Abstract
Despite significant improvements during the past 3 decades, cardiovascular disease remains a leading worldwide health epidemic. The recent identification of a fascinating group of mediators known as long noncoding RNAs (lncRNAs) has provided a wealth of new biology to explore for cardiovascular risk mitigation. lncRNAs are expressed in a highly context-specific fashion, and multiple lines of evidence implicated them in diverse biological processes. Indeed, abnormalities of lncRNAs have been directly linked with human ailments, including cardiovascular biology and disease. Of particular interest to the cardiovascular research community, dysregulation in lncRNA regulatory circuits have been associated with cardiac pathological hypertrophy, vascular disease, cell fate programming and development, atherosclerosis, dyslipidemia, and metabolic syndrome. Although techniques in interrogating noncoding RNAs are rapidly evolving, a major challenge in studying lncRNAs remains navigating through multiple technical constraints. In this review, we provide a road map for lncRNA discovery and interrogation in biological systems relevant to cardiovascular disease and highlight approaches to decipher their modes of action.
Keywords: computational biology, genetics, heart diseases, risk factors, RNA, untranslated
Despite substantial progress in understanding disease mechanisms and the approval of multiple new therapies, for the first time in 3 decades, we are witnessing an increase in overall cardiovascular deaths.1 Recently, the discovery of an attractive group of molecules known as long noncoding RNAs (lncRNAs) has introduced a new catalog of diagnostic and therapeutic opportunities for cardiovascular risk mitigation. For half a century, the principle that proteins are the primary protagonists of cellar functions has dominated the molecular biology landscape. It has long been appreciated that classes of RNAs, such as transfer RNAs and ribosomal RNAs, have infrastructural importance without encoding for proteins.2–5 Thomas Cech and Sidney Altman shared the noble prize in chemistry for their discovery of biological properties of ribozymes, specialized RNAs that catalyze biochemical reactions.6 However, the notion that RNAs that biochemically resemble mRNA, yet do not template protein, serve important functions had gone largely untested for many years.
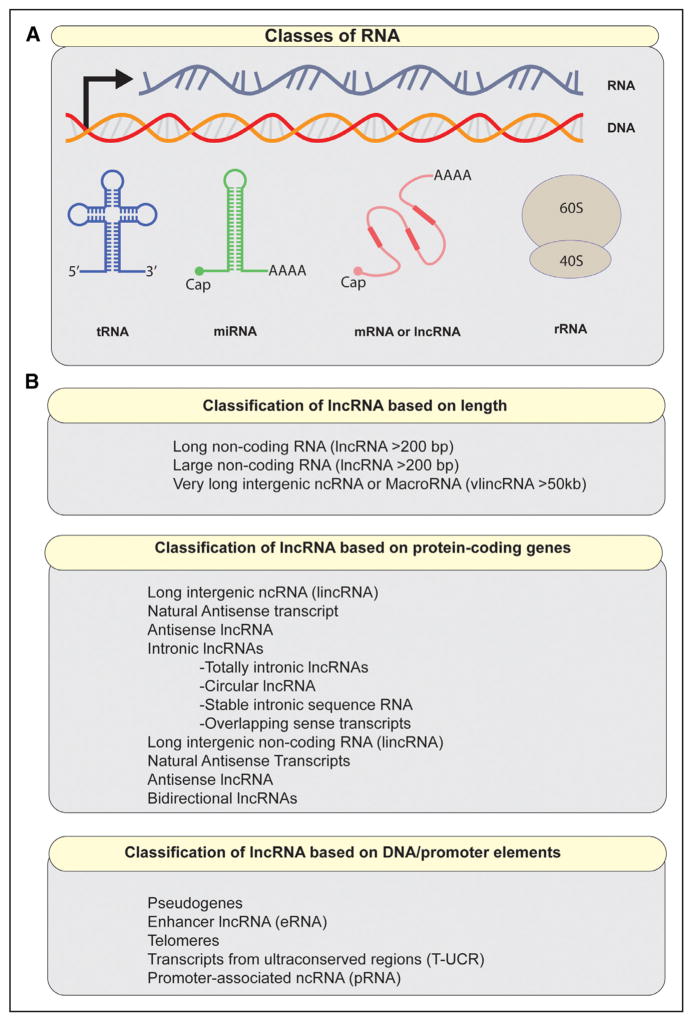
A major turning point in our understanding of the complex role of RNAs in genome regulation came with the sequencing of the human genome. Surprisingly, protein-coding genes account for only 2% of the human genome, despite the fact that >90% of the genome is actively transcribed.7–9 This apparent discrepancy contradicted the presumed efficiency by which mammalian cells were believed to operate. After all, why would living organisms waste so much of the cellular machinery and resources on a process of little biological consequence? A barrage of subsequent studies revealed that this pervasive transcriptional pattern is in fact biologically relevant, thus catalyzing interest in lncRNAs as players in both physiology and disease.10–12 Today lncRNAs are considered important contributors to the diverse RNA community existing within living cells (Figure 1A). However, the substantial overlap between subtypes of lncRNAs presents a challenge to a uniform and consistent classification scheme. The vast majority of definitions describing lncRNAs and related transcripts is shown in Figure 1B.
Figure 1. Categories of RNA in mammalian cells.
A, A schematic of major classes of RNA. Noncoding RNAs may resemble mRNA structurally, but some can have distinctive features, such as lack of polyA tail. B, Classification of noncoding RNAs >200 bp. qPCR indicates quantitative polymerase chain reaction.
