present study was provided by the Hubei Gosign Bio‐feed, Wuhan, China. The percentage of amino acid of YC was shown in Table S2. Experimental diets were prepared by a commercial feed from Haid Feed, (Guangzhou, China) (approximately of 33.66% protein, 8.99% lipid and dry matter basis) supplemented with different levels [0% as Control, 8% (Treat1), 10% (Treat2), 12% (Treat3) and 16% (Treat4)] of YC (Fig. S8). After homogenization, the mixture was pelleted using a grinder (4.5 mm diameter) with 30% water incorporation, and then dried at room temperature and stored in −20 °C.
Fish rearing conditions
A total of 500 healthy grass carp (Ctenopharyngodon idella) (100 ± 5 g) were obtained from Bairong fish breeding base of Huazhong Agricultural University (HZAU). The fish were acclimated to the laboratory rearing conditions for 2 weeks. During this period, a commercial pellets diet was fed twice a day. Thereafter, fish were randomly distributed into fifteen 100‐l tanks (30 fish per tank and triplicate tanks per treatment) and connected to a water‐recirculating system kept at 26 ± 1.5 °C, pH 7 ± 0.5. All tanks were cleaned every 3 days with a 30% water renewal. Each experimental diet was fed to apparent satiety twice a day (09:00 and 16:00 hours) for 10 weeks. Any uneaten feed was collected in 1 h after feeding and dried at 70 °C.
Sample collection and growth performances
All the experimental procedures involving fish were performed in accordance with the guidelines of National Institute of Health Guide for the Care and Use of Laboratory Animals and approved by the Research Ethics Committee of Huazhong Agricultural University.
To determine whether the YC dietary treatment promotes the growth of grass carp, at the end of the feeding trial, the total number, individual body weight and length of fish from each tank were measured. After a 24‐h fasting period, all fish were euthanized with an overdose of anaesthetic and then sacrificed to obtain samples. The SR, GR, FCR and CF were determined and calculated according to the method as described by Dawood et al., (2016). Gain rate was calculated as [weight gain (g)/start weight (g)]. Feed conversion ratio was calculated as [feed eaten (g)/weight gained (g)]. Condition factor was calculated as 100 × [live body weight (g)/length (cm)].
Liver and serum biochemical parameters
Blood was collected from the caudal vein using a 1 ml syringe. Serum samples were obtained by centrifugation at 3000× g for 15 min at 4 °C to collect serum and maintained at −80 °C for further analysis. Liver was aseptically collected and immediately homogenized with 10 volumes of cold PBS buffer (pH 6.8). The homogenate was centrifuged at 12 000× g for 20 min at 4 °C and the supernatant was divided into four Eppendorf tubes and then stored at −80 °C until analysis. To explore whether different concentrations of YC dietary treatments influence the biochemical parameters of liver and serum, the activities of total superoxide dismutase (T‐SOD), total antioxidant capacity (T‐AOC), glutamate pyruvate transaminase (GPT) and glutamate oxalate transaminase (GOT) enzymes were determined using assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China) according to the manufacturer's protocols.
DNA extraction for gut content
To avoid transient bacteria, the whole intestinal tract of individual fish was dissected with sterile instruments and washed in 70% ethanol and sterile water. The gut content from the midgut to hindgut region was squeezed out and mixed thoroughly, then collected into sterilized tubes and immediately stored at liquid nitrogen. Bacterial genomic DNA extraction was carried out from 100 mg of each sample with a QIAamp DNA Stool Mini Kit (Qiagen, Valencia, CA, USA) following the manufacturer's protocol. The concentration of each DNA sample was quantified using NanoDrop ND‐2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and diluted to the same concentration for subsequent PCR amplification.