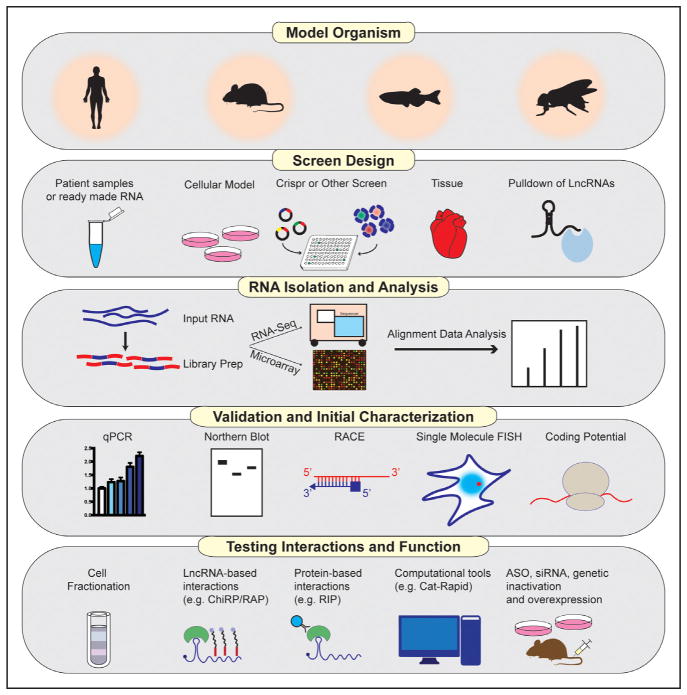
To date, most studies on lncRNAs have focused on their roles in development and differentiation, and many of these findings have had implications for cancer biology. More recently, evidence has begun to emerge that lncRNAs may also participate in diverse additional biological processes and that abnormalities of lncRNAs may be linked to human disease. Cardiovascular biology and disease is one such emerging area of interest. For example, the lncRNA ANRIL was identified in multiple human genome-wide association studies as the most robust predictor of atherosclerotic risk and cardiovascular events.13,14 The precise molecular mechanisms of ANRIL, as is the case with vast majority of lncRNAs, remain poorly understood. The past few years have witnessed a rapid increase in number of studies addressing the roles of lncRNAs in cardiovascular disease. lncRNAs have now been associated with diverse cardiovascular conditions and associated risk factors, including pathological hypertrophy and development, vascular disease, atherosclerosis, dyslipidemia, and metabolic syndrome. A major challenge in studying lncRNAs remains navigating through multiple technical constraints. In this review, we provide a road map for lncRNA discovery and interrogation in biological systems relevant to cardiovascular disease and highlight approaches to decipher their modes of action (Figure 2).
Figure 2. Pipeline of long noncoding RNA (lncRNA) discovery and characterization.
ASO indicates antisense oligonucleotide; ChiRP, chromatin isolation by RNA purification; qPCR, quantitative polymerase chain reaction; RAP, RNA antisense purification; and RIP, RNA immunoprecipitation.
Screening for lncRNAs in Biological Systems
Genome-wide transcriptomic approaches form the foundation of screening for lncRNAs, whether examining mammalian cells, tissues, or serum samples. Novel computational approaches using multiple pipelines can easily identify lncRNAs from RNA-seq data. In general, the data can be analyzed using 2 broad approaches: de novo assembly methods using packages such as Bridger,15 SOAP2,16 Oases,17 or Trinity,18 or alignment against a known standard of defined lncRNAs. The latter approach is increasingly used because lncRNA annotations are rapidly improving. Although knowledge of accompanying chromatin signatures that define active transcription can be helpful, it is not required to identify lncRNAs using sequencing. A subset of lncRNAs showing an element of mammalian conservation is marked by H3K4me3 (trimethylation of lysine 4 of histone H3) at its promoter and H3K36me3 along the transcribed region.19,20 This signature is often referred to as K4k36. Although chromatin features can be helpful in defining lncRNAs, there is ongoing debate as to what distinguishes lncRNAs from other noncoding transcriptional units, such as enhancer RNAs. A summary of these features is provided in Table 1.
Table 1.
Features Distinguishing Long Noncoding eRNAs
| Features | Protein-Coding RNA | LncRNA | eRNA |
|---|---|---|---|
| H3K4me1 | Low | Moderate | High |
| H3K4me3 | High | High | Low |
| H3K27ac | High | Moderate–high | High |
| H3K36me3 | High | High | Low |
| Tissue specific | No | Common | Common |
| Conservation | High | Moderate | Low |
| Splicing | Yes | Common | Not common |
| PolyA tail | Yes | Common | Not common |
| Stability | High | Moderate–high | Low |
eRNA indicates enhancer RNA; and lncRNA, long noncoding RNA.
In general, enhancer RNAs are thought to be unstable, unspliced, nonpolyadenylated, and less abundant than lncRNAs; however, these definitions use arbitrary cutoffs. For practical purposes, there may be some overlap between enhancer RNAs and lncRNAs transcribed from an enhancer element. Some have proposed the use of the chromatin ratio of H3K4me1/H3K4me3 to define enhancers. Although this is one of the defining features that distinguish lncRNAs from other transcriptional units, the sole use of chromatin-binding patterns to define transcriptional elements has limitations.21,22
Although RNA-seq is the most commonly used high-throughput method for RNA detection, other approaches can also be used, including (1) direct RNA sequencing, which involves sequencing of native RNA bypassing library preparation23; (2) cap-assisted gene expression sequencing, which profiles RNAs with 5′ cap and has the advantage of accurately mapping the 5′ transcript end24; (3) serial analysis of gene expression and paired-end tag–expression methods targeting polyA tail25,26; and (4) GrRO-seq—a nuclear run-on assay coupled to deep sequencing to asses nascent transcription.27,28 More recently, multiple commercially available profiling microarrays have been produced that contain probes for human or mouse protein-coding and noncoding transcripts.29–31 The use of microarrays has the advantage of being rapid and efficient and may be comparable with RNA-seq. In our previous experience, differing results can emerge from the 2 methods, although highly enriched lncRNAs were identified by both.32 Similarly, other studies have found the use of RNA-Seq to be superior in detecting low-abundance transcripts, isoform differentiation, and identifying genetic variants.33 Several CRISPRi (CRISPR interference)-based libraries have also been developed to tease out functional roles for lncRNAs.34,35 A recent study targeted >16000 lncRNAs in diverse cell lines, including iPSC cells and identified lncRNAs required for robust cellular growth.34 In addition, several groups have used structural approaches to screen for lncRNAs interacting with transcription factors. For example, Huang et al36 identified the lncRNA Rmrp as important interacting factor in TH17 (T-helper 17) responses by screening for lncRNAs directly interacting with RORγt (RAR-related orphan receptor gamma). Similarly, the cross-linking immunoprecipitation and RNA immunoprecipitation techniques have been used to systematically identify key RNA interactions of binding proteins.37,38
Several important caveats should be considered when designing a screen for lncRNAs. Many lncRNAs are expressed at low levels. As a group, they have a 10-fold lower median expression level than their protein-coding counterparts, so sequencing depth is an important consideration when interrogating noncoding transcripts.39 It is often desirable to retain the strand orientation of RNA sequences to differentiate anti-sense noncoding transcripts, so directional RNA libraries can be prepared. Single-cell sequencing approaches are increasingly being used, but a downside to their use is the need for extensive amplification, which can lead to background noise.
In addition to their low abundance levels, lncRNAs exhibit striking tissue-specific expression patterns, and, therefore, many lncRNAs can be missed unless the correct context and conditions are carefully considered.34,39,40 Many lncRNAs have polyA tails but other types of noncoding transcripts, such as eRNAs, do not, which will influence the optimal library preparation method for sequencing. Finally, it should be noted that computational approaches for analyzing differential expression of assembled transcripts vary substantially. Setting a threshold for lncRNA RPKM (reads per kilobase of transcript per million mapped reads) or FPKM (fragments per kilobase of transcript per million mapped reads) is a somewhat arbitrary decision that can markedly influence the number of identified transcripts. The appropriate number of replicates to use for RNA-seq experiments depends on several factors, including variability in measurements, reproducibility of library preparation, and biological variation.41 Tools such as Scotty can assist in the design of RNA-seq experiments ensuring adequate statistical power while balancing costs.42
Approach to lncRNA Validation
Validation of differentially regulated lncRNAs is an important step before in-depth functional investigation. Quantitative polymerase chain reaction should be used to confirm screen results and differentiate various isoforms that may exist. A particular challenge in studying lncRNAs is that the annotation of many noncoding transcripts is suboptimal and varies depending on genome build. Consequently, careful examination of aligned reads against available annotations for a given lncRNA can provide clues to transcript sequence. RACE (rapid amplification of cDNA ends) experiments can be used to further define the transcript ends and ensure that the lncRNA is indeed produced independent of neighboring genes. Several available databases provide helpful integrative tools and up-to-date annotations, such as NONCODE43 and LNCipedia.44
Defining the copy number for a given lncRNA is an important step that often gives clues to the mechanism of action. Several published studies provide detailed protocols on how to perform this step using real-time polymerase chain reaction.45,46 In addition, RNA-based high-resolution imaging modalities can further validate the abundance and localization of lncRNAs while fostering hypotheses about their potential biological actions. Single-molecule RNA FISH (RNA fluorescent in situ hybridization)—a technique based on multiple complimentary fluorescently tagged oligonucleotides that hybridize to an ln-cRNA of interest—may provide insight into patterns of subcellular localization.47 For example, some lncRNAs, such as Xist, are localized in the nucleus to tight foci, whereas others, including MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), show a more speckled appearance.48 In addition, single-molecule RNA FISH allows for the use of multiple-color localization validation, which improves precision. Several recent studies used this approach along with DNA colocalization to define the contributions of lncRNAs to ribosomal DNA differential expression.49 Probes for single-molecule FISH can be designed according to Raj Laboratory protocol.47 Another alternative is RNAscope—a novel multiplex nucleic acid in situ hybridization technology that has been used to image lncRNAs.50 One caveat relevant to the application of these techniques in cardiovascular disease is that successful imaging can be highly cell-type dependent. Therefore, judicious titration of fixation settings, protease treatment, and hybridization conditions is often required. From our experience, human macrophage cell lines can be particularly challenging to image because of harsh denaturing conditions required.
To Code or Not to Code
lncRNAs are defined as noncoding transcripts >200 bp. A somewhat challenging aspect of this definition is teasing out what constitutes a coding transcript. This is particularly important in light of multiple recent studies relevant to cardiovascular biology showing that annotated lncRNAs may produce micropeptides.51,52 The conserved peptide myoregulin is encoded from a putative lncRNA and has been shown to control muscle relaxation by regulating calcium uptake in the sarcoplasmic reticulum.51 Bioactive peptides can be remarkably small. For example, the open reading frame DWORF—an enhancer of SERCA (sarcoendoplasmic reticulum calcium transport ATPase) activity—is only 34 amino acids long.52 Table 2 provides a summary of methods that may facilitate the identification of peptide production from lncRNAs.
Table 2.
Approaches Used to Investigate the Coding Potential of a Long Noncoding RNA
| Methods | Advantages | Disadvantages |
|---|---|---|
| Bioinformatic tools (eg, CPC, CPAT, and CNCI) | Rapid and easy-to-use web interface for many | Output considered probability rather than definitive evidence |
| In vitro transcription and translation assays | Low hands on time | Somewhat artificial overexpression system |
| Genetic modification, including knockin of a protein tag | Allows for testing of endogenous expression | Time consuming |
| Ribosome profiling | Allows for genome- wide interrogation and resolution mapping of binding sites | Signal-to-noise ratio and other technical concerns have been questioned |
| Mass spectrometry | Definitive | Tedious and requires judicious titration of conditions |
CNCI indicates Coding-Non-Coding Index; CPAT, coding-potential assessment tool; and CPC, coding-potential calculator.
A common approach is to use prediction models that rely on multiple transcript features, including phylogenic conservation, open reading frame length coverage, alignment features, and codon substitution bias, to predict protein-coding potential. Examples of such tools include Coding-Potential Calculator,53 Coding-Potential Assessment Tool,54 and Coding-Non-Coding Index.55 Easy-to-use web-based interfaces are now available for many coding-potential calculator tools. Nevertheless, such tools alone cannot definitively prove the absence of protein-coding ability. Therefore, additional investigative studies are often required.
Coupled in vitro transcription and translation assays may be informative. Genetic modifications that introduce a protein tag at the end of an open reading frame have also been shown to be a valuable tool.51,52 In addition, cellular fractionation analysis can show where an RNA is localized within cells. Ribosome footprinting is a technique that provides a snapshot of the translatome.56 Intriguingly, multiple footprinting studies provide evidence that lncRNAs do not encode for proteins, while others have questioned these results.57–59 Publically available databases archive results on lncRNAs associating with ribosomes in various cell types.60 Finally, it should be noted that the presence of protein product does not exclude the possibility that the transcript itself may have biological activities uncoupled from the encoded polypeptide. For example, recent intriguing work showed that a radiation-induced lncRNA counteracts the activity of a protein produced from the same gene locus.61
Interrogating Your lncRNA Interactome
As mentioned above, a helpful step in defining the range of interactions for a given lncRNA is figuring out its localization. Predominantly cytoplasmic lncRNAs may function through multiple mechanisms, including influencing the stability of an mRNA, affecting translation initiation, acting as competing endogenous RNAs, or influencing post-translational modification. On the contrary, nuclear lncRNAs may influence transcriptional outputs through multiple mechanisms, including epigenetic modifications, interactions with transcription factors, and affecting mRNA processing or export.62
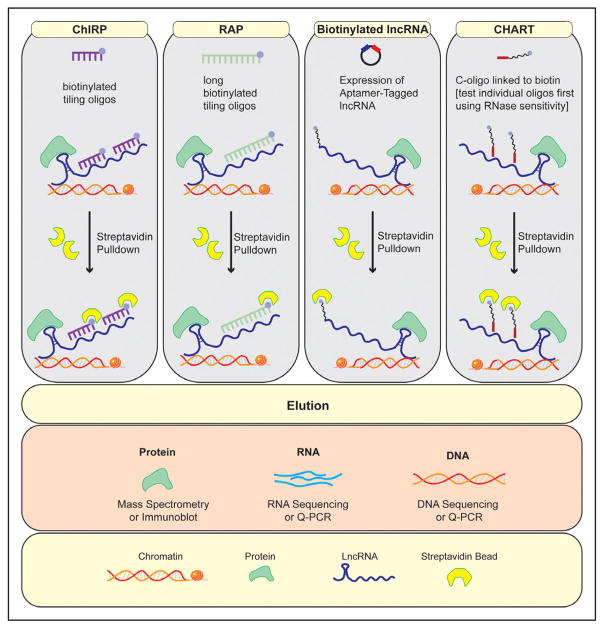
Several techniques have recently been developed that can help to characterize lncRNA interactions (Figure 3). The chromatin isolation by RNA purification (ChIRP) described by Chu et al63,64 was the first of series of chromatin affinity techniques developed to define the repertoire of lncRNA interactions, and the approach has been validated by multiple subsequent studies.32 ChIRP uses tiling complimentary anti-sense oligos to capture an lncRNA of interest and probe its interactions. The method can define genome-binding sites for a given lncRNA and, when coupled with mass spectroscopy, allows for the identification of interacting protein partners. ChIRP has been used to reveal complex interactions of Xist, including its association with hnRNP K and Spen (Spen family transcriptional repressor), which are required for gene silencing.65 ChIRP can even be used to interrogate RNA–RNA interactions, although this has been reported less frequently.63
Figure 3. Summary of techniques used to interrogate a long noncoding RNA’s (lncRNA) interactome.