Amplification, sequencing, assemblage and determination of taxonomic units of fragments
To examine the microbial communities of each sample, a total of 43 libraries, corresponding to nine control, nine Treat1, eight Treat2, eight Treat3 and nine Treat4 were constructed and sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA) with 2 × 250 bp kits following the manufacturer's instructions. The bacterial V3‐V4 hypervariable region of the 16S ribosomal gene was amplified using the following primers: 319F: 5′‐ACTCCTACGGGAGGCAGCAG‐3′) and 806R: 5′‐GGACTACHVGGGTWTCTAAT‐3′ according to the previously described methods (Fadrosh et al., 2014). All control and YC‐treated samples were included in the same sequencing run. All sequences have been deposited in the NCBI's Sequence Read Archive (SRA accession number to be provided upon acceptance).
FLASH (Magoč and Salzberg, 2011) was used to merge the overlapping paired‐end reads into single longer reads covering the full 16S rRNA V3‐V4. UCHIME (Edgar et al., 2011) was used to identify possible chimeric sequences and the unique sequences were aligned using the SILVA database (Pruesse et al., 2007). The filtered and trimmed high‐quality reads were further processed to analyse the microbial community composition. Briefly, the UCLUST (Edgar, 2010) with 97% similar was used to cluster of nearly identified reads as an OTU, then representative sequences of each OTU were aligned to the most recently available version of the bacterial database Green genes using PyNAST (Caporaso et al., 2010). Taxonomic assignment was achieved using RDP Classifier (Wang et al., 2007), clustering the sequences at 97% similarity with a confidence threshold of 0.80. The relative abundance of OTUs and microbial composition in different taxonomic levels was calculated and analysed for each group.
Parameter calculation and statistical analysis on microbiota diversity
Alpha‐diversity metrics including observed OTUs, rarefaction curve, ACE (Eckburg et al., 2006), Chao1 (Chao, 1984) and Shannon's diversity (Shannon, 1948) index were calculated on rarefied OTU tables with Mothur version v.1.30 (Schloss et al., 2009). The overall microbiome dissimilarities among all gut content samples were ordinated using the weighted and unweighted UniFrac distance metrics (Lozupone and Knight, 2005) and visualized with principal coordinates analysis (PCoA) by the R package phyloseq. Discriminatory analysis of the gut microbiome features among control and YC‐treated groups at various taxonomic ranks was performed with LEfSe (http://huttenhower.sph.harvard.edu/galaxy) (Segata et al., 2011) with alpha value < 0.05 for the factorial Kruskal–Wallis test among classes and threshold on the logarithmic LDA score for discriminative features > 2. To predict the differences of metagenome functional profile of the gut microbiota between control and YC‐treated groups, PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) (Langille et al., 2013) was applied using GreenGenes database. We then binned the identified KOs into functional categories under a given subsystem hierarchy at level II and level III derived from KEGG (Kyoto encyclopedia of genes and genomes) modules (Ogata et al., 1999). The output file was further analysed using the software package STAMP (Statistical Analysis of Metagenomic Profiles) (Parks et al., 2014). STAMP implements the Welch's t‐test for comparing profiles organized into two groups and the ANOVA with post hoc tests (Tukey–Kramer) for comparing three or more groups of profiles. Statistical comparison of two groups was performed using a Student's t‐test. Data sets that involved more than two groups were assessed by one‐way analysis of variance (ANOVA) with Tukey's multiple comparison followed by Newman–Keuls post hoc tests. All data are shown as means ± SEM unless otherwise indicated. Values were considered statistically significant when P < 0.05. The statistical analyses were performed with SPSS 19.0 (SPSS, Chicago, IL, USA).
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Fig. S1. Rarefaction analyses of gut samples with different YC dietary treatments.
Fig. S2. The relative abundance of gut microbiota in individual grass carp at phylum level.
Fig. S3. Relative abundance of four taxa at phylum level significantly associated with YC treatments.
Fig. S4. Relative abundance of six taxa at genus level significantly associated with YC treatments.
Fig. S5. Differences in the gut microbiota of grass carp between control and YC dietary Treat3.
Fig. S6. Venn diagrams showing compartmental core microbiota OTU distributions in grass carp.