CHART indicates capture hybridization analysis of RNA targets; ChiRP, chromatin isolation by RNA purification; and RAP, RNA antisense purification.
Based on the same principle of biotin-tagged oligonucleotide retrieval, RNA antisense purification (RAP)66 and capture hybridization analysis of RNA targets67 are 2 other techniques used to investigate the binding patterns and include specific cross-linking protocols and probe design. For example, RAP uses longer probes (≈90 base pair designed with 5′ biotin tags), whereas ChIRP uses 20 nucleotide probes with 3′ tags. Capture hybridization analysis of RNA targets also uses 20 mer olignonucleotides, although it first tests individual olignonucleotides that target accessible regions of a lncRNA using RNase sensitivity. The use of modified protocols that combine elements of multiple designs has been reported to be successful in some cases. For example, we have used a longer RAP probe design with ChIRP in the past. Although nuclear enrichment of samples may be beneficial for interrogating chromatin interactions, it is not necessarily required for any of the above techniques. The use of biotin mimetic lncRNA can also be used as an alternative to the above approaches. In this case, an aptamer sequence (usually 60 bp) is cloned within the expression vector. For example, Zhao et al68 used this approach to identify an interaction between the lncRNA blnc1 and the transcription factor EBF2 (early B-cell factor 2) that drives thermogenic gene stimulation.
There are several important considerations when applying lncRNA interaction methods discussed above. An advantage of ChIRP is that it allows for the retrieval of the endogenous ln-cRNA and can be used with cultured cells or tissues. A major limitation is the need to optimize signal-to-noise ratio. It is conceivable that a series of probes may bind directly to DNA elements, which may result in the identification of false-positive binding sites. This is particularly important for lncRNAs that have prevalent repeats, such as LINE (long interspersed nuclear elements) sequences. The use of genetic inactivation controls and RNase treatment before ChIRP can facilitate data interpretation. dCHIRP is a variant that requires prior knowledge of RNA interacting domains.69 Capture hybridization analysis of RNA targets is more tedious because it involved testing individual probes for retrieval efficiency, but in theory, this method can reduce background noise. Exogenous overexpression can provide evidence for specificity but carries the limitation of interrogating an artificially expressed transcript rather than an endogenous lncRNA. In addition, exogenous tags may interfere with RNA structure.
Although unbiased mass spec approaches have provided valuable insights into lncRNA interactions, a few important technical and interpretive constraints should be considered. First, the use of the above methods coupled with mass spec usually require substantial starting material—typically 200 to 250 million cells per condition. The use of SILAC (stable isotope labeling with amino acids in cell culture) medium is recommended as part of the RAP-MS protocol, but not ChIRP-MS. The inclusion of stringent controls is key to successful chromatin affinity mass spectrometry. We recommend having 2 sets of retrieval probes and 2 sets of control probes (lacZ) as outlined by Chu et al63 to glean direct interactions from background proteins. As is the case with any mass spectrometry experiment, confirmation of interactions and validation of downstream functional effects is key to substantiating lncRNA protein effects. Precise molecular domains important for lncRNA-binding effects can be challenging to predict, but recent evidence used deletion mutants of a lncRNA to map domains required for molecular interactions.46 Work by Atianand et al showed that lncRNA-EPS gates inflammatory gene activation through interactions with the transcription factor hnRNPL (heterogeneous nuclear ribo-nucleoprotein L). The authors used a series of lncRNA-EPS mutants to show that hnRNPL interacted with a distal 3′ nucleotide end of lncRNA-EPS and that ectopic expression of this region of the RNA was sufficient to inhibit inflammation.46 A more detailed discussion of RNA structural analysis is included below. Because many lncRNAs have been shown to influence chromatin dynamics, ATAC-seq analysis of loss- or gain-of-function models can provide clues to enriched domains and cooperative interactions of lncRNAs.70
Structural Analysis of lncRNAs
Deciphering the structure of a specific RNA can give important clues to function and substantiate putative molecular interactions. Computational modeling based on base pairing and physiochemical energetics forms the cornerstone of ln-cRNA structural analysis. Several RNA-modeling platforms are readily available, including JAR3D,71 CMfinder,72 and RNAMotifscan.73 These packages are also useful when comparing across species because RNA 3D structure may be preserved, despite sequence variation. Another stand-alone computational platform is catRAPID (fast predictions of RNA and protein interactions and domains at the Center for Genomic Regulation, Barcelona, Catalonia), which estimates the binding propensity of protein-RNA pairs using computational algorithms to model base-pair structure, hydrogen bonding, and van der Waals forces.74 This powerful omics approach can predict RNA-binding motifs in polypeptides and compute the likelihood of a specific RNA–protein interaction. Similarly, RPI-bind is another theoretical framework that predicts RNA-binding regions on proteins, protein-binding regions on RNA, and their interactions.75
Most recently, multiple techniques that allow for direct mapping of RNA structure using RNA or protein baits have been developed. For example, RAP-RNA is a modification of RAP that uses in vivo cross-linking, antisense oligonucleotides (ASOs) directed against endogenous RNA, and high-throughput RNA sequencing.66 This method was used to investigate the molecular interactions of 2 RNAs: MALAT1 and U1.76 Similarly, Proximity ligation and CLASH (cross-linking ligation and sequencing of hybrids) can be used to interrogate RNA structure.77
Recent work from Howard Chang group described a novel method that identifies base-pairing interactions in living cells at high resolution. PARIS (psoralen analysis of RNA interactions and structures) uses in vivo cross-linking, 2-dimensional purification and proximity ligation to provide high-confidence molecular structure, and interactions of lncRNAs.78 For example, the authors used this approach to reveal the architecture of Xist, including the molecular basis for its interaction with the RNA-binding protein SPEN—a required component of gene repression.79
Approaches to Perturbing lncRNAs
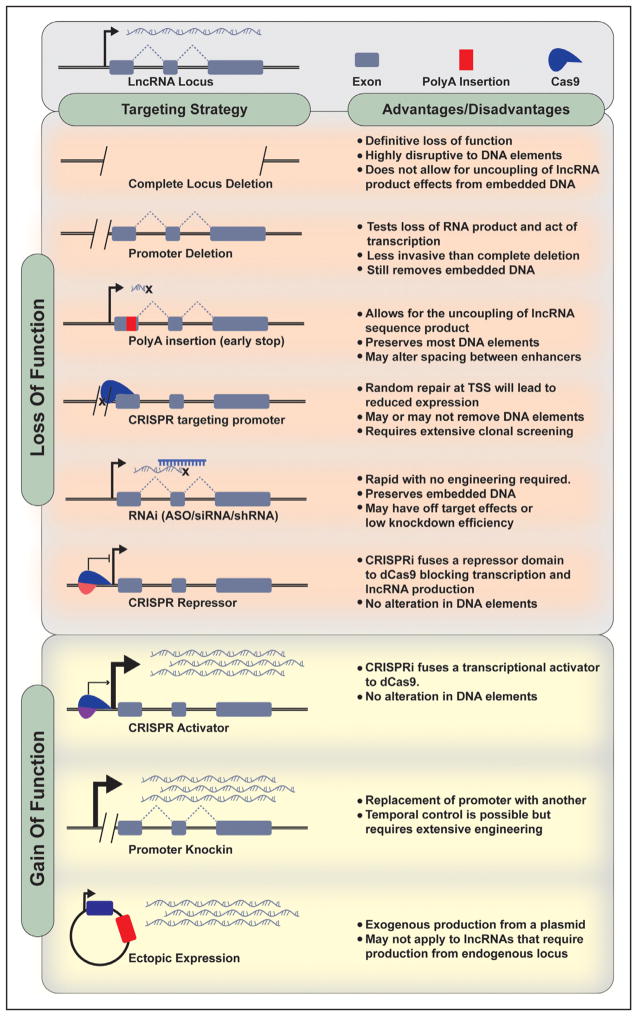
Knockdown or overexpression studies of lncRNAs can be some of the most frustrating or rewarding experiments during a quest to characterize a given lncRNA. Multiple different approaches can be used in perturbing specific lncRNAs (Figure 4). Many lncRNAs regulate transcriptional outputs in cis and, therefore, do not function in exogenous overexpression studies, even with appropriate vector modification that renders the expressed transcript identical to endogenously derived one. A few studies have tried to overcome this constraint by using the λN –Gal4 system to enhance the overexpression of lncRNA in cis.22,80 In addition, the use of RNAi (RNA interference) has notable limitations for the study of lncRNAs. For one, many lncRNAs are expressed at low levels and are predominantly localized in the nucleus, which can limit the efficacy of knockdown.81 In addition, RNAi requires machinery, which can produce off-target effects.
Figure 4. Long noncoding RNA (lncRNA) loss and gain function strategies.
ASO indicates antisense oligonucleotide; and TSS, transcription start site.
More recently, ASOs with appropriate modifications, such as a methylene bridge between the 2′ oxygen and the 4′ carbon (LNAs [locked nucleic acid]), have become readily available.82 In addition, newer generation ASOs allow spatial control of target delivery. For example, triantennary N-acetyl galactosamine has potent affinity for liver.83 Because there are currently >40 human diseases being investigated with ASO-based therapeutics, including cardiometabolic diseases, it is likely that we will witness continued improvements in ASO design, including versatile delivery, higher potency and specificity, and reduced toxicity.84 An advantage of ASO reagents is that they do not require transfection reagents and can be used in vivo without the need to package into an appropriate delivery vector.
Several important caveats should be considered when using ASOs to induce a loss-of-function model. First, it is important to screen multiple ASO or LNA target sites because lncRNAs can form secondary structures that may substantially influence knockdown efficiency depending on site. UNAfold is an example of software analysis that predicts secondary structure in lncRNAs and can assist with targeting sequence design.85 In addition, longer lncRNAs can in theory exhibit retained functional lncRNA fragments even with seemingly efficient knockdown.
Genetic manipulation is rapidly becoming the gold standard in lncRNA perturbation experiments. Although zinc finger86 and Talen87 technologies have been used, CRISPR-CAS (clustered regularly interspaced short palindromic repeats–CRISPR-associated protein)–based modification of lncRNA expression has dominated in recent years.34,35 Advantages of CRISPR over RNAi include improvement in the degree of loss-of-function, improved specificity, and the ability to modulate gene expression in cis.88 CRISPR activation has also been used. Recent work described CRISPR-display, which deploys functional RNA domains to specific genome loci.89 A detailed discussion of approaches of using CRISPR to study lncRNAs in vivo is included below.
Recent work from the Lander laboratory has provided important framework for understanding the contributions of lncRNAs in gene regulation, as well as strategies for manipulating lncRNAs.90 By using CRISPR-based loss-of-function approaches in mouse cell lines, the authors showed that ≈50% of lncRNAs influence the expression of neighboring protein-coding genes. More strikingly, these effects were largely independent of the lncRNA sequence. Of course, protein-coding promoters may influence the expression of neighboring genes as well, so this issue is not distinct to noncoding genes. A CRISPR activation screen from the same group also found 11 lncRNAs that mediate resistance to BRAF (B-Raf proto-oncogene, serine/threonine kinase) inhibitors and that most of them seem to function by influencing their local transcriptional environment.91 These results highlight the fact that neighboring gene regulation by lncRNAs may be more common and more complex than thought previously. It is clear that multiple approaches may be required to glean the mechanism of action for a given lncRNA and assess its potential contributions to local gene regulation.
Knockout Mouse Generation
Although multiple lines of evidence implicate lncRNAs in a range of developmental processes and diseases, few lncRNA loss-of-function studies have yielded robust phenotypes or demonstrated that proposed mechanisms are operative in vivo.92 Because many lncRNAs are thought to function in dose compensation responses, it is certainly conceivable that their effects could be offset by compensatory mechanisms in loss-of-function animal models. However, this limitation should not preclude the study of lncRNAs in vivo. Animal models can provide invaluable insight into the physiological role of an lncRNA and its contribution to disease. However, because lncRNAs by definition do not code for proteins, one cannot simply target an open reading frame or excise a small fraction of the transcript because it may not necessarily render the transcript inactive. Targeting design is thus critical.