Fig. S7. Mean proportion and their differences in predicted functional metagenomes of the gut microbiota among YC dietary treatments.
Fig. S8. Schematic representation of experimental design.
Table S1. Detailed fish gut sample information.
Table S2. The percentage of amino acid in yeast cultures.
Acknowledgements
This work was financially supported by the Central Public‐interest Scientific Institution Basal Research Fund, CAFS (2017HY‐XKQ0205), the Fundament Research Funds for the Central Universities (2662015PY019), the National Science Foundation for Post‐doctoral Scientists of China (2016M600600) and the project of the International Scientific and Technology Cooperation Program of Wuhan City (2015030809020365).
Microbial Biotechnology (2018) 11(3), 551–565
Funding InformationThis work was financially supported by the Central Public‐interest Scientific Institution Basal Research Fund, CAFS (2017HY‐XKQ0205), the Fundament Research Funds for the Central Universities (2662015PY019), the National Science Foundation for Post‐doctoral Scientists of China (2016M600600) and the project of the International Scientific and Technology Cooperation Program of Wuhan City (2015030809020365).
References
- Aron‐Wisnewsky, J. , and Clément, K. (2015) The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol 12: 169–181. [DOI] [PubMed] [Google Scholar]
- Baquero, F. , Martínez, J.L. , and Cantón, R. (2008) Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol 19: 260–265. [DOI] [PubMed] [Google Scholar]
- Berto, R.S. , Pereira, G.V. , Mouriño, J.L.P. , Martins, M.L. , and Fracalossi, D.M. (2016) Yeast extract on growth, nutrient utilization and haemato‐immunological responses of Nile tilapia. Aquac Res 47: 2650–2660. [Google Scholar]
- Blanton, L.V. , Charbonneau, M.R. , Salih, T. , Barratt, M.J. , Venkatesh, S. , Ilkaveya, O. , et al (2016) Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351: aad3311–aad3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick, D.I. , Snowberg, L.K. , Hirsch, P.E. , Lauber, C.L. , Org, E. , Parks, B. , et al (2014) Individual diet has sex‐dependent effects on vertebrate gut microbiota. Nat Commun 5: 4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello, F.C. (2006) Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol 8: 1137–1144. [DOI] [PubMed] [Google Scholar]
- Caporaso, J.G. , Bittinger, K. , Bushman, F.D. , Desantis, T.Z. , Andersen, G.L. , and Knight, R. (2010) PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carda‐Diéguez, M. , Mira, A. , and Fouz, B. (2014) Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets. FEMS Microbiol Ecol 87: 451–459. [DOI] [PubMed] [Google Scholar]
- Chao, A. (1984) Nonparametric estimation of the Scandinavian Journal of Statistics. Scand J Stat 11: 265–270. [Google Scholar]
- Dawood, M.A.O. , and Koshio, S. (2016) Recent advances in the role of probiotics and prebiotics in carp aquaculture: a review. Aquaculture 454: 243–251. [Google Scholar]
- Dawood, M.A.O. , Koshio, S. , Ishikawa, M. , Yokoyama, S. , El Basuini, M.F. , Hossain, M.S. , et al (2016) Effects of dietary supplementation of Lactobacillus rhamnosus or/and Lactococcus lactis on the growth, gut microbiota and immune responses of red sea bream. Pagrus major. Fish Shellfish Immunol 49: 275–285. [DOI] [PubMed] [Google Scholar]
- De Maesschalck, C. , Eeckhaut, V. , Maertens, L. , De Lange, L. , Marchal, L. , Nezer, C. , et al (2015) Effects of Xylo‐oligosaccharides on broiler chicken performance and microbiota. Appl Environ Microbiol 81: 5880–5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schryver, P. , and Vadstein, O. (2014) Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J 8: 2360–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitroglou, A. , Merrifield, D.L. , Spring, P. , Sweetman, J. , Moate, R. , and Davies, S.J. (2010) Effects of mannan oligosaccharide (MOS) supplementation on growth performance, feed utilisation, intestinal histology and gut microbiota of gilthead sea bream (Sparus aurata). Aquaculture 300: 182–188. [Google Scholar]