The idea that lncRNAs can act as epigenetic modifiers is now supported by a wealth of studies.2,92 Many lncRNAs function in cis, and, although some have transcript-specific actions, it is often challenging to uncouple RNA-specific effects from cis-acting genome elements. Indeed, this idea is supported by experimental evidence showing that ln-cRNA promoters can regulate their neighboring protein-coding genes.90,93 This certainly has important implications for knockout mouse generation. The use of zinc finger or CRISPR-based approaches that delete substantial portion of the genomic loci may be impacting cis-acting elements. More recently, the use of an early polyA termination sequence has been used to halt transcription.86,94 An elegant study from the Olson laboratory used this approach to generate a knockout of the lncRNA upperhand showing that blocking upperhand transcription abolished Hand2 expression and led to right ventricular morphological defects and lethality.94 This approach in theory minimally disrupts local genetic architecture. However, it should be noted that altering the grammar of genome topology even without influencing enhancer elements can also impact local transcriptional outputs. As outlined by Levine et al,95 changing the spacing between enhancers while keeping enhancer structure intact dramatically influenced eve stripe development in drosophila. Therefore, genome inactivation strategies that seemingly offer minimal disruption to coding or noncoding genes may still have a profound effect on local cis-acting elements. Approaches that allow spatiotemporal control of gene expression have been applied to lncRNAs. For example, Srivastava et al96 have taken advantage of lox-P strategies to determine the role of the H19 noncoding locus in Igf2 imprinting. Chang et al97 used flox recombination to disrupt the lncRNA Hotair in a context-specific fashion.
Although current technology has some utility in the study of lncRNA, it is only offering good enough solutions. With further technical advances, ideal lncRNA loss-of-function models may be on the horizon. For example, CRISPRi is a versatile platform that is extending initial CRISPR approaches in promising directions. A modified catalytically dead cas9 can block gene transcription without modifying genome elements in an RNA guide-dependent manner.98 The same approach can be used to activate transcription by fusing a transcriptional activator to cas9.99
Conservation Across Species
A fundamental premise in biological systems is that conservation equals importance. However, lack of conservation should not necessarily invoke a lack of function. It is well established that the majority of lncRNAs do not show evidence of strong sequence conservation. This suggests that nature exerts variable evolutionary pressure between mRNA and lncRNAs. Indeed, systemic interrogation shows that lncRNA exons evolve faster than exons of protein-coding genes.20,100 Several studies showed elements of conservation of lncRNAs across species whether it be location, sequence, or structure conservation.101 For example, myocardial infarction-associated transcript, also known as Gomafu, is an lncRNA associated with increased risk of myocardial infarction in Genome-Wide Association Studies and shows sequence conservation with rodents.102,103 Several other lncRNAs, such as HOTAIR,104 Xist,105 and Evf2,106 exhibit clear functional roles in various species, yet exhibit poor mammalian sequence conservation.107 Interestingly, previous work comparing human and zebra fish lncRNAs showed functional resemblance, despite sequence dissimilarity.108 Additionally, it should be noted the presence of sequence conservation may not always predict functions in other species. Malat1 is a moderately conserved and highly abundant lncRNA that seems to have different roles in mice and humans.109
Identifying an orthologue of an lncRNA can be a daunting task, but sequence similarity is only one component of cross-species comparative analysis. For example, some lncRNAs may show location or promoter conservation.108 In addition, examination of secondary structure of lncRNAs can provide important clues. Several databases automate the retrieval of homologs across species, including PLAR,110 lncRNAdb,111 and NONCODE.43
New Emerging Themes in RNA Biology
The identification of lncRNAs as functional units represent only one facet of the current revolution in RNA biology. An additional layer of gene expression control has now become apparent with the recognition of endogenous RNA modifications, which can serve as powerful on/off switches of biological function. These modifications can occur in both mRNA and ln-cRNAs. For example, several studies have shown that the RNA modification m6A contributes dramatically to RNA function, including influencing transcriptional repression by Xist.112 The same RNA signature can exert powerful effects on progenitor cell specificity and function.113 More recently, Werner et al114 described a unique class of lncRNAs based on their chromatin-binding features known as cheRNAs. The CheRNA HIDALGO (hemin-induced cheRNA downstream of fetal hemoglobin) stimulates the fetal HBG1 gene during erythroid differentiation by promoting contacts to a downstream enhancer. Finally, multiple lines of evidence have implicated circular RNA in cardiovascular disease. Circular RNA can be exon or intron produced and are circularized by joining the 3′ and 5′ ends. Just like lncRNAs, they can have diverse functional mechanisms, including regulating transcription, splicing and processing, and acting as sponges. An example is the circular RNA ANRIL (antisense noncoding RNA in the INK4 locus), which affects atherogenesis by acting as a scaffold to prevent ribosomal RNA function.115 Although circular RNAs can be challenging to interrogate because they are not captured by poly-based sequencing methods, their study has filled a substantial gap in mechanisms orchestrating cardiovascular disease.116 Thus, epigenetic modification is no longer a term that is restricted to DNA/chromatin interrogation. Continued exploration of post-transactional RNA modifications will likely yield novel insight into biological disease mechanisms in cardiovascular disease.
Future Perspective and Conclusions
The discovery of lncRNAs has had a transformative impact on our understanding of disease-regulatory circuits and catalyzed the development of technical advances in molecular interrogation techniques. As more and more lncRNAs are linked to cardiovascular physiology, important questions remained to be addressed before we can fully harness the therapeutic potential of these novel biological modulators. For one, how lncRNAs are targeted to specific genetic loci remains poorly understood. Indeed, the temporal–spatial specificity of lncRNA effects is particularly mystifying given that a large number appear to interact with highly promiscuous factors, such as PRC2 (polycomb repressive complex 2).117 The biochemical and structural basis for why some lncRNAs that resemble mRNAs are retained in the nucleus is also mysterious. Finally, an important bottom line question is: can we leverage the diagnostic or therapeutic potential of lncRNA to advance human health? Although this has been an elusive goal, promising translational efforts are not far behind.
Acknowledgments
We apologize to many of our colleagues whose work was not included because of space constraints.
Sources of Funding
This work was supported by grants HL066088 and HL128822 from the National Heart, Lung, and Blood Institute. Additional support was provided by National Institutes of Health grant 5U54GM114833 and the Burroughs Wellcome Fund Career Awards for Medical Scientists.
Nonstandard Abbreviations and Acronyms
- ASO
antisense oligonucleotide
- ChIRP
chromatin isolation by RNA purification
- H3K4me3
trimethylation of lysine 4 of histone H3
- lncRNA
long noncoding RNA
- RAP
RNA antisense purification
Footnotes
Disclosures
None.