- Dong, C. , and Wang, J. (2013) Immunostimulatory effects of dietary fructooligosaccharides on red swamp crayfish, Procambarus clarkii (Girard). Aquac Res 44: 1416–1424. [Google Scholar]
- Eckburg, P.B. , Bik, E.M. , Bernstein, C.N. , Purdom, E. , Dethlefsen, L. , Sargent, M. , et al (2006) Diversity of the Human Intestinal Microbial Flora. Science 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar, R.C. , Haas, B.J. , Clemente, J.C. , Quince, C. , and Knight, R. (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmiller, J.J. , Hamilton, M.J. , Staley, C. , Sadowsky, M.J. , and Sorensen, P.W. (2016) Environment shapes the fecal microbiome of invasive carp species. Microbiome 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essa, M.A. , Mabrouk, H.A. , Mohamed, R.A. , and Michael, F.R. (2011) Evaluating different additive levels of yeast, Saccharomyces cerevisiae, on the growth and production performances of a hybrid of two populations of Egyptian African catfish, Clarias gariepinus . Aquaculture 320: 137–141. [Google Scholar]
- Everard, A. , Lazarevic, V. , Derrien, M. , Girard, M. , Muccioli, G.M. , Neyrinck, A.M. , et al (2011) Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet‐induced leptin‐resistant mice. Diabetes 60: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadrosh, D.W. , Ma, B. , Gajer, P. , Sengamalay, N. , Ott, S. , Brotman, R.M. , and Ravel, J. (2014) An improved dual‐indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO Fisheries and Aquaculture Department (2016) FAO yearbook fishery and aquaculture statistics 2014. Rome, Italy: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Forberg, T. , Sjulstad, E.B. , Bakke, I. , Olsen, Y. , Hagiwara, A. , Sakakura, Y. , and Vadstein, O. (2016) Correlation between microbiota and growth in Mangrove Killifish (Kryptolebias marmoratus) and Atlantic cod (Gadus morhua). Sci Rep 6: 21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. , Zhang, H.J. , Yu, S.H. , Wu, S.G. , Yoon, I. , Quigley, J. , et al (2008) Effects of yeast culture in broiler diets on performance and immunomodulatory functions. Poult Sci 87: 1377–1384. [DOI] [PubMed] [Google Scholar]
- Geurts, L. , Neyrinck, A.M. , Delzenne, N.M. , Knauf, C. , and Cani, P.D. (2014) Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Benef Microbes 5: 3–17. [DOI] [PubMed] [Google Scholar]
- Giatsis, C. , Sipkema, D. , Smidt, H. , Verreth, J. , and Verdegem, M. (2014) The Colonization dynamics of the gut microbiota in tilapia larvae. PLoS One 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri, S.S. , Sukumaran, V. , and Oviya, M. (2013) Potential probiotic Lactobacillus plantarum VSG3 improves the growth, immunity, and disease resistance of tropical freshwater fish. Labeo rohita. Fish Shellfish Immunol 34: 660–666. [DOI] [PubMed] [Google Scholar]
- Grimm, P. , Julliand, V. , Philippeau, C. , and Sadet‐Bourgeteau, S. (2016) Effect of yeast supplementation on hindgut microbiota and digestibility of horses subjected to an abrupt change of hays. Livest Sci 186: 34–40. [Google Scholar]
- Guerreiro, I. , Serra, C.R. , Enes, P. , Couto, A. , Salvador, A. , Costas, B. , and Oliva‐Teles, A. (2016) Effect of short chain fructooligosaccharides (scFOS) on immunological status and gut microbiota of gilthead sea bream (Sparus aurata) reared at two temperatures. Fish Shellfish Immunol 49: 122–131. [DOI] [PubMed] [Google Scholar]
- Hanning, I. , and Diaz‐Sanchez, S. (2015) The functionality of the gastrointestinal microbiome in non‐human animals. Microbiome 3: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L. , Ran, C. , He, S. , Ren, P. , Hu, J. , Zhao, X. , and Zhou, Z. (2015) Effects of dietary Saccharomyces cerevisiae culture or live cells with Bacillus amyloliquefaciens spores on growth performance, gut mucosal morphology, hsp70 gene expression, and disease resistance of juvenile common carp (Cyprinus carpio). Aquaculture 438: 33–38. [Google Scholar]
- Johnson, L. , Walton, G. , Psichas, A. , Frost, G. , Gibson, G. , and Barraclough, T. (2015) Prebiotics modulate the effects of antibiotics on gut microbial diversity and functioning in vitro . Nutrients 7: 4480–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille, M.G.I. , Zaneveld, J. , Caporaso, J.G. , McDonald, D. , Knights, D. , Reyes, J.A. , et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazado, C.C. , and Caipang, C.M.A. (2014) Mucosal immunity and probiotics in fish. Fish Shellfish Immunol 39: 78–89. [DOI] [PubMed] [Google Scholar]
- Li, M. , Monaco, M.H. , Wang, M. , Comstock, S.S. , Kuhlenschmidt, T.B. , Fahey, G.C. Jr , et al (2014) Human milk oligosaccharides shorten rotavirus‐induced diarrhea and modulate piglet mucosal immunity and colonic microbiota. ISME J 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Guo, X. , Gooneratne, R. , Lai, R. , Zeng, C. , Zhan, F. , and Wang, W. (2016) Supplementary information : the gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci Rep 6: 24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn, M.S. , McGinnity, P. , Dionne, M. , Letourneau, J. , Thonier, F. , Carvalho, G.R. , et al (2016) The biogeography of the atlantic salmon (Salmo salar) gut microbiome. ISME J 10: 1280–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C. , and Knight, R. (2005) UniFrac : a New Phylogenetic Method for Comparing Microbial Communities UniFrac : a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoč, T. , and Salzberg, S.L. (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake, S. , Ngugi, D.K. , and Stingl, U. (2015) Diet strongly influences the gut microbiota of surgeonfishes. Mol Ecol 24: 656–672. [DOI] [PubMed] [Google Scholar]
- Mosoni, P. , Chaucheyras‐Durand, F. , Béra‐Maillet, C. , and Forano, E. (2007) Quantification by real‐time PCR of cellulolytic bacteria in the rumen of sheep after supplementation of a forage diet with readily fermentable carbohydrates: effect of a yeast additive. J Appl Microbiol 103: 2676–2685. [DOI] [PubMed] [Google Scholar]
- Narrowe, A.B. , Albuthi‐Lantz, M. , Smith, E.P. , Bower, K.J. , Roane, T.M. , Vajda, A.M. , and Miller, C.S. (2015) Perturbation and restoration of the fathead minnow gut microbiome after low‐level triclosan exposure. Microbiome 3: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak, S.K. (2010) Probiotics and immunity: a fish perspective. Fish Shellfish Immunol 29: 2–14. [DOI] [PubMed] [Google Scholar]
- Ogata, H. , Goto, S. , Sato, K. , Fujibuchi, W. , Bono, H. , and Kanehisa, M. (1999) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.H. , Lee, S.I. , and Ricke, S.C. (2016) Microbial populations in naked neck chicken ceca raised on pasture flock fed commercial yeast cell wall prebiotics via an Illumina MiSeq platform. PLoS One 11: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks, D.H. , Tyson, G.W. , Hugenholtz, P. , and Beiko, R.G. (2014) STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30: 3123–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemiers, S. , Pinheiro, I. , Verhelst, A. , Van Den Abbeele, P. , Maignien, L. , Laukens, D. , et al (2013) A dried yeast fermentate selectively modulates both the luminal and mucosal gut microbiota and protects against inflammation, as studied in an integrated in vitro approach. J Agric Food Chem 61: 9380–9392. [DOI] [PubMed] [Google Scholar]