References
- 1.Heron M, Anderson RN. Changes in the leading cause of death: recent patterns in heart disease and cancer mortality. NCHS Data Brief. 2016;254:1–8. [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul J, Duerksen JD. Chromatin-associated RNA content of heterochromatin and euchromatin. Mol Cell Biochem. 1975;9:9–16. doi: 10.1007/BF01731728. [DOI] [PubMed] [Google Scholar]
- 4.Noller HF. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS. The central role of RNA in human development and cognition. FEBS Lett. 2011;585:1600–1616. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat Rev Mol Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 7.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birney E, Stamatoyannopoulos JA, Dutta A, et al. ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute; Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katayama S, Tomaru Y, Kasukawa T, et al. RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 10.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa FF. Non-coding RNAs: new players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- 12.Maass PG, Luft FC, Bähring S. Long non-coding RNA in health and disease. J Mol Med (Berl) 2014;92:337–346. doi: 10.1007/s00109-014-1131-8. [DOI] [PubMed] [Google Scholar]
- 13.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature. 2011;470:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broadbent HM, Peden JF, Lorkowski S, et al. PROCARDIS Consortium. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008;17:806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 15.Chang Z, Li G, Liu J, Zhang Y, Ashby C, Liu D, Cramer CL, Huang X. Bridger: a new framework for de novo transcriptome assembly using RNA-seq data. Genome Biol. 2015;16:30. doi: 10.1186/s13059-015-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 17.Schulz MH, Zerbino DR, Vingron M, Birney E. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 22.Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, Oh S, Kim HS, Glass CK, Rosenfeld MG. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozsolak F, Platt AR, Jones DR, Reifenberger JG, Sass LE, McInerney P, Thompson JF, Bowers J, Jarosz M, Milos PM. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi H, Kato S, Murata M, Carninci P. CAGE (cap analysis of gene expression): a protocol for the detection of promoter and transcriptional networks. Methods Mol Biol. 2012;786:181–200. doi: 10.1007/978-1-61779-292-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philippe N, Bou Samra E, Boureux A, Mancheron A, Rufflé F, Bai Q, De Vos J, Rivals E, Commes T. Combining DGE and RNA-sequencing data to identify new polyA+ non-coding transcripts in the human genome. Nucleic Acids Res. 2014;42:2820–2832. doi: 10.1093/nar/gkt1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fullwood MJ, Wei CL, Liu ET, Ruan Y. Next-generation DNA sequencing of paired-end tags (PET) for transcriptome and genome analyses. Genome Res. 2009;19:521–532. doi: 10.1101/gr.074906.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardini A. Global run-on sequencing (GRO-Seq) Methods Mol Biol. 2017;1468:111–120. doi: 10.1007/978-1-4939-4035-6_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi Y, Shang J. Long noncoding RNA expression profiling using array-star LncRNA microarrays. Methods Mol Biol. 2016;1402:43–61. doi: 10.1007/978-1-4939-3378-5_6. [DOI] [PubMed] [Google Scholar]
- 30.Chen C, Li Z, Yang Y, Xiang T, Song W, Liu S. Microarray expression profiling of dysregulated long non-coding RNAs in triple-negative breast cancer. Cancer Biol Ther. 2015;16:856–865. doi: 10.1080/15384047.2015.1040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu D, Fang C, Li X, Geng Y, Li R, Wu C, Jiang J, Wu C. Predictive analysis of long non-coding RNA expression profiles in diffuse large B-cell lymphoma. Oncotarget. 2017;8:23228–23236. doi: 10.18632/oncotarget.15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sallam T, Jones MC, Gilliland T, Zhang L, Wu X, Eskin A, Sandhu J, Casero D, Vallim TQ, Hong C, Katz M, Lee R, Whitelegge J, Tontonoz P. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534:124–128. doi: 10.1038/nature17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao S, Fung-Leung WP, Bittner A, Ngo K, Liu X. Comparison of RNA-Seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, Mandegar MA, Olvera MP, Gilbert LA, Conklin BR, Chang HY, Weissman JS, Lim DA. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science. 2017;355:aah7111. doi: 10.1126/science.aah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, Yuan P, Brown M, Liu XS, Wei W. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang W, Thomas B, Flynn RA, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528:517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Kashi K, Henderson L, Bonetti A, Carninci P. Discovery and functional analysis of lncRNAs: methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta. 2016;1859:3–15. doi: 10.1016/j.bbagrm.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 38.Guil S, Soler M, Portela A, Carrère J, Fonalleras E, Gómez A, Villanueva A, Esteller M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat Struct Mol Biol. 2012;19:664–670. doi: 10.1038/nsmb.2315. [DOI] [PubMed] [Google Scholar]
- 39.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsoi LC, Iyer MK, Stuart PE, Swindell WR, Gudjonsson JE, Tejasvi T, Sarkar MK, Li B, Ding J, Voorhees JJ, Kang HM, Nair RP, Chinnaiyan AM, Abecasis GR, Elder JT. Analysis of long non-coding RNAs highlights tissue-specific expression patterns and epigenetic profiles in normal and psoriatic skin. Genome Biol. 2015;16:24. doi: 10.1186/s13059-014-0570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, Mortazavi A. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busby MA, Stewart C, Miller CA, Grzeda KR, Marth GT. Scotty: a web tool for designing RNA-Seq experiments to measure differential gene expression. Bioinformatics. 2013;29:656–657. doi: 10.1093/bioinformatics/btt015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y, Li H, Fang S, Kang Y, Wu W, Hao Y, Li Z, Bu D, Sun N, Zhang MQ, Chen R. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016;44:D203–D208. doi: 10.1093/nar/gkv1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, Mestdagh P. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4364. doi: 10.1093/nar/gkv295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kotzin JJ, Spencer SP, McCright SJ, et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature. 2016;537:239–243. doi: 10.1038/nature19346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Atianand MK, Hu W, Satpathy AT, et al. A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell. 2016;165:1672–1685. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batish M, Raj A, Tyagi S. Single molecule imaging of RNA in situ. Methods Mol Biol. 2011;714:3–13. doi: 10.1007/978-1-61779-005-8_1. [DOI] [PubMed] [Google Scholar]
- 48.Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, Dong R, Yang L, Chen LL. SLERT regulates DDX21 rings associated with Pol I transcription. Cell. 2017;169:664–678e16. doi: 10.1016/j.cell.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Wang MX, Su N, Wang LC, Wu X, Bui S, Nielsen A, Vo HT, Nguyen N, Luo Y, Ma XJ. Rnascope for in situ detection of transcriptionally active human papillomavirus in head and neck squamous cell carcinoma. J VisExp. 2014:85. doi: 10.3791/51426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, Olson EN. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, Cannon SC, Houser SR, Bassel-Duby R, Olson EN. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Park HJ, Dasari S, Wang SQ, Kocher JP, Li W. Cpat: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013;41:e166. doi: 10.1093/nar/gkt646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carlevaro-Fita J, Rahim A, Guigó R, Vardy LA, Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016;22:867–882. doi: 10.1261/rna.053561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Essers PB, Nonnekens J, Goos YJ, Betist MC, Viester MD, Mossink B, Lansu N, Korswagen HC, Jelier R, Brenkman AB, MacInnes AW. A long noncoding rna on the ribosome is required for lifespan extension. Cell Rep. 2015 doi: 10.1016/j.celrep.2014.12.029. pii:S2211–1247(14)01058-4. [DOI] [PubMed] [Google Scholar]