- Pourabedin, M. , Guan, L. , and Zhao, X. (2015) Xylo‐oligosaccharides and virginiamycin differentially modulate gut microbial composition in chickens. Microbiome 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse, E. , Quast, C. , Knittel, K. , Fuchs, B.M. , Ludwig, W. , Peplies, J. , and Glöckner, F.O. (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, C. , Huang, L. , Liu, Z. , Xu, L. , Yang, Y. , Tacon, P. , et al (2015) A comparison of the beneficial effects of live and heat‐inactivated baker's yeast on Nile tilapia: suggestions on the role and function of the secretory metabolites released from the yeast. PLoS One 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss, P.D. , Westcott, S.L. , Ryabin, T. , Hall, J.R. , Hartmann, M. , Hollister, E.B. , et al (2009) Introducing mothur: open‐source, platform‐independent, community‐supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata, N. , Izard, J. , Waldron, L. , Gevers, D. , Miropolsky, L. , Garrett, W.S. , and Huttenhower, C. (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, C.E. (1948) A mathematical theory of communication. Bell Syst Tech J 5: 3. [Google Scholar]
- Shen, Y.B. , Piao, X.S. , Kim, S.W. , Wang, L. , Liu, P. , Yoon, I. , and Zhen, Y.G. (2009) Effects of yeast culture supplementation on growth performance, intestinal health, and immune response of nursery pigs. J Anim Sci 87: 2614–2624. [DOI] [PubMed] [Google Scholar]
- Stanley, D. , Hughes, R.J. , Geier, M.S. , and Moore, R.J. (2016) Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, Z.W. , Burns, A.R. , Stagaman, K. , Wong, S. , Rawls, J.F. , Guillemin, K. , and Bohannan, B.J.M. (2015) The composition of the zebrafish intestinal microbial community varies across development. ISME J 10: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullam, K.E. , Rubin, B.E. , Dalton, C.M. , Kilham, S.S. , Flecker, A.S. , and Russell, J.A. (2015) Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. ISME J 9: 1508–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli, V. , and Bäckhed, F. (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489: 242–249. [DOI] [PubMed] [Google Scholar]
- Tuohy, K. , Rouzaud, G. , Bruck, W. , and Gibson, G. (2005) Modulation of the human gut microflora towards improved health using prebiotics ‐ assessment of efficacy. Curr Pharm Des 11: 75–90. [DOI] [PubMed] [Google Scholar]
- Viaud, S. , Saccheri, F. , Mignot, G. , Yamazaki, T. , Daillere, R. , Hannani, D. , et al (2013) The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342: 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Garrity, G.M. , Tiedje, J.M. , and Cole, J.R. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, L. , Amberg, J. , Chapman, D. , Gaikowski, M. , and Liu, W.T. (2014) Fish gut microbiota analysis differentiates physiology and behaviour of invasive Asian carp and indigenous American fish. ISME J 8: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Rarefaction analyses of gut samples with different YC dietary treatments.
Fig. S2. The relative abundance of gut microbiota in individual grass carp at phylum level.
Fig. S3. Relative abundance of four taxa at phylum level significantly associated with YC treatments.
Fig. S4. Relative abundance of six taxa at genus level significantly associated with YC treatments.
Fig. S5. Differences in the gut microbiota of grass carp between control and YC dietary Treat3.
Fig. S6. Venn diagrams showing compartmental core microbiota OTU distributions in grass carp.
Fig. S7. Mean proportion and their differences in predicted functional metagenomes of the gut microbiota among YC dietary treatments.
Fig. S8. Schematic representation of experimental design.
Table S1. Detailed fish gut sample information.
Table S2. The percentage of amino acid in yeast cultures.