- 60.Michel AM, Fox G, Kiran MA, De Bo C, O’Connor PB, Heaphy SM, Mullan JP, Donohue CA, Higgins DG, Baranov PV. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 2014;42:D859–D864. doi: 10.1093/nar/gkt1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williamson L, Saponaro M, Boeing S, East P, Mitter R, Kantidakis T, Kelly GP, Lobley A, Walker J, Spencer-Dene B, Howell M, Stewart A, Svejstrup JQ. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell. 2017;168:843–855e13. doi: 10.1016/j.cell.2017.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J VisExp. 2012:61. doi: 10.3791/3912. pii: 3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Engreitz J, Lander ES, Guttman M. RNA antisense purification (RAP) for mapping RNA interactions with chromatin. Methods Mol Biol. 2015;1262:183–197. doi: 10.1007/978-1-4939-2253-6_11. [DOI] [PubMed] [Google Scholar]
- 67.Simon MD, Wang CI, Kharchenko PV, West JA, Chapman BA, Alekseyenko AA, Borowsky ML, Kuroda MI, Kingston RE. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci USA. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quinn JJ, Ilik IA, Qu K, Georgiev P, Chu C, Akhtar A, Chang HY. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat Biotechnol. 2014;32:933–940. doi: 10.1038/nbt.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roll J, Zirbel CL, Sweeney B, Petrov AI, Leontis N. JAR3D Webserver: scoring and aligning RNA loop sequences to known 3D motifs. Nucleic Acids Res. 2016;44:W320–W327. doi: 10.1093/nar/gkw453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yao Z, Weinberg Z, Ruzzo WL. CMfinder–a covariance model based RNA motif finding algorithm. Bioinformatics. 2006;22:445–452. doi: 10.1093/bioinformatics/btk008. [DOI] [PubMed] [Google Scholar]
- 73.Zhong C, Tang H, Zhang S. RNAMotifScan: automatic identification of RNA structural motifs using secondary structural alignment. Nucleic Acids Res. 2010;38:e176. doi: 10.1093/nar/gkq672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bellucci M, Agostini F, Masin M, Tartaglia GG. Predicting protein associations with long noncoding RNAs. Nat Methods. 2011;8:444–445. doi: 10.1038/nmeth.1611. [DOI] [PubMed] [Google Scholar]
- 75.Luo J, Liu L, Venkateswaran S, Song Q, Zhou X. RPI-Bind: a structure-based method for accurate identification of RNA-protein binding sites. Sci Rep. 2017;7:614. doi: 10.1038/s41598-017-00795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals rna-rna interactions in yeast. Proc Natl Acad Sci USA. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol. 2017;24:197–204. doi: 10.1038/nsmb.3370. [DOI] [PubMed] [Google Scholar]
- 79.Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, Mesirov JP, Cech TR, Chang HY. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell. 2016;165:1267–1279. doi: 10.1016/j.cell.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–877. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sarma K, Levasseur P, Aristarkhov A, Lee JT. Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA. 2010;107:22196–22201. doi: 10.1073/pnas.1009785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prakash TP, Graham MJ, Yu J, et al. Targeted delivery of antisense oligo-nucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014;42:8796–8807. doi: 10.1093/nar/gku531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Graham MJ, Lee RG, Brandt TA, et al. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N Engl J Med. 2017;377:222–232. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- 85.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 86.Gutschner T, Baas M, Diederichs S. Noncoding RNA gene silencing through genomic integration of RNA destabilizing elements using zinc finger nucleases. Genome Res. 2011;21:1944–1954. doi: 10.1101/gr.122358.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Luo D, Zhao H, Zhu Z, Hu W, Cheng CH. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS One. 2013;8:e76387. doi: 10.1371/journal.pone.0076387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goyal A, Myacheva K, Groß M, Klingenberg M, Duran Arque B, Diederichs S. Challenges of CRISPR/Cas9 applications for long non-coding RNA genes. Nucleic Acids Res. 2017;45:e12. doi: 10.1093/nar/gkw883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by ln-cRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, Tseng YY, Yoon CH, Boehm JS, Lander ES, Zhang F. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548:343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 93.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, Olson EN. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fukaya T, Lim B, Levine M. Enhancer control of transcriptional bursting. Cell. 2016;166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Srivastava M, Hsieh S, Grinberg A, Williams-Simons L, Huang SP, Pfeifer K. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting regulatory region upstream of H19. Genes Dev. 2000;14:1186–1195. [PMC free article] [PubMed] [Google Scholar]
- 97.Li L, Liu B, Wapinski OL, Tsai MC, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA, Helms JA, Chang HY. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grützner F, Kaessmann H. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 101.Ulitsky I. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 2016;17:601–614. doi: 10.1038/nrg.2016.85. [DOI] [PubMed] [Google Scholar]
- 102.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, Saito S, Nakamura Y, Tanaka T. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- 103.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 104.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Okamoto I, Arnaud D, Le Baccon P, Otte AP, Disteche CM, Avner P, Heard E. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- 106.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chodroff RA, Goodstadt L, Sirey TM, Oliver PL, Davies KE, Green ED, Molnar Z, Ponting CP. Long noncoding RNA genes: conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11:R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30:121–123. doi: 10.1016/j.tig.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 110.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11:1110–1122. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Quek XC, Thomson DW, Maag JL, Bartonicek N, Signal B, Clark MB, Gloss BS, Dinger ME. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, Lv J, Heng J, Ding Y, Xue Y, Lu X, Xiao W, Yang YG, Liu F. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549:273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 114.Werner MS, Sullivan MA, Shah RN, Nadadur RD, Grzybowski AT, Galat V, Moskowitz IP, Ruthenburg AJ. Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat Struct Mol Biol. 2017;24:596–603. doi: 10.1038/nsmb.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang H, Yang J, Yang J, Fan Z, Yang C. Circular RNAs: novel rising stars in cardiovascular disease research. Int J Cardiol. 2016;202:726–727. doi: 10.1016/j.ijcard.2015.10.051. [DOI] [PubMed] [Google Scholar]
- 117.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]